Version Changes
Revised. Amendments from Version 1
We have updated the figures to amend some issues with proofing. We have added in some details of other excellent resources for SARS-CoV-2 international surveillance. Over on cov-lineages.org (which has had a facelift since time of publishing), we have also added in a resources page (https://cov-lineages.org/resources.html) that points the user to both internally developed and externally developed resources for SARS-CoV-2 lineage and variant tracking. Figures 1 and 2 along with title were also updated.
Abstract
Late in 2020, two genetically-distinct clusters of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with mutations of biological concern were reported, one in the United Kingdom and one in South Africa. Using a combination of data from routine surveillance, genomic sequencing and international travel we track the international dispersal of lineages B.1.1.7 and B.1.351 (variant 501Y-V2). We account for potential biases in genomic surveillance efforts by including passenger volumes from location of where the lineage was first reported, London and South Africa respectively. Using the software tool grinch (global report investigating novel coronavirus haplotypes), we track the international spread of lineages of concern with automated daily reports, Further, we have built a custom tracking website (cov-lineages.org/global_report.html) which hosts this daily report and will continue to include novel SARS-CoV-2 lineages of concern as they are detected.
Keywords: genomic surveillance, air travel, SARS-CoV-2, genomics, genome sequencing, virus, surveillance, pandemic, B.1.1.7, B.1.351, N501Y, coronavirus, sequencing, genomic epidemiology
Introduction
In December 2020, routine genomic surveillance in the United Kingdom (UK) 1 reported a new and genetically distinct phylogenetic cluster of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (variant VOC202012/01, lineage B.1.1.7). Preliminary analysis suggests that this lineage carries an unusually large number of genetic changes 2. The earliest known cases of B.1.1.7 were sampled in southern England in late September 2020, and by December the lineage had spread to most UK regions and was growing rapidly 3. In October 2020, a separate SARS-CoV-2 cluster (variant 501Y.V2, lineage B.1.351), which carried a different constellation of genetic changes, was detected by the Network for Genomic Surveillance in South Africa 4, 5. Both lineages carry mutations, especially in the virus spike protein, that may affect virus function, and both appear to have grown rapidly in relative frequency since their discovery. Early analyses of the spatial spread of SARS-CoV-2 highlights the potential for rapid virus dissemination through national and international travel 6, 7. Therefore continued genomic monitoring of lineages of concern is required.
To facilitate tracking of these lineages on an international scale, we developed a software tool grinch (global report investigating novel coronavirus haplotypes) that collates SARS-CoV-2 genomic data and epidemiological metadata. Resources such as grinch on cov-lineages.org can inform public health bodies and institutions around the world. Other excellent resources to track lineages and variants are available, including covariants.org, which tracks the spread of SARS-CoV-2 variants of interest, and outbreak.info, which gathers multiple sources of genetic and epidemiological data to track lineages. We include a non-exhaustive list of resources for tracking SARS-CoV-2 at https://cov-lineages.org/resources.html.
Methods
To better characterise the international distribution of lineages B.1.1.7 and B.1.351 we sourced SARS-CoV-2 sequences from GISAID 8, 9 and assigned lineages using pangolin (v2.1.6, https://github.com/cov-lineages/pangolin), which implements the nomenclature scheme described in Rambaut et al., 10. Genomes are assigned lineage B.1.1.7 if they exhibit at least 5 of the 17 mutations inferred to have arisen on the phylogenetic branch immediately ancestral to the cluster ( Table 1) 2; or to B.1.351 if they exhibit at least 5 of 9 lineage-associated mutations ( Table 1) 5. Lineage count and frequency data have been calculated daily using grinch. Using International Air Transport Association (IATA) travel data from October 2020, available through bluedot.global, we aggregated and collated the passenger volumes from international airports in London and South Africa to international destinations on same booking. Destinations with more than 5,000 passengers from London and more than 300 passengers from South Africa during the month of October are displayed on the cov-lineages.org website and in the underlying data for this publication 11. grinch, with custom python modules that make use of geopandas v0.9, matplotlib v3.2 and seaborn v0.10, combines this information and produces reports with descriptive tables and figures that can be found at https://cov-lineages.org/global_report.html.
