Highlights
-
•
Long non coding RNAs are involved in the regulation of multiple cellular processes.
-
•
Cancer stemness and escape from immunological anti-cancer mechanisms are important mechanisms of resistance to anti-cancer agents and are pivotal in controlling cancer development and metastases.
-
•
Long non coding RNAs have deep effects on the immune-modulation and on the control of cancer stem cells.
-
•
Several pathways involved in immunological escape and cancer stemness are modulated by long non coding RNAs.
-
•
Targeting long non coding RNAs is a potential new strategy to control tumor development and metastases.
Abstract
In recent years, it has been evidenced that the human transcriptome includes several types of non-coding RNAs (ncRNAs) that are mainly involved in the regulation of different cellular processes. Among ncRNAs, long-non-coding RNAs (lncRNAs) are defined as longer than 200 nucleotides and have been shown to be involved in several physiological and pathological events, including immune system regulation and cancer. Cancer stem cells (CSCs) are defined as a population of cancer cells that possess characteristics, such as resistance to standard treatments, cancer initiation, ability to undergo epithelial-to-mesenchymal transition, and the ability to invade, spread, and generate metastases. The cancer microenvironment, together with genetic and epigenetic factors, is fundamental for CSC maintenance and tumor growth and progression. Unsurprisingly, lncRNAs have been involved in both CSC biology and cancer progression, prognosis and recurrence. Here we review the most recent literature on IncRNAs involvement in CSC biology and function.
Graphical abstract
Introduction
Long non‐coding RNAs
The presence of a wide number of non-coding RNAs (ncRNAs) significantly contributes to the complexity of the human transcriptome. These molecules form a heterogeneous class of RNAs lacking protein-coding functions. NcRNAs play a key role in regulating transcription and post-transcriptional processes, including X chromosome inactivation, epigenetic regulation, genomic imprinting, and mRNA splicing [1]. According to their size, they can be divided into long (lncRNAs, >200 nt), and small ncRNAs (18–200 nt) [2]. Alternatively, they can be classified as housekeeping ncRNAs, such as ribosomal or transfer RNAs, and regulatory ncRNAs, including microRNAs (miRNAs) and lncRNAs. LncRNAs can be transcribed from intergenic, promoter regions, or antisense into annotated protein-coding genes [1]. Furthermore, lncRNAs can also be produced from transcriptional pseudogenes or mitochondrial genes [3]. Interestingly, several cancer genes, acting as oncosuppressors, generate long antisense ncRNAs [4]. Like mRNAs, most lncRNAs are transcribed by the RNA polymerase II and undergo 5′ capping, polyadenylation, and splicing, including the generation of different isoforms by alternative splicing. LncRNAs can be localized in the nucleus and the cytosol. The main function of lncRNAs is to regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels [5]. Although lncRNAs can belong to more than one archetype, four main molecular mechanisms (archetypes) have been described and by which lncRNAs exert their function. [6]. Firstly, archetype I lncRNAs can act as molecular signals that indicate certain transcriptional activities that are associated with their transcription at a specific location, time, or developmental stage, and in response to distinct stimuli. Secondly, lncRNAs, belonging to archetype II, function as decoys that bind and titrate away proteins, e.g., transcription factors, or miRNAs, thereby negatively regulating their effectors. Thirdly, archetype III lncRNAs act as guides that bind proteins and direct the ribonucleoprotein complex to target genes. Lastly, archetype IV lncRNAs can function as scaffolds that assemble multiple proteins in a complex, such as the stabilization of signaling complexes or direct histone modifications [6,7].
Cancer stem cells and resistance to cancer therapies
Aggressive cancers are characterized by their resistance to therapies, which result in cancer recurrence, poor prognosis, and reduced survival. The underlying cellular and molecular mechanisms of this aggressiveness are subject to intense investigations [8,9]. As part of these efforts, several studies formulated the hypothesis that cancer resistance and recurrence are associated with the existence of highly aggressive cancer cell populations [10,11]. These were termed cancer stem cells (CSCs) due to their abilities to self-renew and to generate differentiated cancer progenies, features that are associated with stemness. Consequently, CSCs can re-initiate, maintain, and promote cancer progression. How CSCs are generated is subject to debate; however, it is thought that oncogenic mutations that result in oncogenic transformations of embryonic and/or tissue-specific stem and progenitor cells are at the heart of CSC emergence [12], [13], [14].
Although cancer therapies efficiently destroy most cancer cells, CSCs escape their effects via several cellular mechanisms [11,15]. For instance, chemotherapies that target highly dividing cells, have limited effects on CSCs that are characterized by slow division or quiescence [16], [17], [18]. CSCs resistance to drugs is also associated with the expression of high levels of ATP-binding cassette (ABC) transporters. These membrane proteins mediate transmembrane transport of various substrates and contribute to the elimination of drugs from cells [18]. Another important mechanism of resistance is related to increased expression of aldehyde dehydrogenases (ALDHs) in CSCs. This superfamily of enzymes is involved in the detoxification of endogenous and exogenous aldehyde substrates by catalyzing the oxidation of aldehydes to carboxylic acids [19,20]. Several studies have shown that elevated levels of ALDH expression correlate with worse prognosis in cancer patients [21], [22], [23].
The resistance of CSCs to apoptosis has also been demonstrated by several studies [24]. For instance, glioma and leukemia CSCs resist extrinsic receptor-mediated cell death by downregulating Fas and Fas-L expression [25], [26], [27], [28], [29], [30]. They also express higher levels of apoptotic inhibitors, such as the cellular Fas‐associated death domain‐like IL‐1β‐converting enzyme‐inhibitory protein (c‐FLIP) [30], [31], [32], [33], [34]. Besides, high levels of expression of the pro-survival protein B‐cell lymphoma-2 have also been found in CSCs from different tumors [35], [36], [37], [38]. Finally, CSCs possess another survival advantage through their capacity to timely activate the DNA damage sensor and repair machinery [39]. CSC resistance to radiotherapy, which causes DNA damage, has been well documented in the literature [40,41]. All these observations suggest that CSCs possess an enhanced DNA damage repair ability that protects them from chemotherapy- and radiotherapy-mediated apoptosis.
CSC niche
The CSC microenvironment or niche is a complex hypoxic environment that comprises a variety of cells such as stromal, immune and epithelial cells, and a network of extracellular macromolecules that support cells within the extracellular matrix (ECM). This niche is essential for CSC-mediated cell invasion, metastasis, and chemoresistance [42]. Cancer-associated fibroblasts (CAFs) offer a mechanical supportive role for CSCs through fibrillary collagen production. However, this is not their only function within the CSC niche as they also secrete cytokines such as CXC chemokine ligand (CXCL)12 and growth factors, including vascular endothelial growth factor (VEGF), hepatocyte growth factor (HGF), and Platelet-Derived Growth factor (PDGF), which significantly promote CSC proliferation, invasion, and metastasis [43], [44], [45]. Furthermore, they also secrete transforming growth factor-beta 1 (TGF-β1), a key player in CSC-mediated epithelial-mesenchymal transition (EMT), that is at the origin of invasive and metastatic processes [46]. Endothelial cells (ECs), together with perivascular cells, are the building blocks of vessels, that ensure the constant supply of nutrients for CSCs metabolism and the recruitment of immune cells such as T regulatory cells (Tregs) that promote immune suppression [47]. ECs are stimulated by angiogenic factors, such as VEGF, which increases tumor vascularization, growth, and metastasis [48]. Besides their involvement in tumor angiogenesis, they also secrete cytokines such as interleukin (IL)−3, granulocyte-colony stimulating factor (G-CSF), granulocyte-macrophage-CSF (GM-CSF), IL-1, IL-6, VEGF-A, and basic fibroblast growth factor (bFGF) that promote and maintain CSC self-renewal, and CSC-mediated cancer progression [49], [50], [51]. The CSC niche is a highly inflammatory environment, that enhances tumor proliferation, invasion, and metastasis [47]. For instance, tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs) secrete TGF-β which contributes to EMT, invasion, and metastasis [52], [53], [54]. TAM-secreted TGF-β recruits Tregs to the CSC niche. MDSCs that secrete TGF-β, but also cytokines, recruit T helper 17 cells to promote their immunosuppressive activity [47]. Other important cellular components of the CSC niche are mesenchymal stem cells (MSCs). These multipotent stromal cells can differentiate into various cell types, including osteocytes, adipocytes, and chondrocytes, and migrate to chronic inflammatory sites such as cancer, where they secrete TGF-β and contribute to EMT and metastasis [55]. MSCs also secrete VEGF, macrophage inflammatory protein-2 (MIP-2), TGF-β1, and the pro-inflammatory cytokines IL-6, and IL-8, which contribute to the colonization process of secondary cancer sites in metastatic breast, prostate, gastric and lung cancers [56], [57], [58], [59], [60]. Finally, due to their multipotent nature, MSCs may be precursors of CAFs that further contribute to the cellular heterogenicity in the CSC microenvironment and its metastatic potential [61], [62], [63].
