Abstract.
Cholera is a severe acute, highly transmissible diarrheal disease which affects many low- and middle-income countries. Outbreaks of cholera are confirmed using microbiological culture, and additional cases during the outbreak are generally identified based on clinical case definitions, rather than laboratory confirmation. Many low-resource areas where cholera occurs lack the capacity to perform culture in an expeditious manner. A simple, reliable, and low-cost rapid diagnostic test (RDT) would improve identification of cases allowing rapid response to outbreaks. Several commercial RDTs are available for cholera testing with two lines to detect either serotypes O1 and O139; however, issues with sensitivity and specificity have not been optimal with these bivalent tests. Here, we report an evaluation of a new commercially available cholera dipstick test which detects only serotype O1. In both laboratory and field studies in Kenya, we demonstrate high sensitivity (97.5%), specificity (100%), and positive predictive value (100%) of this new RDT targeting only serogroup O1. This is the first field evaluation for the new Crystal VC-O1 RDT; however, with these high-performance metrics, this RDT could significantly improve cholera outbreak detection and improve surveillance for better understanding of cholera disease burden.
INTRODUCTION
Cholera is a diarrheal disease caused by the bacterium Vibrio cholerae. It typically occurs in low- and middle-income countries in settings that lack access to clean water and proper hygiene and sanitation. Although confirmation of the etiologic agent is not necessary for treatment of the disease, confirmation of the diagnosis is vital to identify and respond to outbreaks and to better understand the epidemiology of the disease. Unfortunately, cholera confirmation is often challenging because the disease tends to occur in resource-constrained settings; these areas often lack sophisticated microbiology laboratories with skilled personnel. When these laboratories are available, the classical culture method requires serological and biochemical tests which require several days before a report can be issued. Increasingly, a PCR is being used to confirm the diagnosis1 and can be useful for certain situations, but this assay has not been widely used because of lack of PCR equipment, reagents, and trained staff.
In 2017, the Global Task Force on Cholera Control (GTFCC) led by the WHO announced their “Roadmap to End Cholera by 2030,” and a key element of the road map is the ability to rapidly respond to outbreaks.2 A reliable rapid diagnostic test (RDT) that could be used in low-resource settings would greatly facilitate this goal. Several commercial RDTs are available and are being used during outbreaks or for surveillance.1,3–8 These tests are portable and can be performed in nearly any environment. However, the currently available tests have demonstrated a wide range of sensitivities and specificities and their accuracy and reliability can vary based on the user and situation.9,10 Because of the lack of confidence in current cholera RDTs, public health officials have been reluctant to declare an outbreak based solely on results of an RDT; this can result in delays in response to an outbreak.11
Crystal VC is an RDT (Arkray Healthcare Pvt. Ltd, Gujarat, India) that tests for both serogroups O1 and O139. However, specificity for this test has been problematic, requiring confirmation of the diagnosis by culture because false-positive reactions,12 especially for serotype O139,6,13–16 have occurred. Because of this issue, the Centers for Disease Control advises, “it is recommended that fecal specimens that test positive for V. cholerae O1 and/or O139 by the Crystal® VC dipstick always be confirmed using traditional culture-based methods suitable for the isolation and identification of V. cholerae.”17 Theoretically, the ability to detect serotype O139 could be of benefit; however, cholera due to V. cholerae serotype O139 has never been detected in Africa and is now very rarely identified in the South Asia and China during the last two decades.18–20
A new version of the RDT was recently developed and is marketed as Crystal VC O1 (Arkray Healthcare Pvt. Ltd). This new version tests solely for V. cholerae O1 with the intention to improve the accuracy of the assay for serotype O1. Thus, this RDT eliminates concerns for false O139 positives with the potential to increase specificity. If a cholera RDT is developed and demonstrates high sensitivity and specificity, this will greatly change the landscape of cholera detection and control as well as our understanding of true burden of disease. In this study, we conducted laboratory-based testing of a Crystal VC O1 in comparison to the original Crystal VC. Subsequently, we conducted a field evaluation of the test during outbreaks in Kenya from 2018 to 2019 to evaluate the RDT for sensitivity and specificity in comparison to both standard microbiological culture and PCR.
METHODS
Ethics.
