Abstract
Purpose:
There is an increasing availability of large imaging cohorts (such as through The Cancer Imaging Archive (TCIA)) for computational model development and imaging research. To ensure development of generalizable computerized models, there is a need to quickly determine relative quality differences in these cohorts, especially when considering MRI datasets which can exhibit wide variations in image appearance. The purpose of this study is to present a quantitative quality control tool, MRQy, to help interrogate MR imaging datasets for: (a) site- or scanner-specific variations in image resolution or image contrast, and (b) imaging artifacts such as noise or inhomogeneity; which need correction prior to model development.
Methods:
Unlike existing imaging quality control tools, MRQy has been generalized to work with images from any body region to efficiently extract a series of quality measures (e.g. noise ratios, variation metrics) and MR image metadata (e.g. voxel resolution and image dimensions). MRQy also offers a specialized HTML5 based front-end designed for real-time filtering and trend visualization of quality measures.
Results:
MRQy was used to evaluate (a) n=133 brain MRIs from TCIA (7 sites), and (b) n=104 rectal MRIs (3 local sites). MRQy measures revealed significant site-specific variations in both cohorts, indicating potential batch effects. Before processing, MRQy measures could be used to identify each of the 7 sites within the TCIA cohort with 87.5%, 86.4%, 90%, 93%, 90%, 60%, and 92.9% accuracy and the 3 sites within the rectal cohort with 91%, 82.8%, and 88.9% accuracy; using unsupervised clustering. After processing, none of the sites could be distinctively clustered via MRQy measures in either cohort; suggesting that batch effects had been largely accounted for. Marked differences in specific MRQy measures were also able to identify outlier MRI datasets that needed to be corrected for common acquisition artifacts.
Conclusions:
MRQy is designed to be a standalone, unsupervised tool that can be efficiently run on a standard desktop computer. It has been made freely accessible and open-source at http://github.com/ccipd/MRQy for community use and feedback.
I. Introduction
The development of public repositories such as The Cancer Imaging Archive (TCIA) have enabled significant advances in machine and deep learning approaches via radiographic imaging for a variety of oncological applications1. A key aspect to utilizing TCIA cohorts for reliable model development and optimization of computational imaging tools is to curate datasets with minimal to no artifacts; implying they are relatively homogeneous in appearance2. Evaluating variations and relative image quality between cohorts can be critical to determine whether a radiomics or deep learning model that was trained on one cohort will perform reliably on a different cohort2; which can be impacted by:
Site- and scanner-specific variations which can cause poor reproducibility of machine learning models between cohorts and sites. Beyond just differences in image acquisition parameters such as echo and repetition times, there may also be systematic occurrences of technical variations (e.g. voxel resolution or fields-of-view) in subsets of a cohort i.e. batch effects3.
Presence of imaging artifacts which adversely affect the relative quality of clinical MR imaging scans within a cohort and significantly degrade model performance4. Issues such as magnetic field inhomogeneity, aliasing, motion, ringing, or noise5 can cause datasets to harbor anomalies that need correction prior to analysis.
Identification of batch effects and image artifacts in individual MRI datasets (known as image quality assessment) may be performed by experts, however, visual inspection is known to lack sensitivity to subtle variations between MR images6. Subjective quality ratings are also not precise enough to rigorously curate MRI cohorts6 due to inter-rater variability. Additionally, it may be infeasible to laboriously and manually assess the quality of individual imaging datasets in large public repositories such as TCIA. This underscores the need for automated quality control (QC) tools for MRI scans, defined in this context as quantitative approaches that can be used to identify ranges of acceptability for MR imaging datasets7. Importantly, QC allows a user to efficiently identify when a quality measurement falls outside a user-specified range or tolerance, so that an appropriate corrective action can be taken8. QC tools also need to work with minimal to no user intervention while enabling reliable curation of artifact-free and congruent data cohorts for further computational analysis and model development.
Efforts towards automated quality control of MRI datasets has led to development of different Image Quality Metrics (IQMs)9 as well as a Quality Assessment Protocol (QAP)10. However, most studies evaluating IQMs (e.g. noise ratios, energy ratios, entropy values) have focused on supervised prediction of whether a brain MRI should be accepted or excluded, as defined by experts9. IQMs have also been implemented in tools such as MRIQC6 which is a supervised quality classification tool for predicting expert quality ratings for brain MRIs. Other quality control and prediction tools for brain MRIs include the FreeSurfer-specific Qoala-T11 and LABQA2GO12; where the latter outputs an HTML report of image quality measures. However, as summarized in Table 1, none of these tools6,11,12,13,14 are readily generalizable to MRIs of body regions other than the brain, or provide an interactive front-end that can be used to easily interrogate quality issues and batch effects that may be present in large-scale imaging cohorts.
Table 1:
Feature comparison of open-source MRI quality control tools
| Attribute | MRIQC6 | Qoala-T11 | LABQA2GO12 | MRIQC-WebAPI13 | Mindcontrol14 | VisualQC | MRQy |
|---|---|---|---|---|---|---|---|
| Body part | brain | brain | brain | brain | brain | brain | all |
| Supporting raw data (.dcm,.nii) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Supporting processed data (.mha) | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ |
| Supporting modular plugins | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ |
| Supporting phantom data | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✓ |
| Interactive user interface | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ |
| Batch effects identification | ✓ | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ |
| Measurements | 14 | 185 | - | 14 | 3+9 | - | 13+10 |
In this work, we present a technical overview of MRQy, a new open-source quality control tool for MR imaging data. MRQy builds on the digital pathology quality control tool, HistoQC15, and has been optimized for analyzing large-scale MRI cohorts through development of the following modules: (i) automatic foreground detection for any MR image from any body region, from which (ii) a series of imaging-specific metadata and quality measures10 are extracted, generalized to work with any structural MR sequence, in order to (iii) compute representations that capture relevant MR image quality trends in a data cohort. These are presented within a specialized HTML5-based front-end which can be used to: (a) interrogate trends in per-site and per-scanner variations in a multi-site setting, (b) identify which specific image artifacts are present in which MRI scans in a data cohort, and (c) curate together cohorts of MRI datasets which are consistent, of sufficient image quality, and fall within user-specified ranges for computational model development. We will demonstrate the usage of MRQy via a representative publicly available brain MRI cohort from TCIA as well as an in-house multi-site rectal MRI cohort.