Table 1. Defining mutations for lineages of interest.
| Lineage | Defining mutations |
|---|---|
| B.1.1.7 | orf1ab:T1001I; orf1ab:A1708D; orf1ab:I2230T; del:11288:9; del:21765:6; del:21991:3; S:N501Y;
S:A570D; S:P681H; S:T716I; S:S982A; S:D1118H; Orf8:Q27*; Orf8:R52I; Orf8:Y73C; N:D3L; N: S235F |
| B.1.351/501Y-V2 | E:P71L; N:T205I; orf1a:K1655N; S:D80A; S:D215G; S:K417N; S:E484K; S:N501Y; S:E484K |
Implementation
All of the code underlying this daily lineage tracking web-report can be found at GitHub and Zenodo 12. grinch is a python-based tool, the analysis pipeline of which is built on a snakemake backbone 13. Every 24 hours a scheduled cron 14 task runs on our local servers. We download the latest data from GISAID and deduplicate based on sequence names. The sequences are assigned their most likely lineage using pangolin’s latest version and model files. All processed metadata is available and maintained on the cov-lineages.org GitHub repository. To run grinch, the user must have access to a GISAID direct download key and a password and provide these within a configuration file for use. The command used to run grinch is grinch -i grinch_config.yaml, using the config file provided at doi: 10.5281/zenodo.4640379 15.
Operation
Most users will not run grinch themselves, instead all information and useful descriptive figures are provided daily on the web report. Users can navigate to cov-lineages.org in a web browser of choice to view the latest daily report.
Results and discussion
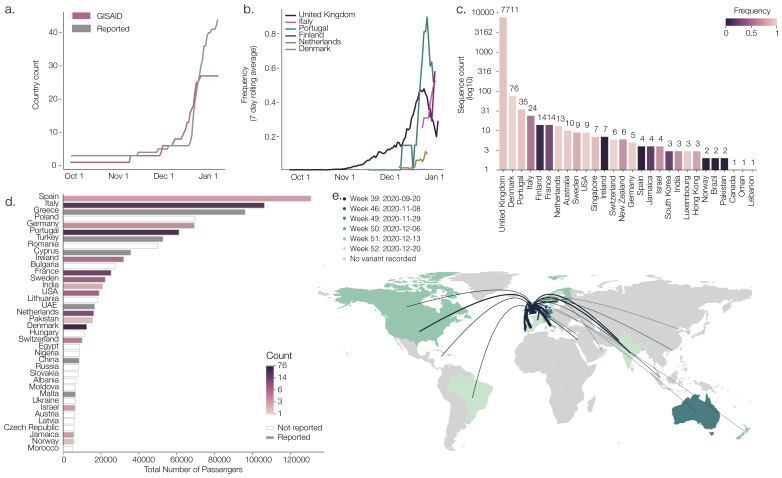
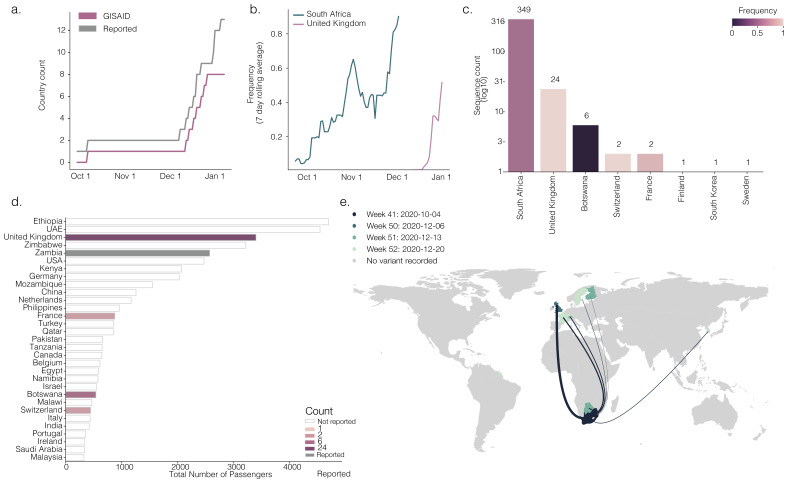
As of 7th Jan 2021, 45 countries had reported the presence of B.1.1.7 and 13 countries had reported B.1.351/501Y.V2. B.1.1.7 and B.1.351 genome sequences were available for 28 and 8 countries, respectively ( Figure 1a, b, c) 11. Although some countries report increases in the relative frequency of B.1.1.7, genome sequencing efforts vary considerably. Potential targeting of sequencing towards travelers from the UK could bias frequency estimates upwards ( Figure 1b, c) and differing genome sharing policies and delays may also skew reporting estimates. The time between the initial collection date of a new variant sample in a country and the first availability of a corresponding virus genome on GISAID was, on average, 12 days (range 1–71).