Ethical approval to conduct the study was obtained from the AMREF Health Africa Ethics and Scientific Review Committee (ESRC P552/2018) and the Johns Hopkins Bloomberg School of Public Health Institutional Review Board (IRB00009067). Outbreaks were responded to according to Ministry of Health Protocol, by the Ministry of Health personnel. Specimens were collected through the Ministry of Health’s disease surveillance program. Patients suspected of having cholera gave informed consent for participation in the study (including parental permission/child assent) before enrollment in the study. Personal identifiers were removed upon enrollment.
Study site.
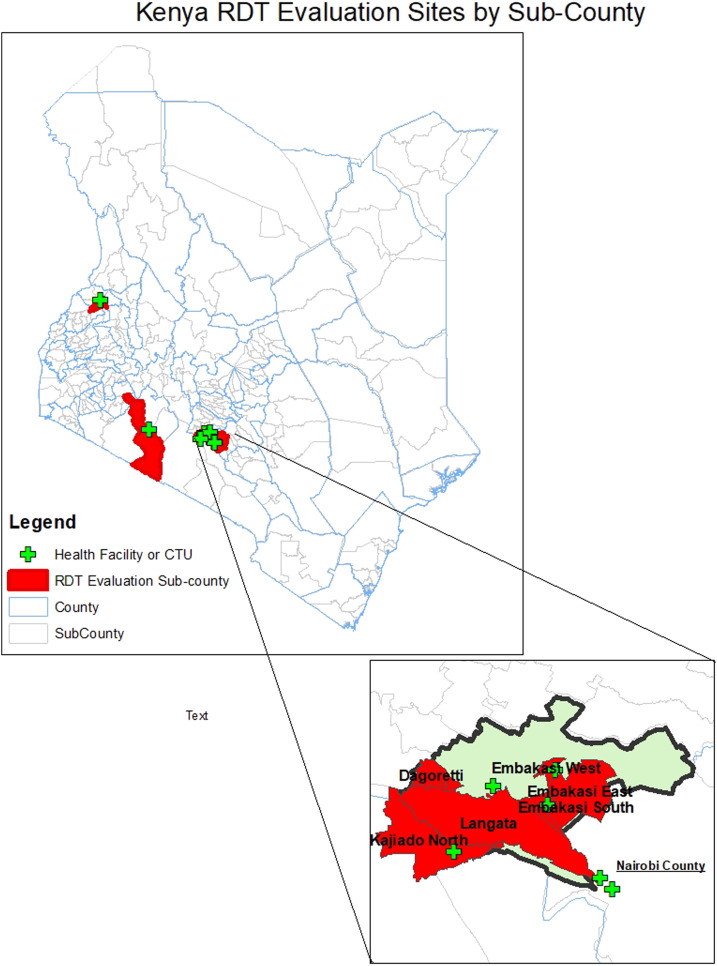
Crystal VC-O1 was evaluated, in comparison to Crystal VC, for specificity and cross-reactivity in the Enteric Laboratory of the International Health Department of the Johns Hopkins Bloomberg School of Public Health, Baltimore, MD. Field evaluations were conducted in the Republic of Kenya where cholera is endemic and has been reported regularly since 1971.21 A cholera outbreak beginning in December 201422 has challenged the Kenyan health system and continued through the evaluation of this RDT. Stool samples were collected from December 2018 to May 2019 at cholera treatment centers (CTCs) and healthcare facilities throughout Kenya (Figure 1). Laboratory testing was completed at the National Public Health Laboratory, Ministry of Health, Kenya.
Figure 1.
Map of rapid diagnostic test (RDT) evaluation sites.
Study Procedures.
Specificity and cross-reactivity testing, Baltimore, MD.
Stool was collected from a healthy individual at three different time points and stored at 4°C until testing. The stool was suspended in sterile, nuclease-free distilled water mixed by vortexing until getting a homogenous mixture at a ratio of 1 g of stool to 3 mL of water.