II. Materials and Methods
The major components of the MRQy tool are illustrated in Figure 1, which can be sub-divided into three specific modules: Input, Backend Processing, and Front-end Visualization.
Figure 1:
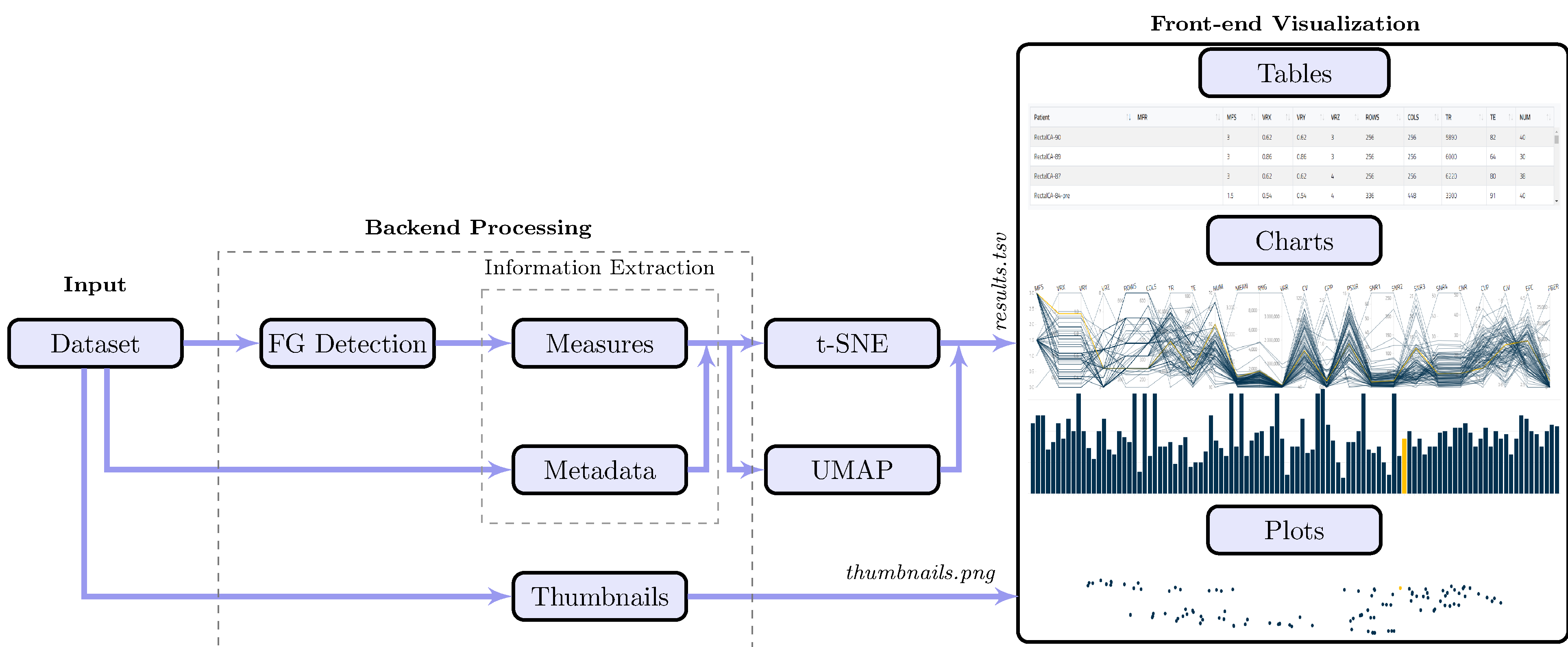
Schematic for overall MRQy workflow and major components.
II.A. Input
To ensure MRQy supported popular file formats for storing radiographic data (e.g. .dcm, .nii, .mha, and .ima), the packages medpy and pydicom libraries were used. MRQy iteratively parses either a single directory input (containing files) or resources through a directory of directories, in order to read in the image volume as well as parse image metadata.
II.B. Backend Processing
Foreground Detection:
As the MRI volume input to MRQy could be acquired from any body region, it is crucial to first identify the primary area within the volume from which quality measures are to be calculated. An in-house image processing algorithm was developed to efficiently and automatically extract and separate the background (outside the body) from the foreground (primary region of interest); further details are provided in the Supplementary Materials. Representative results of foreground detection are illustrated in Figure 2 for MRIs of the brain and the rectum to demonstrate the generalizability of the algorithm across different body regions, including in MR images with minimal background (i.e. "zoomed" acquisitions or those with sufficient phase oversampling, see Figure 2(e)).
Figure 2:
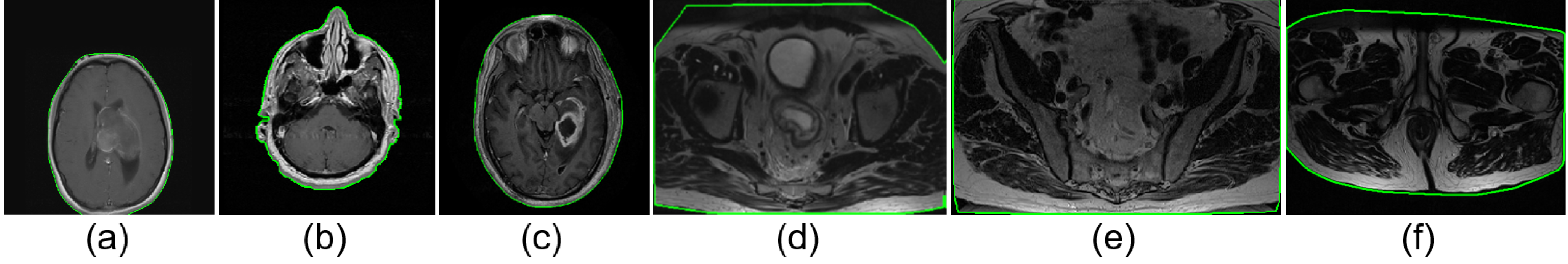
Representative foreground detection results with the foreground outlined in green for representative (a), (b), (c) brain MRI sections, (d), (e), (f) rectal MRI sections. Region outside the green outline is considered background. Note accurate delineation of outer boundary despite visually apparent shadowing and noise artifacts on all MRIs, regardless of body region being imaged.