Cancer stemness, EMT, and tumor progression
CSCs can re-initiate tumorigenesis by generating a new tumor via their capacity to self-renew and to generate differentiated cancer progenies. However, their high invasive and metastatic potential involves a cellular process known as EMT (epithelial-mesenchymal transition). In this process, epithelial cells that are connected via lateral cell-cell contacts such as desmosomes, adherence, tight, and gap junctions, trans-differentiate into mesenchymal-like cells via the loss of these lateral junctions and expression of molecules associated with the mesenchymal phenotype [64]. The resulting cells, that are no longer bound to each other, can begin to invade adjacent tissues, which potentially leads to metastatic spread of cancer cells to distant organs. TGF-β that is secreted by MSCs, CAFs, and immune cells, is an essential player in CSC invasion mediated by EMT [65]. Upon activation of TGF-β receptors, a series of signaling cascades involving the downstream Smad factors are initiated and result in the transcriptional activation of target genes that are implicated in EMT such as SNAIL, bHLH and ZEB transcription factors [65,66]. Consequently, these events lead to the expression of mesenchymal proteins, such as N-cadherin, Fibronectin, and matrix metalloproteases (MMPs) that are involved in cancer cell invasion [66]. Although TGF-β signaling seems to play a leading role in EMT, other signaling pathways, such as the WNT, Notch, and Hedgehog (HH) pathways, also participate in EMT. For instance, WNT signaling via the β-catenin/glycogen synthase kinase-3β (GSK3β) pathway promotes EMT by enhancing the transcriptional activation of lymphoid enhancer-binding factor 1 (LEF1) and T cell factor (TCF), that are involved in gene expression programs that favor EMT [67]. Finally, sonic HH signaling has been shown to increase EMT and invasion by enhancing SNAIL1 expression in carcinomas, while Notch signaling promotes EMT through the Notch intracellular domain that directly activates SNAIL2 expression [68,69].
Involvement of lncRNAs in the CSC niche, chemoresistance, EMT, and metastasis
LncRNAs are aberrantly expressed in various human cancer types and play an important role in tumorigenesis, cancer progression, metastatic spread, and drug resistance [70], [71], [72]. A study that analyzed the expression of over 10,000 lncRNAs and in more than 1000 tumor samples across 4 cancer types, discovered that lncRNAs are associated with tumor subtypes and clinical prognoses [73], [74], [75]. Since lncRNAs are expressed in a tissue- and cancer-specific manners, they are considered potential diagnostic and prognostic biomarkers and promising therapeutic targets [70,76].
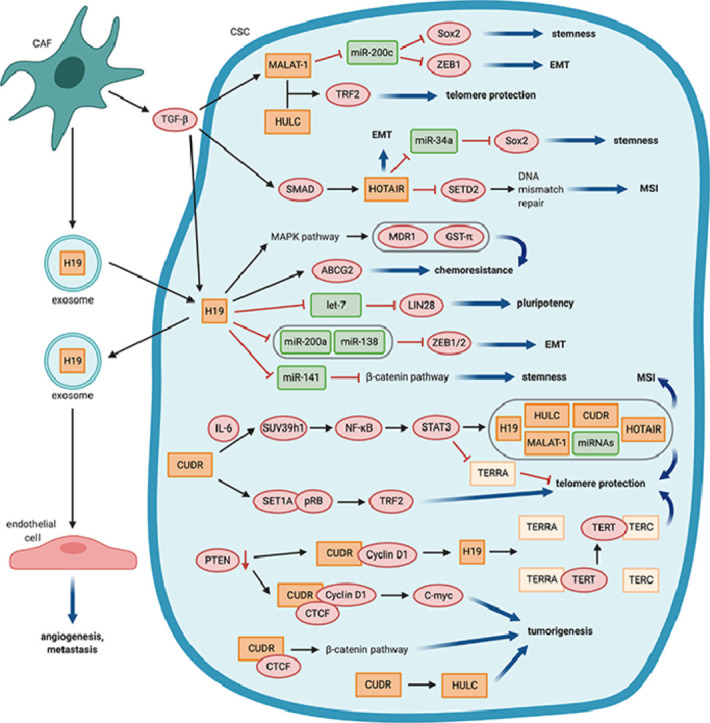
LncRNAs involvement in metastasis is attributed to the regulation of EMT which is intricately linked to CSCs. Recently, several lncRNAs were found to be involved in the WNT/β-Catenin, HH, Notch, and TGF-β signaling pathways, where they regulate the transcription of factors involved in stemness induction and maintenance, such as Sox2, NANOG, and Oct4, and in EMT, such as ZEB1 and ZEB2. In the following, we provide examples on the role of lncRNAs in the regulation of stemness, EMT, and CSC chemoresistance and on their influence on CSC interactions with the tumor microenvironment (Fig. 1, table 1).
Fig. 1.
LncRNAs control main CSC features, LncRNAs (orange) regulate e.g. stemness, epithelial-mesenchymal transition (EMT), chemoresistance and tumorigenesis via interactions with miRNAs (green), proteins (red) or other lncRNAs. In addition, they are involved in the communication of CSCs with the tumor microenvironment. EMT: Epithelial to mesenchymal transition, MSI: Microsatellite instability.
Table 1.
Key characteristics of CSCs are regulated by lncRNAs.
| Enhancement of … | Mechanism of action | LncRNAs | References |
| Stemness | Sponging miRNAs targeting Sox2 | - HOTAIR - MALAT-1 - Sox2ot - linc-DYNC2H1–4 - ROR |
82 (Deng 2017) 100–102 (Pa 2017, Zhuo 2018, Lu 2014) 130 (Li 2018) 129 (Gao 2017) 131 (Hou 2014) |
| Fatty acid oxidation | - MACC1-AS1 | 133 (He 2019) | |
| Pluripotency | Sponging miRNA targeting LIN28 | - H19 - linc-DYNC2H1–4 - ROR |
104, 106 (Peng 2017, Yu 2007) 129 (Gao 2017) New ref |
| EMT | Sponging miRNAs targeting ZEB1/2 | - MALAT-1 - H19 - XIST - lncATB - linc-DYNC2H1–4 - ROR |
99,101 (, Zhuo 2018) 107 (Liang 2015) 127 (Xu 2018) 128 (Yuan 2014) 129 (Gao 2017) 131 (Hou 2014) |
| Unknown | - HOTAIR | 84, 86 (Dou 2016, Lu 2017) | |
| Tumorigenesis | Telomere protection | - MALAT-1 + HULC - CUDR (+ IL-6) - CUDR-induced H19 |
103 (Lu 2014) 122 (Zheng 2017) 124 (Pu 2015) |
| Microsatellite instability | - HOTAIR - CUDR |
85 (Li 2015) 122 (Zheng 2017) |
|
| Chemoresistance | Upregulation of ABC transporters | - H19 | 111 (Bauderlique-Le Roy 2015) |
| Fatty acid oxidation | - MACC1-AS1 | 133 (He 2019) |
Hox transcript antisense intergenic RNA (HOTAIR)
The expression of the HOTAIR is associated with advanced tumor stage, metastasis, and poor prognosis in various human cancer types [77], [78], [79]. Its pro-metastatic function is attributed to HOTAIR-induced retargeting of the polycomb repressive complex 2 (PRC2) and epigenetic silencing of metastasis-repressor-genes [80,81]. HOTAIR expression was enhanced in enriched CSC populations of breast (MCF-7, MDA-MB-231, MCF-10a, and HCC1954) and colon (HT29, DLD1) cancer cell lines compared to non-CSC populations [82,83]. In different cancer stem cell types derived from cell lines such as breast (MCF-7, MDA-MB-231), colon (LoVo), liver (Huh7), ovarian cancer (SKOV3) and oral squamous cell carcinoma (Ca9–22, GNM), HOTAIR overexpression increased stemness- and EMT-associated characteristics, such as proliferation, colony formation, self-renewal, migration, invasion, expression of the pluripotent stem cell markers and E-cadherin downregulation in vitro [82,[84], [85], [86], [87]]. In vivo, tumorigenicity and metastasis were also enhanced [84,85,88,89]. Depending on the publication, CSCs were either enriched via sphere formation or via magnetic (MACS) or fluorescence-activated cell sorting (FACS) sorting of cells expressing cancer type-specific CSC markers, e.g. CD133 or CD44 [82], [83], [84], [85], [86], [87].
A few studies provided insights into the mechanism by which HOTAIR regulate the function of CSCs. In colon and breast CSCs, TGF-β1 that is secreted by CAFs, induced HOTAIR expression through Smad2/3/4, leading to increased EMT and metastasis of breast cancer cells in vivo [83,89]. In breast CSCs, the self-renewal ability was, at least partly, due to HOTAIR inhibition of miR-34a transcription, which targets Sox2, an important transcription factor involved in the maintenance of stem cell self-renewal [82]. HOTAIR also promoted tumorigenesis of liver CSCs by down-regulating SETD2, that ultimately led to impaired MMR and microsatellite instability [85].
