Abstract
Novel coronavirus 2019 (COVID-19) has spread rapidly around the world and is threatening the health and lives of people worldwide. Early detection of COVID-19 positive patients and timely isolation of the patients are essential to prevent its spread. Chest X-ray images of COVID-19 patients often show the characteristics of multifocality, bilateral hairy glass turbidity, patchy network turbidity, etc. It is crucial to design a method to automatically identify COVID-19 from chest X-ray images to help diagnosis and prognosis. Existing studies for the classification of COVID-19 rarely consider the role of attention mechanisms on the classification of chest X-ray images and fail to capture the cross-channel and cross-spatial interrelationships in multiple scopes. This paper proposes a multi-kernel-size spatial-channel attention method to detect COVID-19 from chest X-ray images. Our proposed method consists of three stages. The first stage is feature extraction. The second stage contains two parallel multi-kernel-size attention modules: multi-kernel-size spatial attention and multi-kernel-size channel attention. The two modules capture the cross-channel and cross-spatial interrelationships in multiple scopes using multiple 1D and 2D convolutional kernels of different sizes to obtain channel and spatial attention feature maps. The third stage is the classification module. We integrate the chest X-ray images from three public datasets: COVID-19 Chest X-ray Dataset Initiative, ActualMed COVID-19 Chest X-ray Dataset Initiative, and COVID-19 radiography database for evaluation. Experimental results demonstrate that the proposed method improves the performance of COVID-19 detection and achieves an accuracy of 98.2%.
Keywords: Deep learning, Attention, Coronavirus, X-ray images, Multi-scale
1. Introduction
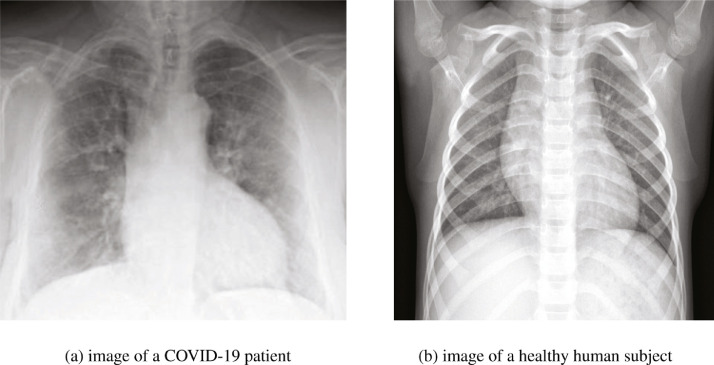
Novel coronavirus 2019 (COVID-19) emerged and spread rapidly worldwide in late 2019 [1]. It poses a great threat and challenge to human beings around the world. To date, more than 50 million people have cumulatively been infected with COVID-19, with a death toll of upwards of 1.3 million in the world. Early detection and isolation of the infected patients are an effective way to prevent the spread of the COVID-19. Conventional methods for COVID-19 detection are unable to meet the needs of the rapidly increasing number of infected patients, and the number of medical specialists became insufficient to meet the demand of healthcare professionals [2]. There is an urgent need for an effective, automatic COVID-19 screening method. Images such as X-ray have been used for the diagnosis of the COVID-19 and an example is shown in Fig. 1 . Fig. 1(a) shows an X-ray image with hazy opacity caused by COVID-19 infection. Fig. 1(b) shows an X-ray image of a healthy subject, in which the chest area is clear.
Fig. 1.
Examples of chest X-ray image. The X-ray image of a patient with COVID-19 infection depicts hazy opacity and fuzziness near the lung region. The lung region of a healthy subject appears to be clear and sharp.
Deep leaning methods have demonstrated great success in many computer vision tasks [3]. Research has been conducted on using deep learning methods to analyze medical images to assist diagnosis such as breast cancer detection [4], [5], skin cancer classification [6], [7], lung disease detection from chest X-ray images [8], fundus image segmentation [9], [10], lung segmentation [11], and magnetic resonance image synthesis [12], [13]. The exceptional results obtained by deep learning techniques excited researchers to explore deep learning for COVID-19 detection. Apostolopoulos et al. [14] applied deep neural network models, e.g.VGG-Net, to classify COVID-19. Ozturk et al. [15] proposed a deep method DarkCovidNet for detecting COVID-19. The model uses fewer convolutional layers and kernels. However, the performance of these methods is far from satisfactory. The features extracted could be out of the lung region that also appears fuzzy, e.g., certain soft tissues. It is, hence, necessary to develop a method to extract features from regions of great potential for classification.
Attention mechanism has been developed for classification and segmentation tasks of images to improve the performance of deep neural networks. The SE-Net [16] applied channel attention to address the interrelationships between channels in the feature map. Woo et al. [17] extended SE-Net by adding a spatial attention module to integrate the relationships between locations across spaces. The ECA-Net proposed by Wang et al. [18] uses a convolutional kernel with adaptively changed the size to learn relationships across channels. These attention mechanisms have achieved good results in processing natural images. However, when the modules for spatial attention and channel attention are executed sequentially, it is likely that important features presented by X-ray images are degraded that leads to inferior performance. Moreover, the kernel with fixed size fails to capture the features of different scales and properties.
In this paper, we propose a deep learning method for processing X-ray chest images to assist COVID-19 detection. Our method extracts features by introducing attention and variable kernel size. The main contribution of this paper is the multi-kernel-size, spatial-channel attention method (MKSC) to analyze chest X-ray images for COVID-19 detection. Our proposed method integrates a feature extraction module, a multi-kernel-size attention module, and a classification module. We use X-ray images from three public datasets for evaluation, which is most comprehensive to our best knowledge.
