Summary
Identification of rare variants associated with schizophrenia has proven challenging due to genetic heterogeneity, which is reduced in founder populations. In samples from the Ashkenazi Jewish population, we report that schizophrenia cases had greater frequency of novel missense or loss of function (MisLoF) ultra-rare variants (URVs) compared to controls, and MisLoF URV burden was inversely correlated with polygenic risk scores in cases. Characterizing 141 “case-only” genes (MisLoF URVs in ≥ 3 cases with none in controls), the cadherin gene set was associated with schizophrenia. We report a recurrent case mutation in PCDHA3 which results in formation of cytoplasmic aggregates and failure to engage in homophilic interactions on the plasma membrane in cultured cells. Modeling purifying selection, we demonstrate that deleterious URVs are greatly over-represented in the Ashkenazi population, yielding enhanced power for association studies. Identification of the cadherin/protocadherin family as risk genes helps specify the synaptic abnormalities central to schizophrenia.
eTOC
Lencz et al. demonstrate the Ashkenazi Jewish population has enhanced power for genetic discovery in schizophrenia. Cases had excess missense or loss-of-function ultra-rare variants, enriched in cadherins and neurodevelopmental genes. A recurrent case mutation in PCDHA3 results in formation of cytoplasmic aggregates and failure to engage in membrane homophilic interactions.
Introduction
Twin studies and other family-based designs have long demonstrated that schizophrenia (SCZ) is highly heritable (h2≈.6-.85) (Hilker et al., 2018; McGue et al., 1983; Sullivan et al., 2003). While large-scale genome-wide association studies (GWAS) have discovered increasing numbers of common (minor allele frequency > 1%) variants associated with illness (Lam et al., 2019; Pardiñas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020), the cumulative effect of such variants accounts for only about a third of the total heritability of SCZ (Lee et al., 2012; Loh et al., 2015). It is therefore likely that rare genetic variants contribute substantially to the heritability of SCZ (Ganna et al., 2018; Purcell et al., 2014), and such rare variants might have considerably higher effect sizes (odds ratios) relative to common variants (Sullivan et al., 2012). For example, several rare (frequency<<1% in the general population) copy number variants have been reliably associated with SCZ, with odds ratios ranging from 5–20 or higher (Marshall et al., 2017).
Identification of rare single nucleotide variants (SNVs) associated with SCZ has proven difficult for several reasons: 1) SCZ is marked by a high degree of locus heterogeneity due to the large “mutational target” (i.e., damage to many different genes can increase risk for the phenotype) (Gratten et al., 2014); 2) at any given gene, a variety of different alleles may have deleterious effects (allelic heterogeneity) (Li & Leal, 2009); 3) deleterious rare variants are generally driven to extremely low frequencies due to purifying selection (Kryukov et al., 2007); and 4) the background rate of benign rare variation across the population is very high (Tennessen et al., 2012). To date, only very large international consortia efforts have identified any schizophrenia-associated SNVs. The largest such effort, the Schizophrenia Exome Sequencing Meta-analysis (SCHEMA) consortium identified only 10 exome-wide significant genes with a sample size of 25,000 cases and nearly 100,000 controls, (Singh et al., 2020).
One approach to enhance power in rare variant studies is to examine unusual populations marked by a strong, (relatively) recent founder effect; such populations are enriched for deleterious rare variants due to inefficient purifying selection (Locke et al., 2019; Wang et al., 2014). For example, the Ashkenazi Jewish (AJ) population, currently numbering more than 10 million individuals worldwide, effectively derives from a mere ~300 founders approximately 750 years ago (Carmi et al., 2014; Palamara et al., 2012). While the AJ population is well known to be enriched for deleterious variants leading to rare recessive disorders (Baskovich et al., 2016), AJ also demonstrate a 10-fold elevated frequency of high-penetrance risk variants for common complex disease, such as the LRRK2 p.G2019S allele associated with Parkinson’s disease (Ozelius et al., 2006) and the BRCA1 c.66_67AG allele associated with breast cancer (Friedman et al., 1995). Importantly, a recent large-scale (n>5,000) sequencing study of AJ individuals demonstrated that this enrichment is widespread across the exome, with approximately one-third of all protein-coding alleles demonstrating frequencies in AJ that were an order of magnitude greater than the maximum frequency in any well-characterized outbred population (Rivas et al., 2018).
In the present study, we examined rates of protein-altering ultra-rare variants (URVs) in AJ cases with schizophrenia compared with AJ controls. Based on prior research (Genovese et al., 2016; Gulsuner et al., 2020; Nguyen et al., 2017; Purcell et al., 2014), we hypothesized that cases would be enriched for rare deleterious variants. We also examined the relationship between URV burden and polygenic risk scores derived from common variant GWAS, to test the hypothesis that these would be inversely correlated in schizophrenia cases as predicted under the additive model. We sought to replicate prior findings that schizophrenia URVs would be over-represented in genes expressed at the neuronal synapse and during neurodevelopment, and to extend these results to additional categories of genes that might be detectable due to the greater frequency of rare variants observed in the AJ population. Additionally, we attempted to replicate the schizophrenia risk genes identified by the SCHEMA consortium, and we sought evidence of individual risk variants that could be observed multiple times across our dataset and SCHEMA. For one such variant, we performed in vitro experiments to identify potential molecular mechanisms by which risk for illness might be conferred. Finally, we modelled the process of purifying selection in a rapidly expanding, bottlenecked population, in order to quantify the relative power of AJ for rare variant discovery.
Results
Greater Rates of Novel Exonic Variants in Cases
After QC procedures, a total of 786 SCZ cases and 463 controls were available for final analysis. Groups did not significantly differ in total number of variants called in their whole genome (~3.68M) or exome (~49K) (Table 1). Called variants were then filtered for novelty against all variants observed in the TOPMED and gnomAD (v2.1.1, non-neuro) datasets. This variant filtration procedure was performed equally to both cases and controls in our dataset, and therefore the number of URVs per genome are not a function of asymmetrical sample size; notably, cases and controls did not significantly differ on total number of novel variants observed genome-wide (~5K). However, cases had significantly more novel exonic variants, exclusively limited to singletons (17.76±6.24 vs 15.44±6.42, p=6.13×10−10; Table 1).
Table 1.
Variant counts in cases and controls.
| Per Case (n = 786) | s.d. | Per Control (n=463) | s.d. | Logistic regression | Linear regression | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p-value | excess variant per case | 95% CI | p-value | ||||||
| Post-QC | variants per genome | 3676280 | 17055 | 3676451 | 18899 | 1 | [1.000, 1.000] | 0.97 | −15.51 | [−2060, 2029] | 0.99 |
| variants per exome | 49437 | 356 | 49454 | 387 | 1 | [1.000, 1.000] | 0.67 | −8.62 | [−51, 33] | 0.69 | |
| Post-filtering on gnomAD (non-neuro) and TOPMed | novel variants per genome | 5028 | 427 | 5034 | 322 | 1 | [1.000, 1.000] | 0.89 | −3.15 | [−48, 42] | 0.89 |
| novel variants per exome | 30.64 | 6.14 | 28.66 | 7.15 | 1.045 | [1.027, 1.064] | 1.53E-06 | 1.99 | [1.20, 2.79] | 1.04E-06 | |
| novel non-singleton per exome | 12.88 | 2.7 | 13.23 | 3.01 | 0.958 | [0.920, 0.998] | 3.78E-02 | −0.34 | [−0.67, −0.02] | 3.76E-02 | |
| novel singleton (URV) per exome | 17.76 | 6.24 | 15.44 | 6.42 | 1.067 | [1.045, 1.090] | 1.33E-09 | 2.34 | [1.61, 3.06] | 4.38E-10 | |
| non-MisLoF URV | 9.86 | 4.13 | 8.81 | 3.97 | 1.07 | [1.037, 1.103] | 1.66E-05 | 1.05 | [0.58, 1.52] | 1.30E-05 | |
| MisLoF URV | 7.9 | 3.54 | 6.63 | 3.51 | 1.116 | [1.077, 1.156] | 1.74E-09 | 1.29 | [0.88, 1.69] | 7.46E-10 | |
Against this backdrop, cases and controls were compared on the number of missense or loss of function (MisLoF) variants within the exome, in two ways: variant-based tests and gene-based tests. First, at the variant level, cases manifest a significantly elevated rate of novel MisLoF URVs (Table 1, last row). Cases also demonstrated an elevated rate of novel non-MisLoF URVs (Table 1, second-to-last row); however, even compared to this background elevation of non-MisLoF URVs, there was a significantly elevated proportion of exonic variants classified as MisLoF in cases (Fisher’s exact p=0.034). Next, we examined case-control differences in MisLoF vs. non-MisLoF URVs at the gene level, as follows: For each class of URV (MisLoF or non-MisLoF), we compared the number of genes hit by one or more such URVs in cases with no such hits in controls (“case-only” genes) to the number of genes hit by one or more such URVs in controls with no such hits in cases (“control-only” genes), using the formula:. As shown in Figure 1, this ratio was much greater (i.e., more case-only genes than control-only genes) for MisLoF URVs relative to non-MisLoF URVs (p=0.0001 by permutation test). Note that, for this analysis, we down-sampled the number of cases to match the number of controls, and iterated across 10,000 permutations.
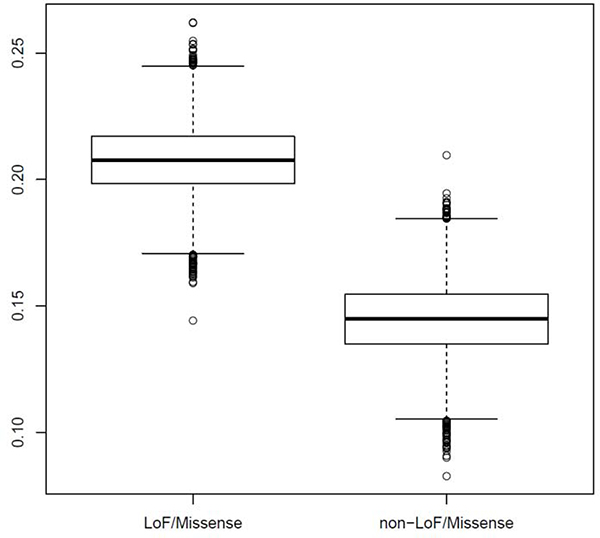
Figure 1: More Genes are hit by missense or loss-of-function ultra-rare variants (URVs) in Cases Relative to Controls.
Y-axis denotes the degree of elevation of “case-only” genes to “control-only” genes, defined by the ratio . Boxplot at left displays this ratio for missense or loss-of-function URVs; there are many more genes in which missense or loss-of-function URVs are observed in cases only, but no controls (ratio≈.205). Boxplot at right displays this ratio for all other URVs in the exome calling interval (including synonymous exonic, flanking intronic variants, and UTRs); while there are more genes in which such URVs are observed in cases only relative to controls only, the ratio (≈.145) is much smaller than that observed for missense and loss-of-function URVs (empirical p=0.0001; note that, for this analysis, we down-sampled the number of cases to match the number of controls, and iterated across 10,000 permutations).
