Highlights
-
•
The study first reports the regulation of LINC01215 on methylation of RUNX3 promoter.
-
•
LINC01215 is highly expressed while RUNX3 is reciprocal in EOC.
-
•
LINC01215 overexpression promotes methylation of RUNX3 and reduces its expression.
-
•
LINC01215 silencing suppresses LNM and EMT of EOC.
-
•
This study may provide a new therapeutic target for EOC.
Keywords: Long non-coding RNA LINC01215, Runt-related transcription factor 3, Epithelial ovarian cancer, Epithelial-mesenchymal transition, Lymph node metastasis, Invasion, Metastasis
Abstract
Epithelial ovarian cancer (EOC) still remains the most lethal gynaecological malignancy in women, despite the recent progress in the management, including surgery and chemotherapy. According to the microarray data of the GSE18520 and GSE54388 datasets, LINC01215 was identified as an upregulated long noncoding RNA (lncRNA) in EOC. Therefore, this study aimed to figure out the involvement of LINC01215 in the progression of EOC. RT-qPCR was conducted to select the EOC cell line with the highest expression of LINC01215. Methylation of RUNX3 was then examined in EOC cells by MS-PCR. Furthermore, the interaction between LINC01215 and methylation-related proteins was revealed according to the results of RIP and RNA pull down assays. Subsequently, the involvement of LINC01215 and RUNX3 in regulating biological behaviors of EOC cells was investigated. Finally, the effects of the ectopic expression of LINC01215 and RUNX3 on the tumor formation and lymph node metastasis (LNM) of EOC cells were assessed in the xenograft tumors of nude mice. Overexpressing LINC01215 contributed to downregulated levels of RUNX3, as demonstrated by the recruitment of methylation-related proteins. Silencing of LINC01215 elevated the expression of RUNX3, thus suppressing cell proliferation, migration, invasion and EMT and decreasing the expressions of MMP-2, MMP-9 and Vimentin, but increased the expression of E-cadherin. The tumor growth and LNM were suppressed by downregulated levels of LINC01215 through inducing the expression of RUNX3. Collectively, the down-regulating LINC01215 could upregulate the expression of RUNX3 by promoting its methylation, thus suppressing EOC cell proliferation, migration and invasion, EMT, tumor growth and LNM.
Introduction
Ovarian cancer is the most common fatal cancer in women with the highest mortality rate among all gynecologic malignancies. Specifically, epithelial ovarian cancer (EOC) ranks the top 5 of the most common type of cancers [1,2]. Some advanced methods of early detection have been used for diagnosis and long-term cancer survival rate is relatively satisfactory; however, a five-year survival rate of 10 - 30% still exists among patients with ovarian cancer with presenting findings at thee advanced stage [3]. Additionally, although the outcome of primary surgery and chemotherapy is optimistic for patients diagnosed with early-stage ovarian cancer, the recurrence commonly arises in patients at late stages and available salvaging therapies have been shown to be ineffective [4]. There are a larger majority of factors that contribute to the poor prognosis as well as recurrence of EOC, such as lack of effective markers for diagnosis or prognosis and treatment protocols [5]. Therefore, it is imperative to identify the regulatory mechanisms on EOC progression and develop new therapeutic targets for EOC treatment.
Long non-coding RNAs (lncRNAs), a novel class of noncoding RNAs that are longer than 200 nucleotides without protein-coding potential, play key regulatory roles in carcinogenesis and cancer metastasis [6,7]. Aberrant expression of lncRNAs has been noted in gastric cancer (GC), hepatocellular carcinoma and renal clear cell carcinoma [8], [9], [10]. Poor prognosis and promotion of tumor metastasis caused by the overexpression of particular lncRNAs in EOC have been reported by several studies [11], [12], [13]. Runt-related transcription factor 3 (RUNX3) is a classical tumor suppressor gene for GC, whose reduced expression level is associated with decreased survival rates of patients with hepatocellular carcinoma [14]. A higher RUNX3 gene promoter hyper-methylation rate is found in GC tumor tissues compared with that in the normal tissues [15]. Promotion of the methylation of the CpG islands in the promoter region of RUNX3 has been suggested to downregulate the expression of RUNX3 [16]. Moreover, the repression of RUNX3 modulated by hyper-methylation is closely linked to unsatisfactory clinical outcomes in Type II EOC [17]. Therefore, we hypothesized that LINC01215 is involved in the progression of EOC, especially epithelial-mesenchymal transition (EMT) and lymph node metastasis (LNM), and the mechanisms by which LINC01215 controlled the expression of RUNX3 were explored in the present study.
Materials and methods
Ethics statement
The study was conducted after the approval of the Ethics Committee of The Second Affiliated Hospital of Harbin Medical University (ethical approval number: SYDW2021-037). All experimental procedures were carried out in strict accordance with the Guide for Care and Use of Laboratory Animals provided by the US National Institute of Health.
Microarray-based gene expression profiling analysis
Ovarian cancer-related gene expression datasets (GSE18520 and GSE54388), as well as probe and annotation files, were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). GSE18520 dataset includes 10 normal samples and 53 ovarian cancer samples [18]. The GSE54388 dataset includes 6 normal samples and 16 ovarian cancer samples [19]. Affy package of the R software was used for background correction and normalization for the two datasets [20]. We then employed cluster analysis to investigate the change of gene expression patterns between the normal samples and ovarian cancer samples using ‘edgeR’ package of the R software [21]. Next, non-specific filtration for the relevant gene expression data was performed to screen out differentially expressed mRNAs using a combination of linear models-empirical Bayes in the Limma package and conventional t-test [22].