The rest of the paper is organized as follows. Section 2 presents the related work of the deep learning method for medical image classification and attention mechanism. Section 3 describes our proposed method in detail. Section 4 discusses the experimental results and a comparison study. Section 5 concludes this paper with a summary and future work.
2. Related Work
Deep learning methods have been developed and applied to medical images in recent years [19]. Esteva et al. [6] applied a deep neural network for skin lesion classification. The network was trained using a dataset with 129,450 images of 2032 diseases and demonstrated much improved performance. Tan et al. [9] used a 10-layer convolutional neural network to segment and discriminate exudates, hemorrhages, and micro-aneurysms. The input image is normalized before segmentation and the network is trained in two stages to improve the performance. On average, their network on 30,275,903 effective points achieved a sensitivity of 0.8758 and 0.7158 for exudates and dark lesions, respectively, on the CLEOPATRA database. It also achieved a sensitivity of 0.6257 and 0.4606 for hemorrhages and micro-aneurysms, respectively. Antony et al. [20] presented the investigations and results of feature learning using convolutional neural networks to automatically assess knee osteoarthritis (OA) severity and the associated clinical and diagnostic features of knee OA from radiographs (X-ray images). They also demonstrated that feature learning in a supervised manner was more effective than using conventional handcrafted features for automatic detection of knee joints and fine-grained knee OA image classification.
Deep learning has also been applied in processing medical images of the chest such as breast disease detection [4], [5], [21], lung segmentation [11], detection and classification of pulmonary nodules [22], etc. Celik et al. [4] proposed an invasive ductal carcinoma (IDC) detection method based on deep transfer learning using pre-trained CNN models to classify IDC and non-IDC image patches. The method obtained a balanced accuracy of 91.57% and an F-score of 94.11% in the balanced group on the image blocks extracted from full-slide images. Zhu et al. [22] proposed DeepLung, a fully automated lung computed tomography (CT) cancer diagnosis system consisting of two parts, nodule detection to determine the location of candidate nodules and classification to classify candidate nodules as benign or malignant. Specifically, a 3D fast regional with convolutional neural network (R-CNN) was designed to detect nodules and efficiently learn nodule features using a 3D two-channel module and a U-net-like encoder-decoder structure. The system achieved good results on nodule identification in the LIDC-IDRI dataset. Zhu et al. [21] proposed end-to-end training of a deep multi-instance network to classify mammogram X-ray images without annotated labels and verified the robustness of the proposed deep network on the INbreast dataset.
The successful application of deep learning in the diagnosis of various diseases prompts scholars to try to use deep learning techniques to process COVID-19 medical images for the diagnosis and detection of COVID-19. Ozturk et al. [15] proposed a deep method DarkCovidNet for the detection of COVID-19, and the method was used as a classifier for the You Only Look Once (YOLO) real-time object detection system based on Darknet-19. They implemented 17 convolutional layers and introduced different filtering on each layer. Hemdan et al. [23] proposed a deep learning framework called COVIDx-Net which includes seven different architectures of deep convolutional neural network methods, such as modified Visual Geometry Group Network (VGG19) and the second version of Google MobileNet. Each of the seven deep neural network methods was able to analyze the normalized intensities of the X-ray image to classify the patient as either a negative or positive COVID-19 case. Abbas et al. [24] proposed a deep convolutional neural network called Decomposition, Transfer and Synthesis (DeTraC) for the classification of chest X-ray images of COVID-19. DeTraC could handle any irregularity in an image dataset by studying the class boundaries of the image dataset using a class decomposition mechanism. DeTraC achieved the accuracy of 95.12% (97.91% sensitivity and 91.87% specificity). Wang et al. [25] introduced COVID-Net, a deep convolutional neural network designed to detect COVID-19 cases from chest X-ray images. COVID-Net uses a lightweight residual projection-expansion-projection-extension (PEPX) network architecture design pattern, where the PEPX network consists of convolutional layers with different convolutional kernel sizes.
Attention plays a very important role in human perception, and the human visual system has a special function of being able to selectively focus on specific parts of the whole scene. Therefore, researchers propose attention mechanisms in the field of deep learning to improve the performance of convolutional neural networks in image classification and segmentation tasks. Hu et al. [16] introduced a compact squeeze and excitation module to exploit the relationships between channels. In this module, global average pooling is used to pool features to obtain channel descriptors, and two fully connected layers are used to capture the relationships across channels. Woo et al. [17] proposed Convolutional Block Attention Module (CBAM), a simple yet effective attention module for feed-forward convolutional neural networks. Given an intermediate feature map, the module sequentially infers attention maps along two separate dimensions, i.e. channel and space, and the attention maps are then multiplied to the input feature map for adaptive feature refinement. Wang et al. [18] proposed an effective channel attention module (ECA) that involves only a small number of parameters but achieves significant performance gains by profiling SE-Net. The authors concluded that avoiding dimensionality reduction is important for learning channel attention and appropriate cross-channel interrelationships significantly reduce model complexity while maintaining performance.