Information Extraction:
Two major types of information were extracted from each MRI volume, broadly categorized as Metadata and Measurements. These were saved into a tab-separated file for further analysis via the specialized front-end interface or commonly available analytical tools (e.g., R, MATLAB, Python, Excel). Metadata were directly extracted from file headers (see Table 2, rows 1–10), and can include user-specified tags and header fields (see Supplementary Materials). Measurements were implemented from a survey of the medical imaging literature6,10 for detecting specific artifacts in MRI scans (Table 2, rows 10–23).
Table 2:
Summary table of metadata and quality measures extracted within MRQy
| Type | Metric | Description* |
|---|---|---|
| MFR | manufacturer name from the file header | |
| MFS | magnetic filed strength from the file header | |
| VRX | voxel resolution in x plane | |
| Metadata | VRY | voxel resolution in y plane |
| VRZ | voxel resolution in z plane | |
| ROWS | rows value of the volume | |
| COLS | columns value of the volume | |
| TR | repetition time value of the volume | |
| TE | echo time value of the volume | |
| NUM | number of slice images in each volume | |
| MEAN | mean of the foreground | |
| RNG | range of the foreground (m2 = max(F) − min(F)) | |
| VAR | variance of the foreground | |
| CV | coefficient of variation of the foreground for shadowing and inhomogeneity artifacts | |
| CPP | contrast per pixel: mean of the foreground filtered by a 3 × 3 2D Laplacian kernel for shadowing artifacts | |
| PSNR | peak signal to noise ratio of the foreground (, f2 is a 5 × 5 median filter) | |
| SNR1 | foreground standard deviation (SD) divided by background SD5 | |
| Measurements | SNR2 | mean of the foreground patch divided by background SD6. |
| SNR3 | foreground patch SD divided by the centered foreground patch SD | |
| SNR4 | mean of the foreground patch divided by mean of the background patch | |
| CNR | contrast to noise ratio for shadowing and noise artifacts5: mean of the foreground and background patches difference divided by background patch SD | |
| CVP | coefficient of variation of the foreground patch for shading artifacts: foreground patch SD divided by foreground patch mean | |
| CJV | coefficient of joint variation between the foreground and background for aliasing and inhomogeneity artifacts16 | |
| EFC | entropy focus criterion for motion artifacts6: | |
| FBER | foreground-background energy ratio for ringing artifacts10: | |
All the computed measurements (numbered m1 – m15) are average values over the entire volume, which calculated for every single slice separately. Variables M, N, F, B, FP, BP stand for slice width size, slice height size, foreground image intensity values, background image intensity values, foreground patch, and background patch respectively. Operators μ,σ,σ2, median stand for mean, standard deviation (SD), and variance measures respectively. Foreground and background patches are random 5 × 5 square patches of the foreground and background images respectively. Measurements SNR1-SNR4 are all signal to noise ratio with different definitions.
II.C. Front-end Visualization
The interactive user-interface of MRQy was built as a locally hosted HTML5/Javascript file (compatible with web browsers such as Google Chrome, Firefox, or Chromium); specifically designed for real-time analytics, data filtering, and interactive visualization by the end-user. The goal was to allow end-users to easily investigate trends in site or scanner variations within an MRI cohort, as well as identify those scans that require additional processing due to the presence of artifacts. The MRQy interface splits into 3 major sections, all of which are inter-connected. Any individual section can also be disabled or re-enabled by the end-user to provide a fully customizable interface.
Tables:
Extracted metadata and measures appear in separate tables within the interface. Each table has sortable columns to easily view outliers in numeric values. Any incomplete metadata values are displayed as "NA" (as well as being ignored in subsequent visualizations). Information can easily be copied out of the tables, which are also fully configurable including allowing for removal of specific subjects or specific columns.
Charts:
Two interactive trend visualizations were implemented. The parallel coordinate (PC) chart is a multivariate visualization tool which has been shown to be effective for understanding trends within multi-variate datasets17. For each MRI volume, a polyline is plotted (i.e. an unbroken line segment) which connected vertices of patient measurements on the parallel axes; i.e the vertex position on each axis corresponds to the value of the point for that specific metadata field or measure. The bar chart involves plotting a single bar per MRI volume for a selected variable (either metadata or measures). This provides an alternative approach to evaluating individual variables, where outliers would be markedly taller or shorter than the remainder of the cohort.
Scatter Plots:
To examine how the MRI volumes in a cohort relate to one another as well as to examine site- or scanner-specific trends, 2 different "embeddings" were computed based on the t-SNE18 (using the sklearn.manifold Python package) and UMAP19 (using the umap-learn Python package) algorithms within the Python backend. Both these algorithms take as an input all 23 measures for each patient and output a 2-dimensional embedding space (visualized as a scatter plot) where pairwise as well as global distances between patients are preserved.
II.D. Experimental Design
II.D.1. TCGA-GBM cohort
The Cancer Genome Atlas Glioblastoma Multiforme (TCGA-GBM) is the largest available dataset of brain MRI data from the Cancer Imaging Archive (TCIA)20, and comprises scans from n=259 subjects. This study was limited to subjects for whom a post-contrast T1-weighted MR image in the axial plane was available, resulting in a cohort of n=133 T1-POST MRI scans accrued from 7 different sites. As these MRI scans were acquired under different environmental conditions and using different scanner equipment and imaging protocols, this cohort includes typical data variations and image artifacts that may be observed in a TCIA dataset21. All 133 T1-POST studies were downloaded as DICOM files from TCIA.
