Verma et al. present an experimental approach for generation of ILC2 memory for allergic asthma. scRNA-seq and ATAC-seq identify a set of preparedness- and repression-inducing genes with memory formation. Their approach demonstrates a critical role for Fhl2 and ICOS in ILC2 memory.
Abstract
Repetitive exposure of Rag1−/− mice to the Alternaria allergen extract generated a form of memory that elicited an asthma-like response upon a subthreshold recall challenge 3–15 wk later. This memory was associated with lung ICOS+ST2+ ILC2s. Genetic, pharmacologic, and antibody-mediated inhibition and adoptive transfer established an essential role for ILC2s in memory-driven asthma. ATAC-seq demonstrated a distinct epigenetic landscape of memory ILC2s and identified Bach2 and AP1 (JunD and Fosl2) motifs as major drivers of altered gene accessibility. scRNA-seq, gene knockout, and signaling studies suggest that repetitive allergenic stress induces a gene repression program involving Nr4a2, Zeb1, Bach2, and JunD and a preparedness program involving Fhl2, FosB, Stat6, Srebf2, and MPP7 in memory ILC2s. A mutually regulated balance between these two programs establishes and maintains memory. The preparedness program (e.g., Fhl2) can be activated with a subthreshold cognate stimulation, which down-regulates repressors and activates effector pathways to elicit the memory-driven phenotype.
Introduction
Type 2 innate lymphoid cells (ILC2s) represent a rare lymphoid cell population that is rich in type 2 cytokines such as IL4, IL5, IL9, and IL13 (reviewed in Klose and Artis, 2016; Spits and Di Santo, 2011). ILC2s play an important role in host defense, allergic inflammation, tissue regeneration, and homeostasis. They are primarily activated by endogenous factors such as cytokines, hormones, neurotransmitters, and lipid mediators (Klose et al., 2017; Oliphant et al., 2014; Wallrapp et al., 2017). Unlike T cells, ILC2s do not express antigen-specific receptors. However, ILC2s internalize antigens, present them to T cells, and facilitate type 2 T helper cell (Th2) differentiation (Halim et al., 2016). Whether they have the ability to generate memory for the internalized antigens and respond to a recall challenge is incompletely understood.
We reported the establishment of a mouse model of asthma wherein airway hyperreactivity and other features of asthma persisted unabated for >6 mo after cessation of allergen exposure (Christianson et al., 2015). ILC2s, but not T or B cells, were essential for persistence of asthma in that model. Adoptive transfer of ILC2s from the allergen-treated chronic asthma model but not from saline (Sal)-treated control mice established chronic asthma in naive recipient mice. Those results indicated the presence of a “memory-like” property of ILC2s from the chronic asthma model. Although ILC2s do not express antigen-specific receptors, they express pattern recognition receptors (PRRs). The PRRs recognize pathogen-associated molecular patterns (PAMPs), which are associated with many allergens. PRR stimulation induces trained immunity (a form of memory) in macrophages (Saeed et al., 2014). Thus, allergen-associated PAMPs could induce trained immunity-like memory in ILC2s. ILC2s have recently been shown to generate long-lasting memory after exposure to IL33 and allergens such as papain. This was associated with increased ILC2s in the lung and mediastinal lymph nodes (Martinez-Gonzalez et al., 2016). Aspergillus-primed ILC2s responded more vigorously to papain and were more efficient in promoting Th2 cell differentiation. In this paper, we study the molecular and epigenetic mechanisms of development of ILC2 memory and its relevance for asthma.
Results
Generation of allergen-induced memory in Rag1−/− mice
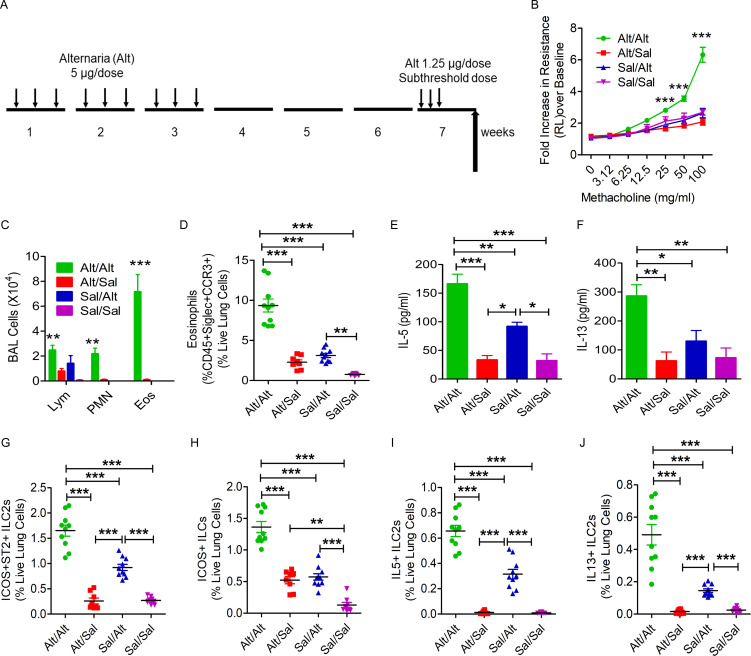
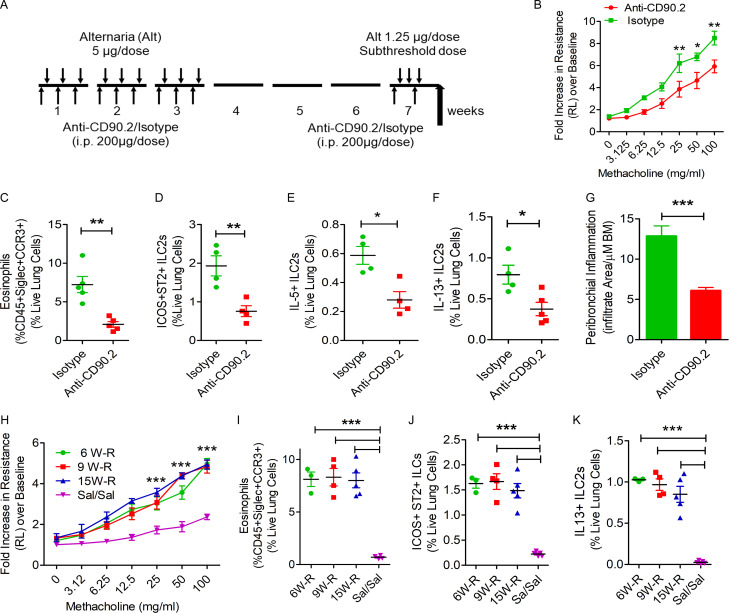
The capacity of ILCs to generate allergen-specific memory and respond to a subthreshold recall challenge is not known. To address this question, we designed the following experiment. We sensitized Rag1−/− female mice intranasally with the Alternaria (Alt) allergen extract (5 µg/dose) or Sal on three alternate days a week for three consecutive weeks (Fig. 1 A). We ceased the exposure for 3 wk and then performed a recall challenge with a predetermined subthreshold dose (1.25 µg, one quarter of the sensitization dose, which did not elicit airway hyperreactivity) of Alt or Sal in week 7 for three subsequent days. We measured airway hyperreactivity to inhaled methacholine 3 d after the last recall dose and analyzed bronchoalveolar lavage (BAL) and lung tissue for inflammatory cell influx and cytokine production. Sensitization of Rag1−/− mice to Alt followed by a recall challenge with a subthreshold dose of Alt (Alt/Alt) but not Sal (Alt/Sal) induced airway hyperreactivity (increase in airway resistance as measured by flexiVent; Fig. 1 B) and eosinophilic influx in BAL and lung (Fig. 1, C and D) and increased IL5 and IL13 in BAL (Fig. 1, E and F), mild airway inflammation (histological examination of H&E staining of the lung tissue; Fig. S1, A and B) and peribronchial collagen deposition (Fig. S1, C and D). The Sal/Alt group had mildly elevated IL5. The control Sal/Sal group did not have any response. Flow cytometry (FCM) analysis of lung cells showed an increase in ICOS+ST2+, ICOS+, IL5+, and IL13+ ILC2s (CD45+CD25+ lung cells) in the Alt/Alt group as compared with other control groups (Fig. 1, G–J). The Sal/Alt group had mildly elevated ICOS+ST2+ and ICOS+, IL5+, and IL13+ ILC2s as compared with the Alt/Sal and Sal/Sal groups. The Alt/Sal group had elevated ICOS+ ILC2s as compared with the Sal/Sal group. The gating strategy for the flow analyses is shown in Fig. S1, E–G. The gated ILC2 population did not have any contamination with TCRγδ+, Ly6G+/Ly6C+, NKp46+, CD11B+, NK1.1+, or FcεRI+ cells (Fig. S2 A). We repeated this experiment using a group of male mice, which are known to have reduced numbers of ILC2s as compared with female mice (Cephus et al., 2017). We observed a similar increase in asthma-like response to a subthreshold dose of Alt in male mice (data not shown). The results suggest that airway innate cells are capable of developing a memory for environmental allergens and generate an asthma-like response to a subthreshold recall challenge.
Figure 1.
Establishment of a memory-driven asthma model in Rag1−/− mice. (A) A schematic diagram of the timeline of allergen exposure, recall challenge, and performance of experiments. Groups of Rag1−/− female mice were intranasally exposed to Alt (5 µg in 20 µl of Sal/dose) or Sal three alternate d/wk in weeks 1–3 and then rested for 3 wk. The mice had a recall challenge in week 7 with a subthreshold dose (1.25 µg/dose) of Alt on three consecutive days and then were examined for airway hyperreactivity and immunological alterations 3 d later. (B) Increase in lung resistance over baseline (as measured by flexiVent) in response to increasing doses of inhaled methacholine in Alt/Alt, Alt/Sal, Sal/Alt, and Sal/Sal groups. ***, P < 0.0001 versus control groups, two-way ANOVA, n = 10/group. (C) Differential leukocyte counts of BAL. **, P < 0.001; ***, P < 0.0001, two-way ANOVA, n = 10/group. Eos, eosinophil; Lym, lymphocyte; PMN, polymorphonuclear cell. (D) Eosinophils (CD45+Siglec8+CCR3+ cells) in the lung from the study groups. Blood-depleted lung tissue from the study mice was digested with collagenase, and the single-cell suspension from the lung digest was stained and analyzed for eosinophils by FCM. ***, P < 0.0001, one-way ANOVA, n = 10/group. (E and F) IL5 and IL13 (measured by ELISA) in BAL. ***, P < 0.0001, n = 10/group, one-way ANOVA. (G–J) ICOS+ST2+, ICOS+, IL5+, and IL13+ ILC2s in the lung. Single-cell suspensions from the lung, processed as described above, were stained and analyzed for ICOS+ST2+, ICOS+, IL5+, and IL13+ ILC2s (CD45+CD25+ lung cells) by FCM. ***, P < 0.0001, n = 10/group, one-way ANOVA.
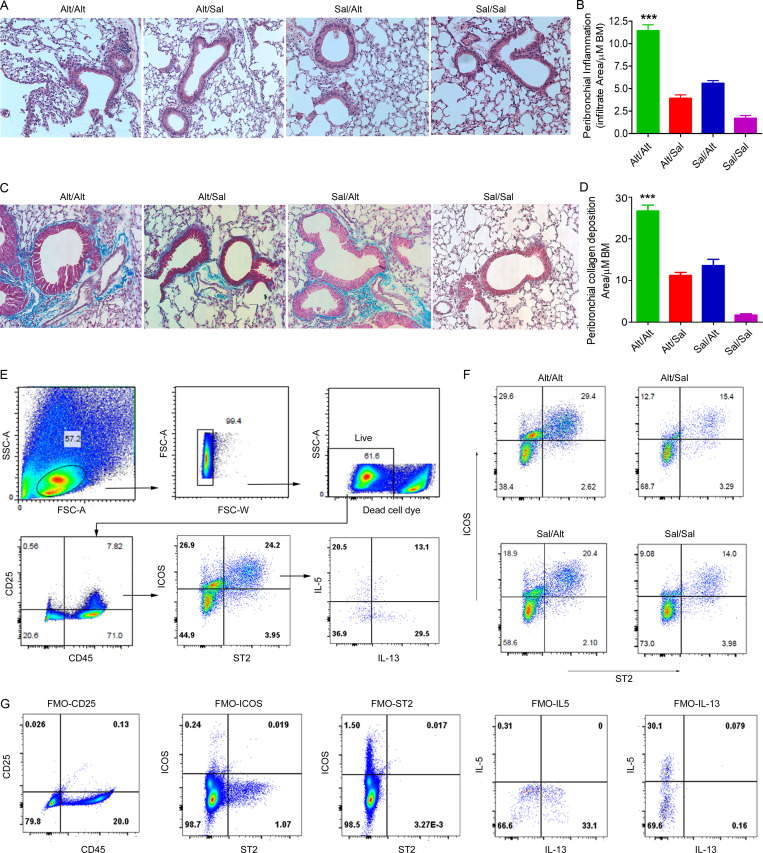
Figure S1.
Representative histological images and gating strategy for flow cytometry. (A–D) Histological changes in the memory model. (A and B) H&E staining and morphometric quantification of the lung tissue from the experimental groups: Sal/Alt, Alt/Sal, Alt/Alt, and Sal/Sal. ***, P < 0.0001. (C and D) Peribronchial collagen deposition and its morphometric quantification from the study groups as described previously (Christianson et al., 2015). ***, P < 0.0001 as compared with other groups, one-way ANOVA, n = 10 per group. Scale bars are 100 µm, and the images were acquired at 20×. BM, bone marrow. (E–G) FCM analysis of lung ILCs from the study groups. Blood-depleted mouse lungs from the study groups were digested with collagenase, and single cells were stained with antibodies against CD45, CD25, ICOS, ST2, IL5, and IL13. Following gating live single cells, they were further gated for CD45+CD25+ cells and then analyzed for ICOS+ST2+, IL5+ICOS+ST2+, and IL13+ICOS+ST2+ cells. (F) Representative flow cytograms showing ICOS+ST2+ cells from the four study groups. (G) Fluorescence minus one (FMO) for the studied molecules used to develop the gating strategy. FSC, forward scatter; SSC, side scatter.
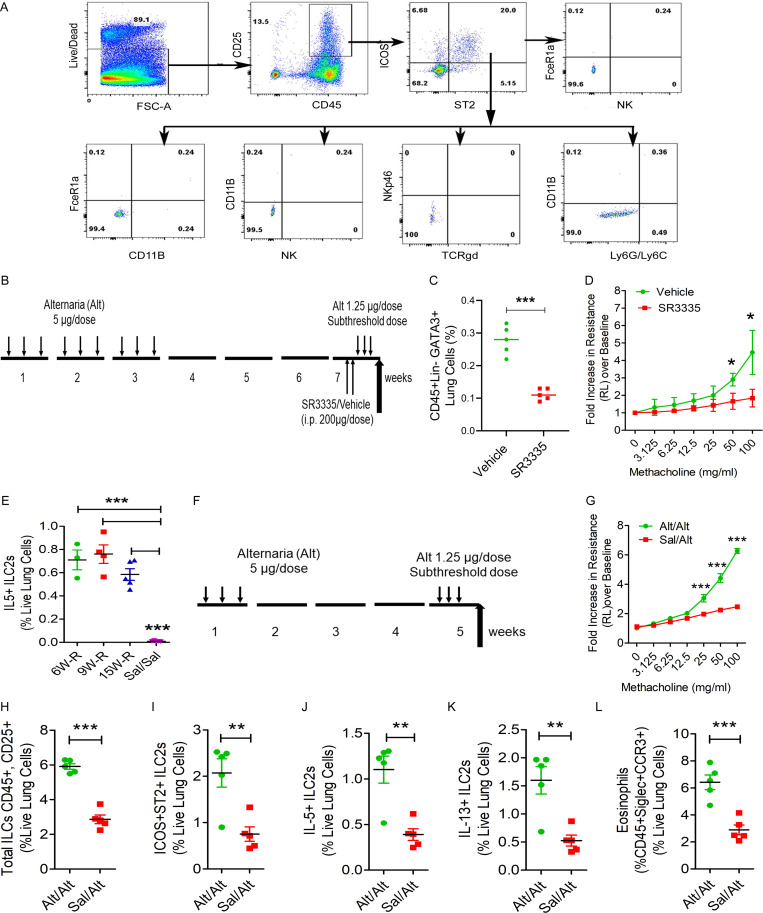
Figure S2.