Cell culture
The ovarian cancer cell lines A2780, HO8910, SKOV-3, ES-2 and HO8910pm as well as normal ovarian cells IOSE80 were purchased from Shanghai Suer Biotech Co., Ltd., (Shanghai, China). After cell resuscitation, A2780, HO8910, SKOV-3 and normal ovarian cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco) in an incubator (Thermo Fisher Scientific Inc., Waltham, MA, USA) with a saturated humidity atmosphere of 5% CO2 at 37 °C. When cells reached a confluence of 80% with good adherence, cells were detached with 0.25% trypsin and subcultured at a ratio of 1: 3. Then cells in the logarithmic growth phase were collected for subsequent experiments.
Cell grouping and transfection
The HO8910pm cells in the logarithmic growth phase were selected for transfection. The cells were assigned into 6 groups: blank (cells without any treatment), negative control (NC) (cells transfected with empty plasmids), overexpressed (oe)LINC01215 (cells transfected with LINC01215 overexpressed plasmids), short hairpin RNA (sh)LINC01215 (cells transfected with LINC01215 interference plasmids), shRUNX3 (cells transfected with RUNX3 interference plasmids), and oeLINC01215 + shRUNX3 (cells co-transfected with LINC01215 overexpressed plasmids and RUNX3 interference plasmids) groups. The HO8910pm cells were seeded in a 12-well plate at a density of 1 × 105 cells/mL. When cells reached a confluence of 50–70%, 800 μL serum-free medium was added into the wells. Next, oeLINC01215, shLINC01215, shRUNX3 or oeLINC01215 + shRUNX3 was mixed with lipo2000 solution (11668027, Thermo Fisher Scientific), and the mixture was added into the 12-well plate. The medium was replaced with a fresh new medium after being cultured for 6 h. Cells were collected after being left to transfect for 48 h. RNA and protein contents were extracted for subsequent experiments.
RNA-binding protein immunoprecipitation (RIP) assay
The binding ability of LINC01215 to DNA methyltransferase 1 (DNMT1), DNMT3A and DNMT3B proteins was detected using an RIP kit (Millipore, MA, USA). The HO8910pm cells were collected and washed with pre-cooled PBS, and the supernatant was discarded. The cells were later left to lyse with an equal volume of radioimmunoprecipitation (RIPA) lysis buffer (P0013B, Beyotime Biotechnology Co., Shanghai, China) in an ice bath for 5 min, and centrifuged at 4°C (14000 rpm) for 10 min, and the supernatant was extracted. A portion of the cell extract served as the Input Control, and the other portion was incubated with antibodies for co-precipitation. 50 μL of beads, which were re-suspended in 100 μL RIP wash buffer, were incubated with 5 μg corresponding antibodies for 30 min at room temperature. The following antibodies: rabbit anti-human DNMT1 (1: 100, ab13537), rabbit anti-human DNMT3A (1: 100, ab2850) and rabbit anti-human DNMT3B (1: 100, ab2851). Rabbit anti-human IgG (1: 100, ab109489) were used as the negative control. Aforementioned antibodies were purchased from Abcam (Cambridge, MA, USA). The magnetic bead-antibody complex was washed, re-suspended in 900 μL RIP wash buffer and incubated with 100 μL cell lysate at 4 °C overnight. The samples were then placed on magnetic pedestals in order to collect magnetic beads-protein complexes. After that, samples and Inputs were detached using proteinase K buffer to extract RNA for reverse transcription quantitative polymerase chain reaction (RT-qPCR).
RNA pull-down assay
According to the instructions provided by the Magnetic RNA-Protein Pull-Down Kit (Pierce, Rockford, IL, USA), 1 μg biotin-labeled RNA was mixed with 500 μL structure buffer, incubated at 95 °C for 2 min, and then ice-bathed for 3 min. 50 μL magnetic beads suspension was added and incubated overnight at 4 °C. After the supernatant was centrifuged at 3000 rpm for 3 min, it was discarded. Subsequently, the precipitate was rinsed thrice with 500 μL RIP wash buffer, and incubated with 10 μL cell lysate at room temperature for 1 h. The bead-RNA-protein mixture was centrifuged at a low speed with the supernatant recycled, and washed 3 times with 500 μL RIP wash buffer. Next, 10 μL cell lysate supernatant was used as the protein Input. After protein concentration was measured, western blot analysis was performed to determine the level of protein expression.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 Kit (WH1199, Shanghai Well Biotech Co., Ltd., Shanghai, China) was applied for cell proliferation detection. The HO8910pm cells were seeded in a 96-well plate with 5 × 103 cells/100 μL per well. After incubation for 24, 48 and 72 h, the supernatant was discarded and 10 μL CCK-8 solution was added to each well, followed by being incubated for an additional 2 h at 37 °C. Next, the optical density (OD) value at the wavelength of 450 nm was measured using a Multiskan FC microplate reader (51119080, Thermo Fisher Scientific). The proliferation rate (%) = [(OD control group - OD experimental group)/OD control group] × 100%. Three replicates were set for each well, and the mean value was calculated. The experiment was repeated 3 times.
Scratch test
Horizontal-lines across the well behind 6-well plates were drawn with a marker pen (interval: 0.8 cm, at least 5 lines across each well). The HO8910pm cells were seeded in a 6-well plate, and the original medium was replaced with 3 mL medium containing 10% FBS and antibiotic. After cells were cultured for 4 h, they were further cultured in complete medium containing serum for 24 h. The scratches through the cell monolayer were vertically made using a sterile micropipette tip (200 µL). The plate was subsequently washed 3 times with PBS to remove the exfoliated cells. Finally, cells were incubated in a serum-free medium at 37 °C with 5% CO2. Images were acquired at the 0th hour and the 24th hour using a microscope to determine the cell migration distance. The experiment was repeated 3 times.