These results showed how one of the most well-studied lncRNAs, HOTAIR, regulates stemness and EMT in CSCs, and provided insights into its role in CSCs-mediated metastasis.
Metastasis‐associated lung adenocarcinoma transcript‐1 (MALAT‐1)
Metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) is highly expressed in various human malignancies, where it enhances invasion and metastasis [90], [91], [92], [93], [94], [95]. MALAT-1 is also overexpressed glioma stem cells (SHG139S), and breast (MCF-7–derived), and pancreatic (e.g., CD133+ FACS-sorted CFPAC-1 and ASPC-1) CSCs. It promoted CSC characteristics, such as proliferation, self-renewal, colony formation, migration, and invasion in vitro [88,92,96,97] and enhanced tumorigenesis of pancreatic cancer cells in vivo [88]. MALAT-1 knockdown reduced the expression of stem cell markers, such as Sox2 [88,96,97], and drug resistance to gemcitabine [88]. Mechanistically, MALAT-1 may function as a sponge for miRNAs. For instance, by binding to miR-200c it increased the expression of ZEB1, an important EMT transcription factor [98], [99], [100], [101]. MiR-200c was also shown to target Sox2, which may explain the upregulation of Sox2 by MALAT-1 [102]. Sponging of miRNAs that regulate stemness and EMT is a common mode of action of lncRNAs in CSCs. Another interesting mechanism that has been described for MALAT-1 and that involves its cooperation with the lncRNA highly upregulated in liver cancer (HULC), has been shown to enhance the proliferation of liver CSCs in vitro and in vivo [103]. This was due to an increased expression and post-translational modification of telomere repeat-binding factor 2 (TRF2), which is an important protein for telomeres’ maintenance and protection. MALAT-1 and HULC formed a telomeric complex with TRF2, thereby protecting the telomeres. Also, TRF2 increased TERC expression and thus enhanced telomerase activity [103].
H19
H19 is strongly expressed by various cancer types, including breast and glioblastoma CSCs stem-like cells [104,105]. H19 was shown to be important for the maintenance of CSC characteristics in breast cancer cells in vitro and in vivo [104]. Mechanistically, it functioned as a competing endogenous RNA sponging miRNA let-7, which resulted in elevated levels of LIN28, a let-7 target, and an important pluripotency factor [104,106]. Besides, H19 acted as a molecular sponge for miR-200a, miR-138, and miR-141, which negatively regulated cancer stemness [107], [108], [109], [110]. In colorectal cancer cells, H19 sponging of miR-200a and miR-138 led to de-repression of ZEB1 and ZEB2 and thus EMT induction as it was measured by a reduced expression of the epithelial marker E-cadherin, an increased expression of the mesenchymal marker Vimentin, an enhanced migration in vitro and tumorigenesis in vivo [107]. Prostate cancer RWPE-1 stem-like cells, expressing higher H19 levels compared to the parent population, showed increased resistance to arsenic and higher mRNA levels of ABCG2, an ABC transporter conferring multi-drug resistance [111,112]. Moreover, in CD133+ FACS-sorted liver CSCs, H19 has been shown to activate the MAPK/ERK signaling pathway, which increased the expression of MDR1 and GST-π, thus likely conveying chemoresistance, while H19 silencing had the opposite effect [113].
Interestingly, H19 packaged into exosomes could be transferred from CAFs to SW480 and HCT116 colorectal cancer cells. H19 expression levels were measured in exosomes and cancer cells following cancer cells cultures with CAF-conditioned media, isolated exosomes from CAFs and CAFs [114]. In the recipient cells, H19 promoted stemness by repressing miR-141 and thereby activating the β-catenin pathway, and by enhancing resistance to oxaliplatin in vitro and in vivo [114]. Furthermore, CD90+ FACS-sorted Huh7 liver CSC-like cells were recently shown to secrete H19-containing exosomes, resulting in an increased angiogenic and pro-metastatic phenotype of endothelial cells in vitro [115].
CUDR and UCA1
Cancer up-regulated drug-resistant (CUDR) or Urothelial cancer associated 1 (UCA1) represent different isoforms of the same transcript and are upregulated in various cancers, including liver, colorectal, and bladder cancers [116], [117], [118]. Moreover, UCA1 was shown to enhance the chemoresistance of squamous carcinoma and bladder cancer cells to doxorubicin and etoposide, and cisplatin and gemcitabine, respectively, in vitro [119], [120], [121]. Zheng et al. showed that CUDR was able to trigger a malignant transformation of human embryonic stem cell-derived hepatocyte-like stem cells in cooperation with IL-6. CUDR and IL-6 increased the expression and mRNA stability of SUV39h1, a histone methyltransferase that catalyses histone 3 lysine 9 tri-methylation (H3K4me3). This histone mark promoted nuclear factor κB (NF-κB) expression and phosphorylation under inflammatory conditions, which in turn increased signal transducer and activator of transcription 3 (STAT3) expression and phosphorylation. Phosphorylated STAT3 bound to the promoters of miRNAs and lncRNAs, including CUDR, HOTAIR, MALAT-1, HULC, and H19, enhanced their expression, while also decreasing the expression of telomeric repeat-containing RNA (TERRA) [122], a non-coding RNA involved in maintaining telomeres’ structure [123]. The altered expression of these non-coding RNAs leads to increased telomerase activity, telomere length, and microsatellite instability [122]. The same research group also described a similar mechanism, by which a decrease in phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a well-known tumor suppressor gene, may promote the binding of CUDR to CyclinD1 in CD133+ CD44+ CD24+ EpCAM+ liver CSCs [124]. CUDR-CyclinD1 increased H19 expression by binding to its promoter region. Overexpressed H19 enhanced the interplay between TERT and TERC and decreased the binding of TERT to TERRA, which resulted in increased telomerase activity and telomere length [124]. Additionally, a complex consisting of CUDR, CyclinD1, and CTCF induced the overexpression of the C-myc oncogene [124]. This group also showed that CUDR increased HULC expression in human embryonic stem cells that were differentiated into hepatocyte-like cells by inhibiting the methylation of its promoter and enhanced β-catenin expression through the CUDR-CTCF complex. HULC and β-catenin activity were essential for the oncogenic activity of CUDR [125]. They also found that CUDR enhanced TRF2 expression via the SET1A-pRB complex, leading to telomere extension [126].
LncRNAs play a crucial role in the induction and maintenance of CSCs
LncRNAs play a crucial role in the induction and maintenance of CSCs in different cancer types. They influence stemness and EMT on many different levels, often by increasing Sox2 and ZEB1/2 expression levels. A well-described mechanism by which lncRNAs upregulate the expression of these transcription factors is by sponging miRNAs that target these same factors. In the above corresponding paragraphs, we provided some examples associated with the function of HOTAIR, MALAT-1, and H19. Furthermore, other lncRNAs, including XIST [127], lncATB [128], linc-DYNC2H1–4 [129], Sox2ot [130] and ROR [131] exert their function in a similar fashion. Another mode of action of lncRNAs is to promote the activation of signaling pathways that are important for CSCs, such as the activation of β-catenin signaling by H19, CUDR, and HOTTIP [132]. Moreover, lncRNAs exert an important role in telomere protection and elongation as described above for MALAT-1 and HULC and CUDR-induced H19. These examples also show that lncRNAs can regulate each other, such as in the case of CUDR, which induces the expression of various other lncRNAs or by direct interaction such as for MALAT-1 and HULC. lncRNAs can also influence the chemoresistance of CSCs. H19 and MACC1-AS1 promote chemoresistance by enhancing the expression of ABC transporters [133], while LET, MEG3, and GAS5 counteract chemoresistance [134], [135], [136]. Gemcitabine-induced TGF-β1 signaling in bladder cancer cells and xenografts decreased the levels of the lncRNA LET, resulting in low miR-145 levels and leading to enhanced stemness and gemcitabine resistance [134]. MiR-145, a known tumor suppressor, represses pluripotency, and reduces drug resistance by targeting MRP1 [137], [138], [139].
LncRNAs are also important mediators and target of the interaction of CSCs with the tumor microenvironment. The expressions of Several lncRNAs, including HOTAIR, MALAT-1 [90], H19 [140], MACC1-AS1 [133], lncATB [128], ZEB2NAT [46], and hPVT1 [141], are upregulated in cancer stem cells by TGF-β1. This latter can be secreted by various cell types in the tumor microenvironment, including CAFs, M2 macrophages, MDSCs, and cancer (stem) cells themselves [142]. It is known to induce stemness [143,144], EMT [54,144], and metastasis [145] in later stages of carcinogenesis. For example, Deng et al. showed that TGF-β1 that is secreted by mesenchymal stem cells contributes to the upregulation of MACC1-AS1 in gastric cancer cells, which enhances stemness and chemoresistance mediated by sponging miR-145–5p [133]. Also, lncRNAs, such H19 and Sox2ot, that are packaged into exosomes, facilitate the communications between CSCs and other cell types in the tumor microenvironment and can enhance EMT and stemness [130].