In addition to the original cohort (as downloaded from TCIA), a processed version of the TCGA-GBM cohort was obtained from a publicly accessible release21,22. Briefly, processed MR volumes had been made available as NIFTI files after undergoing the following steps: (1) re-orientation to the left-posterior-superior coordinate system, (2) co-registration to the T1w anatomical template of SRI24 Multi-Channel Normal Adult Human Brain Atlas via affine registration, (3) resampling to 1 mm3 voxel resolution, (4) skull-stripping, (5) denoising using a low-level image processing smoothing filter, and intensity standardization to an image distribution template.
II.D.2. In-house rectal cancer cohort
This retrospectively curated dataset comprised 104 patients accrued from three different institutions including the Cleveland Clinic (CC, n=60), University Hospitals (UH, n=35), and the Cleveland Veterans Affairs (VA, n = 9) (further details available in [ 23]). For all patients, a T2-weighted turbo spin-echo MRI sequence had been acquired from patients diagnosed with rectal adenocarcinoma. While all scans were anonymized DICOM files, they had been acquired with different sequence parameters and using different scanner equipment; thus making the cohort an exemplar of a retrospectively accrued multi-site cohort.
Both the original scans as well as processed scans in this cohort were utilized, where in the latter case all MRI datasets had undergone the following sequence of operations: (1) linear resampling to a consistent voxel resolution of 0:781 × 0:781 × 4:0 mm, (2) N4ITK bias field correction, and (3) image intensity normalization with respect to a muscle region. The processed images were available as MHA files.
II.D.3. Evaluation
The MRQy front-end was used to assess each of the two cohorts for batch effects as well as image quality artifacts. In order to determine how congruent and consistent the cohorts were as a result of processing, the output of MRQy was compared between the original unprocessed and the processed scans (i.e. after applying artifact correction and normalization operations). All analysis was conducted using Python 3.7.4 for backend processing and Google Chrome 80.0.3987.149 for interacting with the front-end on a PC with Intel(R) Core(TM) i7 CPU 930 (3.60 GHz), 32 GB RAM, and a 64 bit Windows 10 operating system.
The performance of MRQy measures in identifying batch effects in each cohort without any a priori knowledge was evaluated via consensus clustering24 of all 23 measures using the ConsensusClusterPlus package in R. Hierarchical consensus clustering (with k = 7 for TCGA-GBM and k = 3 for the rectal cohort) was performed using Pearson distance and 1000 iterations, including 80% random dataset resampling between iterations. The result was visualized as a consensus cluster heatmap where the shading indicated the frequency with which a pair of patients was clustered together. Cluster overlap accuracy was then computed by first identifying which cluster corresponded to which site (based on precision/recall values) and then calculating what fraction of the datasets from each site had been correctly clustered together in consensus clustering.