MisLoF URVs Are Inversely Correlated with Common Variant Polygenic Risk Score in Cases
Next, we tested the liability threshold model of schizophrenia (Gottesman and Shields, 1967; Kendler, 2015; McGue et al., 1983; Smeland et al., 2020), which suggests that genetic risk factors are largely additive; if true, then it would be expected that cases with a greater URV burden would require a lower burden of common risk variants, as indexed by GWAS-derived polygenic risk score (PRS). By contrast, no relationship between PRS and URV would be expected in controls. For each subject, PRS was calculated based on the large-scale schizophrenia GWAS reported by the Psychiatric Genomics Consortium (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020), excluding our own Ashkenazi cohort, as described in Methods. The p-value threshold (PT) for calculating the PRS from the GWAS summary statistics was determined by optimizing R2 for the comparison of AJ cases and controls. At PT = 0.00725, the Nagelkerke R2 for PRS of the cases vs. controls was optimized at 0.15 on the observed scale (OR = 2.21 for 1 unit of standardized PRS, p < 2×10−16), consistent with previous estimates (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020). Using this threshold, and controlling for sex and the first five principal components derived from the GWAS data, there was a significant inverse relationship between PRS and total number of MisLoF URVs in cases (β=−0.0232, s.e.=0.0095, p=0.014), but not in controls (β=−0.0005, s.e.=0.0126, p=0.967). Results were substantively unchanged when a different PRS threshold was used, based on the optimal level (PT = 0.05) reported in the original GWAS (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020); there remained a significant inverse relationship between PRS and URV for cases (β=−0.0207, s.e.=0.0096, p=0.031), but not controls (β=−0.0031, s.e.=0.0124, p=0.805).
Replication of Previously Identified Schizophrenia Risk Genes
Given that cases demonstrated significant elevation in genes carrying MisLoF URVs, we next sought to characterize the genes that were hit by MisLoF URVs in cases only, with none observed in controls. As shown in Table S1a, eight genes had Case ≥ 5 and Control = 0 MisLoF URVs. Notably, one of these was SETD1A, a methyltransferase gene that was the first to reach genome-wide significance in a schizophrenia rare variant study (Singh et al., 2016). We next tested the set of 9 autosomal exome-wide-significant schizophrenia genes identified by the SCHEMA consortium (Singh et al., 2020) for overlap with 141 “case-only” genes in which ≥ 3 AJ cases in our dataset had MisLoF URVs, with none found in our AJ controls (full list provided in Table S1a). Three of 9 autosomal exome-wide significant SCHEMA genes (SETD1A, TRIO, and XPO7) were among the 141 case-only genes, a 47-fold over-representation relative to chance (hypergeometric test p = 2.89×10−5). Results remained significant when permutation tests controlling for gene size (see Methods) were performed (empirical p=1.7×10−3, see also Table S2). Similar results were obtained examining overlap of our 141 case-only genes with the set of 29 autosomal genes that met the criteria of FDR<.05 in SCHEMA; in addition to the three genes above, STAG1 was also shared between our case-only list and SCHEMA (4/29 genes; hypergeometric p=5.12×10−5; empirical p=8.5×10−3 using permutations controlling for gene size).
In addition to evidence that our dataset provided support for genes identified as significant in SCHEMA, we also used the broader SCHEMA dataset to provide supporting evidence for the genes on our case-only list. We observed that a total of 13 of our case-only genes had nominal (p<.05) support within the SCHEMA dataset, which represents a significant enrichment relative to chance (hypergeometric p=0.012; empirical p<1×10−4 using permutations controlling for gene size). These 13 genes include BSN, CACNA1E, DNAJC14, HMGCR, KIAA0586, MACF1, SPAG5, TOP2B, and WFS1, in addition to the four genes noted in the paragraph above. Additionally, we examined the distribution of p-values in SCHEMA for the 141 case-only genes we identified, as compared against all other genes. As shown in Figure S1, there is a distinct enrichment for our case-only genes compared to all other genes, and the lambda values are 1.14 and 0.73, respectively.
Within our set of 141 case-only genes, there was no evidence of oligogenic effect; the distribution of cases with >1 MisLoF variant in this set of genes are as expected under the null for multinomial distributions (multinomial goodness of fit p=0.3825)(Resin, 2020). Moreover, the distribution of multiply “hit” cases did not significantly differ (Χ2(4)=3.15, p=0.53) from the distribution of controls with >1 MisLoF variant in a similarly-sized set of 148 control-only genes (defined as genes in which ≥2 AJ controls had MisLoF URVs with none found in our AJ cases; Table S1b).
We compared the 347 cases who carried at least one MisLoF URV in the 141 case-only genes to the 439 cases who were non-carriers of such URVs on three clinical variables available in our dataset: 1) age of onset; 2) severity of course (defined as continuous illness without episodes of full or partial remission); and 3) treatment resistance (defined using prescription of clozapine as a proxy for treatment resistance (Ruderfer et al., 2016)). There was no significant difference between case carriers and case non-carriers on age of onset (24.3 years vs 24.2 years; t=0.27, p=0.79); there was no significant difference in designation of course as “continuous” (46.8% vs 42.7%, χ2=1.10, p=0.29); and there was no significant difference in treatment-resistance as suggested by prescription of clozapine (20.6% vs 19.4%, χ2=0.18, p=0.67). As a test of sensitivity of our clinical measures, we confirmed a significant relationship between treatment-resistance and age of onset; patients who were prescribed clozapine (regardless of URV status) had an earlier age of onset compared to patients not prescribed clozapine (22.5 vs 24.7 years of age; t=−2.94, p=0.003), as has been consistently reported in prior studies (Smart et al., 2019).
Case-only Genes are Enriched for Known Neurodevelopmental Genes, Synaptic Genes, and Cadherins
We utilized gene set analyses to characterize our 141 case-only genes and 148 control-only genes. First, we tested sets of genes selected a priori based on prior literature; specifically, previous case-control exome studies in schizophrenia have identified: 1) overlaps with other developmental brain disorders (DBD) including autism spectrum disorder (ASD) and intellectual disability (ID); 2) gene sets representing critical synaptic and/or neurodevelopmental functions such as binding partners of FRMP, RBFOX, and CELF4; and 3) constrained genes (i.e., genes with far fewer missense and/or loss of function variants than average, presumably due to purifying selection) (Genovese et al., 2016; Gonzalez-Mantilla et al., 2016; Gulsuner et al., 2020; Nguyen et al., 2017; Purcell et al., 2014). As shown in Table 2, each of these gene sets demonstrated significant (by hypergeometric test) overlap with the case-only gene lists; moreover, the difference in enrichment between the case-only and control-only gene sets was statistically significant in all cases. Permutation tests accounting for gene size demonstrated similar results, with the exception that the overlap with ASD/ID gene sets were no longer significant, and the SynaptomeDB gene set was marginal (p=.0514) (Table S2). Moreover, the size-matched permutation tests demonstrate that the genes on the control-only list were significantly under-represented amongst many of the key gene sets of interest, as indicated by empirical p-values > 0.95 (i.e., fewer than 5% of all gene-size-matched permuted gene lists had less overlap with the target gene set than the control-only list itself).
Table 2.
A priori gene set analysis (see also Table S2).
| Genes in Gene set | Case-only overlap | Control-only overlap | P(Case) | P(control) | OR | X2 p-value | |
|---|---|---|---|---|---|---|---|
| DBD (Gonzalez-Mantilla et al., 2016) | 214 | 11 | 1 | 3.95×10−7 | 8.01×10−1 | 12.44 | 2.40×10−3 |
| ASD (Satterstrom et al., 2020) | 102 | 4 | 0 | 6.13×10−3 | 1.00 | Inf | 3.91×10−2 |
| ASD_ID_DD (Coe et al., 2019; Satterstrom et al., 2020) | 274 | 8 | 2 | 7.93×10−4 | 6.10×10−1 | 4.39 | 4.45×10−2 |
| SynaptomeDB (Pirooznia et al., 2012) | 1828 | 26 | 15 | 4.88×10−4 | 3.93×10−1 | 2.00 | 4.31×10−2 |
| CELF4 (Wagnon et al., 2012) | 2504 | 41 | 23 | 1.70×10−7 | 1.74×10−1 | 2.23 | 5.60×10−3 |
| FMRP (Darnell et al., 2011) | 1210 | 35 | 19 | 6.15×10−13 | 1.74×10−3 | 2.24 | 8.98×10−3 |
| RBFOX2 (Weyn-Vanhentenryck et al., 2014) | 2911 | 48 | 27 | 7.10×10−9 | 1.37×10−1 | 2.31 | 2.19×10−3 |
| RBFOX1/3 (Weyn-Vanhentenryck et al., 2014) | 3255 | 50 | 27 | 3.18×10−8 | 3.10×10−1 | 2.46 | 9.35×10−4 |
| Missense-constrained (Samocha et al., 2014) | 961 | 23 | 8 | 3.13×10−7 | 4.30×10−1 | 3.41 | 2.74×10−3 |
| LoF-constrained (Karczewski et al., 2020) | 3264 | 62 | 35 | 1.35×10−14 | 1.54×10−2 | 2.53 | 2.55×10−4 |
| PGC3 broad (PGC3, 2020) | 426 | 3 | 4 | 5.88×10−1 | 3.95e-01 | 0.78 | 7.51×10−1 |
| PGC3 prioritized (PGC3, 2020) | 111 | 1 | 0 | 5.49×10−1 | 1.00 | Inf | 3.05×10−1 |
| PsychEncode DEG (Gandal et al., 2018) | 3953 | 35 | 36 | 9.31×10−2 | 1.12×10−1 | 1.03 | 9.22×10−1 |
| PsychEncode TWAS (Wang et al., 2018) | 301 | 4 | 1 | 1.68×10−1 | 8.98×10−1 | 4.29 | 1.59×10−1 |
By contrast, there was no significant overlap of the case-only list with genes identified in the most recent GWAS of the Psychiatric Genomics Consortium (using either broad or narrow criteria for prioritizing genes within a GWAS locus) (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020), nor was there any significant overlap with schizophrenia risk genes identified in two reports from the PsychEncode Consortium: 1) 301 genes identified by a transcriptome-wide association study (TWAS) derived from GWAS results (Wang et al., 2018); and 2) 3,953 differentially expressed genes (DEGs) identified in postmortem brain samples of patients with schizophrenia compared to postmortem samples from controls (Gandal et al., 2018). Because these results were non-significant, they were not examined further with permutation testing.