Current research on deep learning has achieved some good results in processing natural and X-ray images. However, existing studies for the classification of COVID-19 rarely consider the role of attentional mechanisms on the classification of X-ray images and fail to capture the cross-channel and cross-spatial relationship in multiple scopes. COVID-19 X-ray images have the characteristics of multifocality, bilateral hairy glass turbidity, patchy network turbidity, etc. The existing work on attention mechanisms either considers only one of spatial and channel attention or causes damage to certain features when computing attention. For example, when the modules for computing spatial attention and computing channel attention are executed sequentially, the input of computing the second attention is the output of the first attention module, and the output is a feature map to which attention weights have been added to the high-level feature map. When this sequential execution of computing attention is used on chest X-ray images of COVID-19 patients, it is likely that important features presented by X-ray images, such as patchy network turbidity, will be destroyed and the neural network will not achieve the expected results. Moreover, the existing methods only consider the relationship within a single-sized kernel when computing the relationship across channels or space using convolution, and do not integrate the relationship within multiple sized kernels. In this paper, we tackle the problem of COVID-19 diagnosis using deep learning-based analysis of X-ray chest images and propose a multi-kernel-size spatial-channel attention method (MKSC) to identify COVID-19.
3. Multi-Kernel-Size Spatial-Channel Attention Method
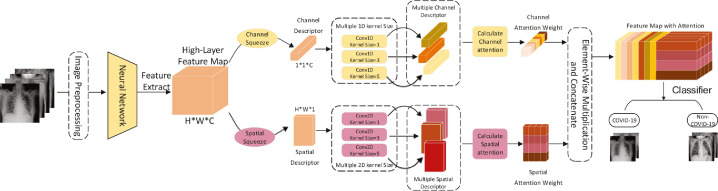
Fig. 2 illustrates our proposed multi-kernel-size spatial-channel attention method (MKSC) that includes three stages. The first stage is feature extraction. We first pre-process the X-ray images in the dataset, adjusting them all to 224 by 224 pixels, and then input them to the convolutional neural network for feature extraction to obtain the high-level feature map. The second stage is the multi-kernel-size attention module which contains two multi-kernel-size attention mechanisms: multi-kernel-size spatial attention and multi-kernel-size channel attention. Using the high-level feature maps obtained from the first stage as the input of the second stage, we suppress the shadow and skeletal noise features and enhance the pathological features of the chest X-ray images through the multi-kernel-size attention mechanism to obtain the channel and spatial attention feature maps, respectively. Furthermore, the two mechanisms capture the cross-channel and cross-spatial interrelationships in multiple scopes using multiple 1D and 2D convolutional kernels of different sizes to obtain channel and spatial attention feature maps. The multi-kernel-size attention module can obtain attention weights by using large-size convolutional kernels for the extraction of dispersed features and small-size convolutional kernels for the extraction of concentrated features, respectively. The third stage is the classification module in which we combine the two attentional feature maps output from the second stage as the input of the third stage, and then classify them by fully connected layers and softmax. The method is expressed as follows:
| (1) |
| (2) |
| (3) |
where and represent the channel and spatial attention feature maps, respectively. represents the concatenation of two obtained attention feature maps and . is the final attention feature map. denotes the channel attention weights computed from the input feature maps . is the channel element-wise multiplication which means multiplying the number of weight coefficients in by in channel direction, respectively. denotes the spatial attention weights computed from the input feature map . denotes the spatial element-wise multiplication which means multiplying the number of weight coefficients in by in spatial direction, respectively.
Fig. 2.
Diagram of the proposed MKSC method.
3.1. Multi-kernel-size channel attention module
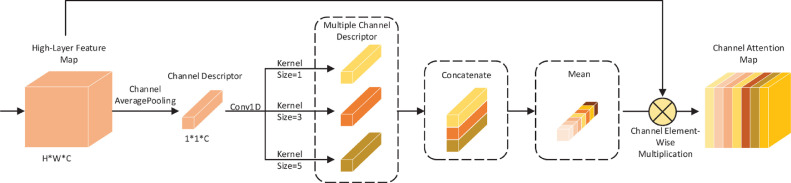
The channel attention module extracts the relationships between feature planes generated by different convolutional kernels in a feature map. The relationships between cross-channel in different ranges are different, so we propose multi-kernel-size channel attention shown in Fig. 3 to integrate the relationships between cross-channel in multiple ranges. The output feature map of the highest layer of the feature extraction part of method MKSC is . We consider the value of each dimension of the feature map and the impact of cross-channel range on the cross-channel relationship, and we choose of 1, 3, and 5, respectively. indicates that we only care about the impact of each channel without considering the cross-channel relationships. means that we consider the relationships between each channel and the adjacent channels.
Fig. 3.
Diagram of multi-kernel-size channel attention method.
In order to take full advantage of the relationship between the feature planes generated by each convolution kernel, we first squeeze the global spatial information into a channel descriptor. That is, the global averaging pooling is performed for each plane of the high-level feature map, which can be expressed as
| (4) |
where denotes the channel descriptor obtained by squeezing, is the spatial dimension of the feature map, and represents each pixel point in the feature plane.
We compute the relationships across channels using different sets of three coefficient matrices respectively and calculate the channel attention weights for a given channel descriptor . The channel attention can be learned as
| (5) |
where , and denote the coefficient matrices used in the calculation of channel attention. The coefficient matrices are shown below, when is set as 1, 3, and 5, respectively.
| (6) |
| (7) |
| (8) |
We implement the above strategy using 1D convolution, such that the process above can be updated with the end-to-end training of the neural network. Therefore, the process becomes as follows. Firstly, we squeeze each feature plane into a channel descriptor using global averaging pooling for the high-level feature map extracted in the first stage. Secondly, Conv1D operations are performed on the obtained channel descriptor using convolutional kernels of size 1, 3, and 5 to obtain three different channel descriptors , , and , respectively. Thirdly, the three channel descriptors are averaged and the final channel attention weight is computed by the sigmoid activation function. Finally, we perform channel element-wise multiplication operation on the obtained attention weights and the original high-level feature map to obtain the attention feature map in the channel dimension. The whole calculation process can be summarized as follows
| (9) |
| (10) |
| (11) |
where . denotes the global average pooling operation for each feature plane of the input feature map. , and denote the one-dimensional convolution operation with kernel sizes of 1, 3, and 5, respectively. The funciton is the average of the three-channel descriptors in the spatial dimension, and the dimension of the output of is . represents the sigmoid activation function, and denotes the channel element-wise multiplication operation.