III. Results
III.A. Assessment of TCGA-GBM datasets for batch effects and imaging artifacts
Analysis via MRQy took 93.5 minutes to process all 133 datasets in the original TCGA-GBM cohort (≈ 42s/dataset) and 101.6 minutes (≈ 46s/dataset) for the processed cohort. As an exemplar, we describe usage of the MRQy front-end interface (Figure 3(a)) to identify imaging artifacts. The PC chart of the CJV measure (Figure 3(b), unprocessed cohort mean CJV=1:30 ± 0:06) shows the presence of a distinct outlier (highlighted in orange + orange arrow, CJV=1.64). Further visualizing representative images (Figure 3(c)) from this dataset depicts a distinct shading artifact compared to a different dataset from the cohort ((Figure 3(c), cyan arrow in PC chart, CJV=1.2). Based on the formulation of the CJV measure16, this dataset may require bias correction or intensity normalization prior to computational analysis. Examining the PC chart of the CJV measure of processed scans (Figure 3(f), cohort mean CJV=0:605 ± 0:05) reveals no distinct outliers (the highlighted dataset from Figure 3(b) has CJV=0.604 after processing). Figure 3(g) and (h) depict processed images corresponding to those in Figure 3(c) and (d) respectively, which confirm the processing steps have accounted for the intensity artifacts previously identified.
Figure 3:
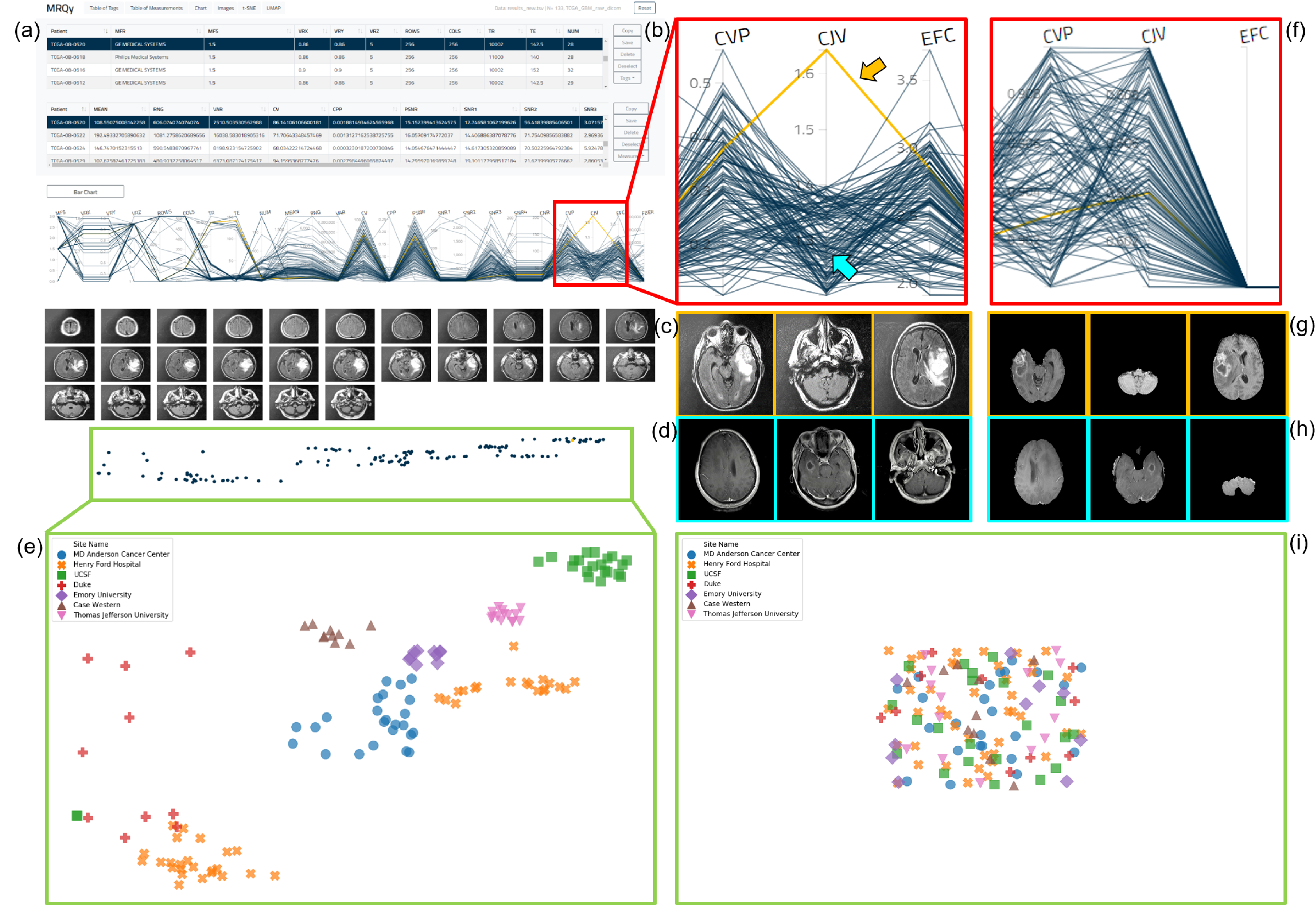
(a) MRQy front-end interface for interrogating TCGA-GBM cohort before processing. (b) Outlier dataset identified on PC chart for the CJV measure found to exhibit shading artifact on (c) representative images, especially when compared to (d) a different dataset. (e) Scatter plot revealing presence of site-specific batch effects in this cohort before processing (colored symbols corresponding to different sites appear in site-specific clusters). (f) corresponds to (b) after data processing. (g) Processed images corresponding to (c). (h) Processed images corresponding to (d). (i) Scatter plot of the processed data fall with in a single merged cluster.
Significant site-specific variations are seen to be present in the embedding scatter plot overlaid with different colored symbols for each site (Figure 3(e)). Each of the 7 sites appear as a distinct cluster, suggesting batch effects that need to be corrected for prior to model development.
After processing, the embedding scatter plot reveals no distinctive clusters corresponding to any site, where the colored symbols in Figure 3(i) fall within a single merged cluster; suggesting that the processing steps may have successfully accounted for site-specific variations.
III.B. Evaluation of multi-site rectal cancer cohort via MRQy