Next, we examined enrichment of our case-only gene list across all GO categories and Panther protein classes (Ashburner et al., 2000; Gene Ontology Consortium, 2019; Mi et al., 2019; The Gene Ontology Consortium, 2017), as well as synaptic components using annotations from SYNGO (Koopmans et al., 2019). As shown in Table 3, the 141 case-only genes were significantly enriched for biological processes related to cell adhesion, and specifically to the cadherin class of proteins. It is noteworthy that the enrichment for cadherins is driven by three of the four FAT atypical cadherins (FAT1, FAT2, and FAT4), all in different chromosomal regions, appearing on our case-only list (as did their key interacting gene, DCHS2); specific mutations observed in these genes are listed in Table S3. By contrast, none of the 18 genes in the cadherin gene set were observed on the list of 148 control-only genes. SYNGO analysis demonstrated that enrichment extended across both presynaptic and postsynaptic genes (Table S4). By contrast, the 148 control-only genes showed no synaptic enrichment (all q-value>.75), no enriched GO categories (all FDR>.05), and only one enriched Panther protein class, in a very small gene set (Hsp90 family chaperone, overlap of 3/9 genes; p=6.37×10−5, FDR=1.24×10−2).
Table 3.
Hypothesis-free gene set analysis (see also Table S2).
| GO bioloqical process | #genes | overlap | expected | Fold Enrichment | raw P value | FDR |
|---|---|---|---|---|---|---|
| homophilic cell adhesion via plasma membrane adhesion molecules | 167 | 10 | 1.12 | 8.92 | 3.14E-07 | 4.98E-03 |
| ↳cell-cell adhesion via plasma-membrane adhesion molecules | 256 | 10 | 1.72 | 5.82 | 1.22E-05 | 4.84E-02 |
| ↳cell adhesion | 947 | 19 | 6.36 | 2.99 | 2.32E-05 | 6.15E-02 |
| ↳biological adhesion | 953 | 20 | 6.40 | 3.13 | 7.29E-06 | 3.85E-02 |
| neurogenesis | 1401 | 24 | 9.41 | 2.55 | 2.33E-05 | 5.29E-02 |
| ↳developmental process | 5765 | 62 | 38.71 | 1.60 | 2.64E-05 | 4.66E-02 |
| ↳nervous system development | 2203 | 32 | 14.79 | 2.16 | 2.51 E-05 | 4.97E-02 |
| ↳system development | 4317 | 53 | 28.99 | 1.83 | 3.22E-06 | 2.55E-02 |
| ↳multicellular organism development | 4906 | 56 | 32.94 | 1.70 | 1.47E-05 | 4.66E-02 |
| ↳multicellular organismal process | 6985 | 71 | 46.90 | 1.51 | 3.10E-05 | 4.92E-02 |
| PANTHER protein class | ||||||
| cadherin | 18 | 4 | .12 | 33.10 | 1.26E-05 | 2.46E-03 |
| ↳cell adhesion molecule | 90 | 5 | .60 | 8.27 | 4.40E-04 | 2.14E-02 |
| intermediate filament binding protein | 15 | 3 | .10 | 29.79 | 2.20E-04 | 2.15E-02 |
| ↳intermediate filament | 15 | 3 | .10 | 29.79 | 2.20E-04 | 1.43E-02 |
| extracellular matrix protein | 166 | 6 | 1.11 | 5.38 | 1.05E-03 | 4.11E-02 |
As shown in Table S2, the lead categories remained significant in the size-matched permutation procedure, and the control-only list was significantly under-represented in several of these gene sets as well. Additionally, we ran the GO biological processes and PANTHER protein class analyses on 100 permuted gene lists, size-matched to the case-only list. For the PANTHER protein class analyses, only 1 out of 100 permutations generated a p-value stronger than the one we reported for the cadherin gene set; moreover, 81/100 permutations resulted in no significant (FDR-corrected) protein classes at all. For the GO biological process analyses, permutation testing was somewhat more equivocal; twelve of the 100 permutations generated p-values stronger than our top gene set (homophilic cell adhesion via plasma membrane adhesion molecules), although 67/100 permutations resulted in no FDR-significant gene sets.
A Damaging URV in PCDHA3 Is Observed Recurrently in Ashkenazi Schizophrenia Cases
The foregoing analyses examined singleton URVs only, which have been the primary focus of exome studies in schizophrenia to date; indeed, the SCHEMA dataset of case-only variants contains ~95% singletons, and <1% of all case-only variants in SCHEMA are observed 3 or more times. However, we hypothesized that the AJ population would be more likely than non-founder populations to retain and propagate multiple copies of deleterious variants. Consequently, we merged exome data from our cohort with the AJ schizophrenia cases (n=869) and controls (n=2415) from SCHEMA, in order to identify individual URVs that were observed ≥3 times in cases (i.e., at least ~1/1000 allele frequency in the 3,310 AJ case chromosomes available). For further filtering, we utilized exome data from an additional 1,587 AJ controls from a separate study of longevity. As shown in Table 4, 17 MisLoF URVs were observed in ≥3 cases and zero controls (nominal p<.05 by Fisher’s exact test). Notably, the most common variant (observed 5 times, case frequency = .15%) is a putatively damaging (CADD score = 23.4) missense variant (chr5:140182458:A/G; p.Asn559Ser) in PCDHA3, part of the protocadherin gene cluster on chromosome 5. Intriguingly, two of the three patients in our own dataset who carried this variant had early-onset schizophrenia (ages at first diagnosis for the three patients were 12, 13, and 22, respectively), and one of these two was prescribed clozapine. Of course, these clinical findings must be considered preliminary due to the very small number of patients involved, although the enrichment of very early-onset cases is statistically significant, given that only 19 other cases in the full cohort had age of onset ≤ 13 (Fisher’s exact p=0.002).
Table 4.
Recurrent (≥3 observations) MisLoF URVs in Ashkenazi cases.
| N Cases | Gene | Chr | Position | Ref | Alt | Impact | Coding Pos | Protein Pos | AA Change |
|---|---|---|---|---|---|---|---|---|---|
| 5 | PCDHA3 | 5 | 140182458 | A | G | missense | 1676 | 559 | N/S |
| 4 | ACBD6 | 1 | 180382593 | T | G | missense | 481 | 161 | N/H |
| 4 | IGF1R | 15 | 99251324 | C | T | missense | 628 | 210 | R/C |
| 3 | ATAD3C | 1 | 1389777 | CA | C | frameshift | 276 | 92 | T/X |
| 3 | ACOX3 | 4 | 8412048 | G | A | missense | 578 | 193 | A/V |
| 3 | FIGNL1 | 7 | 50513662 | G | A | missense | 1324 | 442 | P/S |
| 3 | CEP104 | 1 | 3740011 | T | C | missense | 2480 | 827 | H/R |
| 3 | UBR4 | 1 | 19426131 | A | G | missense | 13262 | 4421 | V/A |
| 3 | KLHL30 | 2 | 239049577 | T | C | missense | 182 | 61 | M/T |
| 3 | TBX18 | 6 | 85457687 | G | C | missense | 860 | 297 | S/C |
| 3 | CACNA2D1 | 7 | 81599254 | A | C | missense | 2287 | 763 | F/V |
| 3 | TENM4 | 11 | 78369465 | A | T | missense | 7948 | 2650 | S/T |
| 3 | TNFRSF1A | 12 | 6440026 | C | A | missense | 618 | 206 | E/D |
| 3 | BCAT1 | 12 | 25002856 | T | C | missense | 574 | 192 | K/E |
| 3 | LCP1 | 13 | 46722531 | C | T | missense | 934 | 312 | E/K |
| 3 | PCSK2 | 20 | 17240934 | A | T | missense | 227 | 76 | K/M |
| 3 | PTK6 | 20 | 62164958 | A | G | missense | 616 | 206 | F/L |
In Vitro Characterization of the PCDHA3 Variant
The PCDHA3 gene is a member of the clustered protocadherin gene family, which is comprised of three gene clusters: α β and γ which are stochastically expressed in individual neurons, generating a cell surface “barcode” required for neuronal self-recognition (Canzio and Maniatis, 2019; Mountoufaris et al., 2018; Rubinstein et al., 2017; Wu and Jia, 2020). Individual PCDH isoforms from all three gene clusters form nearly random cis-dimers (αβ, αγ, β/γ β/β γ/γ − but not α/α), which assemble on the cell surface, and engage in highly specific homophilic interactions with cis-dimers on apposing plasma membranes of neurites, which leads to self-recognition and neurite repulsion (Goodman et al., 2017; Goodman et al., 2016; Rubinstein et al., 2015; Thu et al., 2014). Mouse studies have shown that mutations in the PCDH gene cluster can result in neural circuit deficits and behavioral phenotypes associated with neuropsychiatric or neurodevelopmental disorders (Chen et al., 2017; Katori et al., 2009, 2017; Mountoufaris et al., 2017).
The p.Asn559Ser URV in PCDHA3 is part of the DxND motif within the EC5 domain, and lies within a calcium-coordinating residue which is highly conserved across all clustered protocadherin proteins. The EC5 domain is required for cis-dimerization, and PCDHα isoforms must dimerize with either PCDHβ or γ isoforms in order to localize on the plasma membrane (Goodman et al., 2017; Thu et al., 2014). To investigate the possibility that the p.Asn559Ser URV affects membrane localization, we examined the localization of mCherry-tagged PCDHα3 wildtype and PCDHα3(N559S) mutant isoforms in non-neuronal K562 cells when co-expressed with the PCDHγC3ΔEC1 carrier isoform (Thu et al., 2014). These cells, which do not endogenously express PCDH isoforms, have been used extensively to study PCDH homophilic interactions in prior work (Thu et al., 2014); notably, this prior work has demonstrated the most robust effects for PCDH isoforms lacking the intracellular domain (ICD) of the protein. In the present report, we show primary results for isoforms without the ICD in Figure 2, and comparable results for full-length isoforms in Figure S2; results did not substantively differ according to expression of the ICD, but were somewhat more robust for isoforms lacking the ICD.
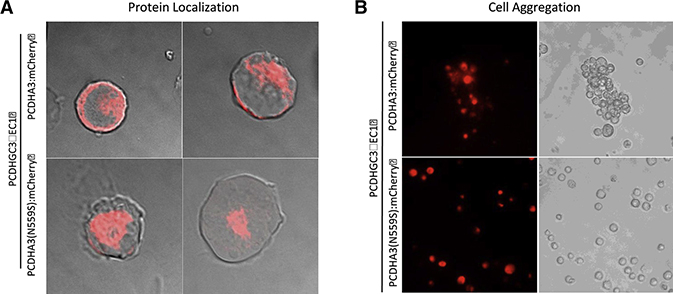
Figure 2: PCDHA3 p.Asn559Ser variant causes mis-localization of PCDHα3 protein and disrupts homophilic aggregation in vitro.
a) Expression of wildtype PCDHα3 or PCDHα3(N559S) mCherry fusion proteins (both excluding the intracellular domain), along with PcdhγC3ΔEC1 carrier protein, in non-neuronal K562 cells to assess cell surface localization. Wildtype mCherry-labeled PCDHα3 protein localized primarily to the cell surface, although some mCherry signal could also be detected in the cytoplasm. By contrast, the PCDHα3(N559S) protein was not detected at the cell surface; rather, it localized to the cytoplasm. b) Cell aggregation assay in K562 cells expressing either wildtype PCDHα3 or PCDHα3(N559S) mCherry fusion proteins (both excluding the intracellular domain), along with PcdhγC3ΔEC1 carrier protein to assess PCDH homophilic interaction. K562 cells expressing the wildtype PCDHα3 protein formed homophilic cell-cell aggregates while those expressing PCDHα3(N559S) proteins failed to aggregate, remaining as individual, non-aggregating cells in suspension.