Flow cytometry of hematopoietic cells, RORα inhibition, and a short memory-induction protocol. (A) Representative flow cytograms examining TCRγδ+, Ly6G+/Ly6C+, NKp46+, CD11B+, NK1.1, and FcεRI+ cells in ICOS+ST2+ cells from the five study groups. (B–D) Effect of the RORα inverse agonist SR3335 on ILC2s. A schematic diagram of the timeline of RORα inverse agonist SR3335 in memory (B), reduced GATA3+ ILC2s (C), and airway hyperreactivity (D) in the memory model. *, P < 0.01; ***, P < 0.0001, unpaired t test, n = 5/group. (E) IL5+ ILC2s from the persistence of memory experiment. ***, P < 0.0001, one-way ANOVA, n = 3–5/group. (F–L) A short exposure model for memory-driven asthma in Rag1−/− mice. (F) A schematic diagram of the timeline of allergen exposure and recall challenge. Groups of Rag1−/− female mice were intranasally exposed to Alt (5 µg in 20 µl of Sal/dose) or Sal on alternate days for 3 d in 1 wk and then rested for 3 wk. The mice had a recall challenge in week 5 with a subthreshold dose (1.25 µg/dose) of Alt on three consecutive days and then were examined for airway hyperreactivity and immunological alterations. (G–L) Lung resistance over baseline (as measured by flexiVent) in response to increasing doses of inhaled methacholine (G), total ILCs (H), ICOS+ST2+ ILCs (I), IL5+ ILC2s (J), and IL13+ ILC2s (K) and eosinophils (L) in lung digest from the Alt/Alt and Sal/Alt study groups. **, P < 0.001; *** P < 0.0001, unpaired t test, n = 5/group.
Role of natural killer (NK) cells in the memory response
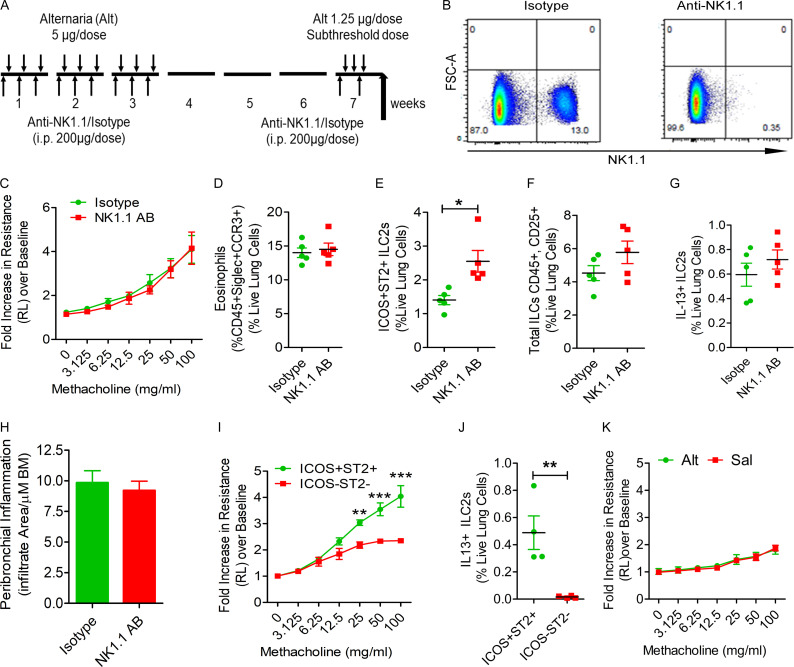
Rag1−/− mice lack T and B cells but have ILCs and NK cells. To determine which of these innate immune cells are involved in the memory response, we treated mice with an anti-NK1.1 antibody that was previously demonstrated to deplete NK cells (Manners et al., 2014). Pretreatment with the anti-NK1.1 antibody completely depleted NK cells from the lungs obtained from Rag1−/− mice (Fig. 2, A and B). However, this treatment did not affect the development of airway hyperreactivity and eosinophilic inflammation in response to the subthreshold Alt challenge (Fig. 2, C and D). It did increase the number of ICOS+ST2+ ILC2s in the lung (Fig. 2 E) but did not affect the total number of ILCs and IL13+ ILC2s and airway inflammation (Fig. 2, F–H).
Figure 2.
The role of NK cells and ILCs in memory-driven asthma. (A–C) Role of NK cells and ILCs in memory-driven asthma. (A) A schematic diagram of the timeline of antibody and allergen exposure and recall challenge experiments. An aliquot of 200 µg of an anti-NK1.1 antibody was administered i.p. to Rag1−/− female mice 1 d before each Alt dose during the exposure phase, 1 d before the recall challenges, and 1 h before the last recall challenge. (B–D) A representative flow cytogram (B) for NK cells in the lung digest in NK1.1 antibody– and control antibody–treated mice, airway hyperreactivity (C) and lung eosinophils (CD45+SiglecF+ CCR3+ cells; D). (E–H) Lung ILC2s (CD45+NK1.1−CD25+ICOS+ST2+ cells; E), total ILCs (CD45+CD25+ ILCs; F), IL13+ ILC2s (CD45+CD25+ICOS+ST2+ cells; G) and airway inflammation (H) in the memory model after depletion of NK cells. *, P < 0.01, n = 5, unpaired t test. FSC, forward scatter. (I and J) Induction of asthma by adoptive transfer of memory ILCs. CD45+NK1.1−CD25+ICOS+ST2+ (ICOS+ST2+) and CD45+NK1.1−CD25+ICOS−ST2− (double-negative) ILCs were obtained from Alt/Alt-treated mice. 50,000 cells were intravenously transferred to Rag2−/−:γc−/− mice. 1 d after the transfer, the recipient mice were challenged with Alt for three consecutive days and examined for airway hyperreactivity (lung resistance in response to methacholine) and IL13+ ILC2s 24 h later. **, P < 0.001; ***, P < 0.0001 (two-way ANOVA and unpaired t test, n = 4/group). (K) Naive Rag2−/−:γc−/− mice were challenged with Alt or Sal for three consecutive days and examined for airway hyperreactivity (lung resistance in response to methacholine). Two-way ANOVA, n = 5/group.
Role of ILCs in the memory response
To address if ICOS+ST2+ ILC2s are independently capable of a memory response, we sensitized Rag1−/− mice intranasally with Alt three times a week for 3 wk as described in Fig. 1 A. We FACS-sorted ICOS+ST2+ ILC2s (CD45+CD25+NK1.1−ICOS+ST2+) and ICOS−ST2− ILCs (CD45+CD25+NK1.1−ICOS−ST2−, referred to as double negative) from the lung digest and transferred them i.v. to Rag2−/−:γc−/− mice. After 24 h, we challenged the recipient mice with the subthreshold dose of Alt for three consecutive days and measured airway hyperreactivity 24 h later. ICOS+ST2+ ILC2 recipient, but not double-negative ILC recipient, Rag2−/−:γc−/− mice developed airway hyperreactivity in response to Alt (Fig. 2 I). The ICOS+ST2+ ILC2 recipients also showed increased numbers of IL13+ ILC2s in the lung (Fig. 2 J). In control experiments, we treated naive and nontransferred Rag2−/−:γc−/− mice with Alt and Sal (as a negative control) the same way as the ILC2 recipient mice. Alt and Sal treatment failed to induced airway hyperreactivity in naive Rag2−/−:γc−/− mice (Fig. 2 K)
ILCs as well as a subgroup of NK cells express CD90. Therefore, we used an anti-CD90 antibody to rule out the role of ILCs in memory formation (Fig. 3 A). An anti-CD90 antibody has previously been used to deplete ILCs in Rag1−/− mice (Monticelli et al., 2011). In our memory model, an anti-CD90.2 antibody but not an isotype control antibody inhibited airway hyperreactivity and eosinophilic inflammation in response to subthreshold Alt challenge (Fig. 3, B and C). There was a concomitant decrease in ICOS+ST2+ ILC2s (Fig. 2 D), IL5+ ILC2s, and IL13+ ILC2s and airway inflammation (Fig. 3, E–G).
Figure 3.
Effect of anti-CD90 antibody-mediated depletion of ILCs and duration of memory. (A) A timeline for anti-CD90.2 administration. (B–G) Effect of depletion ILCs by treatment with an anti-CD90 antibody (isotype antibody as a control) on airway hyperreactivity (B), lung eosinophils (C), ICOS+ST2+ ILC2s (D), IL5+ (E), IL13+ (F), and airway inflammation (G). BM, bone marrow. ***, P < 0.0001; **, P < 0.001; *, P < 0.01, unpaired t test in Rag1−/− mice sensitized and challenged with Alt. (H–K) The duration of persistence of memory. Rag1−/− mice were exposed to Alt as per Fig. 1 A and then had a recall (R) challenge with Alt in week (W) 6, 9, or 15 (labeled as 6, 9, or 15 W-R, respectively). A control group was exposed to Sal and challenged with Sal in week 15 (Sal/Sal). The mice had measurements of airway hyperreactivity (H; two-way ANOVA), lung eosinophils (I), CD45+CD25+ICOS+ST2+ ILCs (J), and IL13+ ILC2s (CD45+CD25+ICOS+ST2+; K) as described above. ***, P < 0.0001 versus Sal/Sal (one-way ANOVA, n = 3–5/group).
ILC2s require the transcription factor retinoic acid–related orphan receptor α (RORα) for maturation and survival (Wong et al., 2012). Recently, the RORα inverse agonist SR3335 was shown to deplete ILC2s in mice (Ito et al., 2017). We treated Rag1−/− mice from the memory model with two doses (200 µg/dose, i.p.) of this compound before the recall challenge (Fig. S2 B). SR3335 significantly reduced the number of GATA3+ ILC2s in mice, which was associated with a concomitant decline in airway hyperreactivity (Fig. S2, C and D). The results in aggregate suggest that ILC2s were sufficient and nonredundant for induction of memory-driven asthma in our model.
Persistence of memory and the duration of allergen exposure for memory induction
Next, we examined the duration of ILC2 memory. We sensitized mice intranasally three times a week for three consecutive weeks as per Fig. 1 A and examined airway resistance in response to subthreshold Alt challenge in weeks 6, 9, and 15. We observed persistence of airway hyperreactivity at all three time points (Fig. 3 H). This was associated with eosinophilic infiltrates (Fig. 3 I), and increased ICOS+ST2+ ILC2s, IL13+ ILC2s (Fig. 3, J and K), and IL5+ ILC2s (Fig. S2 E) in the lung. There was a decreasing trend in the number of lung ILC2s over time.
In initial experiments, we gave mice a total of nine exposures (3 d/wk for 3 wk) for memory induction. In another set of experiments, we gave Rag1−/− mice a total of three exposures of Alt (5 µg/dose) or Sal (3 d/wk for 1 wk) and then performed the subthreshold (1.25 µg/dose) recall challenge 3 wk later. We observed robust induction of airway hyperreactivity and increases in total and ICOS+ST2+ ILC2s, IL5+ and IL13+ ILC2s, and eosinophilic influx upon the recall challenge (Fig. S2, F–L). The results suggest that just three allergen exposures were sufficient to induce memory.
ICOS and IL33 are essential for ILC memory
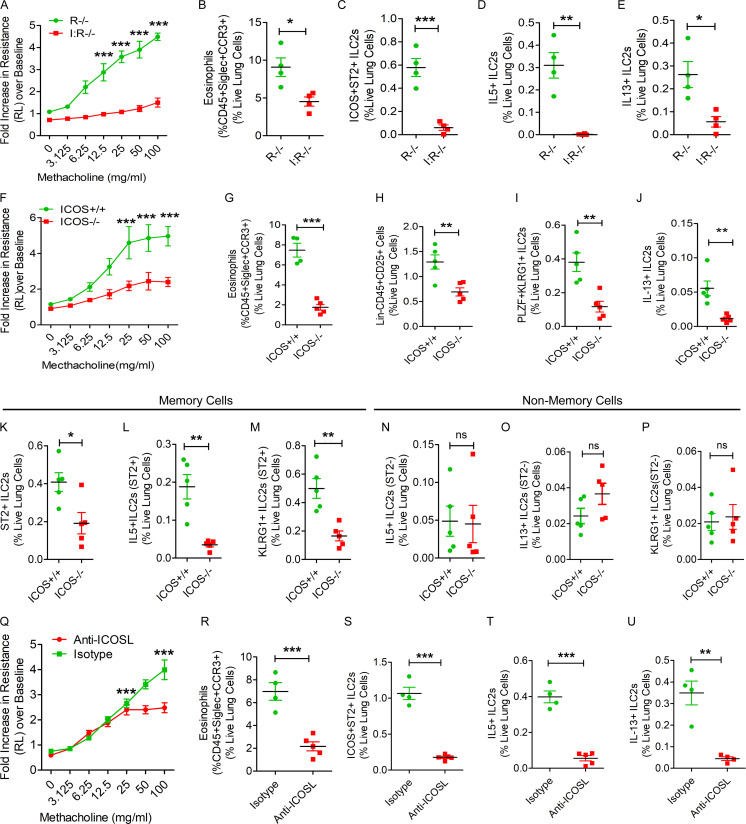
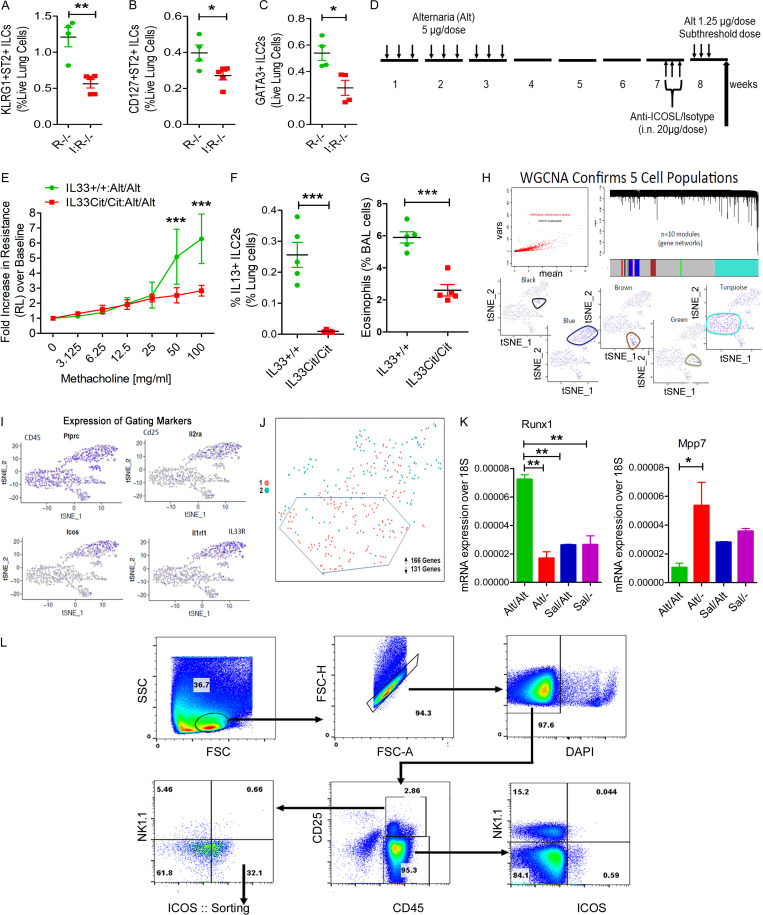
ICOS+ST2+ but not ICOS−ST2− innate immune cells from sensitized Rag1−/− mice mediated memory-driven asthma when adoptively transferred to Rag2−/−:γc−/− mice (Fig. 2 I), suggesting an important role for ICOS and/or ST2 in memory. Next, we examined ILC2 memory generation in ICOS−/−:Rag1−/− mice. These mice had reduced memory-driven airway hyperreactivity and eosinophilic inflammation and near-absent ICOS+ST2+, IL5+, and IL13+ ILC2s in the lung as compared with Rag1−/− mice (Fig. 4, A–E). In addition, ICOS−/−:Rag1−/− mice had reduced numbers of KLRG1+ST2+, CD127+ST2+, and GATA3+ ILC2s (Fig. S3, A–C). We also studied ICOS−/− mice, which express both adaptive and innate immune cells. ICOS−/− mice failed to induce airway hyperreactivity and eosinophilic inflammation and had reduced total ILCs, PLZF+KLRG1+ ILC2s, and IL13+ ILC2s (Fig. 3, F–J). To determine whether the germline deletion of the ICOS gene affects memory versus nonmemory cells, we compared the frequency of ST2+ and ST2− cells as representatives of memory ILC2s and nonmemory ILCs, and then analyzed the expression of the activation marker KLRG1 and type 2 cytokines IL5 and IL13 to memory cell–associated markers ST2 and KLRG1. The frequency of ST2+ memory ILC2s was decreased in ICOS−/− mice (Fig. 4 K). The expression of KLRG1, IL5, and IL13 was impaired in ST2+ memory ILC2s but not ST2− nonmemory ILCs (Fig. 4, L–P). To further substantiate the role of ICOS, we treated Rag1−/− mice with a neutralizing anti-ICOSL antibody before the recall challenge (Fig. S3 D). The anti-ICOSL antibody inhibited airway hyperreactivity and eosinophilic inflammation (Fig. 4, Q and R) and the generation of ICOS+ST2+ ILC2s, IL5+ ILC2s, and IL13+ ILC2s (Fig. 4, S–U). The results, in aggregate, indicate a critical role for ICOS in ILC memory generation.
