Highlights
-
•
CD276 is homogeneously overexpressed in ESCC and EAC.
-
•
CD276-directed CAR-T cells demonstrate remarkable anti-tumor effects in ESCC PDX model.
-
•
CD276-targeting CAR-T cells are successfully generated with patients T cells and show potent cytotoxicity against autologous tumor cells.
Keywords: Esophageal cancer, CD276, CAR-T cell, Co-stimulation, Patient-derived xenograft
Abbreviations: BLI, bioluminescence imaging; CAR, chimeric antigen receptor; CSC, cancer stem cell; CRISPR, clustered regularly interspaced short palindromic repeats; EAC, esophageal adenocarcinoma; ELISA, enzyme-linked immunosorbent assay; ESCC, esophagus squamous cell carcinoma; FFluc, firefly luciferase; IFN-γ, interferon-γ; IHC, immunohistochemistry; IL-2, interleukin-2; PDX, patient-derived xenograft
Abstract
Esophageal cancer, including esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), has a poor prognosis and limited therapeutic options. Chimeric antigen receptor (CAR)-T cells represent a potential ESCC treatment. In this study, we examined CD276 expression in healthy and esophageal tumor tissues and explored the tumoricidal potential of CD276-targeting CAR-T cells in ESCC. CD276 was strongly and homogenously expressed in ESCC and EAC tumor lesions but mildly in healthy tissues, representing a good target for CAR-T cell therapy. We generated CD276-directed CAR-T cells with a humanized antigen-recognizing domain and CD28 or 4–1BB co-stimulation. CD276-specific CAR-T cells efficiently killed ESCC tumor cells in an antigen-dependent manner both in vitro and in vivo. In patient-derived xenograft models, CAR-T cells induced tumor regression and extended mouse survival. In addition, CAR-T cells generated from patient T cells demonstrated potent cytotoxicity against autologous tumor cells. Our study indicates that CD276 is an attractive target for ESCC therapy, and CD276-targeting CAR-T cells are worth testing in ESCC clinical trials.
Introduction
The global incidence and mortality rate of esophageal cancer ranks the seventh and sixth [1], respectively, representing a significant human health risk. Esophageal cancer presents with two major histopathological forms, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), with ESCC accounting for 90% of esophageal cancer cases [2,3]. Although clinical regimens, including surgery, chemotherapy, and radiotherapy, have improved prognosis, the 5-year survival rate of patients with esophageal cancer is ≤ 20% [4,5]. Hence, developing more effective therapeutic strategies would be critical.
Chimeric antigen receptor (CAR) T cells represent a promising therapeutic strategy for malignant diseases [6]. CAR transgenes endow T cells with potent cytotoxicity against target cells in an antigen-dependent manner. CD19-specific CAR-T cells have demonstrated significant efficacy against hematologic malignancies owing to the prevalent expression of the target antigen in malignant cells [6]. In solid tumors, CAR-T cells targeting the uniformly overexpressed antigens in tumor lesions also show promising potential in preclinical and clinical settings [7,8]. However, limited efforts have been made to explore the potential of CAR-T cell therapy in esophageal cancer despite the high prevalence of the disease. Therefore, it remains unclear whether suitable antigens for CAR-T cell therapy would be available for esophageal cancer.
CD276 (also called B7-H3) is a transmembrane protein of the B7 family [9]. CD276 preferentially overexpressed in various tumors [10], [11], [12], [13] and has been explored as the target of CAR-T cells against solid tumors, including ovarian cancer, pancreatic adenocarcinoma, and brain tumors [14,15]. CD276 is reportedly overexpressed in esophageal cancer [10]. However, it remains unclear whether CD276 is extensively overexpressed in esophageal cancer and if it would be suitable for CAR-T cell therapy in patients with such malignancy. In this study, we focused on ESCC because of its predominant prevalence over EAC, explored CD276 expression in malignant and healthy tissues, and compared the anti-tumor effects of CD276-directed CAR-T cells with CD28 or 4–1BB co-stimulation.
Materials and methods
Cell lines
Human ESCC cell lines, including EC109, KYSE150, and TE-1, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human ESCC cell lines KYSE450, KYSE510, and TE-7 were maintained in our laboratory. In addition, 293T cells were obtained from the Cell Bank of the Chinese Academy of Sciences. The cells were cultured in Dulbecco's Modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone), 1% penicillin and streptomycin (Gibco, Grand Island, NY), and anti-mycoplasma agent Plasmocin (Invivogen, San Diego, CA). Firefly luciferase (FFluc)-expressing cells were established using lentiviral transduction.
Primary blood and tissue samples
Peripheral blood samples from healthy donors were supplied by the Henan Red Cross Center (China) in the form of buffy coats. Peripheral blood was collected from patients using ethylenediaminetetraacetic acid vacuum tubes. Malignant and marginal tissues were collected during surgery from patients with ESCC at the First Affiliated Hospital of Zhengzhou University. The study protocol was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all patients.
Immunohistochemistry (IHC) of tumor and healthy tissues
Prepared tissue microarrays (TMAs) including samples from patients diagnosed with ESCC or EAC and healthy tissue samples were obtained from Outdo Biotech Co. LTD (Shanghai, China). IHC tests were performed as previously described [16]. The samples were incubated with anti-human CD276 antibody (1:4000; R&D Systems, Minneapolis, MN) and horseradish peroxidase-linked secondary antibody (Dako, Glostrup, Denmark) and stained with 3,3ʹ-diaminobenzidine substrate (Dako). Next, the samples were counterstained with Mayer's hematoxylin. The staining results were independently evaluated by two experienced observers. Staining intensities (0 = none, 1 = low, 2 = moderate, and 3 = high) and the percentage of the stained cells were determined to calculate H scores according to the following formula: H score = (% at 1) × 1 + (% at 2) × 2 + (% at 3) × 3. Images were obtained using a BX53 microscope (Olympus, Tokyo, Japan).
Public sequencing data analysis
The web server Gene Expression Profiling Interactive Analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn) [17] was used to analyze CD276 mRNA expression in malignant and non-malignant tissues. The Cancer Genome Atlas (TCGA) esophageal tumor tissue (including ESCC and EAC) sequencing data and matched TCGA healthy and GTEx data were compared. Analysis of variance (ANOVA) was used to determine statistically significant differences between the different sample groups.
Fluorescence-activated cell sorting (FACS) analysis and purification
Specific fluorochrome-conjugated antibodies against human CD3, CD62L, CD45RA, CD69, CD107a, CD276, IFN-γ, and EpCAM (BioLegend, San Diego, CA, USA) were used for phenotyping. We also used anti-human CD45 antibody and fixable viability dyes (Thermo Fisher Scientific, Waltham, MA, USA). FACS analysis was performed using a FACSCanto II cytometer with BD Diva software (BD Biosciences, San Jose, CA, USA) unless otherwise indicated. For each sample, at least 10,000 events were acquired, and the data were further analyzed using the FlowJo version 10 (BD Biosciences). For sorting, the cells were purified with a Moflo XDP sorter (Beckman Coulter, Indianapolis, IN, USA). The purity of the sorted samples was >95%.
