Abstract
Single-cell sequencing approaches offer exploration of tissue architecture at unprecedented resolution. These tools are especially powerful when deconvoluting highly specialized microenvironments, such as stem cell (SC) niches. Here, we review single-cell studies that map the cellular and transcriptional makeup of stem and progenitor niches and discuss how these high-resolution analyses fundamentally advance our understanding of how niche factors shape SC biology and activity. In-depth characterization of the blueprint of SC-niche crosstalk, as well as understanding how it becomes dysregulated, will undoubtedly inform the development of more efficient therapies for malignancies and other pathologies.
Every process in our body is mediated by highly orchestrated cellular cross-talk. Elucidating how cellular dialog is established, maintained, and altered by stress represents a major challenge in modern biology. Most mammalian tissues contain rare adult stem cells (SCs) that support organ homeostasis and regeneration. SC function is dependent on unique molecular cues and, as such, SCs are maintained in cellular milieus, termed SC niches. A variety of cell types serve as building blocks of these specialized compartments and are tailored to regulate SC quiescence, self-renewal, survival, and differentiation.
The concept of the SC niche was proposed in the late 1970s (Schofield, 1978). Current understanding of SC niche composition is drawn from studies utilizing elegant imaging approaches (Voog and Jones, 2010) and genetic tools (Crane et al., 2017; Hanoun and Frenette, 2013). More recently, with the advancement of single-cell genomics, simultaneous profiling of thousands of individual cells has become widely accessible and applied to a variety of fields (Kolodziejczyk et al., 2015). Resulting datasets afford a high-resolution glimpse into cellular heterogeneity, molecular topology, and functional plasticity of the tissues. Discerning the cellular identity of niche components and their specific contribution to SC maintenance is central to unravelling the mechanisms governing tissue development, homeostasis, and regeneration. Furthermore, web resources facilitate sharing and exploration of these datasets by the scientific community, taking the acquired knowledge beyond the confinement of published manuscripts (Tanay and Regev, 2017). Here, we review key studies utilizing single-cell transcriptomics to elucidate the molecular architecture of bone, thymus, gut, lung, and central nervous system SC and progenitor niches in mice and men (Figure 1). Specifically, we discuss emerging insights into epithelial, mesenchymal, and immune components of SC microenvironments and their functional plasticity. We place single-cell studies in the context of previous research and discuss unresolved questions and future directions in the field.
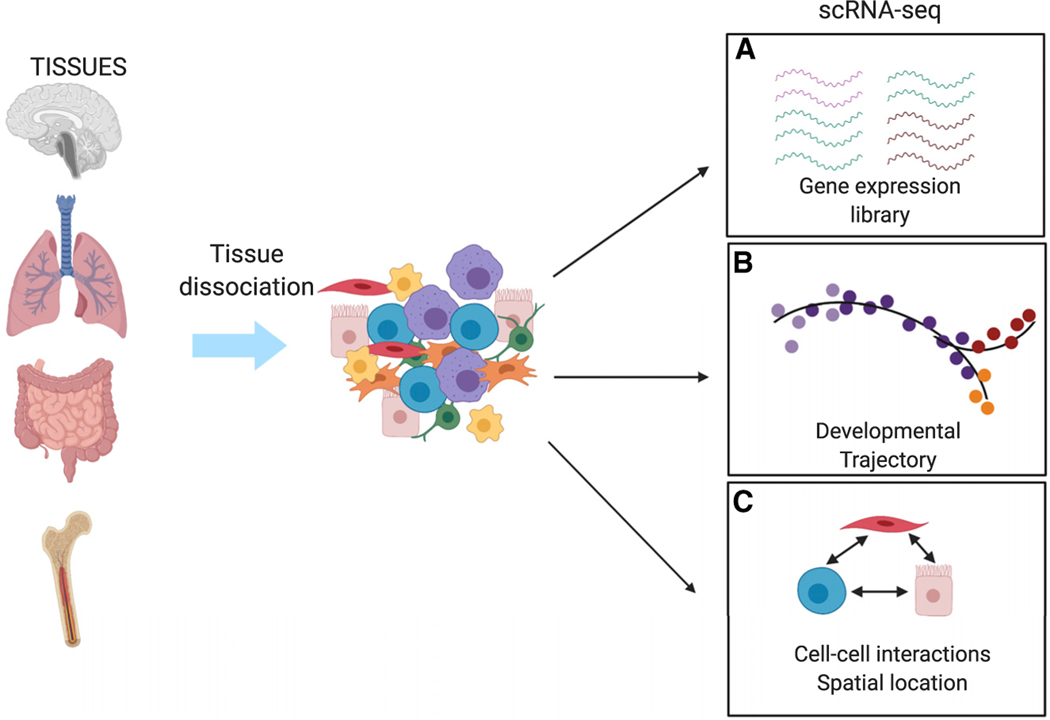
Figure 1. Single Cell Methodologies.
For single-cell analysis, tissues are dissociated into single cells via mechanical digestion, enzymatic digestion, or a combination of both. Various pipelines allow for multidimentional analysis of single cell transcriptomes.
(A) Transcripts originating from single cells are used to generate single-cell gene-expression libraries.
(B) Reconstruction of developmental trajectory infers the relationship between progenitors and differentiated cells.
(C) Transcripts from single cells within a tissue can be used to infer receptor-ligand interactions, leading to discoveries of novel interactions between different cell types.
Hematopoietic Niches
Bone Marrow
Every second, our body produces millions of blood and immune cells. This process, termed hematopoiesis, is vital to oxygen transport, immune defense, and tissue remodeling. The hematopoietic system can adapt rapidly and precisely to varying perturbations such as inflammation and cytopenia. Hematopoietic dysregulation has devastating consequences, including anemia, chronic infections, and tumorigenesis. In mammals, continuous production of the different blood lineages is sustained by a rare population of self-renewing hematopoietic stem cells (HSCs) (Crane et al., 2017; Morrison and Scadden, 2014; Wei and Frenette, 2018). HSCs arise during embryonic development and initiate hematopoiesis in specific fetal niches before relocating to the bone postnatally (Medvinsky et al., 2011). The bone marrow (BM) microenvironment supports differentiation of all hematopoietic lineages with the notable exception of T-lymphocytes that differentiate in the thymus (Abramson and Anderson, 2017). To generate a broad repertoire of hematopoietic lineages, HSCs give rise to an array of oligopotent progenitors, which extensively proliferate and progressively differentiate into mature cells.
A variety of BM cell populations, including endothelial cells (ECs) and mesenchymal stromal cells, support the generation of blood cells. Discovery of the specificity of SLAM family markers to identify HSCs led to the finding that HSCs reside near sinusoidal vascular cells, suggesting that HSCs might be maintained in a perivascular niche, comprised of endothelial and perivascular cells (Kiel et al., 2005). Vascular endothelial and perivascular cells are the source of key niche factors, including SDF-1 (Cxcl12) and SCF (Kitl), required for HSC maintenance. The Cxcl12-Cxcr4 axis serves as a molecular anchor for HSC retention in the BM and plays a critical role in maintaining HSC quiescence (Ding and Morrison, 2013; Greenbaum et al., 2013). Conditional deletion of these factors in the vascular or perivascular compartment results in HSC depletion or mobilization (Ding et al., 2012).
Endothelial niches.
BM endothelial cells (BMECs) form an extensive network of blood vessels and exhibit diverse morphology, potentially representing functionally distinct niches. Branched networks of sinusoidal capillaries (sECs) constitute the majority of BMECs, whereas BM arteriole vessels (aECs) contain comparably few side branches and are longitudinally aligned along the diaphysis. A series of studies based on cell-surface-marker expression and functional characteristics identified two subtypes of BM sECs. CD31highEMCNhigh H-vessels, found in the metaphysis, are organized as vessel columns, whereas CD31lowEMCNlow L-vessels account for the majority of the sECs and are arranged into highly branched capillary networks throughout the BM cavity. These findings highlight heterogeneity within sECs and, given distinct hypoxia status, the presence of distinct metabolic environments in the bone (Kusumbe et al., 2014).
While the important role of vascular cells in HSC maintenance is broadly accepted, the degree of transcriptional heterogeneity of the BM vascular compartment has been hard to define. Furthermore, the preferential localization of quiescent HSCs in relation to sinusoidal versus arteriolar spaces at steady state and after stress remains controversial (Acar et al., 2015; Itkin et al., 2016; Kunisaki et al., 2013). scRNA-seq analysis of BMECs identified two major, transcriptionally distinct subpopulations: Ly6ahigh arteriolar and Stab2high sinusoidal cells. Baryawno et al. identified an additional endosteal BMEC population characterized by Vwf expression (Baccin et al., 2020; Baryawno et al., 2019; Tikhonova et al., 2019).
scRNA-seq analysis confirmed that all BMECs express broad endothelial markers, such as Pecam1, Cdh5, Kdr, Emcn, and Tek. sECs express high levels of Icam1, which supports HSC quiescence (Liu et al., 2018), and angiogenin (Ang), reported to promote stemness in hematopoietic stem progenitor cells (HSPCs) while stimulating proliferation of myeloid progenitors (Goncalves et al., 2016). aECs, in contrast, express lower levels of Icam1 and have upregulated expression of Icam2, suggesting diverse adhesion roles for these compartments. Within BMECs, the arteriolar cluster is enriched in Kitl and Cxcl12 expression. Kitl expressed by the aECs, but not the sEC compartment, resulted in significant reduction in percentage and absolute numbers of long-term HSCs. After 5-FU-mediated myeloablation, HSCs from animals with aEC-specific deletion of SCF had impaired recovery and hematopoietic reconstitution capacity, indicating that aEC-derived SCF significantly contributes to HSC maintenance at steady state and regeneration after myeloablation (Yue et al., 2016). On the other hand, tracking quiescent HSCs in live animals revealed their preferential proximity to both sECs and the endosteal surface (Christodoulou et al., 2020). Therefore, the exact identity of BMECs that maintain quiescent HSCs is a matter of ongoing research.