LncRNAs impact in clinical settings
Several lncRNAs have been found to be expressed in different types of cancer and these expressions correlated with patients’ diagnosis, prognosis, cancer risk and recurrence, and were identified as potential therapeutic targets [146]. Many of these processes are also associated with CSCs’ resistance, and they are associated with poor prognosis; they can also be dormant and develop new tumor recurrence after many years or generate metastases as described before.
Several lncRNAs that we have described to be involved in CSC biology are also associated with those clinical parameters. For example, increased H19 has been associated with reduced disease-free survival from first biopsy to first recurrence in bladder cancer patients. Similarly, increased MALAT-1 has been found to be an independent prognostic marker for recurrence in hepatocellular carcinoma in 112 HCC cases, while HOTAIR can predict recurrence in small-cell lung cancer, urothelial carcinoma, colon cancer, and cervical cancer [146]. MALAT- 1 was also associated with overall survival and advanced tumor stages in cervical cancer and recent data suggest its role as a prognostic marker in many cancer entities such as breast and multiple myeloma [147]. One more example is CCAT1, which increase has been associated with colon cancer patients' clinical stage (higher in stage III/IV than I/II), lymph node metastasis and survival time after surgery [148]. These represent only few examples of the impact of lncRNAs on cancer progression in patients and their prognosis. Based on this evidence, some strategies to target LncRNA has been developed at least in preclinical model. Among these, antisense oligonucleotide (ASO) represents a quite effective tool to be used in clinical settings. ASOs are short DNA sequences complementary to RNA of interest. The typical ASO design to target lncRNA is as a gapmer. This short oligonucleotides consist of RNA-based flanking sequences and an internal DNA “gap” region. Thys binds to complementary RNA and promote the degradation of the RNA/DNA heteroduplex by RNase. An example of this application is given by a gapmer designed to target MALAT-1. In a preclinical model of breast cancer where MALAT-1 is known to drive cancer progression and metastasis, two different ASO were administered resulting in reduced tumor growth, cyst formation and differentiation [149]. Similar results were obtained in a lung cancer model [150]. An additional technology that is very promising for the targeting of lncRNA is CRSPR/Cas9 as it can act on the promoter of specific lncRNA at the genomic level and can be also activated only in cancer cells. A genome-wide CRSPRi strategy evidenced the possibility to target as many as 16.000 lncRNA promoters [151]. Knockout of MALAT-1 and NEAT1 obtained by CRISPR technology inhibited the metastases in different preclinical models [152]. In an animal model of gastric cancer targeting of the lncRNA-GMAN by CRISPR technology suppress metastasis formation and dramatically increased overall survival.
The identification of novel cancer biomarkers and consequently the development of new therapeutic target candidates might lead to more personalized, more effective and less toxic treatments for cancer. In this context, a promising source of response-to-therapy markers is represented by LncRNAs. Among the ncRNAs, LncRNAs have been widely studied as cancer biomarkers because of their stability in formalin-fixed paraffin embedded (FFPE) tissues and in blood circulation. Altered LncRNA expression has been found in several types of cancers and contributes to therapy resistance. [146], [147] Moreover, LncRNAs are critical regulator of immune response and therefore a role as therapeutic targets and/or response predictors to immunotherapy in cancers could be expected. [153] Several studies have demonstrated the importance of lncRNAs as biomarkers to predict the efficacy of conventional chemotherapy. Due to the high stability in FFPE tissues, prognostic or predictive LncRNAs can be detected by using diagnostic biopsies as a source of cancer material; therefore, LncRNA analysis could be rapidly and easily performed during the standard diagnostics procedures. Recently, LncRNA detection in body fluids (e.g., serum and plasma) proved to be a scarcely invasive source of prognostic biomarkers. The opportunity to monitor specific ncRNA levels in serum or plasma in order to get an insight into the responsiveness of tumor cells, before starting a new therapeutic regimen or when the therapy is ongoing, is extremely appealing. Moreover, it is emerging the possibility to efficiently deliver RNA (including ncRNAs) in nanoparticles in vivo in human cells and the recent emergence of COVID-19 pandemic have pushed the scientists and pharmaceutical companies in developing vaccines based on the delivery of viral RNA in pegylated SNALPs. These strategies have been demonstrated to be safe and active and have disclosed a new scenario of intervention based on the therapeutic use of RNAs in human diseases including genetic illnesses and cancer.
How lncRNAs might additionally drive immune ignorance as a hallmark of cancer and cancer stem cells in particular
Immune escape is a hallmark of cancer and a prerequisite for cancer dissemination and progression [153]. Therefore, It is unsurprising, that cancer stem cells have been reported to orchestrate major immune suppressive mechanisms within the tumor microenvironment [154]. While individual mechanisms have been reported for a given cancer entity, it is unclear if these examples are proprietary to CSCs, which is more likely and universally applicable. For instance, in glioblastoma, the downregulation of the antigen-presentation machinery (MHC-I and -II) is reported to dampen immunogenicity and accelerate immune escape [155]. Similarly, immune checkpoint molecules, such as PD-L1 or Galectin-3, are strongly expressed by CSCs and suppress T cell activity [156], while CSCs release macrophage migration inhibitory factor (MIF) to promote arginase 1 expression and T cell suppression through MDSCs [157]. In colon cancer, the production of IL-4 from CSCs has been described to act both in an autocrine loop and to suppress T cell function [158]. In breast cancer, NK cell activatory ligands, MICA and MICB, are downregulated on CSCs, while PD-L1 and CD47 are upregulated to prevent T and myeloid cell activity [159,160]. Lastly, in pancreatic cancer, CSCs are an abundant source of TGF-β, which in addition to its autocrine functions, is an effective suppressor of T cell function [161]. This non-exhaustive list illustrates that immune suppression is an important part of the self-renewing potential of CSCs. It is highly likely, that more mechanisms that were previously described in cancer cells, apply to CSCs.
Under homeostatic and disease conditions, lncRNAs are important regulators of immune function [162]. Specifically, in cancer, different lncRNAs have been reported to boost or dampen immune function [163]. For example, ncRNA-RB1 promotes immunogenic cell death upon chemotherapy application through calreticulin exposure, thus promoting the induction of specific immune responses [164]. TP73-AS1 promotes HMGB1 production in hepatocellular carcinoma, and thereby, maintains tumor-driving inflammation [165]. Lnc-sox5 drives the production of IDO1 and subsequently T cell suppression and protection from specific immune responses [166]. AFAP1-AS1 is highly expressed in nasopharyngeal carcinoma and seems to correlate with PD-1 expression and T cell exhaustion [167]. Along these lines, the various mechanisms illustrate the relevance of lncRNAs in immune escape and different cancer types. For instance, it has been reported that lncRNA TUC339 can be released in exosomes from HCC cells and taken up by myeloid cells. Upon uptake, TUC339 dampens the pro-inflammatory functions of myeloid cells, such as antigen uptake, cytokine production, and migration [168]. As illustrated, CSCs are abundant sources of lncRNAs that regulate major hallmarks of CSCs. It is tempting to speculate that lncRNAs, packed in exosomes, might be used by CSCs to drive immune exhaustion and suppression in cancer. It is of course important to realize that this might not be the only mechanism by which CSCs can influence the immune system. This might also happen, as outlined above, via membrane-bound or even soluble factors. In other words, CSCs do recapitulate mechanisms usually ascribed to immune-privileged organs of the body to shield these from immune effector cells. Any successful immunotherapeutic strategy will need to obtain access and retain activity in the presence of CSCs for a successful therapeutic outcome.
Conclusions and outlook
CSCs significantly contribute to cancer resistance to treatments and cancer following therapy and lncRNAs are important mechanistic contributors to this process. Meanwhile, lncRNAs are significantly involved in the control of immune responses. While this review sheds light on the interplay of these processes, it is also important to stress, that a substantial number of contributions, is made by analogy from other systems. While it is likely that if a given lncRNA has a physiological immune regulatory role, this role will be conserved in the context of CSCs, but the formal proof is mostly lacking. With the rise of immunotherapies as part of the standard of care, there is a growing need to understand how CSCs, in general, and lncRNAs, in particular, might contribute to the primary and acquired resistance to immunotherapeutic modalities. This knowledge will constitute an important prerequisite for the design and development of more efficient immunotherapies.
CRediT authorship contribution statement
Melanie Schwerdtfeger: Investigation, Methodology, Software. Vincenzo Desiderio: Conceptualization, Investigation, Methodology. Sebastian Kobold: Conceptualization, Formal analysis, Funding acquisition, Resources, Supervision. Tarik Regad: Conceptualization, Investigation, Methodology, Project administration, Resources, Software, Supervision. Silvia Zappavigna: Formal analysis, Funding acquisition, Investigation, Methodology, Visualization. Michele Caraglia: Conceptualization, Investigation, Methodology, Supervision, Validation, Visualization.