Lentivirus and CAR-T cell generation
The CD276-recognizing domain was derived from the humanized antibody 8H9S3.3, [18] whereas the CD19-recognizing fragment was derived from the FMC63 antibody [19]. The CAR sequence, including antigen-recognizing and other domains (As shown in Fig. 2B), was synthesized by Sangon Biotech (Shanghai, China). The CAR-coding DNA fragments were then cloned into the pCDH-EF1α-GFP lentiviral vector (System Biosciences, Palo Alto, CA, USA). Lentiviruses were produced using 293T cells. Purified human CD3+ T cells were activated with anti-CD3/CD28 Dynabeads (Thermo Fisher Scientific) in the presence of IL-2 (100 IU/mL; PeproTech, Rocky Hill, NJ, USA) for 48 h. Next, the T cells were infected with lentivirus by centrifugation at 1000 × g for 2 h. The transduced T cells were then expanded in RPMI-1640 with 5% FBS and IL-2 (100 IU/mL) for 12 days. For the in vivo experiments, CAR-T cells were purified using a cell sorter according to GFP expression (purity >95%).
Fig. 2.
Antigen-dependent cytotoxicity of CAR-T cells. (A) FACS analysis of CD276 expression on ESCC cell lines. (B) Structures of CAR used. (C-E) T cells from healthy donors were transduced with CAR via lentivirus and expanded. Then numbers of CAR-T cells were counted at indicated intervals (C). On day 12, CAR transduction efficacy (D) and memory differentiation (E) were determined by FACS. t-test was performed. (F) BLI detection of the viability of tumor cells incubated with indicated CAR-T cells at the listed E/T ratios without exogenous cytokines. (G) Effector cytokines secretion of CAR-T cells at E/T = 1 without exogenous cytokines. (H and I) The FFluc-expressing TE-7 cell line was engineered to express CD276 via lentiviral transduction. Cells were sorted based on CD276 levels and single clones were expanded (H). Then FFluc+ TE-7 (wildtype) and the derivate CD276+ clones were co-cultured with CAR-T cells at E/T = 1 without exogeneous cytokines. Tumor cell viability was detected by BLI after 24 h (I). Data represent assays using CAR-T cells produced from five donors. *, P < 0.05.
Bioluminescence-based cytotoxic analysis
FFluc+ tumor cells (2 × 104) were incubated with CAR-T cells at various effector/target (E/T) ratios for 24 h without exogenous cytokines in 96-well plates. The tumor cell viability was determined by bioluminescence imaging (BLI) using the Lumina II in vivo imaging system (PerkinElmer, Hopkinton, MA).
Enzyme-linked immunosorbent assay (ELISA)
The CAR-T cells (1 × 105) were co-cultured with tumor cells at E/T = 1 without exogenous cytokines for 24 h in 96-well plates. The supernatants were probed for effector cytokines (IFN-γ and IL-2) using specific ELISA kits (R&D Systems) according to the manufacturer's instructions.
CRISPR-mediated CD276 knockout
Gene editing of KYSE150 cells was performed as previously described [20]. pSpCas9(BB)−2A-GFP (PX458) (Addgene plasmid #48,138) was kindly gifted by Feng Zhang. The guide RNA (gRNA; 5ʹ-CTGGTGCACAGTTTCACCGA-3ʹ) targeting CD276 was cloned into a PX458 plasmid, followed by transfection into KYSE150 cells. After 48 h, single clones were obtained using a FACS sorter. CD276 deletion was confirmed by Sanger sequencing.
Cancer stem cell (CSC) enrichment
CSCs were prepared under sphere culture conditions according to our previously described protocol [21]. Briefly, ESCC cells were seeded onto 24-well ultra-low attachment plates (Corning Costar, Corning, NY) at a density of 5 × 103 cells per well and then suspended in DMEM/F12 medium (Thermo Fisher Scientific) containing 4 μg/mL heparin (Sigma Aldrich, St. Louis, MO, USA), B27 (1:50, Gibco), 20 ng/mL EGF, 20 ng/mL fibroblast growth factor (PeproTech), 100 IU/mL penicillin, and100 μg/mL streptomycin. Seven days later, spheres were dissociated with Accutase (Sigma Aldrich) and centrifuged at 150 × g for 5 min. The sedimented cells were then resuspended and used for subsequent assays.
Animal experiments
Female NSG mice (6–8-week old; Vital River Laboratories Co., Ltd., Beijing, China) were used to construct cell line- and patient-derived xenografts (PDXs). Tumor cells suspended in PBS were subcutaneously inoculated into the right flank. Low-passaged PDX tissues were implanted subcutaneously in the back. The CAR-T cells were injected intravenously. Mice were monitored daily. Tumor growth was monitored by BLI or by measuring tumor diameters [22] with calipers (tumor volume = [(length × (width)[2]]/2). Mice were euthanized once the BLI intensity (p/s) exceeded 1.2 × 1011, the tumor volume reached 1200 mm3, or signs of discomfort emerged. Animal experimental protocols were approved by the Institute Ethics Committee of the First Affiliated Hospital of Zhengzhou University, and all procedures strictly followed the approved protocols.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism version 8 software (GraphPad Software Inc., La Jolla, CA, USA). Data are represented as the means ± standard deviation, representing at least three independent experiments. Unpaired and nonparametric t-tests were used to measure the differences between the groups. ANOVA was used to determine the significant differences between the samples for multiple group comparisons. Kaplan-Meier curves were used to demonstrate survival. Log-rank test was used to analyze statistical significance. Differences were considered statistically significant at P-values <0.05.