Figure 4.
Importance of ICOS for memory-driven asthma. (A–E) ICOS−/−:Rag1−/− (I:R−/−) and Rag1−/− (R−/−) mice were subjected to the Alt-induced memory-driven asthma protocol as per Fig. 1 A. Airway hyperreactivity (A) was measured by flexiVent, two-way ANOVA. Lung eosinophils (CD45+SiglecF+CCR3+ cells; B), ST2+, IL5+, and IL13+ ILC2s (CD45+NK1.1−CD25+ICOS+ST2+) cells were measured by FCM (C–E). *, P < 0.01; **, P < 0.001; ***, P < 0.0001, unpaired t test, n = 4/group. (F–J) ICOS−/− and littermate ICOS+/+ mice were subjected to the Alt-induced memory-driven asthma protocol as per Fig. 1 A. Airway hyperreactivity was measured by flexiVent (F). Lung eosinophils, ILCs (Lin−CD45+CD25+), ILC2s (Lin−CD45+CD25+PLZF+KLRG1+), and IL13+ ILC2s (CD45+Lin−CD25+PLZF+KLRG1+) cells were measured by FCM (G–J). *, P < 0.01; **, P < 0.001; ***, P < 0.0001; ns, not significant; two-way ANOVA. (K–P) Comparison of memory (ST2+) and nonmemory (ST2−) ILCs between ICOS+/+ and ICOS−/− mice. The frequency of ST2 + ILC2s (K) and KLRG1+ IL13+ and IL5+ ILC2s in ST2+ and ST2− ILCs (L–P). *, P < 0.01; **, P < 0.001; ***, P < 0.0001, unpaired t test, n = 4–5/group. (Q–U) Rag1−/− mice were intranasally pretreated with an anti-ICOSL antibody or an isotype control antibody before recall challenge. Airway hyperreactivity (Q) was measured by flexiVent, two-way ANOVA. Lung eosinophils (CD45+SiglecF+CCR3+ cells; R), ICOS+ST2+ ILCs (CD45+CD25+; S), and IL5+, IL13+ ILC2s (CD45+CD25+ICOS+ST2+; T and U) in the lung digest were measured by FCM. ***, P < 0.001, unpaired t test, n = 4–5/group.
Figure S3.
Role of ICOS and IL33 in memory and gating strategy for scRNA-seq. (A–C) Importance of ICOS for memory-associated asthma. ICOS−/−:Rag1−/− (I:R−/−) and Rag1−/− (R−/−) mice were subjected to the Alt-induced memory-driven asthma protocol as per Fig. 1 A. KLRG1+ST2+, CD127+ ST2+, and GATA3+ ILC2s (CD45+NK1.1−CD25+) cells were measured by FCM. *, P < 0.01; **, P < 0.001; unpaired t test, n = 4/group. (D) A schematic diagram of the timeline of anti-ICOSL/isotype antibody exposure. (E–G) Role of IL33 in memory formation. IL33Cit/Cit and IL33+/+ mice were subjected to the Alt/Alt or Sal/Sal exposure and recall protocol as per Fig. 1 A. Airway hyperreactivity (E), lung IL13+ ILC2s (Lin−CD45+CD25+NK1.1−ICOS+ST2+; F), and BAL eosinophils (G) were measured. ***, P < 0.0001, n = 5/group, unpaired t test. (H) scRNA-seq of lung CD45+ cells. Lung CD45+ cells were sorted following digestion of blood-depleted lungs with collagenase. Cells from each sample were subjected to scRNA-seq by Wafergen iCell8 protocol followed by Illumina Hi-seq sequencing. t-SNE coordinate clustering of cells with WGCNA network module eigengene expression overlaid for modules highly expressed in each particular cluster. Based on genes contained in the cell discriminatory modules, we defined the clusters as macrophages (turquoise), NK cells (black), dendritic cells (green), endothelial cells (brown), and ILC2s (blue). (I) The expression profile of CD45, Il2ra, Icos, and il1rl1 in each cell cluster is shown. (J) Partial separation of memory ILCs (Alt/− and Alt/Alt in red) from control ILCs (Sal/− and Sal/Alt in turquoise) by t-SNE analysis. A total of 161 and 136 genes were up- and down-regulated, respectively, in memory ILCs. (K) Expression of mRNA for Runx1 and Mpp7 in ILCs from the memory models. *, P < 0.01; **, P < 0.001, n = 5/group, one-way ANOVA. (L) The gating and sorting strategy for isolation of CD45+NK1.1−CD25+ICOS+ cells for ATAC-seq. FSC, forward scatter; SSC, side scatter.
Alt induces IL33 release from airway cells (Snelgrove et al., 2014). To investigate the role of IL33, we used IL33−/− mice that had the cDNA for citrine knocked in (IL33Cit/Cit) the IL33 locus (Hardman et al., 2013). IL33−/− (IL33Cit/Cit) mice had absent memory-driven airway hyperreactivity and IL13+ ILC2s (Fig. S3, E and F) and negligible numbers of eosinophils in the lung (Fig. S3, G). The results underscore a critical role for IL33 and IL33-producing cells in Alt-induced memory.
scRNA-seq identifies ILC memory-related genes
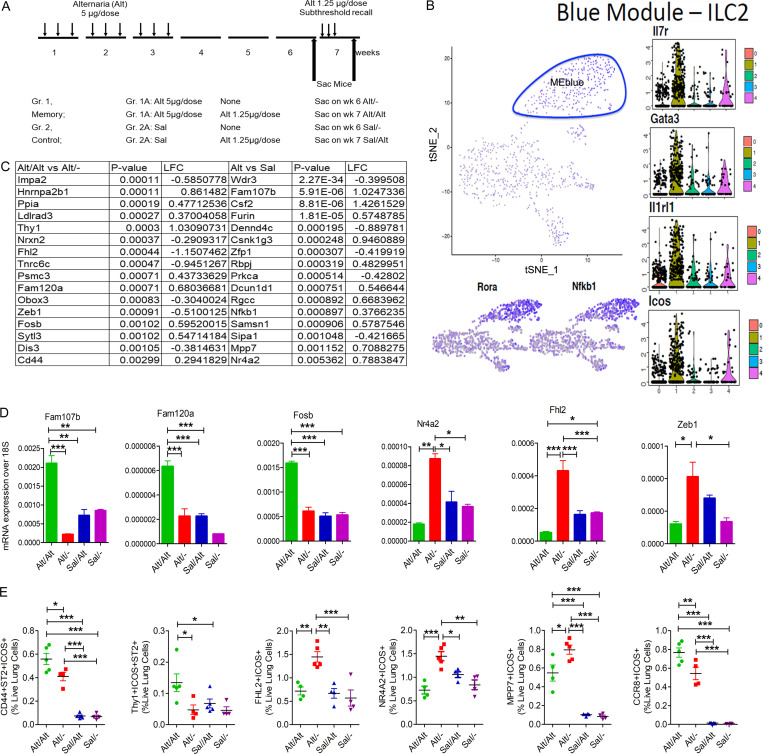
To identify memory-related genes in ILCs, we performed scRNA-seq of lung CD45+ cells from Alt- and Sal-sensitized Rag1−/− mice at two different time points: before and after the subthreshold recall challenge (four experimental groups). The experimental design for the RNA-seq study is shown in Fig. 5 A. Shared nearest neighbor clustering of gene expression from 871 cells identified five distinct cell clusters, which colocalized with t-distributed stochastic neighbor embedding (t-SNE) coordinate clustering (Fig. 5 B). To determine expression profiles most characteristic of cells in each cluster, we performed weighted gene coexpression network analysis (WGCNA) on the most variant genes in the single-cell dataset and then determined network modules that were differentially expressed in each cluster (Fig. S3 H). This analysis found that each of these clusters highly expressed a particular module containing genes characteristic of macrophages (turquoise), NK cells (black), dendritic cells (green), endothelial cells (brown), and ILC2s (blue; Fig. S3 H and Fig. 5 B). The ILC2 population expressed the ILC2-specific genes shown in Fig. 5 B: Rorα, Nfkb1 (expression shown in feature plots in the bottom panel), Il7r, Gata3, Il1rl1, and Icos (expression shown in military green color violin plots, #1). The preferential expression of CD25, Icos, and Il1rl1 in ILC2s as compared with other populations is shown in Fig. S3 I. We identified 166 and 131 genes up- and down-regulated in ILC2s from group 1 (memory group, pink color) versus group 2 (control group, teal color) in a genome-wide differential expression analysis. Coordinate clustering of these DEGs by t-SNE analysis resulted in partial separation of memory and control ILC2s (Fig. S3 J). Fig. 5 C shows top genes that were differentially expressed between group 1B (Alt/Alt) and group 1A (Alt/−), and between group 1 (Alt memory) and group 2 (Sal control). We confirmed the differential expression of Fam107b, Fam120a, Fosb, Nr4a2, Fhl2, Zeb1 (Fig. 5 D), Runx1, and MPP7 (Fig. S3 K) in sorted ILCs (CD45+CD25+ICOS+NK1.1− cells) from the memory model by quantitative PCR (qPCR). We observed two different expression patterns. The expression of Fam107b, Fam120a, Fosb, and Runx1 was low in the memory model before recall but increased after recall. In contrast, the expression of Nr4a2, Fhl2, Zeb1, and MPP7 was increased before recall but declined after recall. The increase in Nr4a2, Zeb1, Fhl2, and MPP7 in ILC2s, which likely occurred during repetitive Alt exposure and was sustained through the rest period until recall, may have relevance for memory formation. We confirmed decreased expression of CD44 (a T cell memory-related gene), Thy1 (an inhibitory surface molecule on lymphoid cells), and CCR8 (a type 2 cell maturation marker; Fig. 5 E) before recall and their increase after recall in ILCs by FCM. Similarly, the increased expression of Fhl2, Nr4a2, and Mpp7 (Fig. 5 E) in CD45+CD25+ICOS+ lung cells before recall and their decrease after recall were confirmed by FCM.
Figure 5.
scRNA-seq of lung hematopoietic cells. (A) A schematic showing study design and timeline for scRNA of lung cells from the memory model. (B) A t-SNE feature plot showing clustering of all CD45+ lung cells with mean expression of the WGCNA blue module, which contains genes characteristic ILC2s. The ILC2 cluster (circled) expressed Il7r, Gata3, Il1rl1, and Icos (violin plots) and Rora and Nfkb1 (feature plots). (C) Log fold-change (LFC) of selected top 16 genes from the memory group 1B versus 1A (after and before recall) and Group1 (memory) versus Group 2 (Sal control). (D) qPCR analysis of mRNA expression of select genes identified by scRNA-seq in sorted ILCs from the lung tissue obtained from the study groups. *, P < 0.01; **, P < 0.001; ***, P < 0.0001, one-way ANOVA, n = 5/group. (E) FCM analysis of expression of CD44 and Thy1 on ICOS+ST2+ ILCs (CD45+CD25+ cells); and the expression of FHL2+, NR4A2 MPP7, and CCR8 in ICOS+ ILCs (CD45+CD25+) in the lung before and after recall. *, P < 0.01; **, P < 0.001; ***, P < 0.0001, n = 4–5/group, one-way ANOVA. Gr., group; Sac, sacrifice.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) demonstrates a unique chromatin landscape in memory ILCs from Rag1−/− mice
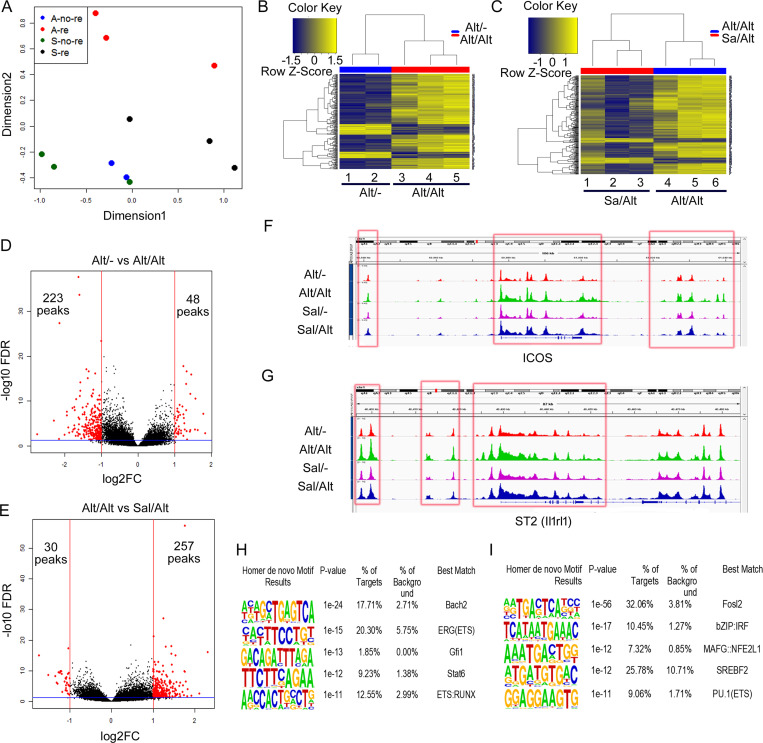
scRNA-seq showed that CD25 (Il2ra) and ICOS were highly expressed by ILCs (Figs. S3 I and 5 B). We FACS-sorted lung CD45+NK1.1−CD25+ICOS+ cells from the four study groups as described for the scRNA-seq study (Fig. 5 A) and used them for ATAC-seq. The gating and sorting strategy for isolation of CD45+NK1.1−CD25+ICOS+ cells is shown in Fig. S3 L. Principal component analysis (PCA) of the ATAC-seq data demonstrated a clear separation representing a unique epigenetic landscape for each group (Fig. 6 A). Moreover, unsupervised clustering of differentially accessible peaks from the ATAC-seq data demonstrated unique chromatin signatures of ILCs from Alt/− versus Alt/Alt groups (Fig. 6 B). Similarly, unique chromatin signatures exist between Alt/Alt versus Sal/Alt (Fig. 6 C). The chromatin signatures of Alt/Alt and Alt/− memory ILCs were also distinct from those of Sal/− ILCs (Fig. S4, A and B). The unique chromatin signature of memory ILCs can also be visualized via volcano plots (Fig. 6, D and E) illustrating differentially accessible peaks between Alt/− and Alt/Alt and between Alt/Alt and Sal/Alt. Alt/− memory ILCs displayed altered chromatin accessibility, with 223 less-accessible peaks and 48 more-accessible peaks by log2FC of 1 (twofold change) and a false discovery rate (FDR) of 0.05 as compared with Alt/Alt memory ILCs. Similarly, Alt/Alt memory ILCs displayed altered chromatin accessibility with 30 less-accessible peaks and 257 more-accessible peaks as compared with Sal/Alt ILCs. The majority of DNA accessibility peaks in all study groups was localized to intergenic and intronic regions, as opposed to promoter regions (Fig. S4, C and D). Fig. 6, F and G, illustrates chromatin accessibility peaks proximal to two representative memory ILC-associated genes: ICOS and ST2 (il1rl1). Both genes were differentially expressed based on the scRNA-seq analysis and simultaneously displayed differentially accessible ATAC-seq peaks proximal to the gene body. The ATAC-seq peaks proximal to each gene locus were characterized by increased chromatin accessibility after recall in memory ILCs (red box).
Figure 6.
ATAC-seq of lung ILCs (sorted CD45+NK1.1−CD25+ICOS+) from the memory models. As per Fig. 4 A. (A) PCA of ATAC data from the study groups: Alt exposure and no recall (Alt/−), Alt exposure and recall (Alt/Alt), Sal exposure and no recall (Sal/−), and Sal exposure and recall (Sal/Alt). n = 3 for all study groups except Alt/− (n = 2). (B and C) Heatmaps of differentially expressed DNA accessibility peaks between Alt/− versus Sal/− (B) and Alt/Alt versus Sal/Alt (C). (D and E) Volcano plots of differentially expressed DNA accessibility peaks between Alt/− versus Alt/Alt (D) and Alt/Alt versus Sal/Alt (E). FC, fold-change. (F and G) DNA accessibility tracing of two representative genes: Icos (F) and ST2 (il1rl1; G), which were differentially expressed both in ATAC-seq and scRNA-seq. (H and I) Top de novo motifs by HOMER analysis distinguishing Alt/− from Alt/Alt (H) and Alt/Alt from Sal/Alt (I).