Transwell assay
Matrigel (Becton, Dickinson and Company, NJ, USA) was diluted with pre-cooled serum-free medium at a ratio of 1: 1 and added to the apical chambers of Transwell (Corning Glass Works, Corning, NY, USA) (50 µL/well) and incubated at 37 °C to polymerize Matrigel into gel. The cells of each group were deprived of nutrients with a medium containing 1% FBS for 24 h, then harvested and resuspended in serum-free medium with a density of 5 × 106 cells/mL. 100 μL cell suspension was placed in the apical chambers, while 800 μL cell culture medium containing 10% FBS was added to the basolateral chambers. Subsequently, the cells were incubated at 37 °C with 5% CO2 for 24 h. Afterwards, cells that failed to migrate were gently wiped off with a cotton swab, while the migrated cells were fixed with 4% paraformaldehyde, stained with crystal violet, and immersed in water to return blue in color. The stained migrated cells were photographed and counted under an inverted microscope (× 100) by randomly selecting 5 visual fields according to the five-spot sampling method. The mean value was obtained and the experiment was repeated 3 times.
Flow cytometry
After HO8910pm cells were transfected of 48 h, they were detached with ethylene diamine tetraacetic acid (EDTA)-free trypsin and collected. After washing with precooled PBS for three times, cells were centrifuged again with the supernatant discarded. Annexin-V-FITC, propidium iodide (PI) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer were used to prepare Annexin-V-FITC/PI dye with the proportion of 1: 2: 50 by following the instructions of Annexin-V-fluorescein isothiocyanate (FITC) cell apoptosis kit (C1065, Beyotime Biotechnology Co., Shanghai, China). 100 μL Annexin-V-FITC/PI dye was used to re-suspend 1 × 106 cells, followed by oscillating and mixing. After incubation for 15 min at room temperature, another 1 mL HEPES buffer was added and fully mixed. Finally, cell apoptosis was detected by flow cytometry at the excitation wavelengths of 525 nm and 620 nm.
RT-qPCR
Total RNA content was extracted from cells by following through the instructions of Trizol (Invitrogen, Carlsbad, CA, USA). The primers of LINC01215, RUNX3, matrix metalloprotein-2 (MMP-2), MMP-9, E-cadherin, and Vimentin were designed and synthesized by AuGCT Biotechnology Co., Ltd., (Beijing, China) (Table S1). Afterwards, RNA was reversely transcribed into cDNA in a PCR amplifier (T100, Bio-Rad Laboratories, Shanghai, China). The StepOnePlus qPCR instrument (StepOne Plus, ABI Company, Oyster Bay, NY, USA) and SYBR Green fluorescent dye method (163795-75-3, Shanghai JiNing ShiYe Co., Ltd., Shanghai, China) were applied for RT-qPCR. U6 was used as the internal reference for LINC01215 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference for the other target genes. The relative expression of the target gene was calculated using the 2-ΔΔCt method [23].
Methylation-specific PCR (MS-PCR)
The primers for RUNX3 methylation and for RUNX3 non-methylation were designed and synthesized by AuGCT Biotechnology Co., (Beijing, China) (Table S2). After HO8910pm cells were detached with proteinase K, the cellular DNA was extracted with chloroform and its concentration was measured with a spectrophotometer. The DNA in the lymphocyte cells served as a positive control. The above extracted DNA was treated with sodium bisulfite to obtain sulfurized DNA. MS-PCR reaction system (25 μL) consisted of 12.5 μL 2 × FMSP buffer, 0.5 μL forward primers, 0.5 μL reverse primers, 2 μL sulfurized DNA and 9.5 μL ddH2O. Reaction conditions included pre-denaturation at 95°C for 15 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s and finally at 72°C for 10 min. The PCR products were subjected to agarose gel electrophoresis and the bands were visualized under ultraviolet light.
Western blot analysis
Total protein content was extracted from cells and separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After that, the protein was transferred onto the nitrocellulose membrane (LC2005, Thermo Fisher Scientific). The membrane was blocked in 5% skim milk powder at room temperature for 2 h and washed 3 times with Tris-buffered saline-Tween (TBST) (10 min each time). Next, the membrane was incubated with the following primary rabbit anti-human polyclonal antibodies at 4°C overnight: RUNX3 (ab92336, 1:5000), MMP-2 (ab92536, 1:1000), MMP-9 (ab73734, 1:1000), E-cadherin (ab40772, 1:10000), Vimentin (ab92547, 1:1000), DNMT1 (ab13537, 1:1000), DNMT3A (ab2850, 1:1000), DNMT3B (ab2851, 1:1000) and GAPDH (ab181602, 1:5000). The membrane was incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (ab97051) at room temperature for 1 hour after being washed with TBST 3 times at room temperature (5 min each time). All above-mentioned antibodies were purchased from Abcam. After three rinses with TBST on a shaking table at room temperature, 5 min per time, the membrane was immersed in enhanced chemiluminescence (ECL) reaction solution (BM101, Biomiga, San Diego, CA, USA) at room temperature for 1 minute. Finally, the membrane was exposed to X-ray in the dark to develop final fixation results. GAPDH was used as an internal protein reference and the ratio of the gray value of the target band to that of the internal reference band was used as the relative expression of the protein.
Xenograft tumor in nude mice
A total of 30 female nude mice (BALB/c-nu/nu) in specific pathogen-free (SPF) grade (aged 4 weeks old and weighing 14 - 16 grams) were purchased from the Institute of Laboratory Animals in Chinese Academy of Medical Sciences (Shanghai, China). The nude mice were randomly assigned into 6 groups (n = 5): blank, NC, oeLINC01215, shLINC01215, shRUNX3 and oeLINC01215 + shRUNX3 groups (mice injected with cells treated with blank, NC, oeLINC01215, shLINC01215, shRUNX3 and both oeLINC01215 and shRUNX3 plasmids, respectively). Each group of nude mice was injected with the corresponding transfected cells, and a single cell suspension (5 × 107 cells/mL) was injected subcutaneously into the back. The maximum diameter (L) and the minimum diameter (W) of the tumor were measured with a Vernier caliper, and the tumor volume was calculated according to the formula: V (mm3) = W2 × L × 0.52 [24]. The tumor volume and weight of nude mice were measured every week. 35 days later, the nude mice were euthanized by cervical dislocation after CO2 inhalation. Then, after removal of the xenograft tumors and resection of the largest surface of the tumor tissue without any necrosis, the larger part was fixed in 4% neutral formaldehyde solution overnight. The tumor tissues were embedded with paraffin at an interval of 50 μm. Finally, 10 slices were continuously sectioned with 5 μm thickness and stained with HE. The total number of tumor metastasis was recorded under the guidance of a microscope with the tumor volume measured.