Results
Overexpression of CD276 in ESCC and EAC tumor tissues
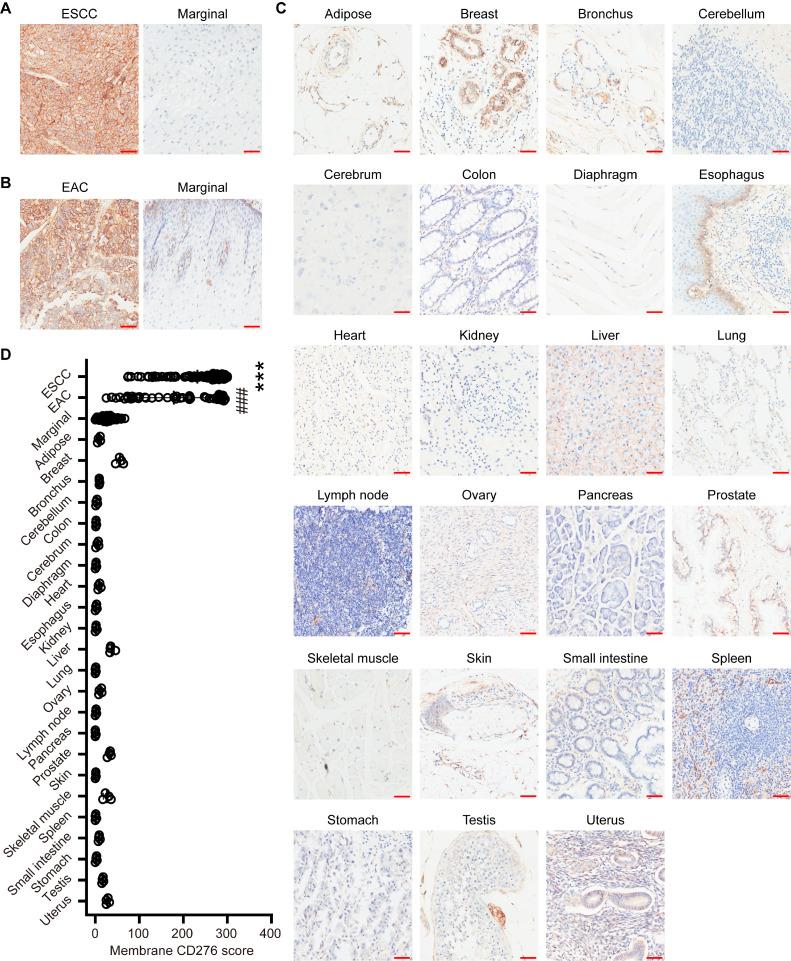
To analyze the expression pattern of CD276, TMAs of ESCC or EAC were stained by IHC (Fig. 1A and B). Among a total of 137 samples, including 91 ESCC and 46 EAC, all tissues demonstrated strong membrane staining of CD276, with more than 73% tumor cells having 2+ and 3+ staining intensities (Table 1 and Supplementary Figure 1). Importantly, in 86.8% of the ESCC samples (79/91) and 60.87% of EAC samples (28/46), the ratio of the CD276-positive tumor cells exceeded 80%. Next, we examined CD276 expression in marginal and healthy tissues. In contrast to malignant samples, the marginal and healthy samples exhibited low levels of CD276 in the limited cells (Fig. 1A-C; Table 1), with significantly lower H scores (P < 0.05; Fig. 1D). The public sequencing data also showed that the CD276 mRNA expression was remarkably higher in esophageal cancer lesions than in marginal and healthy esophageal tissues (P < 0.05; Supplementary Figure 2).
Fig. 1.
CD276 expression in esophagus carcinomas and healthy tissues. (A) IHC analysis of CD276 expression in malignant and healthy tissues from patients with ESCC (n = 91). (B) CD276 expression in EAC. IHC detection in malignant (n = 46) and marginal lesions (n = 21) from patients with EAC. (C) IHC of CD276 in 23 types of healthy tissues from four subjects. Scale bar, 50 μm. (D) ANOVA test of CD276 scores among healthy, marginal and malignant tissues. ***, P < 0.005; ###, P < 0.005.
Table 1.
Membrane expression of CD276 by IHC.
| Tissue type | # Samples stained | % Positive | % Intensity |
|||
|---|---|---|---|---|---|---|
| 0 | 1+ | 2+ | 3+ | |||
| ESCC | 91 | 90.22 | 9.78 | 9.93 | 18.69 | 61.59 |
| EAC | 46 | 76.20 | 23.80 | 16.24 | 17.67 | 42.28 |
| Marginal | 112 | 16.63 | 83.37 | 8.68 | 6.06 | 1.89 |
| Adipose | 4 | 7.25 | 92.75 | 5.75 | 1.50 | 0.00 |
| Breast | 4 | 30.00 | 70.00 | 2.00 | 26.75 | 0.25 |
| Bronchus | 4 | 6.75 | 93.25 | 4.25 | 2.50 | 0.00 |
| Cerebellum | 4 | 2.50 | 97.50 | 2.00 | 0.50 | 0.00 |
| Cerebrum | 4 | 1.50 | 98.50 | 1.25 | 0.25 | 0.00 |
| Colon | 4 | 4.00 | 96.00 | 3.00 | 1.00 | 0.00 |
| Diaphragm | 4 | 1.50 | 98.50 | 1.50 | 0.00 | 0.00 |
| Esophagus | 4 | 8.50 | 91.50 | 8.00 | 0.50 | 0.00 |
| Heart | 4 | 1.50 | 98.50 | 1.00 | 0.50 | 0.00 |
| Kidney | 4 | 1.50 | 98.50 | 1.25 | 0.25 | 0.00 |
| Liver | 4 | 34.00 | 66.00 | 31.00 | 3.00 | 0.00 |
| Lung | 4 | 0.75 | 99.25 | 0.75 | 0.00 | 0.00 |
| Lymphnode | 4 | 10.25 | 89.75 | 9.00 | 1.25 | 0.00 |
| Ovary | 4 | 1.50 | 98.50 | 1.50 | 0.00 | 0.00 |
| Pancreas | 4 | 0.50 | 99.50 | 0.50 | 0.00 | 0.00 |
| Prostate | 4 | 25.25 | 74.75 | 18.25 | 7.00 | 0.00 |
| Skeletal muscle | 4 | 1.00 | 99.00 | 1.00 | 0.00 | 0.00 |
| Skin | 4 | 19.75 | 80.25 | 12.75 | 7.00 | 0.00 |
| Small intestine | 4 | 1.00 | 99.00 | 1.00 | 0.00 | 0.00 |
| Spleen | 4 | 5.25 | 94.75 | 3.25 | 2.75 | 0.00 |
| Stomach | 4 | 2.25 | 97.75 | 2.25 | 0.00 | 0.00 |
| Testis | 4 | 15.00 | 85.00 | 13.00 | 2.00 | 0.00 |
| Uterus | 4 | 25.75 | 74.25 | 23.75 | 2.00 | 0.00 |
Characterization of CD276-directed CAR-T cells
CD276 expression was assessed in the ESCC cell lines. In agreement with the primary samples, most ESCC cell lines exhibited high levels of CD276 expression (Fig. 2A). However, TE-7 exhibited very faint CD276 staining (Fig. 2A), reflecting the heterogeneous antigen expression in the tumor. Next, CD276-specific CAR vectors with CD28 (CAR276.28ζ) or 4–1BB (CAR276.BBζ) co-stimulation were constructed (Fig. 2B). CD19-directed CAR was used as a control (CAR19.BBζ). After lentiviral CAR transduction, the engineered T cells proliferated exponentially with minimally different kinetics (Fig. 2C). On day 12, the transduction efficacy and memory differentiation of CD276-directed CAR-T cells were evaluated. CAR276.28ζ and CAR276.BBζ cells had comparable transduction efficacy (Fig. 2D), and the latter displayed a larger proportion of central memory cells than that in the former (P < 0.05; Fig. 2E). Next, we tested the cytotoxicity of CD276-specific CAR-T cells. CAR276.28ζ and CAR276.BBζ cells demonstrated potent and comparable cytolytic capacity and effector cytokine secretion in antigen-specific tumor cells (Fig. 2F and G). To confirm the antigen-dependent cytotoxicity of CAR-T cells, we generated CD276-deficient KYSE150 cells (KYSE150-CD276KO) using the CRISPR/cas9 system (Fig. 2A). As expected, CD276-directed CAR-T cells maintained their resting state when encountering CD276-negative derivate cells (Fig. 2F and G). Next, we assessed how the density of CD276 in target cells affected the tumoricidal activity of CAR-T cells by generating single-cell clones of tumor cells expressing different levels of CD276 (Fig. 2H). As shown in Fig. 2I, tumor cells expressing low levels of CD276 induced less effective cytotoxicity of CD276-direceted CAR-T cells (P < 0.05).