Strains of Vibrio were cultured to create test combinations of Vibrio strains in the spiked healthy stool. The strains used in this study included strains of Vibrio cholerae O1 Ogawa, Vibrio cholerae O1 Inaba, V. cholerae O139, nontoxigenic V. cholerae, and V. cholerae non-O1, non-0139 (Supplemental Table 1). Frozen stocks were revived via direct streaking on thiosulfate citrate bile salt sucrose (TCBS; Becton Dickinson and Co., Franklin Lakes, NJ) agar followed by overnight incubation at 37°C. A sterile cotton swab was used to touch 10 colonies from the plate and resuspend in 3 mL (mL) of PBS. And, 0.1 mL of the suspension is used to subculture lawns of the bacteria on Luria agar overnight at 37°C. The bacterial lawn is harvested with sterile PBS. The optical density of the bacterial suspension was determined by measurement at 600 nm, and the bacterial–stool mixture was diluted to a final concentration of approximately 1 × 108 bacteria per milliliter. The number of colony-forming units (CFU) in the inoculum was confirmed by titrating and plating on Luria agar plates.23 Bacterial–stool suspensions were created for nontoxigenic V. cholerae, V. cholerae non-O1, non-O139, V. cholerae O1 Ogawa, V. cholerae O1 Inaba, and V. cholerae O139 bacteria.
Specificity testing was performed by testing combinations of strain-spiked stool samples to validate specificity to V. cholerae O1. Testing was performed in triplicate with Crystal VC and Crystal VC-O1 RDTs. Cross-reactivity testing was performed using one spiked sample per specified combination. Before testing, each sample was vortexed for an additional 30 seconds. The samples were tested using the RDTs following the manufacturers’ instructions for testing fecal samples.24
Field evaluation study procedures, Kenya.
Patients of any age who presented to the health facility seeking care for acute watery diarrhea were eligible for inclusion in the study. No other inclusion or exclusion criteria were applied. A fresh stool sample, and basic clinical and demographic characteristics were collected from patients who consented to participate. Information collected included, but was not limited to, location of the health facility, age, gender, and dehydration status (Table 3). Standard medical care was provided by the healthcare facility, and participation in the study did not affect the care provided. Fresh stool specimens were immediately tested using the Crystal VC O1 RDT according to manufacturers’ instructions. In brief, approximately two drops of liquid stool were transferred to the reagent vial included in the Crystal VC/Crystal VC-O1 kit. For solid stools, a small portion of stool was collected with the sampling stick and then transferred to the reagent vial and shaken to mix. From the mixed reagent vial, four drops were then transferred to the provided test tube. A dipstick was removed from its sealed packaging and then placed into the liquid stool mixture such that only the absorbent tip was immersed. The strip was removed after 15 and before 30 minutes and read by two independent technicians, and results were recorded separately. The dipsticks were photographed to preserve the appearance of the dipstick. A positive result appears as two pink lines on the dipstick, the upper line being the control band and the lower line being the band specific to serogroup O1.
Table 3.
Participant demographics and clinical status
| Variable | RDT positive (n = 79) | RDT negative (n = 148) | P-value | |
|---|---|---|---|---|
| Gender (%) | Male | 50 (42.4) | 68 (57.6) | 0.013 |
| Female | 29 (25.9) | 83 (74.1) | ||
| Mean age (years) (±SD) | 28.1 (14.86) | 24.5 (14.6) | 0.1276 | |
| Age category (years) | 0–4 (%) | 7 (25) | 21 (75) | 0.499 |
| 5–14 | 6 (34.6) | 13 (68.4) | ||
| ≥ 15 | 66 (36.1) | 117 (63.9) | ||
| Type of diarrhea | Bloody | 0 (0) | 1 (100) | < 0.001 |
| Mucus | 15 (16.1) | 78 (83.9) | ||
| Watery | 64 (47.4) | 71 (52.6) | ||
| Dehydration status (%) | Plan A (none) | 1 (1.2) | 81 (98.7) | < 0.001 |
| Plan B (moderate) | 36 (39.6) | 55 (60.4) | ||
| Plan C (severe) | 42 (73.7) | 15 (26.3) | ||
| IV Rehydration (%) | No | 0 (0) | 82(100) | < 0.001 |
| Yes | 79 (54.1) | 67(45.9) | ||
An aliquot of the stool samples was preserved used for standard microbiological methods. Prepackaged Cary-Blair tubes were labeled with the specimen ID, the supplied swab was inserted into the stool sample and subsequently used to inoculate the media. All specimens were cultured within 3 days of collection. The microbiology laboratory recorded the results of the microbiological culture tests independent of the results of the rapid test.
