Abstract
The ST6GAL1 sialyltransferase, which adds α2–6 linked sialic acids to N-glycosylated proteins, is overexpressed in a wide range of human malignancies. Recent studies have established the importance of ST6GAL1 in promoting tumor cell behaviors such as invasion, resistance to cell stress and chemoresistance. Furthermore, ST6GAL1 activity has been implicated in imparting cancer stem cell characteristics. However, despite the burgeoning interest in the role of ST6GAL1 in the phenotypic features of tumor cells, insufficient attention has been paid to the molecular mechanisms responsible for ST6GAL1 upregulation during neoplastic transformation. Evidence suggests that these mechanisms are multifactorial, encompassing genetic, epigenetic, transcriptional and posttranslational regulation. The purpose of this review is to summarize current knowledge regarding the molecular events that drive enriched ST6GAL1 expression in cancer cells.
Keywords: cancer, gene regulation, sialic acid, ST6GAL1
Introduction
Aberrant cell surface glycosylation constitutes one of the well-known hallmarks of a cancer cell (Pinho and Reis 2015; Taniguchi and Kizuka 2015; Varki et al. 2015; Munkley and Elliott 2016; Rodrigues et al. 2018). Among the many tumor-associated alterations in glycan structure, increased surface sialylation has been reported for at least three decades (Harduin-Lepers et al. 2012; Schultz et al. 2012; Dall'Olio et al. 2014; Pearce and Laubli 2016; Bhide and Colley 2017; Rodrigues and Macauley 2018). Enhanced tumor cell sialylation typically results from the overexpression of select sialyltransferases and/or the downregulation of sialidases such as Neu1 or Neu4 (Dall'Olio et al. 2014; Pearce and Laubli 2016; Vajaria et al. 2016). While increases in both α2–3 and α2–6 sialylation are common in cancer cells (Harduin-Lepers et al. 2012; Dall'Olio et al. 2014), some studies have described a selective enrichment in the α2–6 sialic acid linkage (Sata et al. 1991; Sethi et al. 2015). Tumor cells display elevated levels of α2–6 sialic acids on O-linked, and N-linked, glycans. For example, the sialylTn antigen is an α2–6 sialylated O-linked tumor glycan generated, in part, by the ST6GalNAc family of sialyltransferases (Ju et al. 2013; Munkley 2016). The principal sialyltransferase responsible for adding α2–6 sialic acids to N-linked tumor glycans is ST6GAL1 (Lu and Gu 2015; Garnham et al. 2019). A second enzyme, ST6GAL2, can also α2–6 sialylate N-glycans; however, this enzyme seems to be primarily expressed in the brain (Takashima et al. 2002; Lehoux et al. 2010). ST6GAL1 is overexpressed in a plethora of human malignancies, and an extensive literature has established a crucial role for ST6GAL1 in directing cell behaviors that propel tumor initiation and progression (Dall'Olio 2000; Lu and Gu 2015; Garnham et al. 2019). While much is known about ST6GAL1’s functional contribution to tumor cell characteristics, there is currently a dearth of information regarding the upstream mechanisms that control ST6GAL1 expression. Accordingly, the focus of this review is on the molecular events that mediate alterations in ST6GAL1 expression during neoplastic development.
ST6GAL1 is upregulated in many cancers, and high levels of ST6GAL1 correlate with a poor prognosis
The ST6GAL1 enzyme was initially purified by the Paulson laboratory (Weinstein et al. 1982) and the rat and human cDNAs were cloned a few years later (Weinstein et al. 1987; Grundmann et al. 1990). Around that same time, increases in ST6GAL1 expression and tumor cell α2–6 sialylation were reported in colon cancer (Dall'Olio et al. 1989; Sata et al. 1991; Dall'Olio and Trere 1993). Subsequent studies confirmed ST6GAL1 upregulation in colon carcinoma, as well as in many other epithelial malignancies including breast, ovarian, pancreatic, cervical, gastric, liver and prostate cancer (Recchi et al. 1998; Gretschel et al. 2003; Hebbar et al. 2003; Dall'Olio et al. 2004; Wang et al. 2004; Vazquez-Martin et al. 2005; Wang et al. 2005; Swindall et al. 2013; Munkley et al. 2016; Schultz et al. 2016; Wei et al. 2016; Hsieh et al. 2017; Cui et al. 2018; Wichert et al. 2018). Overexpression of ST6GAL1 has also been observed in some myeloid leukemias and brain tumors (Skacel et al. 1991; Kaneko et al. 1996; Mondal et al. 2010). High levels of ST6GAL1 are associated with a more advanced tumor grade, metastatic progression, lymphovascular invasion and reduced progression-free and overall survival (Recchi et al. 1998; Lise et al. 2000; Schultz et al. 2016; Wei et al. 2016; Agrawal et al. 2017; Hsieh et al. 2017; Cui et al. 2018; Wichert et al. 2018). Additionally, numerous cell culture and animal studies have attested to ST6GAL1’s role in conferring a malignant cell phenotype. Literature describing ST6GAL1’s tumor-promoting activity has been widely reviewed elsewhere and will not be a major focus of this article (Dall'Olio and Chiricolo 2001; Harduin-Lepers et al. 2012; Park and Lee 2013; Lu and Gu 2015; Bhide and Colley 2017; Garnham et al. 2019; Li and Ding 2019). However, in brief, ST6GAL1 sialylates key surface glycoproteins that regulate oncogenic signaling networks, for example, select adhesion molecules and growth factor receptors (Lu and Gu 2015; Garnham et al. 2019). In turn, this promotes tumor cell behaviors such as migration/invasion, epithelial to mesenchymal transition (EMT), and resistance to a wide range of cytotoxic stimuli including chemotherapy drugs, radiation, death receptor ligands, serum deprivation, hypoxia and apoptotic galectins (Seales et al. 2005; Lee et al. 2008; Lee et al. 2010; Swindall and Bellis 2011; Zhuo and Bellis 2011; Park et al. 2012; Schultz et al. 2013; Lu et al. 2014; Schultz et al. 2016; Wei et al. 2016; Britain et al. 2017; Hsieh et al. 2017; Chakraborty et al. 2018; Holdbrooks et al. 2018; Jones et al. 2018). In tandem with intrinsic effects on tumor cell signaling and phenotype, surface α2–6 sialic acids serve as ligands for a subset of inhibitory Siglec receptors, implicating ST6GAL1 in dampening antitumor immunity (Barenwaldt and Laubli 2019). On the other hand, tumor-suppressive functions for ST6GAL1 have been reported for certain types of cancer cells. ST6GAL1 expression is downregulated in advanced bladder cancer (Antony et al. 2014), and Moskal’s group has shown that forced ST6GAL1 overexpression in the U373 glioblastoma cell line decreases cell invasion, chemoresistance and in vivo tumor growth (Yamamoto et al. 1997; Yamamoto et al. 2001; Dawson et al. 2004). A similar inhibitory effect of ST6GAL1 on cell invasion and tumor growth has been documented for some colon cancer cell lines (Chiricolo et al. 2006; Park et al. 2012; Jung et al. 2016; Zhou et al. 2019). As well, ST6GAL1-mediated sialylation appears to inhibit VEGF-independent angiogenesis (Croci et al. 2014). While further studies will be needed to resolve these disparate findings, there is evidence that ST6GAL1 can have cell-type-specific activity (Venturi et al. 2019).