3.2. Multi-kernel-size spatial attention module
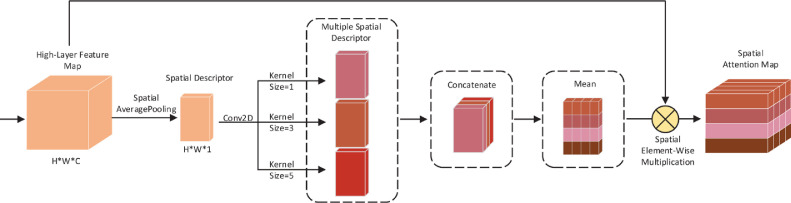
In addition to considering the interrelationships between channels, the proposed network model MKSC tries to capture the relationships between pixel points or local receptive fields by integrating the internal spatial relationships of the feature maps produced by each convolutional kernel under a certain channel. Specifically, we design a spatial attention module that also takes the feature map generated at the top layer of the feature extraction section as the input and obtain the spatial attention map via the multi-kernel-size spatial attention module, as shown in Fig. 4 . The spatial attention module and the channel attention module are mutually supportive and can better capture the impact and interaction of the two attention mechanisms. Our proposed spatial attention module includes averaging pooling operations different from the existing operations in that the spatial attention module averages each pixel point along the channel dimension. The average pooling operation is as follows:
| (12) |
where denotes the spatial descriptor obtained by squeezing, is the number of channels in the feature map , and represents the local pixel value of each channel at a specific spatial location.
Fig. 4.
Diagram of multi-kernel-size spatial attention module.
We use three sets of coefficient matrices of different sizes to compute the relationship between cross-spatial locations and calculate the spatial attention weights for a given spatial descriptor . The spatial attention can be learned as follows:
| (13) |
where , and denote the coefficient matrices used to compute spatial attention when is 1, 3, and 5, respectively. The coefficient matrices are shown below:
| (14) |
| (15) |
We implement the process above using 2D convolution, such that the process can be updated with the end-to-end training of the neural network. Firstly, we squeeze the feature information of pixels at each spatial location of the high-level feature map extracted in the first stage into a representative descriptor. We then aggregate all descriptors into a single spatial descriptor . Secondly, we use 2D convolution operations to learn the relationships between pixels as well as between local receptive fields, such that the network is able to better learn the relationships across spaces. Since the interrelationships in different cross-space range sizes are different, we also choose the convolution kernel sizes of (1,1), (3,3), and (5,5) for learning the relationships in different cross-space range sizes. We then obtain the three spatial descriptors , , and . The final spatial attention weights are computed by the sigmoid activation function after averaging the three descriptors. Finally, we perform spatial element-wise multiplication operation on the obtained attention weights and the original high-level feature map to get the attention weight map in the spatial dimension. The whole calculation process of spatial attention is as follows:
| (16) |
| (17) |
| (18) |
where . denotes the global average pooling operation over the spatial dimension of the input feature map. , , and denote the convolution with kernel sizes of (1,1), (3,3), and (5,5), respectively. The function is the average of the three-spatial descriptors in the channel dimension, and the dimension of the output of is . represents the sigmoid activation function, and denotes the element-wise multiplication.
4. Experimental Results
4.1. Datasets and Settings
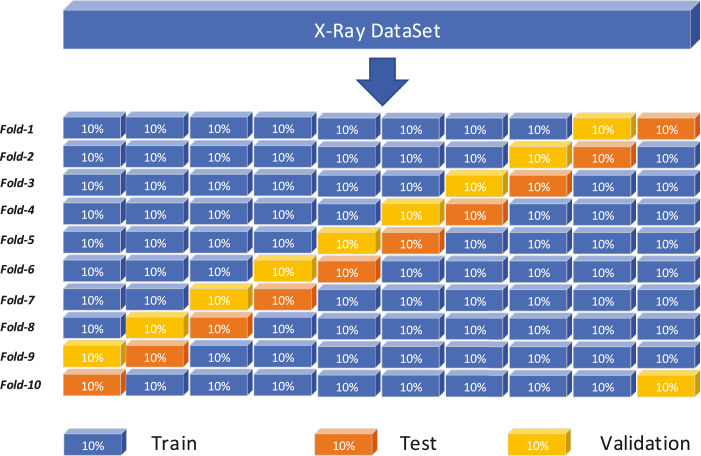
We integrate the chest X-ray images from 3 public datasets: COVID-19 Chest X-ray Dataset Initiative [26], ActualMed COVID-19 Chest X-ray Dataset Initiative [27], and COVID-19 radiography database [28]. Our dataset consists of 500 COVID-19 X-ray images and 500 non-COVID-19 X-ray images. All images are resized to pixels. We use cross-validation to evaluate the proposed MKSC by first dividing the dataset into ten random folds, with each consisting of 100 randomly selected COVID-19 X-ray images and non-COVID-19 X-ray images. We then use each fold as a validation set and a test set, respectively, as shown in Fig. 5 . We perform 30 experiments on each fold and take the average of the results as the final results. We set the initial learning rate as . When the validation accuracy does not grow three times consecutively, the learning rate drops to half of the previous rate. The number of iterations is 30.