Figure S4.
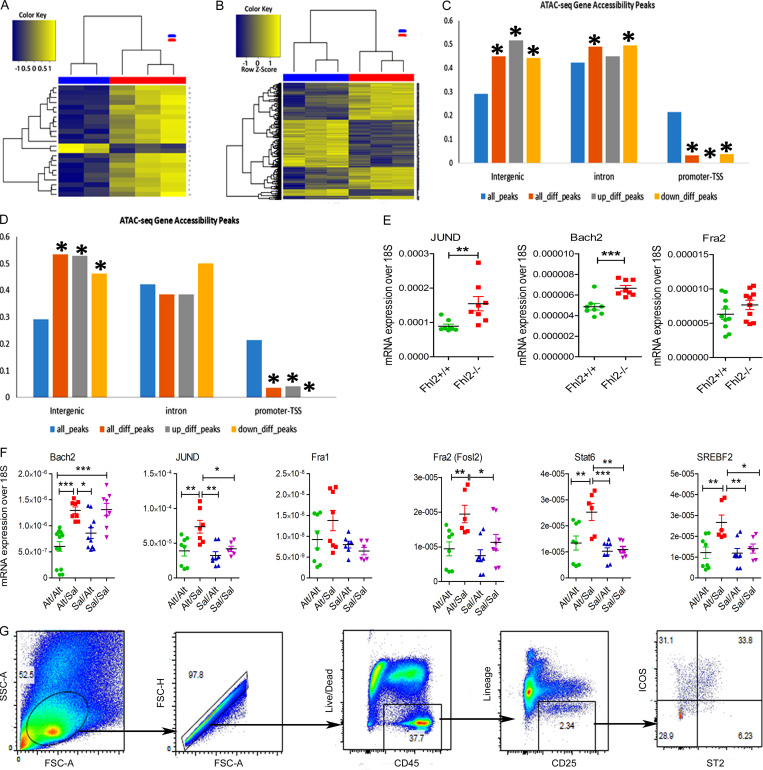
ATAC-seq and gating strategy for ILCs. (A–D) Heatmaps of differentially expressed DNA accessibility peaks between Alt/− (blue) versus Sal/− (red; A) and Alt/Alt (blue) versus Sal/− (red; B). Distribution of ATAC-seq gene accessibility peaks among intergenic, intronic and promoter-TSS as compared Alt/− versus Sal/− (C) and Alt/Alt versus Alt/− (D). *, Statistical significance. (E) Expression of mRNA for JunD, Bach2 and Fra2 (Fosl2) in the lung from Alt-sensitized and recalled Fhl2+/+ and Fhl2−/− mice. **, P < 0.001; ***, P < 0.001, unpaired t test. (F) The expression of mRNA (qPCR) for the top motif genes (Bach2, JunD, Fra1, Fosl2, Stat6, and Srebf2) in the lung tissue from study groups. *, P < 0.01; **, P < 0.001; ***, P < 0.001, n = 8–10/group; **, P < 0.001; ***, P < 0.0001, n = 5/group, one-way ANOVA. (G) The gating strategy for FCM for lung cells from the wild-type and Fhl2−/− mice. FSC, forward scatter; SSC, side scatter.
Given the unique chromatin signatures defining memory ILCs, we next investigated the molecular programs regulating memory formation via enrichment analysis for transcription factor binding motifs in the ATAC-seq data. The top de novo motifs for memory ILCs from Alt/− versus Alt/Alt groups were Bach2, ERG(ETS), Gfi1, STAT6, and ETS-RUNX (Fig. 6 H). The top de novo motifs for memory ILCs from Alt/Alt versus Sal/Alt groups were Fosl2, IRF, MAFG, SREBF2, and PU.1 (Fig. 6 I). Most relevant de novo motifs for Alt/− versus Sal/− and Sal/− versus Alt/Alt as well as hypergeometric optimization of motif enrichment (HOMER) known motifs for Alt/− versus Alt/Alt, Alt/Alt versus Sal/Alt, Alt/− versus Sal/−, and Sal/− versus Alt/Alt are shown in Table S1 and Table S2. Some of the top motifs included JunD (Alt/− versus Sal/−), Fosl1/Fra1 (Alt/− versus Alt/Alt and Alt/Alt versus Sal/Alt), and Batf, Runx1, and Stat6 (Sal/− versus Alt/Alt). We examined mRNA for the top motif genes in the lung tissue by qPCR (Fig. S4 F). The mRNA for JunD, Fosl2 (Fra2), Stat6, and Srebf2 (encodes for sterol regulatory element-binding factor 2) was increased in the lung tissue from Alt/Sal as compared with other groups. Their levels decreased after Alt recall (Alt/Alt). Bach2 mRNA decreased after Alt challenge in Alt/Alt and Sal/Alt. JunD and Fosl2 are components of the AP1 transcription factor and function as repressors (Meixner et al., 2004) as well as activators (Ruppert et al., 2012; Stelekati et al., 2018) in T cells. Stat6 is a key transcription factor for type 2 cells (Foster, 1999). Srebf2 (encodes for SREBP2) is essential for lipid metabolism during CD8 T cell clonal expansion (Kidani et al., 2013). Bach2 is a master repressor of cytokine genes and is involved in T cell memory (Roychoudhuri et al., 2016). The enriched occurrence of these DNA-binding motifs in differential peaks suggests an important role for these molecules in regulating the molecular program governing ILC2 memory.
Fhl2 is one of the top up-regulated genes in memory ILCs
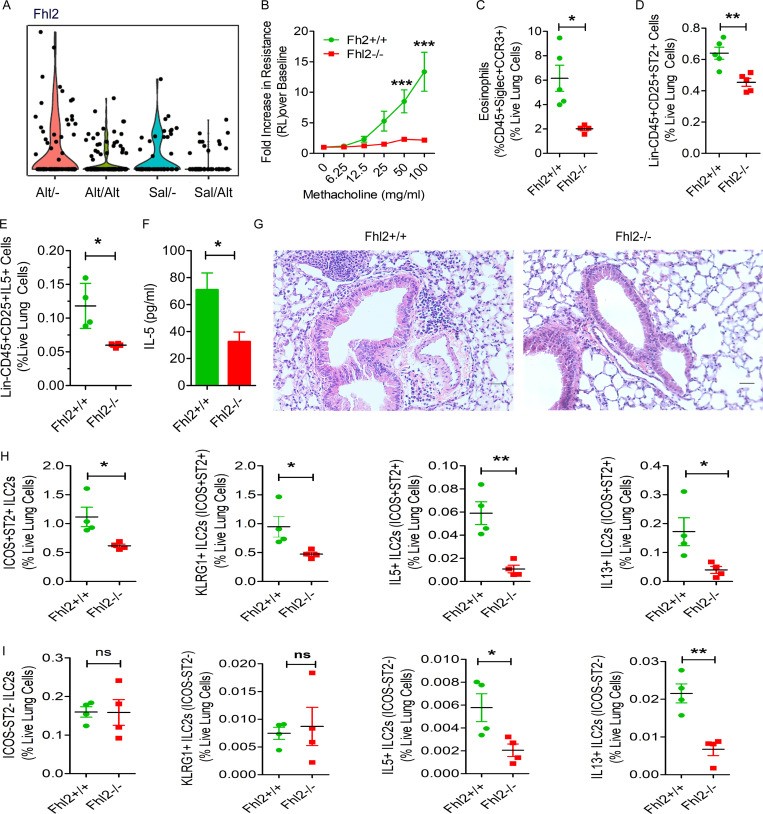
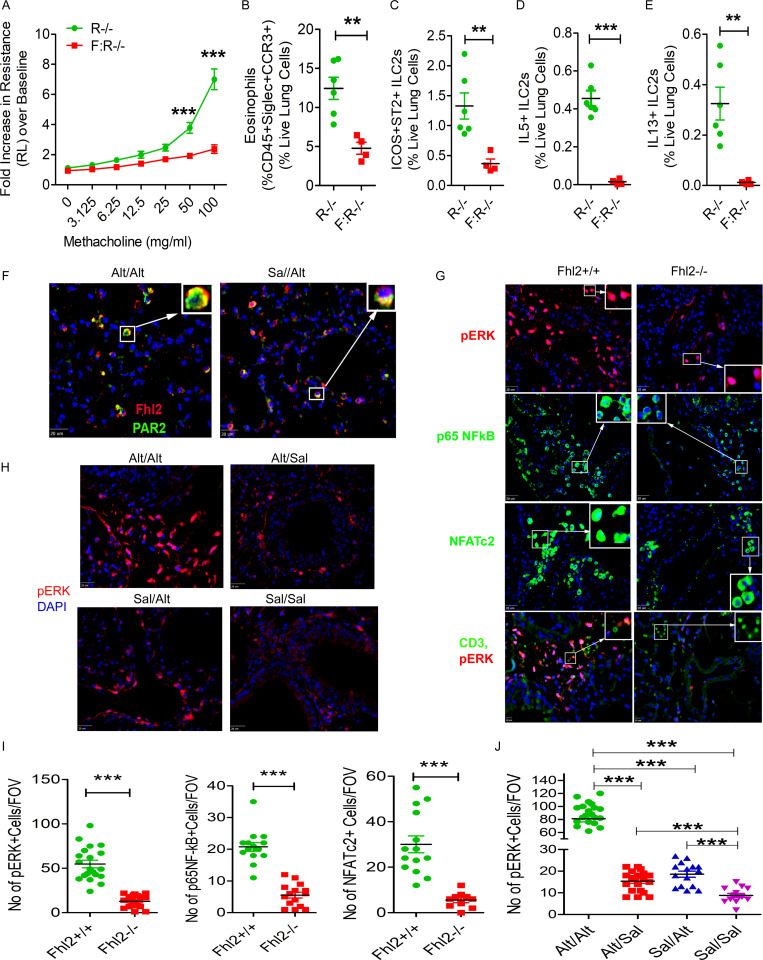
qPCR of memory ILCs validated up-regulation of four molecules: Nr4a2, Fhl2, Mpp7, and Zeb1. The Immgen database shows that Mpp7 and Zeb1 are expressed in all lymphoid cells. In contrast, Nr4a2 and Fhl2 are mostly expressed in ILCs, NK cells, and virally infected CD8 T cells. Because of its high expression in ILCs from the memory model (Fig. 7 A), we examined the role of Fhl2 in memory-driven asthma. We confirmed the expression of Fhl2 in mouse lung ILCs (Fig. 5 E). Fhl2 knockout (KO) B6 mice were unable to mount airway hyperreactivity (Fig. 6 B) upon Alt recall. This confirms a previous report on the essential role of Fhl2 in asthma (Kurakula et al., 2015). Fhl2 KO mice had reduced numbers of eosinophils and ST2+ and IL5+ ILC2s in the lung, IL5 in BAL (Fig. 7, C–F), and airway inflammation (Fig. 7 G). To assess the role of FHL2 in activation of memory versus nonmemory cells, we studied ICOS+ST2+ ILC2s as memory cells and ICOS−ST2− as nonmemory cells and examined their frequency and expression of KLRG1 (as an activation marker) and type 2 cytokines. The frequency of ICOS+ST2+ memory ILC2s and their expression of KLRG1 and type 2 cytokines were significantly lower in Fhl2−/− mice (Fig. 7, H and I). The frequency of KLRG1+ and type 2 cytokine+ cells was 10-fold lower in ICOS−ST2− ILCs. The expression of type 2 cytokines but not KLRG1 was lower in Fhl2−/− mice. The FCM gating strategy for wild-type and Fhl2 KO mice is shown in Fig. S4 G. The role of Fhl2 in ILC2 memory was also studied using Fhl2−/−;Rag1−/− mice. Similar to Fhl2−/− mice, Fhl2−/− × Rag1−/− mice failed to develop airway hyperreactivity, eosinophilic inflammation, ICOS+ST2+, and IL5+ and IL13+ ILC2s as compared with Rag1−/− mice (Fig. 8, A–E).
Figure 7.
The effect of Fhl2 gene deletion on memory-driven asthma. (A) The expression pattern (violin plots) of Fhl2 in ILC2s from the study groups that were examined by the single cells RNA-seq. Fhl2−/− and littermate Fhl2+/+ mice were subjected to the Alt-induced memory-driven asthma protocol as per Fig. 1 A. (B–G) Airway hyperreactivity (B; two-way ANOVA), lung eosinophils (CD45+SiglecF+CCR3+ cells; C), Lin−CD45+CD25+ST2+ cells (D), and IL5+ ILCs (E) were measured by FCM and IL5 in BAL by ELISA (F). Representative histological images of H&E staining of the lung tissue from Fhl2+/+ and Fhl2−/− mice from the memory model (n = 5/group; G). Scale bars are 100 µm and the images were acquired at 20×. (H and I) Comparison of memory (ICOS+ST2+) and nonmemory (ICOS−ST2−) ILCs between Fhl2+/+ and Fhl2−/− mice. The frequency of ICOS+ST2+ memory ILCs and the expression of KLRG1, IL5, and IL13 in ICOS+ST2+ memory ILCs and ICOS−ST2− nonmemory ILCs were measured by FCM (H and I). *, P < 0.01; **, P < 0.001; ***, P < 0.0001, unpaired t test, n = 4–5/group.
Figure 8.
Importance of Fhl2 in memory-associated signaling. (A–E) Fhl2−/−;Rag1−/− (F:R−/−) mice were sensitized and challenged with Alt and Sal as per Fig. 1 A and then examined for airway hyperreactivity and immunological changes. The increase in lung resistance over baseline (as measured by flexiVent) in response to increasing doses of inhaled methacholine (A), lung eosinophils (B), ICOS+ST2+ ILCs (C), IL5+ ILC2s (D), and IL13+ ILC2s (E). **, P < 0.001; ***, P < 0.001, unpaired t test, n = 5/group. (F) Double immunostaining of the lung tissue from the Alt/Sal and Sal/Sal study group for Fhl2 (red) and PAR2 (green). The insets show a higher magnification of cells displaying colocalization (yellow color; n = 5 mice/group). Scale bars are 20 µm. (G) Immunofluorescence staining of the lung tissue from Fhl2+/+ and Fhl2−/− B6 mice from the memory model for pERK1/2 (red) and p65NFkB and NFATc2 (green). The insets show nuclear localization of NF-κB and NFAT under a higher magnification. Scale bars are 20 µm. The last panel shows costaining of pERK1/2 (red) with CD3 (green). Scale bars are 10 µm. (H) Immunofluorescence staining of the lung tissue from the memory models from Rag1−/− mice for pERK1/2 (red). All sections were counterstained with DAPI (blue), and images were taken at 40×. Scale bars are 20 µm. (I and J) Quantification of pERK+, and nuclear NF-kB+ and NFAT+ cells in the lung tissue from Fhl2+/+ and Fhl2−/− mice (***, P < 0.001, unpaired t test; I) and the quantification for pERK1/2 in memory models from Rag1−/− mice (J). One-way ANOVA. ***, P < 0.0001. FOV, field of view.
Next, we examined the signaling mechanism by which Fhl2 contributed to memory-driven activation of ILCs. Double immunostaining of the lung tissues showed colocalization of Fhl2 with PAR2 in inflammatory cells (Fig. 8 F). We immunostained the lung tissue from the memory model for three major signaling pathways that are relevant for the type 2 immune response. These pathways included pERK1/2, p65NFkB, and NFATc2. There was strong staining of pERK1/2 and nuclear staining of p65NFkB and NFATc2 in peribronchial inflammatory cells from the wild-type mice (Fig. 8, G and I). This staining was significantly reduced in Fhl2−/− mice. pERK1/2 staining was primarily associated with CD3− infiltrating cells (Fig. 8 G, bottom panel). We confirmed increased pERK1/2 staining in peribronchial inflammatory cells from the memory model in Rag1−/− mice (Fig. 8, H and J). The ERK1/2 signaling pathway regulates AP1, and the latter antagonizes Bach2. Since Fhl2 positively regulated ERK1/2 signaling, we examined the expression of mRNA for Bach2 and the AP1 transcription factors JunD and Fra2 (Fosl2) in the lung tissue. The expression of JunD and Bach2 but not Fra2 was increased in Fhl2−/− mice, suggesting that Fhl2 negatively regulated mRNA for the memory-associated repressors JunD and Bach2 (Fig. S4 E).