In addition to the RDT and microbiological culture, two drops of the stool specimen were spotted and dried on a Whatman 903 Protein Saver Card (10534612 903; Sigma-Aldrich, St. Louis, MO; catalog no. WHA10534612). The filter paper specimen was then stored in a plastic bag for PCR testing for V. cholerae at Johns Hopkins University.1,25 The laboratories were blind to the results of tests completed in separate laboratories.
Laboratory methods.
Microbiological stool culture.
Swabs in specimen-labeled Cary-Blair tubes were used to streak TCBS plates according to published protocol.26
Multiplex PCR.
Stool samples preserved on Whatman 903 Protein Saver cards were shipped to the laboratory at Johns Hopkins University. DNA was extracted from filter papers according to previously published methods.1 In brief, the stool-spotted filter paper was excised and rehydrated in 1 mL of sterile 1X PBS for 10 minutes, centrifuged (14,000 × g for 2 minutes), and the supernatant discarded. Then, 1 mL of sterile 1X PBS was added to the sample tube and immediately centrifuged, and the supernatant was discarded. Subsequently, 140 µL of 2% (w/v) Chelex-100 (Bio-Rad, Hercules, CA) was added to each sample. And then, 50 µL of sterile water was added to each sample, followed by heating at 100°C for 8 minutes. The samples were then centrifuged, and the supernatant was transferred to a clean microcentrifuge tube and stored at −20°C until PCR processing. Conventional PCR was performed targeting the ompW and cholera toxin A gene (ctxA) genes, as previous described.25 The Nandi multiplex targets 1) the outer membrane protein (OmpW), a gene conserved in the V. cholerae genome; and 2) the ctxA. The PCR was performed, as published, using Terra PCR Direct Polymerase (Takara, Mountain View, CA).
Statistical methods.
A true cholera case was defined as a clinically suspected case with either a positive culture or PCR result for V. cholerae O1. A true-negative case was defined as a suspected case that was negative by culture and PCR for V. cholerae O1. The sensitivity, specificity, positive predictive values (PPVs) and negative predictive values (NPVs) were calculated using the Stata command diagti for both laboratory evaluations and field evaluations. The analysis was conducted in Stata 16 (Stata Corporation, College Station, TX). The kappa coefficient was calculated in Stata using the kap command to estimate the agreement between the gold standards (culture and/or PCR) and the RDT. Chi-square and T-test analyses were used evaluate clinical characteristics of study participants.
RESULTS
Laboratory evaluation.
Table 1 demonstrates the sensitivity of both the original Crystal VC RDT and the new Crystal VC O1 RDT. In three evaluations of stool spiked with nontoxigenic O1, non-O1/non-O139, or O139 Vibrio strains, both RDTs accurately displayed positive results for samples spiked with O1 strains, and negative results for the non-O1/non-O139 strains. For the O139 spiked stools, the Crystal VC O1 showed a negative result, whereas the Crystal VC RDT was positive for O139.
Table 1.
Laboratory evaluation of Crystal VC and Crystal VC O1
| Serotype | Trial | Crystal VC O1 | Crystal VC | |
|---|---|---|---|---|
| O1 line | O1 line | O139 line | ||
| Nontoxigenic V. cholerae O1 | 1 | + | + | – |
| 2 | + | + | – | |
| 3 | + | + | – | |
| V. cholerae (non-O1 and non-O139) | 1 | – | – | – |
| 2 | – | – | – | |
| 3 | – | – | – | |
| V. cholerae O139 | 1 | – | – | + |
| 2 | – | – | + | |
| 3 | – | – | + | |
V. cholerae = Vibrio cholerae. Results from stool spiking of various strains of V. cholerae for both Crystal VC and Crystal VC O1 rapid tests. Three separate stools were used for three independent assays, and stools were spiked with a single strain for each assay.