CD276 reportedly exists in both membrane-bound and soluble forms [23]. Therefore, we speculated whether soluble CD276 would activate or inhibit the cytotoxicity of antigen-specific CAR-T cells. However, even in the presence of high concentrations of the soluble antigen, the cytolytic potency of CD276-specific CAR-T cells was not impeded or enhanced (Supplementary Figure 3), diminishing the concern that CAR-T cell activity could be affected by the soluble CD276 form.
CD276-directed CAR-T cells efficiently kill ESCC stem cells
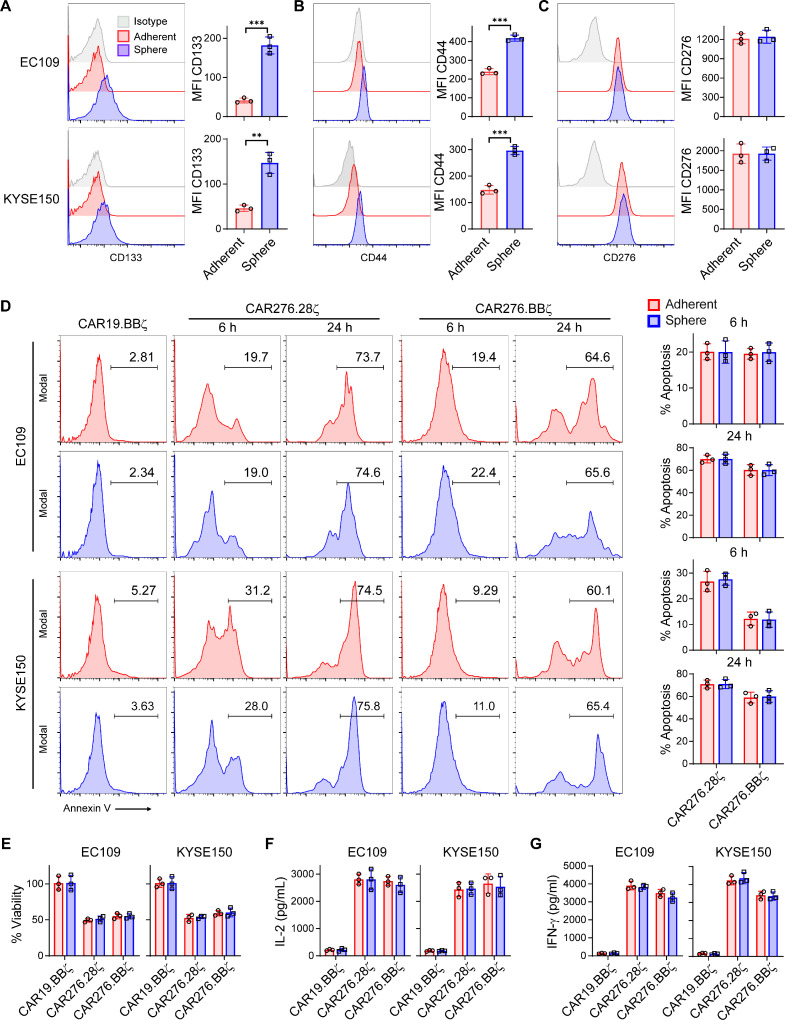
CSCs mediate the resistance to conventional and T cell-based immunotherapeutic approaches [24,25], Hence, the capacity of CAR-T cells to kill CSCs is critically important for therapeutic success. Our previous study showed that ESCC stem cells enhanced therapeutic resistance and metastatic capacity [21]. We speculated whether ESCC stem cells would resist CAR-T cell attack. Using our established protocol [21], CSCs were enriched under sphere culture conditions. Compared with adherently cultured tumor cells, CSCs demonstrated enhanced CSC marker [3] CD133 and CD44 expressions (P < 0.05; Fig. 3A and B). However, the surface CD276 expression was comparable between the tumor cells (adherent) and CSCs (sphere) (Fig. 3C). To verify if CSCs could be eliminated as efficiently as tumor cells, the spheres or adherent tumor cells were dissociated into single cells and then incubated with CAR-T cells. In the co-culture assay, indistinguishable apoptosis levels were induced by CAR-T cells between adherent and sphere ESCC cells (Fig. 3D). The BLI assay showed that CAR-T cells equally reduced tumor cell and CSC viability (Fig. 3E). In good agreement with these results, CD276-directed CAR-T cells produced comparable amounts of effector cytokines upon ESCC tumor cell or CSC engagement (Fig. 3F and G). These results suggest that CD276-specific CAR-T cells could efficiently kill ESCC stem cells.
Fig. 3.
CAR-T cells kill cancer stem cells. Single cell suspension of adherently cultured ESCC tumor cells (adherent) and sphere-derived CSCs (sphere) was separately prepared. (A-C) Surface expressions of CD133 (A), CD44 (B), and CD276 (C) were detected via FACS in tumor cells (adherent) and CSCs (sphere), respectively. Mean fluorescence intensity (MFI) of the target molecules was compared with t-test. (D) Tumor cells (adherent) and CSCs (sphere) were suspended in DMEM/F12 media and added into ultra-low attachment plate, respectively. Then CAR-T cells were added at E/T = 1 without exogenous cytokines. At 6 h and 24 h, apoptosis of tumor cells (EpCAM+CD3−) was determined by Annexin V staining. (E) FFluc+ tumor cells were incubated with indicated CAR-T cells at E/T = 1 without exogenous cytokines for 24 h. Then cell viabilities were calculated relatively to CAR19.28ζ group according to BLI intensity. (F and G) ELISA of effector cytokines from CAR-T cells incubated with adherent or sphere tumor cells. t-test was performed in D-G. Data represent 3 independent repeats. **, P < 0.01; ***, P < 0.005.
