Abstract
Acute Kidney Injury (AKI) is a common manifestation of COVID-19 and several cases have been reported in the setting of the high-risk APOL1 genotype (common genetic variants). This increases the likelihood that African American people with the high-risk genotype APOL1 are at increased risk for kidney disease in the COVID-19 environment. Single-nucleotide polymorphisms (SNPs) are found in various microRNAs (miRNAs) and target genes change the miRNA activity that leads to different diseases. Evidence has shown that SNPs increase/decrease the effectiveness of the interaction between miRNAs and disease-related target genes. The aim of this study is not only to identify miRSNPs on the APOL1 gene and SNPs in miRNA genes targeting 3′UTR but also to evaluate the effect of these gene variations in kidney patients and their association with SARS-COV-2 infection. In 3′UTR of the APOL1 gene, we detected 96 miRNA binding sites and 35 different SNPs with 10 different online software in the binding sites of the miRNA (in silico). Also we studied gene expression of patients and control samples by using qRT-PCR (in vitro). In silico study, the binding site of miR-6741-3p on APOL1 has two SNPs (rs1288875001, G > C; rs1452517383, A > C) on APOL1 3′UTR, and its genomic sequence is the same nucleotide as rs1288875001. Similarly, two other SNPs (rs1142591, T > A; rs376326225, G > A) were identified in the binding sites of miR-6741-3p at the first position. Here, the miRSNP (rs1288875001) in APOL1 3′UTR and SNP (rs376326225) in the miR-6741-3p genomic sequence are cross-matched in the same binding region. In vitro study, the relative expression levels were calculated by the 2−ΔΔCt method & Mann–Whitney U test. The expression of APOL1 gene was different in chronic kidney patients along with COVID-19. By these results, APOL1 expression was found lower in patients than healthy (p < 0.05) in kidney patients along with COVID-19. In addition, miR-6741-3p targets many APOL1-related genes (TLR7, SLC6A19, IL-6,10,18, chemokine (C–C motif) ligand 5, SWT1, NFYB, BRF1, HES2, NFYB, MED12L, MAFG, GTF2H5, TRAF3, angiotensin II receptor-associated protein, PRSS23) by evaluating online software in the binding sites of the miR-6741-3p. miR-6741-3p has not previously shown any association with kidney diseases and SARS-COV-2 infection. It assures that APOL1 can have a significant consequence in kidney-associated diseases by different pathways. Henceforth, this study represents and demonstrates an effective association between miR-6741-3p and kidney diseases, i.e., collapsing glomerulopathy, chronic kidney disease (CKD), acute kidney injury (AKI), and tubulointerstitial lesions susceptibility to SARS-COV-2 infection via in silico and in vitro exploration and recommended to have better insight.
Background
Coronaviruses (CoV) are serious health problems linked to enteric, respiratory, hepatic, and central nervous diseases in humans and animals. Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) were identified as the source of epidemics in 2002 and 2012 with rates of high mortality due to severe respiratory syndrome (Chang et al. 2019; Wilde et al. 2017). Respiratory pneumonia (COVID-19) caused by Severe acute respiratory syndrome 2 (SARS-CoV-2) has been detected in Wuhan, where China has spread worldwide since December 2019.
Recently, two cases of focal segmental glomeruloscleroses (FSGS) and tubulointerstitial lesions have been reported in sub-Saharan African ancestry patients with COVID-19 and APOL1 polymorphism. They also highlighted the potentially important role of the risk alleles APOL1 G1 and G2 in the formation of collapsed FSGS linked to SARS-CoV-2 in humans (Couturier et al. 2020). Larsen et al. reported CG variants of the APOL1 gene associated with COVID-19 in kidney disease patients (Larsen et al. 2019). CG has not been reported in outbreaks in China and Europe, perhaps the high-risk APOL1 genotypes are only available in sub-Saharan African ancestry populations (Larsen et al. 2019; Peleg et al. 2020). High-risk genotypes have been reported in 10% to 15% of African American individuals. Given the expected attack rates of 50% to 80% of COVID-19 in the general population, a significant portion of the West African population is the COVID-19 epidemic in America and sub-Saharan African ancestry (Larsen et al. 2019; Peleg et al. 2020).
Numerous studies have reported that miRNAs act not only as signatures of tissue expression and function, but also as potential biomarkers that play an important role in regulating the pathophysiology of diseases (Macha and M, Seshacharyulu P, Ram Krishn S, Pai P, Rachagani S, Jain M, K Batra S 2014). In viral infections, host antiviral miRNAs play an important role in regulating the immune response to viral infection caused by the viral substance. It appears that many known human miRNAs can target viral genes and their functions, such as replication, translation, and interference with expression (Bátkai and Thum 2012; Bronze-da-Rocha 2014; Song et al. 2014). Irregularity of miRNAs expression plays an important role in the development of kidney disease susceptibility to SARS-COV-2 infection (Demirci and Adan 2020; Mallick et al. 2009). Some structural variations of human APOL1 and miRNA genes targeting 3′UTRs have been identified, characterized by lower binding affinity with a pointed viral protein, which has potential protective effects (Kontaraki et al. 2014).
