Abstract
While functional studies of long noncoding RNAs (lncRNAs) have mostly focused on how they influence disease diagnosis and prognosis, the pharmacogenomic relevance of lncRNAs remains largely unknown. Here, we test the hypothesis that the expression of a lncRNA, grow arrest-specific 5 (GAS5) can be a biomarker for docetaxel response in castration resistant prostate cancer (CRPC) using both prostate cancer (PCa) cell lines and CRPC patient datasets. Our results suggest that lower GAS5 expression is associated with docetaxel resistance in both PCa cell lines and CRPC patients. Further experiments also suggest that GAS5 is downregulated in docetaxel resistant CRPC cell lines, which reinforces its potential as a biomarker for docetaxel response. To examine the underlying biological mechanisms, we transiently knockdown GAS5 expression in PCa cell lines and then subject the cells to docetaxel treatment overtime. We did not observe a decrease in docetaxel induced growth inhibition or apoptosis in the siRNA treated cells. The findings suggest that there is no direct causal relationship between change in GAS5 expression and docetaxel response. Subsequently, we explored the indirect regulation among GAS5, ATP binding cassette subfamily B member 1 (ABCB1), and docetaxel sensitivity. We showed that transient knockdown GAS5 did not lead to significant changes in ABCB1 expression. Therefore, we rule out the hypothesis that GAS5 directly down regulate ABCB1 that lead to docetaxel sensitivity. In conclusion, our work suggests that GAS5 can serve as a predictive biomarker for docetaxel response in CRPC; however, the exact mechanism behind the observed correlation remain to be elucidated.
Keywords: long noncoding (lnc) RNA, GAS5, castration-resistant prostate cancer (CRPC), docetaxel, drug resistance, ABCB1
Introduction
Long noncoding RNAs (lncRNAs) are defined as transcripts greater than 200 nucleotides with no protein-coding capacities. Constituting the majority of the non-coding transcriptome (1), lncRNAs are shown to be involved in essential biological processes at both transcriptional and posttranscriptional levels including but not limited to chromosome silencing, RNA processing, and protein-RNA interactions (2, 3). Most recently, emerging evidence has shown that certain lncRNAs play important roles in human cancers by functioning as either oncogenes or tumor suppressor genes (4–6).
The growth arrest-specific arrest 5 (GAS5) lncRNA was originally identified by Schneider et al. as being preferentially expressed in grow-arrested cells (7). Later studies have suggested GAS5 as a tumor suppressor gene in various types of cancer through inhibiting proliferation, invasion and promoting apoptosis (8–11). In addition to its potential suppressor role in tumor growth, GAS5 has also been shown to be associated with the response of several anticancer agents such as docetaxel, doxorubicin, and tamoxifen (12–14). A recent lncRNA study that systematically surveyed the pharmacologic role of lncRNAs shows that the expression of GAS5 correlates with the sensitivity of over 100 anti-cancer drugs in a collection of pan-cancer cell lines (15). This work suggests that GAS5 may be a master biomarker for the response to chemotherapeutics in various cancer types.
Prostate cancer (PCa) is the most commonly diagnosed cancer in men, and it consists of about 8% of the total cancer deaths in male population in the US (16). 10- 50% of all PCa patients will advance to castration resistant prostate cancer (CRPC) within three years of diagnosis and CRPC accounts for nearly all PCa mortality (17). For CRPC patients, docetaxel is recommended clinically as the first-line treatment (18). Unfortunately, patients’ response to docetaxel varies and there is no clinically actionable biomarker to predict its efficacy in CRPC patients (19).
In this study, we set out to examine the value of GAS5 expression in predicting docetaxel response in CRPC. Furthermore, after confirming the correlative nature of GAS5 expression and docetaxel response, we explore the potential underlying biology based on a hypothesis that GAS5 achieves its docetaxel sensitizing role by affecting ABCB1, whose expression level was shown to be upregulated in CRPC in previous studies (20, 21).
Materials and Methods
Data Acquisition
The transcriptome profiles (in the form of RPKM) of cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle/data, release date 01/02/2019). In vitro drug response data were obtained from the Cancer Therapeutics Response Portal (CTRPv2, https://ocg.cancer.gov/programs/ctd2/data-portal/, CTRPv2.1_2016). Transcriptome profiles and in vitro drug response data were also obtained from an independent high throughput cancer cell line drug screening dataset: Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/) as the training dataset for docetaxel sensitivity imputations in PCa patients. In addition, a total of 3 PCa clinical studies were evaluated including 1) The Cancer Genome Atlas (TCGA) prostate cancer, with RNA-seq data (in the form of FPKM) and clinical phenotypes obtained from the University of California, Santa Cruz Xena browser (https://xenabrowser.net/datapages/, Release 18.0); 2) The Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) trial, with RNA-seq data (in the form of RPKM) clinical phenotypes obtained from dbGaP (https://www.ncbi.nlm.nih.gov/gap/), study ID: phs001141.v1.p1; 3) Stand Up To Cancer (SU2C) prostate cancer study, with RNA-seq data (in the form of RPKM) and clinical phenotypes obtained from cBioPortal (https://www.cbioportal.org/). Gene expression data were normalized using the log2(FPKM + 1) or log2(RPKM + 1) method.