Hematoxylin-eosin (HE) staining
The sections of lymphatic tissue in nude mice were fixed with 4% paraformaldehyde solution for 24 h. The routine dehydration process was conducted with conventional gradient alcohol (ethanol concentration of 70%, 80%, 90%, 95% and 100%) for 1 minute each time. The tissues were later cleared with xylene twice (each for 5 min), immersed in wax, embedded in paraffin, and then sliced into 4 μm sections (partial sections were prepared for immunohistochemistry). The paraffin-embedded sections were routinely dewaxed in water, stained with hematoxylin for 4 min and washed. Hydrochloric acid and ethanol were used for differentiation for 10 s. The slices were rinsed for 5 min, and then placed back in ammonia solution for 10 min to revert to blue color. Eosin was used to stain the slices for 2 min, followed by gradient ethanol dehydration (1 minute each time) and clearing with xylene for 2 times (1 min each time). Next, the slides were sealed with neutral gum. Finally, the pathological changes of the tumor tissues were observed under the guidance of an optical microscope (CSW-PH50, Shenzhen Coosway Optical Instrument Co., Ltd., Shenzhen, China).
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was employed for statistical analysis. The measurement data were expressed as the mean ± standard deviation. The data comparison among multiple groups was performed by one-way analysis of variance (ANOVA), followed by Tukey's post hoc test. Data comparison at different time points was conducted by repeated measures of ANOVA, followed by Bonferroni post hoc test. The difference was considered to be statistically significant at p < 0.05.
Results
LINC01215 is highly expressed in EOC
Based on the data analysis for GSE18520 dataset, 2872 differentially expressed genes were screened, including 1741 downregulated genes and 1131 upregulated genes; while for GSE54388 dataset, 1368 differentially expressed genes were screened, including 743 downregulated genes and 625 upregulated genes. We subsequently conducted hierarchical cluster analysis on these screened genes, which accurately distinguished the top 10 differentially expressed mRNAs in ovarian cancer in the two datasets. Also, LINC01215 was found to be highly expressed in EOC tissues in comparison to that of the normal tissues (Fig. 1A, B). Next, RT-qPCR was performed to determine the expression of LINC01215 in EOC cell lines A2780, HO8910, SKOV-3, ES-2, HO8910pm and the normal ovarian IOSE80 cell lines. As depicted in Fig. 1C, the HO8910pm cell line exhibited the highest expression of LINC01215 among these five ECO cell lines (p < 0.05) compared with that of the IOSE80 cell line. Thus, the HO8910pm cell line was selected for the subsequent experiment.
Fig. 1.
LINC01215 is predicted to be highly expressed in EOC tissues and its highest expression was found in HO8910pm EOC cell line determined by RT-qPCR.
Notes: Panel A, a heat map of differentially expressed lncRNAs in the GSE18520 dataset; Panel B, LINC01215 was highly expressed in EOC tissues in comparison to that of the normal tissues, based on the heat map of differentially expressed lncRNAs in the GSE54388 dataset; Panel C, the HO8910pm cell line exhibited the highest expression of LINC01215 among the five EOC cell lines (A2780, HO8910, SKOV-3, ES-2, HO8910pm) compared with that of the human normal IOSE80 cell line; EOC, epithelial ovarian cancer; RT-qPCR, reverse transcription quantitative polymerase chain reaction; *, p < 0.05 vs. the IOSE80 cells; #, p < 0.05 vs. the HO8910pm cells. Data were shown as the mean ± standard deviation of three technical replicates and a t-test was used for data comparison between two groups.
LINC01215 promotes the methylation of RUNX3 promoter by recruiting methylation proteins
Bioinformatics analysis showed that LINC01215 was located in the nucleus (Fig. 2A). The relationship between LINC01215 and methylation-related proteins DNMT1, DNMT3A and DNMT3B, together with the binding relationship with RUNX3, were explored by RNA pull down and RIP assay. As illustrated in Fig. 2B, C, the LINC01215 could recruit DNMT1, DNMT3A and DNMT3B and then bind to the promoter region of RUNX3. Moreover, high levels of methylation activity of RUNX3 in HO8910pm cells was detected by MS-PCR (Fig. 2D). These findings demonstrated that the LINC01215 could promote the methylation of RUNX3 promoter region through the recruitment of methylation-related proteins DNMT1, DNMT3A and DNMT3B.
Fig. 2.
LINC01215 enhances RUNX3 promoter methylation by recruiting methylation-related proteins.