In this study, we focus on the relationship among the 3′UTR of APOL1 and miRNAs target sites associated with kidney diseases susceptibility to COVID-19 and aim to facilitate the exploration of new therapeutic medicine to control kidney diseases, i.e., collapsing glomerulopathy, chronic kidney disease (CKD), acute kidney injury (AKI), and tubulointerstitial lesions and decrease susceptibility to SARS-COV-2 infection.
Material and methods
Screening of miRNA targeting 3′UTR of APOL1 gene
Ten unique online databases including DIANA Tools, miRTarBase, Miranda, TargetScan, miRDB, miRecords, miRcode, miRO, miRWalk, and MiCosm were used for the detection of miRNA targeting the 3′UTR of the APOL1 gene, and then we followed several algorithms and calculations below methodologies provided (Ergun and Oztuzcu 2014; Safdar et al. 2020). Accordingly, the scanning of all miRNA targets can be performed without missing any of them using the multiple databases below.
DIANA tools
To obtain estimates about in-silico–miRNA–mRNA interactions, a server used DIANA-microT containing specific databases, so that each communication and further estimation of each target site for estimation of miRNA–target gene interactions. The 3′UTR and coding regions have a positive and negative set of miRNA recognition elements, so these positive and negative clusters are locations where this tool operates. A fantastic increase in sensitivity was observed by DIANA-microT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php) compared to experimental proteomic efficiency (Paraskevopoulou et al. 2013; Vlachos et al. 2012).
miRTarBase
miRTarbase not only provides fifty thousand miRNA–target interactions to scan research articles related to functional miRNA studies, but also provides interaction information for manual review of appropriate literature after investigation of text consistency. Commonly, experiments confirm Western blot, microarray, a reporter assay, next-generation sequencing, and obtained miRNA–target interactions (MTIs). Compared to pre-built databases, the most current and greatest amount of data are provided by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) (Hsu et al. 2014).
MiRanda
In a comprehensive assessment, miRanda competes with other target prediction databases, not only because of the specification of the target genes, but also the downregulation prediction at the translational or transcriptional level. Experimental identification of a large number of unprotected and non-canonical areas is determined by this database (http://www.microrna.org/microrna/home.do) (Betel et al. 2010).
TargetScan
To estimate the computational targets of miRNAs, filtration of the protected 8 and 7 occurrences per region matching the seed portion of all miRNAs, and then another prediction of unprotected sites, target scanning was used. The context and scores of places in mammals make calculations, and these calculations also help align the target productivity estimates. The ordering of forecasts is also made with target probabilities in protected areas (http://www.targetscan.org/vert_72/) (Bommer et al. 2007).
miRDB
miRDB is an online analytical database tool used to predict miRNA targets and explain their functions. Another bioinformatics tool, miR target, was used to predict all targets of miRDB created after high-output sequencing research by studying thousands of miRNA–target interactions. MiRNA target binding has some well-known common features. These features are used to predict the target of miRNA by the machine learning method (http://www.mirdb.org/). miRDB provides miRNA targets of five species; mouse, human, rat, chicken, and dog. The latest update of this software makes it easy for the user to predict customized target sequences (Wong and Wang 2015).
miRecords
miRecords is a source of miRNA–target interaction in animals. This online database tool is divided into two components. The main component consists of authenticated targets and a high inferiority database, and these databases use experimental miRNA targets after certain literature improvements. Another component of MiRecords (http://c1.accurascience.com/miRecords/) is to estimate targets, which are the assimilation of expected miRNA targets produced by 11 verified miRNA target computation series. Since April 27, 2013, this software’s authenticated targets component has been recording approximately 2705 interactions between 644 miRNAs of 9 animal species and 1901 target genes (Wang et al. 2016). Among these records, it was handled from 2028 “low input” trials. The predicted targets of miRNA target estimation tools include HedefScan/TargertScanS, MiInspector, miRanda, MirTarget2, miTarget, NBmiRTar, RNA hybrid, NBmiRTar, PicTar, PITA, RNA22, and DIANA-miT.
miRcode
miRcode (http://www.mircode.org/) contains more than 10,000 non-encoding RNA genes. The miRNA target estimates in miRcode provided by “full transcriptome” are based on extensive GENCODE gene detailing. This tool also includes coding genes and these coding genes also contain unusual regions such as 5′UTR and CDS. The names and definitions in the MiRNA family are consistent with TargetScan. Vertebrate species form the basis of evaluation for the protection of the site (Jeggari et al. 2012).