With the advent of The Cancer Genome Atlas (TCGA), studies linking ST6GAL1 mRNA expression to patient outcomes have greatly expanded. Results from these investigations have been mixed, with some reports describing reduced survival for patients with high ST6GAL1 mRNA levels (Wichert et al. 2018), and others suggesting no association (Venturi et al. 2019). However, there are some caveats to using TCGA transcriptomics data to infer cancer-associated changes in ST6GAL1 expression. First, it has been noted that most glycogenes are expressed in low abundance, and therefore, transcript levels do not accurately correspond with protein expression (Thu and Mahal 2020). Second, ST6GAL1 is expressed in most immune cell populations, and thus, mRNA levels quantified from whole tumor homogenates represent the combined tumor/immune cell pool. This can confound comparisons of differential ST6GAL1 mRNA expression across tumor grades or stages, given that immune cells can, in some malignancies, constitute a significant proportion of the total tumor cellularity (Kim and Bae 2016). In pancreatic ductal adenocarcinoma, tumor cells comprise only 5–20% of the cells within a malignant lesion (Wood and Hruban 2012). Furthermore, correlations between bulk tumor RNA levels and patient survival are problematic for immune-expressed genes like ST6GAL1 because the type of tumor-infiltrating immune cell has a significant effect on prognosis (Grivennikov et al. 2010; Schnell et al. 2018). The presence of cells such as cytotoxic T lymphocytes and Natural Killer cells usually corresponds with a better prognosis, whereas T regulatory cells and myeloid-derived suppressor cells portend an unfavorable outcome (all of these immune cell types are ST6GAL1 positive). Hence, if a large proportion of the bulk tumor mRNA is derived from the immune compartment, then ST6GAL1 mRNA levels within this immune pool can influence interpretations of disease progression, survival and other parameters. Newer technologies such as single cell RNA sequencing may address this issue in the future, enabling a more definitive view of how tumor cell-specific ST6GAL1 levels correlate with phenotypic characteristics such as metastatic potential and chemosensitivity.
Immunohistochemistry (IHC) offers an alternative approach for analyzing ST6GAL1 expression in tumors. This technique can distinguish between tumor and immune/stromal cells and also offer information regarding ST6GAL1 expression in distinct regions of a tumor (e.g., hypoxic areas and invasive margins). Historically, IHC analyses of ST6GAL1 were limited by the lack of effective antibodies; however, numerous commercial antibodies have been developed in recent years. Our group extensively tested many of these and identified an antibody that is reliable for IHC (Swindall et al. 2013; Schultz et al. 2016) (as a side note, some antibodies previously used for IHC were not actually generated against ST6GAL1, such as the LN1 monoclonal antibody). Using IHC, we confirmed that ST6GAL1 is overexpressed in many epithelial cancers (Swindall et al. 2013; Schultz et al. 2016), and subsequently, we and others showed that high ST6GAL1 protein levels correlate with reduced patient survival (Liu et al. 2014; Schultz et al. 2016; Wei et al. 2016; Hsieh et al. 2017). To demonstrate differential expression of ST6GAL1 protein, images of IHC-stained normal and malignant pancreatic tissues are shown in Figure 1. No detectable ST6GAL1 expression is found in normal human pancreatic acinar cells, which are thought to comprise the cell of origin for most epithelial pancreatic cancers (Figure 1A). Interestingly, a subset of cells within the islets stains positively for ST6GAL1 (Figure 1A). Co-staining for ST6GAL1 and insulin in the murine pancreas revealed that ST6GAL1 is expressed in the β cells of the islets (Figure 1B). Specificity of the antibody is verified by the lack of ST6GAL1 staining in the islets of St6gal1 knockout (KO) mice (Figure 1B). In contrast to the negligible ST6GAL1 expression in normal acinar cells, ST6GAL1 is robustly expressed in the malignant epithelium. In ductal-like malignant lesions, ST6GAL1 expression is detected on the luminal side of the tumor cell nuclei, consistent with Golgi localization (Noda et al. 2018) (Figure 1C). In nonpolarized pancreatic cancer cells, ST6GAL1 staining often presents as multiple punctae (Figure 1D, arrowheads), which aligns with a long-standing literature describing Golgi disruption in tumor cells (Kellokumpu et al. 2002). In immune cells, ST6GAL1 typically stains as a single spot adjacent to the nucleus (Figure 1D, arrows). Tumor cells can be distinguished from immune/stromal cells by IHC because they have large, amorphous nuclei with prominent nucleoli. To confirm that ST6GAL1 is expressed in both tumor and immune populations, tissues were co-stained for ST6GAL1, a tumor-marker, Sox9 (Figure 1E, arrowheads), and an immune marker, CD45 (Figure 1E, arrows). These examples of ST6GAL1 staining patterns are provided to illustrate that by using IHC, one can selectively quantify tumor cell-specific expression of ST6GAL1. This enables analyses of: (i) ST6GAL1 levels in tumor vs. normal epithelium; (ii) differences in ST6GAL1 abundance across tumor stages and (iii) the relationship between the degree of ST6GAL1 expression and clinical outcomes such as metastatic progression, response to chemotherapy, and progression-free and overall survival.
Fig. 1.
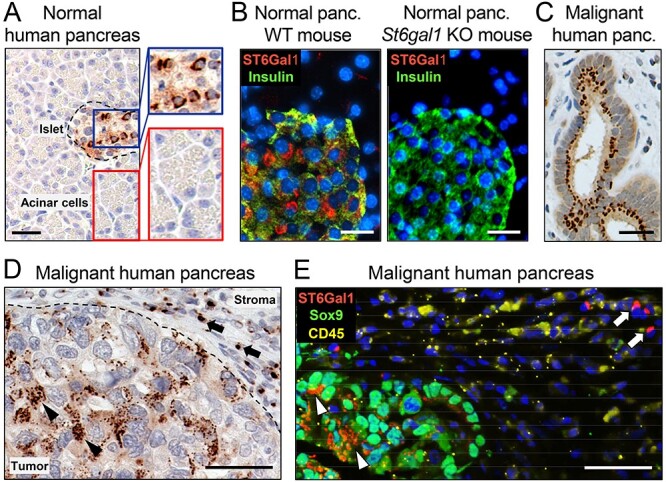
ST6GAL1 expression in normal and malignant pancreatic tissues. ST6GAL1 protein expression was evaluated by IHC in pancreatic tissues using a well-validated antibody, as in our prior studies (Swindall et al. 2013; Schultz et al. 2016). (A) No detectable ST6Gal-I expression was observed in normal pancreatic acinar cells (red inset); however, a subset of cells within the islets (blue inset) expresses ST6GAL1. Scale bar = 50 μM. (B) Left panel: co-staining for ST6GAL1 (red) and insulin (green) in wild-type mice confirms that ST6GAL1 is expressed in the β cells of the islets. Right panel, no ST6GAL1-positive cells are detected in the islets of St6gal1 KO mice, verifying specificity of the antibody. Scale bars = 25 μM. (C) In ductal-like malignant lesions, ST6GAL1expression is found on the luminal side of the nucleus, consistent with Golgi localization. Scale bar = 50 μM. (D) In non-ductal-like malignant lesions, ST6GAL1 staining often presents as multiple punctae within cancer cells (arrowheads), whereas staining in immune/stromal cells typically appears as a single spot adjacent to the nucleus (arrows). Scale bar = 50 μM. (E) ST6GAL1 (red) is co-expressed with Sox9 (green), a known tumor cell marker (arrowheads). ST6GAL1 is also found in immune cells (arrows), illustrated by co-staining with the immune marker CD45 (yellow). Scale bar = 50 μM. This figure is available in black and white in print and in colour at Glycobiology online.
ST6Gal-I is upregulated in cancer cells through gene amplification
Despite substantial evidence that ST6GAL1 is upregulated during carcinogenesis, the mechanisms that direct increased ST6GAL1 expression remain poorly defined. ST6GAL1 appears to be regulated through diverse conduits including genetic and epigenetic alterations, increased transcription and posttranscriptional/posttranslational processes (Harduin-Lepers et al. 2012). Among these diverse mechanisms, TCGA databases of genetic modifications in tumors suggest that one prominent event occurring in malignant cells is an increase in ST6GAL1 gene copy number. As reported, ST6GAL1 is pervasively amplified across many types of human cancers, but rarely deleted (Dorsett et al. 2019). Mutations in ST6GAL1 also occur, but these are less common than ST6GAL1 copy number gains (Figure 2). Within individual cancer cohorts, the percentage of tumors harboring ST6GAL1 gene amplifications can be quite high. For instance, a gain in ST6GAL1 copy number is found in 30–40% of lung squamous cell carcinomas and ~25% of ovarian serous adenocarcinomas (Figure 2 and Dorsett et al. 2019). The human ST6GAL1 gene lies within the well-known “3q26” amplicon, which spans from 3q26–29 (Wang et al. 2013; Fields et al. 2016; Davidson and Shanks 2017). This amplicon is one of the most frequently amplified chromosomal segments in epithelial cancers (Sonoda et al. 1997; Wang et al. 2013; Fields et al. 2016; Davidson and Shanks 2017). There are a number of well-known tumor-driver genes within this amplicon, including SOX2, ECT2, PRKCI (PKCι) and PIK3CA (catalytic subunit of PI3K) (Fields et al. 2016). A wealth of research has focused on the potency of this amplicon in promoting neoplastic development, chemoresistance and a more aggressive, stem-like tumor cell phenotype (Sonoda et al. 1997; Qian and Massion 2008; Wang et al. 2013; Fields et al. 2016; Davidson and Shanks 2017); however, the presence of ST6GAL1 within this amplicon has gone unnoticed. While it is possible that ST6GAL1 amplification is a bystander event, studies in animal models strongly support a causal role for ST6GAL1 in tumor initiation and progression (Schultz et al. 2016; Hsieh et al. 2017; Zhao et al. 2017). It is tempting to speculate that some of the tumorigenic activity commonly attributed to other genes within the 3q26 amplicon may be mediated, at least in part, by ST6GAL1.