While the spatial distribution of HSCs in relation to vascular subsets continues to be debated and cannot be resolved with transcriptomics alone, scRNA-seq analysis has significantly improved our understanding of the transcriptional landscape of BMECs. scRNA-seq analysis has revealed two major, transcriptionally distinct BMEC subpopulations: arterioles and sinusoids. Notably, scRNA-seq data are a snapshot of a point in time and likely do not reflect the full extent of BMEC complexity. Future studies throughout different developmental stages and conditions, coupled, for example, with mass cytometry (CyTOF)based single-cell protein analysis (Severe et al., 2019) and spatial transcriptomic approaches, will further inform our understanding of the BM vascular architecture.
Mesenchymal niches.
HSC localization in proximity of BMECs directed the field’s interest to the perivascular cells that wrap around blood vessels throughout the BM. BM perivascular cells contain mesenchymal stem and progenitor cells (MSPCs) that have the potential to differentiate into osteoblasts, adipocytes, and chondrocytes (Kfoury and Scadden, 2015). Considering their multi-lineage differentiation potential, the phenotypic markers of BM mesenchymal stem cells (MSCs) have been of great interest. Earlier studies proposed a BM Pdgfra+Sca-1+ population to contain MSCs (Houlihan et al., 2012). Subsequent reports, however, revealed functional heterogeneity within Pdgfra+Sca-1+ cells. Taking advantage of transgenic labeling of Leptin-receptor-expressing (Lepr+) mesenchymal populations, enriched for MSC activity (Yue et al., 2016; Zhou et al., 2014), our group identified two transcriptionally similar but distinct sub-populations of quiescent MSPC: adipogenesisand osteogenesis-primed cells. While Lepr-Cre-driven lineage tracing labels both progenitor groups, adipo-primed cells express significantly higher levels of Lepr transcript compared to the osteo-primed subset. Fate mapping indicates that osteo-primed cells are the progeny of the Leprhigh adipo-primed population. Immunofluorescence revealed that adipo-primed cells co-localize with sECs in the bone metaphysis, whereas osteo-primed cells reside adjacent to aECs. Coupling laser capture microdissection of fixed BM sections with bulkRNA-seq, and then integrating resulting signatures with BM scRNA-seq, Baccin et al. independently confirmed the spatial distribution of mesenchymal populations and additionally highlighted the presence of osteo-primed cells in nonvascular spaces (Baccin et al., 2020). By applying pseudotime ordering tools to BM stroma single-cell data, Wolock et al. inferred the gene-expression trajectories of mesenchymal progenitors as they differentiate (Wolock, 2019). RNAVelocity identified Foxc1+Cbln1+Clec2d+ cells that also express Adipoq as the strongest “source” cell state, and pre-adipocytes, pro-osteoblasts, and pro-chondrocytes probably serve as end state. Importantly, functional assessment of in vitro stem cell capacity indicated that adipo-primed MSPCs are significantly enriched in fibroblastic colony-forming units (CFU-F) and account for the CFU-F activity of the total perivascular population. Finally, adipo-primed MSPCs strongly correlate with the gene signature of human BM MSCs (CD45CD271+) (Ghazanfari et al., 2016), further suggesting that early mesenchymal progenitors reside within the adipo-primed mesenchymal subset (Tikhonova et al., 2019).
scRNA-seq analysis of BM vascular, perivascular, and osteo compartments revealed adipo-primed Lepr+ MSPC to be a major source of pro-hematopoietic factors in the BM. In addition to Cxcl12 (Ding and Morrison, 2013), Kitl (Ding and Morrison, 2013), and ll7 (Cordeiro Gomes et al., 2016), Lepr+ adipo-primed cells expressed ll15, ll34, Csf1, Bmp4, Ccl19, and Ccl2. To gain a broader understanding of BM stroma, Baryawno et al. profiled all non-hematopoietic (Ter119−/CD71−/Lin−) cells in the BM, identifying previously unappreciated subsets of fibroblasts, which also constitute a considerable reservoir of different pro-hematopoietic factors (Baryawno et al., 2019). However, the precise location and function of BM fibroblasts in the context of HSPC homeostasis or disease remain to be defined.
Functional plasticity of BM microenvironment.
During steadystate hematopoiesis, blood and immune cells are replaced in a relatively constant manner. After stress such as infection, inflammation, chemotherapy, or blood loss, LT-HSCs start rapidly dividing and sharply skew their differentiation toward the myeloid compartment, a process termed emergency hematopoiesis. This augmented myeloid output provides rapid protection against the threat of pathogens, blood loss, and tissue damage. Importantly, the emergency hematopoietic program is transient, and HSCs return to their quiescent state. The resolution of this response is essential to limit HSC exhaustion, restore the production of the diverse blood lineages, and limit immunopathologies (Pietras, 2017; Pietras et al., 2016; Schuettpelz et al., 2014). Chemotherapy induces widespread cellular stress throughout the BM, ablating cycling hematopoietic progenitor cells and driving HSC differentiation (Mauch et al., 1995). Despite the mostly quiescent status of BM stroma, myeloablation affects vasculature and perivascular subsets and causes a dramatic increase in frequencies of BM adipocytes. After myeloablation, adipo-primed Lepr+ cells sharply upregulate adipogenic signature genes, while osteo-primed perivascular progenitors downregulate osteogenesis-associated genes (Tikhonova et al., 2019). These findings highlight the transcriptional plasticity of MSPCs in response to stress and are consistent with the previous work reporting expansion of BM adipocytes after irradiation or myeloblation (Zhou et al., 2017).
The role of the microenvironment in the onset and development of hematopoietic malignancies has been gaining attention, due to the potential for therapeutic interventions to mitigate the development of a chemo-resistant leukemic niche. In a murine model of acute myeloid leukemia (AML), Baryawno et al. identified significant shifts in both the cellular composition and the transcriptional landscape of BM stroma. Leukemic development triggers impaired osteogenesis and vascular remodeling, also reported in AML patients. Key HSPC factors such as Cxcl12, Kitl, ll7, Csf1, and Vcam1 were downregulated in MSPCs, which is consistent with the idea that AML clones remodel their environment to disadvantage normal hematopoiesis (Baryawno et al., 2019). Future functional studies will explore the safety and feasibility of targeting the leukemic microenvironment to halt AML progression and restore hematopoiesis.
Single-cell analyses have systematically mapped the transcriptional landscape of the BM microenvironment, identifying novel cellular fractions and resolving cellular sources of pro-hematopoietic growth factors, chemokines, and membrane-bound ligands. These efforts illuminate our understanding of the BM niche (Figure 2) and offer novel lines of inquiry. The proteomic architecture of BM vasculature, spaciotemporal analysis of HSCs and HSPCs (Christodoulou et al., 2020), and precise cellular identity of BM MSCs remain areas of active investigation.
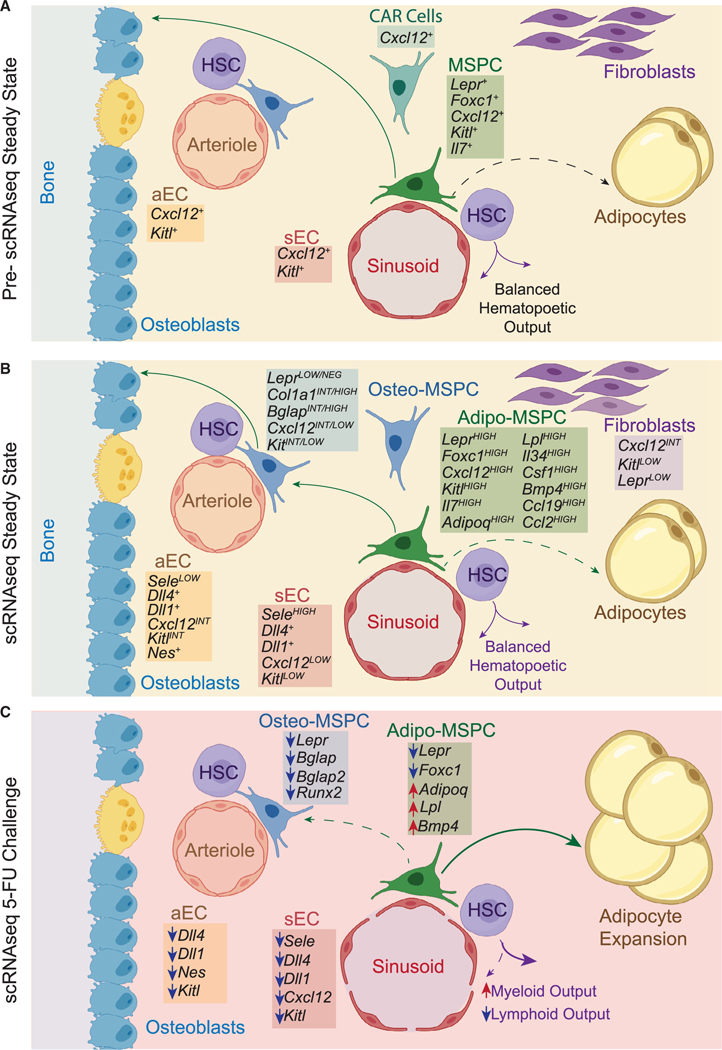
Figure 2. Bone Marrow Niches.
(A) Prior to scRNA-seq studies, BM vascular and perivascular Lepr+ cells were identified as the key populations constituting the HSC niche. The degree of the heterogeneity within those cellular subsets was unknown.
(B) scRNA-seq of BM vascular endothelial and perivascular Lepr+ cells resolved cellular composition of these populations and defined expression patterns of pro-hematopoietic growth factors, chemokines, and membrane-bound ligands.
(C) Changes in gene expressionin BM vascular endothelial and perivascula Lepr+ cells following acute chemotherapy treatment.