Notes: Panel A, the location of LINC01215 was predicted to be in the nucleus by bioinformatics method; Panel B&C, the LINC01215 could recruit DNMT1, DNMT3A and DNMT3B determined by RNA pull down RIP assays; Panel D, HO8910pm cells exhibited great methylation of RUNX3 analyzed by MS-PCR. *, p < 0.05 vs. IgG;
LINC01215 upregulation or RUNX3 silencing promotes EOC cell proliferation
First, western blot analysis confirmed the silencing efficiency of shRUNX3-1, shRUNX3-2 and shRUNX3-3, as shown by the decreased expression of RUNX3 in cells after being treated with shRUNX3-1, shRUNX3-2 or shRUNX3-3. In particular, shRUNX3-2 showed the most superior silencing efficiency and was thus used in the subsequent experiments. At the same time, western blot analysis results also validated the efficiency of RUNX3 overexpression (Fig. 3A). In addition, RT-qPCR results showed the successful transfection of shLINC01215-1, shLINC01215-2 and shLINC01215-3, of which, shLINC01215-1 had the best silencing efficiency and was therefore used for the subsequent experiments. The successful transfection of oeLINC01215 was also confirmed by RT-qPCR (Fig. 3B). CCK-8 assay results revealed that on the 1st day, the proliferative ability did not show any difference among these groups. The proliferative ability on the 2nd and 3rd day was significantly higher in cells treated with oeLINC01215 plasmids, with shRUNX3 plasmids, and with both oeLINC01215 and shRUNX3 plasmids compared with cells without any treatment; on the other hand, the shLINC01215 group showed a complete opposite outcome. On the 2nd and 3rd day, relative to cells with both oeLINC01215 and shRUNX3 plasmids, decreased proliferative abilities were observed in cells treated with oeLINC01215 plasmids, shRUNX3 plasmids or oeLINC01215 + oeRUNX3 plasmids. The comparison among cells treated with oeLINC01215 plasmids and cells with shRUNX3 plasmids revealed no significantly different proliferative changes (Fig. 3C). These data suggested that overexpressing LINC01215 or RUNX3 silencing could stimulate EOC cell proliferation.
Fig. 3.
LINC01215 upregulation and RUNX3 silencing can both promote HO8910pm cell proliferation.
Notes: Panel A, transfection efficiency of shRUNX3-1, shRUNX3-2 and shRUNX3-3 and oeRUNX3 confirmed by western blot analysis; Panel B, transfection efficiency of shLINC01215-1, shLINC01215-2, shLINC01215-3 and oeLINC01215 confirmed by RT-qPCR; Panel C, cell proliferation detected by CCK-8 assay. *, p < 0.05 vs. the blank group; #, p < 0.05 vs. the oeLINC01215 + shRUNX3 treatment; NC, negative control; oeLINC01215, LINC01215 overexpression; shRUNX3, short hairpin RNA targeting runt-related transcription factor 3. Data were shown as the mean ± standard deviation of three technical replicates. One-way ANOVA was used for data comparisons among multiple groups.
LINC01215 overexpression or RUNX3 silencing induces EOC cell migration and invasion
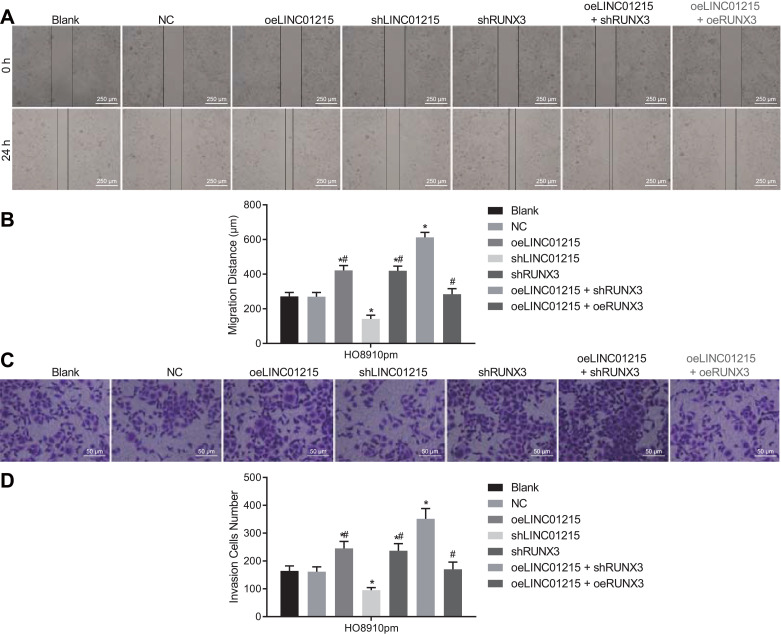
The migrative and invasive abilities of EOC cell after transfection were evaluated by scratch test and Transwell assay, respectively. As presented in Fig. 4, oeLINC01215, shRUNX3 and oeLINC01215 + shRUNX3 treatment resulted in an increased cell migration distance and more invasive cells (p < 0.05) in contrast to cells not receiving any treatment. Cells transfected with the shLINC01215 displayed shorter cell migration distance and fewer invasive cells (p < 0.05) compared with the cells transfected with oeLINC01215 and shRUNX3, while there was no distinct difference between the cells treated with oeLINC01215 and with shRUNX3 (p > 0.05). However, cell migration distance and invasive cells were decreased in cells treated with oeLINC01215, shRUNX3 or oeLINC01215 + oeRUNX3 relative to treatment with both oeLINC01215 and shRUNX3 (p < 0.05). Above all, overexpressing LINC01215 and silencing RUNX3 could contribute to EOC cell migration and invasion.
Fig. 4.
HO8910pm cell migration and invasion were induced by either LINC01215 overexpression or RUNX3 silencing.
Notes: Panel A, the cell migrative ability determined by scratch test (40 ×); Panel B, the cell migration distance of each group; Panel C, the cell invasive ability according to the results of Transwell assay (200 ×); Panel D, the number of invasive cells in each group; *, p < 0.05 vs. the blank group; #, p < 0.05 vs. the oeLINC01215 + shRUNX3 co-treatment; NC, negative control; oeLINC01215, LINC01215 overexpression; shRUNX3, short hairpin RNA targeting runt-related transcription factor 3. Data were shown as the mean ± standard deviation of three technical replicates. One-way ANOVA was used for data comparisons among multiple groups.