Fig. 2.
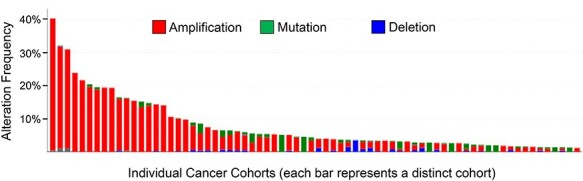
Gene amplification of ST6GAL1 in multiple human cancers. TCGA databases were evaluated using cBioportal (Cerami et al. 2012) for ST6GAL1 mutations or changes in ST6GAL1 copy number. Amplifications are denoted in red, mutations in green and deletions in blue. Each vertical bar represents a distinct cancer cohort, with the first three bars on the left representing lung squamous cell carcinomas, and the fourth bar, ovarian serous adenocarcinoma. Included in the figure are 15 different cancer types showing ST6GAL1 amplification. Original data are available from the TCGA website: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga This figure is available in black and white in print and in colour at Glycobiology online.
Transcription factors that promote ST6GAL1 expression
In addition to copy number alterations, ST6GAL1 expression is regulated through transcriptional mechanisms, both in cancer and normal cells (O'Hanlon et al. 1989; Paulson et al. 1989). Transcriptional regulation of ST6GAL1 is complex, encompassing the use of distinct promoters and alternative splicing events. This facilitates the expression of ST6GAL1 in a cell-type and tissue-specific manner. ST6GAL1 levels can vary dramatically in accordance with cell differentiation state or in response to specific stimuli (Dall'Olio 2000). There are multiple ST6GAL1 mRNA isoforms, driven by at least four different promoters (Svensson et al. 1990; Wang et al. 1990; Wen et al. 1992; Dall'Olio et al. 2004). The main promoters for ST6GAL1 are designated as P1, P2 and P3 (Figure 3). P1 is selectively utilized within the liver; transcription from P2 is restricted to B cells and P3 is a ubiquitous promoter (Xu et al. 2003; Dall'Olio et al. 2004; Lu and Gu 2015). A fourth promoter, P4, is only active in the mammary gland during lactation (Dalziel et al. 2001). Each of these promoters directs the expression of an mRNA species with a distinct 5′ untranslated region (UTR). The ST6GAL1 coding sequence and the 3’UTR are well-conserved among the isoforms, with the exception of isoform 1b, which encodes a soluble ST6GAL1 enzyme lacking the transmembrane domain.
Fig. 3.
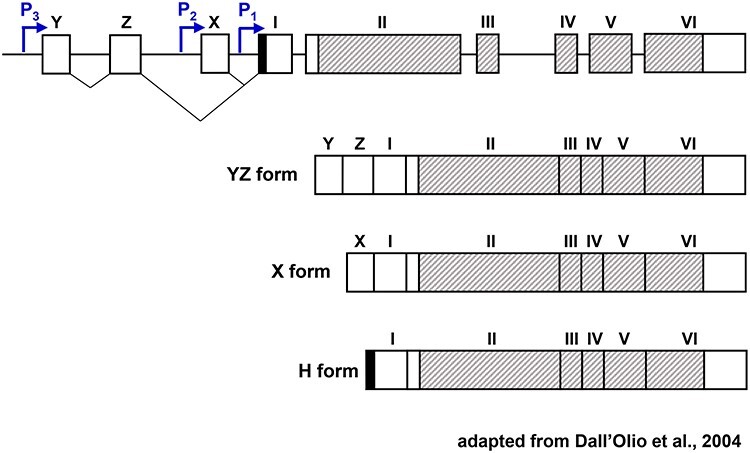
Promoters used to transcribe ST6GAL1 and their associated mRNA isoforms. Schematic representation of the genomic organization of ST6GAL1 and the three major ST6GAL1 transcripts. The three ST6GAL1 mRNA isoforms share exons I–VI. The shaded region denotes the coding sequence, which begins in Exon II and spans through part of Exon VI. The P3-driven ST6GAL1 isoform (“YZ form”) includes the untranslated exons Y and Z, whereas the P2-driven isoform (“X form”) includes the untranslated X exon. The P1-driven isoform, referred to as the “H form”, has a short sequence (solid) directly upstream of exon I. This figure was adapted from a manuscript by Dall'Olio et al. (2004). This figure is available in black and white in print and in colour at Glycobiology online.
In epithelial cancer cells, the most prevalent ST6GAL1 isoforms are transcribed from either the P1 or P3 promoter (Dall'Olio et al. 2000; Xu et al. 2003; Milflores-Flores et al. 2012). While both of these promoters have numerous consensus sequences for transcription factor binding, very few studies have focused on identifying the specific factors that mediate ST6GAL1 transcription. Xu et al. reported that the HNF1 transcription factor binds the ST6GAL1 P1 promoter to upregulate ST6GAL1 in colon cancer cells (Xu et al. 2003). HNF1 is also responsible for the transcription of the P1-driven isoform in nonmalignant hepatocytes (Svensson et al. 1992). Contrarily, the ST6GAL1 P3 promoter is preferentially utilized in ovarian cancer cells (Dorsett et al. 2019). In this study, it was shown that the stem cell transcription factor, Sox2, binds to regions proximal to the P3 promoter and consequently induces both ST6GAL1 expression and cell surface α2–6 sialylation (Dorsett et al. 2019). These findings may explain, in part, the strong association between ST6GAL1 and stem-like cancer cells, known as cancer stem cells (CSCs) or tumor-initiating cells. Multiple studies have demonstrated that ST6GAL1 activity confers all of the hallmark characteristics of a CSC including expression of CSC markers, self-renewal capability, tumor-initiating potential in limiting dilution assays, invasiveness and chemoresistance (Zhu et al. 2001; Christie et al. 2008; Schultz et al. 2013; Swindall et al. 2013; Chen et al. 2016; Schultz et al. 2016; Wei et al. 2016; Zhang et al. 2016; Britain et al. 2018; Chakraborty et al. 2018; Cui et al. 2018).
Notably, ST6GAL1 expression is also high in some nonmalignant stem and progenitor cell populations, as well as stem cell-associated tissue niches (Swindall et al. 2013; Dorsett et al. 2019; Alexander et al. 2020; Verge et al. 2020). ST6GAL1 expression is likewise enriched in a range of induced pluripotent stem cell (iPSC) lines, as compared with somatic cell lines (Wang et al. 2015). The transduction of somatic cells with the four Yamanaka factors (one of which is Sox2) to induce pluripotency causes a marked upregulation in ST6GAL1 (Tateno et al. 2011; Swindall et al. 2013; Wang et al. 2015). This is accompanied by dramatic changes in the surface glycome, with somatic cells expressing mostly α2–3 sialylation, and iPSCs, predominantly α2–6 sialylation (Hasehira et al. 2012). A functional role for ST6GAL1 in cellular reprogramming was confirmed by experiments showing that the transition to pluripotency is impeded by shRNA-mediated knockdown of ST6GAL1 (Wang et al. 2015). Contrasting with enriched expression in stem and cancer cells, levels of ST6GAL1 in differentiated epithelial cells are variable. ST6GAL1 protein expression is very low in tissues such as ovarian surface epithelium and pancreatic acini (Schultz et al. 2016), whereas hepatocytes, a specialized type of epithelia, constitutively express a considerable amount of ST6GAL1 (Bhide and Colley 2017). The molecular processes that repress the expression of ST6GAL1 in certain differentiated cell types are even less well-defined than those that turn on ST6GAL1 expression in cancer cells, but likely involve promoter methylation, at least in part. While this review is focused on mechanisms regulating ST6GAL1 expression during carcinogenesis, it is worth noting that the ST6GAL1 P3 promoter has a CpG island that is hypermethylated in accordance with cell differentiation state. As an example, Kaburagi et al. reported that ST6GAL1 downregulation during the differentiation of pre-adipocytes into adipocytes occurs through P3 promoter methylation (Kaburagi et al. 2017).