The chromatin landscape of memory ILC2s from wild-type mice
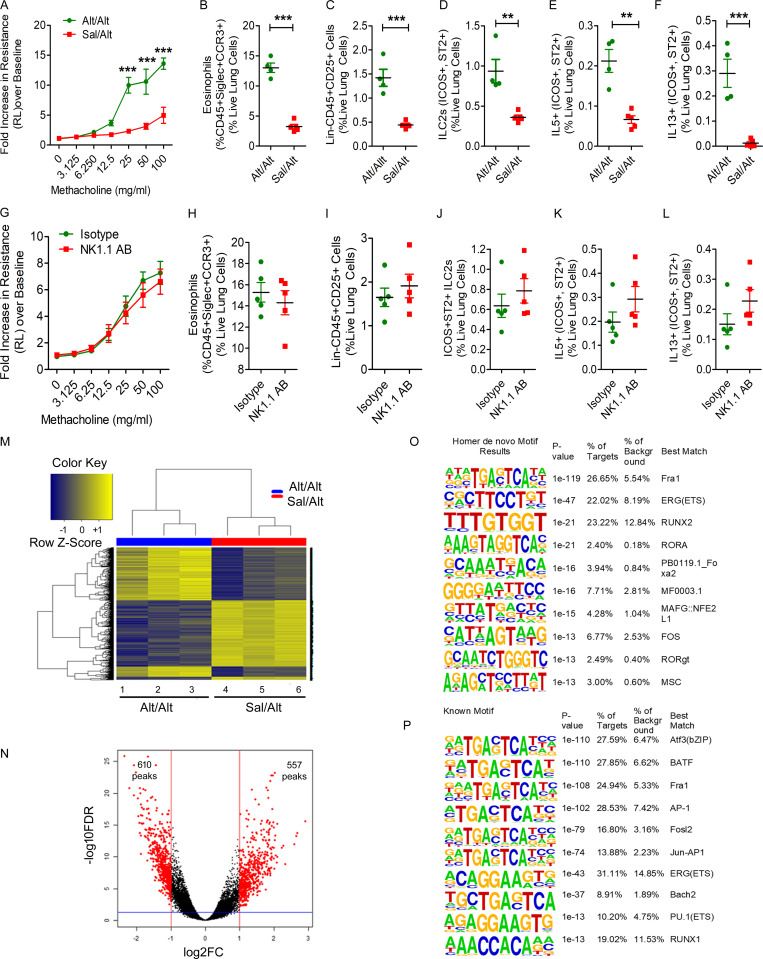
To validate the ATAC-seq findings from the Rag1−/− mice that are devoid of T and B cells, we studied two groups of wild-type B6 mice as per the Alt/Alt and Sal/Alt protocol (Fig. 5 A). Like Rag1−/− mice, the wild-type mice from the Alt/Alt but not Sal/Alt group developed memory-driven airway hyperreactivity and eosinophilic inflammation (Fig. 9, A and B). This was associated with an increase in total ILCs (Lin−CD45+CD25+), memory ILC2s (ICOS+ST2+ ILC2s), and type 2 cytokine–positive ILC2s (Fig. 9, C–F). Antibody-mediated depletion of NK cells did not affect the foregoing parameters in wild-type mice (Fig. 9, G–L). We performed ATAC-seq on FACS-sorted lung CD45+Lin−NK1.1−CD25+ICOS+ cells as per Fig. 6. Unsupervised clustering of differentially accessible peaks from the ATAC-seq data illustrated unique chromatin signatures of ILCs from Alt/Alt versus Sal/Alt groups (Fig. 9 M). The differentially accessible peaks between Alt/Alt and Sal/Alt representing the unique chromatin signature of memory ILCs is shown as a volcano plot in Fig. 9 N. Alt/Alt memory ILCs displayed altered chromatin accessibility, with 610 less accessible peaks and 557 more accessible peaks by log2FC of 1 (twofold change) and an FDR of 0.05 as compared with Sal/Alt ILCs. The top de novo motifs for Alt/Alt memory ILCs from the wild-type mice were Fra1 (AP1), ERG(ETS), and RUNX2, whereas the top known motifs were Atf3, BATF, and Fra1 (all belong to the AP1 family; Fig. 9, O and P). The results suggest that select motifs, AP1, ERG, and RUNX2, dominate transcriptional regulation of memory ILC2s from both wild-type and Rag1−/− mice.
Figure 9.
Studies of ILC2 memory in wild-type mice. (A–F) Wild-type mice (C57BL/6) mice were subjected to the Alt-induced memory-driven asthma protocol as per Fig. 1 A. Airway hyperreactivity, as assessed by lung resistance to inhaled methacholine, was measured by flexiVent (A). Lung eosinophils (CD45+SiglecF+CCR3+ cells; B), total ILCs (Lin−CD45+CD25+; C), ICOS+ST2+ ILC2s (D), and IL5+ and IL13+ ILC2s (CD45+NK1.1−CD25+ICOS+ST2+; E and F) cells were measured by FCM. **, P < 0.001; ***, P < 0.0001, unpaired t test, n = 4 or 5/group. (G–L) The role of NK cells in memory-driven asthma in wild-type mice. The protocol for NK cells depletion and allergen challenge in wild-type mice was similar to that described in Fig. 2 A. We measured airway hyperreactivity (G), lung eosinophils (CD45+SiglecF+CCR3+ cells; H), total ILCs (Lin−CD45+CD25+; I), ILC2s (CD45+NK1.1−CD25+ICOS+ST2+ cells; J), and IL5+ and IL13+ ILC2s (CD45+NK1.1−CD25+ICOS+ST2+; K and L). Unpaired t test, n = 5/group. (M–P) Comparison of the heatmaps of differentially expressed DNA accessibility peaks between the Alt/Alt and Sal/Alt groups (M). Volcano plots of differentially expressed DNA accessibility peaks from the Alt/Alt and Sal/Alt groups (N). Top de novo and known motifs by HOMER analysis distinguishing Alt/Alt from Sal/Alt (O and P). FC, fold-change.
Human memory ILCs in asthma
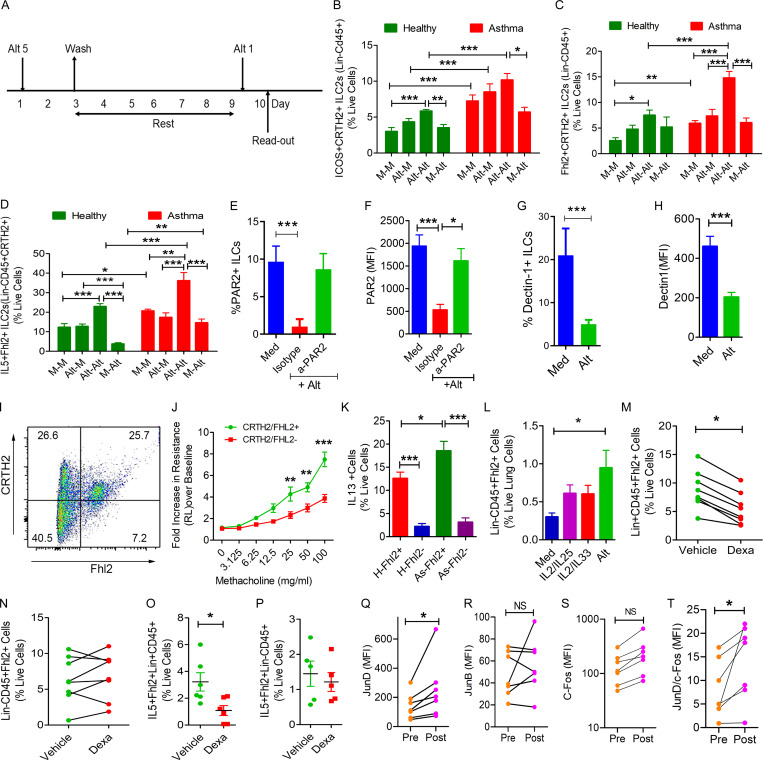
We developed an in vitro model of memory induction in human ILCs (Fig. 10 A) similar to what was developed for monocytes (Cheng et al., 2014; Saeed et al., 2014). In this model, we exposed peripheral blood mononuclear cells (PBMCs) to 5 µg/ml Alt or medium for 2 d, washed to remove the allergen, and cultured in the media alone. After 6 d of rest, we reexposed the cells to a subthreshold dose (one fifth the original dose), 1 µg/ml Alt or medium, and measured the frequency of ICOS+CRTH2+, Fhl2+CRTH2+, and IL5+Fhl2+CRTH2+ ILC2s (Lin− cells) by FCM. We studied 8 healthy and 10 Alt-sensitive allergic asthma patients. We observed that the frequency of ICOS+CRTH2+ and Fhl2+CRTH2+ memory ILC2s and their expression of IL5 was increased under medium-treated conditions in asthma as compared with healthy controls (Fig. 10, B–D). Prior treatment of cells with Alt 6 d before the recall challenge increased the frequency of memory ILC2s and their cytokine expression in both healthy subjects and asthma patients, although the increase was significantly higher in asthma patients.
Figure 10.
Role of Fhl2 in human memory ILC2s. (A) A schematic of in vitro memory induction in human blood ILCs. (B–D) Human PBMCs from 8 healthy (H) and 10 allergic asthmatic (As) patients were treated with medium (M) or Alt (5 µg/ml) for 48 h. After washing (3×) and culturing in medium for 6 d, the cells were rechallenged with medium or a subthreshold dose of Alt (1 µg/ml) for 24 h ICOS+ (B) and FHL2+ (C) and IL5+ (D) ILC2s (Lin−CD45+CRTH2+) were analyzed by FCM. One- and two-way ANOVA. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (E–H) Isolated blood Lin− cells were cultured in medium alone (Med) or with Alt (5 µg/ml). An anti-PAR2 antibody or an isotype control antibody were added to select cultures (E). After 24 h, the cells were analyzed for PAR2 (E and F) or dectin-1 (G and H) by FCM (n = 3). *, P < 0.01; ***, P < 0.0001, one-way ANOVA or unpaired t test. (I and J) Single cells were isolated from a healthy human lung digest and analyzed for Fhl2 in CRTH2+ ILCs (CD45+Lin−; I). CRTH2+Fhl2+ and CRTH2−Fhl2− ILCs (CD45+Lin−) lung cells (100,000) were adoptively transferred to Rag2−/−:γc−/− mice. The mice were challenged with Alt (5 µg/dose) for three consecutive days, and airway hyperreactivity was measured 48 h later. **, P < 0.001; ***, P < 0.0001, paired t test. n = 3/group, two-way ANOVA (J). (K) Comparison of IL13 expression by Fhl2+ and Fhl2− ILC2s (CD45+Lin−CRTH2+) from 8 healthy (H) and 10 asthma (As) patients, two-way ANOVA. *, P < 0.01; ***, P < 0.0001. (L) Human lung–derived mononuclear cells were cultured with medium, IL2/IL25, IL2/IL33 (10 ng/ml each), or Alt (5 µg/ml) for 10 d and then examined for Fhl2 expression by FCM (n = 6; *, P < 0.05), one-way ANOVA. (M–P) The frequency of Fhl2+ and IL5+Fhl2+ cells in Lin+ and Lin− cell populations in PBMCs that were cultured with dexamethasone (Dexa; 10−7 M) or vehicle (ethanol) for 3 d. Mann–Whitney U test. *, P < 0.01. (Q–T) Effect of repetitive allergen injections on memory-related AP1 genes. PBMCs were isolated from seven allergic patients before and after rush allergen immunotherapy and stained ex vivo for c-Fos, JunD, and JunB in Lin− cells. *, P < 0.01.
Alt-associated PAMPs activate PAR2 and dectin-1
The Alt allergen extract has multiple PAMPs including a serine protease and a β-glucan. The foregoing PAMPs activate PAR2 and dectin-1, respectively. We examined the direct effect of Alt on PAR2 and dectin-1 using human blood Lin− cells. For these experiments, we isolated Lin− cells from PBMCs by negative selection. Culture of human blood Lin− cells with Alt reduced the frequency of PAR2+ and dectin-1+ cells and the expression of PAR2 (Fig. 10, E and F) and dectin-1 (Fig. 10, G and H), which is suggestive of ligand-specific desensitization and receptor down-regulation (Rajagopal and Shenoy, 2018). Anti-PAR2 blocking antibody prevented the Alt-induced receptor down-regulation. The results suggest that Alt directly acts on human ILCs through PAR2 and dectin-1. We recognize that these studies are preliminary as PAR2 can heterodimerize with other PAR receptors and transduce the Alt-associated protease signaling.
Fhl2+ human ILCs in asthma
We examined the expression and function of Fhl2 in human ILCs. Human lung-derived CRTH2+ ILCs (CD45+Lin−FcεRI−CD25+) expressed Fhl2 (Fig. 10 I). We FACS-sorted human lung Fhl2+CRTH2+ and Fhl2−CRTH2− ILCs and adoptively transferred them to Rag2−/−:γc−/− mice. 1 d later, the mice were intranasally exposed to Alt for three consecutive days and examined for airway hyperreactivity to methacholine (measurement of lung resistance by flexiVent) 48 h later. Fhl2+CRTH2+ but not Fhl2−CRTH2− ILCs induced airway hyperreactivity upon adoptive transfer (Fig. 10 J). We compared IL13 expression between Fhl2+ memory and Fhl2− nonmemory ILCs from 8 healthy and 10 asthma patients. IL13 expression was significantly higher in Fhl2+ ILCs (Fig. 10 K). Culture of human lung mononuclear cells with Alt but not with cytokines (IL2, IL25, or IL33) up-regulated Fhl2 in CD45+Lin−cells (Fig. 10 G).
Next, we examined the disease relevance of Fhl2+ memory ILCs for asthma. Severe refractory asthma is often associated with steroid resistance. We asked if Fhl2+ ILCs were steroid resistant. We isolated PBMCs, cultured them with dexamethasone (10−7 M) or vehicle (ethanol) for 3 d, and examined the frequency of Fhl2+Lin− and Fhl2+Lin+ cells. Dexamethasone reduced the frequency of Fhl2+Lin+ cells but failed to inhibit Fhl2+Lin− cells (Fig. 10, M and N). Likewise, dexamethasone inhibited IL5 expression in Fhl2+Lin+ cells but not in Fhl2+Lin− cells (Fig. 10, O and P). The results suggest that Fhl2+Lin− cells were relatively steroid resistant and, thus, could contribute to the development of steroid-resistant asthma.
JunD expression increases after repetitive allergen injections in allergic patients
The AP1 motif was one of the top DNA-binding motifs in ILCs after repetitive exposure of mice to Alt (Fig. 6 I; Fig. 9, O and P; and Table S2). Allergen immunotherapy, especially the so-called rush immunotherapy in allergic patients, partially mimics the repetitive allergen exposure to mice in our model. The rush immunotherapy protocol involves subcutaneous administration of increasing doses of the sensitizing allergens every 30 min for 6 h/d for two consecutive days (Horst et al., 1990). We examined the expression of JunD and two other AP1 family members, JunB and c-Fos (Fig. 10, Q–S), in ILCs (Lin− PBMCs) by FCM before and after repetitive allergen injections in seven patients. JunD but not JunB or c-Fos showed a significant increase in its expression (mean fluorescence intensity). JunD is a negative regulator of lymphocyte function (Meixner et al., 2004), and JunB (Li et al., 1999) and c-Fos (Agrawal et al., 2003) are positive regulators. The ratio of JunD over c-Fos in the same patients significantly increased after immunotherapy (Fig. 10 T). The results show that repetitive allergen exposure increases the expression of JunD in human ILCs.
Discussion
Upon repetitive exposure to Alt, airway cells including ILC2s developed a form of memory, which could be recalled with a subthreshold dose of Alt 3–15 wk later. A reduction in activation threshold is characteristic of cellular memory (Pihlgren et al., 1996). This memory was associated with increased type 2 cytokine production, eosinophilic influx, and airway hyperreactivity and with a concomitant increase in ICOS+ST2+ ILC2s. ILC2s were sufficient for the memory-driven asthma phenotype, as (a) their adoptive transfer induced asthma; (b) their depletion by anti-ICOSL and anti-CD90 antibodies and by genetic ablation of ICOS blocked asthma; (c) their inhibition by RORα antagonism failed to induce asthma; and (d) the depletion of NK cells did not affect asthma. We previously observed memory generation by ILC2s in a chronic asthma model in genetically unmodified B6 mice (Christianson et al., 2015). In this chronic asthma model, airway hyperreactivity persisted >6 mo after cessation of the allergen exposure. Allergen-experienced ILC2s from the chronic asthma model, but not Sal-experienced ILC2s, induced chronic asthma following adoptive transfer. Finally, we present the human disease relevance of memory ILC2s by demonstrating increased frequency of ICOS+ and Fhl2+ memory ILC2s in asthma and steroid resistance of Fhl2+ memory ILCs.