Figure 1.
a) The cumulative number of countries with reports of lineage B.1.1.7 (grey line) and cumulative number of genomes of B.1.1.7 deposited in GISAID. b) Rolling seven-day average of the proportion of B.1.1.7 genomes in countries with more than ten sequences of the variant, and with more than ten days between the first B.1.1.7 sequence and the most recent one compared to all sampled genomes in that country. c) Number of sequences (log10) per country. Colour indicates the proportion of sequences that are classified as lineage B.1.1.7. d) Number of air travellers from major international London airports (Heathrow, Gatwick, Luton, City, Stansted, Southend) during October 2020. Colour indicates the number of sampled genomes of lineage B.1.1.7. Reported refers to countries that we found media reports stating there had been sequences of that particular lineage, but for which there were no sequences on GISAID. This is distinct from ‘not reported’ where there were no records found of that lineage in a given country. e) Map of international flights from major international London airports to countries with B.1.1.7 sequences. Colours indicate the date of earliest detection of B.1.1.7. in each country. The width of the lines indicates the number of flights. International Air Transport Association data used here account for ~90% of passenger travel itineraries on commercial flights, excluding transportation via unscheduled charter flights (the remainder is modelled using market intelligence). Data shown represents origin-destination journeys during October 2020. Routes to countries that have not yet detected B.1.1.7 and deposited data on GISAID are not included.
The number of B.1.1.7 and B.1.351/501Y.V2 genome sequences reported in each country is a consequence of (i) the intensity of local genomic surveillance; (ii) the level of concern about new variant introductions; (iii) the volume of international travel among affected countries, and (iv) the amount of local transmission following the introduction of lineage from elsewhere. To explore these factors, we analysed the most recent available IATA travel data (October 2020). We collated the total number of origin-to-destination air journeys between major London international airports and each country. The calculation was repeated for journeys originating in all international South African airports. We focussed on London and South Africa as they are the locations with the first reports and highest reported prevalence of lineages B.1.1.7 and B.1.351 respectively 2, 5. However, due to low SARS-CoV-2 genomic surveillance in many locations, we cannot reject the hypotheses that these lineages initially originated elsewhere. Figure 1d shows destinations receiving >5,000 travellers in October 2020 from the UK ( Figure 2 shows destinations receiving >300 travellers from South Africa).
Figure 2.
a) Shows the cumulative number of countries with reports of lineage B.1.351 (black line) and cumulative number of genomes of B.1.351 deposited in GISAID. b) Rolling seven-day average of the proportion of B.1.351 genomes in countries with more than ten sequences of the variant, and with more than ten days between the first B.1.351 sequence and the most recent one compared to all sampled genomes in that country. c) Number of sequences (log10) per country. Colour indicates the proportion of sequences that are classified as lineage B.1.351 d) Number of air travellers from South Africa during October 2020. Colour indicates the number of sampled genomes of lineage B.1.351. Not reported refers to a given country having no record of B.1.351, and reported refers to countries that we found media reports but that country had no SARS-CoV-2 genomes shared on GISAID at that time. e) Map of international flights to countries with B.1.351 sequences. Colours indicate the date of earliest detection of B.1.351 in each country. The width of the lines indicates the number of flights. International Air Transport Association data used here account for ~90% of passenger travel itineraries on commercial flights, excluding transportation via unscheduled charter flights (the remainder is modelled using market intelligence). Data shown represents origin-destination journeys during October 2020. Routes to countries that have not yet detected B.1.351 and deposited data on GISAID are not included. >300 travellers from South Africa).