Treatment Response of Docetaxel in PCa Cell Lines and PCa Patients
To evaluate the relationship between GAS5 expression and docetaxel sensitivity, we first obtained GAS5 expression from RNAseq data in CCLE, PROMOTE, and SU2C. The docetaxel response data were generated from both in vitro and in vivo sources. For in vitro assessment, the area under the docetaxel dose-response curve (AUC) parameter from all cancer cell lines was obtained from CTRPv2. To assess the docetaxel response in PCa patients, we imputed docetaxel drug sensitivity score (DSS) using “pRRophetic” package, a computational tool that has been shown to accurately impute patient tumor response to various anti-cancer agents (22). More specifically, in this study, we built the relationship matrix using the transcriptome and measured docetaxel response data from GDSC before imputing docetaxel sensitivity score in PCa patients. We used GDSC instead of CPRPv2 as the training dataset is for the reason that the docetaxel concentration range screened in GDSC is more clinically relevant than that in CTRPv2. Pearson’s correlation coefficient was calculated between the GAS5 expression and docetaxel response as represented by either measured docetaxel AUC in cancer cell lines or docetaxel DSS in PCa patients.
Expression Correlation Between GAS5 and ABCB1
Again, the expression levels of GAS5 and ABCB1 were obtained from both cancer cell lines (CCLE) and PCa patients (TCGA-PRAD, PROMOTE, SU2C) RNA-seq data. Pearson’s correlation coefficient was calculated between the expression of GAS5 and ABCB1 in cancer cell lines and PCa patients.
Cell Culture and Reagents
We obtained the human PCa cell line DU145 from ATCC (https://www.atcc.org/) and another PCa cell line, R1D567 from the Dehm laboratory (23). Both cell lines were cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific). Cells were maintained in a humidified atmosphere of 5% CO2 at 37°C and were observed periodically to confirm morphology.
Docetaxel Resistant Cell Lines Development
Docetaxel resistant prostate cancer cell lines were established over 3 months by chronically exposing the parent cell lines to stepwise increasing concentrations of docetaxel. In brief, the cell lines were initially exposed to docetaxel at a concentration of 2×IC50 of the respective parent cell lines for 72 hours. Then, they were cultured under the regular medium and continuously monitored under the microscope until colonies formed. The cell lines were resuspended into a new T25 flask and the treatment cycle was repeated with the same concentration of docetaxel when the cell lines reached 50% confluency. As cells displayed resistance to docetaxel, the concentration was subsequently increased to 3×IC50, 4×IC50, and the treatment cycle was repeated as described above. The resistance was determined by the decrease of cell death during exposure to docetaxel. Control cells were maintained under the same percentage of the vehicle (DMSO) as resistant cell lines. All cell lines were within a passage number of 20.
Knock Down GAS5 through siRNA
R1D567 cells were plated in 6-well plates under a seeding density of 2 × 105 cells/well. Cells were reversely transfected with Lincode GAS5 Control Pool (Human) using DharmaFECT reagent 2 according to the manufacturer-supplied protocol. Transfection reagent was removed after 48 hours due to potential cytotoxicity. The final siRNA concentration was 25 nM and the final concentration of DharmaFECT reagent 2 was 0.1%. Four different siRNAs were employed for GAS5 knockdown (catalog ID: D-001310-1): GAUGGAGUCUCAUGGCACA, UGGAUGACUUGCUUGGGUA, AGGUAUGGAGAGUCGGCUU, AGGCAGACCUGUUAUCCUA. Scramble control was transfected with negative control siRNA (catalog ID: D-001320-10): UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA,UGGUUUACAUGUUUUCCUA.
Real-Time Reverse Transcriptase PCR
We obtained total RNA from cultured cells using the Quick-RNA MiniPrep Plus Kit (Zymo Research, Irvine, CA). The NanoDrop ND-8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) was used for RNA quantification. cDNA was synthase using The High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA). The Sso-Advanced Universal SYBR Green SuperMix (Bio-Rad, Hercules, CA) and the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) were used to conduct real-time PCR analyses under manufacturer’s protocol. The PCR primers used to amplify target gene/lncRNA and housekeeping genes are as follows: ABCB1 Forward 5’ – GATGCTGGTGTTTGGAGAAATG, ABCB1 Reverse 5’ - GCCTATCTCCTGTCGCATTATAG, GAS5 Forward 5’ – TGGATGACTTGCTTGGGTAAG, GAS5 Reverse 5’ – TAACAGGTCTGCCTGCATTT, GAPDH Forward 5’- GAACATCATCCCTGCCTCTAC -3’, GAPDH Reverse 5’- CCTGCTTCACCACCTTCTT -3’. All results were normalized with the expression of GAPDH. Expression results were quantified using the ΔΔCt method relative to the scramble control.
Determine Docetaxel Response Through Growth Inhibition and Apoptosis Assay After GAS5 Knockdown
R1D567 cells were trypsinized, harvested, counterstained with Hoechst 33342 Fluorescent Stain (Thermo Scientific, Pierce Biotechnology, Rockford, IL) and resuspended in full growth media to 1x105 cells per mL prior to plating and reverse transfection with siRNAs as previously described. Cells were plated in 96-well microplates (Thermo Scientific) using a seeding density of 1x104 cells per well and allowed to attach and transfect with appropriate siRNA for 48 hours. Following incubation, cells were treated with different concentrations of docetaxel ranging from 1nM to 16nM. Apoptosis was kinetically measured over 72 hours using the CellEvent™ Caspase-3/7 Green Detection Reagent (Invitrogen, Life Technologies, Carlsbad, CA) following manufacturer’s protocol. Cell culture plates were transferred by the BioSpa 8 to a Cytation™ 1 Cell Imaging Multi-Mode Reader (BioTek Instruments, Winooski, VT) every six hours, and images were captured in the DAPI, GFP, and brightfield channels. Automatic background flattening parameters were used to remove background fluorescence from the GFP and DAPI channels. Object masking thresholds were then set to identify each cell for counting. Total cell counts per well were obtained using the DAPI fluorescence intensity > 2000 as threshold. Apoptotic cell count was obtained as a subpopulation of the total cell count using the GFP fluorescence intensity > 2000 as threshold. Results are reported as a mean and standard deviation of two independent biological experiments, each containing three technical replicates for each experimental condition.