As shown in Table 2, the Crystal VC–O1 RDT yielded a positive result with both the Inaba and Ogawa strains when combined with different Vibrio strains (non-O1/non-O139, nontoxigenic O1, and O139).These results demonstrate ability of the tests to accurately identify V. cholerae O1 without cross-reactivity from other Vibrio species that may be present in the sample. Similarly, when evaluated for cross-reactivity, the Crystal VC RDT accurately showed a positive result for O1 for all stool aliquots containing V. cholerae O1 and accurately showed a positive result for O139 for all stool aliquots containing V. cholerae O139. No false-positive results were observed for either test.
Table 2.
Laboratory evaluation of cross-reactivity
| Vibrio cholerae serotype | Crystal VC O1 | Crystal VC | ||
|---|---|---|---|---|
| Strain 1 | Strain 2 | O1 line | O1 line | O139 line |
| O1 Inaba | O1 (nontoxigenic) | + | + | – |
| O1 Inaba | non-O1 and non-O139 | + | + | – |
| O1 Ogawa | O1 (nontoxigenic) | + | + | – |
| O1 Ogawa | non-O1 and non-O139 | + | + | – |
| O139 | O1 (nontoxigenic) | + | + | + |
| O139 | 2non-O1 and non-O139 | – | – | + |
Results from stool spiking of two strains of Vibrio cholerae to evaluate cross-reactivity of Crystal VC and Crystal VC O1 rapid tests.
Field evaluation.
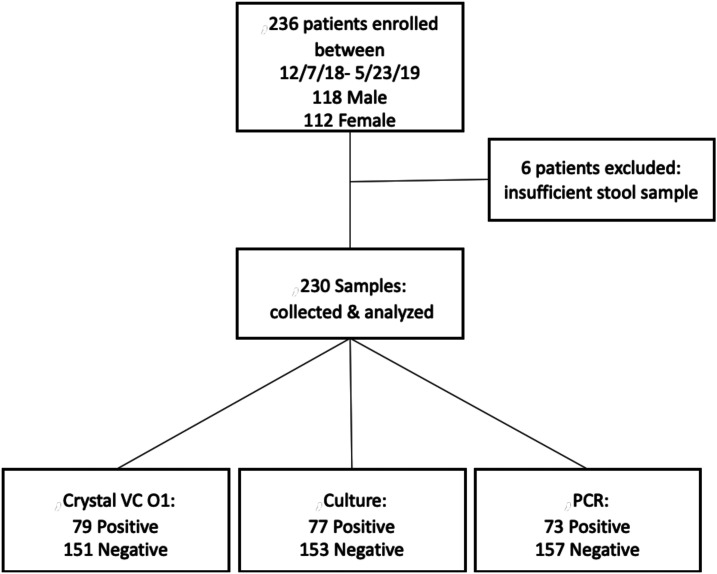
From December 2018 to May 2019, a total of 236 patients were enrolled, from which 230 had specimens suitable for inclusions in the study. The participants presented at health facilities in nine subcounties, representing five counties: 3-Mavoko (Machakos); 10-Dagoretti North, 17-Embakasi East, 6-Embakasi South, 93-Embakasi West, 5-Langata, (Nairobi); 11-Kaijado North (Kaijado); 36-Narok South (Narok); and 49-Kimini (Trans-Nzoia). There were 112 female and 118 male participants that ranged in age from infant to 64 years old, with a mean age of 25. The demographic and clinical characteristics of the participants in the study is shown in Table 3. A flow chart summarizing the results of the different tests with these specimens is shown on Figure 2.
Figure 2.
Study flow chart.
Of the 230 stool samples obtained for this analysis, 77 were positive for V. cholerae by culture methods, 79 were positive by RDT and 73 were positive by PCR. All culture-positive samples were also RDT positive. Two samples were RDT positive and PCR positive but culture negative. The comparison of the culture and PCR results with RDT, sensitivity, specificity, and positive and negative predictive values is shown in Table 4. As noted, the two specimens that were culture negative by culture but RDT positive were found positive by PCR, and thus considered positive. Two additional specimens were positive by PCR but negative by culture and RDT. Eight specimens were negative by PCR but positive by culture and RDT.
Table 4.