As mentioned previously, Martinez-Gonzalez et al. (2016) reported the generation of memory ILC2s. The authors showed that allergen- and IL33-experienced ILC2s persisted for months and responded more robustly to a different allergen. Their results suggested an antigen-nonspecific memory of prior activation. This is more akin to acquisition of fitness through training. This has also been called trained immunity (Netea et al., 2016). Martinez-Gonzalez et al. (2016) examined differential gene expression in their IL33-elicited memory ILC2s. There was an increased expression of IL25R that persisted for 4 mo. We did not see any increase in IL25R in Alt-elicited memory ILC2s, suggesting that the pathways involved in memory formation are different in these two experimental models.
In scRNA-seq of memory ILC2s, we observed two different gene expression patterns. The expression of Fam107b, Fam120a, Fosb, and Runx1 was low in the memory model before recall but increased after recall. The increased expression of Fam120a, Fosb, and Runx1 after recall was of interest. Fam120a is a multifunctional scaffold protein involved in (a) activation of Src family kinases and the PI-3K-AKT-mTOR pathway (Tanaka et al., 2009); (b) interaction with argonaut-2 and rescue of >2,000 mRNAs from argonaut-mediated degradation (Kelly et al., 2019); and (c) interaction with IL13ra2, which protects cells from oxidative stress–induced apoptosis (Bartolomé et al., 2015). Fosb is a component of the AP1 complex, a major transcriptional activator (Hasenfuss et al., 2014). Runx1 directs ILC differentiation and function by activating the enhancer for promyelocytic leukemia zinc finger (PLZF; Mao et al., 2017). In contrast to the foregoing molecules, the expression of Nr4a2, Fhl2, Zeb1, and MPP7 was increased before recall but declined after recall. Nr4a2 (nuclear receptor subfamily 4 member a2) and Zeb1 (zinc finger E box-binding homeobox 1) are repressors. Nr4a2 is involved in memory formation in the brain (McNulty et al., 2012), CD8 T cell exhaustion (Chen et al., 2019), and maintenance of the anti-inflammatory function of regulatory T cells (Sekiya et al., 2011). Zeb1 is involved in memory formation in T cells (Guan et al., 2018). Fhl2 (four and half lim domain containing 2) is a multifunctional scaffold protein, which interacts with >30 molecules: transcriptional activators and repressors in the nucleus and receptors and signaling molecules in the cytosol (Johannessen et al., 2006). Fhl2 interacts with ID2 and PLZF, genes involved in ILC2 development, and ERK2 as well as its downstream effectors, Fos and Jun, which are components of the transcription factor AP1. MPP7 (membrane palmitoylated protein 7) interacts with the transcription factor cAMP response element-binding protein, regulates Angiomotin and YAP1, and regulates the competence of stem cells to expand and self-renew (Li and Fan, 2017).
ATAC-seq analysis revealed a unique chromatin landscape of memory ILC2s. PCA showed clear separation of memory ILC2s from nonmemory ILC2s. Unsupervised clustering demonstrated a unique chromatin signature specific to each type of memory ILC, indicating the potential for distinct molecular pathways regulating memory formation versus activation. TF motif analysis of the ATAC-seq data identified the motifs for AP1 (Fra1, Fosl2, JunD, Atf3, and Batf), ERG, RUNX2, Bach2, and STAT6 as the top DNA-binding motifs of memory ILC2s from Rag1−/− and wild-type mice. The AP1 motif was the top motif in memory NK cells (Lau et al., 2018) and CD8 T cells (Roychoudhuri et al., 2016). The results suggest that memory formation in ILC2s, NK cells, and T cells involves similar chromatin and molecular processes. The AP1 heterodimer (sometimes homodimers) can be composed of proteins from four families: Fos (c-Fos, Fosb, Fra1, and Fosl2), Jun (c-Jun, JunB, and JunD), ATF (ATF2, ATF3, and BATF1-3), and MAF (Hasenfuss et al., 2014; Murphy et al., 2013). Most AP1 molecules function as transcriptional activators for lymphocytes (Agrawal et al., 2003; Mondino et al., 1996; Murphy et al., 2013). Fosl2 (Fra2) regulates the expression of the memory-associated ICOS gene in Th2 cells (Watanabe et al., 2012). Interestingly, JunD (Meixner et al., 2004) and Fosl2 (Ciofani et al., 2012) can also function as repressors in a context-dependent manner. Bach2 promotes memory formation in CD4 and CD8 T cells and prevents effector cell differentiation (Roychoudhuri et al., 2016; Roychoudhuri et al., 2013). The AP1 DNA-binding motif has a near complete overlap with that of Bach2, implying that an excess of one outcompetes the other (Roychoudhuri et al., 2016).
Fhl2 is one of the top up-regulated genes before recall in memory ILC2s. Fhl2 was also the top up-regulated gene in β-glucan–activated macrophages (Walachowski et al., 2017). Adoptive transfer and KO studies showed that Fhl2 was essential and sufficient for memory-driven asthma. Fhl2+ ILC2s were superior to Fhl2− ILC2s in expression of KLRG1 and type 2 cytokines. Fhl2 interacts with c-Fos (Morlon and Sassone-Corsi, 2003) and Nr4a1 (Kurakula et al., 2011) in the nucleus and with ERK2, TRAF6, and calcineurin (Tran et al., 2016) in the cytosol. We show that Fhl2 interacts with PAR2 on the cell membrane. KO mouse studies suggest that Fhl2 positively regulates activation of ERK1/2, nuclear expression of NFAT, and to a lower extent, NF-κB in airway inflammatory cells. The activation of ERK1/2 is important, as it induces and/or phosphorylates many AP1 factors including Fosb, Fra1, and Fosl2 (Hasenfuss et al., 2014; Murphy et al., 2013). In contrast to the signaling activators, the repressors JunD and Bach2 are negatively regulated by Fhl2. We believe that the loss of Fhl2 led to defective AP1 generation, heightened gene repression, and consequently, impaired memory-driven asthma in Fhl2 KO mice.
Based on our results, we believe that repetitive allergen exposure is perceived by the airways as chronic stress. To cope with this stress, the airway cells including ILCs establish two mutually regulated programs: a gene repression program and a preparedness program. The gene repression program includes repressors such as Nr4a2, Zeb1, JunD, and Bach2. The preparedness program includes transcriptional and signaling activators such as Fhl2, FosB, STAT6, MPP7, and Srebf2. Many of these genes were up-regulated, and others (Bach2 and JunD) were actively engaged in memory ILC2s. The repressors mark and repress previously activated genes, which constitutes the molecular/epigenetic mechanism of ILC memory. The repressors are regulated by the preparedness-associated molecules. For instance, Fhl2 negatively regulates JunD and Bach2. ERK1/2 positively regulates Nr4a2 (Jacobsen et al., 2008), whereas Nr4a2 inhibits ERK1/2 signaling through up-regulation of ERK1/2 phosphatases DUSP2 and DUSP14 (Ashraf et al., 2019). These results suggest that a mutually regulated balance between the repression and the preparedness programs is necessary to generate and maintain memory. The preparedness program is primed but not activated. Its activation upon a recall allergen challenge down-regulates the repressors Bach2 and JunD and activates the ERK1/2-AP1 and STAT6 pathways to elicit the memory-driven asthma phenotype.
Materials and methods
Mouse studies
The animal protocol for this study was approved by the National Jewish Health institutional animal care and use committee. We used Rag1−/− (B6.129S7-Rag1tm1Mom/J), ICOS−/− (B6.129P2-Icos tm1Mak/J), Rag2−/−:γc−/− (C;129S4-Rag2tm1.1FlvIl2rgtm1.1Flv/J), and CLEC7A−/− (B6.129S6-Clec7a tm1Gdb/J) mice, also known as Dectin-1−/− (all from the Jackson Laboratory); Fhl2−/− (gift from J. Chen, University of California, San Diego, San Diego, CA) and IL33Cit/Cit knock-in (gift from Dr. Andrew McKenzie, University of Cambridge, Cambridge, UK) mice; and ICOS−/− × Rag1−/− and Fhl2−/− × Rag1−/− mice. All the mutant mouse strains except Rag2−/−:γc−/− and IL33Cit/Cit were on B6 background. We used littermate or C57BL/6 (as identified in the text) as wild-type controls.
Mucosal sensitization of mice
The allergen extracts used in this experiment were Alt (Alternaria alternata). Mice were sensitized to allergens as shown in Fig. 1. Briefly, Alt (5 µg/dose) was administered in a 20 µl volume intranasally on alternate days three times a week for three consecutive weeks unless otherwise stated. The mice were then rested for 3 wk. In week 7, they were given a recall challenge with a subthreshold dose of the sensitizing allergen on three consecutive days. The mice were sacrificed 3 d later. The subthreshold recall dose was 1.25 µg/dose.
Antibody, pharmacologic, and genetic interventions
For ICOS blockade experiments, an anti-ICOSL antibody (#107410; BioLegend; 20 µg/20 µl in Sal) was delivered intranasally for three consecutive days 3 d before analysis in week 8. An isotype rat IgG2a (#400544; BioLegend) antibody was used as a control. For NK1.1 and ILCs cell depletion, we used as anti-NK1.1(#BE0036, clone PK136; Bio X Cell) and anti-CD90.2 (#BE0066, clone 30H12; Bio X Cell), respectively. Mouse IgG2a (#BE0085, clone C1.18.4; Bio X Cell) and rat IgG2b (#BE0090, clone LTF-2; Bio X Cell) antibodies were used as isotype controls, respectively. For ILC2 depletion in vivo, we treated mice with SR3335 (200 µg/dose, i.p.) or vehicle (ethanol diluted with Sal; final ethanol concentration 20 µl/dose) on two consecutive days before the recall challenge.
Airway hyperreactivity measurement
The measurement of airway hyperreactivity in response to methacholine by flexiVent was described in detail previously (Verma et al., 2018). Briefly, mice were anesthetized with ketamine (180 mg/kg), xylazine (9 mg/kg), and acepromazine (4 mg/kg). After loss of footpad pinch reflex, a tracheotomy was performed, and the mouse was attached via an 18-gauge cannula to a small-animal ventilator with a computer-controlled piston (flexiVent Fx; Scireq). After performing initial calibrations (cylinder pressure channel and nebulizer calibration), we conducted dynamic tube calibration and used a default program called QuickPrime 3 (v7) for measurement of airway resistance in response to methacholine. This program uses prime perturbations, which are a family of complex forced oscillation perturbations at a frequency greater than and less than the subject’s ventilation frequency (1–20.5 Hz). The amplitude of the oscillatory signal is preset to a volume that is slightly smaller than the subject’s tidal volume (0.2 ml). Volume and pressure signal are recorded during a measurement, and the flow signal is derived from the volume. The foregoing allows calculation of Newtonian resistance, tissue damping, tissue elastance, and hysteresivity. Resistance measurements were taken to establish the baseline for total lung resistance and at each methacholine dose. Group averages were expressed as the fold increase over baseline resistance (mean ± SEM; Verma et al., 2018).
Histology and imaging
Paraffin-embedded lungs were sectioned and stained with H&E for morphometric analysis and Mason’s trichrome for collagen deposition. Images were acquired on a Nikon Eclipse TE2000-U microscope using 20× dry lenses at room temperature through a Diagnostics Instruments camera model #4.2 using Spot software 5.0. H&E sections were mounted using Permount medium. Images were adjusted for brightness and contrast to improve viewing (Verma et al., 2018)
Thin-section immunofluorescence microscopy
Paraffin-embedded lung sections (4-µm thickness) were used for this study. Images were taken under polarized light using an upright dry under 40× objective. Tissues were deparaffinized and, after antigen retrieval, permeabilized with 0.4% Triton X-100. Tissues were blocked with 10% BSA and maintained in PBS + 5% BSA + 0.4% Triton X-100 throughout antibody treatments. Primary antibodies were incubated at 4°C overnight, and secondary antibodies were incubated for 1 h at room temperature. Primary antibodies used (1:200 dilution) included rabbit anti-phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; #9101S; Cell Signaling Technology); rabbit anti-NF-κB p65 (#8242; Cell Signaling Technology); mouse anti-NFATC2 (#sc-7296, Santa Cruz Biotechnology) mouse anti-CD3 (#sc-7296; Santa Cruz Biotechnology); rabbit anti-Fhl2 (#ab12328; Abcam); and mouse anti-PAR2 (SAM11, #817201; BioLegend). Goat anti-rabbit IgG-A594 or IgG-A488 was used as secondary antibody (1:200 dilution at room temperature). DAPI was used for nuclear counterstaining. ProLong Gold antifade reagent was used as mounting media. Mounted slides were examined with a Leica DM6000 B. Slide Book 6 (3i) was used for analysis and capturing images.
Lung digestion for isolation of single-cell preparations
Mouse lungs were perfused with Sal and then subjected to mechanical mincing followed by digestion at 37°C for 45 min in RPMI with 10% FBS, 1% penicillin/streptomycin, and collagenase type I (1 mg/ml; #LS004197; Worthington) as described previously (Verma et al., 2018). Isolated cell suspensions were agitated at room temperature for 10 min in RPMI with 100 U/ml DNase I before filtration through 40-µm filters and red blood cell lysis. Single-cell suspensions were either subsequently cultured in RPMI with 10% FBS and 1% penicillin/streptomycin at 37°C in a CO2 incubator overnight or used immediately for staining for FCM and analysis, depending on the experiment and schedules (Verma et al., 2018).
FCM analyses of ILCs, eosinophils, neutrophils, and other cells
Most of the fluorophore-conjugated antibodies used for FCM were purchased from BioLegend. Others were from eBioscience or R&D, unless otherwise stated. The mouse lung single-cell suspension was blocked with an anti-FcR blocking reagent (#130-092-575; Miltenyi Biotec) before staining. ILC cells were stained with FITC-labeled anti-CD45.2 (clone 104), Pacific Blue–labeled lineage marker antibodies (CD3, Ly-6G/Ly-6C, CD11b, CD45R/B220, TER-119/erythroid cells, and FcεRI), PerCP-Cy5.5–conjugated anti-CD25 (clone PC61.5; eBioscience), PE-Cy–labeled anti-NK1.1 (#108714; BioLegend), Pacific Blue anti-mouse NK-1.1 antibody (#108722; BioLegend), allophycocyanin- or Brilliant Violet 421–labeled anti-IL5 (TRFK5), Alexa Fluor 647–labeled anti-PAR 2 (#FAB3949R; R&D), and PE-Cy7–labeled anti-IL13 (clone eBio13A; eBioscience). Lung ILCs from Rag1 KO, Rag2−/−γc−/−, and DKO (Rag1KO × ST2 KO) mice were initially characterized with PE-conjugated anti-ST2 (#FAB10041P-100UG; R&D) and Brilliant Violet 510–conjugated anti-ICOS (CD278; clone C398.4) antibodies. For detection of surface FHL2 on live cells, we first applied FcR blocking reagent, followed by an unconjugated anti-FHL2 antibody (#MBS584089, clone F4B2-B11; Mybiosource) and then F488- or Pacific Blue–conjugated mouse IgG1 secondary antibody. After three washes, we used labeled antibodies for other markers. For detection of intracellular Fhl2, we treated cells with FcR block, fixed cells with 4% paraformaldehyde, and then applied Perm/Wash Buffer (BD). After this step, we incubated cells with the anti-Fhl2 antibody followed by the secondary antibody. After three washes, we applied labeled antibodies against other markers. For eosinophil and neutrophil: PerCP/Cy5.5 anti-mouse Ly-6G (#127616; clone1A8; BioLegend), PE/Cy7 anti-mouse CD11c (#117318; clone N418; BioLegend), Brilliant Violet 421 anti-mouse/human CD11b (#101236; clone M1/70; BioLegend), PE anti-mouse CD193 (#144506; clone J073E5; BioLegend), Alexa Fluor 647 rat anti-mouse Siglec-F (#562680; clone E50-2440; BD PharMingen). PerCP-eFluor 710 anti-GATA3 antibody (#46-9966-42; eBioscience), Alexa Fluor 488 anti-mouse TCR γ/δ antibody, Alexa Fluor 700 anti-mouse CD45.2 antibody, Brilliant Violet 605 anti-mouse NK-1.1 antibody, APC/Cy7 anti-mouse NKp46 antibody, and PerCP/Cy5 anti-mouse Ly6C and Ly6G. Staining for other molecules (when applicable) was done subsequent to the staining for Fhl2. Fixable Viability Dye eFluor 780 (#65-0865-14; eBioscience) and Zombie Aqua Fixable Viability Kit (#423102; BioLegend) were used for detection of cell viability. Stained cells were analyzed using the LSRFortessa cell analyzer (BD). Flow data were analyzed with FlowJo v10.0.7 software (TreeStar).