Cell viability after docetaxel treatment was also measured after knocking down GAS5 with siRNAs as previously described. R1D567 (5x103 cells per well) and DU145 cells (3x103 cells per well) were plated in 96-well microplates (Thermo Scientific) and allowed to attach and transfect with appropriate siRNA for 24 hours. Cells were then exposed to various concentrations of docetaxel for 72 hours. Cell viability was measured using WST-1 assay [(Roche Applied. Science, Penzberg, Upper Bavaria, Germany) following the manufacturer’s protocol. Absorbance at the 450nm wavelength was assessed using the Synergy HTX Multi-Mode Plate Reader (BioTek, Winooski, VT)].
Statistical Analysis and Software
All imputations, predictions, and statistical analyses were performed in the R statistical computing environment. The drug sensitivity curves were plotted and analyzed to obtain the IC50 values using GraphPad Prism 7.0. A p value of < 0.05 was considered statistically significant.
Results
GAS5 as a Biomarker for Docetaxel Sensitivity in CRPC
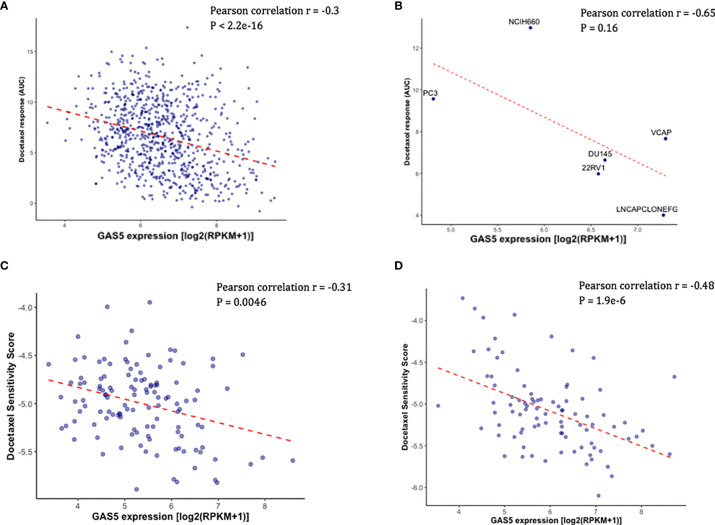
Using the existing in vitro docetaxel sensitivity screening dataset (CTRPv2) and cancer cell line RNA-seq dataset (CCLE), we first assessed the correlation between the expression of GAS5 and the measured docetaxel sensitivity across 811 cancer cell lines. As expected, a significant negative correlation of GAS5 expression and the AUC of docetaxel has been observed, with higher docetaxel sensitivity in higher GAS5 expressed cells (Pearson correlation coefficient r = -0.3, P < 2.2 x 10-16) (Figure 1A). The same directional effect was also observed in a small collection of 6 PCa cell lines available in CTRPv2 (Pearson correlation coefficient r = -0.65, P = 0.16) (Figure 1B). To evaluate the GAS5 and docetaxel response relationship in CRPC patients, we obtained the GAS5 expression from two independent CRPC patient datasets: SU2C and PROMOTE. In both studies, higher expression levels of GAS5 correlated with higher docetaxel sensitivity (as indicated with higher imputed docetaxel response) in prostate cancer patients (SU2C: Pearson correlation coefficient r = -0.31, P = 0.0046, PROMOTE: Pearson correlation coefficient r = -0.48, P < 1.9 x 10-6 (Figures 1C, D). These results from both cancer cell lines and PCa patients suggest the potential of GAS5 expression as a predictive biomarker for docetaxel sensitivity in prostate cancer.
Figure 1.
Higher GAS5 expression was associated with higher docetaxel sensitivity in cancer cell lines and CRPC patients. Lower imputed docetaxel sensitivity score indicates higher sensitivity in CRPC patients. (A) Scatter plot showing docetaxel response (AUC) (Y-axis) and GAS5 expression in 811 cancer cell lines in CTRPv2. (B) Scatter plot showing docetaxel response (AUC) (Y-axis) and GAS5 expression in 6 PCa cell lines. Name of the cell line was identified above the corresponding point. (C) Scatter plot showing imputed docetaxel sensitivity score (Y-axis) and measured GAS5 expression in 117 CRPC patients in SU2C clinical study. (D) Scatter plot showing imputed docetaxel sensitivity score (Y-axis) and measured GAS5 expression in 91 CRPC patients in PROMOTE trial.