Fig. 5.
Schematic representation of training and validation scheme employed in the 10-fold cross-validation procedure.
We use a cross-validation method to evaluate the performance of the proposed MKSC, and we can get the four expected results of true positive (TP), true negative (TN), false positive (FP), and false-negative (FN). True positive (TP) means that a COVID-19 patient is classified as COVID-19. True negative (TN) indicates that a non-COVID-19 person is identified to be non-COVID-19. False-positive (FP) means that a non-COVID-19 person is classified as COVID-19. False-negative (FN) indicates that a COVID-19 patient is identified as non-COVID-19. The four expected results can be expressed by the confusion matrix shown in Table 1 .
Table 1.
Components of the confusion matrix.
| Predicted Positive | Predicted Negative | |
|---|---|---|
| Actual Positive | True Positive(TP) | False Negative(FN) |
| Actual Negative | False Positive(FP) | True Negative(TN) |
Based on the confusion matrix, we calculate the 5 performance metrics commonly used in deep learning classification tasks to evaluate the performance of MKSC: accuracy, precision, recall (sensitivity), specificity, and F1-score.
4.2. Results and Discussion
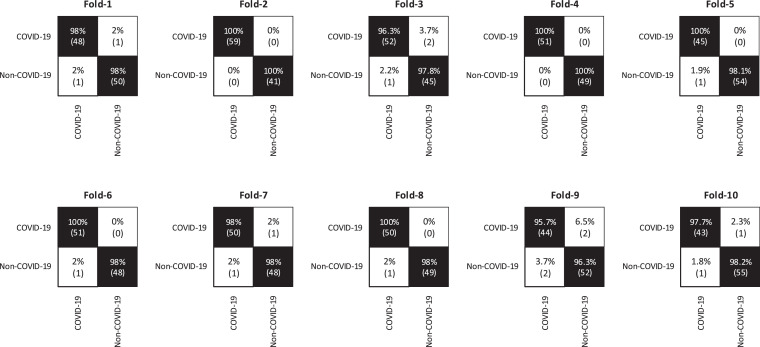
We conduct the experiments and get the confusion matrices shown in Fig. 6 . We can see that our proposed MKSC has high recognition accuracy for both COVID-19 and non-COVID-19 X-ray images. In most cases, TP and TN are above 98%, which shows that the proposed MKSC can correctly classify samples. In the following experiments, we take the average of the results in the confusion matrices in all the folds as the final results.
Fig. 6.
The 10-fold confusion matrix results for the binary classification task.
We compare the proposed MKSC method with VGG-Net [14], DarkCovidNet [15] and ResNet18 [29], which are the state of arts used for COVID-19 detection, and VGG-Net with the attention modules of SpatialAttention, SE-Net [16], CBAM [17], and ECA-Net [18], which are the state-of-art attention mechanisms in processing natural images.
Table 2 shows the accuracy of each fold and the average accuracy, and our proposed MKSC method achieves better performance than all the other methods. MKSC outperforms VGG-Net, DarkCovidNet, and ResNet18 which are without attention. These methods try to enhance classification accuracy by using different depths of convolutional layers. The results demonstrate that the attention mechanisms used in MKSC enable the neural network to know better where and what to focus on, and hence obtain higher accuracy.
Table 2.
Accuracy (%) and standard deviation of different methods.
| ResNet18 | VGG-Net | DarkCovidNet | SE-Net | ECA-Net | Spatia | CBAM | ||
|---|---|---|---|---|---|---|---|---|
| [29] | [14] | [15] | [16] | [18] | Attention | [17] | MKSC | |
| Fold-1 | 96.7 | 97.0 | 98.0 | 97.0 | 98.0 | 96.3 | 95.3 | 98.0 |
| Fold-2 | 97.3 | 96.7 | 98.7 | 98.7 | 98.3 | 99.0 | 99.0 | 99.0 |
| Fold-3 | 95.0 | 96.0 | 97.3 | 97.0 | 95.0 | 97.3 | 97.3 | 97.3 |
| Fold-4 | 95.3 | 97.3 | 97.0 | 95.3 | 98.3 | 96.7 | 96.3 | 99.0 |
| Fold-5 | 98.3 | 96.0 | 98.3 | 98.3 | 98.7 | 97.0 | 97.3 | 98.7 |
| Fold-6 | 96.7 | 97.3 | 96.0 | 97.3 | 99.3 | 99.0 | 99.7 | 99.7 |
| Fold-7 | 96.0 | 96.0 | 99.0 | 98.0 | 97.7 | 97.7 | 98.0 | 97.7 |
| Fold-8 | 95.3 | 97.0 | 98.0 | 97.0 | 97.0 | 97.7 | 98.3 | 98.0 |
| Fold-9 | 95.0 | 95.3 | 95.0 | 95.0 | 95.0 | 96.0 | 95.3 | 96.3 |
| Fold-10 | 94.0 | 95.3 | 96.0 | 95.0 | 96.7 | 95.3 | 95.7 | 98.0 |
| Average | 95.96 | 96.39 | 97.33 | 96.93 | 97.4 | 97.2 | 97.22 | 98.17 |
| STD | 1.29 | 0.73 | 1.25 | 1.3 | 1.4 | 1.15 | 1.47 | 0.92 |
MKSC obtains better results than methods SE-Net, ECA-Net, and Spatial Attention. SE-Net and ECA-Net introduce attention in the channel domain, while SpatialAttention computes the attention in the space domain. Table 2 shows that SE-Net, ECA-Net, and SpatialAttention achieve better performance than VGG-Net and DarkCovidNet without attention, illustrating attention can promote the classification performance. However, our proposed MKSC method outperforms the three attention mechanisms, since MKSC jointly considers channel and spatial attention. During the training of the neural network, the attention mechanisms in MKSC not only tell the network where and what to focus on but also suppress the shadows and skeletal noises of the images that are not helpful for the classification. In addition, the convolution operation in MKSC extracts both cross-channel and cross-spatial features. MKSC captures the cross-channel and cross-spatial interrelationships in multiple ranges using multiple 1D and 2D convolutional kernels of different sizes to obtain channel and spatial attention feature maps. That is, MKSC avoids only considering the relationship between either the channels or the spaces.