Sensitivities and specificities of Crystal VC O1 compared with culture or PCR in a field evaluation
| Culture or PCR positive | Culture or PCR negative | Culture positive | Culture negative | PCR positive | PCR negative | |
|---|---|---|---|---|---|---|
| RDT positive | 79 | 0 | 77 | 2 | 71 | 8 |
| RDT negative | 2 | 149 | 0 | 151 | 2 | 149 |
| Total | 81 | 149 | 77 | 153 | 79 | 151 |
| Sensitivity | 97.5% (91.4–99.7) | – | – | – | – | |
| Specificity | 100% (97.6–100) | – | – | – | – | |
| Positive predictive value | 100% (95.4–100) | – | – | – | – | |
| Negative predictive value | 98.7% (95.0–99.7) | – | – | – | – | |
The RDT was positive for 79 of 81 samples found positive by either test, as shown in Table 4. Applying this definition to our results, the sensitivity of the RDT test was 97.5%, and specificity was 100%, the positive predictive value was 100%, and the negative predictive value was 98.7%. There were no false-positive results using the RDT. The overall agreement between Crystal VC-O1 and culture was kappa = 0.9806, and between Crystal VC-O1 and PCR was Kappa = 0.9018.
DISCUSSION
Laboratory-based evaluations for sensitivity and cross-reactivity of Crystal VC O1 demonstrated high sensitivity and specificity. There was no evidence of interference to the RDT performance for either O1 Inaba or O1 Ogawa strains when evaluated in the presence of other Vibrio strains. Experiments evaluating different strains reproducibly confirmed that the RDT accurately yielded a positive result for O1 when tested with a nontoxigenic O1 strain and a negative result when tested with a non-O1/non-O139 strain. In addition, the Crystal VC test yielded a positive result for O139 for stools spiked with O139.
The current price of the kit is approximately $2 USD, similar to the original Crystal VC RDT. It is of similar ease of use as the original Crystal VC RDT, with comparisons to the use of a pregnancy test. The aspects of RDT use which remain critical include diluting the specimen in the sample buffer, ensuring the RDT is not submerged past the line of demarcation on the RDT and ensuring the test is read 15–30 minutes from the time it was started. The RDT kit includes all necessary supplies for the RDT test itself, but sample collection cups for the patient must be supplied. Furthermore, if testing after enrichment in alkaline peptone water (APW) is desired, the APW will be needed.27
The field evaluation corroborated the laboratory evaluation, demonstrating that Crystal VC-O1 yielded high sensitivity (%) and specificity when used to identify V. cholerae O1–positive patients presenting with watery diarrhea. When comparing the results of the three cholera detection methods used (RDT, culture, and PCR), most of the samples in the field evaluation were concordant. Among the few samples in which there were discordant results, two samples were positive by RDT and PCR but negative by culture. Because the samples were found to be positive by PCR, which is more sensitive, suggesting that the RDT result was correct and the culture was falsely negative. In addition, two samples were positive only by PCR but negative by culture and RDTs. Because PCR is more sensitive, it is likely that these samples contained a very low concentration of V. cholerae and/or that the participant had taken antibiotics before reporting to the health facility, causing the V. cholerae to be undetectable by the other two tests. Finally, there were eight samples that were positive by culture and negative by PCR; this was unexpected, given the high sensitivity of the PCR. Four of these samples came from a single cluster of cases. To further investigate these results, we will conduct additional studies to investigate this finding. This is likely due to an inadequate amount of stool placed on the filter paper. In all eight of these samples, the RDT and culture results were in agreement, so the RDT results were considered accurate in accordance with our definition of a true cholera case. Although additional studies are needed to confirm these results with this new RDT in different settings, our study team responded to outbreaks in different regions of Kenya and were able to use the RDT in the various field settings, maintaining its high performance. Using the manufacturers’ instructions, the Crystal VC-O1 RDT meets the minimum and the desired performance recommendations set forth by the GTFCC for cholera RDTs.28
Studies evaluating the original Crystal VC in comparison to culture and PCR report sensitivities in the range from 65.6 to 99% and specificities from 49.2% to 95.7%.5,6,9,12,29,30 This field evaluation found sensitivity and specificities at the higher end of this range for both sensitivity and, particularly, specificity. In most of the previous studies, direct specimen testing via RDTs shows higher sensitivity and lower specificity, whereas in our Crystal VC-O1 evaluation, we found extremely high sensitivity and specificity. In fact, the Crystal VC-O1 evaluation reported higher specificities than those in any of the previous studies evaluating Crystal VC. Given the historical lack of specificity with using Crystal VC directly, the performance metrics found in this evaluation may significantly impact cholera RDT use.