Analysis via MRQy took 51.8 minutes to process all 104 original datasets in rectal cohort (≈ 30s/dataset) and 60.28 minutes (≈ 35s/dataset) for the processed cohort. Within the MRQy front-end (Figure 4(a) for unprocessed data and Figure 4(d), for processed data), the bar chart of PSNR values (Figure 4(b)), unprocessed cohort mean PSNR=11:17 ± 2:38) helps quickly identify a specific dataset with very poor PSNR (indicated via orange arrow, PSNR=0.2). PSNR thus appears to accurately characterize noise artifacts in MR images as designed6, suggesting this dataset requires denoising prior to analysis. In the processed cohort, the PSNR value for this dataset (Figure 4(e), orange arrow, PSNR=18.4) is markedly higher as well as appears more consistent with the remainder of the cohort (processed cohort mean PSNR=17:94 ± 1:42).
Figure 4:
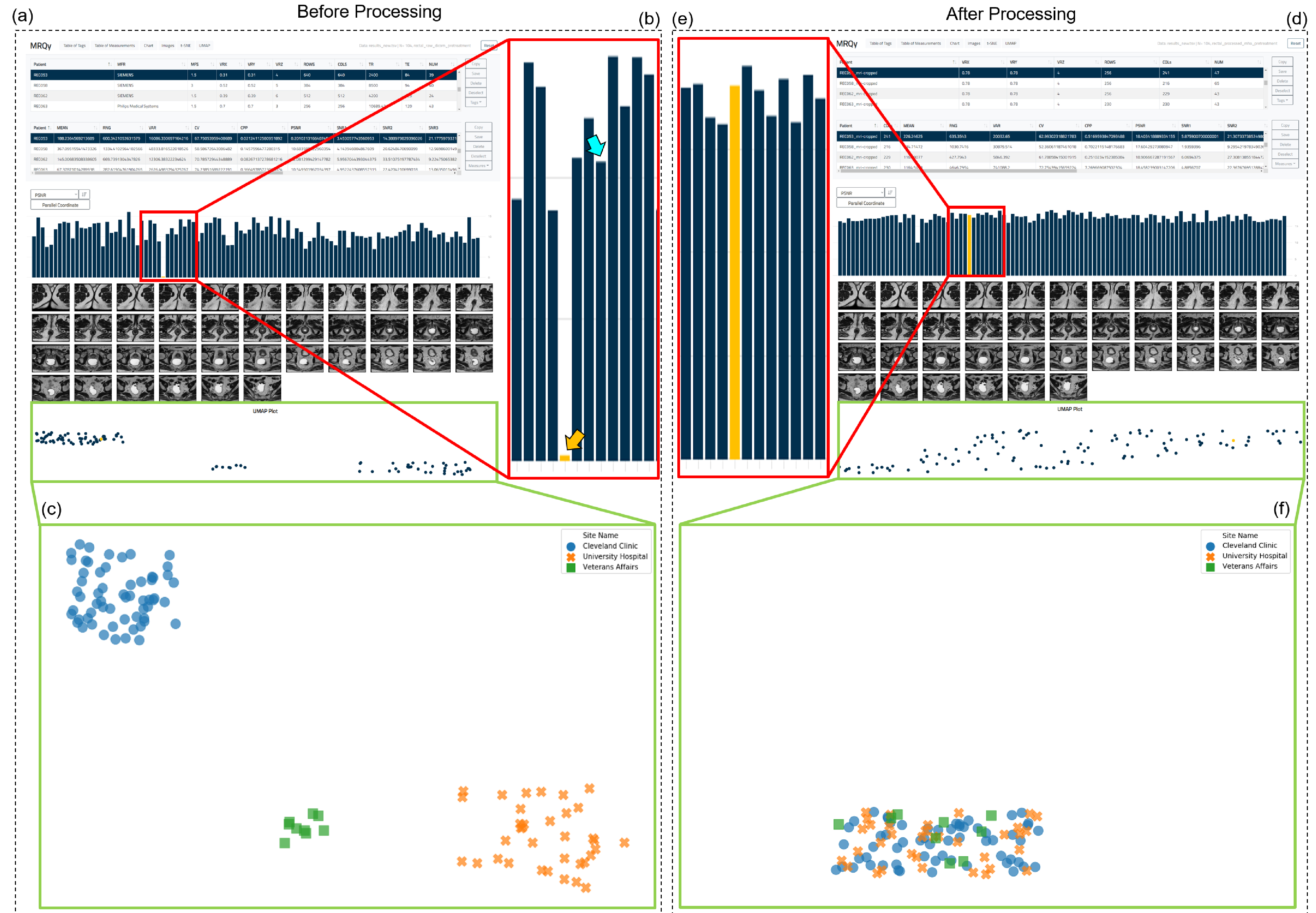
(a) MRQy front-end interface for interrogating rectal cohort before processing. (b) Outlier dataset identified on the bar chart for the PSNR measure found to exhibit subtle noise artifacts. (c) Scatter plot reveals presence of site-specific clusters in this cohort before processing (colored symbols corresponding to different sites appear in site-specific clusters). (d) MRQy front-end interface showing results using processed data. (e) Improved PSNR value of dataset highlighted in (b) after processing, also more consistent with remainder of cohort. (f) Scatter plot a single merged cluster for the cohort, suggesting batch effects and site-specific variations may have been accounted for.
Marked batch effects are seen to be present in the scatter plot in Figure 4(e) as each of the colored symbols (corresponding to different sites in the unprocessed cohort) appear in distinct clusters. By contrast, the scatter plot of the processed cohort in Figure 4(f) reveals a single merged cluster suggesting that site-specific differences have been accounted for.
III.C. Quantitative evaluation of batch effects before and after processing
Figure 5 (a) and (b) visualizes the clustergrams for each cohort (based on unprocessed data) obtained via consensus clustering of the 23 measures. For the TCGA-GBM cohort, the cluster overlap accuracy of MRQy measures with respect to each of the 7 sites is 87.5%, 86.4%, 90%, 93%, 90%, 60%, and 92.9% respectively. Similarly, MRQy measures are found to cluster each of the 3 sites in the rectal cohort with an overlap accuracy of 91%, 82.8%, and 88.9%, respectively. The strong co-clustering of datasets from the same site as as well as relatively high clustering accuracy of datasets within site-specific groupings are suggestive of batch effects in both cohorts.
Figure 5:
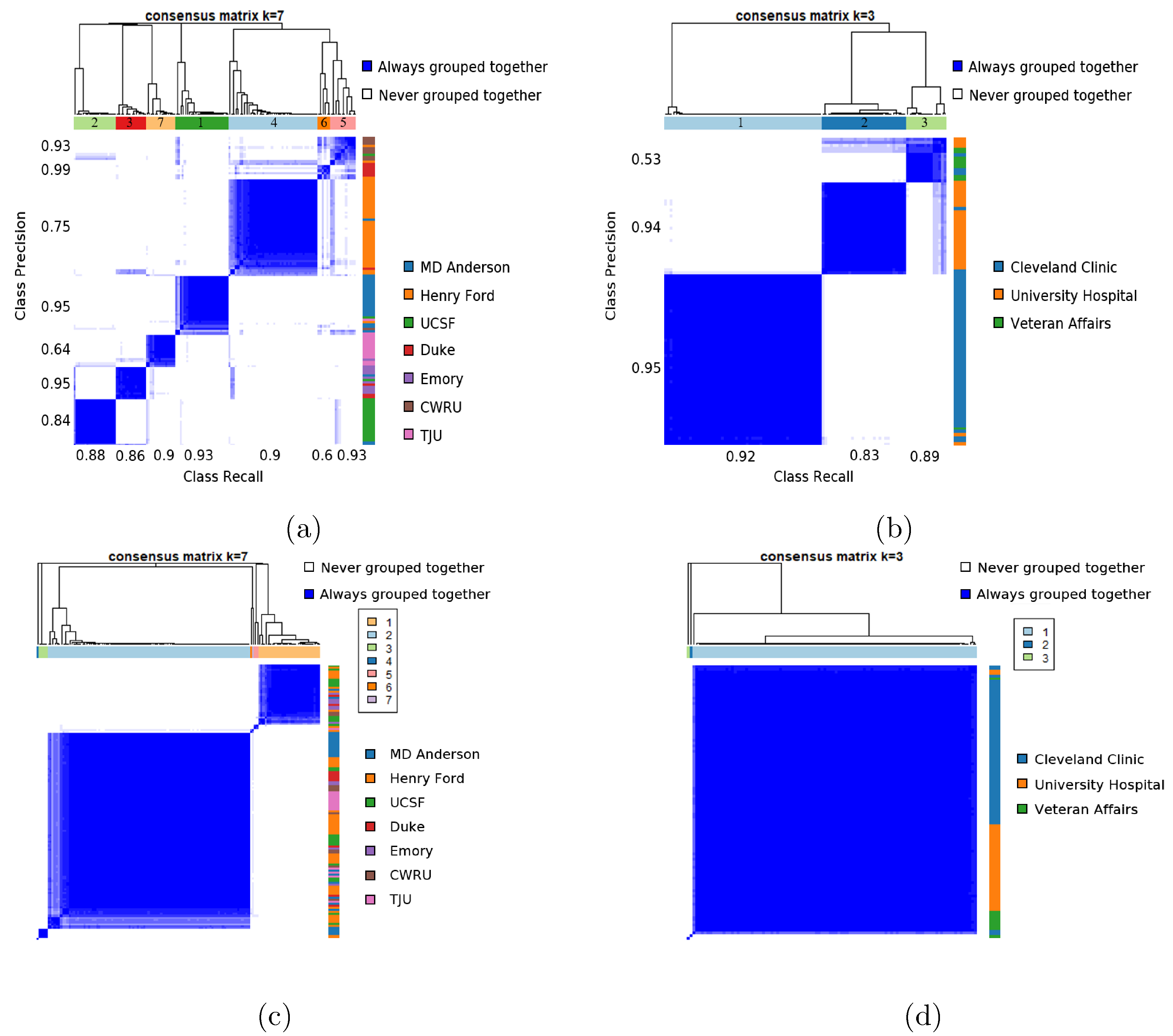
Consensus clustegram of all 23 MRQy measures computed for (a) unprocessed TCGA-GBM cohort, (b) unprocessed rectal cohort, (c) processed TCGA-GBM cohort, and (d) processed rectal cohort. In each plot, the colorbar along the right corresponds indicates which site each dataset belongs to (legend alongside) while the colorbar at the top is the cluster label obtained via consensus clustering. For unprocessed data, precision and recall values for each cluster in identifying a specific site are also noted along the left and bottom of the clustergram, respectively.