Table 2 also shows that the MKSC method achieves better performance than CBAM. CBAM uses a hybrid attention mechanism by computing spatial and channel attention in sequence. That is, the input of computing the second attention is the output of the first attention module. Note that the output of the first attention module is a feature map that has already increased attention weights on the high-level feature map, which is likely to destroy the important features presented by the images and make the neural network fail to achieve the expected results. The attention mechanism in MKSC computes the spatial and channel attention in parallel, and the obtained attention maps are concatenated to obtain the final attention feature map. In this way, the important features of the images are kept. In addition, CBAM only considers the relationships between cross-channel and cross-space within a single range when computing attention. In contrast, MKSC introduces a multi-kernel-size channel attention module and a multi-kernel-size spatial attention module to integrate the cross-channel and cross-spatial relationships in multiple ranges.
As presented in Table 2, ECA-Net achieves the second highest accuracy but has a high standard deviation, which indicates large performance fluctuation. The accuracy of VGG-Net is low, but its degree of performance is the least. Our model achieves the greatest average accuracy (98.17%) with a small standard deviation, which demonstrates superior performance and robustness.
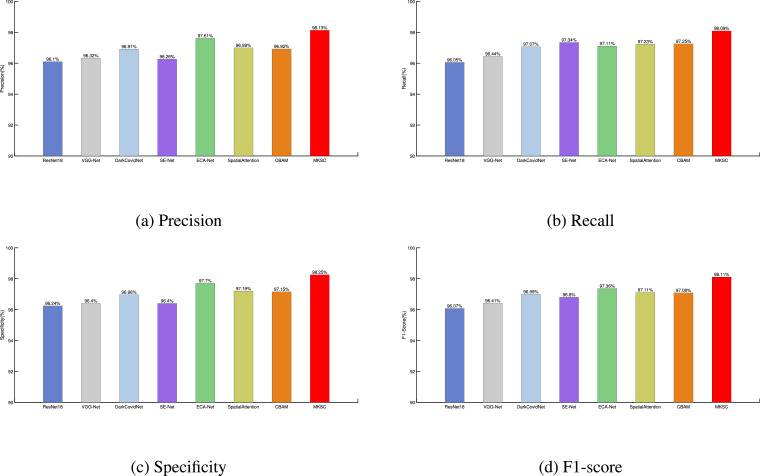
In the following, we compute the precision, recall, specificity, and F1-score of each method via confusion matrices. The performance of the methods in terms of the four metrics is shown in Fig. 7 . It can be observed from the figures that the proposed MKSC method improves the performance of precision, recall, specificity, and F1-score over the existing methods. The values of the four metrics are all above 98%, indicating that MKSC identifies COVID-19 and non-COVID-19 effectively. For the COVID-19 X-ray images with the characteristics of multifocality, bilateral hairy glass turbidity, patchy network turbidity, etc., the parallel multi-kernel-size spatial and the multi-kernel-size channel attention module in MKSC can suppress the shadows and skeletal noises of the images and enhance the pathological features of the chest X-ray images.
Fig. 7.
A comparison of COVID-19 classification performance using different methods.