Thymus
The cardinal feature of adaptive immunity is the ability to discriminate between self and non-self antigens. Lymphoid progenitors originate in the BM and migrate to the thymus, where T cell development takes place. To prevent autoimmunity, T-lymphocytes are selected based on their ability to recognize self-antigens with low affinity. Thymic selection is marked by two consecutive stages: positive selection for thymocytes with functional TCRs that takes place in the thymic cortex (Klein et al., 2009) and negative selection of self-reactive T cells in the thymic medulla (Kappler et al., 1987; Kisielow et al., 1988). A variety of antigen-presenting cells (APCs) – cortical thymic epithelial cells (cTECs), medullary thymic epithelial cells (mTECs), and dendritic cells (DCs) – play critical roles in thymic selection (Cheng and Anderson, 2018), shaping the T cell repertoire. Thymic APCs have unique properties in the expression, processing, and presentation of self-antigens specialized for thymic selection (Bornstein et al., 2018; Miller et al., 2018).
cTECs express unique machineries for protein degradation, contributing to self-antigen presentation and positive selection of thymocytes (Kondo et al., 2017). mTECs mediate central tolerance by presenting a broad range of tissue-restricted self-antigens, a process termed promiscuous gene expression (PGE). Autoimmune regulator (AIRE) is a transcription factor that promotes the expression of tissue-restricted antigens (TRAs) in the thymic medulla (Anderson et al., 2005). Autoreactive T cells that recognize TSAs with high affinity undergo negative selection, preventing the release of autoreactive T cells into the periphery. Individuals harboring single AIRE point mutations on only one allele have increased susceptibility to autoimmune diseases (Anderson and Su, 2016).
Epithelial heterogeneity.
scRNA-seq studies substantially refined our understanding of the thymic microenvironment. Kernfeld et al. analyzed 3,341 epithelial cells isolated from murine thymi during embryonic organogenesis at different time points. This analysis revealed three medullary and four cortical subsets. Whereas mTEC populations were transcriptionally distinct, cTEC subsets fell along a transcriptional continuum. Relative frequencies of TEC populations, as well as their transcriptional landscape, varied considerably over the course of development. Later embryonic stages, for example, were marked by upregulation of MHC-II genes (H2aa, Cd74) in all TEC subsets. This study captured heterogeneity and dynamic changes in the transcriptional architecture of the epithelial compartment over the course of thymic development (Kernfeld et al., 2018).
To resolve TEC heterogeneity post-development, Bornstein et al. examined 2,341 epithelial cells sorted from 4–8-weekold murine thymi. They identified a single cortical and four medullary epithelial subsets (mTEC I-IV). mTEC populations displayed distinct transcriptional characteristics and various levels of PGE. mTEC I cells expressed Itga6 and Ly6a, associated with putative TEC progenitors. AireHIGH mTEC II cells were characterized by canonical mTEC markers and expressed the highest number of variable and uncorrelated genes, which are associated with PGE, among medullary epithelial subsets. High levels of PGE were also detected in AireLOW mTEC III cells. Notably, Aire-deficient mTEC II and III subsets had virtually no “stochastic gene expression,” confirming the key role of Aire in PGE. mTEC IV cells, despite shared lineage with mTEC II and III cells, lacked expression of canonical medullary markers as well as “stochastic gene expression,” suggesting functions outside of negative selection. Surprisingly, mTEC IV cells express genes associated with gut chemosensory tuft-cells, such as Sox9, Trpm5, Pou2f3, and cytokine, ll25, typically associated with epithelial barrier tissues (Bornstein et al., 2018).
A study by Miller et al., employing an inducible Aire-lineage tracing model, also found tuft-like mTECs in adult mice. By using a combination of low-input bulk RNA-seq and single-cell approaches, the authors established that 18 out of 20 tuft-specific markers are shared between gut-resident tuft cells and tuft-like mTECs. Functional studies from both groups revealed that animals deficient in Pou2f3, a master regulator of tuft cells, had a complete loss of tuft-like mTEC cells. Pou2f3 knock-out studies revealed that tuft-like mTECs establish tolerance to antigens they express, control the pool-size of thymic ILC2s, and contribute to development and intra-thymic function of type 2 invariant natural killer T (NKT2) cells, unconventional “innate-like” T lymphocytes implicated in regulating immunity to a diverse range of infections (Miller et al., 2018). Although thymic cells bearing tuft-cell-like morphology and expressing canonical taste transduction genes have been reported prior to these studies (Panneck et al., 2014; Soultanova et al., 2015), single-cell approaches have enabled characterization of the previously unappreciated transcriptional and functional features of this intriguing subset (Figure 3).
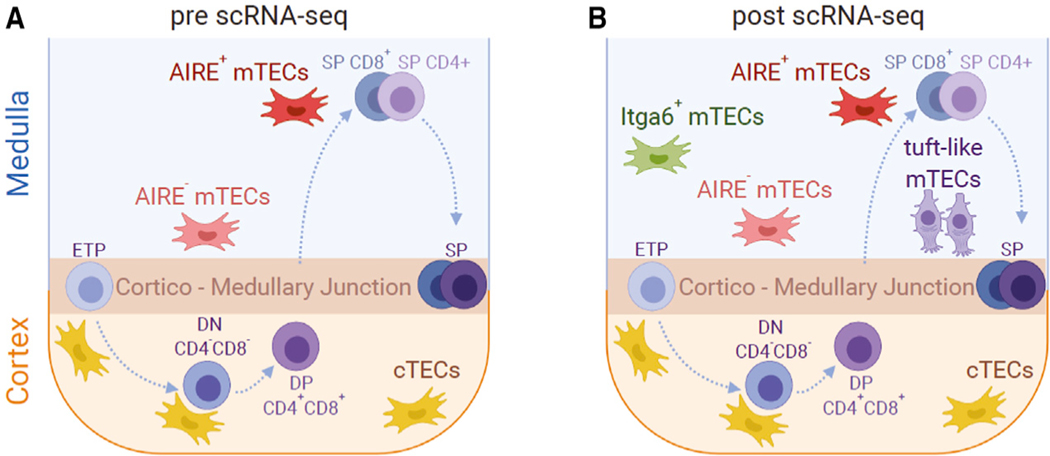
Figure 3. Thymic Niches.
(A) The major role of the cTEC and mTEC populations inT-cell development has been long established. However, the degree of cellular diversity within those populations was not well defined.
(B) scRNA-seq revealed significant molecular and epigenetic heterogeneity within the mTEC compartment, highlighting an mTEC lineage with molecular characteristics of gut chemosensory epithelial tuft cells (tuft-like mTECs).
Our understanding of T cell selection is shaped largely by studies in animal models. Adaptive responses in humans are significantly more complex and command in-depth understanding of the human thymus. In a tour-de-force work, Park et al. performed scRNA-seq on 255,901 cells from developing, pediatric, and adult thymi. This extensive dataset includes differentiating T cells, resident immune subsets (B cells, natural killer [NK] cells, innate lymphoid cells [ILCs], macrophages, monocytes, and DCs), as well as epithelial, vascular smooth muscle, endothelial, lymphatic, ECs, and fibroblasts. Notably, scRNA-seq identified previously uncharacterized thymic fibroblasts that were further split into two subtypes, F1 and F2. Single-molecule fluorescence in situ hybridization (smFISH) spatially placed these subsets in perilobular (F1) and interlobular (F2) areas. Human TEC populations were annotated and compared to their mouse counterparts (Bornstein et al., 2018), revealing cTECs and mTEC I-III cells to be largely conserved across these species. Low frequencies of tuft-like mTEC IV cells were also detected in human thymi, but in contrast to mice, POU2F3 expression was not specific to human tuft-like mTECs. In addition to the thymic microenvironment, Park et al. characterized T cell developmental trajectories, combined with TCR repertoire analysis. We encourage the readers to independently explore these fascinating datasets (Park et al., 2020).
Collectively, these studies conducted in-depth analyses of the complex thymic compartments and uncovered novel cellular layers in the organization of the thymic niche. The effects of malignant transformation and infection have not been examined so far with scRNA-seq. These will be critical areas of focus as the field moves forward, shedding light on the thymic niche under conditions of stress.
Barrier Tissues
Barrier tissues in the mammalian body – intestine, lungs, skin, and liver – represent the organism’s first line of defense against environmental insults. These specialized milieus are composed of myriad cell types and are host to commensal microbiota. Highly orchestrated interplay between tissue resident populations maintains homeostasis, shapes responses to sudden changes in the environment, and repairs tissue damage. In this section, we will discuss advances in understanding the intestinal and lung SC niches, brought to light by single-cell sequencing studies, and review topics where our current understanding is still lacking.
Intestine
Unlike the BM and thymic niches, where elegant experimental work is complemented by in-depth characterization by scRNA mapping of stromal compartments, current knowledge of the intestinal SC (ISC) microenvironment is based mainly on mouse models and ex vivo organoid cultures, and, to date, understanding of the complex heterogeneity of the intestinal niche is in its initial stages. The intestinal tract consists of epithelial, immune, mesenchymal, and vascular cells, and neurons. Although the epithelial and mesenchymal components of the intestinal niche have largely been described, and the immune component of the niche has been brought to light, the molecular function of other important subpopulations is yet to be elucidated.
The intestinal epithelium consists of enterocytes, Paneth, goblet, tuft, and neuroendocrine cells that are constantly generated by highly-proliferative Lgr5+ ISCs (Sato et al., 2009). Paneth cells act as an epithelial niche, supporting Lgr5+ cell proliferation (Sato et al., 2011). In contrast to the clear hierarchy of the epithelium, contribution of other cells in the intestinal tract to epithelial maintenance is largely unknown. Mesenchymal cells have been shown to play an important role in maintaining intestinal integrity and contribute to epithelial differentiation (Valenta et al., 2016). However, the heterogeneity of the intestinal mesenchymal compartment has complicated identification of specific populations contributing to ISC niches.
Mesenchymal niches.