Docetaxel Resistant Prostate Cancer Cell Lines Exhibit Decreased GAS5 Expression
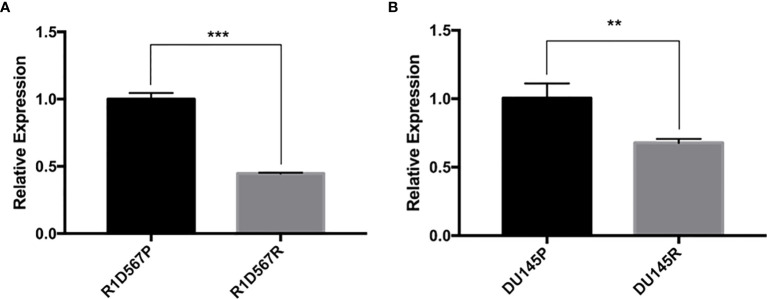
Two PCa cell lines (DU145 and R1D567) were used as the CRPC models for the experimental validation. DU145 is widely used as a CRPC cell line model as it is insensitive to anti-hormonal treatment. R1D567 is a genetically engineered cell line derived from a R1AD1 prostate cancer cell line, which was isolated from a CRPC patient. The exons 5-7 were deleted from the androgen receptor gene in R1AD1 using TALEN to establish R1D567. As a result, R1D567 cells are androgen independence (23). We established docetaxel resistant cell lines (DU145R and R1D567R) by chronically exposing parent cell lines to increasing concentrations of docetaxel. The resulting resistant cell lines have the following features: DU145R tolerates 2 × IC50 (2 x 6 nM) docetaxel treatment and R1D567R tolerates 4 × IC50 (4 x 3 nM) docetaxel. GAS5 expression was measured in both docetaxel resistant cell lines and their corresponding DMSO treated control cell lines. In DU145R, GAS5 was downregulated by nearly 40% compared to the DU145 control cell line (P = 0.0068) (Figure 2A). Similarly, compared to the parent R1D567 DMSO treated cell line, GAS5 was significantly downregulated by over 50% in R1D567R (P = 0.0001) (Figure 2B). These results provided additional support for the role of GAS5 in docetaxel sensitivity in CRPC.
Figure 2.
Docetaxel resistant PCa cell lines exhibit lower GAS5 expression when compared to parent docetaxel sensitive PCa cell lines. The expression was measured using real-time RT-PCR and was normalized to GAPDH expression. (A) Comparison of relative GAS5 expression levels in R1D567 parental line (R1D567P, black bar) and docetaxel resistant line (R1D567R, grey bar). (B) Comparison of relative GAS5 expression levels in DU145 parental line (DU145P, black bar) and docetaxel resistant line (DU145R, grey bar). **P < 0.01, ***P < 0.001.
Transient GAS5 Knockdown Did Not Decrease Percentage Apoptosis or Increase Percentage of Viable Cells After Docetaxel Treatment
To evaluate whether there is a causal relationship between GAS5 expression level and docetaxel sensitivity in CRPC, we transiently knocked down GAS5 using siRNA pool in R1D567 before treating the cells with different concentrations of docetaxel. According to qPCR results, GAS5 knockdown efficacy was greater than 75% at 48h, 72h, and 96h (Supplementary Figure 1). Then, we assessed cell proliferation in both GAS5 knockdown and controlled R1D567 cells. If lower GAS5 expression led to docetaxel resistance, we would observe a higher proliferation rate in GAS5 knockdown group. However, our results showed that there was no difference in cell proliferation between the two groups (Supplementary Figure 2). Since docetaxel exerts its cytotoxic effect mainly through inducing cell apoptosis, we then investigated whether knocking down GAS5 would change percentage apoptosis under docetaxel treatment. We did not observe a decrease in percentage apoptosis under docetaxel treatment after knocking down GAS5 (Supplementary Figure 3). In fact, we found a slight but significant increase in percentage apoptosis in the siRNA treated group after exposing to 2-4nM of docetaxel. The cell proliferation and apoptosis results together suggest that there is no short-term causal relationship between GAS5 expression and docetaxel sensitivity.
GAS5 Expression Negatively Correlated With ABCB1 Expression in PCa Patients
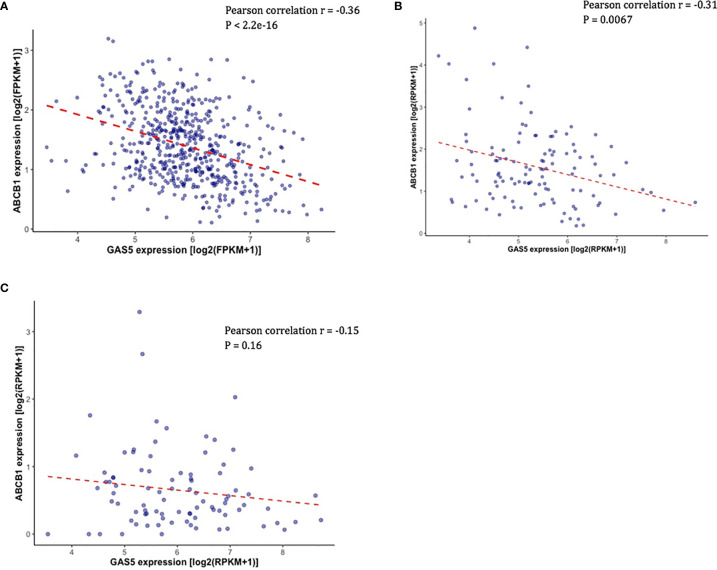
Overexpression of ABCB1 is known to be associated with docetaxel resistance in CRPC cell lines, and knockdown of ABCB1 has been shown to be able to re-sensitized resistant cell lines to docetaxel (20). Given that ABCB1 expression is functionally associated with docetaxel resistance in CRPC, we sought to determine if there is a link between the expression of GAS5 and ABCB1. In TCGA-PRAD, the expression of ABCB1 was found to be negatively correlated with GAS5 expression (Pearson correlation coefficient r = -0.36, P < 2.2 x 10-16) (Figure 3A). In addition, the same directional correlations were observed in CRPC patients in SU2C and PROMOTE datasets (SU2C Pearson correlation coefficient r = -0.31, P = 0.0067, PROMOTE Pearson correlation coefficient r = -0.15, P = 0.16) (Figures 3B, C).