Upregulated LINC01215 or downregulated RUNX3 inhibits EOC cell apoptosis
Flow cytometry assay was conducted to explore how LINC01215 and RUNX3 regulated the apoptosis rates of EOC cells. Firstly, the results of Annexin V-FITC/PI double staining (Fig. 5A and B) depicted that treatment of oeLINC01215, shRUNX3 and oeLINC01215 + shRUNX3 induced markedly lower cell apoptosis rates in EOC relative to no treatment (p < 0.05). However, we found that the apoptosis rate of cells treated with shLINC01215 was lower than that of cells treated with oeLINC01215 and shRUNX3 (p < 0.05), while the difference between the oeLINC01215 treatment and the shRUNX3 treatment was not distinct (p > 0.05). In contrast to the co-treatment of oeLINC01215 and shRUNX3, a higher cell apoptosis rate was noted following treatment with oeLINC01215, shRUNX3 or oeLINC01215 + oeRUNX3. Therefore, it was concluded that upregulated LINC01215 or downregulated RUNX3 inhibited the apoptosis rates of EOC cells.
Fig. 5.
HO8910pm cell apoptosis was inhibited through upregulated LINC01215 or downregulated RUNX3 evaluated by flow cytometry.
Notes: Panel A, HO8910pm cells in the scatter plots; Panel B, the cell apoptosis rate of each group; *, p < 0.05 vs. the blank group; #, p < 0.05 vs. the oeLINC01215 + shRUNX3 co-treatment; NC, negative control; oeLINC01215, LINC01215 overexpression; shRUNX3, short hairpin RNA targeting runt-related transcription factor 3. Data were shown as the mean ± standard deviation of three technical replicates. One-way ANOVA was used for data comparisons among multiple groups.
LINC01215 silencing inhibits EMT by upregulating RUNX3
RT-qPCR and western blot analysis were applied to determine the following expressions of E-cadherin, MMP-2, MMP-9 and Vimentin, all being EMT-associated markers. As revealed in Fig. 6A–C, depletion of RUNX3 and/or overexpression of LINC01215 markedly inhibited the expressions of RUNX3 and Vimentin, but promoted the following expressions of LINC01215, MMP-2, MMP-9 and Vimentin in cells, compared to cells receiving no treatment, while opposite results were observed in cells treated with shLINC01215. The treatment with oeLINC01215, shRUNX3 or oeLINC01215 + oeRUNX3 declined the expressions of LINC01215, MMP-2, MMP-9 and Vimentin (p < 0.05), but elevated the expressions of RUNX3 and Vimentin relative to the oeLINC01215 + shRUNX3 treatment (p < 0.05). No significant difference was observed in the mRNA and protein expressions of LINC01215, RUNX3 or EMT-associated markers (p > 0.05) compared with cells treated with oeLINC01215 and cells treated with shRUNX3. Taken together, the present findings confirmed that the inhibition of EMT could be achieved through LINC01215 silencing which upregulated the expression of RUNX3.
Fig. 6.
EMT of HO8910pm cells was suppressed by LINC01215 silencing and upregulation of RUNX3.
Notes: Panel A, RT-qPCR was used to determine the relative expressions of LINC01215, RUNX3, and EMT-related genes (MMP-2, MMP-9, E-cadherin and Vimentin); Panel B&C, the relative protein expressions of RUNX3 and EMT-related genes (MMP-2, MMP-9, E-cadherin and Vimentin) accessed by western blot analysis; *, p < 0.05 vs. the blank group; #, p < 0.05 vs. the oeLINC01215 + shRUNX3 co-treatment; NC, negative control; RUNX3, runt-related transcription factor 3; EMT, epithelial-mesenchymal transition; RT-qPCR, reverse transcription quantitative polymerase chain reaction. Data were shown as the mean ± standard deviation of three technical replicates. One-way ANOVA was used for data comparisons among multiple groups.
LINC01215 promotes tumor growth and LNM in nude mice by downregulating RUNX3
To verify the roles of LINC01215 and RUNX3 on tumor growth and LNM, xenograft tumor was established in nude mice and the tumor volume was monitored every week. After 5 weeks, the xenograft tumors were extracted from the nude mice and were subsequently used for HE staining (Fig. 7). The accelerated tumor growth, larger tumor volume, and increased tumor metastasis were observed in mice inoculated with cells transfected with the oeLINC01215 and shRUNX3 plasmids, while the shLINC01215 treatment showed opposite outcomes. The tumor extracted from mice co-treated with oeLINC01215 and shRUNX3 exhibited the quickest growth rate, the largest tumor volume and the largest number of tumor metastasis (all p < 0.05). According to the results of HE staining on the lymphatic tissues, there were massive metastatic tumor cells resulted from oeLINC01215 and shRUNX3, which demonstrated the promotion of LNM (p < 0.05), while the inhibition of LNM revealed no metastatic tumor cells in the shLINC01215 group. The largest number of tumor cells metastasized to the lymph nodes was induced by the oeLINC01215 + shRUNX3 treatment (p < 0.05). The results provided evidence that the tumor growth, as well as LNM, were augmented by the overexpression of LINC01215, which could reduce RUNX3.
Fig. 7.
Tumor growth and LNM in nude mice were promoted by LINC01215 overexpression through RUNX3 downregulation.
Notes: Panel A, the tumor growth of xenograft tumor in nude mice; Panel B, quantitative analysis for the number of tumor metastasis in mice in each group; Panel C, HE staining of mouse lymphatic tissue sections (200 ×); Panel D, the tumor volume of mice in each group in the 1st, 2nd, 3rd and 4th week; *, p < 0.05 vs. the blank group; NC, negative control; RUNX3, runt-related transcription factor 3; LNM, lymph node metastasis; HE, hematoxylin-eosin. Data were shown as the mean ± standard deviation of three technical replicates. Repeated measures of ANOVA were used to analyze the data at different time points (n = 5).
Discussion
EOC is a type of malignancy that poses a serious threat to women, and patients diagnosed at a late-stage usually has a 5-year survival rate of less than 30% [25]. As mounting research has focused on the function of lncRNAs in various cancers, an increasing number of lncRNAs have been identified to play a crucial role in the development of EOC. In this study, we strived to figure out the mechanism by which LINC01215 affects the progression of EOC. We demonstrated that LINC01215 was highly expressed in EOC cells and promoted invasive, migrative, proliferative, EMT, LNM, tumor growth while inhibiting EOC cell apoptosis through inducing RUNX3 promoter methylation.