Acknowledgments
SK is supported by grants from the international doctoral program “i-Target” funded by the Elite Network of Bavaria, the Melanoma Research Alliance (grant numbers N269626, 409510), the Marie-Sklodowska-Curie “Training Network for the Immunotherapy of Cancer (IMMUTRAIN)” and the Marie-Sklodowska-Curie “Training Network for Optimizing Adoptive T cell Therapy (T-OP), both funded by the H2020 program of the European Union, the Else Kröner-Fresenius-Stiftung, the German Cancer Aid, the Ernst-Jung-Stiftung, the Bundesministerium für Bildung und Forschung VIP+ grant ONKATTRACT, the European Research Council Starting Grant (grant number 756017), the Deutsche Forschungsgemeinschaft (DFG), the José Carreras Foundation, the Hector Foundation, and the Fritz-Bender Foundation. The figures were created with Biorender.com.
References
- 1.Mercer T., Mattick J. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2021;20:300–307. doi: 10.1038/nsmb.2480. n.d. [DOI] [PubMed] [Google Scholar]
- 2.Eidem T.M., Kugel J.F., Goodrich J.A. Noncoding RNAs: regulators of the Mammalian Transcription Machinery. J. Mol. Biol. 2016;428:2652–2659. doi: 10.1016/j.jmb.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rackham O., Shearwood A.M.J., Mercer T.R., Davies S.M.K., Mattick J.S., Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louro R., El-Jundi T., Nakaya H.I., Reis E.M., Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D., Xiong M., Xu C., Xiang P., Zhong X. Long noncoding RNAs: an overview. Methods Mol. Biol. 2016:287–295. doi: 10.1007/978-1-4939-3378-5_22. [DOI] [PubMed] [Google Scholar]
- 6.Wang K.C., Chang H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butova R., Vychytilova-Faltejskova P., Souckova A., Sevcikova S., Hajek R. Long non-coding RNAs in multiple myeloma. Noncoding RNA. 2019;5 doi: 10.3390/ncrna5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regad T. Tissue-specific cancer stem cells: reality or a mirage? Transl. Med. Reports. 2017:1. doi: 10.4081/tmr.6535. [DOI] [Google Scholar]
- 9.Gottesman M.M., Lavi O., Hall M.D., Gillet J.-.P. Toward a Better Understanding of the Complexity of Cancer Drug Resistance. Annu. Rev. Pharmacol. Toxicol. 2016;56:85–102. doi: 10.1146/annurev-pharmtox-010715-103111. [DOI] [PubMed] [Google Scholar]
- 10.Oskarsson T., Batlle E., Massagué J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J. Cancer stem cells and chemoresistance: the smartest survives the raid. Pharmacol. Ther. 2016;160:145–158. doi: 10.1016/j.pharmthera.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afify S.M., Seno M. Conversion of stem cells to cancer stem cells: undercurrent of cancer initiation. Cancers (Basel) 2019;11 doi: 10.3390/cancers11030345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atashzar M.R., Baharlou R., Karami J., Abdollahi H., Rezaei R., Pourramezan F., Zoljalali Moghaddam S.H. Cancer stem cells: a review from origin to therapeutic implications. J. Cell. Physiol. 2020;235:790–803. doi: 10.1002/jcp.29044. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Atkinson K., Zhang T. Combination of chemotherapy and cancer stem cell targeting agents: preclinical and clinical studies. Cancer Lett. 2017;396:103–109. doi: 10.1016/j.canlet.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Vidal S.J., Rodriguez-Bravo V., Galsky M., Cordon-Cardo C., Domingo-Domenech J. Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene. 2014;33:4451–4463. doi: 10.1038/onc.2013.411. [DOI] [PubMed] [Google Scholar]
- 17.Ajani J.A., Song S., Hochster H.S., Steinberg I.B. Cancer stem cells: the promise and the potential. Semin. Oncol. 2015;42:S3–S17. doi: 10.1053/j.seminoncol.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Abdullah L.N., Chow E.K.-H. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013;2 doi: 10.1186/2001-1326-2-3. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikawa M., Impraim C.C., Wang G., Yoshidas A. 1983. Isolation and Characterization of Aldehyde Dehydrogenase Isozymes from Usual and Atypical Human Livers*.http://www.jbc.org/ [PubMed] [Google Scholar]
- 20.Sládek N.E. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J. Biochem. Mol. Toxicol. 2003;17:7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 21.Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S., Schott A., Hayes D., Birnbaum D., Wicha M.S., Dontu G. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald T.L., Rangan S., Dobbs L., Starr S., Sigounas G. The impact of Aldehyde dehydrogenase 1 expression on prognosis for metastatic colon cancer. J. Surg. Res. 2014;192:82–89. doi: 10.1016/j.jss.2014.05.054. [DOI] [PubMed] [Google Scholar]
- 23.Vogler T., Kriegl L., Horst D., Engel J., Sagebiel S., Schäffauer A.J., Kirchner T., Jung A. The expression pattern of aldehyde dehydrogenase 1 (ALDH1) is an independent prognostic marker for low survival in colorectal tumors. Exp. Mol. Pathol. 2012;92:111–117. doi: 10.1016/j.yexmp.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Li T., Su Y., Mei Y., Leng Q., Leng B., Liu Z., Stass S.A., Jiang F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients outcome. Lab. Investig. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Scadden D.T. Harnessing the apoptotic programs in cancer stem-like cells. EMBO Rep. 2015;16:1084–1098. doi: 10.15252/embr.201439675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costello R.T., Mallet F., Gaugler B., Sainty D., Arnoulet C., Gastaut J.-.A., Olive D. Human Acute Myeloid Leukemia CD34 /CD38 Progenitor Cells Have Decreased Sensitivity to Chemotherapy and Fas-induced Apoptosis. Reduced Immunogenicity, and Impaired Dendritic Cell Transformation Capacities. 2000;1 [PubMed] [Google Scholar]
- 27.Weller M., Frei K., Groscurth P., Krammer P.H., Yonekawa Y., Fontana A. Anti-Fas/APO-1 antibody-mediated apoptosis of cultured human glioma cells: induction and modulation of sensitivity by cytokines. J. Clin. Invest. 1994;94:954–964. doi: 10.1172/JCI117462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frei K., Ambar B., Adachi N., Yonekawa Y., Fontana A. Ex vivo malignant glioma cells are sensitive to Fas (CD95/APO-1) ligand- mediated apoptosis. J. Neuroimmunol. 1998;87:105–113. doi: 10.1016/S0165-5728(98)00065-4. [DOI] [PubMed] [Google Scholar]
- 29.Tao J., Qiu B., Zhang D., Wang Y. Expression levels of Fas/Fas-L mRNA in human brain glioma stem cells. Mol. Med. Rep. 2012;5:1202–1206. doi: 10.3892/mmr.2012.791. [DOI] [PubMed] [Google Scholar]
- 30.G. Nter Eisele, P. Roth, K. Hasenbach, S. Aulwurm, F. Wolpert, G. Tabatabai, W. Wick, M. Weller, APO010, a Synthetic Hexameric CD95 ligand, Induces Human Glioma Cell Death in Vitro and in Vivo, (n.d.). doi:10.1093/neuonc/noq176. [DOI] [PMC free article] [PubMed]
- 31.Dong L.F., Freeman R., Liu J., Zobaiova R., Marin-Hernandez A., Stantic M., Rohlena J., Valis K., Rodriguez-Enriquez S., Butcher B., Goodwin J., Brunk U.T., Witting P.K., Moreno-Sanchez R., Scheffler I.E., Raiph S.J., Neuzil J. Suppression of tumor growth in vivo by the mitocan α-tocopheryl succinate requires respiratory complex II. Clin. Cancer Res. 2009;15:1593–1600. doi: 10.1158/1078-0432.CCR-08-2439. [DOI] [PubMed] [Google Scholar]
- 32.Zobalova R., McDermott L., Stantic M., Prokopova K., Dong L.F., Neuzil J. CD133-positive cells are resistant to TRAIL due to up-regulation of FLIP. Biochem. Biophys. Res. Commun. 2008;373:567–571. doi: 10.1016/j.bbrc.2008.06.073. [DOI] [PubMed] [Google Scholar]
- 33.Ding L., Yuan C., Wei F., Wang G., Zhang J., Bellail A.C., Zhang Z., Olson J.J., Hao C. Cisplatin restores TRAIL apoptotic pathway in glioblastoma-derived stem cells through up-regulation of DR5 and down-regulation of c-flip. Cancer Invest. 2011;29:511–520. doi: 10.3109/07357907.2011.605412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutton A., Young L.S., Murray P.G. The role of cellular flice inhibitory protein (c-FLIP) in the pathogenesis and treatment of cancer. Expert Opin. Ther. Targets. 2006;10:27–35. doi: 10.1517/14728222.10.1.27. [DOI] [PubMed] [Google Scholar]
- 35.Shirley S., Micheau O. Targeting c-FLIP in cancer. Cancer Lett. 2013;332:141–150. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Lauria F., Raspadori D., Rondelli D., Ventura M., Fiacchini M., Visani G., Forconi F., Tura S. High bcl-2 expression in acute myeloid leukemia cells correlates with CD34 positivity and complete remission rate. Leukemia. 1997;11:2075–2078. doi: 10.1038/sj.leu.2400854. [DOI] [PubMed] [Google Scholar]
- 37.Tóthová E., Fricova M., Stecová N., Kafková A., Elbertová A. High expression of Bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Neoplasma. 2002;49:141–144. [PubMed] [Google Scholar]