Figure 3.
Validation of the correlation between the expression of GAS5 and the expression of ABCB1 in PCa patient datasets. (A) Scatter plot showing the expression level of ABCB1 (Y-axis) and the expression level of GAS5 in PCa patients in TCGA. (B) Scatter plot showing the expression level of ABCB1 (Y-axis) and the expression level of GAS5 in PCa patients in SU2C. (C) Scatter plot showing the expression level of ABCB1 (Y-axis) and the expression level of GAS5 in PCa patients in PRMOTE.
ABCB1 Was Found to Be Upregulated in Both Docetaxel Resistant PCa Cell Lines and Predicted Poor Docetaxel Responders
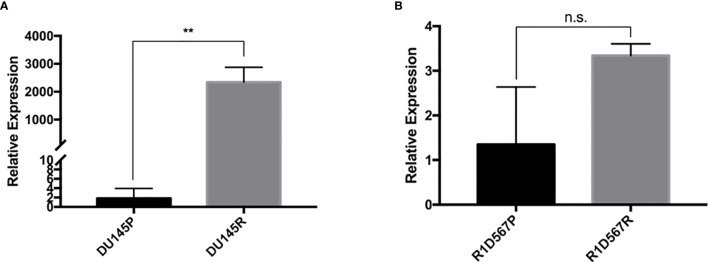
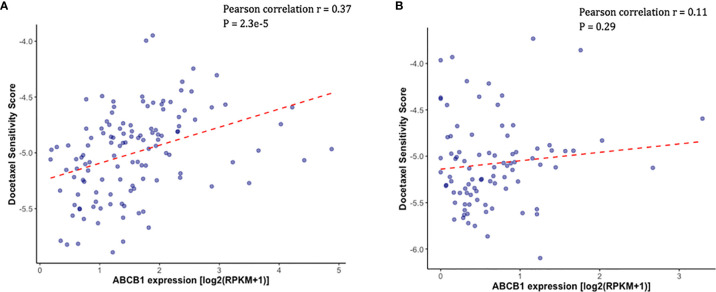
Docetaxel is a well-established substrate of ABCB1. We measured ABCB1 expression level in both docetaxel resistant and control cell lines. In DU145R, ABCB1 was significantly upregulated compared to the corresponding DMSO treated control cell line (P = 0.0031) (Figure 4A). Similarly, although not statistically significant, upregulation of ABCB1 was also observed in R1D567R compared to the control (P = 0.1) (Figure 4B). Furthermore, we observed a negative correlation between ABCB1 expression and imputed docetaxel sensitivity in two CRPC trials. In SU2C, higher ABCB1 expression was found to be correlated with lower predicted docetaxel sensitivity (Pearson correlation coefficient r = 0.37, P = 2.3 x 10-5) (Figure 5A). Similar direction was also found in PROMOTE trial (Pearson correlation coefficient r = 0.11, P = 0.29) (Figure 5B).
Figure 4.
In vitro validation of ABCB1 dysregulation in docetaxel response in CRPC cell lines. The expression was measured using real-time RT-PCR and was normalized to GAPDH expression. (A) Comparison of relative ABCB1 expression levels in DU145 parental line (black bar) and docetaxel resistant line (grey bar). (B) Comparison of relative ABCB1 expression levels in R1D567 parental line (black bar) and docetaxel resistant line (grey bar). **P < 0.01, n.s., not significant.
Figure 5.
Higher measured ABCB1 expression was correlated with higher predicted docetaxel sensitivity score (indicating lower sensitivity to docetaxel) in two CRPC trials. (A) Scatter plot showing the predicted docetaxel sensitivity score (Y-axis) and the expression level of ABCB1 (X-axis) in PCa patients in SU2C trial. (B) Scatter plot showing the predicted docetaxel sensitivity score (Y-axis) and the expression level of ABCB1 (X-axis) in PCa patients in PROMOTE trial.
Transient GAS5 Knockdown Did Not Change ABCB1 Expression in CRPC Cell Lines
Since we did not observe a direct effect of GAS5 knockdown to docetaxel response, we hypothesized that GAS5 may indirectly affect docetaxel response through its regulation on ABCB1 gene. We performed GAS5 knockdown experiments in R1D567 using a pooled siRNA targeting GAS5. The expression level of ABCB1 was measured 48h, 72h, and 96h after GAS5 knockdown. We expected that knocking down GAS5 would result in ABCB1 upregulation. However, qPCR results showed that the expression level of ABCB1 was not significantly changed after GAS5 being knocked down (Supplementary Figure 4).