In keeping with a putative function in stem-like cells, ST6GAL1 activity promotes EMT in several cell models (Lu et al. 2014; Britain et al. 2020). The process of EMT is central to the acquisition of metastatic potential, and EMT also serves as a mechanism for generating CSCs (Ye and Weinberg 2015). In a TGFβ-induced EMT model, Lu et al. determined that ST6GAL1 was the only sialylation-related enzyme to show significant upregulation in response to TGFβ, as compared with 10 other sialyltransferases, the CMP-sialic acid transporter and the four Neu family member sialidases (Lu et al. 2014). Furthermore, knockdown of ST6GAL1 expression inhibited TGFβ-induced EMT, confirming ST6GAL1’s role in promoting mesenchymal cell properties. In this same model, ST6GAL1 upregulation was dependent upon the SP-1 transcription factor. Transcriptional regulation of ST6GAL1 by SP-1 has been observed in several cancer types. SP-1 activates transcription from the ST6GAL1 P3 promoter in HL60 leukemia cells (Taniguchi et al. 1998), whereas the ST6GAL1 P1 promoter is utilized by SP-1 in HepG2 hepatocellular carcinoma cells (Milflores-Flores et al. 2012).
Regulation of ST6GAL1 by epigenetic mechanisms
A variety of investigations have demonstrated that cancer cells exhibit changes in ST6GAL1 DNA methylation. Gene methylation can have either positive or negative effects on gene expression, depending upon proximity to the transcriptional start site, as well as other factors (Laurent et al. 2010; Ronneberg et al. 2011). Hypermethylation of ST6GAL1 has been reported in breast, glioblastoma and bladder cancer (Antony et al. 2014; Fleischer et al. 2014; Kroes and Moskal 2016), whereas ST6GAL1 is hypomethylated in lung cancer (Vojta et al. 2016). In glioblastoma and bladder cancer cells, hypermethylation occurs within the CpG island of the P3 promoter, and this corresponds with decreased expression of ST6GAL1 (Antony et al. 2014; Kroes and Moskal 2016).
ST6GAL1 expression is also regulated by the activity of microRNAs (miRs). The ST6GAL1 3’UTR has many consensus sequences for binding of diverse miRs, although to date, only a few studies have shown direct interaction of miRs with the ST6GAL1 transcript. Han et al. demonstrated that miR-9 is more highly expressed in nonmetastatic vs. metastatic hepatocellular carcinoma cells, and forced expression of miR-9 in the metastatic line suppressed ST6GAL1 expression and cell invasive capability (Han et al. 2018). Minami et al. (2013) reported that the inhibition of ST6GAL1 expression by miR-199a promoted oncogenic signaling because the loss of α2–6 sialylation on Nectin-like molecule 2 (Necl2) ablated its ability to inhibit the ErbB2/ErbB3 complex. In breast cancer cells, repression of ST6GAL1 expression by miR-214-3p led to enhanced breast cancer cell migration and invasion (Tao et al. 2019). Finally, miR-200, a tumor suppressor miR that inhibits EMT, was shown to reduce ST6GAL1 expression, although it was not determined whether this mechanism was direct or indirect (Schliekelman et al. 2011). This latter study employed a broad proteomics screen to ascertain that metastatic lung cancer cells with low miR-200 levels had high ST6GAL1 expression (along with other proteins) and a mesenchymal phenotype, as compared with nonmetastatic cells. Forced expression of miR-200 reduced ST6GAL1 expression and reverted EMT.
Posttranscriptional and posttranslational regulation of ST6GAL1
Other than miR-dependent mechanisms that hinder the translation of ST6GAL1, not much is known about posttranscriptional regulation. On the other hand, there is substantial evidence of posttranslational regulation of ST6GAL1 through mechanisms including oligomerization, subcellular localization and proteolytic processing. Many years ago, the Colley laboratory examined the activity of two ST6GAL1 isoforms, STtyr and STcys, which have a single amino acid difference in the catalytic domain (Chen et al. 2000; Bhide and Colley 2017). It was shown that the STcys form is particularly prone to oligomerization, and this, in turn, facilitates ST6GAL1 retention within the Golgi (Chen et al. 2000). Golgi localization of ST6GAL1 is also enhanced by its association with GOLPH3, a phosphoprotein which is upregulated in cancer, and has oncogenic activity (Welch and Munro 2019). In a study by Gu’s group, ST6GAL1 was identified as a direct binding partner for GOLPH3 (Isaji et al. 2014). The formation of the GOLPH3/ST6GAL1 complex was critical for ST6GAL1-mediated sialylation of the β1 integrin and β1 integrin-driven cell migration (Isaji et al. 2014). Counterbalancing mechanisms that retain ST6GAL1 within the Golgi, the proteolytic cleavage and release of ST6GAL1 from the Golgi is also biologically important. ST6GAL1 is cleaved by the BACE1 β-secretase, leading to the secretion of ST6GAL1 (Kitazume et al. 2001). Secreted ST6GAL1 is enzymatically active in the extracellular milieu, if provided with a source of CMP-sialic acid. Work from the Lau laboratory provided a seminal advance by determining that platelets release CMP-sialic acid and that this source of sialic acid can be utilized by extrinsic ST6GAL1 to sialylate the surface of nearby cells (Lee et al. 2014). Although not yet tested, this mechanism could be operative within the tumor microenvironment, given that thrombosis is a common feature of malignant tissues. In addition to secreted, soluble ST6GAL1, both cleaved and membrane-bound forms of ST6GAL1 are found within tumor-derived extracellular vesicles (EVs) including exosomes (Zhang et al. 2019). The presence of active ST6GAL1 in the extracellular fluid or tumor-derived EVs points to the intriguing possibility that the ST6GAL1 released from one tumor cell could mediate the sialylation, and therefore, phenotypic remodeling, of adjacent tumor or stromal cells.
Oncogenic signaling pathways that induce ST6GAL1 expression
As another knowledge gap, limited research has been devoted to identifying intracellular signaling networks that regulate ST6GAL1 expression. Such research would be beneficial for predicting ST6GAL1 levels in tumor cell subsets that harbor genetic and/or transcriptomic alterations in known cancer-associated pathways such as ras, PI3K/AKT, etc. This information would enhance ST6GAL1’s potential to serve as a biomarker or prognostic indicator. While studies in this area are scant, the ras pathway has long been observed to induce ST6GAL1 expression. Activating mutations in ras constitute one of the most prevalent oncogenic events, occurring in ~25% of all human cancers (Hobbs et al. 2016) and ranging up to 93% in malignancies such as pancreatic cancer (Lennerz and Stenzinger 2015). Nearly 30 years ago, it was observed that ST6GAL1 expression was elevated in various fibroblast cell lines following transfection with the ras oncogene (Easton et al. 1991; Le Marer et al. 1992; Vandamme et al. 1992). This finding was later corroborated in epithelial cancer cell models (Seales et al. 2003). All three of the ras isoforms, H-, N- and K-ras, stimulate ST6Gal-I expression (Easton et al. 1991; Le Marer et al. 1992; Vandamme et al. 1992; Seales et al. 2003; Dalziel et al. 2004). Piller’s group further showed that oncogenic ras signals through a RalGEF pathway to induce the expression of the P3-driven ST6GAL1 isoform (Dalziel et al. 2004). This same study quantified the expression of multiple α2–6 and α2–3 sialyltransferases in ras-transfected cells and determined that ST6GAL1 was the principal enzyme upregulated by oncogenic ras.