Potent anti-ESCC effects of CD276-specific CAR-T cells in vivo
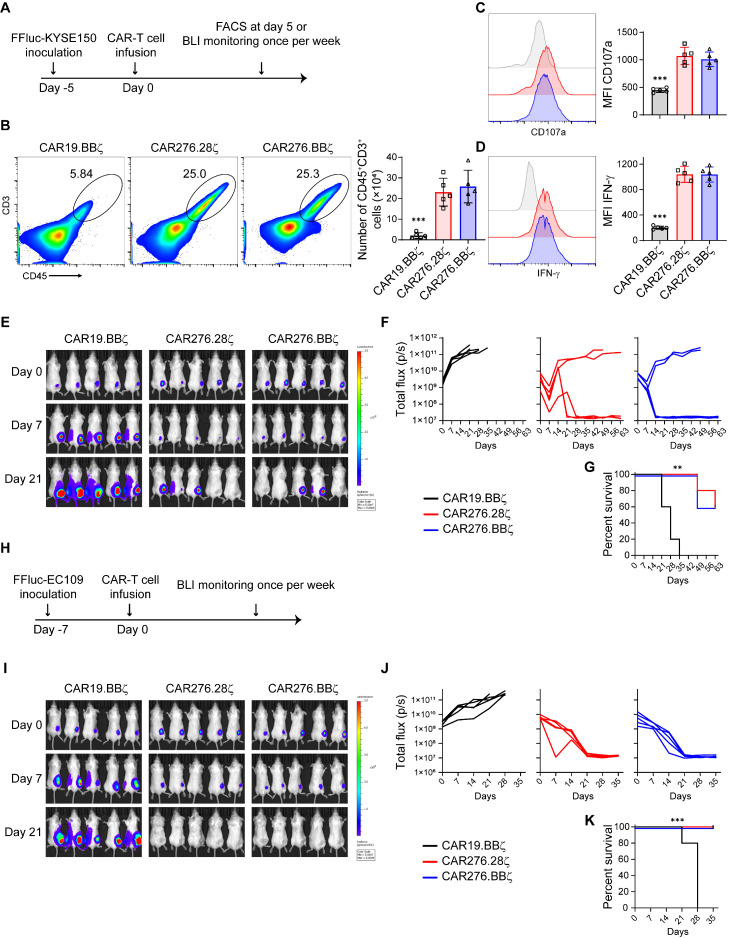
As CD276-specific CAR-T cells could efficiently eliminate ESCC cells in vitro, we evaluated the anti-tumor activity of the engineered T cells in vivo. Immune-deficient NSG mice bearing FFluc+ KYSE150 xenografts were administered with a single injection of CAR-T cells (Fig. 4A). After 5 days, we assessed the tumoral accumulation and activation of CAR-T cells (Supplementary Figure 4A and Fig. 4B-D). In contrast to CD19-specific CAR-T cells, CD276-directed CAR-T cells efficiently expanded (P < 0.05; Fig. 4B). CD276-specific CAR-T cells expressed higher levels of the degranulation marker CD107a and effector molecule IFN-γ than these in the CAR19.BBζ cells (P < 0.05; Fig. 4C and D), indicating that the cytotoxic function of specific CAR-T cells was upregulated. The enhanced accumulation and cytotoxicity suggest that CD276-directed CAR-T cells recognize ESCC cells and then activate in vivo. The differences in expansion and activation between the CAR276.28ζ and CAR276.BBζ cells were negligible (Fig. 4B-D). However, CAR276.BBζ cells exhibited more central memory cells than their CAR276.28ζ counterparts (P < 0.05; Supplementary Figure 4B). The BLI assay showed that both CAR276.28ζ and CAR276.BBζ cells efficiently delayed tumor growth compared with the CAR19.BBζ-treated group (Fig. 4E and F). Accordingly, mouse survival was improved by the CD276-directed CAR-T cells (P < 0.05; Fig. 4G). In the CAR276.28ζ and CAR276.BBζ groups, 60% of the mice reached long-term complete regression (Fig. 4E-G). Intriguingly, CAR-T cells were still circulating in the blood of these mice with complete remission even on day 63 after T cell infusion, although CAR276.BBζ cells were more frequently observed than CAR276.28ζ cells (P < 0.05; Supplementary Figure 4C). Next, the anti-ESCC effect of CD276-directed CAR-T cells was tested in an EC109 xenograft model (Fig. 4H). Both CAR276.28ζ and CAR276.BBζ cells inhibited tumor growth and extended mouse survival (P < 0.05; Fig. 4I-K). These observations suggest that CD276-directed CAR-T cells could eradicate ESCC tumors in vivo.
Fig. 4.
Anti-ESCC effects of CAR-T cells in vivo. (A-G) Anti-ESCC effects of CAR-T cells in KYSE150 xenograft model. NSG mice were inoculated subcutaneously with 1 × 106 FFluc+ KYSE150 cells at day −5 and given a single injection of purified 2 × 106 CAR-T cells via tail veins at day 0 (A). Another 5 days later, FACS was performed to determine expansion (B), just as well as CD107a (C) and IFN-γ (D) expressions of tumor-infiltrating CAR-T cells (n = 5 in each group). Meanwhile, tumor growth was measured by bioluminescence photometry and flux values (photons per second) were calculated using Living Image software weekly (E and F). Mouse survival was recorded (G). Each group had 5 mice. (H-K) Anti-ESCC effects of CAR-T cells in the EC109 xenograft model. NSG mice were inoculated subcutaneously with 5 × 106 FFluc+EC109 cells at day −7 and given 2 × 106 CAR-T cells once via tail veins at day 0 (5 mice per group) (H). Then tumor growth was measured by bioluminescence photometry weekly (I-J). Survival of mice was recorded (K). Survival curves were compared by Log-rank test following Bonferroni correction. **, P < 0.01; ***, P < 0.005.