The intestinal epithelium is organized in crypt-villus units, whereby epithelial cells migrate from the base of the crypt and along the villus axis as they differentiate. This relies on opposing gradients of Wnt and BMP signaling (Gehart and Clevers, 2019). The Wnt pathway is active in the base of the crypt and required for stem-cell maintenance, whereas the BMP pathway is active in the villi and mediates progenitor differentiation. In the small intestine, Paneth cells express Wnt ligand Wnt3a, Notch ligands, and EGF required for ISC maintenance and proliferation (Sato et al., 2011). In addition to Paneth cells, mesenchymal cells line the intestinal epithelium and have been shown to express key factors required for ISC maintenance. Mesenchymal cells are thought to be the main source of Wnt ligands in the colon, where there are no Paneth cells, and are important for maintaining epithelial architecture also in the small intestine. In the small intestine, Gli1+ mesenchymal cells residing in the crypt area produce Wnt2b (Valenta et al., 2016), and blockade of Wnt secretion by these cells leads to loss of crypt architecture. Furthermore, in mice with impaired epithelial Wnt secretion, the Gli1+ mesenchymal compartment is enlarged, suggesting these cells compensate for a dysfunctional epithelial niche in the small intestine. In the colon, where Wnt is not produced by epithelial cells, Wnt production by Gli1+ mesenchymal cells is essential for maintaining colonic crypts. scRNA-seq of colonic Gli1+ cells revealed the presence of eight clusters, representing eight transcriptionally different sub-populations, two of which express Wnt2b and other Wnt ligands. In parallel to these studies, a subset of mesenchymal cells, Foxl1+ telocytes, lining the entire length of the intestinal epithelium from small intestine to colon, were also shown to express Wnt ligands Wnt2b and Wnt5a. Blockade of Wnt secretion in these cells lead to loss of epithelial structures throughout the intestine (Shoshkes-Carmel et al., 2018). scRNA-seq of mesenchymal cells also demonstrated overlap between Gli1 and Foxl1 mRNA expression (Degirmenci et al., 2018), indicating that they mark the same cell population. Indeed, Madison et al. have shown that Gli proteins bind the Foxl1 promoter in the intestine (Madison et al., 2009), providing a mechanistic link between these two markers. While parts of the murine colonic niche have been elucidated at high resolution by single-cell transcriptomics, a full census of the colonic and small-intestinal niche is still lacking.
The role of the mesenchymal niche in maintenance of the colonic epithelium appears to be conserved in humans. A scRNA-seq census of human mesenchymal cells has revealed a subpopulation expressing non-canonical Wnt ligands, WNT5A and WNT5B, as well as BMP2 and BMP5, residing in close proximity to the epithelium (Kinchen et al., 2018). However, it is not clear if Wnt-secreting cells in humans are the same Gli1+/ Foxl1+ cells identified in murine models. Importantly, Kinchen et al. also characterized unique markers for each mesenchymal subpopulation, providing important tools for further isolation and characterization of the human mesenchymal niche (Kinchen et al., 2018). Functional validation of these markers could help resolve the degree of conservation between the human and murine niche.
Immune niches.
Cytokines produced by T cells are another important component of the intestinal niche. Surprisingly, scRNA-seq analysis revealed that a subset of Lgr5+ ISCs in the small intestine express the MHC class-II antigen presentation machinery, previously thought to be exclusive to antigen-presenting cells in the immune system. Cytokines secreted from different T helper (Th) subsets influence ISC trajectories, and MHC-II-bearing ISCs play an important role in resolving infection (Biton et al., 2018). Cytokines secreted by T cells influence differentiation of epithelial cells in response to injury; while acute bacterial infection induces Th1 cytokines and leads to an increase in enterocyte and Paneth cell frequency, helminth infections induce Th2 cytokines, leading to an increase in goblet and tuft cell frequency. These in vivo observations were supported by elegant organoid experiments, where addition of Th1 or Th2 cytokines skewed organoid differentiation toward enterocytes or goblet and tuft cells, respectively. This is in line with the established role of these cells in response to bacterial or helminth infection: Paneth cells produce anti-microbial peptides (Bevins and Salzman, 2011), whereas tuft cells secrete the epithelial cytokine IL-25 and enhance Th2 immune responses (von Moltke et al., 2016). Furthermore, interactions with regulatory T cells (Tregs) control ISC numbers, and depletion of Tregs led to expansion of proliferating Lgr5+ ISCs. Furthermore, complete T cell depletion, or deletion of MHC-II in ISCs, leads to an increase in ISC numbers (Biton et al., 2018). Thus, immune cells constitute an important part of the small-intestinal niche (Figure 4), and it would be interesting to examine their role in other barrier tissue niches. It is tempting to speculate that immune cells play a similar role in regulating the colonic niche, which is more susceptible to inflammatory bowel disease (IBD) and where exposure to commensal bacteria is more prevalent.
Figure 4. Intestinal Niches.
(A) Before the advent of scRNA-Seq, knowledge of the intestinal niche was limited. In the intestine, Lgr5+ISCs lie adjacent to Paneth cells that secrete Wnt and EGF ligands that are crucial for ISC maintenance. In addition, Wnt ligands are secreted by the mesenchymal compartment, but the cellular composition of this compartment was unknown.
(B) Single-cell analysis of the intestinal epithelium has shed light on many components of the niche. In the small intestine, a subset of Lgr5+ ISCs express MHC-II and interact with Tregs that control the ISC pool.
(C) During helminth infection, epithelial cell populations change, giving rise to an increase in goblet and tuft cells. MHCII-expressing ISC subsets increase innumber and mediate these changes via interaction with TH2 cells.
(D) Prior to scRNA-seq studies, the mesenchymal niche in the colon was thought to be the major source of Wnt ligands, but its composition was largely unknown.(E) In the mesenchymal compartment, eight subpopulations of mesenchymal cells (denoted by different colors) were identified by scRNA-Seq, which also highlighted the overlap between Foxl1+ and Gli1+ cells that secrete Wnt ligands.
(F) Inflammatory bowel disease (IBD) remodels the intestinal epithelium. scRNA-Seq identified expansion of inflammatory mesenchymal cells in IBD patients and loss of Wnt-secreting mesenchymal cells, as well as expansion of microfold cells.
Functional plasticity.
The intestinal epithelium has remarkable plasticity and can regenerate following acute injury, such as irradiation or bacterial infection. In mice, damage to the intestinal epithelium leads to remodeling of the mesenchymal niche, with expansion of an inflammatory subset of mesenchymal cells. The mesenchymal niche is also altered in inflammatory bowel disease (IBD) patients, with emergence of mesenchymal cells expressing MHC-II genes, chemokines CCL19 and CCL21, and proinflammatory cytokine ll33, coinciding with depletion of Wnt-expressing niche cells (Kinchen et al., 2018). This depletion might account for the structural remodeling of the intestinal epithelium in IBD, and indeed, in murine models, depletion of Wnt signals from mesenchymal cells leads to loss of epithelial structure (Degirmenci et al., 2018; Shoshkes-Carmel et al., 2018). Considering the role of T cell-derived cytokines in dictating intestinal differentiation, it is possible that this mesenchymal cluster acts as an inflammatory niche in IBD. scRNA-seq of colonic epithelial samples from IBD patients has revealed epithelial composition changes, including depletion of pHsensing cells, enrichment of microfold cells (specialized cells with a role in antigen sampling), and changes in Wnt regulatory factors in ISCs (Parikh et al., 2019; Smillie et al., 2019). The functional significance of these changes in disease progression remains to be determined and might pave the way for novel therapeutic strategies for IBD.
Lung
The lung epithelium is subdivided into conducting airways, responsible for oxygen transport to the lungs, and respiratory epithelium, where gas exchange takes place. These key functions are enabled by specialized structures and cell types. Within the lung, the two epithelial compartments originate from two distinct SC subpopulations; in the conducting airways, epithelial SCs are a subset of basal cells (Rock et al., 2009), whereas in the respiratory epithelium, type 2 alveolar cells contain the SC fraction (Barkauskas et al., 2013). Single-cell analysis of different lung compartments, in both murine and human tissue, has revealed important insights into lung cell hierarchy and niche structure, highlighting novel cell types with important implications in lung function and pathology.
Conducting and respiratory airways.
The conducting airway epithelium consists of basal, secretory, and ciliated cells, as well as rare pulmonary neuroendocrine cells and brush cells (Rock et al., 2009). Lineage-tracing studies combined with single-cell sequencing analysis revealed a hierarchy of differentiation, originating with basal cells and giving rise to different subsets of secretory populations, including tuft and goblet cells and ciliated cells (Montoro et al., 2018). Likewise, in a census of human lung epithelium by Vieira Braga and colleagues, pseudotime analysis inferred a differentiation trajectory starting with basal cells and bifurcating into secretory and ciliated populations (Vieira Braga et al., 2019). An independent study of human lung revealed that 5% of basal cells are actively proliferating, whereas 29% of the basal compartment expressed lineage-restricted markers, indicating ongoing differentiation (Travaglini et al., 2019). Importantly, the human lung was shown to contain 17 unique cell types that were not identified in mouse models, including novel stromal and vascular populations, as well as novel epithelial populations (Travaglini et al., 2019). These findings highlight the need for comprehensive molecular atlases of different tissues, which will allow for better modeling of development and disease. Furthermore, these results emphasize that special care should be taken when inferring human cell function using murine models, and where possible, findings in mouse models should be validated using human samples.
In the respiratory epithelium, type 1 alveolar cells (AT1) mediate gas exchange, whereas type 2 alveolar cells (AT2) secrete surfactant. A small subset of AT2 cells act as respiratory SCs (Barkauskas et al., 2013), giving rise to all alveolar cells in the respiratory epithelium, yet the signals distinguishing AT2 SCs from bulk AT2 cells are not fully understood. Travaglini et al. identified two clusters of AT2 cells. One expresses canonical AT2 genes, as well as Wnt and Hedgehog inhibitors and cell cycle inhibitors, indicating these are mature AT2 cells. The second cluster expressed Wnt pathway genes, including Wnt ligand WNT5A, Wnt receptor LRP5, and Wnt transcription factor TCF4. These cells may correspond to the Wnt-active AT2 SCs identified in mice, yet experimental evidence for this is lacking (Travaglini et. al, 2019).
Mesenchymal niches.