We found that the wildtype PCDHα3 protein localized primarily to the surface of the K562 cells, although the mCherry signal could also be detected in the cytoplasm (Figure 2a; Figure S2a). By contrast, the PCDHα3(N559S) protein was not detected at the cell surface. Rather, it localized to the cytoplasm, suggesting that the PCDHα3 URV may disrupt dimerization with the PCDHγC3 carrier isoform, which is required for plasma membrane localization, and thus fails to be transported to the cell surface. Alternatively, the amino acid substitution may destabilize the protein causing it to form aggregates in the cytoplasm.
To determine whether the mis-localization of the PCDHα3(N559S) prevents the protein from engaging in the homophilic interactions required for self-recognition, we performed a cell aggregation assay in K562 cells expressing either the mCherry-tagged wild type PCDHα3 or the PCDHα3(N559S) mutant isoform along with the PCDHγC3ΔEC1 carrier isoform. As shown in Figure 2b (and Figure S2b), K562 cells expressing the wildtype PCDHα3 protein formed homophilic aggregates (consistent with previous findings (Thu et al., 2014)), while those expressing PCDHα3(N559S) proteins failed to form aggregates, remaining as individual, non-aggregating cells in suspension. These results indicate that the PCDHα3 URV identified in schizophrenia cases may prevent formation of cis-dimers, which are required for membrane localization, homophilic interactions, and proper self-avoidance. Thus, the absence of Pcdhα heterodimerization may interfere with normal cell surface lattice formation, potentially resulting in neural circuit deficits in individuals bearing the p.Asn559Ser variant.
Damaging Rare Variants Escape Purifying Selection in a Founder Population
Given that we were able to detect numerous recurrent case-only variants, despite our relatively small sample size compared to SCHEMA, we sought to model the parameters affecting the persistence of deleterious alleles in a founder population. We initiated a series of simulations based on our prior estimates of the size (N=300) and timing (30 generations ago) of the Ashkenazi bottleneck (Carmi et al., 2014; Palamara et al., 2012), and population-based estimates (Power et al., 2013) of reduced fecundity in schizophrenia (fecundity ratio ~0.5 for females and ~0.25 for males). We then generated 10,000 simulations for each of a series of variations on these parameters (Table S5) in order to model the odds of a deleterious variant, present in a single individual at the time of the bottleneck, escaping extinction to persist in the present AJ population. In addition to the parameters noted above, simulations were performed as a function of penetrance for schizophrenia. As shown in Figure 3a, between 30–50% of such variants escape extinction within the range of penetrance expected, given the genetic architecture of the disorder (Sullivan et al., 2012).
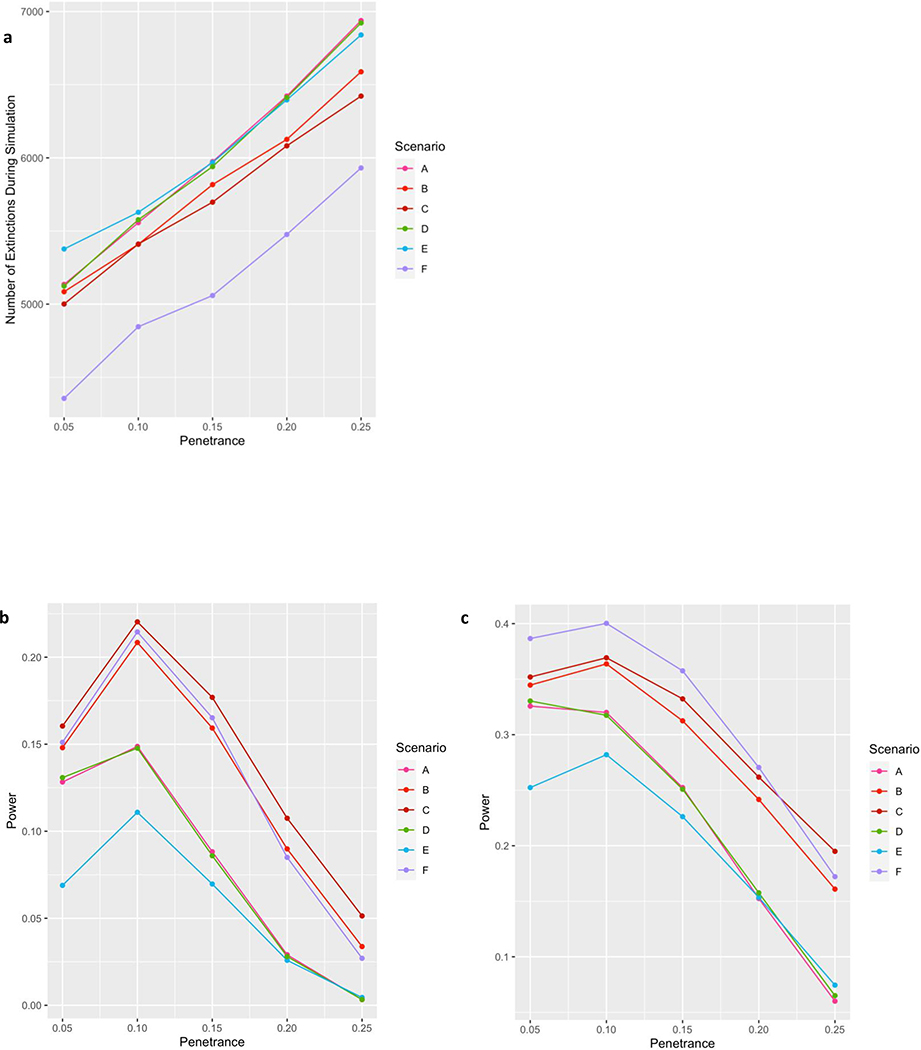
Figure 3: Damaging Rare Variants Escape Purifying Selection in a Founder Population.
a) Under a range of scenarios consistent with known population and disease parameters, as many as half of all damaging variants remain in a rapidly expanding founder population. Scenario A represents best estimates of the size and timing of the AJ population bottleneck based on our prior work (Carmi et al., 2014; Palamara et al. 2012), and effects of schizophrenia on fecundity based on the work of Power et al. (2013); other scenarios (detailed in Table S5) test sensitivity of the model to variations in fecundity effects (Scenarios B and C), size of the bottleneck (Scenario D), and number of generations since the bottleneck (Scenarios E and F). Y-axis denotes number of extinctions out of 10,000 simulations for each condition. b) Given rough estimates of the AJ population today (~10M) and the prevalence of schizophrenia (~1%), the power of our discovery cohort combined with the SCHEMA AJ cohort (N=1637 cases and 2878 controls) to detect a given variant at exome-wide significance generally ranged between 5% and 20%. Notably, while power tends to decrease at higher levels of penetrance due to increased variant extinction (as shown in panel a), there also tends to be a decrease in power at the lowest level of penetrance due to an increased frequency of these variants appearing in controls. c) For a slightly larger study (N=5000 cases and 9000 controls) power would be ~20–40% to detect any individual variant in the range of realistic penetrance, resulting in >80% power to detect at least one variant, assuming there are at least 7 such variants circulating in the population.
Based on these results, and given rough estimates of the AJ population today (~10M) and the prevalence of schizophrenia (~1%), we could then estimate the total number of case and control carriers expected in the contemporary AJ population for each scenario. These calculations allowed us to determine the power of our discovery cohort combined with the SCHEMA AJ cohort (total N=1637 cases and 2878 controls) to detect a given variant at exome-wide significance (Figure 3b), which generally ranged between 5% and 20%. However, we also calculated that a slightly larger study, of 5000 cases and 9000 controls, would have power of ~20–40% to detect any individual variant in the range of realistic penetrance (Figure 3c), and would therefore have >80% power to detect at least one variant, assuming there are at least 7 such variants circulating in the population (i.e., even if only 5% of our case-only list were true positives). Such an assumption is likely to be extremely conservative, given the estimated mutational target of 1000 genes or more (Nguyen et al., 2017; Purcell et al., 2014), the replication of the SCHEMA results in our dataset, the significant findings documented in Table 2, and the long list of variants at greater than doubleton frequency documented in Table 4.
Discussion
The present study demonstrates the enhanced power available to genetic studies performed in populations enriched for rare variants, consistent with recent work in schizophrenia (Gulsuner et al., 2020) and other phenotypes (Locke et al., 2019; Rivas et al., 2018; Selvan et al., 2020). We further reduced background heterogeneity by utilizing a strict filter against all variants reported in non-neuropsychiatric samples across the two largest publicly available sequencing datasets, gnomAD (Karczewski et al., 2020) and TOPMED (The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program., 2018). Thus, despite relatively modest sample sizes, the present study was able to replicate several previously identified schizophrenia-associated genes (SETD1A, TRIO, XPO7) (Singh et al., 2016, 2020) and gene sets (synaptic, DBD-related, and constrained genes) (Genovese et al., 2016; Gonzalez-Mantilla et al., 2016; Gulsuner et al., 2020; Nguyen et al., 2017; Purcell et al., 2014), with these analyses serving as a positive control for our approach. Beyond these replications, we were also able to make several additional discoveries, as described below.
First, we identified several gene sets associated with schizophrenia that have not been reported in previous studies. The strongest statistical signal was observed for cell adhesion processes, especially the cadherin family genes (Table 3). Cadherins form calcium-dependent adherence junctions at the synapse and are involved in both neuronal migration and mature synaptic activity (Friedman et al., 2015). Surprisingly, cadherins have not received much attention in the schizophrenia genetics literature, despite the considerable recent focus on both calcium activity and synaptic proteins (Nanou and Catterall, 2018). While there are more than 100 different proteins in the cadherin superfamily (Friedman et al., 2015), it is noteworthy that three of the four FAT atypical cadherins, all in different chromosomal regions, appeared on our case-only list (as did their key interacting gene, DCHS2). These genes are specifically involved in regulating microtubule polarity, thereby directing cellular migration in the developing nervous system (Avilés and Goodrich, 2017; Fulford and McNeill, 2020). Homozygous mutations in FAT4 cause Van Maldergem syndrome, a recessive intellectual disability marked by periventricular neuronal heterotopia (Cappello et al., 2013), while mutations in FAT1 have been observed in autism (Cukier et al., 2014).