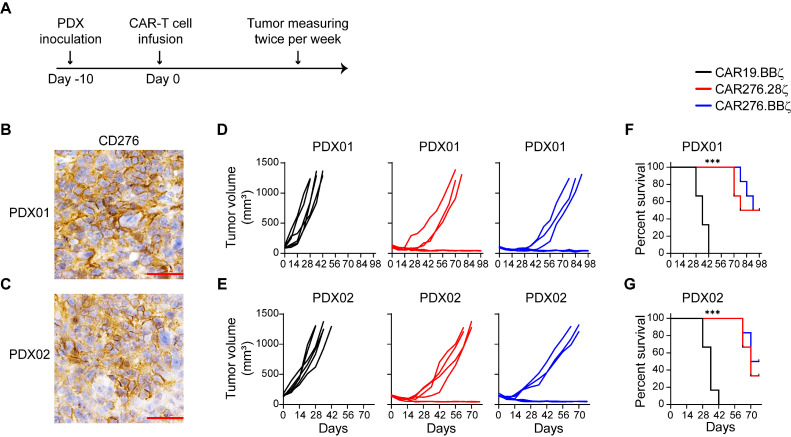
Potent CAR-T cell cytotoxicity against the ESCC PDX tumors
The PDX model better reflects the clinical scenarios in solid tumors [26]. To test the clinical applicability of CD276-directed CAR-T cells, their anti-tumor activity was tested in PDX tumors (Fig. 5A). CD276-expressing ESCC tumor tissues were implanted into NSG mice (Fig. 5B). When PDX tumors were established, CAR-T cells were injected. Compared with the CAR19.BBζ group, the CAR276.28ζ and CAR276.BBζ cells significantly suppressed tumor growth and improved mouse survival (P < 0.05; Fig. 5D and F). In addition, the anti-ESCC effect of CD276-directed CAR-T cells was tested in PDX tumors derived from another ESCC patient (Fig. 5C). As expected, compared with the CD19.BBζ-modified T cells, the CD276-targeting CAR-T cells significantly suppressed tumor growth and extended survival (P < 0.05; Fig. 5E and G).
Fig. 5.
CAR-T cells efficiently eradicate PDX tumors. (A) Schema of experiments. On day −10, ESCC tumor tissues from 2 patients were transplanted into NSG mice at the back, respectively (6 mice/group). On day 0, a total of 2 × 106 CAR-T cells generated from healthy donors were injected through the tail vein. (B and C) CD276 expression in PDX tissues derived from 2 patients with ESCC. Scale bar, 50 μm. (D and E) Tumors were measured with a caliper and the volumes were calculated twice per week. (F and G) Mouse survival was recorded. Survival curves were compared by Log-rank test following Bonferroni correction. ***, P < 0.005.
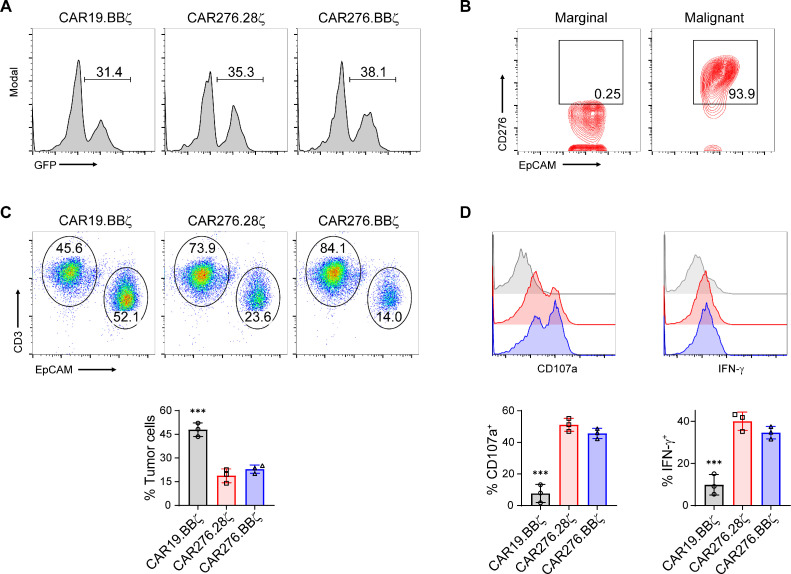
Patient CAR-T cells can eliminate autologous tumor cells
To further test the clinical feasibility of the developed technique, we generated CD276-directed CAR-T cells using T cells from ESCC patients and examined their cytotoxicity against primary tumor cells. As shown in Fig. 6A, patient T cell-derived CAR-T cells were successfully generated. Tumor cells of patients with ESCC had significantly stronger CD276 expression than that in epithelial cells in marginal tissues (Fig. 6B). The tumor cells were then purified and co-incubated with autologous T cells. The CAR19.BBζ cells failed to eliminate the primary tumor cells in the co-incubation system (Fig. 6C and Supplementary Figure 5). In contrast, CD276-targeting CAR-T cells efficiently eliminated autologous tumor cells (Fig. 6C and Supplementary Figure 5). The FACS assay showed that CD276-specific CAR-T cells but not CAR19.BBζ cells expressed high levels of effector molecules (P < 0.05; Fig. 6D), suggesting the CAR-dependent activation of primary T cells.
Fig. 6.
Tumoricidal function of CAR-modified T cells from patients with ESCC. (A) Peripheral T cells isolated from patients with ESCC were transduced and CAR expression was detected 12 days later. (B) Expression of CD276 on healthy epithelial cells or tumor cells from patients with ESCC. (C and D) Primary tumor cells (EpCAM+CD3−) were purified and co-cultured with indicated CAR-T cells (EpCAM−CD3+) at E/T = 1 without exogenous cytokines overnight. Ratios of the residual tumor and T cells were determined by FACS (C). T cells were further analyzed for CD107a and IFN-γ expressions (D). ANOVA tests were performed. Data represent independent experiments with samples from three patients. ***, P < 0.005.
Discussion
The cytotoxicity of CAR-T cells is antigen dependent. However, solid tumors exhibit high inter- and intra-tumoral heterogeneity, resulting in remarkable differences in antigen expression, largely restricting CAR-T cell efficacy [27]. This heterogeneity could also be observed in ESCC [28]. Therefore, identifying widely overexpressed antigens in ESCC is crucial for maximizing CAR-T cell efficacy. However, despite its high prevalence and poor prognosis, our knowledge of suitable ESCC targets remains limited.
CD276 is a transmembrane protein that is an attractive target for CAR-T cells. Previous reports showed that CD276 expression is limited in healthy tissues but prevalent in certain types of solid tumors [13,29]. In ESCC, CD276 is reportedly overexpressed extensively [13]. In good agreement with these findings, our results showed that CD276 expression was upregulated and exhibited extensive coverage of the tumor cells in ESCC. Our study also showed that CD276 is prevalent in EAC. We presented the largest currently available esophageal cancer screen, including the major histopathological forms, ESCC and EAC, for CD276 expression. Our data suggest that CD276 is ubiquitously overexpressed in the majority of tumor tissues of patients with esophageal cancer, especially in case of ESCC. Compared with CD276, the other potential targets, including EGFR and Her2, were only overexpressed in less than 25% of patients with esophageal cancers and demonstrated highly heterogeneous expression [30], [31], [32]. Hence, targeting CD276 is more suitable for esophageal cancer and might reduce therapeutic failure due to antigen heterogeneity.