More recently, work by Johansson et al. implicated another key downstream effector of ras, the B-raf kinase, in regulating ST6Gal-I expression in melanoma cells (Johansson et al. 2007). This group identified a gene signature that could reliably predict whether a tumor had activating mutations in BRAF. ST6GAL1 was one of these signature genes (and the only glycosyltransferase) consistently upregulated in melanoma cells with mutated BRAF. Taken together, these studies provide strong evidence pointing to the ras pathway as a critical signaling node responsible for ST6GAL1 overexpression in cancer cells. However, the specific molecular players that induce ST6GAL1 expression downstream of ras remain to be elucidated. While Piller’s group confirmed that ras stimulates ST6GAL1 transcription, the specific transcription factor mediating this event was not determined. Moreover, ras signaling may also modulate ST6GAL1 expression through mechanisms independent of transcriptional activation. Given the preponderance of activated ras signaling in most human cancers, studies are needed to define the mechanistic link between ras activation and ST6GAL1 upregulation. Additionally, there is a need to identify other intracellular signaling cascades that may contribute to ST6GAL1 upregulation during malignant transformation.
Conclusions
Cancer biologists have invested significant effort into elucidating mechanisms that control the expression of proteins with oncogenic function. However, studies of molecular pathways that direct the expression of tumor-promoting glycogenes, such as ST6GAL1, have lagged behind. While it is known that ST6GAL1 is dynamically regulated, there is a paucity of research focused on the upstream events that induce or repress ST6GAL1 expression. A comprehensive knowledge of ST6GAL1 regulation would foster a better understanding of ST6GAL1’s function in select cell types, both normal and malignant. Cancer cells are notoriously heterogeneous, and certain populations, such as CSCs, play an inordinate role in cancer progression, recurrence and metastasis. Defining the genetic, epigenetic, transcriptional and posttranscriptional/posttranslational events that promote ST6GAL1 expression is critical for predicting which cancer types, or subclones within tumors, exhibit upregulated ST6GAL1 expression and activity. Additionally, elucidating ST6GAL1 regulatory mechanisms would provide insight into potential therapeutic avenues for suppressing ST6GAL1 expression in cancer cells. The overall goal of this review is to summarize current knowledge regarding the molecular events that modulate ST6GAL1 expression in neoplastic cells and highlight the need for further research into the regulation of ST6GAL1, as well as other glycosyltransferases and glycosidases that play important functional roles in cancer biology.
Funding
National Institutes of Health (CA233581 to S.L.B., CA225177 to S.L.B).
Contributor Information
Kaitlyn A Dorsett, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Michael P Marciel, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Jihye Hwang, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Katherine E Ankenbauer, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Nikita Bhalerao, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Susan L Bellis, Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Abbreviations
CSC, Cancer Stem Cell; EMT, Epithelial to Mesenchymal Transition; EV, Extracellular Vesicle; IHC, Immunohistochemistry; iPSC, inducted Pluripotent Stem Cell; miR, microRNA; ST6GAL1, β-galactoside α-2,6 sialyltransferase 1; ST6GAL2, β-galactoside α-2,6 sialyltransferase 2; TCGA, The Cancer Genome Atlas; TGFβ, Transforming Growth Factor β; TIC, Tumor-Initiating Cells; UTR, Untranslated Region.
References
- Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S, Darvishian F, et al. 2017. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell. 31:804, e807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander KL, Serrano CA, Chakraborty A, Nearing M, Council LN, Riquelme A, Garrido M, Bellis SL, Smythies LE, Smith PD. 2020. Modulation of glycosyltransferase ST6Gal-I in gastric cancer-derived organoids disrupts homeostatic epithelial cell turnover. J Biol Chem. 295:14153–14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony P, Rose M, Heidenreich A, Knuchel R, Gaisa NT, Dahl E. 2014. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer. 14:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenwaldt A, Laubli H. 2019. The sialoglycan-Siglec glyco-immune checkpoint - a target for improving innate and adaptive anti-cancer immunity. Expert Opin Ther Targets. 23:839–853. [DOI] [PubMed] [Google Scholar]
- Bhide GP, Colley KJ. 2017. Sialylation of N-glycans: Mechanism, cellular compartmentalization and function. Histochem Cell Biol. 147:149–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britain CM, Dorsett KA, Bellis SL. 2017. Glycosyltransferase ST6Gal-I protects tumor cells against serum growth factor withdrawal by enhancing survival signaling and proliferative potential. J Biol Chem. 292:4663–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britain CM, Holdbrooks AT, Anderson JC, Willey CD, Bellis SL. 2018. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J Ovarian Res. 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britain CM, Bhalerao N, Silva AD, Chakraborty A, Buchsbaum DJ, Crowley MR, Crossman DK, Edward YJK, Bellis SL. 2020. Glycosyltransferase ST6Gal-I promotes the epithelial to mesenchymal transition in pancreatic cancer cells. J. Biol Chem. Nov 4 epub ahead of priDK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Dorsett KA, Trummell HQ, Yang ES, Oliver PG, Bonner JA, Buchsbaum DJ, Bellis SL. 2018. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J Biol Chem. 293:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ma J, Lazic A, Backovic M, Colley KJ. 2000. Formation of insoluble oligomers correlates with ST6Gal I stable localization in the golgi. J Biol Chem. 275:13819–13826. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang L, Zhao Y, Yuan S, Wu Q, Zhu X, Niang B, Wang S, Zhang J. 2016. ST6Gal-I modulates docetaxel sensitivity in human hepatocarcinoma cells via the p38 MAPK/caspase pathway. Oncotarget. 7:51955–51964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiricolo M, Malagolini N, Bonfiglioli S, Dall'Olio F. 2006. Phenotypic changes induced by expression of beta-galactoside alpha2,6 sialyltransferase I in the human colon cancer cell line SW948. Glycobiology. 16:146–154. [DOI] [PubMed] [Google Scholar]
- Christie DR, Shaikh FM, Lucas JA 4th, Lucas JA 3rd, Bellis SL. 2008. ST6Gal-I expression in ovarian cancer cells promotes an invasive phenotype by altering integrin glycosylation and function. J Ovarian Res. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci DO, Cerliani JP, Dalotto-Moreno T, Mendez-Huergo SP, Mascanfroni ID, Dergan-Dylon S, Toscano MA, Caramelo JJ, Garcia-Vallejo JJ, Ouyang J, et al. 2014. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 156:744–758. [DOI] [PubMed] [Google Scholar]
- Cui H, Yang S, Jiang Y, Li C, Zhao Y, Shi Y, Hao Y, Qian F, Tang B, Yu P. 2018. The glycosyltransferase ST6Gal-I is enriched in cancer stem-like cells in colorectal carcinoma and contributes to their chemo-resistance. Clin Transl Oncol. 20:1175–1184. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F. 2000. The sialyl-alpha2,6-lactosaminyl-structure: Biosynthesis and functional role. Glycoconj J. 17:669–676. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Chiricolo M. 2001. Sialyltransferases in cancer. Glycoconj J. 18:841–850. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Chiricolo M, Ceccarelli C, Minni F, Marrano D, Santini D. 2000. Beta-galactoside alpha2,6 sialyltransferase in human colon cancer: Contribution of multiple transcripts to regulation of enzyme activity and reactivity with Sambucus nigra agglutinin. Int J Cancer. 88:58–65. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Chiricolo M, D'Errico A, Gruppioni E, Altimari A, Fiorentino M, Grigioni WF. 2004. Expression of beta-galactoside alpha2,6 sialyltransferase and of alpha2,6-sialylated glycoconjugates in normal human liver, hepatocarcinoma, and cirrhosis. Glycobiology. 14:39–49. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Malagolini N, di Stefano G, Minni F, Marrano D, Serafini-Cessi F. 1989. Increased CMP-NeuAc:Gal beta 1,4GlcNAc-R alpha 2,6 sialyltransferase activity in human colorectal cancer tissues. Int J Cancer. 44:434–439. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Malagolini N, Trinchera M, Chiricolo M. 2014. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim Biophys Acta. 1840:2752–2764. [DOI] [PubMed] [Google Scholar]