Discussion
Despite the significant advances in cancer treatment, CRPC remains a deadly disease. Docetaxel is a first-line therapy for CRPC. However, the response to this key therapy varies largely between individuals (24). Currently, with additional treatment options becoming available for CRPC patients such as radium 223, sipuleucel, and cabazitaxel, there is an urgent need to identify actionable biomarkers to predict docetaxel sensitivity in CRPC so the patients can be triaged to different therapies if he is not going to respond to docetaxel therapy. In this study, we leveraged existing in vitro and in vivo data for hypothesis generation and performed experimental validation to identify the role of GAS5, a lncRNA, as a predictive biomarker for docetaxel response in CRPC.
As a well-established tumor suppressor gene, GAS5 is shown to exert growth arrest effects in different types of cancer. Luo et al. demonstrated that GAS5 enhances the promoter activity of cyclin-dependent kinase inhibitor 1B (P27Kip1), a regulator of cell cycle, by enhancing the binding of E2F1 to P27Kip1 promotor in prostate cancer (10). Moreover, GAS5 was shown to inhibit pancreatic cancer cell proliferation by inhibiting the transcriptional activity of glucocorticoid receptor (GR) in CD133+ population which is often attributed to tumor metastasis and recurrence (25). Besides acting as a tumor suppressor, GAS5 was also found to confer pharmacogenomic significance in various types of cancer. Yacqub-Usman et al. showed that in androgen-dependent prostate cancer, mTOR inhibitors induced GAS5 upregulation and knocking down GAS5 resulted in resistance to mTOR inhibitors (26). Similarly, Li et al. demonstrated that GAS5 downregulation led to trastuzumab resistance in breast cancer, and the resistance could be alleviated by lapatinib which inhibits PI3K/Akt/mTOR pathway and eventually upregulates GAS5 (27). In this study, we focused on the role of GAS5 in docetaxel response in CRPC. The initial analysis in CTRP dataset suggested that more GAS5 expression was associated with high docetaxel sensitivity in cancer cell lines. Then, after establishing docetaxel resistant cell line models by chronically exposing two CRPC cell lines (DU145 and R1D567) to docetaxel, we found that GAS5 was downregulated by nearly 50% in both docetaxel resistant cell line models. These findings support the predictive value of GAS5 as a docetaxel response biomarker in CRPC.
Beyond the in vitro findings, we also investigated whether the association between GAS5 expression and docetaxel sensitivity can be recapitulated in CRPC patients. By utilizing both RNAseq data and predicted docetaxel sensitivity score, we further demonstrated that higher GAS5 expression correlated with higher predicted docetaxel sensitivity in CRPC patients through analysis of two independent clinical studies (PROMOTE and SU2C). Notably, even if some of the patients in PROMOTE and SU2C received docetaxel as one of their treatments, the RNAseq was obtained before they physically got the drug. Therefore, the predictive role of GAS5 becomes more valuable and clinically actionable since the docetaxel sensitivity can be predicted before the treatment starts.
To test the hypothesis that GAS5 expression is a direct cause of docetaxel response in CRPC, we performed GAS5 knockdown in R1D567 and then the cellular sensitivity to docetaxel (measured through cellular growth inhibition and cell apoptosis after docetaxel exposure) were compared between the GAS5 knockdown and control groups. We found that transient knockdown of GAS5 did not result in docetaxel resistance within 72 hours. This finding is in conflict with a previously reported in vitro study, in which short-term knockdown of GAS5 diminished the cell-killing effects of docetaxel in prostate cancer cells (12). Further examination revealed that the cell line model utilized in the previous study was 22Rv1, a hormone sensitive PCa cell line (28). Therefore, the discrepancy in the observation may be in part due to the model system employed. Our results suggest that there is no direct causal relationship between GAS5 expression and docetaxel response in CRPC cell lines.
We then hypothesized that GAS5 can indirectly affect docetaxel resistance by influencing other coding genes and subsequently lead to docetaxel resistance. To test this hypothesis, we first chose to examine a potential intermediate gene, ABCB1. This gene encodes a major transporter, P-glycoprotein (P-gp), that pumps out foreign substances in an ATP-dependent manner. Our rationale to focus on this gene is that ABCB1 has been known to be related to chemoresistance in many different types of cancer (29–31). Recent studies have shown that ABCB1 could potentially modulate docetaxel resistance in prostate cancer (20, 21). Indeed, we confirmed these literature findings upon examining the correlation between ABCB1 gene expression and imputed docetaxel response scores in both clinical studies. In addition, we found that the expression of ABCB1 was increased in both DU145R and R1D567R docetaxel resistant cell lines. We also observed a negative correlation between the expression of GAS5 and ABCB1 in CRPC patients in both PROMOTE and SU2C trials. These findings suggest that there may be a link either directly or indirectly between GAS5 and ABCB1.
To investigate whether there is a direct interaction between GAS5 and ABCB1, we performed GAS5 knockdown experiments in R1D567 to explore whether transient knockdown of GAS5 would lead to ABCB1 overexpression. To this end, we observed no ABCB1 expression change at 48h, 72h, and 96h after GAS5 knockdown. These results are in contrast to our observations in docetaxel resistant cell models, which were established through chronic docetaxel exposure (months as compared to the siRNA experiments which only go up to 96 h). In the docetaxel resistant cell lines, we found significant decrease in GAS5 expression and significant increase in ABCB1 expression. Taken together, these results suggest that the expression of GAS5 did not directly affect ABCB1 expression. Therefore, we speculate that there is an indirect relationship between GAS5 and ABCB1; and that it will take time for the impact of GAS5 expression to show both at the ABCB1 expression level and for docetaxel response. This theory has support from a study by Xiao Lin et al, in which the authors suggest that the expression and function of lncRNAs may be time-dependent (32).