Considering the different anatomical location and specialized functions performed by different sub-populations of lung epithelial cells, it has been presumed that distinct niche elements regulate each compartment. Indeed, Lee et al., using genetic lineage tracing, identified two subpopulations of lung mesenchymal cells, distinguished by expression of Wnt-pathway receptors Lgr5 and Lgr6 (Lee et al., 2017). Lgr5 and Lgr6 are well-known markers of intestinal SCs and skin progenitors. In the adult lung, however, their expression is restricted to mesenchymal populations. Whereas Lgr6+ cells are found in proximity to both bronchiolar and alveolar cells, Lgr5+ cells are predominantly found in proximity to alveolar cells. scRNA-seq of Lgr5- and Lgr6-expressing lung mesenchymal cells revealed five distinct mesenchymal subpopulations, including one cluster enriched for Pdgfra, Wnt2, and Fgf10 – all of which are key extrinsic factors for lung epithelial maintenance. Whereas Lgr5- and Lgr6-expressing cell subsets express common factors, the role of each subset is restricted to specific lung compartments. Specifically, Lgr6+ mesenchymal cells support organoid formation from club cells via production of Fgf10. In sharp contrast to their role in airway epithelium maintenance, Lgr6+ cells reduced alveolar organoid formation, whereas Lgr5+ cells increased alveolar organoid output, indicating spatial regulation of lung epithelium by these distinct mesenchymal elements. Interestingly, some Lgr6+ cells were found to express Lgr5, suggesting a developmental relationship between these subsets. However, the full composition and developmental trajectory of the lung mesenchymal niche needs to be further elucidated to clarify this link.
In a complementary study of the murine lung, Zepp et al. show that AT2 cells reside in close proximity to Pdgfra-expressing fibroblasts, which can promote AT2 cell self-renewal and differentiation into AT1 cells in ex vivo cultures (Zepp et al., 2017). scRNA-seq of mesenchymal cells, combined with elegant use of signaling reporters, allowed comprehensive mapping of the murine respiratory niche compartment, integrating transcriptional profiles with spatial locations. Mesenchymal cells were divided into five distinct subsets on the basis of expression of Axin2, Wnt2, and Pdgfra. Of these, mesenchymal cells expressing both Axin2 and dgfra were found in closest proximity to AT2 alveolar cells, and, in ex vivo experiments, were shown to preferentially support organoid generation and differentiation of AT2 cells to AT1 cells compared to other mesenchymal lineages. Similarly, an examination of the transcriptional landscape of alveolar fibroblasts revealed a mesenchymal subset expressing Pdgfra and Wnt5a. Imaging analysis indicated that Wnt5a-expressing fibroblasts lie in close proximity to AT2 cells, identifying a single Wnt5a-expressing fibroblast adjacent to each AT2 stem cell (Nabhan et al., 2018). These findings suggest a niche model where Wnt secretion by subepithelial fibroblasts supports alveolar SC function in the respiratory epithelium. However, the relationship between Lgr5- and Lgr6-expressing mesenchymal cells and Pdgfra-expressing fibroblasts remains obscure, and it is not clear if these are markers for the same population, or if they represent different subsets of the mesenchymal niche. Further characterization of the cellular components of the lung niche is required to identify factors that govern the spatial specification of lung function.
Functional plasticity.
The lungs are susceptible to several chronic diseases, which can be induced either by inherited mutations or by exposure to environmental insults. Pathologic changes in the lung epithelium, due to chronic disease, are likely to be accompanied by changes in the niche, highlighting the need for in-depth understanding of niche architecture. Because the lung is a relatively quiescent tissue (Peng et al., 2015), studies of lung SC function largely depend on models of injury to trigger regeneration. Although these models are somewhat artificial, they can provide important insight into the molecular pathways involved in epithelial repair. Injury models have revealed signaling pathways and cytokines important for lung regeneration, but their cellular sources remain unclear.
In humans, scRNA-seq of lung tissue from pulmonary fibrosis (PF) patients and from healthy lung donors revealed significant changes in epithelial structure in PF patients, including expansion of conducting epithelial cells and decrease in alveolar epithelial cells (Habermann et al., 2020). A novel epithelial subtype, expressing disease-associated extracellular matrix components, was identified specifically in PF patients from all disease subtypes. Trajectory reconstruction inferred that these cells arise from transitional AT2 cells, which can originate from either Scgb3a2+ secretory cells or from AT2 cells. Notably, these cells were not identified in murine models of fibrosis, highlighting the need for in-depth characterization of human disease. Lungs from PF patients were also enriched in myofibroblasts, as well as a novel Has1+ fibroblast population, which expressed high levels of stress-response genes and type 2 cytokines. These data demonstrate that epithelial remodeling in PF is accompanied by niche remodeling and suggest that targeting of niche components may enable restoration of healthy tissue architecture.
Chronic inflammation induced by asthma also significantly remodels the lung. scRNA-seq of lung biopsies from patients with chronic asthma (Vieira Braga et al., 2019) revealed an increase in CD4+ T cells, as well as changes in the differentiation trajectory of the lung epithelium, assessed by pseudotime analysis. In healthy lungs, basal cells differentiated into a secretory lineage, consisting mostly of club cells, and a ciliated lineage, whereas in asthma patients, the secretory lineage consisted of a mixture of club and goblet cells, and the ciliated lineage was dominated by mucous ciliated cells, which were not found in healthy donors. Furthermore, examination of receptor-ligand expression patterns demonstrated that the interactome of the asthmatic lung was drastically altered. In healthy lungs, the interaction landscape is dominated by epithelial and mesenchymal cells interacting with each other or with CD4+ T cells. In contrast, in asthmatic lungs the communication landscape was dominated by CD4+ Th2 cells, which interacted with all other cell types in the lung. This was accompanied by reduced interactions between epithelial and mesenchymal cells.
Understanding the molecular landscape of tissues in health and disease is key to developing novel therapies. Indeed, scRNA-seq of lung epithelium has revealed that the CFTR gene, implicated in cystic fibrosis (CF), is expressed by a novel cell type, pulmonary ionocytes (Montoro et al., 2018; Plasschaert et al., 2018). This finding has significant implications for our understanding of CF pathogenesis and is sure to influence the clinical outcome of CF patients.
Although advances in single-cell sequencing technologies have enabled significant progress in understanding the architecture of barrier tissue, much remains to be discovered. The plasticity of the mesenchymal niche in response to different types of injury requires crosstalk between all niche components, and this remains to be elucidated. Furthermore, it is unclear how changes in the epithelial compartment are sensed in the niche. Implementation of newly developed spatial sequencing methods may shed light on the interplay of all niche components, by revealing their location relative to epithelial cells. Furthermore, different functions of specialized tissue areas are supported by corresponding niche populations, which may also be revealed by spatial sequencing. Exposure to pathogenic cues, whether bacterial, viral, or environmental, that can also change according to their location within the tissue probably influences niche function. Finally, analysis of other cell types, such as neurons and vascular cells, which play a significant role in the bone and neurogenic niches, might reveal novel contributors to the barrier tissue niche.
Central Nervous System
Neurogenesis, the process by which neural stem cells (NSCs) differentiate into mature neurons and glia, predominantly takes place during embryonic development and drives the formation of the central nervous system (CNS). Although animal studies have found strong evidence for adult neurogenesis, its prevalence in the human brain is actively debated (Paredes et al., 2018; Petrik and Encinas, 2019; Sorrells et al., 2018). In this section, we discuss studies utilizing single-cell transcriptomics to examine various aspects of adult neurogenesis, including NSC differentiation, niche architecture, and age-associated inflammation.
NSCs are defined by their ability to differentiate into neurons, astrocytes, and oligodendrocytes. In rodents, adult neurogenesis maintains normal brain function throughout life and is restricted to two major sites, the ventricular-subventricular zone (V-SVZ) and the subgranular zone (SGZ). In the V-SVZ, NSCs, type B1 cells, reside along the walls of the lateral ventricle and generate neural progenitors that migrate to the olfactory bulb. B1 cells give rise to periglomerular and granule neuronal cells, which are required for the maintenance of fine odor discrimination (Figure 5A). In the SGZ, radial astrocytes in the ease (AD) (Moreno-Jiménez et al., 2019; dentate gyrus of the hippocampus give rise to granule neurons, which are required for memory and learning (Bjornsson et al., 2015; Kriegstein and Alvarez-Buylla, 2009; Zhao et al., 2008). In humans, abundant immature neurons were identified in healthy hippocampus, suggesting neuronal regeneration in the postnatal human brain. Interestingly, these immature neurons were drastically reduced in patients with Alzheimer’s disease (AD) (Moreno-jiménez et al., 2019; Tobin et al., 2019). Yet, unequivocal identification of NSCs and their niche in the adult human brain is still under active research.
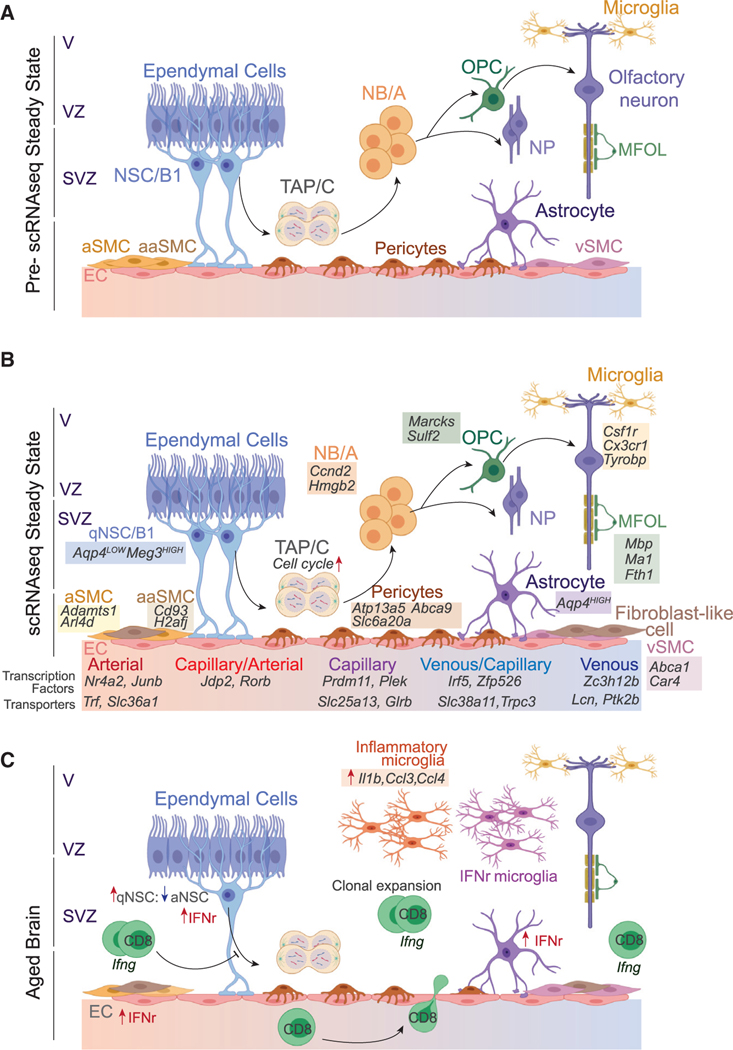
Figure 5. Neurogenic Niches.