Adoptive transfer of ICOS+ST2+ and ICOS−ST2− cells from Rag1 KO mice
Lung cells were isolated from Alt-treated Rag1 KO mice after the last sensitizing exposure (Fig. 1 A) according to the protocol described above. Mouse lung single-cell suspension was blocked with an anti-FcR blocking reagent (#130-092-575; Miltenyi Biotec) before staining. ILCs were stained with FITC-labeled anti-CD45.2 (clone 104), PerCP-Cy5.5–conjugated anti-CD25 (clone PC61.5; eBioscience), Pacific Blue anti-mouse NK-1.1 antibody (#108722; BioLegend), or PE-Cy7–labeled anti-NK1.1 (#108714; BioLegend), anti-ST2 (#FAB10041P-100UG; clone #245707; R&D), and APC-Cy7–labeled anti-ICOS (clone C398.4A) antibodies. Live CD45.2, CD25, and ICOS+ST2+ and ICOS−ST2− cells were sorted on a BD FACSAria Fusion and delivered i.v. (50,000 cells/mice) to Rag2−/−γc−/− mice 24 h after sorting. Intranasal Alt (2.5 µg in 20-µl dose in PBS) was administered on three consecutive days after i.v. transfer of ICOS+ST2+ and ICOS−ST2− cells. The recipient mice were studied on day 6 for measurement of airway hyperreactivity, inflammation, and type 2 cytokine production.
Flow sorting and scRNA-seq
Lung cells were isolated from the Alt-treated Rag1 KO mice as described above. The single-cell suspension was stained with a FITC-labeled anti-CD45.2 (clone 104) antibody. Live CD45+ cells were sorted on a BD FACSAria Fusion. In total, 100,000 cells were sorted from the four study groups for scRNA-seq.
Generation of scRNA-seq library
Flow sorted cells were dispensed into a 5,184-nanowell microchip using the ICELL8 platform (WaferGen Biosystems), from which single cells were selected for library preparation and sequencing. Library preparation was performed using ICell8 single-cell poly-(A)+ transcriptome amplification hands-on workflow (with Triton X-100 for cell lysis), protocol D07-000040-003 rev4. Generated libraries for captured cells were then sequenced as 100-bp single-end reads on an Illumina HiSeq 4000.
scRNA-seq data processing
Generated cDNA reads in FASTQ format were trimmed on 5′ and 3′ ends based on quality (quality score q < 20) using Cutadapt (Martin, 2011). PolyA tails and short reads were removed (<25 bp). We performed trimmed alignment to the mouse genome (GRCm38/mm10) using GSNAP (Wu et al., 2016). Alignment was performed with max-mismatches = 0.05 and in a splice- and single-nucleotide polymorphism–aware fashion using known Ensembl sites. Gene expression was then quantified using HTSeq (Anders et al., 2015) with parameters “stranded = yes,” “mode = intersection-nonempty,” and “t = gene.” The number of unique molecular identifiers (UMIs) for each gene was summed to obtain a UMI count matrix.
We removed mitochondrial, ribosomal, nonexpressed, unannotated, and predicted transcripts to reduce data noise and facilitate interpretability. We then removed 113 cells that expressed <1,000 UMIs, resulting in an expression matrix with 871 single cells and 21,920 genes. The final quality controlled cells had a mean depth of 7,389 UMIs and a mean of 2,830 expressed genes. To account for differences in sequencing depth among cells, we normalized UMI counts by dividing each cell by the sum of its UMIs, multiplying by 10,000, and taking the log of this vector. The log-normalized data were then centered and scaled, and the number of UMIs was regressed from the from the expression matrix. The Seurat R package (Butler et al., 2018) was used to perform data normalization and scaling, as well as downstream dimensionality reduction, clustering, and differential expression analyses. All downstream analysis parameters were kept on default settings unless specified otherwise.
scRNA-seq dimensionality reduction and clustering
To focus analysis to the most informative genes, the relationship between each gene’s mean expression and its dispersion was modeled. 2,739 outlier genes in this mean–variance relationship model were selected (based on Z-score, default Seurat parameters) as the “most variant” genes. This subset of high variance genes was then used to compute PCA. The first four principal components were selected based the amount of relative variance they explained (i.e., the “elbow principle”). To reduce dimensionality further, t-SNE was performed (perplexity = 50) on the first four principal components. Cell cluster detection was performed using the first four principal components passed as features to the Louvain algorithm (k-nearest neighbors = 30). Seven clusters were initially detected, however, downstream analysis strongly suggested that these clusters in fact represented five cell types, and three of these clusters were combined into a single cluster. This resulted in five distinct post hoc cell clusters.
WGCNA
To detect coexpressed gene networks and refine the cluster detection of the Louvain algorithm, we performed WGCNA (Langfelder and Horvath, 2008). Briefly, the WGCNA method measures pairwise Pearson correlations among all expressed genes in the dataset, constructs a gene network based on these correlations, calculates topological overlap among genes in the network, and then, by carrying out hierarchical clustering of a distance metric based on the topological overlap matrix, identifies discrete clusters of genes. We set the soft-thresholding power to 9, which best maximized the number of connections in the gene network, while still meeting the scale-free network assumptions of the method, and we retained the direction of correlations among indirect connections in the network (i.e., analysis was signed). In addition, minimum network size was set to 20, and maxCoreScatter minGap and cutHeight were calculated according to the 90% quantile of the tree height. For each of the 10 gene networks (modules) detected from this analysis, a principal component–based summary metric (the module “eigengene”) was calculated for each cell and module.
Differential expression analysis
Differential expression for each gene between the detected cell clusters was performed using the likelihood ratio test for count bimodality. We limited each comparison to genes exhibiting both an estimated log fold-change >0.25 and detectable expression in >10% of cells in one of the two clusters being compared. We corrected for multiple hypothesis testing by calculating FDR-adjusted P values. Genes were considered differentially expressed when FDR < 0.05. Within the ILC2 cell cluster, effect sizes among the treatment groups were much smaller than the cluster-level differences. Thus, we assessed treatment-level differential expression using a Tobit test and reported uncorrected P values.
Real-time PCR
Total RNA was isolated from frozen lung samples using Trizol (Invitrogen). cDNA was synthesized using the Verso cDNA synthesis kit (#AB-1453/B; Thermo Fisher Scientific) according to manufacturer’s instructions as described previously (Verma et al., 2018). Gene-specific PCR products were amplified using the qPCR SYBR Green Rox mix (#AB-4162/B; Thermo Fisher Scientific) and primers outlined in the online repository (Table S3). Primers were designed using the Applied Biosystems 7000 Sequence Detection System software. The levels of target gene expression were normalized to 18S expression using the 2−ΔCt method, where Ct = threshold count. The primer list is given in Table S3.
Flow sorting and ATAC-seq
Lung cells were isolated from the Alt-treated Rag1 KO mice as described above. The single-cell suspension was stained with FITC-labeled anti-CD45.2 (clone 104), PerCP-Cy5.5-conjugated anti-CD25 (clone PC61.5; eBioscience), PE-Cy7–labeled anti-NK1.1 (#108714; BioLegend), and APC-Cy7–labeled anti-ICOS (clone C398.4A) antibodies. Live (DAPI-negative) CD45.2+NK1.1−CD25+ICOS+ cells were sorted on the Synergy SY3200 cell sorter. We sorted 50,000 cells/sample, and each group had three replicate samples except the allergen no-recall group, which had two replicate samples for ATAC-seq.
Omni-ATAC-seq
50,000 viable cells were resuspended in 50 µl lysis buffer (0.01% Digitonin, 0.1% NP-40, 0.1% Tween-20, 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, and 3 mM MgCl2) and incubated on ice for 3 min. Lysis buffer was washed out by adding 1 ml resuspending buffer (0.1% Tween-20, 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, and 3 mM MgCl2) and inverting tubes three times. Nuclei were centrifuged at 500 g for 10 min at 4°C. All supernatant was carefully aspirated. Transposase reaction was performed with a Nextera DNA Library Preparation kit from Illumina. Transposed DNA was isolated with a MiniElute PCR Purification Kit from Qiagen. Libraries were made by PCR. Real-time PCR was performed to determine PCR cycle number by calculating one-third of maximum fluorescent intensity. Primers are listed in (Buenrostro et al., 2015) and synthesized by Integrated DNA technologies (Coralville, IA). All libraries were sequenced using 2 × 50-bp reads on a Hiseq 2500 from Illumina (Buenrostro et al., 2015; Corces et al., 2017)
Bioinformatic analysis of omni-ATAC-seq
ATAC-seq reads were trimmed of adapters and poor sequence by skewer v0.2.2 (Jiang et al., 2014), and quality control was assessed with fastqc 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). BAM files were generated by mapping to the mm10 mouse genome with Bowtie 2 v2.3.2 (Langmead and Salzberg, 2012) with maximum fragment length set to −X 2000. The resulting BAM files were filtered for ChrM, PCR duplicates, and the ATAC-seq Blacklist from Encode (https://www.encodeproject.org/files/ENCFF759PJK) using picardtools v2.12.1 (http://broadinstitute.github.io/picard/) and samtools v1.5 (Li et al., 2009). Peaks were called for each sample using MACS v2.1.1 (Zhang et al., 2008) with –B–SPMR–nomodel–nolambda–call-summits options. A bed file of peak summits ±200 bp was generated using bedtools 2 v2.26.0 (Quinlan and Hall, 2010) slopBed tool and using bedops v2.4.14 (Neph et al., 2012) merged across samples to create a common reference peak list. A count table of the number of fragments intersecting each reference peak was then generated with Bedtools 2 multiBamCov tool for each sample and imported into R v3.4.2, where differential peak calling was performed using EdgeR v3.20.9 (Robinson et al., 2010). Significant differential peaks were determined as those with relative peak fragment counts between different exposed ILC2 groups differing by more than twofold and adjusted P < 0.05. Peaks were annotated by Homer v4.8 (http://homer.ucsd.edu/homer/index.html) annotatePeaks.pl with nearest gene and genomic location. Homer findMotifsGenome.pl was used to determine enriched motifs in the defined peaks of each dataset. Peaks were visualized with the genome using the Integrated Genome Viewer v2.3.97.
ELISA for cytokines
IL5 (#DY405-05; R&D) and IL13 (#DY413-05; R&D) in the BAL fluid were measured by ELISA kits as per manufacturer’s instructions.
Human subject studies
Flow sorting and adoptive transfer of ILCs from human lung
Human lung tissues were obtained from deceased adult organ donors and were procured by National Jewish Health from Donor Alliance. All subjects were free of acute or chronic infections (HIV and HBV/HCV) and cancer. As donors were deceased at the time of tissue collection, this work does not qualify as using human subjects as confirmed by the institutional review board of National Jewish Health. A piece of lung tissue was perfused with Sal to remove residual blood. A single-cell suspension was acquired by mechanical mincing of the lungs followed by digestion at 37°C for 40 min in RPMI with 10% FBS, 1% penicillin/streptomycin, and collagenase type I (1 mg/ml; #LS004197; Worthington) as described previously (Tait Wojno et al., 2015). The cell suspension was incubated with DNase I (Roche; 20 mg/ml) in RPMI at room temperature for 10 min and then filtered through a wire mesh sieve and a 70-mm cell strainer. RBCs were lysed with ACK buffer (Quality). Mononuclear cells from the isolated lung cells were separated by centrifugation on a Ficoll-Paque gradient (GE Healthcare). Isolated mononuclear cells were cultured in RPMI with 10% FBS and 1% penicillin/streptomycin at 37°C in a CO2 incubator overnight with recombinant human IL-2 (10 µg/ml). The next day, the mononuclear cell suspension was depleted of Lin+ cells by negative selection using the Human Lineage Depletion Kit (#130-092-211; Miltenyi Biotech). Collected Lin− cells were first stained with an unconjugated anti-FHL2 (#MBS584089, clone F4B2-B11; Mybiosource) antibody followed by F488-conjugated mouse IgG1 secondary antibody. The cells were then washed and subsequently stained with APC-Cy7–conjugated anti-CRTH2 (BM16), and live CRTH2+Fhl2+/− cells were sorted on a BD FACSAria Fusion and cultured with IL2 in a CO2 incubator overnight. The next daythese cells were administered i.v. (100,000 cells/mice) to Rag2−/−γc−/− mice. Intranasal Alt (5 µg in 20 µl PBS) was administered on three consecutive days after i.v. transfer of CRTH2+Fhl2+ and CRTH2+Fhl2− cells. The recipient mice were studied on day 6 (48 h after the last exposure) for airway hyperreactivity, inflammation, and cytokine production.
Human blood ILC studies
Human subjects
We studied blood ILCs (Lin− cells) from allergic patients (with and without asthma) and healthy donors. Allergic patients were recruited from the outpatient clinics at National Jewish Health. Allergic patients undergoing allergen immunotherapy (rush immunotherapy protocol) were recruited from the National Jewish Health Minimally Invasive Diagnostic Center. Healthy donors were recruited from the blood bank of National Jewish Health. The protocols for studies of lymphoid cells from allergic patients and healthy subjects were approved by the institutional review board. Written informed consent was obtained from each participant.
Isolation of PBMCs, Lin− cells, culture, and FCM for ILC2s
PBMCs were isolated from EDTA-treated blood by density centrifugation with Ficoll-Hypaque. PBMCs were cultured in RPMI 1640 plus 10% FBS in the medium alone or with Alt extract (5 µg/ml) for the indicated period of time as described in Fig. 8 A. Human PBMCs from 9 healthy and 14 allergic asthmatic patients were treated with medium or Alt (5 µg/ml) for 24 h. The cells were then washed three times and placed in the medium alone for 7 d. The cultures were rechallenged with either medium or a subthreshold dose of Alt (1 µg/ml) for 24 h. Human blood Lin− cells were isolated form PBMCs by negative selection using antibody-coated magnetic beads (Miltenyi Biotec and Stem Cell Technologies). Lin− cells were cultured in medium alone or with Alt (5 µg/ml), anti-PAR2 antibody, or an isotype control antibody. In select experiments, Lin− cells were isolated from PBMCs by negative selection using antibody-coated magnetic beads (Lineage Cell Depletion Kit; Miltenyi Biotec). To study the effect of repetitive allergen injections during rush immunotherapy on memory-related AP1 genes, we isolated PBMCs from seven allergic patients before and after rush immunotherapy with multiple allergens when the patient received incremental doses of the allergens every 30 min, 6 h a day, for two consecutive days. After culture, cells were pelleted and stained for FCM as described previously (Christianson et al., 2015; Liu et al., 2018).
FCM staining of cells
Staining for FCM was performed as described previously (Christianson et al., 2015; Liu et al., 2018). We added a labeled FcεRI antibody to the human lineage cocktail (CD3, CD14, CD16, CD19, CD20, and CD56) antibodies (#348805; BioLegend) to eliminate basophils. We stained cell surface markers BV510-conjugated anti-ICOS (#313525, CD278; clone:C398.4; BioLegend), PAR2 (#3949R; R&D), and dectin-1 (#355404; BioLegend) on live cells first. Then, we fixed cells using 4% paraformaldehyde, permeabilized them with 0.1% saponin, and performed staining for intracellular molecules JunD (#sc271938; Santa Cruz Biotechnology), c-Fos (#14609; Cell Signaling Technology), and JunB (#sc-8051; Santa Cruz Biotechnology). FCM was performed on an LSRII (BD Biosciences). The data were analyzed by FlowJo software. Isotype antibody controls and fluorescence minus one were used to develop the gating and data analysis strategies.
Statistical analyses
Statistical analyses were performed and figures were prepared with Prism software (v6; GraphPad). Statistical significance was analyzed by t test or ANOVA for mouse data and Mann–Whitney U test for human data. The data for scRNA-seq and ATAC-seq data are deposited under accession no. GSE172258.