Relatedly, we observed a single missense variant in a protocadherin gene (PCDHA3) at higher rate of recurrence (5 observations) in cases than any other ultra-rare (i.e., not in healthy individuals) variant in the published schizophrenia literature (although it should be noted that one splice acceptor variant in SETD1A appears six times in the SCHEMA database). PCDHA3 is a member of the PCDH gene cluster, which in conjunction with the stochastic expression of PCDHα, β and γ isoforms, generates a “molecular barcode” on the cell surface required for neuronal self-recognition (Canzio and Maniatis, 2019). Individual PCDH proteins form nearly random cis-dimers, which are transported to the plasma membrane, and engage in trans with homophilic partners on the apposing cell surface. Ultimately, the assembly of PCDH cis/trans tetramers on the plasma membrane creates a lattice-like structure, which may be required for repulsion and self-avoidance (Brasch et al., 2019).
The missense variant in the PCDHA3 gene is located in the EC5 domain of the protein, which is critical for cis-dimerization and cell surface localization of PCDHα isoforms (Thu et al., 2014). Consistent with the role of the EC5 domain, we found that the PCDHα3 variant protein failed to localize on the surface of K562 cells, instead accumulating as cytoplasmic aggregates. Such an effect in primary neurons may result in failure of the PCDHα3 variant protein to localize to the plasma membrane, preventing homophilic engagement and disrupting the assembly of the PCDH protein lattice, which would cause deficits in self-avoidance. In patients carrying this variant, these effects would likely have a significant impact on neural circuit formation. For example, previous studies have shown that the PcdhαC2 protein is required for normal serotonergic neuron wiring (Chen et al., 2017; Katori et al., 2009, 2017). Moreover, disruption of PCDHα isoforms is consistent with other findings in schizophrenia; for example, altered expression of protocadherins (including PCDHα3) in schizophrenia has been implicated by a recent transcriptome-wide association study of both prefrontal cortex and hippocampus (Collado-Torres et al., 2019). This observation is supported by functional interrogation of the schizophrenia GWAS locus encompassing the PCDHA gene cluster in hiPSC derived neural cells (Rajarajan et al., 2018). In addition, cortical interneurons derived from induced pluripotent stem cells (iPSCs) of patients with schizophrenia showed reduced PCDHA3 expression compared to similarly derived interneurons from controls (Shao et al., 2019). The latter study further demonstrated that reduced protocadherin expression was associated with deficient synaptic arborization in both rodent and iPSC-derived human interneurons (but not glutamatergic neurons), and that these deficits could be reversed by treatment with an inhibitor of protein kinase C (Shao et al., 2019).
When our samples were combined with Ashkenazi patients from the SCHEMA database, the PCDHA3 missense variant was observed in 0.3% of all Ashkenazi cases in the present study. While unusually high for a schizophrenia-associated URV, this carrier rate is low compared to the ~4% rate observed for the most common BRCA1 founder variant in Ashkenazi breast cancer cases (King et al., 2003), and the ~15% rate of the LRRK2 G2019S variant amongst Ashkenazi patients with Parkinson’s disease (Correia Guedes et al., 2010). These latter disorders have onset in late-life, and therefore susceptibility alleles for these diseases are not under the strong purifying selection affecting genes for schizophrenia (Pardiñas et al., 2018), a disorder which results in markedly reduced fecundity (Power et al., 2013). Nevertheless, our simulations demonstrated the limits of purifying selection in a founder population with a tight bottleneck. One-third to one-half of all damaging variants escape purifying selection, and these variants tend to become surprisingly frequent in the context of a rapidly expanding population, as described previously for the Finnish population (Wang et al., 2014). Consequently, ascertainment of additional samples from founder populations can be a highly cost-effective way of rapidly enhancing power of rare variant studies (Locke et al., 2019).
The overlap of our schizophrenia case-only gene list with gene sets derived from developmental brain disorders was notable, insofar as exome studies in these disorders have been more well-powered than schizophrenia studies to date (Myers et al., 2020). Consequently, the overlapping genes indicated in the first three rows of Table 2 (and especially the first row, which remained significant after permutation testing) have a strong prior probability of association, especially given prior evidence that rare single nucleotide variants (e.g., SETD1A) (Singh et al., 2016) and copy number variants (Kirov, 2015) tend to be shared across schizophrenia and other neurodevelopmental disorders. In the present study, case-only variants in the 11 genes overlapping the prior report of developmental brain disorders (DBD) (Gonzalez-Mantilla et al., 2016) (ASXL3, BIRC6, DIP2A, DST, LAMA2, NSD1, PCDH15, SETBP1, SETD1A, TRIO, WDFY3) were overwhelmingly (10:1 ratio) missense rather than loss of function; by contrast, the DBD list was generated from prior reports of loss of function variation exclusively. Thus, it is possible that our findings represent allelic series at these genes, in which more damaging variants are associated with more severe clinical phenotypes emerging in early childhood (Shohat et al., 2017).
Similarly, we identified 5 cases (and no controls) with novel missense variants in TSC2, a gene in which mutations (primarily loss of function) are known to cause tuberous sclerosis (TS). TS is an autosomal dominant disorder marked by hamartomas across multiple organs, potentially including the brain (Henske et al., 2016). Case reports of psychotic features in TS patients have proliferated for decades (Herkert et al., 1972); a recent survey of a large international cohort of TS patients identified psychosis in 11% of adults (de Vries et al., 2018). Since the affected cases in the present study were not noted in their clinical report to have TS, our results suggest that schizophrenia can be the primary presenting feature of TSC2 mutations.
In the last 15 years, genetic research in schizophrenia has given consistent support to the long-posited liability-threshold model (Gottesman and Shields, 1967; Kendler, 2015; McGue et al., 1983; Smeland et al., 2020), which states that manifestation of illness requires that the additive total of risk factors (including genetic and environmental) crosses an (unknown) threshold. While most schizophrenia research to date has focused on the total burden of common genetic variants, as captured by the polygenic risk score (Lee et al., 2012; van Rheenen et al., 2019; The International Schizophrenia Consortium, 2009), the model suggests that rare variants contribute in the same manner, albeit with much greater individual weighting (penetrance) (Richards et al., 2016). Supporting evidence has come from four very recent studies of schizophrenia patients carrying known, high-penetrance copy number variants (Bergen et al., 2019; Cleynen et al., 2020; Davies et al., 2020; Taniguchi et al., 2020). These studies all show that patients with highly penetrant CNVs have lower common-variant PRS compared to patients not carrying a known CNV, presumably because the CNV has already pushed them closer to the threshold for illness. In the present study, we have demonstrated, for the first time, a similar inverse correlation between common-variant PRS and rare variant burden indexed by missense and loss-of-function single nucleotide changes and small indels. Interestingly, the cases who carried MisLoF URVs in the 141 case-only genes did not show earlier age at onset or more severe course of illness, suggesting that these variants do not generally manifest in a fundamentally different illness relative to other additive risk factors. However, there was initial evidence that the recurrent PCDHA3 mutation may convey risk for an early-onset form of schizophrenia.
This study had several limitations, most notably that sample size was relatively small for a genetic association study; on the other hand, we demonstrated that the AJ population is enriched for rare variants and has substantially greater power than comparably-sized studies of outbred populations. Moreover, we were able to utilize external, well-powered datasets (i.e., SCHEMA and various studies of neurodevelopmental disorders) as validation/replication, and our gene set results for synaptic and constrained genes served as a positive control for our approach. Relatedly, although we utilized whole-genome sequencing to obtain our data, we restricted our analysis to the exome in order to make use of the largest possible set of samples both for purposes of these comparisons and for filtering of variants. Finally, we did not have neuropsychological testing data available for our cases, so we were unable to differentiate the relative contribution of URVs to cognitive deficits in our samples (Singh et al., 2017). Because of the enhanced power available in the Ashkenazi population, future rare variant studies in well-characterized AJ samples can be useful in overcoming a common limitation in large-scale genetic studies of schizophrenia, namely understanding the clinical impact of URVs on phenotypic expression.
STAR Methods
RESOURCE AVAILIBILITY
Lead Contact
Further information and requests for resources and data should be directed to, and will be fulfilled by, the Lead Contact, Todd Lencz, Ph.D. (tlencz@northwell.edu).
Materials Availability
Plasmids generated in this study have been deposited to Addgene (#164616 and #164617)
Data and Code Availability
DNA sequence data for the AJ cohort are available to qualified investigators through the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession code EGAS00001000664
Code generated for this study is available at https://github.com/rrkhan/RareVariantSimulation
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
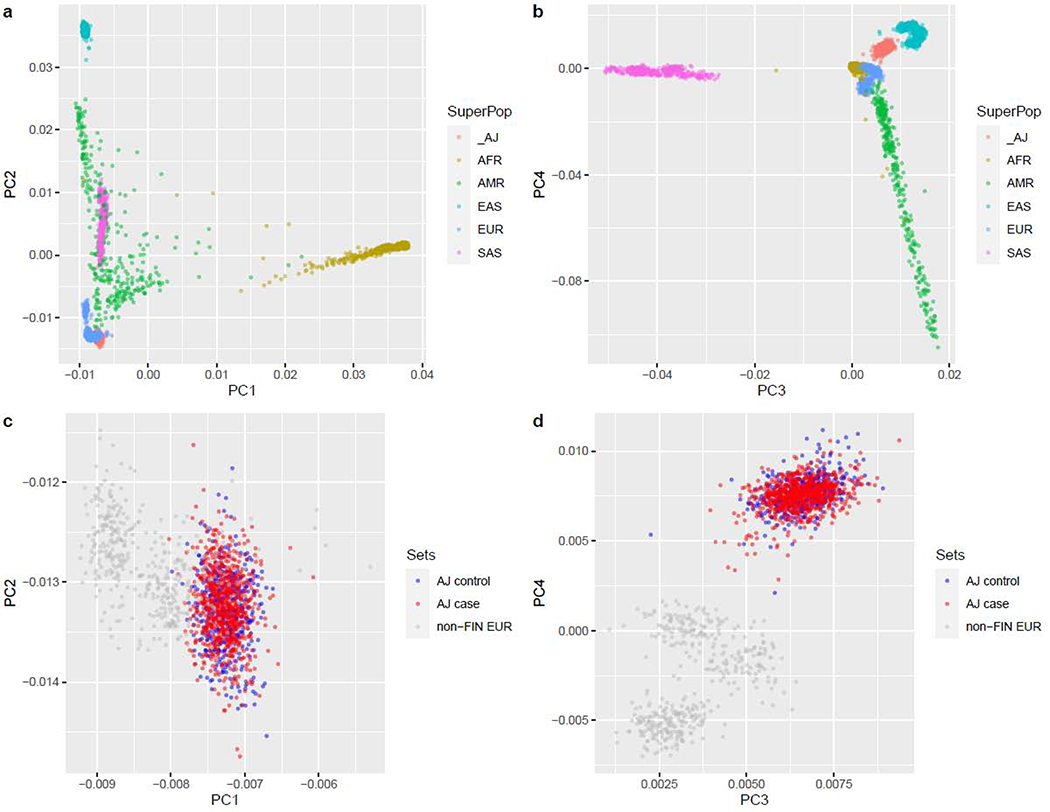
Sequenced samples were derived from subjects described previously from multiple case-control cohorts summarized in Table S6. All subjects were self-reported to be Ashkenazi Jewish, and were only selected for sequencing if ancestry was also verified as AJ by principal components analysis of previously collected SNP array data as described in our prior publications (Atzmon et al., 2010; Guha et al., 2012). Moreover, no outliers were observed in principal components analysis of all included subjects in comparison to reference population data (1000 Genomes Phase 3), with cases and controls overlapping within the AJ cluster (Figure 4). Following the variant calling and quality control steps described below (Method Details), the final sample available for analysis consisted of 1249 subjects (560 female; 689 male).