Corresponding clustergrams for the processed datasets are shown in Figure 5(c) (for TCGA-GBM cohort) and Figure 5(d) (for rectal cohort). After processing, the cluster overlap accuracy of MRQy measures in the TCGA-GBM cohort for each of the 7 sites reduced to 16.67%, 2.33%, 4.55%, 70%, 10%, 20%, and 7.14% respectively. Similarly, the cluster overlap accuracy for each of the 3 sites in the rectal cancer cohort is 1.67%, 0%, and 88.89%, respectively. This suggests that none of the clusters identified via MRQy measures are associated with any specific site after processing, in either cohort.
IV. Discussion
We have presented a technical overview and experimental evaluation of a novel open-source quality control tool for MR imaging data; called MRQy. Using cohorts from 2 different body regions (brain MRIs from TCIA, in-house rectal MRIs), we demonstrated that MRQy can quantify and evaluate (a) site- or scanner-specific variations within an MRI cohort, (b) imaging artifacts that compromise relative image quality of MRI datasets, and (c) how well specific processing pipelines have accounted for artifacts that are present in an given data cohort.
Quality control of MR imaging has long been a topic of active research9,25, but a majority of the resulting tools have been specifically developed for brain MRIs6,11,12,13,14 in order to identify acceptable scans via supervised learning (based on subjective expert quality ratings). By contrast, MRQy represents a unique solution for quality control of MRI datasets that has been designed to work with scans of any body region while running in an efficient and unsupervised fashion. MRQy has been developed by expanding on an initial framework developed as part of a digital pathology quality control tool, HistoQC15, but has been evolved significantly to specialize it for MR imaging data by incorporating imaging-specific processing steps and quality measures. Unlike existing approaches for MRI quality control, MRQy also includes a specialized interactive front-end which can help quickly identify what relative imaging artifacts and batch effects are present in a cohort. While the current work has been limited to demonstrating detection of 2 specific artifacts (shading, noise) via MRQy, we believe this approach will generalize to other artifacts due to the extensive list of extracted quality measures which have been previously validated for this purpose6.
Further development of MRQy has been enabled by its modular design, exemplified by the HTML front-end which is already highly customizable by the end-user. One of our planned functionalities is to enable real-time generation of the embedding scatter plots directly within the MRQy HTML front-end rather than via the backend. Similarly, the Python backend can be easily modified via user-developed plugins to use a different foreground detection algorithm, compute additional quality measures, or even to integrate quality prediction algorithms25 in the future. One of the current limitations of MRQy is that it can only be used for quality control of structural MRI data, but we are working on developing new measures that could allow MRQy to be used to interrogate non-structural MRIs (e.g. diffusion or dynamic contrast-enhanced MRI). Additionally, by modifying the quality measures appropriately, this tool could be adapted for quality control of any radiographic imaging collection (CT, ultrasound, PET, etc.) housed in public repositories such as TCIA.
V. Conclusions
We have presented a technical overview and experimental evaluation of MRQy, a new quality control tool for MR imaging data. MRQy works by computing actionable quality measurements as well as image metadata from any structural MR sequence of any body region via a Python backend, which can be interrogated via a specialized HTML5 front end. MRQy can be executed completely offline and the entire tool requires only a few commands to run in an unsupervised fashion, as well as being designed to be easily extensible and modular. Our initial results successfully demonstrated how MRQy could be used for (a) quantifying site- and scanner-specific batch effects within large multi-site cohorts of MR imaging data (such as TCIA), (b) identifying relative imaging artifacts within MRI datasets which require correction prior to model development, as well as (c) evaluating how well specific processing pipelines have corrected for these issues in a given data cohort. We have publicly released results of our QC analysis of MRI studies from 3 different TCIA collections (https://doi.org/10.7937/K9/TCIA.2020.JHZ2-T694) to assist researchers in this regard. MRQy has been made publicly accessible as an open-source project through GitHub (https://github.com/ccipd/MRQy), and can be downloaded as well as contributed to freely by any end-user.
Supplementary Material
Acknowledgements
The authors thank Drs. Andrei Purysko, Matthew Kalady, Sharon Stein, Rajmohan Paspulati, and Eric Marderstein for making available the multi-institutional rectal MRI cohort used in this study.