(A) Before scRNA-seq, it was known that NSCs (type B1 cells) in the V-SVZ contact multi-ciliated ependymal cells in the ventricular zone and vascular cells of the blood-brain barrier (BBB). Upon receiving activation signals, qNSCs exit quiescence to generate transient amplifying progenitors (TAPs, type C), which then transition to NBs (type A) to make neural progenitors (NPs), including oligodendrocyte progenitor cells (OPCs). NPs migrate to the olfactory bulb and differentiate into neural lineages, including neurons and myelin-forming oligodendrocytes (MFOLs). Microglia are resident neural macrophages that play a neuroprotective role. ECs, connected by tight junctions, line the interior of BBB blood vessels and pericytes, and smooth muscle cells (SMCs) cover the exterior.
(B) scRNA-seq has defined the transcriptional programs of the V-SVZ neurogenic niche cells. NSCs can be separated from astrocytes by differential expression of Aqp4. qNSCs are defined by high expression of glycolysis and lipid metabolism genes, and aNSCs are defined by upregulation of oxidative phosphorylation genes before transitioning to rapidly cycling TAPs. Gene signatures of neurogenic niche cells are shown underneath individual cells.
(C) The aged brain is characterized by decreased number of NSCs and increased NSC quiescence. Neurogenic niches have increased infiltration and clonal expansion of CD8+ effector memory T cells expressing high levels of Ifng. Neurogenic niche cells are enriched for interferon-response signatures (IFNr). Microglia upregulate the inflammatory program.
The lack of definitive NSC-specific markers is a major challenge in the field. For example, GFAP and Nestin, commonly used to identify NSCs, are also expressed on astrocytes and oligodendrocyte progenitor cells (Lim and Alvarez-Buylla, 2016; Silver and Steindler, 2009; Urbán and Guillemot, 2014). In order to comprehensively characterize the V-SVZ compartment, Zywitza et al. performed scRNA-seq on all V-SVZ-residing cells. This analysis underscored close transcriptional resemblance between NSCs and parenchymal astrocytes and revealed that NSCs can be distinguished from astrocytes by low expression of astrocyte-specific Aqp4, as well as high levels of the long, non-coding RNA Meg3 (Zywitza et al., 2018). In the murine hippocampus, Artegiani et al. demonstrated that NSCs do not cluster separately from astrocytes, but can be discerned by astrocyte-specific expression of S100b and Fzd2 (Artegiani et al., 2017). These studies highlight the caveats associated with NSC-specific marker identification, driven by close transcriptional similarities with astrocytes, as well as uneven expression of commonly used markers throughout the NSC compartment. Although the challenge remains, the transcriptional signatures provided by these studies could inform sophisticated reporter models that are required for labeling NSCs in vivo.
scRNA-seq has been fundamental in dissecting the transcriptional dynamics of NSC activation and differentiation into different neurogenic lineages. These studies show that NSC activation and transition into transient-amplifying progenitor cells was accompanied by rapid upregulation of cycling genes and changes in metabolism and protein homeostasis (Artegiani et al., 2017; Leeman et al., 2018; Zywitza et al., 2018). Furthermore, single-cell analysis has unraveled the transcriptional basis of neural progenitor differentiation into oligodendrocyte progenitor cells that transition to myelin-forming mature oligodendrocytes (Artegiani et al., 2017) (Figure 5). Understanding NSC differentiation trajectories carries significant clinical implications, as exemplified by the major contribution of aberrant progenitors to formation of gliomas (Lee et al., 2018).
Neurons.
Neurogenesis depends on exogenous signals such as cytokines, growth factors, and neurotransmitters (Berg et al., 2013; Lim and Alvarez-Buylla, 2016). NSC quiescence is a highly regulated state that preserves the NSC pool and is critical for maintaining continuous neurogenesis in the adult brain. How NSC quiescence is maintained is an active area of research that carries significant clinical implications.
In the V-SVZ, NSCs are localized along the lateral ventricle walls, distal to olfactory bulb inhibitory neurons. NSC proliferation is dependent on the signals of neurotransmitters such as serotonin (5HT). Another neurotransmitter, dopamine, might have dual inhibitory and activating effects on NSCs (Kippin et al., 2005; Lennington et al., 2011). SGZ NSCs, on the other hand, reside in close proximity to excitatory granule neurons and parvalbumin-expressing (PV+) interneurons (Berg et al., 2013). SGZ NSCs are maintained in quiescence by neurotransmitter γ-aminobutyric acid (GABA) released from PV+ interneurons during high circuitry activity (Song et al., 2012). Hippocampal NSC quiescence is also regulated by Wnt signaling, specifically the expression of Wnt inhibitor Sfrp3 by excitatory granule neurons. Jang et al. showed that electroconvulsive stimulation to increase neuronal activity decreased Sfrp3 expression, and this change subsequently upregulated Wnt signaling in NSCs, increasing NSC activation and the formation of immature neurons (Jang et al., 2013).
Shin et al. compared qNSC-enriched genes downregulated during activation with newly upregulated genes after qNSC activation and early neurogenesis. After activation, NSCs downregulated signaling pathways associated with quiescence. Ligands activating these pathways are present in the adult SGZ niche, suggesting that to maintain quiescence, qNSCs actively receive and integrate these signals. Remarkably, NSC activation led to suppression of genes involved in signal transduction of these niche cues, suggesting that aNSCs lose sensitivity to niche signals associated with quiescence. This conceptual leap required a systematic analysis, simultaneously integrating hundreds of parameters (Shin et al., 2015). In a follow-up study, this group investigated the role of Mfge8, a transcript previously found to be enriched in qNSCs compared to the activated state. The authors found that SGZ NSC attrition might also be prevented by high expression of Mfge8, which maintains NSC quiescence through suppression of the mTOR1 pathway (Zhou et al., 2018).
Endothelial niches.
The cardinal feature of the brain vasculature is the blood-brain barrier (BBB). This selective barrier regulates nutrient supply from the blood to the brain, protects the brain from toxins, and prevents immune-cell infiltration. The BBB consists of tightly packed capillaries connected by tight junctions that impede the movement of lipid-insoluble macromolecules out of the blood stream into the brain parenchyma (Baeten and Akassoglou, 2011). Importantly, NSCs have direct contact with ECs that secrete factors to control NSC function (Gómez-Gaviro et al., 2012; Kokovay et al., 2010). Understanding how NSCs integrate vascular signals is an area of ongoing research (Ottone, 2015).
Molecular mechanisms underlying the unique properties of BBB components are of great clinical interest. Vanlandewijck et al. utilized transgenic animal reporters to examine transcriptional dynamics and specialization of brain vasculature at single-cell resolution. The authors show that BBB ECs exhibit a continuum of six transcriptional states across venous, capillary, and arterial zones, and arterial ECs are enriched for transcription factors, and capillary and venous ECs are enriched for transporters (Figure 5) (Vanlandewijck et al., 2018). An independent study investigating tissue-specific heterogeneity of ECs across 11 mouse tissues performed pseudotime analysis of brain ECs, obtaining analogous transcriptional topography. Notably, intratissue EC heterogeneity varied significantly across tissues. Consistent with BBB function, ECs and pericytes highly expressed genes involved in molecular transport, and BBB ECs downregulated transcriptional signatures associated with immune regulation compared to ECs from other peripheral tissues (Kalucka et al., 2020). Highlighting the power of scRNA-seq, this study identified a novel fibroblast-like cell located in the perivascular space and enriched in genes associated with maintenance of the extracellular matrix, including collagens and collagen-modifying enzymes. The functional relevance of these transcriptional signatures for NSCs and neurogenic niche cells remains unclear and will need to be addressed in future functional studies.
Functional plasticity.
Aging is a major risk factor for neurodegenerative disorders. As the brain ages, a number of deleterious processes arise, including abnormal protein deposits, lysosomal dysfunction, DNA damage, and immune dysregulation (Elobeid et al., 2016; Wyss-Coray, 2016). Neurogenic aging in both the SGZ and V-SVZ, as assessed by flow cytometry and single-cell transcriptomic analysis, is marked by a drop in NSC numbers and increased quiescence (Artegiani et al., 2019; Kalamakis et al., 2019; Ximerakis et al., 2019). Kalamakis et al. demonstrated that aged V-SVZ qNSCs were more resistant to injuryinduced activation compared to younger counterparts. Surprisingly, young and aged NSCs had comparable functionality and transcriptional signatures, suggesting microenvironment regulation of activation dynamics. Differential expression analysis in V-SVZ niche cells and NSCs revealed a common enrichment of transcripts related to inflammatory response. Notably, the inflammatory signature was captured with bulk RNA-seq of the individual cell populations, as the observed changes in gene expression were difficult to capture at the single-cell level. This study established a causal link between niche-derived inflammatory signals and increased NSC quiescence. Moreover, this increased quiescence is likely to contribute to the challenge of detecting active neurogenesis in the aged adult human brain at steady state (Kalamakis et al., 2019).