Patients with more severe disease tended to have a higher proportion of positive tests; 73.7% of severely dehydrated patients were RDT positive. However, some patients with moderate to severe dehydration reporting watery diarrhea were RDT and culture negative, suggesting that there are other pathogens able to cause moderate to severe diarrhea even during an outbreak. We did not search for other pathogens in this study, but a likely pathogen is enterotoxigenic Escherichia coli.
There were several limitations in this study. First, there were samples that were positive by culture but negative by PCR. It is suspected that insufficient amount of stool was spotted on the filter paper for preservation. The specimens were not enriched in APW and tested via RDTs because of preservation of the RDT for direct assessment. Such enrichment would have facilitated a full comparison of the RDTs performance metrics in comparison to those required by the Target Product Profile (TPP).28 In addition, we did not explore the 151 negative specimens to understand the pathogens presenting with similar clinical indications. Finally, this evaluation was conducted only within Kenya and for the results to be generalizable to other cholera prone countries, the evaluation must be reproduced in additional settings.
In conclusion, the new Crystal VC-O1 RDT, with a single line for serogroup O1, performed with high sensitivity and high specificity in this study. Importantly, we did not find any false-positive results. If this is confirmed in other studies, this suggests that an outbreak could be confirmed based on a positive Crystal VC-O1 RDT, particularly if specimens from multiple patients are positive. The impact that a cholera RDT with such high sensitivity and specificity can have on cholera surveillance and control is vast; not only would we have an improved understanding of the true cholera burden but also such an RDT would eliminate many of the laboratory challenges surrounding cholera confirmation, expedite outbreak detection and response, improving disease-monitoring methods in endemic and high-risk areas.
Supplemental tables
Acknowledgment:
We thank the Kenyan Ministry of Health for the assistance in conducting this study.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Debes AK, Ateudjieu J, Guenou E, Ebile W, Sonkoua IT, Njimbia AC, Steinwald P, Ram M, Sack DA, 2016. Clinical and environmental surveillance for Vibrio cholerae in resource constrained areas: application during a 1-year surveillance in the far north region of Cameroon. Am J Trop Med Hyg 94: 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legros D, 2018. Global cholera epidemiology: opportunities to reduce the burden of cholera by 2030. J Infect Dis 218: S137–S140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuiyan NA, Qadri F, Faruque AS, Malek MA, Salam MA, Nato F, Fournier JM, Chanteau S, Sack DA, Balakrish Nair G, 2003. Use of dipsticks for rapid diagnosis of cholera caused by Vibrio cholerae O1 and O139 from rectal swabs. J Clin Microbiol 41: 3939–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bwire G, Orach CG, Abdallah D, Debes AK, Kagirita A, Ram M, Sack DA, 2017. Alkaline peptone water enrichment with a dipstick test to quickly detect and monitor cholera outbreaks. BMC Infect Dis 17: 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris JR, et al. 2009. Field evaluation of crystal VC rapid dipstick test for cholera during a cholera outbreak in Guinea-Bissau. Trop Med Int Health 14: 1117–1121. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee P, Ghosh S, Ramamurthy T, Bhattacharya MK, Nandy RK, Takeda Y, Nair GB, Mukhopadhyay AK, 2010. Evaluation of a rapid immunochromatographic dipstick kit for diagnosis of cholera emphasizes its outbreak utility. Jpn J Infect Dis 63: 234–238. [PubMed] [Google Scholar]
- 7.Sinha A, et al. 2012. Evaluation of a rapid dipstick test for identifying cholera cases during the outbreak. Indian J Med Res 135: 523–528. [PMC free article] [PubMed] [Google Scholar]
- 8.Dick MH, Guillerm M, Moussy F, Chaignat CL, 2012. Review of two decades of cholera diagnostics--how far have we really come? PLoS Negl Trop Dis 6: e1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George CM, et al. 2014. Evaluation of enrichment method for the detection of Vibrio cholerae O1 using a rapid dipstick test in Bangladesh. Trop Med Int Health 19: 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denue BA, 2018. Evaluation of a rapid dipstick test (crystal Vc®) for the diagnosis of cholera in Maiduguri, northeastern Nigeria. Arch Med Health Sci 6: 24–27. [Google Scholar]