Screening of miRSNPs
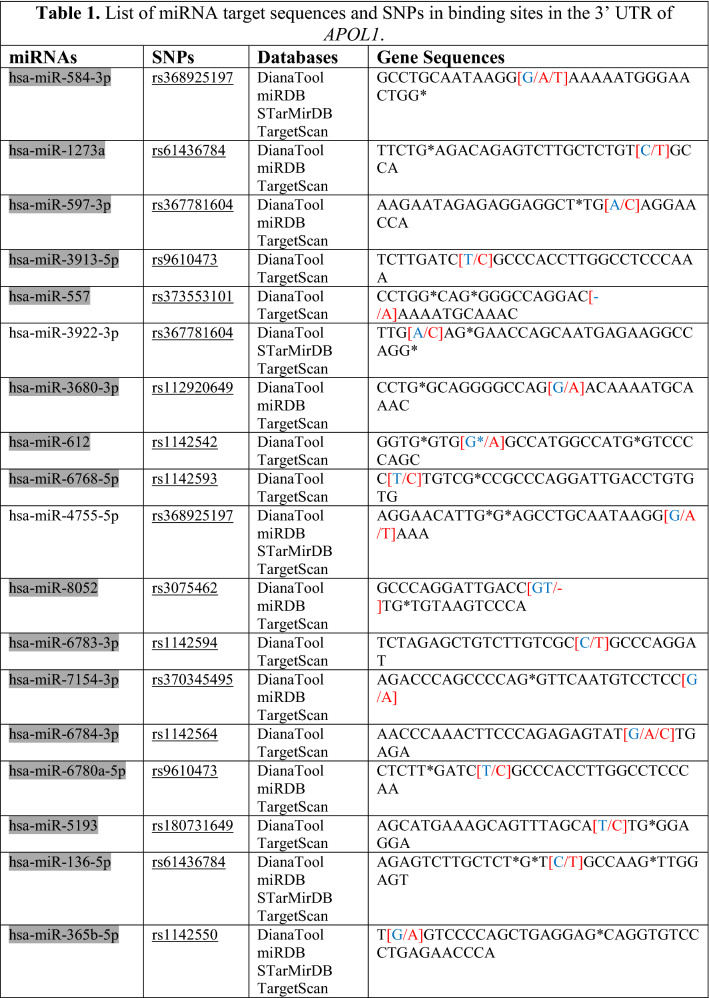
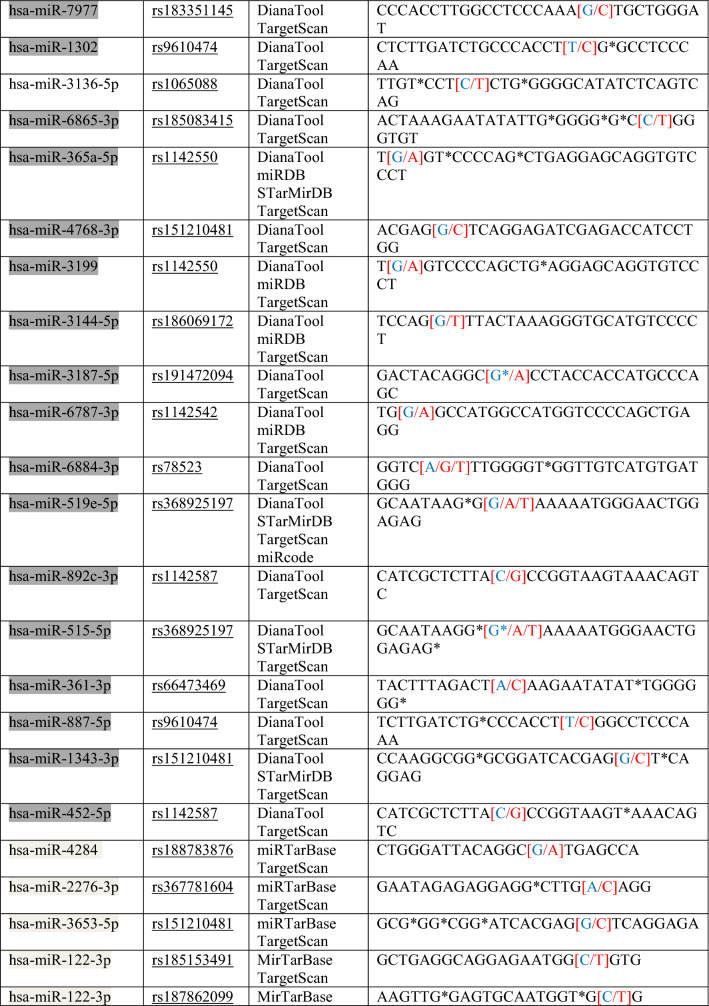
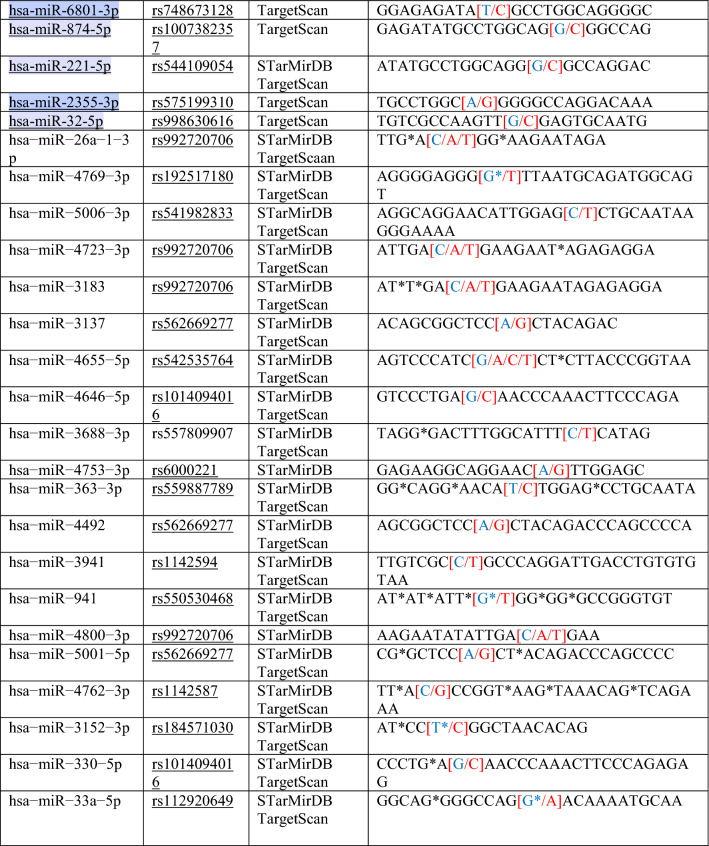
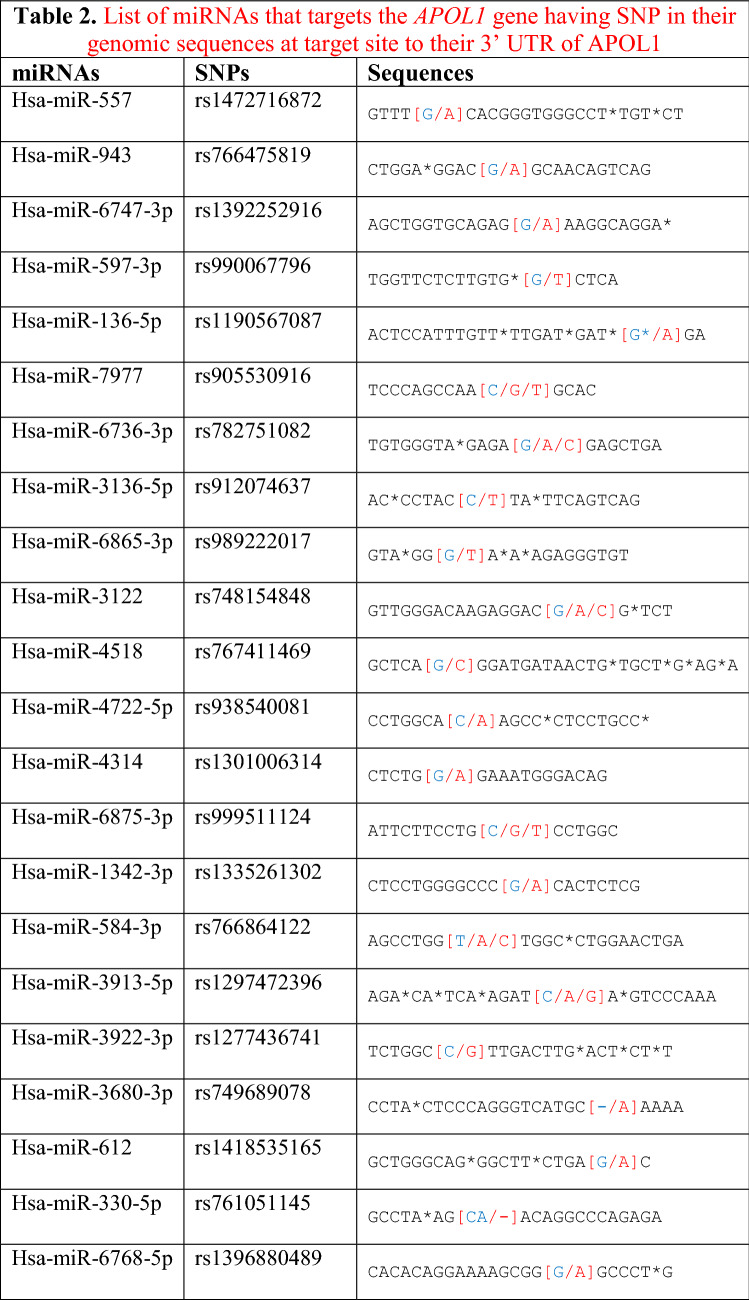
Regions targeted by miRNAs in 3′UTR of the APOL1 gene were taken from these ten databases and a target site list was constructed. Meanwhile, direct matching (A-U, G-C) and wobble matching (G-U) were well thought out in target prediction. GU wobble in seed matching indicates that a G match with a U is allowed instead of C. SNPs in the 3′UTR of APOL1 were obtained from dbSNP build 96 databases (Table 1). SNPs related to APOL1 activity that do not match these matching areas were identified and evaluated. Scanning SNPs of miRNA genes targeting the 3′UTR of APOL1 on the miRNA gene targeting the 3′UTR of APOL1 was scanned from the NCBI database. SNPs in the genes of miRNAs and miRSNPs in the 3′UTR of APOL1 were matched (Table 2). Cross-match SNPs were searched as done by Ergun and Oztuzcu (2014); Safdar et al. (2020). Matched and unmatched SNPs are shown and interpreted for APOL1 activity with kidney diseases that has susceptibility to COVID-19.
Table 1.
List of miRNA target sequences and SNPs in binding sites in the 3′ UTR of APOL1
*Red text means “Change/SNP”, **Blue text means “The particular base that changed into SNPs”
Table 2.