Immune cells play critical roles in regulating the neurogenic niche. Tissue-resident macrophages, termed microglia, comprise 10% of brain cells and maintain the neurogenic niche by performing synaptic remodeling, phagocytosis of dying cells, and maintaining myelin homeostasis (Lannes et al., 2017; Sasaki, 2017; Schafer and Stevens, 2015). Genetic lesions in microglia have been associated with neurodegenerative disease and leukodystrophies (Jonsson et al., 2013; Lambert et al., 2013; Rademakers et al., 2011). scRNA-seq studies found that microglia diversity peaks during early development and decreases in adulthood until onset of aging or perturbation by injury (Hammond et al., 2019). Aging caused a redistribution of microglia states, with marked expansion of subsets enriched for interferon (IFN)-response genes and inflammatory microglia with high expression of ll1b, Ccl4, and Ccl3 (Hammond et al., 2019). In pathological contexts, microglia display a broad range of activation states. However, comparison of transcriptional signatures of microglia from models of multiple sclerosis (MS) and AD to axontract-associated microglia found during brain development revealed a shared transcriptional signature of 12 genes, including Spp1, Lpl, and Apoe (Hammond et al., 2019; Keren-Shaul et al., 2017). Interestingly, these genes correspond to pathways expressed by axon-tract-associated microglia specific to a restricted developmental window, in line with the hypothesis that developmental pathways are re-expressed in brain pathologies, a concept that can be leveraged in the design of precision therapies.
Aging-mediated enrichment of IFN-response genes in microglia raises the question of the cellular source(s) of IFN. Single-cell mass and fluorescence cytometry analysis has demonstrated that aging has a profound effect on the immune landscape in the brain. One of the most striking changes in the aged brain is a ten-fold increase in T cell frequency (Mrdjen et al., 2018). Dulken et al. found infiltration of effector memory CD8+ T cells that expressed high levels of Ifng in the aged murine SVZ niche. V(D)J analysis revealed that brain-infiltrating lymphocytes were clonally expanded, and their TCR repertoire differed from T cells isolated from peripheral blood. Several cell types, including astrocytes, NSCs, ECs, and microglia, in the aged SVZ niche were enriched for an IFNγ response signature. Importantly, T cell-mediated IFNγ production inhibited NSC function, providing a potential mechanism of mediating age-associated decline in NSCs (Dulken et al., 2019). Clonal expansion of CD8+ T cells indicates that infiltrating CD8+ T cells recognize brain-specific antigen(s). Identification of the specific antigens and blocking their recognition would have major therapeutic implications for understanding and treating age-related pathologies.
Although scRNA-seq is an essential tool for transcriptionally profiling individual cells, cellular dissociation leads to loss of spatial context that is essential to understanding the cellular architecture of different regions and connections between brain circuits and neurogenic niche cells. Technological advancement in multiple spatial transcriptomic techniques, such as sequential fluorescence in situ hybridization (seqFISH+) and multiplexed error robust fluorescence in situ hybridization (MERFISH), has facilitated the generation of high-sensitivity transcriptional maps while maintaining topological information of brain regions (Eng et al., 2019; Moffitt et al., 2018; Rodriques et al., 2019). Application of these approaches to SC niches can clarify the spatial resolution of niche components, advancing our understanding of SC-niche crosstalk.
Concluding Remarks
scRNA-seq has significantly informed SC biology by resolving cellular and molecular units of the SC microenvironment. Detection of rare populations is exemplified by the identification of tuft-like medullary epithelial population in the thymus and CFTR+ pulmonary ionocytes in the lung. SC niche elements can be identified through unbiased and focused transcriptional profiling. Clonally expanded IFN-producing CD8+ T-lymphocytes in old neurogenic niches were identified by scRNA-seq analysis of the entire SVZ. In the intestine, on the other hand, MHC-II expression by ISC-facilitated identification of CD4+ T helper cells as a critical component of the intestinal SC niche. Thus, the insights afforded by single-cell transcriptomics extend beyond phenomenology and are largely dictated by the study design.
Single-cell transcriptomic studies are quickly becoming mainstream, yet challenges remain. Due to the low amount of starting material, scRNA-seq is hindered by low capture efficiency and sequencing coverage (Kolodziejczyk et al., 2015). Though sequencing at relatively shallow depth is sufficient for common tasks such as cell-type identification, deep sequencing effectively mitigates technical noise and allows for the dissection of cell-specific transcriptional programs (Zhang et al., 2020). Unfortunately, the choice of sequencing scale and depth is dictated not only by the biological question, but also by experimental cost. Significant expenses associated with single-cell studies impede broad utilization of scRNA-seq.
Human application is perhaps the strongest facet of single-cell analysis. Although SCs and their niches have been extensively studied in animal models, the real power of SC studies lies in generating detailed blueprints of human tissues, which largely remained a mystery before the advent of scRNA-seq. International initiatives such as the Human Cell Atlas and the National Institutes of Health’s Human BioMolecular Atlas Program are ambitious efforts to map all the cells in the human body and build a comprehensive database, enabling data sharing worldwide. These consortiums have already generated detailed maps of several organs, yielding important insights into the biology of thymus, heart, lungs, small intestine, and liver. Although all of these studies could not be included here due to space constraints, this critical effort will guide future research. Importantly, these consortiums have also set tissue and sample preparation standards, which could help limit variability in results from different groups. Open-access data will enable investigators to bridge the gap between basic research and human health. Indeed, an unrestricted data-sharing approach should be applied to every study with single-cell data. To magnify the impact of published research, in addition to depositing raw sequencing data, the manuscript should be accompanied by web-based, interactive, user-friendly resources, enabling independent data exploration. This will facilitate dialog among groups from different fields and speed up scientific advances.
scRNA-seq analysis enabled the deciphering of SC niche architecture; however, in-depth understanding of SC-niche crosstalk requires knowledge of the genome, the chromatin landscape, protein levels, and spatial context of the cell. Emerging experimental platforms profile each of these modalities at the single-cell level and will inevitably gain broad utilization across disciplines. None of the platforms available to date has the capacity to profile each of these parameters simultaneously. Therefore, integration of single-cell data across independent datasets and platforms requires an effective computational strategy, such as LIGER (Welch et al., 2019) and Seurat 3.0 (Stuart et al., 2019). The best-designed studies will integrate multimodal approaches and perform functional validation. As single-cell technology continues to advance, it will markedly transform the day-to-day of laboratory research and shift scientific culture toward open-access. Independently of utilized approaches, the most meaningful discoveries will still be based on the ability to define a biologically relevant question and the careful interpretation of results.
ACKNOWLEDGMENTS
This research was supported by the US National Institutes of Health (RO1CA202025, RO1CA202027, R01CA216421, R01CA173636, and RO1CA242020), the Leukemia & Lymphoma Society (TRP #6580), The Edward P. Evans Foundation, Alex’s Lemonade Stand Foundation for Childhood Cancer, and St. Baldrick’s Cancer Research Foundation (I.A). The work was also supported by the New York State Department of Health (NYSTEM #C32563GG and #C32587GG). A.N.T. was supported by the Leukemia and Lymphoma Society and the Alex’s Lemonade Stand Foundation for Childhood Cancer. We thank Dr. Shane Liddelow, Dr. Gilad Evrony, Dr. Kathryn Hockemeyer, and Dr. Matthew T. Witkowski for their input on the manuscript.
REFERENCES
- Abramson J, and Anderson G. (2017). Thymic Epithelial Cells. Annu. Rev. Immunol. 35, 85–118. [DOI] [PubMed] [Google Scholar]
- Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, and Morrison SJ (2015). Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, and Su MA (2016). AIRE expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol. 16, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, and Mathis D. (2005). The cellular mechanism of Aire control of T cell tolerance. Immunity 23, 227–239. [DOI] [PubMed] [Google Scholar]
- Artegiani B, Lyubimova A, Muraro M, van Es JH, van Oudenaarden A, and Clevers H. (2017). A Single-Cell RNA Sequencing Study Reveals Cellular and Molecular Dynamics of the Hippocampal Neurogenic Niche. Cell Rep. 21, 3271–3284. [DOI] [PubMed] [Google Scholar]
- Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, Lopez-Iglesias C, Postrach D, Dayton T, Oka R, et al. (2019). Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 24, 927–943 e926. [DOI] [PubMed] [Google Scholar]
- Baccin C, Al-Sabah J, Velten L, Helbling PM, Grünschläger F, Hernández-Malmierca P, Nombela-Arrieta C, Steinmetz LM, Trumpp A, and Haas S. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 22, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten KM, and Akassoglou K. (2011). Extracellular matrix and matrix receptors in blood-brain barrier formation and stroke. Dev. Neurobiol. 71, 1018–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BL (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, Kokkaliaris KD, Mercier F, Tabaka M, Hofree M, et al. (2019). A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 177, 1915–1932 e1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DA, Belnoue L, Song H, and Simon A. (2013). Neurotransmitter-mediated control of neurogenesis in the adult vertebrate brain. Development 140, 2548–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins CL, and Salzman NH (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. [DOI] [PubMed] [Google Scholar]
- Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson CS, Apostolopoulou M, Tian Y, and Temple S. (2015). It takes a village: constructing the neurogenic niche. Dev. Cell 32, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Tó th, B., et al. (2018). Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626. [DOI] [PubMed] [Google Scholar]
- Cheng M, and Anderson MS (2018). Thymic tolerance as a key brake on autoimmunity. Nat. Immunol. 19, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou Constantina, Spencer Joel, A, Yeh, Shu-Chi, A, et al. (2020). Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou C, Spencer JA, Yeh SA, Turcotte R, Kokkaliaris KD, Panero R, Ramos A, Guo G, Seyedhassantehrani N, Esipova TV, et al. (2020). Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro Gomes A, Hara T, Lim VY, Herndler-Brandstetter D, Nevius E, Sugiyama T, Tani-Ichi S, Schlenner S, Richie E, Rodewald HR, et al. (2016). Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 45, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane GM, Jeffery E, and Morrison SJ (2017). Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590. [DOI] [PubMed] [Google Scholar]
- Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, and Basler K. (2018). GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453. [DOI] [PubMed] [Google Scholar]
- Ding L, and Morrison SJ (2013). Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, and Morrison SJ (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV, et al. (2019). Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elobeid A, Libard S, Leino M, Popova SN, and Alafuzoff I. (2016). Altered Proteins in the Aging Brain. J. Neuropathol. Exp. Neurol. 75, 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, and Cai L. (2019). Transcriptome-scale superresolved imaging in tissues by RNA seqFISH. Nature 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, and Clevers H. (2019). Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34. [DOI] [PubMed] [Google Scholar]
- Ghazanfari R, Li H, Zacharaki D, Lim HC, and Scheding S. (2016). Human Non-Hematopoietic CD271pos/CD140alow/neg Bone Marrow Stroma Cells Fulfill Stringent Stem Cell Criteria in Serial Transplantations. Stem Cells Dev. 25, 1652–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gaviro MV, Lovell-Badge R, Fernández-Avilés F, and Lara-Pezzi E. (2012). The vascular stem cell niche. J. Cardiovasc. Transl. Res. 5, 618–630. [DOI] [PubMed] [Google Scholar]
- Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, Scadden DT, and Hu GF (2016). Angiogenin Promotes Hematopoietic Regeneration by Dichotomously Regulating Quiescence of Stem and Progenitor Cells. Cell 166, 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, and Link DC (2013). CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung M, Taylor CJ, Jetter C, et al. (2020). Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoun M, and Frenette PS (2013). This niche is a maze; an amazing niche. Cell Stem Cell 12, 391–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan DD, Mabuchi Y, Morikawa S, Niibe K, Araki D, Suzuki S, Okano H, and Matsuzaki Y. (2012). Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-α. Nat. Protoc. 7, 2103–2111. [DOI] [PubMed] [Google Scholar]
- Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, Ledergor G, Jung Y, Milo I, Poulos MG, et al. (2016). Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MH, Bonaguidi MA, Kitabatake Y, Sun J, Song J, Kang E, Jun H, Zhong C, Su Y, Guo JU, et al. (2013). Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. (2013). Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamakis G, Brüne D, Ravichandran S, Bolz J, Fan W, Ziebell F, Stiehl T, Catalá-Martinez F, Kupke J, Zhao S, et al. (2019). Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell 176, 1407–1419.e14. [DOI] [PubMed] [Google Scholar]
- Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. (2020). SingleCell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779 e720. [DOI] [PubMed] [Google Scholar]
- Kappler JW, Roehm N, and Marrack P. (1987). T cell tolerance by clonal elimination in the thymus. Cell 49, 273–280. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, and Maehr R. (2018). A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation. Immunity 48, 1258–270 e1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury Y, and Scadden DT (2015). Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 16, 239–253. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, and Morrison SJ (2005). SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121. [DOI] [PubMed] [Google Scholar]
- Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. (2018). Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 175, 372–386 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin TE, Kapur S, and van der Kooy D. (2005). Dopamine specifically inhibits forebrain neural stem cell proliferation, suggesting a novel effect of antipsychotic drugs. J. Neurosci. 25, 5815–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Blüthmann H, Staerz UD, Steinmetz M, and von Boehmer H. (1988). Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333, 742–746. [DOI] [PubMed] [Google Scholar]
- Klein L, Hinterberger M, Wirnsberger G, and Kyewski B. (2009). Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844. [DOI] [PubMed] [Google Scholar]
- Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, and Temple S. (2010). Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell 7, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, and Teichmann SA (2015). The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620. [DOI] [PubMed] [Google Scholar]
- Kondo K, Takada K, and Takahama Y. (2017). Antigen processing and presentation in the thymus: implications for T cell repertoire selection. Curr. Opin. Immunol. 46, 53–57. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. (2013). Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe AP, Ramasamy SK, and Adams RH (2014). Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature 507, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. ; European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannes N, Eppler E, Etemad S, Yotovski P, and Filgueira L. (2017). Microglia at center stage: a comprehensive review about the versatile and unique residential macrophages of the central nervous system. Oncotarget 8, 114393–114413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um J-Y, Kim WK, Lee J-K, Park J, et al. (2018). Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560, 243–247. [DOI] [PubMed] [Google Scholar]
- Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, Dulken BW, Zhao X, Yeo RW, Ho TT, et al. (2018). Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennington JB, Pope S, Goodheart AE, Drozdowicz L, Daniels SB, Salamone JD, and Conover JC (2011). Midbrain dopamine neurons associated with reward processing innervate the neurogenic subventricular zone. J. Neurosci. 31, 13078–13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, and Alvarez-Buylla A. (2016). The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb. Perspect. Biol. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Zhang SY, Chen YY, Shi K, Zou B, Liu J, Yang Q, Jiang H, Wei L, Li CZ, et al. (2018). ICAM-1 Deficiency in the Bone Marrow Niche Impairs Quiescence and Repopulation of Hematopoietic Stem Cells. Stem Cell Reports 11, 258–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, McKenna LB, Dolson D, Epstein DJ, and Kaestner KH (2009). FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J. Biol. Chem. 284, 5936–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch P, Constine L, Greenberger J, Knospe W, Sullivan J, Liesveld JL, and Deeg HJ (1995). Hematopoietic stem cell compartment: acute and late effects of radiation therapy and chemotherapy. Int. J. Radiat. Oncol. Biol. Phys. 31, 1319–1339. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, and Taoudi S. (2011). Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031. [DOI] [PubMed] [Google Scholar]
- Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, et al. (2018). Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, and Zhuang X. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Jiménez EP, Flor-García M, Terreros-Roncal J, Rábano A, Cafini F, Pallas-Bazarra N, Ávila J, and Llorens-Martín M. (2019). Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, and Scadden DT (2014). The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, Lelios I, Heppner FL, Kipnis J, Merkler D, et al. (2018). High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48, 599. [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottone Christina, et al. (2015). Multifaceted Control of Adult SVZ Neurogenesis by the Vascular Niche. Cell Cycle 14, 2222–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneck AR, Rafiq A, Schütz B, Soultanova A, Deckmann K, Chubanov V, Gudermann T, Weihe E, Krasteva-Christ G, Grau V, et al. (2014). Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res. 358, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF, Sorrells SF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, et al. (2018). Does Adult Neurogenesis Persist in the Human Hippocampus? Cell Stem Cell 23, 780–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, et al. (2019). Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55. [DOI] [PubMed] [Google Scholar]
- Park JE, Botting RA, Domínguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, and Morrisey EE (2015). Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik D, and Encinas JM (2019). Perspective: Of Mice and Men - How Widespread Is Adult Neurogenesis? Front. Neurosci. 13, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM (2017). Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood 130, 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, Lakshminarasimhan R, Chin CP, Techner JM, Will B, et al. (2016). Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, et al. (2011). Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 44, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BL (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, and Macosko EZ (2019). Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. (2017). Microglia and brain macrophages: An update. Neuropathology 37, 452–464. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, and Clevers H. (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, and Stevens B. (2015). Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb. Perspect. Biol. 7, a020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. (1978). The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4, 7–25. [PubMed] [Google Scholar]
- Schuettpelz LG, Borgerding JN, Christopher MJ, Gopalan PK, Romine MP, Herman AC, Woloszynek JR, Greenbaum AM, and Link DC (2014). G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia 28, 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe N, Karabacak NM, Gustafsson K, Baryawno N, Courties G, Kfoury Y, Kokkaliaris KD, Rhee C, Lee D, Scadden EW, et al. (2019). Stress-Induced Changes in Bone Marrow Stromal Cell Populations Revealed through Single-Cell Protein Expression Mapping. Cell Stem Cell 25, 570–583 e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, and Song H. (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DJ, and Steindler DA (2009). Common astrocytic programs during brain development, injury and cancer. Trends Neurosci. 32, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2019). Intraand Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730 e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhong C, Bonaguidi MA, Sun GJ, Hsu D, Gu Y, Meletis K, Huang ZJ, Ge S, Enikolopov G, et al. (2012). Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanova A, Voigt A, Chubanov V, Gudermann T, Meyerhof W, Boehm U, and Kummer W. (2015). Cholinergic chemosensory cells of the thymic medulla express the bitter receptor Tas2r131. Int. Immunopharmacol. 29, 143–147. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, and Regev A. (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, et al. (2019). The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin MK, Musaraca K, Disouky A, Shetti A, Bheri A, Honer WG, Kim N, Dawe RJ, Bennett DA, Arfanakis K, and Lazarov O. (2019). Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer’s Disease Patients. Cell Stem Cell 24, 974–982.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, et al. (2019). A molecular cell atlas of the human lung from single cell RNA sequencing. bioRxiv. 10.1101/742320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán N, and Guillemot F. (2014). Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 8, 396, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantu C, Aguet M, and Basler K. (2016). Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep. 15, 911–918. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. (2019). A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, and Locksley RM (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voog J, and Jones DL (2010). Stem cells and the niche: a dynamic duo. Cell Stem Cell 6, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, and Frenette PS (2018). Niches for Hematopoietic Stem Cells and Their Progeny. Immunity 48, 632–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, and Macosko EZ (2019). Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887 e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock Samuel, L., et al. (2019). Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Reports 28, 302–311.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T. (2016). Ageing, neurodegeneration and brain rejuvenation. Nature 539, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, et al. (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708. [DOI] [PubMed] [Google Scholar]
- Yue R, Zhou BO, Shimada IS, Zhao Z, and Morrison SJ (2016). Leptin Receptor Promotes Adipogenesis and Reduces Osteogenesis by Regulating Mesenchymal Stromal Cells in Adult Bone Marrow. Cell Stem Cell 18, 782–796. [DOI] [PubMed] [Google Scholar]
- Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, and Morrisey EE (2017). Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MJ, Ntranos V, and Tse D. (2020). Determining sequencing depth in a single-cell RNA-seq experiment. Nat. Commun. 11, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, and Gage FH (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. [DOI] [PubMed] [Google Scholar]
- Zhou BO, Yue R, Murphy MM, Peyer JG, and Morrison SJ (2014). Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Yu H, Yue R, Zhao Z, Rios JJ, Naveiras O, and Morrison SJ (2017). Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 19, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Bond AM, Shade JE, Zhu Y, Davis CO, Wang X, Su Y, Yoon KJ, Phan AT, Chen WJ, et al. (2018). Autocrine Mfge8 Signaling Prevents Developmental Exhaustion of the Adult Neural Stem Cell Pool. Cell Stem Cell 23, 444–452 e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zywitza V, Misios A, Bunatyan L, Willnow TE, and Rajewsky N. (2018). Single-Cell Transcriptomics Characterizes Cell Types in the Subventricular Zone and Uncovers Molecular Defects Impairing Adult Neurogenesis. Cell Rep. 25, 2457–2469.e8. [DOI] [PubMed] [Google Scholar]