There are several limitations to our study. First of all, instead of the measured docetaxel response in prostate cancer patients, we utilized the docetaxel sensitivity score imputed using patients’ RNAseq data to perform the correlation between GAS5 expression and docetaxel response. The reasons behind this decision are 1. The lack of recorded docetaxel response in CRPC patients with measured tumor RNA data; 2. multiple treatment modalities leads to difficulty in attributing survival outcome to any specific treatment agents. It is worth noting that the drug sensitivity imputation model employed in this study has been proven to be able to capture a large proportion of response variability in different clinical trials (22). A second limitation of our study is that, utilizing transient GAS5 knockdown by siRNA, we were unable to track the long-term effects of GAS5 dysregulation in CRPC cell lines. Even though we observe dysregulation of both GAS5 and ABCB1 in DU145R and R1D567R and their correlation in PCa patients, our experimental findings suggest that there is no direct regulatory relationship between GAS5 expression and docetaxel sensitivity nor with ABCB1 expression. Therefore, the exact mechanism behind the GAS5 expression and docetaxel sensitivity correlation is yet to be elucidated.
In summary, we have identified that a lncRNA, GAS5, can potentially serve as a predictive biomarker for docetaxel sensitivity in CRPC. Our findings were validated in both PCa cell lines and PCa patient data. Given the urgent clinical need for biomarkers to predict docetaxel sensitivity in CRPC, we expect future work in this area to compare performance of various new biomarkers, including ours and to explore the underlying biological mechanisms to fully appreciate GAS5 as a biomarker for docetaxel response and promote its clinical application.
Data Availability Statement
The original contributions presented in the study are included in the Date Acquisition section under Materials and Methods. Further inquiries can be directed to the corresponding author.
Author Contributions
RH and YS conceived the idea. YS, AML, and YH designed and performed the experiments. JM and AL imputed docetaxel sensitivity scores in PCa patients. YS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a NIH/NCI grant R01CA204856. YS received a CTSI A-PReP Fellowship. YH receives 3M Fellowship. RH also receives support from a research grant from the Avon Foundation for Women; a NIH/NCI grant R01CA229618; a University of Minnesota Faculty Research development grant and a University of Minnesota Grant-in-Aid award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Aritro Nath at the Department of Medical Oncology and Therapeutics Research at City of Hope for giving suggestions at the early stage of this project. In addition, we thank the generous support provided by the Office of the Vice President for Research, University of Minnesota for a Grant-in-Aid support of purchasing a key equipment to complete our experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.675215/full#supplementary-material
References
- 1. Wang F, Liang R, Soibam B, Yang J, Liu Y. Coregulatory Long Non-Coding RNA and Protein-Coding Genes in Serum Starved Cells. Biochim Biophys Acta - Gene Regul Mech (2019) 1862:824–95. 10.1016/j.bbagrm.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 2. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding Rnas. Cell (2018) 172:393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell (2011) 43:904–14. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gutschner T, Diederichs S. The Hallmarks of Cancer: A Long non-Coding RNA Point of View. RNA Biol (2012) 9:703–19. 10.4161/rna.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitra SA, Mitra AP, Triche TJ. A Central Role for Long Non-Coding RNA in Cancer. Front Genet (2012) 3:17. 10.3389/fgene.2012.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang Y, Ling A, Pareek S, Huang RS. Oncogene or Tumor Suppressor? Long Noncoding RNAs Role in Patient’s Prognosis Varies Depending on Disease Type. Transl Res (2020) 230:98–110. 10.1016/j.trsl.2020.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider C, King RM, Philipson L. Genes Specifically Expressed At Growth Arrest of Mammalian Cells. Cell (1988) 54:787–93. 10.1016/S0092-8674(88)91065-3 [DOI] [PubMed] [Google Scholar]
- 8. Ji J, Dai X, Yeung SCJ, He X. The Role of Long Non-Coding RNA GAS5 in Cancers. Cancer Manag Res (2019) 11:2729–37. 10.2147/CMAR.S189052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a Non-Protein-Coding RNA, Controls Apoptosis and is Downregulated in Breast Cancer. Oncogene (2009) 28:195–208. 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- 10. Luo G, Liu D, Huang C, Wang M, Xiao X, Zeng F, et al. Lncrna GAS5 Inhibits Cellular Proliferation by Targeting P27kip1. Mol Cancer Res (2017) 15:789–99. 10.1158/1541-7786.MCR-16-0331 [DOI] [PubMed] [Google Scholar]