List of miRNAs that targets the APOL1 gene having SNP in their genomic sequences at target site to their 3′ UTR of APOL1
*Red text means ’Change/SNP”, **Blue text means ’The particular base that changed into SNPs”
Samples collection, RNA isolation, and cDNA conversion
A total of 40 samples of nasopharyngeal swabs were collected from Erbil. There were 20 samples of kidney disease as well as COVID-19 patients and 20 were healthy controls. RNA isolation was achieved using a kit (Trizol, Sigma-Aldrich, Life Sciences Technologies) reagent as per the direction of the manufacturer and stored at − 20 C. The cDNA was synthesized using the kit (QuantiTect Reverse Transcription kit, Qiagen) as per the instructions of the manufacturer (Fig. 1).
Fig. 1.
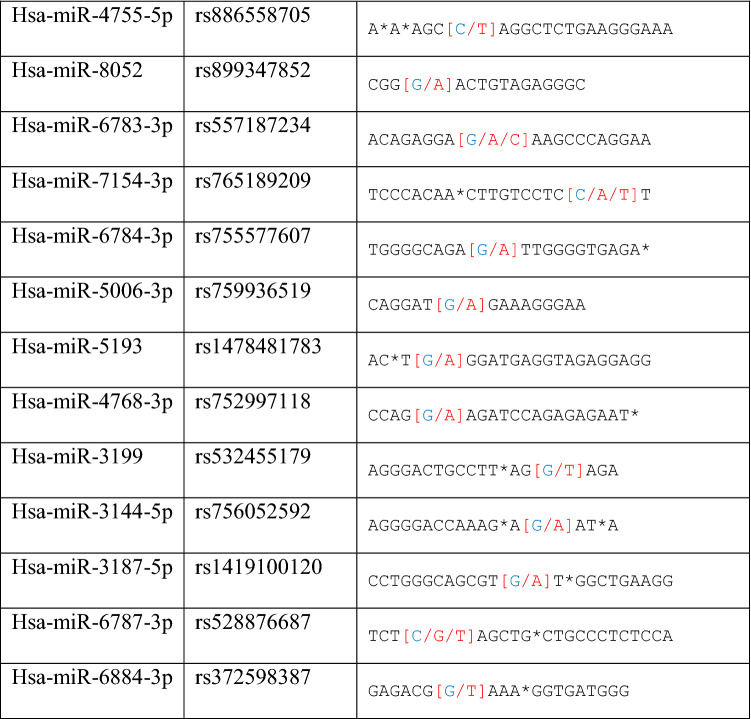
There are four important SNPs at the binding sites of miR-6741-3p and APOL1 genes. Interestingly, miR-6741-3p’s binding site on APOL1 has two SNPs (rs1288875001, G > C; rs1452517383, A > C) on APOL1 3′UTR. Similarly, two other SNPs (rs1142591, T > A; rs376326225, G > A) were identified at the first position binding sites of miR-6741-3p. Here, miRSNP (rs1288875001) at APOL1 3′UTR and SNP (rs376326225) at miR-6741-3p genomic sequence cross-match at the same site of binding region
Relative gene expression using qRT-PCR
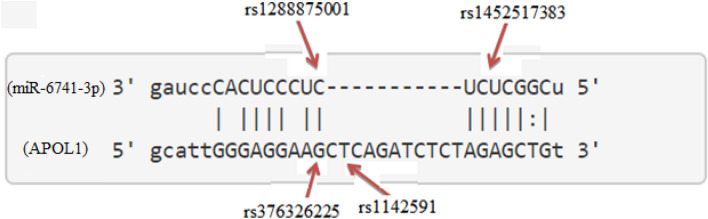
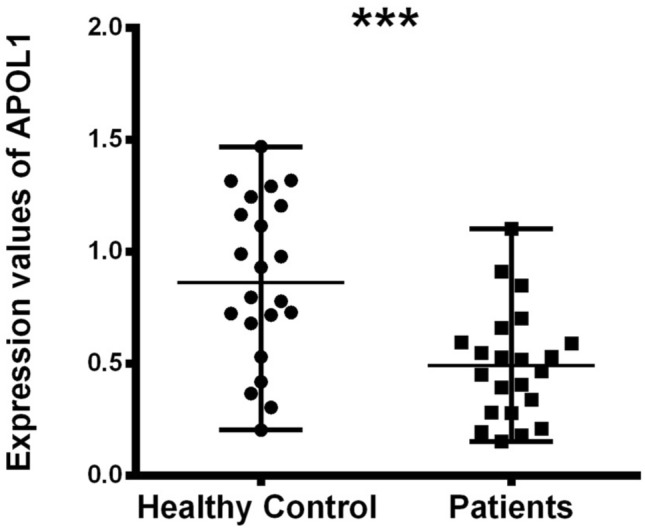
We used Rotor Gene qReal-Time PCR (qRT-PCR) to measure the expression level of miR-6741-3p and APOL1 gene in patient (kidney disease + COVID-19) and healthy subjects. Expression values of APOL1 in healthy and patients subjects are shown in Figs. 2, 3, and Table 3. The relative expression levels were calculated by the 2−ΔΔCt method & Mann–Whitney U test. By the results, APOL1 expression was found statistically lower in the patients than healthy (p < 0.05). As our hypothesis, we thought that miR-6741-3p targeting APOL1 gene may be used to inhibit complications in kidney patients with SARS-COV-2 infection. Because APOL1 gene inhibit partially transcription with miR-6741-3p since miR-6741-3p targets 3′UTR region of APOL1 according to computational analysis.