Of the countries that receive >5,000 travellers from London, 16 have sequenced B.1.1.7. Of the 45 countries that have identified B.1.1.7 (32 in travellers and 13 with local onward transmission), only 6 perform real-time routine genomic surveillance (Denmark, UK, Iceland, The Netherlands, Australia, Sweden), 3 have prioritised sequencing based on S-gene target failure tests 16, 30 primarily targeted sequencing towards arriving travellers from the UK, and there was no information available for 10 (details at https://github.com/cov-lineages/lineages-website/blob/master/_data/). Of the 13 countries that have identified B.1.351 (four with local onward transmission including South Africa), 4 perform routine sequencing (South Africa, UK, Botswana, Australia), 6 target sequencing of travellers, and there was no information available for 3. Consequently, there is no clear relationship between number of sequences reported and flight numbers, but rather reflects the current genomic surveillance effort. For example, in September, the UK sequenced ~13% of its reported cases and Denmark sequenced ~21%. In comparison, Israel sequenced ~0.002% of its cases during the same period 17, 18.
Our study has several limitations. The passenger flight data do not include recent changes to holiday travel, and recent restrictions on travel from the UK and South Africa is not reflected in the mobility data. Further, flight data may not accurately reflect the final destination if multiple tickets are purchased.
The discovery and rapid spread of B.1.1.7 and B.1.351/501Y.V2 highlights the importance of real-time and open data for tracking the spread of SARS-CoV-2 and for informing future public health interventions and travel advice.
Data availability
Underlying data
Zenodo: Accession IDs included in publication Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. https://doi.org/10.5281/zenodo.4642401 9.
This project contains the following underlying data:
-
-
Accession IDs of B.1.1.7 and B.1.351 genome sequences included in report up until January 7 th, 2021. All accession IDs link to data on the GISAID repository, http://doi.org/10.17616/R3Q59F. These data are available under the terms of the GISAID EpiFlu™ Database Access Agreement.
Zenodo: cov-lineages.org website. https://doi.org/10.5281/zenodo.4640140 11.
This project contains the following underlying data:
-
-
Website data archived at time of publication
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Extended data
Zenodo: Supplementary materials with group affiliations for Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. https://doi.org/10.5281/zenodo.4704471 19.
This project contains the following extended data:
-
-
Supplementary materials with group authorship affiliations and full acknowledgements.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Software availability
-
-
Software available from: https://cov-lineages.org/global_report.html
-
-
Source code available from: https://github.com/cov-lineages/grinch
-
-
Archived source code at time of publication:https://doi.org/10.5281/zenodo.464003712; https://doi.org/10.5281/zenodo.464037915
-
-
Licenses:GNU General Public License v3.0; Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Acknowledgements
An earlier version of this article can be found on Virological (url: https://virological.org/t/tracking-the-international-spread-of-sars-cov-2-lineages-b-1-1-7-and-b-1-351-501y-v2/592).
We thank Norelle Sherry, Benjamin Howden and Michelle Sait for their contribution to sequencing in Australia. We also include full acknowledgements and details of group authorships at https://doi.org/10.5281/zenodo.4704471 19. We would also like to extend our gratitude to everyone involved in the global sequencing effort.