- 11. Chen L, Yang H, Yi Z, Jiang L, Li Y, Han Q, et al. LncRNA GAS5 Regulates Redox Balance and Dysregulates the Cell Cycle and Apoptosis in Malignant Melanoma Cells. J Cancer Res Clin Oncol (2019) 145:637–52. 10.1007/s00432-018-2820-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long Non-Coding RNA Gas5 Regulates Apoptosis in Prostate Cancer Cell Lines. Biochim Biophys Acta - Mol Basis Dis (2013) 1832:1613–23. 10.1016/j.bbadis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z, Pan T, Jiang D, Jin L, Geng Y, Feng X, et al. The Lncrna-GAS5/Mir-221-3p/DKK2 Axis Modulates Abcb1-Mediated Adriamycin Resistance of Breast Cancer Via the Wnt/β-Catenin Signaling Pathway. Mol Ther - Nucleic Acids (2020) 19:1434–48. 10.1016/j.omtn.2020.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M, et al. Downregulation of LncRNA Gas5 Confers Tamoxifen Resistance by Activating miR-222 in Breast Cancer. Cancer Lett (2018) 434:1–10. 10.1016/j.canlet.2018.06.039 [DOI] [PubMed] [Google Scholar]
- 15. Nath A, Lau EYT, Lee AM, Geeleher P, Cho WCS, Huang RS. Discovering Long Noncoding RNA Predictors of Anticancer Drug Sensitivity Beyond Protein-Coding Genes. Proc Natl Acad Sci USA (2019) 116:22020–9. 10.1073/pnas.1909998116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin (2017) 67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 17. Kirby M, Hirst C, Crawford ED. Characterising the Castration-Resistant Prostate Cancer Population: A Systematic Review. Int J Clin Pract (2011) 65:1180–92. 10.1111/j.1742-1241.2011.02799.x [DOI] [PubMed] [Google Scholar]
- 18. National Comprehensive Cancer Network . Prostate Cancer (version 2.2021) (2021). Retrieved from https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- 19. Varnai R, Koskinen LM, Mäntylä LE, Szabo I, Fitzgerald LM, Sipeky C. Pharmacogenomic Biomarkers in Docetaxel Treatment of Prostate Cancer: From Discovery to Implementation. Genes (Basel) (2019) 10:599. 10.3390/genes10080599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Y, Liu C, Nadiminty N, Lou W, Tummala R, Evans CP, et al. Inhibition of Abcb1 Expression Overcomes Acquired Docetaxel Resistance in Prostate Cancer. Mol Cancer Ther (2013) 12:1829–36. 10.1158/1535-7163.MCT-13-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lombard AP, Liu C, Armstrong CM, Cucchiara V, Gu X, Lou W, et al. Abcb1 Mediates Cabazitaxel–Docetaxel Cross-Resistance in Advanced Prostate Cancer. Mol Cancer Ther (2017) 16:2257–66. 10.1158/1535-7163.MCT-17-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geeleher P, Cox NJ, Huang RS. Clinical Drug Response Can be Predicted Using Baseline Gene Expression Levels and In Vitro Drug Sensitivity in Cell Lines. Genome Biol (2014) 15:1–12. 10.1158/1538-7445.AM2014-5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KAT, et al. Talen-Engineered AR Gene Rearrangements Reveal Endocrine Uncoupling of Androgen Receptor in Prostate Cancer. Proc Natl Acad Sci USA (2013) 110:17492–7. 10.1073/pnas.1308587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin HM, Castillo L, Mahon KL, Chiam K, Lee BY, Nguyen Q, et al. Circulating microRNAs are Associated With Docetaxel Chemotherapy Outcome in Castration-Resistant Prostate Cancer. Br J Cancer (2014) 110:2462–71. 10.1038/bjc.2014.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma NS, Gnamlin P, Durden B, Gupta VK, Kesh K, Garrido VT, et al. Long non-Coding RNA Gas5 Acts as Proliferation “Brakes” in CD133+ Cells Responsible for Tumor Recurrence. Oncogenesis (2019) 8:68. 10.1038/s41389-019-0177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal Regulation of GAS5 LncRNA Levels and Mtor Inhibitor Action in Prostate Cancer Cells. Prostate (2015) 75:693–705. 10.1002/pros.22952 [DOI] [PubMed] [Google Scholar]
- 27. Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W, et al. Downregulation of LncRNA Gas5 Causes Trastuzumab Resistance in Breast Cancer. Oncotarget (2016) 7:27778–86. 10.18632/oncotarget.8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giatromanolaki A, Fasoulaki V, Kalamida D, Mitrakas A, Kakouratos C, Lialiaris T, et al. CYP17A1 and Androgen-Receptor Expression in Prostate Carcinoma Tissues and Cancer Cell Lines. Curr Urol (2019) 13:157–65. 10.1159/000499276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chou CW, Wang CC, Wu CP, Lin YJ, Lee YC, Cheng YW, et al. Tumor Cycling Hypoxia Induces Chemoresistance in Glioblastoma Multiforme by Upregulating the Expression and Function of ABCB1. Neuro Oncol (2012) 14:1227–38. 10.1093/neuonc/nos195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fultang N, Illendula A, Lin J, Pandey MK, Klase Z, Peethambaran B. Ror1 Regulates Chemoresistance in Breast Cancer Via Modulation of Drug Efflux Pump Abcb1. Sci Rep (2020) 10:1821. 10.1038/s41598-020-58864-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, et al. Abcb1 (Mdr1) Induction Defines a Common Resistance Mechanism in Paclitaxel- and Olaparib-Resistant Ovarian Cancer Cells. Br J Cancer (2016) 115:431–141. 10.1038/bjc.2016.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin X, Lin W, Ku YS, Wong FL, Li MW, Lam HM, et al. Analysis of Soybean Long non-Coding RNAs Reveals a Subset of Small Peptide-Coding Transcripts1[Open]. Plant Physiol (2020) 182:1359–74. 10.1104/PP.19.01324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the Date Acquisition section under Materials and Methods. Further inquiries can be directed to the corresponding author.