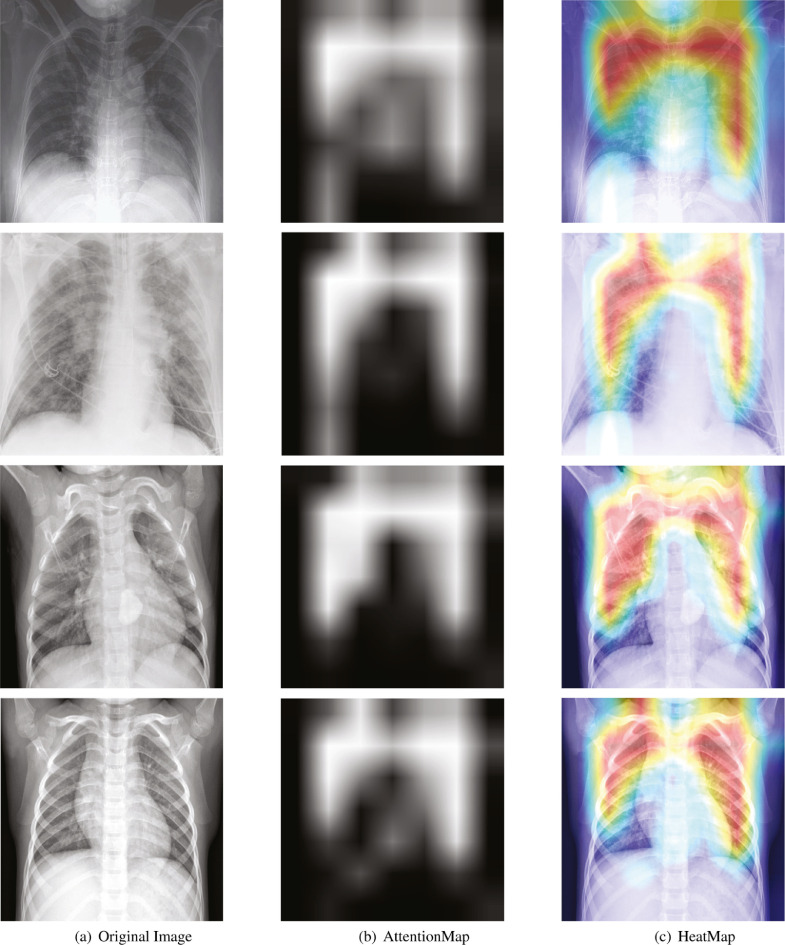
Fig. 8 illustrates the attention maps and heatmaps of COVID-19 and non-COVID-19 images. We can observe the differences between images with COVID-19 infection (top two rows) and without COVID-19 infections (bottom two rows). Images with COVID-19 infection show turbidity and opacity, whereas images without COVID-19 infections are mostly clear and sharp. The attention maps and heatmaps demonstrate that our model focuses mostly on the opaque areas of the images. The attention scatters across the lung region in the images without COVID-19 infections. The experiment result shows that our model identifies different features in images with COVID-19 and without COVID-19 infections.
Fig. 8.
The first column (a): original images; the second column (b): attention maps obtained by our method; the third column (c): corresponding heatmaps. The first two rows show the COVID-19 images, and the last two rows show the non-COVID-19 images.
We add Gaussian noise to the images to evaluate the robustness of the compared methods. The noise distribution follows zero mean and 0.6 standard deviation. Table 3 reports the performance in terms of accuracy, precision, recall, specificity, and F1-score. With added noise, the performance of all models decreases, and the performance of the benchmark models drops significantly. In contrast, the proposed MKSC method achieves the best performance in all aspects. More importantly, the performance of our model decreases the least. For example, the recall of our model remains almost the same, which demonstrates the robustness of our proposed method.
Table 3.
Performance of different methods on the data with Gaussian noise.
| Model | Performance Indicators (%) |
||||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | Sprcificity | F1-Score | |
| ResNet18 | 95.64 | 96.37 | 95.26 | 96.19 | 95.81 |
| VGG-Net | 96.03 | 96.48 | 96.19 | 95.66 | 96.33 |
| DarkCovidNet | 96.32 | 96.39 | 96.21 | 96.85 | 96.30 |
| SE-Net | 95.67 | 95.50 | 95.60 | 95.71 | 95.55 |
| ECA-Net | 96.60 | 97.40 | 95.87 | 97.50 | 96.63 |
| SpatialAttention | 95.80 | 95.50 | 94.84 | 96.50 | 95.17 |
| CBAM | 96.33 | 96.20 | 95.30 | 96.50 | 96.50 |
| MKSC | 97.82 | 97.52 | 98.05 | 97.82 | 97.82 |
5. Conclusions and Future Work
In this paper, we proposed a multi-kernel-size spatial-channel attention method for the automatic detection of COVID-19 based on X-ray images. The method has three stages. The first stage is feature extraction. The second stage contains two parallel multi-kernel-size attention modules: multi-kernel-size spatial attention and multi-kernel-size channel attention. The two mutually supportive attention modules capture the cross-channel and cross-spatial interrelationships in multiple ranges using multiple 1D and 2D convolutional kernels of different sizes to obtain channel and spatial attention feature maps. Both modules learn to focus on the pathological features of the chest X-ray images of COVID-19 patients and suppress the shadow and skeletal features. The third stage is the classification module, which performs the classification via the fully connected layer and softmax. We integrated the chest X-ray images from 3 public datasets: COVID-19 Chest X-ray Dataset Initiative, ActualMed COVID-19 Chest X-ray Dataset Initiative, and COVID-19 radiography database. We conducted experiments on the integrated dataset to validate the proposed MKSC method. Experimental results demonstrated that the proposed method achieved the accuracy of 98.2%, the precision of 98.1%, the recall of 98.1%, the specificity of 98.3%, and the F1-Score of 98.1%, which improved the performance of COVID-19 detection.
Limited by the available data set, our model achieves two-class classification: COVID-19 infection or not. There could be various severities of COVID-19 infection, which are valuable in clinical practices for planning treatments and monitoring recovery. Our future work includes an extension of this method to address stage-wise evaluation of COVID-19 infection as well as integrating data of other modalities and sources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [61806067]; and the National Key Research And Development Plan [2018YFB200050].
Biographies
Yuqi Fan is an Associate Professor at School of Computer Science and Information Engineering at Hefei University of Technology, China. He received both B.S. and M.S. degrees in computer science and engineering from Hefei University of Technology in 1999 and 2003, respectively and received his Ph.D. in Computer Science and Engineering from Wright State University in 2009. His research interests include blockchain, computer networks, cloud computing, and cyber-physical systems.
Jiahao Liu is currently a graduate student in Computer Science at School of Computer and Information Engineering at Hefei University of Technology, Hefei, China. His research interests include deep learning and computer vision.