Initially, our results revealed that the expression of LINC01215 was significantly increased in EOC cells. LINC01215 had the ability to recruit methylation related proteins DNMT1, DNMT3A and DNMT3B, by which way the methylation of RUNX3 promoter was promoted. RUNX3 is controversially discussed in relation to possible oncogenic or tumor suppressive functions [26]. The association between hypermethylation of RUNX3 and the progression of Type II EOC has been previously demonstrated by a study [17]. Our study presented that silencing LINC01215 inhibits cell migrative, invasive and proliferative capabilities, while accelerating apoptosis in EOC by restoring RUNX3 promoter methylation. Besides, the loss of RUNX3 expression can contribute to the inhibition of hepatocellular carcinoma cell apoptosis by which tumorigenesis is induced [27].
Furthermore, through RT-qPCR and western blot analysis, increases in miRNA and protein expressions of MMP-2, MMP-9 and Vimentin along with decreases of E-cadherin demonstrated induced EMT in EOC, which resulted from overexpressed LINC01215 or depletion of RUNX3. EMT is a biological process involving enhanced migrative and invasive abilities, reduced apoptosis, as well as increased extracellular matrix components [27] and is also an important mechanism underlying cellular migrative and invasive capabilities [28]. There was a study that has emphasized the significance of forced expression of E-cadherin in ovarian-specific metastasis [29]. In the process of EMT in EOC, the loss of expression of E-cadherin is conducive to the interaction with endothelial and stromal components [30]. The upregulation of Vimentin is commonly observed in the cells that undergo EMT process as it promotes EMT, thus aggravating tumor malignancy [31]. The association between MMP and EMT has been previously discussed that the MMPs-induced alterations in phenotype and genotype can contribute to tumor progression [32]. The association between upregulation of MMP-9 and the undesirable prognosis of patients with ovarian cancer has been elucidated, suggesting that the repression of MMP-9 might be of significant importance in ovarian cancer [33]. In vivo xenograft tumor in nude mice was implemented to elucidate the inhibitory effect of LINC01215 silencing on tumor growth and LNM.
Conclusions
In summary, our findings helped demonstrated that LINC01215 contributes to the progression of EOC, and that the silencing of LINC01215 may provide new therapeutic insights for the treatment of EOC. The methylation of RUNX3 triggered by LINC01215 overexpression can be reversed by silencing LINC01215, thus contributing to the suppression of invasive, migrative, proliferative capabilities, EMT, LNM and tumor growth in EOC (Fig. 8). However, further studies are warranted to explore the association of LINC01215 with tumorigenesis, metastasis and prognosis and reveal the potential clinical value of LINC01215.
Fig. 8.
A diagram showing the molecular mechanisms of LINC01215 involved in EOC progression by promoting methylation of RUNX3. LINC01215, which is highly expressed in EOC cells, binds to the promoter region of RUNX3 and recruits DNMT1, DNMT3A and DNMT3B proteins to inhibit the transcription of RUNX3, further upregulating the expressions of MMP-2, MMP-9 and Vimentin and downregulating the expression of E-cadherin, therefore promoting proliferation and EMT of EOC cells. EOC, epithelial ovarian cancer; RUNX3, runt-related transcription factor 3; DNMT1, DNA methyltransferase 1; DNMT3A, DNA methyltransferase 3A; DNMT3B, DNA methyltransferase 3B; MMP-2, matrix metalloprotein-2; MMP-9; matrix metalloprotein-9; EMT, epithelial-mesenchymal transition.
Author contributions
L-W, T-S, B-XX, M-SH and C-XW designed the study. L-W and T-S collated the data, carried out data analyses and produced the initial draft of the manuscript. B-XX, M-SH and C-XW contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgements
We would like to give our sincere appreciation to the colleagues for their helpful comments on this article.
Funding
This study was supported by the project of Science and Technology Department in Heilongjiang Province (GC09C406-5) and the National Natural Science Foundation of China (No. 81772274).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101135.
Appendix. Supplementary materials
Data for reference
The datasets generated/analysed during the current study are available.