Research reported in this publication was supported by the National Cancer Institute under award numbers 1U24CA199374–01, R01CA202752–01A1, R01CA208236–01A1, R01CA216579–01A1, R01CA220581–01A1, 1U01CA239055–01, 1F31CA216935–01A1, 1U01CA248226–01, the National Heart, Lung and Blood Institute under award number 1R01HL15127701A1, National Institute for Biomedical Imaging and Bioengineering 1R43EB028736–01, National Center for Research Resources under award number 1C06RR12463–01, the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program (W81XWH-19–1-0668), the Prostate Cancer Research Program (W81XWH-15–1-0558, W81XWH-20–1-0851), the Lung Cancer Research Program (W81XWH-18–1-0440, W81XWH-20–1-0595), and the Peer Reviewed Cancer Research Program (W81XWH-16–1-0329, W81XWH-18–1-0404), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Dana Foundation David Mahoney Neuroimaging Program, the V Foundation Translational Research Award, the Johnson & Johnson WiSTEM2D Research Scholar Award, the Ohio Third Frontier Technology Validation Fund, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Footnotes
Conflicts of Interest
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. In addition he has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol Meyers-Squibb and Merck. Currently he serves on the advisory board of Aiforia Inc. He also has sponsored research agreements with Philips, AstraZeneca and Bristol Meyers-Squibb. His technology has been licensed to Elucid Bioimaging. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc. Dr. Beig is an employee of Tempus Labs, Inc.
References
- 1.Zanfardino M, Pane K, Mirabelli P, Salvatore M, and Franzese M, TCGA-TCIA Impact on Radiogenomics Cancer Research: A Systematic Review, International Journal of Molecular Sciences 20, 6033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu A, Warzel D, Eftekhari A, Kirby JS, Freymann J, Knable J, Sharma A, and Jacobs P, Call for Data Standardization: Lessons Learned and Recommendations in an Imaging Study, JCO Clinical Cancer Informatics 3, 1-–11. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, and Irizarry RA, Tackling the widespread and critical impact of batch effects in high-throughput data, Nature Reviews Genetics 11, 733–739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zwanenburg A, Vallires M, Abdalah MA, Aerts HJ, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, and Bogowicz M, The Image Biomarker Standardization Initiative: standardized quantitative radiomics for high-throughput image-based phenotyping, Radiology, 191145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushberg JT, Seibert JA, Leidholdt EM, and Boone JM, The essential physics of medical imaging, Lippincott Williams & Wilkins, 2011. [Google Scholar]
- 6.Esteban O, Birman D, Schaer M, Koyejo OO, Poldrack RA, and Gorgolewski KJ, MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites, PloS one 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delis H, Christaki K, Healy B, Loreti G, Poli GL, Toroi P, and Meghzifene A, Moving beyond quality control in diagnostic radiology and the role of the clinically qualified medical physicist, Physica Medica 41, 104–108 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Ondategui-Parra S, Quality Management in Radiology: Defining the Parameters, Health Management 8(4), (2008). [Google Scholar]
- 9.Woodard JP and Carley-Spencer MP, No-reference image quality metrics for structural MRI, Neuroinformatics 4, 243–262 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Van Essen DC et al. , The Human Connectome Project: A data acquisition perspective, NeuroImage 62, 2222–2231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klapwijk ET, van de Kamp F, van der Meulen M, Peters S, and Wierenga LM, Qoala-T: A supervised-learning tool for quality control of FreeSurfer segmented MRI data, NeuroImage 189, 116–129 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Vogelbacher C, Bopp MH, Schuster V, Herholz P, Jansen A, and Sommer J, LABQA2GO: A free, easy-to-use toolbox for the quality assessment of magnetic resonance imaging data, Frontiers in neuroscience 13, 688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteban O, Blair RW, Nielson DM, Varada JC, Marrett S, Thomas AG, Poldrack RA, and Gorgolewski KJ, Crowdsourced MRI quality metrics and expert quality annotations for training of humans and machines, Scientific data 6, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshavan A, Datta E, McDonough I, Madan C, Jordan K, and Henry R, Mind-control: A web application for brain segmentation quality control, NeuroImage 170, 365–372 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Janowczyk A, Zuo R, Gilmore H, Feldman M, and Madabhushi A, HistoQC: an open-source quality control tool for digital pathology slides, JCO clinical cancer informatics 3, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui C, Zhou YX, and Narayana P, Fast algorithm for calculation of inhomogeneity gradient in magnetic resonance imaging data, Journal of Magnetic Resonance Imaging 32, 1197–1208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edsall RM, The parallel coordinate plot in action: design and use for geographic visualization, Computational Statistics & Data Analysis 43, 605–619 (2003). [Google Scholar]
- 18.Maaten L. v. d. and Hinton G, Visualizing data using t-SNE, Journal of machine learning research 9, 2579–2605 (2008). [Google Scholar]
- 19.McInnes L, Healy J, and Melville J, Umap: Uniform manifold approximation and projection for dimension reduction, arXiv preprint arXiv:1802.03426 (2018).
- 20.Scarpace L, Mikkelsen T, Cha S, Rao S, Tekchandani S, Gutman D, Saltz J, Erickson B, Pedano N, Flanders A, Barnholtz-Sloan J, Ostrom Q, Barboriak D, and Pierce L, Radiology Data from The Cancer Genome Atlas Glioblastoma Multiforme [TCGA-GBM] collection, The Cancer Imaging Archive, 10.7937/K9/TCIA.2016.RNYFUYE9. [DOI]
- 21.Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, and Davatzikos C, Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features, Scientific data 4, 170117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beig N, Bera K, Prasanna P, Antunes J, Correa R, Singh S, Saeed Bamashmos A, Ismail M, Braman N, Verma R, Hill VB, Statsevych V, Ahluwalia MS, Varadan V, Madabhushi A, and Tiwari P, Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in Glioblastoma, Clinical Cancer Research 26 1866–1876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antunes JT, Ofshteyn A, Bera K, Wang EY, Brady JT, Willis JE, Friedman KA, Marderstein EL, Kalady MF, Stein SL, Purysko AS, Gollamudi Paspulati RJ, Madabhushi A, and Viswanath SE, Radiomic Features of Primary Rectal Cancers on Baseline T2-Weighted MRI Are Associated With Pathologic Complete Response to Neoadjuvant Chemoradiation: A Multisite Study, Journal of Magnetic Resonance Imaging n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monti S, Tamayo P, Mesirov J, and Golub T, Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data, Machine learning 52, 91–118 (2003). [Google Scholar]
- 25.Ding Y, Suffren S, Bellec P, and Lodygensky GA, Supervised machine learning quality control for magnetic resonance artifacts in neonatal data sets, Human brain mapping 40, 1290–1297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