Funding Statement
I.I.B. is supported by the Canadian Institutes of Health Research, COVID-19 Rapid Research Funding Opportunity (02179-000). K.K. is the founder of BlueDot, a social enterprise that develops digital technologies for public health. K.K., A.W., A.T.B. and C.H. are employed at BlueDot. I.I.B. has consulted for BlueDot. T.d.O. and the NGS-SA is funded by the South African Medical Research Council (SAMRC), MRC SHIP and the Department of Science and Innovation (DSI) of South Africa. N.R.F. acknowledges support from a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (204311/Z/16/Z) and a Medical Research Council-São Paulo Research Foundation CADDE partnership award (MR/S0195/1 and FAPESP 18/14389-0). VH was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/M010996/1]. M.U.G.K. acknowledges support from the Branco Weiss Fellowship and EU grant 874850 MOOD. The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. O.G.P. , J.P.M. and M.U.G.K. acknowledge support from the Oxford Martin School. AR acknowledges the support of the Wellcome Trust (Collaborators Award 206298/Z/17/Z – ARTIC network) and the European Research Council (grant agreement no. 725422 – ReservoirDOCS). A.OT is supported by the Wellcome Trust Hosts, Pathogens & Global Health Programme [grant number: grant.203783/Z/16/Z] and Fast Grants [award number: 2236]. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute.TFS acknowledges support from the Deutsche Forschungsgemeinschaft (SFB900, EXC2155 RESIST). SeqCOVID-SPAIN is supported by a grant from the Instituto de Salud Carlos III COV0020/00140.
[version 2; peer review: 3 approved]
References
- 1.COVID-19 Genomics UK (COG-UK) consortiumcontact@cogconsortium.uk: An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe. 2020;1(3):e99–100. 10.1016/S2666-5247(20)30054-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambaut A, Loman N, Pybus O, et al. : Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.2020; published online Dec 18. (accessed Jan 8, 2021). Reference Source [Google Scholar]
- 3.Volz E, Mishra S, Chand M, et al. : Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. bioRxiv. 2021. 10.1101/2020.12.30.20249034 [DOI] [Google Scholar]
- 4.Msomi N, Mlisana K, de Oliveira T: A genomics network established to respond rapidly to public health threats in South Africa. Lancet Microbe. 2020;1(6):e229–30. 10.1016/S2666-5247(20)30116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegally H, Wilkinson E, Giovanetti M, et al. : Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv. 2020. 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- 6.du Plessis L, McCrone JT, Zarebski AE, et al. : Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371(6530):708–712. 10.1126/science.abf2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, du Plessis L, Liu Z, et al. : Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181(5):997–1003.e9. 10.1016/j.cell.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbe S, Buckland-Merrett G: Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall. 2017;1(1):33–46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Toole A: Accession IDs included in publication Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 [Data set]. Zenodo. 2021. 10.5281/zenodo.4642401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rambaut A, Holmes EC, O’Toole Á,, et al. : A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–7. 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Toole A: cov-lineages.org website. Zenodo. 2021. 10.5281/zenodo.4640140 [DOI] [Google Scholar]
- 12.O Toole A, Hill V: grinch. Zenodo. 2021. 10.5281/zenodo.4640037 [DOI] [Google Scholar]
- 13.Mölder F, Jablonski KP, Letcher B, et al. : Sustainable data analysis with Snakemake [version 1; peer review: 1 approved, 1 approved with reservations]. F1000Res. 2021;10:33. 10.12688/f1000research.29032.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reznick L: Using cron and crontab. Sys Admin. 1993;2(4):29–32. [Google Scholar]
- 15.O'Toole A: grinch_config.yaml [Data set]. Zenodo. 2021. 10.5281/zenodo.4640379 [DOI] [Google Scholar]
- 16.Bal A, Destras G, Gaymard A, et al. : Two-step strategy for the identification of SARS-CoV-2 variants co-occurring with spike deletion H69-V70, Lyon, France, August to December 2020. bioRxiv. 2020. 10.1101/2020.11.10.20228528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasell J, Mathieu E, Beltekian D, et al. : A cross-country database of COVID-19 testing. Sci Data. 2020;7(1):345. 10.1038/s41597-020-00688-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong E, Du H, Gardner L: An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–4. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Toole A: Supplementary materials with group affiliations for Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Zenodo. 2021. 10.5281/zenodo.4704471 [DOI] [PMC free article] [PubMed] [Google Scholar]