Figure 4: PCA plots of Ashkenazi Jewish Subjects Compared to Global Populations.
AJ subjects demonstrate clear separation from other groups at the global scale (a,b) and within populations of European ancestry (c,d, gray dots represent CEU, GBR, IBS, and TSI populations from 1000 Genomes). Importantly, no clear distinction can be drawn between AJ cases and controls (c,d, orange and green dots, respectively).
Patients with schizophrenia were recruited from hospitalized inpatients at seven medical centers in Israel as described previously (Guha et al., 2013; Lencz et al., 2013). All diagnoses were assigned after direct interview using a structured clinical interview, a questionnaire with inclusion and exclusion criteria, and cross-references to medical records. The inclusion criteria specified that subjects had to be diagnosed with schizophrenia or schizoaffective disorder by the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV). The exclusion criteria eliminated subjects diagnosed with at least one of the following disorders: psychotic disorder due to a general medical condition, substance-induced psychotic disorder, or any Cluster A (schizotypal, schizoid or paranoid) personality disorder. Controls were taken from several cohorts, primarily those screened for multiple forms of chronic illness (Lencz et al., 2013; Walter et al., 2011), but also including a small number of subjects ascertained for non-psychiatric disorders (Inflammatory Bowel Disease or Dystonia)(Kenny et al., 2012; Risch et al., 2007). Informed consent was obtained from all subjects in accordance with institutional policies and the studies were approved by the corresponding institutional review boards.
Cell lines
For plasmid generation, the variable coding sequence of the PCDHA3 isoform was amplified by PCR from human genomic DNA isolated from HEK293T cells. For isoforms containing the intracellular domain, the constant coding region was amplified from cDNA isolated from SKNSH cells. For the cell aggregation and immunostaining experiments, K562 cells were utilized.
METHOD DETAILS
Sequencing and variant calling pipeline
All samples were sequenced on the Illumina HiSeq X platform, using methods described previously (Lencz et al., 2018). Briefly, genomic DNA was isolated from whole blood and was quantified using PicoGreen, and integrity was assessed using the Fragment Analyzer (Advanced Analytical). Sequencing libraries were prepared using the Illumina TruSeq Nano DNA kit, with 100ng input, and pooled in equimolar amounts (8 samples/pool); a 2.5–3nM pooled library was loaded onto each lane of the patterned flow cell, and clustered on a cBot, generating ~375–400M pass filter 2×150bp reads per flow cell lane. For samples that did not meet 30x mean genome coverage post alignment, additional aliquots of the sequencing libraries were pooled in proportion to the amount of additional reads needed for resequencing.
Upon completion of sequencing runs, bcl files were demultiplexed and quality of sequencing data reviewed using SAV software (Illumina) and FastQC for deviations from expected values with respect to total number of reads, percent reads demultiplexed (>95%), percent clusters pass filter (>55%), base quality by lane and cycle, percent bases >Q30 for read 1 and read 2 (>75%), GC content, and percent N-content. FastQ files were aligned to hg19/GRCh37 using the Burrows-Wheeler Aligner (BWA-MEM v0.78)(Li & Durbin, 2009) and processed using the best-practices pipeline that includes marking of duplicate reads by the use of Picard tools (v1.83, http://picard.sourceforge.net), realignment around indels, and base recalibration via Genome Analysis Toolkit (GATK v3.5)(Van der Auwera et al., 2013). A total of 1,310 samples proceeded to the last steps of joint genotyping and VQSR variant filtering after removal of 10 samples (7 cases, 3 controls) with < 80% 20X read depth coverage of the genome, and removal of 26 duplicate samples (1 case and 25 controls) sequenced as part of QC procedures. All remaining samples were jointly genotyped to generate a multi-sample VCF. Variant Quality Score Recalibration (VQSR) was performed on the multi-sample VCF, and variants were annotated using VCFtools (Danecek et al., 2011). After the GATK pipeline, we also filtered the SNP and small INDEL on LCR regions (Li, 2014) and 1000G masked difficult regions (Auton et al., 2015), since these regions are enriched with calling errors that cannot be filtered effectively by the VQSR model, as we demonstrated previously (Lencz et al., 2018). Furthermore, we masked the genotypes with GQ < 20 as “./.” and filtered variants where <= 80% individuals could not be genotyped confidently. For purposes of downstream case-control analyses, we also removed 1 member of any pair of samples that were related at the first-cousin level or greater (n=27 controls).
Defining ultra-rare variants (URVs) in TAGC samples
To maximize the power of existing reference population databases, we focused our primary analysis on the exome regions (as defined by gnomAD exome calling intervals (Karczewski et al., 2020)). We focused on URVs that were novel singletons in our TAGC cohort, filtering out all variants called in gnomAD (v2.1.1) non-neuro samples of any ethnicity, regardless of their call quality, and filtering out all variants called in TOPMed (The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program., 2018) freeze 5 release (with coordinates lifted over to hg19). We identified a small set of TAGC samples (n = 34; 21 cases / 13 controls) with an excessive number of exonic URVs (>=50), shown as outliers in the distribution (Figure S3). Importantly, these outlier samples also demonstrated excess number of intergenic URVs and were not restricted to any sequencing batch. After filtering these outliers, we ended up with 1,249 samples for the downstream analyses (Table S6).
Primary analyses compared potentially functional (missense or loss of function) to putatively silent (synonymous or other) variants; these were defined as MisLoF and non-MisLoF, respectively. Exonic URVs were classified as loss of function, missense, synonymous, or other (generally intronic bases flanking exons as well as UTR regions), based on their most damaging impact annotated for any transcript. Thus, non-MisLoF variants had no missense or loss of function annotation on any known transcript.
Plasmid Generation
The variable coding sequence of the PCDHA3 isoform was amplified by PCR from human genomic DNA isolated from HEK293T cells. For isoforms including the intracellular domain, the constant coding region was amplified from cDNA isolated from SKNSH cells. HiFi DNA assembly was used to generate a mCherry tagged PCDHα3 protein by inserting the PCR fragments into the KpnI and AgeI restriction sites of a plasmid containing the immediate early promoter of cytomegalovirus with intron element (PCMV-IE) and mCherry sequence previously described (Thu et al., 2014). To generate the PCDHα3(N559S) mutant expression plasmid, a single nucleotide substitution was made using Q5 site directed mutagenesis (NEB) following manufacturer’s instructions. Plasmid sequences were confirmed by Sanger sequencing (Genewiz). Primers used to generate the wildtype PCDHα3 expression construct and perform the site directed mutagenesis are found in Table S7.
Cell Aggregation Assay
K562 cell aggregation assay was performed as previously described (Thu et al., 2014). Briefly, PCDHα proteins require the co-expression of PCDHβ or PCDHγ isoforms to reach the cell surface, wildtype and mutant PCDHA3 expression plasmids were nucleofected together with a PCDHγC3ΔEC1 carrier expression construct into K562 cells (ATCC CCCL243) using the Amaxa 4D- Nucleofector (Lonza) following the manufacturer’s recommended protocol. After 24 hours in culture, the cells were allowed to aggregate for 3–4 hours on a rocker kept inside of the incubator. The cells were then fixed in 4% paraformaldehyde (PFA), washed with PBS, and imaged using an Olympus IX71 microscope.
Immunostaining
K562 cells were nucleofected as described above. Following 24 hours post-nucleofection, the cells were fixed with 4% paraformaldehyde and washed in PBS. Cells were then collected onto poly-D-lysine coated coverslips by centrifugation at 800rcf for 10min. Cells were then mounted onto glass slides and the K562 cells expressing mCherry tagged PCDHα3 isoforms were imaged using an Olympus Fluoview FV1000 confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Common variant polygenic risk score (PRS)
A common variant PRS for schizophrenia was calculated for each subject based on summary statistics from the large-scale schizophrenia GWAS reported by the Psychiatric Genomics Consortium (Schizophrenia Working Group of the Psychiatric Genomics Consortium et al., 2020), excluding our own Ashkenazi cohort. These summary statistics were filtered to include only high-quality SNPs (imputation INFO score ≥ 0.9). Additional filtering was also applied to the genotype data of the AJ samples to maximize quality of the variants used for PRS calculation; variants were only utilized if: 1) ≥99% of the AJ samples were called with high confident genotypes (GQ≥20) at that position; 2) MAF ≥ 1% in the AJ samples; 3) HWE P > 1×10−10.
The PRS calculation was performed using PRSice-2 (v2.3.3) after applying LD clumping (R2 threshold = 0.1 within 250kb) on the AJ genotypes. In order to optimize the p-value threshold (PT) for the primary comparison of interest (PRS vs. URV burden), we examined the effect of different values of PT on the amount of PRS variance explained by case/control status. The largest R2 for the case-control comparison was obtained at PT = 0.00725, and this threshold was then used for the primary regression analysis examining the relationship between PRS and total number of MisLoF URVs, controlling for sex and the loadings of top five genetic principal components derived from genotypes on common SNP sites, were performed separately for cases and controls.