- Dall'Olio F, Trere D. 1993. Expression of alpha 2,6-sialylated sugar chains in normal and neoplastic colon tissues. Detection by digoxigenin-conjugated Sambucus nigra agglutinin. Eur J Histochem. 37:257–265. [PubMed] [Google Scholar]
- Dalziel M, Dall'Olio F, Mungul A, Piller V, Piller F. 2004. Ras oncogene induces beta-galactoside alpha2,6-sialyltransferase (ST6Gal I) via a RalGEF-mediated signal to its housekeeping promoter. Eur J Biochem. 271:3623–3634. [DOI] [PubMed] [Google Scholar]
- Dalziel M, Huang RY, Dall'Olio F, Morris JR, Taylor-Papadimitriou J, Lau JT. 2001. Mouse ST6Gal sialyltransferase gene expression during mammary gland lactation. Glycobiology. 11:407–412. [DOI] [PubMed] [Google Scholar]
- Davidson MA, Shanks EJ. 2017. 3q26-29 amplification in head and neck squamous cell carcinoma: A review of established and prospective oncogenes. FEBS J. 284:2705–2731. [DOI] [PubMed] [Google Scholar]
- Dawson G, Moskal JR, Dawson SA. 2004. Transfection of 2,6 and 2,3-sialyltransferase genes and GlcNAc-transferase genes into human glioma cell line U-373 MG affects glycoconjugate expression and enhances cell death. J Neurochem. 89:1436–1444. [DOI] [PubMed] [Google Scholar]
- Dorsett KA, Jones RB, Ankenbauer KE, Hjelmeland AB, Bellis SL. 2019. Sox2 promotes expression of the ST6Gal-I glycosyltransferase in ovarian cancer cells. J Ovarian Res. 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton EW, Bolscher JG, van den Eijnden DH. 1991. Enzymatic amplification involving glycosyltransferases forms the basis for the increased size of asparagine-linked glycans at the surface of NIH 3T3 cells expressing the N-ras proto-oncogene. J Biol Chem. 266:21674–21680. [PubMed] [Google Scholar]
- Fields AP, Justilien V, Murray NR. 2016. The chromosome 3q26 OncCassette: A multigenic driver of human cancer. Adv Biological Reg. 60:47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer T, Edvardsen H, Solvang HK, Daviaud C, Naume B, Borresen-Dale AL, Kristensen VN, Tost J. 2014. Integrated analysis of high-resolution DNA methylation profiles, gene expression, germline genotypes and clinical end points in breast cancer patients. Int J Cancer. 134:2615–2625. [DOI] [PubMed] [Google Scholar]
- Garnham R, Scott E, Livermore KE, Munkley J. 2019. ST6GAL1: A key player in cancer. Oncol Lett. 18:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretschel S, Haensch W, Schlag PM, Kemmner W. 2003. Clinical relevance of sialyltransferases ST6GAL-I and ST3GAL-III in gastric cancer. Oncology. 65:139–145. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell. 140:883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann U, Nerlich C, Rein T, Zettlmeissl G. 1990. Complete cDNA sequence encoding human beta-galactoside alpha-2,6-sialyltransferase. Nucleic Acids Res. 18:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Liu Y, Fu X, Zhang Q, Huang H, Zhang C, Li W, Zhang J. 2018. miR-9 inhibits the metastatic ability of hepatocellular carcinoma via targeting beta galactoside alpha-2,6-sialyltransferase 1. J Physiol Biochem. 74:491–501. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Krzewinski-Recchi MA, Colomb F, Foulquier F, Groux-Degroote S, Delannoy P. 2012. Sialyltransferases functions in cancers. Front Biosci (Elite Ed). 4:499–515. [DOI] [PubMed] [Google Scholar]
- Hasehira K, Tateno H, Onuma Y, Ito Y, Asashima M, Hirabayashi J. 2012. Structural and quantitative evidence for dynamic glycome shift on production of induced pluripotent stem cells. Mol Cell Proteomics. 11:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar M, Krzewinski-Recchi MA, Hornez L, Verdiere A, Harduin-Lepers A, Bonneterre J, Delannoy P, Peyrat JP. 2003. Prognostic value of tumoral sialyltransferase expression and circulating E-selectin concentrations in node-negative breast cancer patients. Int J Biol Markers. 18:116–122. [DOI] [PubMed] [Google Scholar]
- Hobbs GA, Der CJ, Rossman KL. 2016. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 129:1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdbrooks AT, Britain CM, Bellis SL. 2018. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 293:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CC, Shyr YM, Liao WY, Chen TH, Wang SE, Lu PC, Lin PY, Chen YB, Mao WY, Han HY, et al. 2017. Elevation of beta-galactoside alpha2,6-sialyltransferase 1 in a fructoseresponsive manner promotes pancreatic cancer metastasis. Oncotarget. 8:7691–7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaji T, Im S, Gu W, Wang Y, Hang Q, Lu J, Fukuda T, Hashii N, Takakura D, Kawasaki N, et al. 2014. An oncogenic protein Golgi phosphoprotein 3 up-regulates cell migration via sialylation. J Biol Chem. 289:20694–20705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Pavey S, Hayward N. 2007. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 20:216–221. [DOI] [PubMed] [Google Scholar]
- Jones RB, Dorsett KA, Hjelmeland AB, Bellis SL. 2018. The ST6Gal-I sialyltransferase protects tumor cells against hypoxia by enhancing HIF-1alpha signaling. J Biol Chem. 293:5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Wang Y, Aryal RP, Lehoux SD, Ding X, Kudelka MR, Cutler C, Zeng J, Wang J, Sun X, et al. 2013. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 7:618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YR, Park JJ, Jin YB, Cao YJ, Park MJ, Kim EJ, Lee M. 2016. Silencing of ST6Gal I enhances colorectal cancer metastasis by down-regulating KAI1 via exosome-mediated exportation and thereby rescues integrin signaling. Carcinogenesis. 37:1089–1097. [DOI] [PubMed] [Google Scholar]
- Kaburagi T, Kizuka Y, Kitazume S, Taniguchi N. 2017. The inhibitory role of alpha2,6-sialylation in Adipogenesis. J Biol Chem. 292:2278–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Yamamoto H, Kersey DS, Colley KJ, Leestma JE, Moskal JR. 1996. The expression of gal beta 1,4GlcNAc alpha 2,6 sialyltransferase and alpha 2,6-linked sialoglycoconjugates in human brain tumors. Acta Neuropathol. 91:284–292. [DOI] [PubMed] [Google Scholar]
- Kellokumpu S, Sormunen R, Kellokumpu I. 2002. Abnormal glycosylation and altered Golgi structure in colorectal cancer: Dependence on intra-Golgi pH. FEBS Lett. 516:217–224. [DOI] [PubMed] [Google Scholar]
- Kim J, Bae JS. 2016. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. 2016:6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y. 2001. Alzheimer's beta-secretase, beta-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc Natl Acad Sci U S A. 98:13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes RA, Moskal JR. 2016. The role of DNA methylation in ST6Gal1 expression in gliomas. Glycobiology. 26:1271–1283. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res. 20:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marer N, Laudet V, Svensson EC, Cazlaris H, Van Hille B, Lagrou C, Stehelin D, Montreuil J, Verbert A, Delannoy P. 1992. The c-ha-ras oncogene induces increased expression of beta-galactoside alpha-2, 6-sialyltransferase in rat fibroblast (FR3T3) cells. Glycobiology. 2:49–56. [DOI] [PubMed] [Google Scholar]
- Lee M, Lee HJ, Bae S, Lee YS. 2008. Protein sialylation by sialyltransferase involves radiation resistance. Mol Cancer Res. 6:1316–1325. [DOI] [PubMed] [Google Scholar]
- Lee M, Park JJ, Lee YS. 2010. Adhesion of ST6Gal I-mediated human colon cancer cells to fibronectin contributes to cell survival by integrin beta1-mediated paxillin and AKT activation. Oncol Rep. 23:757–761. [PubMed] [Google Scholar]