- 11.Ngwa MC, et al. 2020. The multi-sectorial emergency response to a cholera outbreak in internally displaced persons camps in Borno state, Nigeria, 2017. BMJ Glob Health 5: e002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley B, et al. 2012. Evaluation of a rapid dipstick (Crystal VC) for the diagnosis of cholera in Zanzibar and a comparison with previous studies. PLoS One 7: e36930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matias WR, Julceus FE, Abelard C, Mayo-Smith LM, Franke MF, Harris JB, Ivers LC, 2017. Laboratory evaluation of immunochromatographic rapid diagnostic tests for cholera in Haiti. PLoS One 12: e0186710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ontweka LN, et al. 2016. Cholera rapid test with enrichment step has diagnostic performance equivalent to culture. PLoS One 11: e0168257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page AL, Alberti KP, Mondonge V, Rauzier J, Quilici ML, Guerin PJ, 2012. Evaluation of a rapid test for the diagnosis of cholera in the absence of a gold standard. PLoS One 7: e37360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha A, et al. 2013. Culture-independent real-time PCR reveals extensive polymicrobial infections in hospitalized diarrhoea cases in Kolkata, India. Clin Microbiol Infect 19: 173–180. [DOI] [PubMed] [Google Scholar]
- 17.CDC , 2020. Cholera–Vibrio cholerae Infection. Available at: https://www.cdc.gov/cholera/diagnosis.html. Accessed September 4, 2020. [Google Scholar]
- 18.Zhang P, Zhou H, Diao B, Li F, Du P, Li J, Kan B, Morris JG, Jr., Wang D, 2014. A molecular surveillance reveals the prevalence of Vibrio cholerae O139 isolates in China from 1993 to 2012. J Clin Microbiol 52: 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdhury F, et al. 2015. Vibrio cholerae serogroup O139: isolation from cholera patients and asymptomatic household family members in Bangladesh between 2013 and 2014. PLoS Negl Trop Dis 9: e0004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization , 2018. Weekly epidemiological record. Cholera, 2017: 489–500. [Google Scholar]
- 21.Mutonga D, Langat D, Mwangi D, Tonui J, Njeru M, Abade A, Irura Z, Njeru I, Dahlke M, 2013. National surveillance data on the epidemiology of cholera in Kenya, 1997–2010. J Infect Dis 208: S55–S61. [DOI] [PubMed] [Google Scholar]
- 22.George G, et al. 2016. Notes from the field: ongoing cholera outbreak - Kenya, 2014-2016. MMWR Morb Mortal Wkly Rep 65: 68–69. [DOI] [PubMed] [Google Scholar]
- 23.Talaat KR, et al. 2020. Oral delivery of Hyperimmune bovine serum antibodies against CS6-expressing enterotoxigenic Escherichia coli as a prophylactic against diarrhea. Gut Microbes 12: 1732852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arkray_Diagnostics , 2020. Crystal VC Test Instructions. Available at: http://www.arkray.co.in/english/upload/docs/Crystal%20VC.pdf. Accessed September 4, 2020. [Google Scholar]
- 25.Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC, 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol 38: 4145–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CDC , 2020. Isolation of Vibrio cholerae from fecal specimens. Available at: https://www.cdc.gov/cholera/laboratory.html. Accessed September 4, 2020. [Google Scholar]
- 27.Debes AK, et al. 2016. Evaluation in Cameroon of a novel, simplified methodology to assist molecular microbiological analysis of V. cholerae in resource-limited settings. PLoS Negl Trop Dis 10: e0004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Global Task Force on Cholera Control , 2017. Target Product Profile (TPP) for the Development of Improved Cholera Rapid Diagnostic Tests. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 29.Sayeed MA, et al. 2018. Development of a new dipstick (Cholkit) for rapid detection of Vibrio cholerae O1 in acute watery diarrheal stools. PLoS Negl Trop Dis 12: e0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Islam MT, et al. 2019. Field evaluation of a locally produced rapid diagnostic test for early detection of cholera in Bangladesh. PLoS Negl Trop Dis 13: e0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.