Fig. 2.
APOL1 expression difference between healthy and patients subjects (nasopharyngeal swab samples) normalized by GAPDH is shown by the dot plot (p < 0.05). Data were expressed as median with range. Data were non-parametric; they were analyzed by Mann–Whitney U test
Fig. 3.
miR-6741-3p expression difference between healthy and patients subjects (nasopharyngeal swab samples) normalized by RNU6B_13 is shown by the dot plot (p < 0.05). Data were expressed as median with range. Data were non-parametric, they were analyzed by Mann–Whitney U test
Table 3.
Comparisons of APOL1 and miR-6741-3p between healthy controls and patients
| Parameters | Healthy Controls (median (IQR)) | Patients (median (IQR)) | p Values |
|---|---|---|---|
| APOL1 | 0.863 (0.643 to 1.216) | 0.493 (0.280 to 0.611) | 0.0003 |
| miR-6741-3p | 0.985 (0.726 to 1.257) | 1.380 (1.188 to 1.600) | < 0.0001 |
Data were expressed as median with range. Data were non-parametric; they were analyzed by Mann–Whitney U test. p < 0.05 was regarded as statistically significant
Results and discussion
Finding binding and targeting sites on MiRNA is expensive and time-consuming. Estimation of miRNA/gene interaction by computational analysis is a valuable tool in a wet lab for experimental transmission. Also, as researchers face difficulties in predicting the undefined basic mechanisms of miRNA targets, finding a comparative figure of miRNA-mediated target binding is a powerful activity because it discourages the trouble of recognizing the potential miRNA–mRNA interaction of millions of miRNA–gene combinations (Huang et al. 2010). In addition, by suppressing mRNAs, various biological functions are targeted by the miRNA, and the representation of miRNA-mediated effects is problematic, even when they are attached to large-scale regulatory systems (Gosline et al. 2016). One of the benefits of existing databases is that it mostly includes miRNA–gene interaction in the 3′UTR regions.
In this study, we used ten different tools for analysis to detect miRSNPs in the APOL1 gene and SNPs on miRNA genes targeting APOL1 with COVID-19-sensitive kidney diseases. Online databases were used to find the 3′UTR of APOL1 gene targeted by miRNAs. Instead of just one database, ten different databases were used, followed by various algorithms and computational approaches. Accordingly, scanning of all miRNA targets can be performed without missing any of them using various databases. So we detected 96 miRNA-binding sites and 35 different SNPs in binding sites of miRNA in 3′UTR of the APOL1 gene (Tables 1, 2). Interestingly, miR-6741-3p’s binding site on APOL1 has two SNPs (rs1288875001, G > C; rs1452517383, A > C) on APOL1 3′UTR, and its genomic sequence has an SNP (rs376326225, G > A) at the same nucleotide with rs1288875001. Similarly, two other SNPs (rs1142591, T > A; rs376326225, G > A) were identified at the first position binding sites of miR-6741-3p. Here, miRSNP (rs1288875001) at APOL1 3′UTR and SNP (rs376326225) at miR-6741-3p genomic sequence cross-match at the same site of binding region (Fig. 1). These variations may assist the entry of SARS-COV-2 and develop infection in patients and damage lungs along with kidneys. Interestingly, some studies explained the mechanisms of the role of miRNAs in kidney diseases such as the overexpression of miR-29c decreased the levels of Spry1 protein because it is a direct target of miR-29c, and activated the Rho-kinase pathway in mice modeling (Long et al. 2011). Previously, some researchers found variants in APOL1 at G1 (rs73885319 and rs609101) and G2 (rs71785313). They also found variants in other related genes which have strong relation with kidney diseases, i.e., rs10854687 (APOL2), rs73641143 (APOL4), and rs4821481 and rs3752462 (MYH9) in kidney patients. Two variants of APOL1 gene at G1 and G2 showed strong association with FSGS. By reduction of G1 it does not show elimination of FSGS but by reduction of both G1 and G2 with respect to G0 shows reduction in FSGS (Genovese et al. 2013; Reidy et al. 2018). In our in vitro study, when we analyze miR-6741-3p and APOL1 expression profiles in healthy and patient subjects (co-infection of kidney disease and COVID-19), miR-6741-3p expression was found higher in patients than healthy according to Fig. 3 (p < 0.05). At the same time, APOL1 expression was found lower in patients than healthy subjects but this difference was not statistically significant according to Fig. 2 (p > 0.05). These results show that miR-6741-3p may have a role on regulation of APOL1 expression because of the fact that miR-6741-3p expression level increases while APOL1 expression level decreases in patients (Table 3). However, at this point, it is not possible to comment APOL1 expression profile upon mechanism.