Online supplemental material
Fig. S1 shows histological changes in the memory model and gating strategy for FCM. Fig. S2 shows representative flow cytograms for TCRγδ+, Ly6G+/Ly6C+, NKp46+, CD11B+, NK1.1, and FcεRI+ cells in ICOS+ST2+ cells; the effect of RORα inverse agonist SR3335 on ILC2s; a short exposure model for memory-driven asthma in Rag1−/− mice; and the importance of ICOS for memory-associated asthma. Fig. S3 shows the role of ICOS and IL33 in memory formation; scRNA-seq; t-SNE coordinate clustering of cells with WGCNA network module eigengene expression overlaid for modules highly expressed in each particular cluster; and the gating and sorting strategy for isolation of CD45+NK1.1−CD25+ICOS+ cells for ATAC-seq. Fig. S4 shows heatmaps of differentially expressed DNA accessibility peaks between Alt/− versus Sal/− and Alt/Alt versus Sal/−; expression of mRNA for motif of Fig. 6, H and I, from Alt-sensitized and recalled Fhl2+/+ and Fhl2−/− mice and all four groups of memory model (Rga1−/−); and the gating strategy for FCM for lung cells from the wild-type and Fhl2−/− mice. Table S1 shows top known motifs by HOMER for all memory groups. Table S2 shows top de novo and known motifs for Alt/− versus Sal/− and Sal/− versus Alt/Alt. Table S3 lists the primer sequences for RT-PCR.
Supplementary Material
shows top known motifs (HOMER) for all memory groups.
shows top de novo and known motifs for Alt/− versus Sal/− and Sal/− versus Alt/Alt.
lists the primer sequences for RT-PCR.
Acknowledgments
We are indebted to Allen Stevens and Christina Kolakowski for the outstanding work with patient recruitment.
The work was supported by National Institutes of Health grants AI102943, AI137970, and HL126895 and grants from the Cohen Family Foundation, NBL Fellowship Foundation, Colorado Technology, and Department of Medicine of National Jewish Health to R. Alam.
Author contributions: M. Verma conducted the majority of the experiments, contributed to the design of select experiments, analyzed and prepared the data, and contributed to preparation of the manuscript. L. Michalec, A. Sripada, K. Sirohi, and D. Verma performed select experiments and helped M. Verma in other mouse experiments. J. McKay and D. Sheth conducted some of the human experiments. N. Dyjack and M.A. Seibold performed scRNA-seq and analyzed the data. J.R. Knapp, T-H. Tu, and B.P. O’Connor performed ATAC-seq and analyzed the data. R. Martin and M.M. Gorska helped with discussion of the study design and review of the manuscript. R. Alam conceived the concept, designed the experiments, analyzed the data, and wrote the manuscript.
References
- Agrawal, S., Agrawal A., Doughty B., Gerwitz A., Blenis J., Van Dyke T., and Pulendran B.. 2003. Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th responses via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J. Immunol. 171:4984–4989. 10.4049/jimmunol.171.10.4984 [DOI] [PubMed] [Google Scholar]
- Anders, S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf, S., Hegazy Y.K., and Harmancey R.. 2019. Nuclear receptor subfamily 4 group A member 2 inhibits activation of ERK signaling and cell growth in response to β-adrenergic stimulation in adult rat cardiomyocytes. Am. J. Physiol. Cell Physiol. 317:C513–C524. 10.1152/ajpcell.00526.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé, R.A., García-Palmero I., Torres S., López-Lucendo M., Balyasnikova I.V., and Casal J.I.. 2015. IL13 Receptor α2 Signaling Requires a Scaffold Protein, FAM120A, to Activate the FAK and PI3K Pathways in Colon Cancer Metastasis. Cancer Res. 75:2434–2444. 10.1158/0008-5472.CAN-14-3650 [DOI] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cephus, J.Y., Stier M.T., Fuseini H., Yung J.A., Toki S., Bloodworth M.H., Zhou W., Goleniewska K., Zhang J., Garon S.L., et al. 2017. Testosterone Attenuates Group 2 Innate Lymphoid Cell-Mediated Airway Inflammation. Cell Rep. 21:2487–2499. 10.1016/j.celrep.2017.10.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., López-Moyado I.F., Seo H., Lio C.J., Hempleman L.J., Sekiya T., Yoshimura A., Scott-Browne J.P., and Rao A.. 2019. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 567:530–534. 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A., et al. 2014. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 345:1250684. 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, C.A., Goplen N.P., Zafar I., Irvin C., Good J.T. Jr., Rollins D.R., Gorentla B., Liu W., Gorska M.M., Chu H., et al. 2015. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J. Allergy Clin. Immunol. 136:59–68.e14. 10.1016/j.jaci.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, P.S. 1999. STAT6: an intracellular target for the inhibition of allergic disease. Clin. Exp. Allergy. 29:12–16. 10.1046/j.1365-2222.1999.00476.x [DOI] [PubMed] [Google Scholar]
- Guan, T., Dominguez C.X., Amezquita R.A., Laidlaw B.J., Cheng J., Henao-Mejia J., Williams A., Flavell R.A., Lu J., and Kaech S.M.. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med. 215:1153–1168. 10.1084/jem.20171352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim, T.Y., Hwang Y.Y., Scanlon S.T., Zaghouani H., Garbi N., Fallon P.G., and McKenzie A.N.. 2016. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 17:57–64. 10.1038/ni.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman, C.S., Panova V., and McKenzie A.N.. 2013. IL-33 citrine reporter mice reveal the temporal and spatial expression of IL-33 during allergic lung inflammation. Eur. J. Immunol. 43:488–498. 10.1002/eji.201242863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenfuss, S.C., Bakiri L., Thomsen M.K., Williams E.G., Auwerx J., and Wagner E.F.. 2014. Regulation of steatohepatitis and PPARγ signaling by distinct AP-1 dimers. Cell Metab. 19:84–95. 10.1016/j.cmet.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst, M., Hejjaoui A., Horst V., Michel F.B., and Bousquet J.. 1990. Double-blind, placebo-controlled rush immunotherapy with a standardized Alternaria extract. J. Allergy Clin. Immunol. 85:460–472. 10.1016/0091-6749(90)90156-X [DOI] [PubMed] [Google Scholar]
- Ito, I., Bhopale K.K., Kobayashi M., Finnerty C.C., Herndon D.N., and Suzuki F.. 2017. Lamina propria group 2 innate lymphoid cells impair the antibacterial defense of burned mice to enterococcal translocation. J. Leukoc. Biol. 102:1451–1460. 10.1189/jlb.4A0517-195R [DOI] [PubMed] [Google Scholar]
- Jacobsen, K.X., MacDonald H., Lemonde S., Daigle M., Grimes D.A., Bulman D.E., and Albert P.R.. 2008. A Nurr1 point mutant, implicated in Parkinson’s disease, uncouples ERK1/2-dependent regulation of tyrosine hydroxylase transcription. Neurobiol Dis. 29:117–122. 10.1016/j.nbd.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Lei R., Ding S.W., and Zhu S.. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 15:182. 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, M., Møller S., Hansen T., Moens U., and Van Ghelue M.. 2006. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell. Mol. Life Sci. 63:268–284. 10.1007/s00018-005-5438-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, T.J., Suzuki H.I., Zamudio J.R., Suzuki M., and Sharp P.A.. 2019. Sequestration of microRNA-mediated target repression by the Ago2-associated RNA-binding protein FAM120A. RNA. 25:1291–1297. 10.1261/rna.071621.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani, Y., Elsaesser H., Hock M.B., Vergnes L., Williams K.J., Argus J.P., Marbois B.N., Komisopoulou E., Wilson E.B., Osborne T.F., et al. 2013. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 14:489–499. 10.1038/ni.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, C.S., and Artis D.. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17:765–774. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- Klose, C.S.N., Mahlakõiv T., Moeller J.B., Rankin L.C., Flamar A.L., Kabata H., Monticelli L.A., Moriyama S., Putzel G.G., Rakhilin N., et al. 2017. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 549:282–286. 10.1038/nature23676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakula, K., van der Wal E., Geerts D., van Tiel C.M., and de Vries C.J.. 2011. FHL2 protein is a novel co-repressor of nuclear receptor Nur77. J. Biol. Chem. 286:44336–44343. 10.1074/jbc.M111.308999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakula, K., Vos M., Logiantara A., Roelofs J.J., Nieuwenhuis M.A., Koppelman G.H., Postma D.S., Brandsma C.A., Sin D.D., Bossé Y., et al. 2015. Deficiency of FHL2 attenuates airway inflammation in mice and genetic variation associates with human bronchial hyper-responsiveness. Allergy. 70:1531–1544. 10.1111/all.12709 [DOI] [PubMed] [Google Scholar]
- Langfelder, P., and Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.M., Adams N.M., Geary C.D., Weizman O.E., Rapp M., Pritykin Y., Leslie C.S., and Sun J.C.. 2018. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 19:963–972. 10.1038/s41590-018-0176-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Tournier C., Davis R.J., and Flavell R.A.. 1999. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18:420–432. 10.1093/emboj/18.2.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Fan C.M.. 2017. A CREB-MPP7-AMOT Regulatory Axis Controls Muscle Stem Cell Expansion and Self-Renewal Competence. Cell Rep. 21:1253–1266. 10.1016/j.celrep.2017.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Verma M., Michalec L., Liu W., Sripada A., Rollins D., Good J., Ito Y., Chu H., Gorska M.M., et al. 2018. Steroid resistance of airway type 2 innate lymphoid cells from patients with severe asthma: The role of thymic stromal lymphopoietin. J. Allergy Clin. Immunol. 141:257–268.e6. 10.1016/j.jaci.2017.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners, S., Alam R., Schwartz D.A., and Gorska M.M.. 2014. A mouse model links asthma susceptibility to prenatal exposure to diesel exhaust. J. Allergy Clin. Immunol. 134:63–72.E7. 10.1016/j.jaci.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, A.P., Ishizuka I.E., Kasal D.N., Mandal M., and Bendelac A.. 2017. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 8:863. 10.1038/s41467-017-00882-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martinez-Gonzalez, I., Mathä L., Steer C.A., Ghaedi M., Poon G.F., and Takei F.. 2016. Allergen-Experienced Group 2 Innate Lymphoid Cells Acquire Memory-like Properties and Enhance Allergic Lung Inflammation. Immunity. 45:198–208. 10.1016/j.immuni.2016.06.017 [DOI] [PubMed] [Google Scholar]
- McNulty, S.E., Barrett R.M., Vogel-Ciernia A., Malvaez M., Hernandez N., Davatolhagh M.F., Matheos D.P., Schiffman A., and Wood M.A.. 2012. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn. Mem. 19:588–592. 10.1101/lm.026385.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner, A., Karreth F., Kenner L., and Wagner E.F.. 2004. JunD regulates lymphocyte proliferation and T helper cell cytokine expression. EMBO J. 23:1325–1335. 10.1038/sj.emboj.7600133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondino, A., Whaley C.D., DeSilva D.R., Li W., Jenkins M.K., and Mueller D.L.. 1996. Defective transcription of the IL-2 gene is associated with impaired expression of c-Fos, FosB, and JunB in anergic T helper 1 cells. J. Immunol. 157:2048–2057. [PubMed] [Google Scholar]
- Monticelli, L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon, A., and Sassone-Corsi P.. 2003. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. USA. 100:3977–3982. 10.1073/pnas.0735923100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, T.L., Tussiwand R., and Murphy K.M.. 2013. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13:499–509. 10.1038/nri3470 [DOI] [PubMed] [Google Scholar]
- Neph, S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S., et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics. 28:1919–1920. 10.1093/bioinformatics/bts277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea, M.G., Joosten L.A.B., Latz E., Mills K.H.G., Natoli G., Stunnenberg H.G., O’Neill L.A.J., and Xavier R.J.. 2016. Trained immunity: A program of innate immune memory in health and disease. Science. 352:aaf1098. 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant, C.J., Hwang Y.Y., Walker J.A., Salimi M., Wong S.H., Brewer J.M., Englezakis A., Barlow J.L., Hams E., Scanlon S.T., et al. 2014. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 41:283–295. 10.1016/j.immuni.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlgren, M., Dubois P.M., Tomkowiak M., Sjögren T., and Marvel J.. 1996. Resting memory CD8+ T cells are hyperreactive to antigenic challenge in vitro. J. Exp. Med. 184:2141–2152. 10.1084/jem.184.6.2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal, S., and Shenoy S.K.. 2018. GPCR desensitization: Acute and prolonged phases. Cell. Signal. 41:9–16. 10.1016/j.cellsig.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri, R., Clever D., Li P., Wakabayashi Y., Quinn K.M., Klebanoff C.A., Ji Y., Sukumar M., Eil R.L., Yu Z., et al. 2016. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17:851–860. 10.1038/ni.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri, R., Hirahara K., Mousavi K., Clever D., Klebanoff C.A., Bonelli M., Sciumè G., Zare H., Vahedi G., Dema B., et al. 2013. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 498:506–510. 10.1038/nature12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert, S.M., Chehtane M., Zhang G., Hu H., Li X., and Khaled A.R.. 2012. JunD/AP-1-mediated gene expression promotes lymphocyte growth dependent on interleukin-7 signal transduction. PLoS One. 7:e32262. 10.1371/journal.pone.0032262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, S., Quintin J., Kerstens H.H., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.C., Ratter J., Berentsen K., van der Ent M.A., et al. 2014. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 345:1251086. 10.1126/science.1251086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya, T., Kashiwagi I., Inoue N., Morita R., Hori S., Waldmann H., Rudensky A.Y., Ichinose H., Metzger D., Chambon P., and Yoshimura A.. 2011. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2:269. 10.1038/ncomms1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelgrove, R.J., Gregory L.G., Peiró T., Akthar S., Campbell G.A., Walker S.A., and Lloyd C.M.. 2014. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J. Allergy Clin. Immunol. 134:583–592.e6. 10.1016/j.jaci.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits, H., and Di Santo J.P.. 2011. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12:21–27. 10.1038/ni.1962 [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Sasaki K., Kamata R., Hoshino Y., Yanagihara K., and Sakai R.. 2009. A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol. Cell. Biol. 29:402–413. 10.1128/MCB.01035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, M.K., Kurakula K., Koenis D.S., and de Vries C.J.. 2016. Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta. 1863:219–228. 10.1016/j.bbamcr.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Verma, M., Liu S., Michalec L., Sripada A., Gorska M.M., and Alam R.. 2018. Experimental asthma persists in IL-33 receptor knockout mice because of the emergence of thymic stromal lymphopoietin-driven IL-9+ and IL-13+ type 2 innate lymphoid cell subpopulations. J. Allergy Clin. Immunol. 142:793–803.e8. 10.1016/j.jaci.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walachowski, S., Tabouret G., Fabre M., and Foucras G.. 2017. Molecular Analysis of a Short-term Model of β-Glucans-Trained Immunity Highlights the Accessory Contribution of GM-CSF in Priming Mouse Macrophages Response. Front. Immunol. 8:1089. 10.3389/fimmu.2017.01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrapp, A., Riesenfeld S.J., Burkett P.R., Abdulnour R.E., Nyman J., Dionne D., Hofree M., Cuoco M.S., Rodman C., Farouq D., et al. 2017. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 549:351–356. 10.1038/nature24029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati, E., Chen Z., Manne S., Kurachi M., Ali M.-A., Lewy K., Cai Z., Nzingha K., McLane L.M., Hope J.L., et al. 2018. Long-term persistence of exhausted CD8 T Cells in chronic infection is regulated by microRNA-155. Cell Rep. 23:2142–2156. 10.1016/j.celrep.2018.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait Wojno, E.D., Monticelli L.A., Tran S.V., Alenghat T., Osborne L.C., Thome J.J., Willis C., Budelsky A., Farber D.L., and Artis D.. 2015. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8:1313–1323. 10.1038/mi.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M., Nakajima S., Ohnuki K., Ogawa S., Yamashita M., Nakayama T., Murakami Y., Tanabe K., and Abe R.. 2012. AP-1 is involved in ICOS gene expression downstream of TCR/CD28 and cytokine receptor signaling. Eur J Immunol. 42:1850–1862. 10.1002/eji.201141897 [DOI] [PubMed] [Google Scholar]
- Wong, S.H., Walker J.A., Jolin H.E., Drynan L.F., Hams E., Camelo A., Barlow J.L., Neill D.R., Panova V., Koch U., et al. 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13:229–236. 10.1038/ni.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.D., Reeder J., Lawrence M., Becker G., and Brauer M.J.. 2016. GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. Methods Mol. Biol. 1418:283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows top known motifs (HOMER) for all memory groups.
shows top de novo and known motifs for Alt/− versus Sal/− and Sal/− versus Alt/Alt.
lists the primer sequences for RT-PCR.