Assigning novel URVs to genes and defining “case-only” and “control-only” genes
Each gene was characterized by the number of cases and the number of controls harboring a novel MisLoF or non-MisLoF variant within it. We focused on genes in which only cases or only controls harbored a MisLoF variant; genes in which both cases and controls were observed to have a given type of URV were excluded from subsequent analyses for that variant type. Of course, it was more likely that a gene would be identified as “case-only” rather than “control-only” for each type of URV due to the unequal numbers of cases relative to controls. Consequently, we controlled for the both effects by utilizing a re-sampling strategy, down-sampling the number of cases to match the number of controls, and iterated 10,000 times. For each iteration for each variant type, the following calculation was performed:
Replication of SCHEMA genes
The SCHEMA consortium lists 10 significant genes using strict exome-wide criteria (p<2.2×10−6) and 32 genes using false discovery rate <.05 (p<7.9×10−5)(Singh et al., 2020). We used the hypergeometric test to statistically compare the overlap between case-only MisLoF genes in our AJ sample and these SCHEMA genes (restricting the comparison to the autosome). Note that we did not simply merge datasets due to differences in sequence acquisition, calling, and quality control procedures. To guard against potential confound by gene size, permutations were performed to match the size distribution of our case-only gene set in the following manner: First, we created a ranked list of all autosomal genes in order of size of the coding sequence, and divided this list into ten bins (deciles). In permutation testing, a gene was randomly sampled from the same decile for each gene in the set to be matched. A total of 10,000 iterations were performed, and the empirical p-value was defined by the proportion of permuted gene sets with overlap ≥ the case-only gene set. If no permutation was observed to meet this criteria, the p-value was reported as <1×10−4.
Gene set analyses
We compared the AJ case-only and control-only MisLoF genes to three categories of gene sets based on prior studies: 1. De novo mutation genes implicated across multiple developmental brain disorders (DBD)(Gonzalez-Mantilla et al., 2016), a large scale autism spectrum disorder(ASD) exome study(Satterstrom et al., 2020), and the integration of the ASD set with a large-scale study of ASD, developmental disorder(DD), and intellectual disability(ID) exome sequencing studies (Coe et al., 2019); 2. Genes known to encode proteins of the synapse aggregated in SynaptomeDB (Pirooznia et al., 2012), and genes regulated by known neuronal RNA-binding proteins, including CELF4 (Wagnon et al., 2012), FMRP (Darnell et al., 2011), RBFOX2 and RBFOX1/RBFOX3 (Weyn-Vanhentenryck et al., 2014); 3. Genes constrained by missense (Samocha et al., 2014) and LoF variants (Karczewski et al., 2020). Since X and Y chromosomes of AJ samples are not included, we adjusted the number of genes in each set and total protein-coding genes (n=19,780; http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database) to autosomal-only to calculate the p-values using hypergeometric tests. We further compared the relative enrichment of AJ case-only vs. control-only MisLoF genes for these gene sets using the chi-squared test based on the 2×2 table of overlap and non-overlap for case-only and control-only. As above, we also performed permutation testing to control for effects of gene size. In addition to the a priori gene sets identified above, we performed hypothesis-free testing of the case-only and control-only lists against all GO categories and PANTHER protein classes available from the Gene Ontology Consortium (www.geneontology.org, GO Ontology database DOI: 10.5281/zenodo.4081749)(Ashburner et al., 2000; Gene Ontology Consortium, 2019; Mi et al., 2019; The Gene Ontology Consortium, 2017), as well as synaptic components (annotated by SYNGO (Koopmans et al., 2019)). To control for gene size, we extended the permutation procedures described above to the significant gene sets identified from the hypothesis-free GO analysis. Moreover, we ran the full set of GO and PANTHER analyses on 100 permuted gene lists, size-matched to the case-only list.
Modeling the effects of purifying selection
We simulated the spread of a single schizophrenia-causing variant in a founder population under a number of empirical conditions. The initial conditions of the simulation assume a single variant carrier in a fixed size population at the time of bottleneck, which we denote as NB. From there, we modeled the growth of the number of carriers and total population size over a set number of G generations until a maximum population size Nmax is achieved. The growth rate R is computed as R = ((Nmax/NB)/G). We generated the number of offspring per individual within a generation using a Poisson branching process, in which the lambda parameter is a function of growth rate R, sex, and case/control status. A healthy individual will have a lambda equal to R, while an individual with schizophrenia will have reduced relative fecundity, differing by sex (estimated at 0.5 for females and 0.25 for males (Power et al., 2013)). Within our simulated population, we track the number of variant carriers for each of several scenarios, in which we test the sensitivity of the model to variations on our initial estimates of population bottleneck size NB, number of generations G, and relative fecundity of schizophrenia cases; we report results as a function of the disease penetrance of the variant. We computed 10,000 simulations for each scenario, calculating the proportion of simulations in which the number of variant carriers goes to zero (extinction) within G generations, and the proportion of simulations in which a variant escapes extinction. We then use the number of variant carriers remaining after G generations in the population to calculate the power of Fisher’s exact test for detecting the variant effect in a study cohort size of a given size.
Supplementary Material
Table S1a: list of 141 “case-only” genes, related to Figure 1.
Table S1b: list of 148 “control-only” genes, related to Figure 1.
Highlights.
Schizophrenia rare variant discovery is enhanced in the Ashkenazi founder population
Ultra-rare variant burden is inversely correlated with polygenic risk scores in cases
Ultra-rare exonic variants in schizophrenia cases are enriched in cadherin genes
A recurrent case mutation in PCDHA3 disrupts homophilic interactions in culture
Acknowledgements
The authors are extremely grateful to Soren Germer, Ph.D. and his team at the New York Genome Center for performing the Illumina sequencing. We acknowledge financial support from the Human Frontier Science Program (SC); NIH research grants AG042188 (GA), DK62429, DK062422, DK092235 (JHC), NS050487, NS060113 (LNC), AG021654, AG027734 (NB), MH089964, MH095458, MH084098 (TL), and CA121852 (computational infrastructure, IPe’er); NSF research grants 08929882 and 0845677 (IPe’er); Rachel and Lewis Rudin Foundation (HE); Northwell Health Foundation (TL); Brain & Behavior Foundation (TL); US-Israel Binational Science Foundation (TL, AD); LUNGevity Foundation (ZHG); New York Crohn’s Disease Foundation (IPeter); Edwin & Caroline Levy and Joseph & Carol Reich (SB); the Parkinson’s Disease Foundation (LNC); the Sharon Levine Corzine Cancer Research Fund (KO); and the Andrew Sabin Family Research Fund (KO).
Footnotes
Declaration of interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, et al. (2010). Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet 86, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilés EC, and Goodrich LV (2017). Configuring a robust nervous system with Fat cadherins. Seminars in Cell & Developmental Biology 69, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskovich B, Hiraki S, Upadhyay K, Meyer P, Carmi S, Barzilai N, Darvasi A, Ozelius L, Peter I, Cho JH, et al. (2016). Expanded genetic screening panel for the Ashkenazi Jewish population. Genet. Med 18, 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen SE, Ploner A, Howrigan D, CNV Analysis Group and the Schizophrenia Working Group of the Psychiatric Genomics Consortium, O’Donovan MC, Smoller JW, Sullivan PF, Sebat J, Neale B, and Kendler KS (2019). Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. Am J Psychiatry 176, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J, Goodman KM, Noble AJ, Rapp M, Mannepalli S, Bahna F, Dandey VP, Bepler T, Berger B, Maniatis T, et al. (2019). Visualization of clustered protocadherin neuronal self-recognition complexes. Nature 569, 280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, and Maniatis T (2019). The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Current Opinion in Neurobiology 59, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Gray MJ, Badouel C, Lange S, Einsiedler M, Srour M, Chitayat D, Hamdan FF, Jenkins ZA, Morgan T, et al. (2013). Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat. Genet 45, 1300–1308. [DOI] [PubMed] [Google Scholar]
- Carmi S, Hui KY, Kochav E, Liu X, Xue J, Grady F, Guha S, Upadhyay K, Ben-Avraham D, Mukherjee S, et al. (2014). Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat Commun 5, 4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Nwakeze CL, Denny CA, O’Keeffe S, Rieger MA, Mountoufaris G, Kirner A, Dougherty JD, Hen R, Wu Q, et al. (2017). Pcdhαc2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science 356, 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Engchuan W, Hestand MS, Heung T, Holleman AM, Johnston HR, Monfeuga T, McDonald-McGinn DM, Gur RE, Morrow BE, et al. (2020). Genetic contributors to risk of schizophrenia in the presence of a 22q11.2 deletion. Molecular Psychiatry 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, Dougherty JD, Lein ES, Hormozdiari F, Bernier RA, et al. (2019). Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nature Genetics 51, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Torres L, Burke EE, Peterson A, Shin J, Straub RE, Rajpurohit A, Semick SA, Ulrich WS, Price AJ, Valencia C, et al. (2019). Regional Heterogeneity in Gene Expression, Regulation, and Coherence in the Frontal Cortex and Hippocampus across Development and Schizophrenia. Neuron 103, 203–216.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia Guedes L, Ferreira JJ, Rosa MM, Coelho M, Bonifati V, and Sampaio C (2010). Worldwide frequency of G2019S LRRK2 mutation in Parkinson’s disease: a systematic review. Parkinsonism Relat. Disord 16, 237–242. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Dueker ND, Slifer SH, Lee JM, Whitehead PL, Lalanne E, Leyva N, Konidari I, Gentry RC, Hulme WF, et al. (2014). Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Molecular Autism 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RW, Fiksinski AM, Breetvelt EJ, Williams NM, Hooper SR, Monfeuga T, Bassett AS, Owen MJ, Gur RE, Morrow BE, et al. (2020). Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nat Med 26, 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LG, Benson DL, and Huntley GW (2015). Cadherin-based transsynaptic networks in establishing and modifying neural connectivity. Curr. Top. Dev. Biol 112, 415–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LS, Szabo CI, Ostermeyer EA, Dowd P, Butler L, Park T, Lee MK, Goode EL, Rowell SE, and King MC (1995). Novel inherited mutations and variable expressivity of BRCA1 alleles, including the founder mutation 185delAG in Ashkenazi Jewish families. Am. J. Hum. Genet 57, 1284–1297. [PMC free article] [PubMed] [Google Scholar]
- Fulford AD, and McNeill H (2020). Fat/Dachsous family cadherins in cell and tissue organisation. Current Opinion in Cell Biology 62, 96–103. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, Bakel H. van, Varghese M, Wang Y, et al. (2018). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, Alfoldi J, Martin AR, Havulinna AS, Byrnes A, et al. (2018). Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am. J. Hum. Genet 102, 1204–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium T (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, Moran JL, Purcell SM, Sklar P, Sullivan PF, et al. (2016). Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci 19, 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, and Martin CL (2016). A Cross-Disorder Method to Identify Novel Candidate Genes for Developmental Brain Disorders. JAMA Psychiatry 73, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsén G, Rittenhouse C, Maniatis T, Honig B, and Shapiro L (2016). Structural Basis of Diverse Homophilic Recognition by Clustered α- and β-Protocadherins. Neuron 90, 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KM, Rubinstein R, Dan H, Bahna F, Mannepalli S, Ahlsén G, Aye Thu C, Sampogna RV, Maniatis T, Honig B, et al. (2017). Protocadherin cis-dimer architecture and recognition unit diversity. Proc Natl Acad Sci U S A 114, E9829–E9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, and Shields J (1967). A polygenic theory of schizophrenia. Proc. Natl. Acad. Sci. U.S.A 58, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Wray NR, Keller MC, and Visscher PM (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat. Neurosci 17, 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Rosenfeld JA, Malhotra AK, Lee AT, Gregersen PK, Kane JM, Pe’er I, Darvasi A, and Lencz T (2012). Implications for health and disease in the genetic signature of the Ashkenazi Jewish population. Genome Biol. 13, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, Magnusson PK, Cormican P, Morris D, Gill M, et al. (2013). Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry 70, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Stein DJ, Susser ES, Sibeko G, Pretorius A, Walsh T, Majara L, Mndini MM, Mqulwana SG, Ntola OA, et al. (2020). Genetics of schizophrenia in the South African Xhosa. Science 367, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske EP, Jóźwiak S, Kingswood JC, Sampson JR, and Thiele EA (2016). Tuberous sclerosis complex. Nat Rev Dis Primers 2, 16035. [DOI] [PubMed] [Google Scholar]