- Lee MM, Nasirikenari M, Manhardt CT, Ashline DJ, Hanneman AJ, Reinhold VN, Lau JT. 2014. Platelets support extracellular sialylation by supplying the sugar donor substrate. J Biol Chem. 289:8742–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoux S, Groux-Degroote S, Cazet A, Dhaenens CM, Maurage CA, Caillet-Boudin ML, Delannoy P, Krzewinski-Recchi MA. 2010. Transcriptional regulation of the human ST6GAL2 gene in cerebral cortex and neuronal cells. Glycoconj J. 27:99–114. [DOI] [PubMed] [Google Scholar]
- Lennerz JK, Stenzinger A. 2015. Allelic ratio of KRAS mutations in pancreatic cancer. Oncologist. 20:e8–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ding J. 2019. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell. 10:550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lise M, Belluco C, Perera SP, Patel R, Thomas P, Ganguly A. 2000. Clinical correlations of alpha2,6-sialyltransferase expression in colorectal cancer patients. Hybridoma. 19:281–286. [DOI] [PubMed] [Google Scholar]
- Liu HO, Wu Q, Liu WS, Liu YD, Fu Q, Zhang WJ, Xu L, Xu JJ. 2014. ST6Gal-I predicts postoperative clinical outcome for patients with localized clear-cell renal cell carcinoma. Asian Pac J Cancer Prev. 15:10217–10223. [DOI] [PubMed] [Google Scholar]
- Lu J, Gu J. 2015. Significance of beta-Galactoside alpha2,6 Sialyltranferase 1 in cancers. Molecules. 20:7509–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Isaji T, Im S, Fukuda T, Hashii N, Takakura D, Kawasaki N, Gu J. 2014. Beta-Galactoside alpha2,6-sialyltranferase 1 promotes transforming growth factor-beta-mediated epithelial-mesenchymal transition. J Biol Chem. 289:34627–34641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milflores-Flores L, Millan-Perez L, Santos-Lopez G, Reyes-Leyva J, Vallejo-Ruiz V. 2012. Characterization of P1 promoter activity of the beta-galactoside alpha2,6-sialyltransferase I gene (siat 1) in cervical and hepatic cancer cell lines. J Biosci. 37:259–267. [DOI] [PubMed] [Google Scholar]
- Minami A, Shimono Y, Mizutani K, Nobutani K, Momose K, Azuma T, Takai Y. 2013. Reduction of the ST6 beta-galactosamide alpha-2,6-sialyltransferase 1 (ST6GAL1)-catalyzed sialylation of nectin-like molecule 2/cell adhesion molecule 1 and enhancement of ErbB2/ErbB3 signaling by microRNA-199a. J Biol Chem. 288:11845–11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Chandra S, Mandal C. 2010. Elevated mRNA level of hST6Gal I and hST3Gal V positively correlates with the high risk of pediatric acute leukemia. Leuk Res. 34:463–470. [DOI] [PubMed] [Google Scholar]
- Munkley J. 2016. The role of Sialyl-Tn in cancer. Int J Mol Sci. 17:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J, Elliott DJ. 2016. Hallmarks of glycosylation in cancer. Oncotarget. 7:35478–35489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J, Vodak D, Livermore KE, James K, Wilson BT, Knight B, McCullagh P, McGrath J, Crundwell M, Harries LW, et al. 2016. Glycosylation is an androgen-regulated process essential for prostate cancer cell viability. EBioMedicine. 8:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Okayama H, Tachibana K, Sakamoto W, Saito K, Thar Min AK, Ashizawa M, Nakajima T, Aoto K, Momma T, et al. 2018. Glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin Cancer Res. 24:4468–4481. [DOI] [PubMed] [Google Scholar]
- O'Hanlon TP, Lau KM, Wang XC, Lau JT. 1989. Tissue-specific expression of beta-galactoside alpha-2,6-sialyltransferase. Transcript heterogeneity predicts a divergent polypeptide. J Biol Chem. 264:17389–17394. [PubMed] [Google Scholar]
- Park JJ, Lee M. 2013. Increasing the alpha 2, 6 sialylation of glycoproteins may contribute to metastatic spread and therapeutic resistance in colorectal cancer. Gut Liver. 7:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Yi JY, Jin YB, Lee YJ, Lee JS, Lee YS, Ko YG, Lee M. 2012. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 83:849–857. [DOI] [PubMed] [Google Scholar]
- Paulson JC, Weinstein J, Schauer A. 1989. Tissue-specific expression of sialyltransferases. J Biol Chem. 264:10931–10934. [PubMed] [Google Scholar]
- Pearce OM, Laubli H. 2016. Sialic acids in cancer biology and immunity. Glycobiology. 26:111–128. [DOI] [PubMed] [Google Scholar]
- Pinho SS, Reis CA. 2015. Glycosylation in cancer: Mechanisms and clinical implications. Nat Rev Cancer. 15:540–555. [DOI] [PubMed] [Google Scholar]
- Qian J, Massion PP. 2008. Role of chromosome 3q amplification in lung cancer. J Thorac Oncol. 3:212–215. [DOI] [PubMed] [Google Scholar]
- Recchi MA, Hebbar M, Hornez L, Harduin-Lepers A, Peyrat JP, Delannoy P. 1998. Multiplex reverse transcription polymerase chain reaction assessment of sialyltransferase expression in human breast cancer. Cancer Res. 58:4066–4070. [PubMed] [Google Scholar]
- Rodrigues E, Macauley MS. 2018. Hypersialylation in cancer: Modulation of inflammation and therapeutic opportunities. Cancers. 10:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JG, Balmana M, Macedo JA, Pocas J, Fernandes A, de-Freitas JCM Jr, Pinho SS, Gomes J, Magalhaes A, Gomes C, et al. 2018. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 333:46–57. [DOI] [PubMed] [Google Scholar]
- Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, Potapenko I, Nebdal D, Daviaud C, Gut I, Bukholm I, et al. 2011. Methylation profiling with a panel of cancer related genes: Association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 5:61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata T, Roth J, Zuber C, Stamm B, Heitz PU. 1991. Expression of alpha 2,6-linked sialic acid residues in neoplastic but not in normal human colonic mucosa. A lectin-gold cytochemical study with Sambucus nigra and Maackia amurensis lectins. Am J Pathol. 139:1435–1448. [PMC free article] [PubMed] [Google Scholar]
- Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, Wong CH, Wang H, Ungewiss C, Ahn YH, et al. 2011. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 71:7670–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell A, Schmidl C, Herr W, Siska PJ. 2018. The peripheral and Intratumoral immune cell landscape in cancer patients: A proxy for tumor biology and a tool for outcome prediction. Biomedicines. 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, Conner MG, Arend RC, Yoon KJ, Klug CA, et al. 2016. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 76:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Swindall AF, Bellis SL. 2012. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 31:501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Swindall AF, Wright JW, Sztul ES, Landen CN, Bellis SL. 2013. ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J Ovarian Res. 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL. 2005. Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65:4645–4652. [DOI] [PubMed] [Google Scholar]
- Seales EC, Jurado GA, Singhal A, Bellis SL. 2003. Ras oncogene directs expression of a differentially sialylated, functionally altered beta1 integrin. Oncogene. 22:7137–7145. [DOI] [PubMed] [Google Scholar]
- Sethi MK, Kim H, Park CK, Baker MS, Paik YK, Packer NH, Hancock WS, Fanayan S, Thaysen-Andersen M. 2015. In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology. 25:1064–1078. [DOI] [PubMed] [Google Scholar]
- Skacel PO, Edwards AJ, Harrison CT, Watkins WM. 1991. Enzymic control of the expression of the X determinant (CD15) in human myeloid cells during maturation: The regulatory role of 6-sialytransferase. Blood. 78:1452–1460. [PubMed] [Google Scholar]
- Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, Testa JR. 1997. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer. 20:320–328. [PubMed] [Google Scholar]
- Svensson EC, Conley PB, Paulson JC. 1992. Regulated expression of alpha 2,6-sialyltransferase by the liver-enriched transcription factors HNF-1, DBP, and LAP. J Biol Chem. 267:3466–3472. [PubMed] [Google Scholar]
- Svensson EC, Soreghan B, Paulson JC. 1990. Organization of the beta-galactoside alpha 2,6-sialyltransferase gene. Evidence for the transcriptional regulation of terminal glycosylation. J Biol Chem. 265:20863–20868. [PubMed] [Google Scholar]