The differential expression between malignant and non-malignant tissues makes CD276 a good immunotherapeutic target. The CD276-targeting antibody or CAR-T cells have demonstrated good safety and efficacy in immune-proficient mouse and non-human primate models [13,15,33]. In these studies, CD276-targeting antibody or CAR-T cells showed tumor-limited toxicity but had negligible effects on healthy tissues and cells, including immune cells [13,15,33]. More importantly, the feasibility of targeting CD276 has been tested in humans. Several clinical trials showed that CD276-targeting antibodies exhibit clinical benefits without obvious side effects [34], [35], [36]. These studies encourage the development of CD276-targeting CAR-T cells for tumor treatment.
The CD276-specific 8H9 antibody has already been used in the treatment of different tumor types for more than 10 years [34,35], which can better guarantee therapeutic safety when used as an antigen binder within CAR-T cells. However, 8H9 is a mouse-sourced antibody that can induce graft-versus-host disease, thereby mitigating therapeutic effectiveness. Hence, we selected the humanized derivative 8H9S3.318 to construct CAR. The 8H9S3.3 antibody has a comparable affinity to CD276 with the parental 8H9 IgG (KD = 9.85 nM vs KD = 8.9 nM for 4Ig-CD276) and the defined recognizing epitope within CD276 [18]. Our results showed that 8H9S3.3 antibody-derived CAR-T cells had potent cytotoxicity and strict antigen dependence. Similarly, Majzner et al. showed that CAR-T cells with antigen binders from antibodies tested in clinical trials exhibit a significant tumoricidal function in an antigen-dependent manner [14]. With the encouraging outcomes of humanized binder-containing CAR-T cells, we plan to screen antibodies against CD276 from large human antibody libraries and develop fully human CAR-T cells.
The co-stimulatory domains within the CAR are also important for optimal therapeutic responses. The incorporated co-stimulation within the CAR-T cells enhances the cytotoxic functions, maintains T cell fitness, and extends persistence [37]. CD28 and 4–1BB are the most extensively used co-stimulation segments [6]. CD28 is believed to upregulate cytotoxicity but limits the persistence of CAR-T cells [38]. In contrast, 4–1BB induces central memory differentiation and improves CAR-T cell persistence [39], hence providing CAR-T cells with better anti-tumor effects [8]. Our study showed that CAR-T cells with CD28 or 4–1BB co-stimulation exhibited comparable tumoricidal functions in vitro and in vivo, although 4–1BB-co-stimulated CAR-T cells preferred central memory differentiation and better persisted in the blood. Similarly, several reports showed that the anti-tumor efficacy is not significantly different between the CAR-T cells with CD28 or 4–1BB co-stimulation [40], [41], [42]. Our study and previous reports underline that the impact of co-stimulation may be context dependent and co-stimulation should be selected according to the observations in preclinical and clinical trials.
Our study highlights CD276-targeting CAR-T cells as a promising therapeutic strategy for ESCC. Nevertheless, several limitations remain to be addressed. First, an orthotopic tumor model was lacking. The trafficking of CAR-T cells into malignant tissues is critical for tumor eradication. It was observed that the suppression of tumor growth by CD276-specific CAR-T cells in some mice was not as efficient as that in other mice. This may be because of the differential trafficking and infiltration of CAR-T cells. Orthotopic models can better evaluate CAR-T cell trafficking and anti-tumor function in situ. Second, the on-target/off-tumor effects in humans remain undetermined. Owing to limited samples, our study and previous studies could not fully reveal the expression pattern of CD276 in healthy cells and tissues. The off-tumor killing of specific CAR-T cells and the resulting effects require careful evaluation in appropriate models, especially in clinical trials. Indeed, several clinical trials (such as NCT04432649, NCT04185038, and NCT04385173) have been conducted to evaluate the safety of CD276-directed CAR-T cells. We plan to conduct a clinical trial to determine whether CD276-specific CAR-T cells are safe and effective in ESCC patients. Third, the impact of the tumor microenvironment, especially immunosuppressive cells and molecules, on CAR-T cell therapy remains unclear. The immunosuppressive microenvironment largely limits CAR-T cell activity and therapeutic efficacy. A more suitable modality other than immune-deficient mice can help elucidate such impacts and would help develop more efficient regimens in combination with immune checkpoint inhibitors or T cell agonists.
In the present study, we identified CD276 as a candidate target in ESCC and explored the tumoricidal efficacy of CD276-directed CAR-T cells. Overall, our findings support CD276 as a good therapeutic target for ESCC, and targeting CD276 via CAR-T cells may provide a new treatment choice for patients with ESCC.
Grant support
This work was supported by the National Key Research and Development Program (2018YFC1313400), National Natural Science Foundation of China (U2004115, U1804281), Key Research and Development Project of Henan Provincial Science and Technology Department (192102310035), and Medical Science and Technology Breakthrough Project of Henan Province (SB201903004).
Author contributions
Yujing Xuan: Investigation; Methodology; Validation; Formal analysis;
Yuqiao Sheng: Investigation; Methodology; Resources; Visualization;
Daiquan Zhang: Investigation; Validation;
Kai Zhang: Investigation;
Zhen Zhang: Investigation;
Yu Ping: Investigation;
Shumin Wang: Investigation;
Xiaojuan Shi: Investigation;
Jingyao Lian: Investigation;
Kangdong Liu: Methodology; Resources; Writing;
Yi Zhang: Funding acquisition; Writing; Supervision;
Feng Li: Formal analysis; Funding acquisition; Project administration; Supervision; Writing.
Author contributions
F.L. conceived and designed the project; Y.X., D.Z., and F.L. the CAR-T cells; Y.S. generated the PDX tumor models and processed the primary samples; Y.X., D.Z., K.Z., Z.Z., Y.P., S.W., X.S., and J.L. performed the in vitro experiments; F.L. and Y.S. prepared the manuscript draft; and K.L. and Y.Z. helped design the project and revised the manuscript. All authors contributed to the interpretation of the results and the preparation of the manuscript.
Declaration of Competing Interest
All authors declare no competing interests.
Acknowledgments
Acknowledgments
We would like to thank Ning Wang and Lijing Zhang for their assistance in the in vivo imaging experiments, Xixi Duan for help with the IHC experiments, and Jing Zhang for assistance with animal and IHC assays.
Data and material availability
All data associated with this study are presented in the paper or the Supplementary Materials. The cell lines and vectors used in this study are available upon request under a material transfer agreement.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101138.
Contributor Information
Kangdong Liu, Email: kdliu@zzu.edu.cn.
Yi Zhang, Email: yizhang@zzu.edu.cn.