- Herkert EE, Wald A, and Romero O (1972). Tuberous sclerosis and schizophrenia. Dis Nerv Syst 33, 439–445. [PubMed] [Google Scholar]
- Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, Nordentoft M, and Glenthøj B (2018). Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol. Psychiatry 83, 492–498. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, and Yagi T (2009). Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci 29, 9137–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori S, Noguchi-Katori Y, Okayama A, Kawamura Y, Luo W, Sakimura K, Hirabayashi T, Iwasato T, and Yagi T (2017). Protocadherin-αC2 is required for diffuse projections of serotonergic axons. Sci Rep 7, 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS (2015). A joint history of the nature of genetic variation and the nature of schizophrenia. Mol Psychiatry 20, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EE, Pe’er I, Karban A, Ozelius L, Mitchell AA, Ng SM, Erazo M, Ostrer H, Abraham C, Abreu MT, et al. (2012). A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet. 8, e1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M-C, Marks JH, Mandell JB, and New York Breast Cancer Study Group (2003). Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646. [DOI] [PubMed] [Google Scholar]
- Kirov G (2015). CNVs in neuropsychiatric disorders. Hum. Mol. Genet 24, R45–49. [DOI] [PubMed] [Google Scholar]
- Koopmans F, van Nierop P, Andres-Alonso M, Byrnes A, Cijsouw T, Coba MP, Cornelisse LN, Farrell RJ, Goldschmidt HL, Howrigan DP, et al. (2019). SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron 103, 217–234.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, and Sunyaev SR (2007). Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet 80, 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Chen C-Y, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown BC, et al. (2019). Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51, 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ), International Schizophrenia Consortium (ISC), Molecular Genetics of Schizophrenia Collaboration (MGS), Sullivan PF, Goddard ME, Keller MC, et al. (2012). Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet 44, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P, John M, Cheng L, Zhang C, Badner JA, et al. (2013). Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat Commun 4, 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Yu J, Palmer C, Carmi S, Ben-Avraham D, Barzilai N, Bressman S, Darvasi A, Cho JH, Clark LN, et al. (2018). High-depth whole genome sequencing of an Ashkenazi Jewish reference panel: enhancing sensitivity, accuracy, and imputation. Hum. Genet 137, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2014). Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Leal SM (2009). Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 5, e1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Steinberg KM, Chiang CWK, Service SK, Havulinna AS, Stell L, Pirinen M, Abel HJ, Chiang CC, Fulton RS, et al. (2019). Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 572, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, Schizophrenia Working Group of Psychiatric Genomics Consortium, de Candia TR, Lee SH, Wray NR, et al. (2015). Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet 47, 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, et al. (2017). Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet 49, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Gottesman II, and Rao DC (1983). The transmission of schizophrenia under a multifactorial threshold model. Am. J. Hum. Genet 35, 1161–1178. [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountoufaris G, Chen WV, Hirabayashi Y, O’Keeffe S, Chevee M, Nwakeze CL, Polleux F, and Maniatis T (2017). Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356, 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, and Maniatis T (2018). Writing, Reading, and Translating the Clustered Protocadherin Cell Surface Recognition Code for Neural Circuit Assembly. Annu Rev Cell Dev Biol 34, 471–493. [DOI] [PubMed] [Google Scholar]
- Myers SM, Challman TD, Bernier R, Bourgeron T, Chung WK, Constantino JN, Eichler EE, Jacquemont S, Miller DT, Mitchell KJ, et al. (2020). Insufficient Evidence for “Autism-Specific” Genes. Am. J. Hum. Genet 106, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanou E, and Catterall WA (2018). Calcium Channels, Synaptic Plasticity, and Neuropsychiatric Disease. Neuron 98, 466–481. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bryois J, Kim A, Dobbyn A, Huckins LM, Munoz-Manchado AB, Ruderfer DM, Genovese G, Fromer M, Xu X, et al. (2017). Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, et al. (2006). LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N. Engl. J. Med 354, 424–425. [DOI] [PubMed] [Google Scholar]
- Palamara PF, Lencz T, Darvasi A, and Pe’er I (2012). Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet 91, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, et al. (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, Goes FS, Potash JB, and Zandi PP (2012). SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics 28, 897–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RA, Kyaga S, Uher R, MacCabe JH, Långström N, Landen M, McGuffin P, Lewis CM, Lichtenstein P, and Svensson AC (2013). Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry 70, 22–30. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kähler A, et al. (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, Powell S, Yashaswini C, LaMarca EA, Kassim B, et al. (2018). Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resin J (2020). A Simple Algorithm for Exact Multinomial Tests. ArXiv:2008.12682 [Stat]. [Google Scholar]
- van Rheenen W, Peyrot WJ, Schork AJ, Lee SH, and Wray NR (2019). Genetic correlations of polygenic disease traits: from theory to practice. Nat. Rev. Genet 20, 567–581. [DOI] [PubMed] [Google Scholar]
- Richards AL, Leonenko G, Walters JT, Kavanagh DH, Rees EG, Evans A, Chambert KD, Moran JL, Goldstein J, Neale BM, et al. (2016). Exome arrays capture polygenic rare variant contributions to schizophrenia. Hum. Mol. Genet 25, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch NJ, Bressman SB, Senthil G, and Ozelius LJ (2007). Intragenic Cis and Trans modification of genetic susceptibility in DYT1 torsion dystonia. Am. J. Hum. Genet 80, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Avila BE, Koskela J, Huang H, Stevens C, Pirinen M, Haritunians T, Neale BM, Kurki M, Ganna A, et al. (2018). Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 14, e1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, Ahlsen G, Chevee M, Halim A, Clausen H, et al. (2015). Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell 163, 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R, Goodman KM, Maniatis T, Shapiro L, and Honig B (2017). Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin Cell Dev Biol 69, 140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Charney AW, Readhead B, Kidd BA, Kähler AK, Kenny PJ, Keiser MJ, Moran JL, Hultman CM, Scott SA, et al. (2016). Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. The Lancet Psychiatry 3, 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. (2014). A framework for the interpretation of de novo mutation in human disease. Nature Genetics 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins R, Grove J, Klei L, et al. (2020). Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–584.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Walters JT, and O’Donovan MC (2020). Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. MedRxiv 2020.09.12.20192922. [Google Scholar]
- Selvan ME, Zauderer MG, Rudin CM, Jones S, Mukherjee S, Offit K, Onel K, Rennert G, Velculescu VE, Lipkin SM, et al. (2020). Inherited rare, deleterious variants in ATM increase lung adenocarcinoma risk. MedRxiv 2020.03.19.20034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Noh H, Bin Kim W, Ni P, Nguyen C, Cote SE, Noyes E, Zhao J, Parsons T, Park JM, et al. (2019). Dysregulated protocadherin-pathway activity as an intrinsic defect in induced pluripotent stem cell-derived cortical interneurons from subjects with schizophrenia. Nat. Neurosci 22, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohat S, Ben-David E, and Shifman S (2017). Varying Intolerance of Gene Pathways to Mutational Classes Explain Genetic Convergence across Neuropsychiatric Disorders. Cell Reports 18, 2217–2227. [DOI] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G, et al. (2016). Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat. Neurosci 19, 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, Rees E, Iyegbe C, Blackwood D, McIntosh AM, et al. (2017). The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet 49, 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Poterba T, Curtis D, Akil H, Eissa MA, Barchas JD, Bass N, Bigdeli TB, Breen G, Bromet EJ, et al. (2020). Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. MedRxiv 2020.09.18.20192815. [Google Scholar]
- Smart SE, Kępińska AP, Murray RM, and MacCabe JH (2019). Predictors of treatment resistant schizophrenia: a systematic review of prospective observational studies. Psychol. Med 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Dale AM, and Andreassen OA (2020). The polygenic architecture of schizophrenia — rethinking pathogenesis and nosology. Nat Rev Neurol. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, and Neale MC (2003). Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, and O’Donovan M (2012). Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi S, Ninomiya K, Kushima I, Saito T, Shimasaki A, Sakusabe T, Momozawa Y, Kubo M, Kamatani Y, Ozaki N, et al. (2020). Polygenic risk scores in schizophrenia with clinically significant copy number variants. Psychiatry Clin. Neurosci 74, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. (2012). Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program. (2018). BRAVO variant browser.
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, and Maniatis T (2014). Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell 158, 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries PJ, Belousova E, Benedik MP, Carter T, Cottin V, Curatolo P, Dahlin M, D’Amato L, d’Augères GB, Ferreira JC, et al. (2018). TSC-associated neuropsychiatric disorders (TAND): findings from the TOSCA natural history study. Orphanet Journal of Rare Diseases 13, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Briese M, Sun W, Mahaffey CL, Curk T, Rot G, Ule J, and Frankel WN (2012). CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function. PLOS Genetics 8, e1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Atzmon G, Demerath EW, Garcia ME, Kaplan RC, Kumari M, Lunetta KL, Milaneschi Y, Tanaka T, Tranah GJ, et al. (2011). A genome-wide association study of aging. Neurobiol. Aging 32, 2109.e15-–28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, et al. (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SR, Agarwala V, Flannick J, Chiang CWK, Altshuler D, GoT2D Consortium, and Hirschhorn JN (2014). Simulation of Finnish population history, guided by empirical genetic data, to assess power of rare-variant tests in Finland. Am. J. Hum. Genet 94, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. (2014). HITS-CLIP and Integrative Modeling Define the Rbfox Splicing-Regulatory Network Linked to Brain Development and Autism. Cell Reports 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, and Jia Z (2020). Wiring the Brain by Clustered Protocadherin Neural Codes. Neurosci Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1a: list of 141 “case-only” genes, related to Figure 1.
Table S1b: list of 148 “control-only” genes, related to Figure 1.
Data Availability Statement
DNA sequence data for the AJ cohort are available to qualified investigators through the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession code EGAS00001000664
Code generated for this study is available at https://github.com/rrkhan/RareVariantSimulation