- 38.Sun Q., Wang Y., Desgrosellier J.S. Combined Bcl-2/Src inhibition synergize to deplete stem-like breast cancer cells. Cancer Lett. 2019;457:40–46. doi: 10.1016/j.canlet.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Campos L., Rouault J.P., Sabido O., Oriol P., Roubi N., Vasselon C., Archimbaud E., Magaud J.P., Guyotat D. High expression of bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Blood. 1993;81:3091–3096. doi: 10.1182/blood.v81.11.3091.3091. [DOI] [PubMed] [Google Scholar]
- 40.Abad E., Graifer D., Lyakhovich A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 2020;474:106–117. doi: 10.1016/j.canlet.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Tsao T., Beretov J., Ni J., Bai X., Bucci J., Graham P., Li Y. Cancer stem cells in prostate cancer radioresistance. Cancer Lett. 2019;465:94–104. doi: 10.1016/j.canlet.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Karimi-Busheri F., Rasouli-Nia A., Mackey J.R., Weinfeld M. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res. 2010;123:1–16. doi: 10.1186/bcr2583. 12 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaccio F., Paino F., Regad T., Papaccio G., Desiderio V., Tirino V. Concise Review: cancer Cells, Cancer Stem Cells, and Mesenchymal Stem Cells: influence in Cancer Development. Stem Cells Transl. Med. 2017;6:2115–2125. doi: 10.1002/sctm.17-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Junttila M.R., De Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 45.Plaks V., Kong N., Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuang J., Lu Q., Shen B., Huang X., Shen L., Zheng X., Huang R., Yan J., Guo H. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura T., Qian B.Z., Pollard J.W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hida K., Maishi N., Annan D.A., Hida Y. Contribution of tumor endothelial cells in cancer progression. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fessler E., Borovski T., Medema J.P. Endothelial cells induce cancer stem cell features in differentiated glioblastoma cells via bFGF. Mol. Cancer. 2015:14. doi: 10.1186/s12943-015-0420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.G P., JB N. Endothelium-derived Factors as Paracrine Mediators of Prostate Cancer Progression. Prostate. 2000;44 doi: 10.1002/1097-0045(20000615)44:1<77::aid-pros10>3.0.co;2-g. 10.1002/1097-0045(20000615)44:1<77::AID−PROS10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 51.Butler J.M., Kobayashi H., Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat. Rev. Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabarcas S.M., Mathews L.A., Farrar W.L. The cancer stem cell niche-there goes the neighborhood? Int. J. Cancer. 2011;129:2315–2327. doi: 10.1002/ijc.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buczek M.E., Miles A.K., Green W., Johnson C., Boocock D.J., Pockley A.G., Rees R.C., Hulman G., Van Schalkwyk G., Parkinson R., Hulman J., Powe D.G., Regad T. Cytoplasmic PML promotes TGF-β-associated epithelial-mesenchymal transition and invasion in prostate cancer. Oncogene. 2016;35:3465–3475. doi: 10.1038/onc.2015.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katsuno Y., Lamouille S., Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 55.Ridge S.M., Sullivan F.J., Glynn S.A. Mesenchymal stem cells: key players in cancer progression. Mol. Cancer. 2017;16:31. doi: 10.1186/s12943-017-0597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karnoub A.E., Dash A.B., Vo A.P., Sullivan A., Brooks M.W., Bell G.W., Richardson A.L., Polyak K., Tubo R., Weinberg R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 57.Corcoran K.E., Trzaska K.A., Fernandes H., Bryan M., Taborga M., Srinivas V., Packman K., Patel P.S., Rameshwar P. Mesenchymal stem cells in early entry of breast cancer into bone marrow. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nabha S.M., Dos Santos E.B., Yamamoto H.A., Belizi A., Dong Z., Meng H., Saliganan A., Sabbota A., Bonfil R.D., Cher M.L. Bone marrow stromal cells enhance prostate cancer cell invasion through type I collagen in an MMP-12 dependent manner. Int. J. Cancer. 2008;122:2482–2490. doi: 10.1002/ijc.23431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duda D.G., Duyverman A.M.M.J., Kohno M., Snuderl M., Steller E.J.A., Fukumura D., Jain R.K. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W., Zhou Y., Yang J., Zhang X., Zhang H., Zhang T., Zhao S., Zheng P., Huo J., Wu H. Gastric cancer-derived mesenchymal stem cells prompt gastric cancer progression through secretion of interleukin-8. J. Exp. Clin. Cancer Res. 2015;34 doi: 10.1186/s13046-015-0172-3. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaeth E.L., Dembinski J.L., Sasser A.K., Watson K., Klopp A., Hall B., Andreeff M., Marini F. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS ONE. 2009:4. doi: 10.1371/journal.pone.0004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra P.J., Mishra P.J., Humeniuk R., Medina D.J., Alexe G., Mesirov J.P., Ganesan S., Glod J.W., Banerjee D. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z., Peng Y., Li Z. GRP78 secreted by tumor cells stimulates differentiation of bone marrow mesenchymal stem cells to cancer-associated fibroblasts. Biochem. Biophys. Res. Commun. 2013;440:558–563. doi: 10.1016/j.bbrc.2013.09.108. [DOI] [PubMed] [Google Scholar]
- 64.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Padua D., Massagué J. Roles of TGFβ in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 66.Massagué J. TGFβ in Cancer, Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niehrs C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 68.Xie M., Zhang L., He C.-.S., Xu F., Liu J.-.L., Hu Z.-.H., Zhao L.-.P., Tian Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J. Cell. Biochem. 2012;113 doi: 10.1002/jcb.24019. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 69.Fendrich V., Waldmann J., Esni F., Ramaswamy A., Mullendore M., Buchholz M., Maitra A., Feldmann G. Snail and Sonic Hedgehog activation in neuroendocrine tumors of the ileum. Endocr. Relat. Cancer. 2007;14:865–874. doi: 10.1677/ERC-07-0108. [DOI] [PubMed] [Google Scholar]
- 70.Meseure D., Drak Alsibai K., Nicolas A., Bieche I., Morillon A., Alsibai K.D., Nicolas A., Bieche I., Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed. Res. Int. 2015 doi: 10.1155/2015/320214. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lv H., Lv G., Han Q., Yang W., Wang H. Noncoding RNAs in liver cancer stem cells: the big impact of little things. Cancer Lett. 2018;418:51–63. doi: 10.1016/j.canlet.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Yan H., Bu P. Non-coding RNAs in cancer stem cells. Cancer Lett. 2018;421:121–126. doi: 10.1016/j.canlet.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 73.Trimarchi T., Bilal E., Ntziachristos P., Fabbri G., Dalla-Favera R., Tsirigos A., Aifantis I. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D., Cao X., Jing X., Wang X., Siddiqui J., Wei J.T., Robinson D., Iyer H.K., Palanisamy N., Maher C.A., Chinnaiyan A.M. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du Z., Fei T., Verhaak R.G.W., Su Z., Zhang Y., Brown M., Chen Y., Liu X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malek E., Jagannathan S., Driscoll J.J. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–8038. doi: 10.18632/oncotarget.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Min S.N., Wei T., Wang X.T., Wu L.L., Yu G.Y. Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: a meta-analysis. Med. 2017;96:e7084. doi: 10.1097/MD.0000000000007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S., Chen S., Yang G., Gu F., Li M., Zhong B., Hu J., Hoffman A., Chen M. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qu X., Alsager S., Zhuo Y., Shan B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90–97. doi: 10.1016/j.canlet.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 80.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., Wang Y., Brzoska P., Kong B., Li R., West R.B., van de Vijver M.J., Sukumar S., Chang H.Y. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., Miyano S., Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 82.Deng J., Yang M., Jiang R., An N., Wang X., Liu B. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0170860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Padua Alves C., Fonseca A.S., Muys B.R., de Barros E.L.B.R., Burger M.C., de Souza J.E., Valente V., Zago M.A., J. Silva W A. Brief report: the lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31:2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 84.Dou J., Ni Y., He X., Wu D., Li M., Wu S., Zhang R., Guo M., Zhao F. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am. J. Transl. Res. 2016;8:98–108. https://www.ncbi.nlm.nih.gov/pubmed/27069543 [PMC free article] [PubMed] [Google Scholar]