Feng Li, Email: lifeng01@msn.com.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rustgi A.K., El-Serag H.B. Esophageal carcinoma. N. Engl. J. Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 3.Liu K., Zhao T., Wang J. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 2019;458:21–28. doi: 10.1016/j.canlet.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H., Zheng R., Guo Y. Cancer survival in China, 2003-2005: a population-based study. Int. J. Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.June C.H., Sadelain M. Chimeric antigen receptor therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C., Wang Z., Yang Z. Phase I escalating-dose trial of CAR-T therapy targeting CEA+ metastatic colorectal cancers. Mol. Ther. 2017;25:1248–1258. doi: 10.1016/j.ymthe.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellegatta S., Savoldo B., Di Ianni N. Constitutive and TNFalpha-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: implications for CAR-T cell therapy. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aao2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suh W.K., Gajewska B.U., Okada H. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 2003;4:899–906. doi: 10.1038/ni967. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Chen J., Xu B. B7-H3 expression associates with tumor invasion and patient's poor survival in human esophageal cancer. Am. J. Transl. Res. 2015;7:2646–2660. [PMC free article] [PubMed] [Google Scholar]
- 11.Chavin G., Sheinin Y., Crispen P.L. Expression of immunosuppresive B7-H3 ligand by hormone-treated prostate cancer tumors and metastases. Clin. Cancer Res. 2009;15:2174–2180. doi: 10.1158/1078-0432.CCR-08-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J.T., Chen C.H., Ku K.L. Glycoprotein B7-H3 overexpression and aberrant glycosylation in oral cancer and immune response. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13057–13062. doi: 10.1073/pnas.1516991112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seaman S., Zhu Z., Saha S. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell. 2017;31:501–515. doi: 10.1016/j.ccell.2017.03.005. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majzner R.G., Theruvath J.L., Nellan A. CAR T cells targeting B7-H3, a pan-cancer antigen, demonstrate potent preclinical activity against pediatric solid tumors and brain tumors. Clin. Cancer Res. 2019;25:2560–2574. doi: 10.1158/1078-0432.CCR-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du H., Hirabayashi K., Ahn S. Antitumor responses in the absence of toxicity in solid tumors by targeting B7-H3 via chimeric antigen receptor T cells. Cancer Cell. 2019;35:221–237. doi: 10.1016/j.ccell.2019.01.002. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F., Jiang Q., Shi K.J. RhoA modulates functional and physical interaction between ROCK1 and ERK1/2 in selenite-induced apoptosis of leukaemia cells. Cell Death. Dis. 2013;4:e708. doi: 10.1038/cddis.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z., Kang B., Li C. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic. Acids. Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed M., Cheng M., Zhao Q. Humanized affinity-matured monoclonal antibody 8H9 has potent antitumor activity and binds to FG loop of tumor antigen B7-H3. J. Biol. Chem. 2015;290:30018–30029. doi: 10.1074/jbc.M115.679852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kochenderfer J.N., Feldman S.A., Zhao Y. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J. Immunother. 2009;32:689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran F.A., Hsu P.D., Wright J. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., Yue D., Chen X. Epigenetic regulation of CD271, a potential cancer stem cell marker associated with chemoresistance and metastatic capacity. Oncol. Rep. 2015;33:425–432. doi: 10.3892/or.2014.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W., Bi C., Credille K.M. Inhibition of tumor growth and metastasis in non-small cell lung cancer by LY2801653, an inhibitor of several oncokinases, including MET. Clin. Cancer Res. 2013;19:5699–5710. doi: 10.1158/1078-0432.CCR-13-1758. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G., Hou J., Shi J. Soluble CD276 (B7-H3) is released from monocytes, dendritic cells and activated T cells and is detectable in normal human serum. Immunology. 2008;123:538–546. doi: 10.1111/j.1365-2567.2007.02723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Tomaso T., Mazzoleni S., Wang E. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res. 2010;16:800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Y., Yang H., Levorse J. Adaptive immune resistance emerges from tumor-initiating stem cells. Cell. 2019;177:1172–1186. doi: 10.1016/j.cell.2019.03.025. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassidy J.W., Caldas C., Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75:2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newick K., O'Brien S., Moon E. CAR T cell therapy for solid tumors. Annu. Rev. Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- 28.Yan T., Cui H., Zhou Y. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat. Commun. 2019;10:1670. doi: 10.1038/s41467-019-09255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zang X., Thompson R.H., Al-Ahmadie H.A. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato H., Arao T., Matsumoto K. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int. J. Oncol. 2013;42:1151–1158. doi: 10.3892/ijo.2013.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fusco N., Bosari S. HER2 aberrations and heterogeneity in cancers of the digestive system: implications for pathologists and gastroenterologists. World J. Gastroenterol. 2016;22:7926–7937. doi: 10.3748/wjg.v22.i35.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., Zhu S.M., Xu X.L. Expression levels of HER2 and MRP1 are not prognostic factors of long-term survival in 829 patients with esophageal squamous cell carcinoma. Oncol. Lett. 2016;11:745–752. doi: 10.3892/ol.2015.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loo D., Alderson R.F., Chen F.Z. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin. Cancer Res. 2012;18:3834–3845. doi: 10.1158/1078-0432.CCR-12-0715. [DOI] [PubMed] [Google Scholar]
- 34.Kramer K., Kushner B.H., Modak S. Compartmental intrathecal radioimmunotherapy: results for treatment for metastatic CNS neuroblastoma. J. Neurooncol. 2010;97:409–418. doi: 10.1007/s11060-009-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer K., Smith M., Souweidane M.M. Safety profile of long-term intraventricular access devices in pediatric patients receiving radioimmunotherapy for central nervous system malignancies. Pediatr. Blood Cancer. 2014;61:1590–1592. doi: 10.1002/pbc.25080. [DOI] [PubMed] [Google Scholar]
- 36.Powderly J., Cote G., Flaherty K. Interim results of an ongoing Phase I, dose escalation study of MGA271 (Fc-optimized humanized anti-B7-H3 monoclonal antibody) in patients with refractory B7-H3-expressing neoplasms or neoplasms whose vasculature expresses B7-H3. J. Immunother. Cancer. 2015;3:O8. [Google Scholar]
- 37.Rafiq S., Hackett C.S., Brentjens R.J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 2020;17:147–167. doi: 10.1038/s41571-019-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z., Condomines M., van der Stegen S.J.C. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell. 2015;28:415–428. doi: 10.1016/j.ccell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawalekar O.U., O'Connor R.S., Fraietta J.A. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. 2016;44:380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Carpenito C., Milone M.C., Hassan R. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3360–3365. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Z., Wei R., Ma Q. In vivo expansion and antitumor activity of coinfused CD28- and 4-1BB-engineered CAR-T cells in patients with B cell leukemia. Mol. Ther. 2018;26:976–985. doi: 10.1016/j.ymthe.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam N., Trinklein N.D., Buelow B. Anti-BCMA chimeric antigen receptors with fully human heavy-chain-only antigen recognition domains. Nat. Commun. 2020;11:283. doi: 10.1038/s41467-019-14119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.