Recently, the researchers have been focused on the genetic variations in APOL1 gene and found its relationship between COVID-19 and kidney diseases, especially collapsing glomerulopathy, chronic kidney disease (CKD), acute kidney injury (AKI), and tubulointerstitial lesions that lead to increase in the rate of COVID-19 in sub-Saharan African ancestry (Couturier et al. 2020; Larsen et al. 2019; Peleg et al. 2020). The relationship between the levels of miR-15b and miR-17 in kidney tissue and acute kidney damage in human and animal models has been reported in terms of expression analysis (Fan et al. 2016). Since gene expression levels are highly tissue specific, it is not clear whether gene expression studies in one tissue can be generalized to a different tissue (Consortium G 2015; Fu et al. 2012; Rodwell et al. 2004). APOL1 variant gene expression in kidney is required for disease induction, as shown by clinical renal transplant studies. So it is suggested for further studies immediately.
In addition, miR-6741-3p targets many APOL1-related genes (TLR7, SLC6A19, IL-6,10,18, chemokine (C–C motif) ligand 5, SWT1, NFYB, BRF1, HES2, NFYB, MED12L, MAFG, GTF2H5, TRAF3, angiotensin II receptor-associated protein, PRSS23) (Table 4) that have direct interaction with kidney diseases which may lead to SARS-COV-2 (Guo et al. 2020; Li et al. 2016; Moreno-Eutimio et al. 2020; Naicker et al. 2020; Vellingiri et al. 2020; Zhang 2020). Formerly, miR-6741-3p had not demonstrated any association towards kidney diseases and SARS-COV-2, but these genes have been studied intensively that explains different pathways for kidney diseases and SARS-COV-2 infection (Couturier et al. 2020; Larsen et al. 2019; Peleg et al. 2020). It assures that APOL1 could have a significant consequence of kidney diseases and its associated diseases through different pathways and could be hoping to be the association between miR-6741-3p and APOL1 gene because their SNPs are located in a splicing site that needs to be investigated in further in vivo studies.
Table 4.
List of kidney diseases-related genes (other than APOL1) targeted miR-6741-3p and associated with kidney disease pathways susceptibility to SARS-CoV-2 infection
| Sr# | Genes | Relationship of genes with Kidney diseases and SARS-COV-2 |
|---|---|---|
| 1 | AGTRAP,PRSS23SWT1, NFYB, BRF1, HES2, NFYB, MED12L, MAFG, GTF2H5 | Viral gene structure variations and human genes relationship with drugs |
| 2 | TLR7 and TRAF3 | SARS Coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6 |
| 3 | SLC6A19 | Functional association of mutant SLC6A19 transporters with APOL1 in the intestine |
| 4 | IL-6, IL-10, IL-18, CCL5 | APOL1 showed enhancement of pro-inflammatory cytokines, i.e., IL6,10,18 and chemokine (C–C motif) ligand 5 |
Conclusion
The single-nucleotide polymorphisms (SNPs) and targets found in various microRNA (miRNA) genes may change the miRNA activity leading to various diseases including cancer and neurodegenerative. Evidence reported that SNPs increase/decrease the effectiveness of the interaction between miRNAs and disease-related target genes. Therefore, there was a need to find the actual mechanisms of miRSNPs on the APOL1 gene and SNPs in miRNA genes targeting 3′UTR to evaluate the effect of these gene variations in kidney diseases and their associated COVID-19 infection. This study advances information by showing that the effects of miRSNPs on the APOL1 gene and SNPs in miRNA genes targeting 3′UTR are related to changes in kidney diseases and their associated SARS-COV-2 infection. After this, a validated in silico and in vitro analysis emerges that this study has better prediction and validation of miRNA targets and that the genomic sequences of SNPs and miRNAs in target regions are of great importance, especially in the 3′UTR region of APOL1 to develop new drugs for COVID-19.
Acknowledgements
We would like to thank Dr. Artak Heboyan and Dr. Qui Thanh Hoai Ta for their help in the analysis of this paper.
Authors’ contributions
All the authors have equally contributed to this research. All authors read and approved the fnal manuscript.
Funding
Departmental funds only, there are no external funds used and no external financial aid has been got.
Data availability
All data analyzed or generated during this study are included in this publication and its additional files.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate.