Ruixuan Yao is currently a graduate student in Computer Science at School of Computer and Information Engineering at Hefei University of Technology, Hefei, China. His research interests include deep learning and computer vision.
Dr. Xiaohui Yuan received a B.S. degree in electrical engineering from the Hefei University of Technology, Hefei, China in 1996 and a Ph.D. degree in computer science from Tulane University, New Orleans, LA, USA in 2004. He is currently an Associate Professor at the Department of Computer Science and Engineering in the University of North Texas. His research interests include computer vision, data mining, machine learning, and artificial intelligence. His research findings have been reported in over one hundred peer-reviewed papers. Dr. Yuan is a recipient of Ralph E. Powe Junior Faculty Enhancement award in 2008 and the Air Force Summer Faculty Fellowship in 2011, 2012, and 2013.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Z., Tang J., Wang Z., Zhang K., Zhang L., Sun Q. Deep learning for image-based cancer detection and diagnosis - A survey. Pattern Recognition. 2018;83:134–149. [Google Scholar]
- 3.Hu Z., Tang J., Zhang P., Jiang J. Deep learning for the identification of bruised apples by fusing 3D deep features for apple grading systems. Mechanical Systems and Signal Processing. 2020;145:106922. [Google Scholar]
- 4.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognition Letters. 2020;133:232–239. [Google Scholar]
- 5.Cruz-Roa A., Basavanhally A., González F., Gilmore H., Feldman M., Ganesan S., Shih N., Tomaszewski J., Madabhushi A. Medical Imaging 2014: Digital Pathology, volume 9041, International Society for Optics and Photonics. SPIE; 2014. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks; pp. 1–15. [Google Scholar]
- 6.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codella N.C., Nguyen Q.-B., Pankanti S., Gutman D.A., Helba B., Halpern A.C., Smith J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM Journal of Research and Development. 2017;61(4/5):5:1–5:15. [Google Scholar]
- 8.Jiang H., Shen F., Gao F., Han W. Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recognition. 2021;113:107825. [Google Scholar]
- 9.Tan J.H., Fujita H., Sivaprasad S., Bhandary S.V., Rao A.K., Chua K.C., Acharya U.R. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Information Sciences. 2017;420:66–76. [Google Scholar]
- 10.Wang L., Gu J., Chen Y., Liang Y., Zhang W., Pu J., Chen H. Automated segmentation of the optic disc from fundus images using an asymmetric deep learning network. Pattern Recognition. 2021;112:107810. doi: 10.1016/j.patcog.2020.107810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souza J.C., Diniz J.O.B., Ferreira J.L., da Silva G.L.F., Silva A.C., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Computer Methods and Programs in Biomedicine. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Yuan X. Medical Imaging 2010: Image Processing. Vol. 7623. International Society for Optics and Photonics; 2010. Segmentation of blurry object by learning from examples; p. 76234G. [Google Scholar]
- 13.Zhang M., Desrosiers C., Zhang C. Atlas-based reconstruction of high performance brain MR data. Pattern Recognition. 2018;76:549–559. [Google Scholar]
- 14.Apostolopoulos I.D., Mpesiana T.A. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J., Shen L., Sun G. Proceedings of the IEEE Conference on Computer Cision and Cattern Cecognition (CVPR) 2018. Squeeze-and-excitation networks; pp. 7132–7141. [Google Scholar]
- 17.Woo S., Park J., Lee J.-Y., Kweon I.S. Proceedings of the European Conference on Computer Vision (ECCV) 2018. CBAM: Convolutional block attention module; pp. 3–19. [Google Scholar]
- 18.Wang Q., Wu B., Zhu P., Li P., Zuo W., Hu Q. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2020. ECA-Net: Efficient channel attention for deep convolutional neural networks; pp. 11531–11539. [Google Scholar]
- 19.Yuan X., Xie L., Abouelenien M. A regularized ensemble framework of deep learning for cancer detection from multi-class, imbalanced training data. Pattern Recognition. 2018;77:160–172. [Google Scholar]
- 20.Antony J., McGuinness K., Moran K., O’Connor N.E. Deep Learners and Deep Learner Descriptors for Medical Applications. Springer International Publishing; 2020. Feature learning to automatically assess radiographic knee osteoarthritis severity; pp. 9–93. [Google Scholar]
- 21.Zhu W., Lou Q., Vang Y.S., Xie X. Medical Image Computing and Computer Assisted Intervention - MICCAI 2017. Springer International Publishing; 2017. Deep multi-instance networks with sparse label assignment for whole mammogram classification; pp. 603–611. [Google Scholar]
- 22.Zhu W., Liu C., Fan W., Xie X. 2018 IEEE Winter Conference on Applications of Computer Vision (WACV) IEEE; 2018. Deeplung: Deep 3D dual path nets for automated pulmonary nodule detection and classification; pp. 673–681. [Google Scholar]
- 23.Hemdan E.E.-D., Shouman M.A., Karar M.E. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055. 2020 [Google Scholar]
- 24.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Applied Intelligence. 2020;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Lin Z.Q., Wong A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Scientific Reports. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.COVID-Net Team, Figure1 COVID-19 chest X-ray data initiative, 2020, Accessed in Oct. 2020.
- 27.C.-N. Team, Actualmed COVID-19 chest X-ray data initiative, 2020, Accessed in Oct. 2020.
- 28.T. Rahman, M. Chowdhury, A. Khandakar, COVID-19 Radiography Database - COVID-19 Chest X-ray Database, 2020, Accessed in Oct. 2020.
- 29.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]