- 85.Li H., An J., Wu M., Zheng Q., Gui X., Li T., Pu H., Lu D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6:27847–27864. doi: 10.18632/oncotarget.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu M.Y., Liao Y.W., Chen P.Y., Hsieh P.L., Fang C.Y., Wu C.Y., Yen M.L., Peng B.Y., Wang D.P., Cheng H.C., Wu C.Z., Shih Y.H., Wang D.J., Yu C.C., Tsai L.L. Targeting LncRNA HOTAIR suppresses cancer stemness and metastasis in oral carcinomas stem cells through modulation of EMT. Oncotarget. 2017;8:98542–98552. doi: 10.18632/oncotarget.21614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J., Chen D., He X., Zhang Y., Shi F., Wu D., Chen J., Zhang Y., Zhao F., Dou J. Downregulated lincRNA HOTAIR expression in ovarian cancer stem cells decreases its tumorgeniesis and metastasis by inhibiting epithelial-mesenchymal transition. Cancer Cell Int. 2015;15 doi: 10.1186/s12935-015-0174-4. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiao F., Hu H., Han T., Yuan C., Wang L., Jin Z., Guo Z., Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren Y., Jia H.H., Xu Y.Q., Zhou X., Zhao X.H., Wang Y.F., Song X., Zhu Z.Y., Sun T., Dou Y., Tian W.P., Zhao X.L., Kang C.S., Mei M. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol. Cancer. 2018;17 doi: 10.1186/s12943-018-0758-4. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 91.Gutschner T., Hammerle M., Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 92.Jiao F., Hu H., Yuan C., Wang L., Jiang W., Jin Z., Guo Z., Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 93.Schmidt L.H., Spieker T., Koschmieder S., Schaffers S., Humberg J., Jungen D., Bulk E., Hascher A., Wittmer D., Marra A., Hillejan L., Wiebe K., Berdel W.E., Wiewrodt R., Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 94.Wu X.S., Wang X.A., Wu W.G., Hu Y.P., Li M.L., Ding Q., Weng H., Shu Y.J., Liu T.Y., Jiang L., Cao Y., Bao R.F., Mu J.S., Tan Z.J., Tao F., Liu Y.B. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol. Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu Y., Zhang X., Hu X., Zhou W., Zhang P., Zhang J., Yang S., Liu Y. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol. Med. 2018;24 doi: 10.1186/s10020-018-0050-5. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han Y., Zhou L., Wu T., Huang Y., Cheng Z., Li X., Sun T., Zhou Y., Du Z. Downregulation of lncRNA-MALAT1 Affects Proliferation and the Expression of Stemness Markers in Glioma Stem Cell Line SHG139S. Cell. Mol. Neurobiol. 2016;36:1097–1107. doi: 10.1007/s10571-015-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeng L., Cen Y., Chen J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 2018;15:2117–2122. doi: 10.3892/ol.2017.7557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liang Z., Chen Y., Zhao Y., Xu C., Zhang A., Zhang Q., Wang D., He J., Hua W., Duan P. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell Res Ther. 2017;8:251. doi: 10.1186/s13287-017-0706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pa M., Naizaer G., Seyiti A., Kuerbang G. Long Noncoding RNA MALAT1 Functions as a Sponge of MiR-200c in Ovarian Cancer. Oncol. Res. 2017 doi: 10.3727/096504017X15049198963076. [DOI] [PubMed] [Google Scholar]
- 101.Zhuo M., Yuan C., Han T., Cui J., Jiao F., Wang L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer. 2018;18:1032. doi: 10.1186/s12885-018-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu Y.X., Yuan L., Xue X.L., Zhou M., Liu Y., Zhang C., Li J.P., Zheng L., Hong M., Li X.N. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin. Cancer Res. 2014;20:2631–2642. doi: 10.1158/1078-0432.CCR-13-2348. [DOI] [PubMed] [Google Scholar]
- 103.Wu M., Lin Z., Li X., Xin X., An J., Zheng Q., Yang Y., Lu D. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci. Rep. 2016;6:36045. doi: 10.1038/srep36045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng F., Li T.T., Wang K.L., Xiao G.Q., Wang J.H., Zhao H.D., Kang Z.J., Fan W.J., Zhu L.L., Li M., Cui B., Zheng F.M., Wang H.J., Lam E.W., Wang B., Xu J., Liu Q. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death. Dis. 2017;8:e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang X., Yan Y., Hu M., Chen X., Wang Y., Dai Y., Wu D., Wang Y., Zhuang Z., Xia H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016;124:129–136. doi: 10.3171/2014.12.JNS1426. [DOI] [PubMed] [Google Scholar]
- 106.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science (80-.) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 107.Liang W.C., Fu W.M., Wong C.W., Wang Y., Wang W.M., Hu G.X., Zhang L., Xiao L.J., Wan D.C., Zhang J.F., Waye M.M. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu C., Liu R., Zhang D., Deng Q., Liu B., Chao H.P., Rycaj K., Takata Y., Lin K., Lu Y., Zhong Y., Krolewski J., Shen J., Tang D.G. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017;8:14270. doi: 10.1038/ncomms14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu N., Zhong L., Zeng J., Zhang X., Yang Q., Liao D., Wang Y., Chen G., Wang Y. Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer. Neoplasma. 2015;62:550–559. doi: 10.4149/neo_2015_066. [DOI] [PubMed] [Google Scholar]
- 110.Yang Q., Wang X., Tang C., Chen X., He J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int. J. Oncol. 2017;50:1801–1809. doi: 10.3892/ijo.2017.3941. [DOI] [PubMed] [Google Scholar]
- 111.Bauderlique-Le Roy H., Vennin C., Brocqueville G., Spruyt N., Adriaenssens E., Bourette R.P. Enrichment of Human Stem-Like Prostate Cells with s-SHIP Promoter Activity Uncovers a Role in Stemness for the Long Noncoding RNA H19. Stem Cells Dev. 2015;24:1252–1262. doi: 10.1089/scd.2014.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.An Y., Ongkeko W.M. ABCG2: the key to chemoresistance in cancer stem cells? Expert Opin. Drug Metab. Toxicol. 2009;5:1529–1542. doi: 10.1517/17425250903228834. [DOI] [PubMed] [Google Scholar]
- 113.Ding K., Liao Y., Gong D., Zhao X., Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018;502:194–201. doi: 10.1016/j.bbrc.2018.05.143. [DOI] [PubMed] [Google Scholar]
- 114.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., Wang Y., Wang T., Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conigliaro A., Costa V., Lo Dico A., Saieva L., Buccheri S., Dieli F., Manno M., Raccosta S., Mancone C., Tripodi M., De Leo G., Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Han Y., Yang Y.N., Yuan H.H., Zhang T.T., Sui H., Wei X.L., Liu L., Huang P., Zhang W.J., Bai Y.X. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 117.Wang F., Ying H.Q., He B.S., Pan Y.Q., Deng Q.W., Sun H.L., Chen J., Liu X., Wang S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang X.S., Zhang Z., Wang H.C., Cai J.L., Xu Q.W., Li M.Q., Chen Y.C., Qian X.P., Lu T.J., Yu L.Z., Zhang Y., Xin D.Q., Na Y.Q., Chen W.F. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 119.Tsang W.P., Wong T.W., Cheung A.H., Co C.N., Kwok T.T. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;12:890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 121.Pan J., Li X., Wu W., Xue M., Hou H., Zhai W., Chen W. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382:64–76. doi: 10.1016/j.canlet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 122.Zheng Q., Lin Z., Li X., Xin X., Wu M., An J., Gui X., Li T., Pu H., Li H., Lu D. Inflammatory cytokine IL6 cooperates with CUDR to aggravate hepatocyte-like stem cells malignant transformation through NF-kappaB signaling. Sci. Rep. 2016;6:36843. doi: 10.1038/srep36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pu H., Zheng Q., Li H., Wu M., An J., Gui X., Li T., Lu D. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget. 2015;6:40775–40798. doi: 10.18632/oncotarget.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gui X., Li H., Li T., Pu H., Lu D. Long Noncoding RNA CUDR Regulates HULC and beta-Catenin to Govern Human Liver Stem Cell Malignant Differentiation. Mol. Ther. 2015;23:1843–1853. doi: 10.1038/mt.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li T., Zheng Q., An J., Wu M., Li H., Gui X., Pu H., Lu D. SET1A Cooperates With CUDR to Promote Liver Cancer Growth and Hepatocyte-like Stem Cell Malignant Transformation Epigenetically. Mol. Ther. 2016;24:261–275. doi: 10.1038/mt.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 127.Xu R., Zhu X., Chen F., Huang C., Ai K., Wu H., Zhang L., Zhao X. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. doi: 10.1186/s12935-018-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C., Wang S.B., Wang Y.Z., Yang Y., Yang N., Zhou W.P., Yang G.S., Sun S.H. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 129.Gao Y., Zhang Z., Li K., Gong L., Yang Q., Huang X., Hong C., Ding M., Yang H. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death. Dis. 2017;8:e2924. doi: 10.1038/cddis.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Z., Jiang P., Li J., Peng M., Zhao X., Zhang X., Chen K., Zhang Y., Liu H., Gan L., Bi H., Zhen P., Zhu J., Li X. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 131.Hou P., Zhao Y., Li Z., Yao R., Ma M., Gao Y., Zhao L., Zhang Y., Huang B., Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death. Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fu Z., Chen C., Zhou Q., Wang Y., Zhao Y., Zhao X., Li W., Zheng S., Ye H., Wang L., He Z., Lin Q., Li Z., Chen R. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 133.He W., Liang B., Wang C., Li S., Zhao Y., Huang Q., Liu Z., Yao Z., Wu Q., Liao W., Zhang S., Liu Y., Xiang Y., Liu J., Shi M. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019 doi: 10.1038/s41388-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhuang J., Shen L., Yang L., Huang X., Lu Q., Cui Y., Zheng X., Zhao X., Zhang D., Huang R., Guo H., Yan J. TGFbeta1 Promotes Gemcitabine Resistance through Regulating the LncRNA-LET/NF90/miR-145 Signaling Axis in Bladder Cancer. Theranostics. 2017;7:3053–3067. doi: 10.7150/thno.19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng R., Fan F.Y., Yi H., Liu F., He G.C., Sun H.P., Su Y. MEG3 affects the progression and chemoresistance of T-cell lymphoblastic lymphoma by suppressing epithelial-mesenchymal transition via the PI3K/mTOR pathway. J. Cell. Biochem. 2018 doi: 10.1002/jcb.28093. [DOI] [PubMed] [Google Scholar]
- 136.Liu B., Wu S., Ma J., Yan S., Xiao Z., Wan L., Zhang F., Shang M., Mao A. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 138.Zhan M., Zhao X., Wang H., Chen W., Xu S., Wang W., Shen H., Huang S., Wang J. miR-145 sensitizes gallbladder cancer to cisplatin by regulating multidrug resistance associated protein 1. Tumour Biol. 2016;37:10553–10562. doi: 10.1007/s13277-016-4957-6. [DOI] [PubMed] [Google Scholar]
- 139.Gao M., Miao L., Liu M., Li C., Yu C., Yan H., Yin Y., Wang Y., Qi X., Ren J. miR-145 sensitizes breast cancer to doxorubicin by targeting multidrug resistance-associated protein-1. Oncotarget. 2016;7:59714–59726. doi: 10.18632/oncotarget.10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matouk I.J., Raveh E., Abu-lail R., Mezan S., Gilon M., Gershtain E., Birman T., Gallula J., Schneider T., Barkali M., Richler C., Fellig Y., Sorin V., Hubert A., Hochberg A., Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 141.Wang F., Yuan J.H., Wang S.B., Yang F., Yuan S.X., Ye C., Yang N., Zhou W.P., Li W.L., Li W., Sun S.H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 142.Ahmadi A., Najafi M., Farhood B., Mortezaee K. Transforming growth factor-beta signaling: tumorigenesis and targeting for cancer therapy. J. Cell. Physiol. 2018 doi: 10.1002/jcp.27955. [DOI] [PubMed] [Google Scholar]
- 143.Chaffer C.L., Marjanovic N.D., Lee T., Bell G., Kleer C.G., Reinhardt F., D’Alessio A.C., Young R.A., Weinberg R.A. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Penuelas S., Anido J., Prieto-Sanchez R.M., Folch G., Barba I., Cuartas I., Garcia-Dorado D., Poca M.A., Sahuquillo J., Baselga J., Seoane J. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 145.Heldin C.H., Vanlandewijck M., Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 146.Gupta S.Chandra, Tripathi Y.Nandan. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int. J. Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 147.Wang Y., Zhang Y., Hu K., Qiu J., Hu Y., Zhou M., Zhang S. Elevated long noncoding RNA MALAT-1 expression is predictive of poor prognosis in patients with breast cancer: a meta-analysis. Biosci. Rep. 2020:40. doi: 10.1042/BSR20200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.He X., Tan X., Wang X., Jin H., Liu L., Ma L., Yu H., Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 149.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L., Brogi E., Egeblad M., Spector D.L. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gutschner T., Hämmerle M., Eißmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Groß M., Zörnig M., MacLeod A.R., Spector D.L., Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y., Mandegar M.A., Olvera M.P., Gilbert L.A., Conklin B.R., Chang H.Y., Weissman J.S., Lim D.A. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science (80-.) 2017:355. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jiang M.-.C., Ni J.-.J., Cui W.-.Y., Wang B.-.Y., Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019;9:1354–1366. http://www.ncbi.nlm.nih.gov/pubmed/31392074 (accessed April 27, 2021) [PMC free article] [PubMed] [Google Scholar]
- 153.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 154.Badrinath N., Yoo S.Y. Recent Advances in Cancer Stem Cell-Targeted Immunotherapy. Cancers (Basel) 2019;11 doi: 10.3390/cancers11030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Di Tomaso T., Mazzoleni S., Wang E., Sovena G., Clavenna D., Franzin A., Mortini P., Ferrone S., Doglioni C., Marincola F.M., Galli R., Parmiani G., Maccalli C. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res. 2010;16:800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei J., Barr J., Kong L.Y., Wang Y., Wu A., Sharma A.K., Gumin J., Henry V., Colman H., Sawaya R., Lang F.F., Heimberger A.B. Glioma-associated cancer-initiating cells induce immunosuppression. Clin. Cancer Res. 2010;16:461–473. doi: 10.1158/1078-0432.CCR-09-1983. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 157.Otvos B., Silver D.J., Mulkearns-Hubert E.E., Alvarado A.G., Turaga S.M., Sorensen M.D., Rayman P., Flavahan W.A., Hale J.S., Stoltz K., Sinyuk M., Wu Q., Jarrar A., Kim S.H., Fox P.L., Nakano I., Rich J.N., Ransohoff R.M., Finke J., Kristensen B.W., Vogelbaum M.A., Lathia J.D. Cancer Stem Cell-Secreted Macrophage Migration Inhibitory Factor Stimulates Myeloid Derived Suppressor Cell Function and Facilitates Glioblastoma Immune Evasion. Stem Cells. 2016;34:2026–2039. doi: 10.1002/stem.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Volonte A., Di Tomaso T., Spinelli M., Todaro M., Sanvito F., Albarello L., Bissolati M., Ghirardelli L., Orsenigo E., Ferrone S., Doglioni C., Stassi G., Dellabona P., Staudacher C., Parmiani G., Maccalli C. Cancer-initiating cells from colorectal cancer patients escape from T cell-mediated immunosurveillance in vitro through membrane-bound IL-4. J. Immunol. 2014;192:523–532. doi: 10.4049/jimmunol.1301342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang B., Wang Q., Wang Z., Jiang J., Yu S.C., Ping Y.F., Yang J., Xu S.L., Ye X.Z., Xu C., Yang L., Qian C., Wang J.M., Cui Y.H., Zhang X., Bian X.W. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014;74:5746–5757. doi: 10.1158/0008-5472.CAN-13-2563. [DOI] [PubMed] [Google Scholar]
- 160.Zhang H., Lu H., Xiang L., Bullen J.W., Zhang C., Samanta D., Gilkes D.M., He J., Semenza G.L. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci U S A. 2015;112:E6215–E6223. doi: 10.1073/pnas.1520032112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 162.Fitzgerald K.A., Caffrey D.R. Long noncoding RNAs in innate and adaptive immunity. Curr. Opin. Immunol. 2014;26:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu W.D., Wang H., He Q.F., Xu Y., Wang X.C. Long noncoding RNAs in cancer-immunity cycle. J. Cell. Physiol. 2018;233:6518–6523. doi: 10.1002/jcp.26568. [DOI] [PubMed] [Google Scholar]
- 164.Musahl A.S., Huang X., Rusakiewicz S., Ntini E., Marsico A., Kroemer G., Kepp O., Orom U.A. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene. 2015;34:5046–5054. doi: 10.1038/onc.2014.424. [DOI] [PubMed] [Google Scholar]
- 165.Li S., Huang Y., Huang Y., Fu Y., Tang D., Kang R., Zhou R., Fan X.G. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J. Exp. Clin. Cancer Res. 2017;36:51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu K., Zhao Z., Liu K., Zhang J., Li G., Wang L. Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell Cycle. 2017;16:1295–1301. doi: 10.1080/15384101.2017.1317416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tang Y., He Y., Shi L., Yang L., Wang J., Lian Y., Fan C., Zhang P., Guo C., Zhang S., Gong Z., Li X., Xiong F., Li X., Li Y., Li G., Xiong W., Zeng Z. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001–39011. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li X., Lei Y., Wu M., Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19102958. [DOI] [PMC free article] [PubMed] [Google Scholar]