- Swindall AF, Bellis SL. 2011. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem. 286:22982–22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindall AF, Londono-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. 2013. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 73:2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Tsuji S, Tsujimoto M. 2002. Characterization of the second type of human beta-galactoside alpha 2,6-sialyltransferase (ST6Gal II), which sialylates Galbeta 1,4GlcNAc structures on oligosaccharides preferentially. Genomic analysis of human sialyltransferase genes. J Biol Chem. 277:45719–45728. [DOI] [PubMed] [Google Scholar]
- Taniguchi A, Higai K, Hasegawa Y, Utsumi K, Matsumoto K. 1998. Differentiation elicits negative regulation of human beta-galactoside alpha2,6-sialyltransferase at the mRNA level in the HL-60 cell line. FEBS Lett. 441:191–194. [DOI] [PubMed] [Google Scholar]
- Taniguchi N, Kizuka Y. 2015. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res. 126:11–51. [DOI] [PubMed] [Google Scholar]
- Tao Y, Zhao Z, Ma J, Dong L, Liang Y, Li S, Mao Y, Li Y, Zhang Y. 2019. MiR-214-3p regulates the viability, invasion, migration and EMT of TNBC cells by targeting ST6GAL1. Cytotechnology. 71:1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Toyota M, Saito S, Onuma Y, Ito Y, Hiemori K, Fukumura M, Matsushima A, Nakanishi M, Ohnuma K, et al. 2011. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J Biol Chem. 286:20345–20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu CT, Mahal LK. 2020. Sweet control: MicroRNA regulation of the Glycome. Biochemistry. 59:3098–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajaria BN, Patel KR, Begum R, Patel PS. 2016. Sialylation: An avenue to target cancer cells. Pathol Oncol Res. 22:443–447. [DOI] [PubMed] [Google Scholar]
- Vandamme V, Cazlaris H, Le Marer N, Laudet V, Lagrou C, Verbert A, Delannoy P. 1992. Comparison of sialyl- and alpha-1,3-galactosyltransferase activity in NIH3T3 cells transformed with ras oncogene: Increased beta-galactoside alpha-2,6-sialyltransferase. Biochimie. 74:89–99. [DOI] [PubMed] [Google Scholar]
- Varki A, Kannagi R, Toole V, Stanley P. 2015. Glycosylation changes in cancer. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of glycobiology. 2nd ed: Cold Spring Harbor (NY). p. 597–609. [Google Scholar]
- Vazquez-Martin C, Gil-Martin E, Fernandez-Briera A. 2005. Elevation of ST6Gal I activity in malignant and transitional tissue in human colorectal cancer. Oncology. 69:436–444. [DOI] [PubMed] [Google Scholar]
- Venturi G, Gomes Ferreira I, Pucci M, Ferracin M, Malagolini N, Chiricolo M, Dall'Olio F. 2019. Impact of sialyltransferase ST6GAL1 overexpression on different colon cancer cell types. Glycobiology. 29:684–695. [DOI] [PubMed] [Google Scholar]
- Verge C, Bouchatal A, Chirat F, Guerardel Y, Maftah A, Petit JM. 2020. Involvement of ST6Gal I-mediated alpha2,6 sialylation in myoblast proliferation and differentiation. FEBS Open Bio. 10:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Samarzija I, Bockor L, Zoldos V. 2016. Glyco-genes change expression in cancer through aberrant methylation. Biochim Biophys Acta. 1860:1776–1785. [DOI] [PubMed] [Google Scholar]
- Wang J, Qian J, Hoeksema MD, Zou Y, Espinosa AV, Rahman SM, Zhang B, Massion PP. 2013. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clin Cancer Res. 19:5580–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PH, Lee WL, Juang CM, Yang YH, Lo WH, Lai CR, Hsieh SL, Yuan CC. 2005. Altered mRNA expressions of sialyltransferases in ovarian cancers. Gynecol Oncol. 99:631–639. [DOI] [PubMed] [Google Scholar]
- Wang PH, Lee WL, Lee YR, Juang CM, Chen YJ, Chao HT, Tsai YC, Yuan CC. 2004. Enhanced expression of alpha 2,6-sialyltransferase ST6Gal I in cervical squamous cell carcinoma. Gynecol Oncol. 93:722–723. [DOI] [PubMed] [Google Scholar]
- Wang X, O'Hanlon TP, Young RF, Lau JT. 1990. Rat beta-galactoside alpha 2,6-sialyltransferase genomic organization: Alternate promoters direct the synthesis of liver and kidney transcripts. Glycobiology. 1:25–31. [DOI] [PubMed] [Google Scholar]
- Wang YC, Stein JW, Lynch CL, Tran HT, Lee CY, Coleman R, Hatch A, Antontsev VG, Chy HS, O'Brien CM, et al. 2015. Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells. Sci Rep. 5:13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A, Fan B, Zhao Y, Zhang H, Wang L, Yu X, Yuan Q, Yang D, Wang S. 2016. ST6Gal-I overexpression facilitates prostate cancer progression via the PI3K/Akt/GSK-3beta/beta-catenin signaling pathway. Oncotarget. 7:65374–65388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J, de Souza-e-Silva U, Paulson JC. 1982. Purification of a gal beta 1 to 4GlcNAc alpha 2 to 6 sialyltransferase and a gal beta 1 to 3(4)GlcNAc alpha 2 to 3 sialyltransferase to homogeneity from rat liver. J Biol Chem. 257:13835–13844. [PubMed] [Google Scholar]
- Weinstein J, Lee EU, McEntee K, Lai PH, Paulson JC. 1987. Primary structure of beta-galactoside alpha 2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J Biol Chem. 262:17735–17743. [PubMed] [Google Scholar]
- Welch LG, Munro S. 2019. A tale of short tails, through thick and thin: Investigating the sorting mechanisms of Golgi enzymes. FEBS Lett. 593:2452–2465. [DOI] [PubMed] [Google Scholar]
- Wen DX, Svensson EC, Paulson JC. 1992. Tissue-specific alternative splicing of the beta-galactoside alpha 2,6-sialyltransferase gene. J Biol Chem. 267:2512–2518. [PubMed] [Google Scholar]
- Wichert B, Milde-Langosch K, Galatenko V, Schmalfeldt B, Oliveira-Ferrer L. 2018. Prognostic role of the sialyltransferase ST6GAL1 in ovarian cancer. Glycobiology. 28:898–903. [DOI] [PubMed] [Google Scholar]
- Wood LD, Hruban RH. 2012. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 18:492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kurusu Y, Takizawa K, Tanaka J, Matsumoto K, Taniguchi A. 2003. Transcriptional regulation of human beta-galactoside alpha2,6-sialyltransferase (hST6Gal I) gene in colon adenocarcinoma cell line. Biochem Biophys Res Commun. 307:1070–1074. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kaneko Y, Rebbaa A, Bremer EG, Moskal JR. 1997. alpha2,6-Sialyltransferase gene transfection into a human glioma cell line (U373 MG) results in decreased invasivity. J Neurochem. 68:2566–2576. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Oviedo A, Sweeley C, Saito T, Moskal JR. 2001. Alpha2,6-sialylation of cell-surface N-glycans inhibits glioma formation in vivo. Cancer Res. 61:6822–6829. [PubMed] [Google Scholar]
- Ye X, Weinberg RA. 2015. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 25:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA, et al. 2019. Transfer of functional cargo in exomeres. Cell Rep. 27:940–954 e946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Pan C, Zhou L, Cai Z, Zhao S, Yu D. 2016. Knockdown of ST6Gal-I increases cisplatin sensitivity in cervical cancer cells. BMC Cancer. 16:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wei A, Zhang H, Chen X, Wang L, Zhang H, Yu X, Yuan Q, Zhang J, Wang S. 2017. alpha2,6-sialylation mediates hepatocellular carcinoma growth in vitro and in vivo by targeting the Wnt/beta-catenin pathway. Oncogenesis. 6:e343. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou L, Zhang S, Zou X, Lu J, Yang X, Xu Z, Shan A, Jia W, Liu F, Yan X, et al. 2019. The beta-galactoside alpha2,6-sialyltranferase 1 (ST6GAL1) inhibits the colorectal cancer metastasis by stabilizing intercellular adhesion molecule-1 via sialylation. Cancer Manag Res. 11:6185–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Srivatana U, Ullah A, Gagneja H, Berenson CS, Lance P. 2001. Suppression of a sialyltransferase by antisense DNA reduces invasiveness of human colon cancer cells in vitro. Biochim Biophys Acta. 1536:148–160. [DOI] [PubMed] [Google Scholar]
- Zhuo Y, Bellis SL. 2011. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 286:5935–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]