References
- 1.Kurman R.J., Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anugraham M., Jacob F., Nixdorf S., Everest-Dass A.V., Heinzelmann-Schwarz V., Packer N.H. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: glycan structures reflect gene expression and DNA methylation status. Mol. Cell. Proteom. 2014;13:2213–2232. doi: 10.1074/mcp.M113.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore L.E., Pfeiffer R.M., Zhang Z., Lu K.H., Fung E.T., Bast R.C., Jr. Proteomic biomarkers in combination with CA 125 for detection of epithelial ovarian cancer using prediagnostic serum samples from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Cancer. 2012;118:91–100. doi: 10.1002/cncr.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayson G.C., Kohn E.C., Kitchener H.C., Ledermann J.A. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 5.Kim H.J., Jeon H.K., Cho Y.J., Park Y.A., Choi J.J., Do I.G., Song S.Y., Lee Y.Y., Choi C.H., Kim T.J., Bae D.S., Lee J.W., Kim B.G. High galectin-1 expression correlates with poor prognosis and is involved in epithelial ovarian cancer proliferation and invasion. Eur. J. Cancer. 2012;48:1914–1921. doi: 10.1016/j.ejca.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Liu S.P., Yang J.X., Cao D.Y., Shen K. Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol. Med. 2013;10:138–141. doi: 10.7497/j.issn.2095-3941.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spizzo R., Almeida M.I., Colombatti A., Calin G.A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang F., Zhang L., Huo X.S., Yuan J.H., Xu D., Yuan S.X., Zhu N., Zhou W.P., Yang G.S., Wang Y.Z., Shang J.L., Gao C.F., Zhang F.R. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 9.Cao W.J., Wu H.L., He B.S., Zhang Y.S., Zhang Z.Y. Analysis of long non-coding RNA expression profiles in gastric cancer. World J. Gastroenterol. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G., Yao W., Wang J., Ma X., Xiao W., Li H., Xia D., Yang Y., Deng K., Xiao H., Wang B., Guo X., Guan W. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J.J., Lin Y.Y., Ye L.C., Ding J.X., Feng W.W., Jin H.Y., Zhang Y., Li Q., Hua K.Q. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol. Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 12.Qiu J.J., Wang Y., Liu Y.L., Zhang Y., Ding J.X., Hua K.Q. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget. 2016;7:32478–32492. doi: 10.18632/oncotarget.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuang D., Zhang X., Hua S., Dong W., Li Z. Long non-coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial-mesenchymal transition. Exp. Mol. Pathol. 2016;101:267–273. doi: 10.1016/j.yexmp.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Shiraha H., Nishina S., Yamamoto K. Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J. Cell. Biochem. 2011;112:745–749. doi: 10.1002/jcb.22973. [DOI] [PubMed] [Google Scholar]
- 15.Xia Y., Zhang M., Zhang X., Liu X. A systematic review and meta-analysis of runt-related transcription factor 3 gene promoter hypermethylation and risk of gastric cancer. J. Cancer Res. Ther. 2014;(10 Suppl):310–313. doi: 10.4103/0973-1482.151539. [DOI] [PubMed] [Google Scholar]
- 16.Rehman A.U., Iqbal M.A., Sattar R.S.A., Saikia S., Kashif M., Ali W.M., Medhi S., Saluja S.S., Husain S.A. Elevated expression of RUNX3 co-expressing with EZH2 in esophageal cancer patients from India. Cancer Cell Int. 2020;20:445. doi: 10.1186/s12935-020-01534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafner N., Steinbach D., Jansen L., Diebolder H., Durst M., Runnebaum I.B. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int. J. Cancer. 2016;138:217–228. doi: 10.1002/ijc.29690. [DOI] [PubMed] [Google Scholar]
- 18.Mok S.C., Bonome T., Vathipadiekal V., Bell A., Johnson M.E., Wong K.K., Park D.C., Hao K., Yip D.K., Donninger H., Ozbun L., Samimi G., Brady J. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung T.L., Leung C.S., Wong K.K., Gutierrez-Hartmann A., Kwong J., Gershenson D.M., Mok S.C. ELF3 is a negative regulator of epithelial-mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:16951–16963. doi: 10.18632/oncotarget.15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita A., Sato J.R., Rodrigues Lde O., Ferreira C.E., Sogayar M.C. Evaluating different methods of microarray data normalization. BMC Bioinform. 2006;7:469. doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Morimoto A., Kannari M., Tsuchida Y., Sasaki S., Saito C., Matsuta T., Maeda T., Akiyama M., Nakamura T., Sakaguchi M., Nameki N., Gonzalez F.J., Inoue Y. An HNF4alpha-microRNA-194/192 signaling axis maintains hepatic cell function. J. Biol. Chem. 2017;292:10574–10585. doi: 10.1074/jbc.M117.785592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim C.J., Terado T., Tambe Y., Mukaisho K.I., Sugihara H., Kawauchi A., Inoue H. Anti-oncogenic activities of cyclin D1b siRNA on human bladder cancer cells via induction of apoptosis and suppression of cancer cell stemness and invasiveness. Int. J. Oncol. 2018;52:231–240. doi: 10.3892/ijo.2017.4194. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y., Yang D.Z., Zhai Y.N., Xue K., Xu F., Gu X.Y., Wang S.M. Microarray expression profiling of long non-coding RNAs in epithelial ovarian cancer. Oncol. Lett. 2017;14:2523–2530. doi: 10.3892/ol.2017.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang L.S., Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29:2605–2615. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi Y., Shiraha H., Nishina S., Tanaka S., Matsubara M., Horiguchi S., Iwamuro M., Takaoka N., Uemura M., Kuwaki K., Hagihara H., Toshimori J., Ohnishi H. Loss of runt-related transcription factor 3 expression leads hepatocellular carcinoma cells to escape apoptosis. BMC Cancer. 2011;11:3. doi: 10.1186/1471-2407-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliev R., Kleinova R., Juracek J., Dolezel J., Ozanova Z., Fedorko M., Pacik D., Svoboda M., Stanik M., Slaby O. Overexpression of long non-coding RNA TUG1 predicts poor prognosis and promotes cancer cell proliferation and migration in high-grade muscle-invasive bladder cancer. Tumour Biol. 2016;37:13385–13390. doi: 10.1007/s13277-016-5177-9. [DOI] [PubMed] [Google Scholar]
- 29.Kuwabara Y., Yamada T., Yamazaki K., Du W.L., Banno K., Aoki D., Sakamoto M. Establishment of an ovarian metastasis model and possible involvement of E-cadherin down-regulation in the metastasis. Cancer Sci. 2008;99:1933–1939. doi: 10.1111/j.1349-7006.2008.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergara D., Merlot B., Lucot J.P., Collinet P., Vinatier D., Fournier I., Salzet M. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291:59–66. doi: 10.1016/j.canlet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Liu C.Y., Lin H.H., Tang M.J., Wang Y.K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6:15966–15983. doi: 10.18632/oncotarget.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlichenko L.S., Radisky D.C. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin. Exp. Metastasis. 2008;25:593–600. doi: 10.1007/s10585-008-9143-9. [DOI] [PubMed] [Google Scholar]
- 33.Li L.N., Zhou X., Gu Y., Yan J. Prognostic value of MMP-9 in ovarian cancer: a meta-analysis. Asian Pac. J. Cancer Prev. 2013;14:4107–4113. doi: 10.7314/apjcp.2013.14.7.4107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated/analysed during the current study are available.