This study was approved by the ethical committee of Salahaddin University-Erbil (File No: 04/03/6120). This study was conducted in compliance with the Declaration of Helsinki (2013 revised Forta Reza) and the Ethical Guidelines for Medical Research Involving Human Subjects (partially revised on 28 February 2017). All patients included in the study provided written, informed consent at the time of enrolment of the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bátkai S, Thum T. MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr Hypertens Rep. 2012;14(1):79–87. doi: 10.1007/s11906-011-0235-6. [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. BioMed Res Int. 2014 doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 2020 doi: 10.1016/j.tmrv.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier A, Ferlicot S, Chevalier K, Guillet M, Essig M, Jauréguiberry S, Collarino R, Dargelos M, Michaut A, Geri G. Indirect effects of severe acute respiratory syndrome coronavirus 2 on the kidney in coronavirus disease patients. Clin Kidney J. 2020 doi: 10.1093/ckj/sfaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde AH, Snijder EJ, Kikkert M, van Hemert MJ. Roles of Host Gene and Non-coding RNA Expression in Virus Infection. Cham: Springer; 2017. Host factors in coronavirus replication; pp. 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci MDS, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun S, Oztuzcu S. Computational analysis of 3′ UTR Region of CASP3 with respect to miRSNPs and SNPs in targetting miRNAs. Comput Biol Chem. 2014;53:235–241. doi: 10.1016/j.compbiolchem.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Fan P-C, Chen C-C, Chen Y-C, Chang Y-S, Chu P-H. MicroRNAs in acute kidney injury. Hum Genom. 2016;10(1):29. doi: 10.1186/s40246-016-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wolfs MG, Deelen P, Westra H-J, Fehrmann RS, Te Meerman GJ, Buurman WA, Rensen SS, Groen HJ, Weersma RK. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8(1):e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Friedman DJ, Pollak MR. APOL1 variants and kidney disease in people of recent African ancestry. Nat Rev Nephrol. 2013;9(4):240. doi: 10.1038/nrneph.2013.34. [DOI] [PubMed] [Google Scholar]
- Gosline SJ, Gurtan AM, JnBaptiste CK, Bosson A, Milani P, Dalin S, Matthews BJ, Yap YS, Sharp PA, Fraenkel E. Elucidating MicroRNA regulatory networks using transcriptional, post-transcriptional, and histone modification measurements. Cell Rep. 2016;14(2):310–319. doi: 10.1016/j.celrep.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D, Jin H-J, Tan K-S, Wang D-Y, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-D, Tseng Y-T, Shrestha S, Lin Y-L, Khaleel A, Chou C-H, Chu C-F, Huang H-Y, Lin C-M, Ho S-Y. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(D1):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zou Q, Song H, Song F, Wang L, Zhang G, Shen X. A study of miRNAs targets prediction and experimental validation. Protein Cell. 2010;1(11):979–986. doi: 10.1007/s13238-010-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontaraki JE, Marketou ME, Zacharis EA, Parthenakis FI, Vardas PE. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: potential markers of target-organ damage. J Am Soc Hypertens. 2014;8(6):368–375. doi: 10.1016/j.jash.2014.03.324. [DOI] [PubMed] [Google Scholar]
- Larsen C, Bourne T, Wilson J. Collapsing glomerulopathy in a patient with coronavirus disease 2019 (COVID-19) Kidney Int Rep. 2020;5(6):935–939. doi: 10.1016/j.ekir.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-W, Wang C-Y, Jou Y-J, Huang S-H, Hsiao L-H, Wan L, Lin Y-J, Kung S-H, Lin C-W. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci. 2016;17(5):678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macha M, Seshacharyulu P, Ram Krishn S, Pai P, Rachagani S, Jain M, Batra S. MicroRNAs (miRNAs) as biomarker (s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 2014;20(33):5287–5297. doi: 10.2174/1381612820666140128213117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE. 2009;4(11):e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Eutimio MA, Lopez-Macias C, Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbess Infect. 2020 doi: 10.1016/j.micinf.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naicker S, Yang C-W, Hwang S-J, Liu B-C, Chen J-H, Jha V. The Novel Coronavirus 2019 epidemic and kidneys. Kidney Int. 2020;97(5):824–828. doi: 10.1016/j.kint.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(W1):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Y, Kudose S, D’Agati V, Siddall E, Ahmad S, Kisselev S, Gharavi A, Canetta P. Acute kidney injury due to collapsing glomerulopathy following COVID-19 infection. Kidney Int Rep. 2020 doi: 10.1016/j.ekir.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy KJ, Hjorten R, Parekh RS. Genetic risk of APOL1 and kidney disease in children and young adults of African ancestry. Curr Opin Pediatr. 2018;30(2):252. doi: 10.1097/MOP.0000000000000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2(12):e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar M, Zaheer S, Khailany RA et al (2020) The relevance of SNPs at 3’UTR region of CASP7 and miR-371b-5p associated diseases: a computational analysis. Cell Biochem Biophys 78:541–557. 10.1007/s12013-020-00941-2 [DOI] [PubMed]
- Song X, Shan D, Chen J, Jing Q. miRNAs and lncRNAs in vascular injury and remodeling. Sci China Life Sci. 2014;57(8):826–835. doi: 10.1007/s11427-014-4698-y. [DOI] [PubMed] [Google Scholar]
- Vellingiri B, Jayaramayya K, Iyer M, Narayanasamy A, Govindasamy V, Giridharan B, Ganesan S, Venugopal A, Venkatesan D, Ganesan H. COVID-19: a promising cure for the global panic. Sci Total Environ. 2020;725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. DIANA miRPath v. 2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(W1):W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang S-T, Wang Y-D, Wu G, Du Y, Qian M-Q, Liang X-G, Elbatreek MH, Yang H-Y, Liu Z-R. Stress-responsive heme oxygenase-1 isoenzyme participates in Toll-like receptor 4-induced inflammation during brain ischemia. NeuroReport. 2016;27(6):445–454. doi: 10.1097/WNR.0000000000000561. [DOI] [PubMed] [Google Scholar]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(D1):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-m, Zhang H. Genetic roadmap for kidney involvement of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) Infection. Clin J Am Soc Nephrol. 2020 doi: 10.2215/CJN.04370420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed or generated during this study are included in this publication and its additional files.