Abstract
Background:
Disseminated tumor cells (DTCs) have been reported in the bone marrow (BM) of patients with localized prostate cancer (PCa). However, the existence of these cells continues to be questioned, and few methods exist for viable DTC isolation. Therefore, we sought to develop novel approaches to identify and, if detected, analyze localized PCa patient DTCs.
Methods:
We used fluorescence-activated cell sorting (FACS) to isolate a putative DTC population, which was negative for CD45, CD235a, alkaline phosphatase, and CD34, and strongly expressed EPCAM. We examined tumor cell content by bulk cell RNA sequencing (RNA-Seq) and whole-exome sequencing after whole genome amplification. We also enriched for BM DTCs with α-EPCAM immunomagnetic beads and performed quantitative reverse trancriptase polymerase chain reaction (qRT-PCR) for PCa markers.
Results:
At a threshold of 4 cells per million BM cells, the putative DTC population was present in 10 of 58 patients (17%) with localized PCa, 4 of 8 patients with metastatic PCa of varying disease control, and 1 of 8 patients with no known cancer, and was positively correlated with patients’ plasma PSA values. RNA-Seq analysis of the putative DTC population collected from samples above (3 patients) and below (5 patients) the threshold of 4 putative DTCs per million showed increased expression of PCa marker genes in 4 of 8 patients with localized PCa, but not the one normal donor who had the putative DTC population present. Whole-exome sequencing also showed the presence of single nucleotide polymorphisms and structural variants in the gene characteristics of PCa in 2 of 3 localized PCa patients. To examine the likely contaminating cell types, we used a myeloid colony formation assay, differential counts of cell smears, and analysis of the RNA-Seq data using the CIBERSORT algorithm, which most strongly suggested the presence of B-cell lineages as a contaminant. Finally, we used EPCAM enrichment and qRT-PCR for PCa markers to estimate DTC prevalence and found evidence of DTCs in 21 of 44 samples (47%).
Conclusion:
These data support the presence of DTCs in the BM of a subset of patients with localized PCa and describe a novel FACS method for isolation and analysis of viable DTCs.
Keywords: disseminated cancer cell, dormancy, flow cytometry, metastasis
1 ∣. INTRODUCTION
Prostate cancer (PCa) is the second most common cause of cancer-related deaths among men in the US.1 Unlike most cancers, late recurrences in PCa are relatively common, with over 20% of recurrences occurring more than 5 years after curative intent radiation or surgery.2 These recurrences are thought to result from early dissemination of PCa cells, which initially exhibit a dormant behavior for months or years, but eventually undergo reactivation and lead to clinical recurrence.3,4 Many of these recurrences may result from distant cells, termed disseminated tumor cells (DTCs) or disseminated cancer cells. To find the reservoir for dormancy and subsequent recurrence, investigators have noted PCa avidity for the bone microenvironment, with 90% of fatal metastatic cases involving bone as one of the metastatic sites.5
Previous investigators have found evidence of DTCs in the bone marrow (BM) from patients with localized PCa using various techniques including RT-PCR for KLK3 (prostate-specific antigen [PSA]), immunocytochemistry for PSA or pan-cytokeratin, and immunomagnetic enrichment and single-cell isolation for EPCAM coupled with immunomagnetic depletion of normal BM cells,4,6-24 as we recently reviewed.25 However, most of these techniques did not allow isolation of viable cells for subsequent messenger RNA (mRNA) analysis. Furthermore, those that were able to isolate viable cells did not have the advantage of next-generation sequencing technologies.4,24 Most recently, investigators from four institutions were unable to detect DTCs in BM from localized PCa patients using four platforms validated for the detection of circulating tumor cells (CTCs) from blood.26
Therefore, to help reconcile these discrepancies in the literature, we designed novel techniques to detect DTCs in BM of patients with localized PCa, and, if present, estimate their frequency (ie, number of putative DTCs per million marrow cells) and prevalence (ie, percentage of localized PCa patients with detectable DTCs). Furthermore, because we ultimately hope to use these techniques to understand the biology of PCa dormancy and recurrence, we desired to isolate viable DTCs so that RNA could be extracted.
Therefore, to detect and isolate viable BM DTCs from localized PCa patients, we designed a fluorescence-activated cell sorting (FACS)-based protocol and chose high expression of EPCAM as our positive marker, combined with negative markers for possibly contaminating BM cell types with no reported expression in PCa cells: CD45 (PTPRC) for hematopoietic cells, CD235a (glycophorin A) for erythroids, alkaline phosphatase (ALPL) for osteoblastic lineage cells, and CD34 for hematopoietic stem and progenitor cells. As an additional estimate of the prevalence of BM DTCs in localized PCa, we also did a simple immunomagnetic enrichment for EPCAM and performed qRT-PCR for expression of prostate markers.
Here, using cell enumeration by flow cytometry, bulk cell RNA sequencing (RNA-Seq), whole genome amplification followed by whole-exome sequencing, and qRT-PCR, we show evidence for the presence of DTCs in the BM of a subset of patients with localized PCa. These cells are rare, with a frequency less than one putative DTC per 105 viable, nucleated BM cells in most patients. Our findings are in agreement with the majority of the literature that does find DTCs to be present in some patients with localized PCa, although at a lower prevalence (% of patients) than some reports, which have been as high as 72% of patients.12 Perhaps more importantly, we have developed methods using standard FACS equipment to isolate viable PCa DTCs. We expect this technical advance to be a boon for investigators studying PCa recurrence.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Patients and sample collection
Patient characteristics are summarized in Table 1. The use of localized PCa samples for complimentary approaches is summarized in Figure 1. All subjects provided written informed consent as part of IRB-approved protocols. All patients with localized PCa (AJCC 8th Edition Stage IIIC or less) underwent radical prostatectomy as their initial treatment for PCa. Any localized PCa patient who received neoadjuvant (before surgery) treatment was excluded from FACS analysis. All samples from patients with the localized disease were collected by BM aspiration at Johns Hopkins University either before their surgery from the posterior superior iliac crest, or at the time of surgery from the pubic bone. Samples were transferred to 7.5 mL EDTA (purple cap) tubes and shipped overnight to The University of Michigan on wet ice. Normal marrow samples were purchased from AllCells (Alameda, CA), drawn from the posterior superior iliac crest of paid donors, transferred to heparinized tubes and shipped overnight on wet ice, or aspirated from vertebrae into EDTA tubes during noncancer spine surgeries at the University of Michigan. Samples from patients with metastatic (all castration resistant) PCa were collected from the University of Michigan by aspiration of the posterior superior iliac crest into EDTA tubes (living donors) or en bloc during bisection of the femur or vertebrae at autopsy as part of the Michigan Legacy Tissue Program (rapid autopsy program). Autopsy specimens were homogenized to release cells for analysis.
TABLE 1.
Patient characteristics
| Criterion | Subjects (n) | |
|---|---|---|
| Localized prostate cancer patients | ||
| Total | … | 72 |
| Age, y | ≤59 | 19 |
| 60 -69 | 39 | |
| ≥70 | 12 | |
| PSA (ng/mL) before prostatectomy | ≤6 | 32 |
| 6.01-10 | 28 | |
| 10.01-20 | 8 | |
| >20 | 2 | |
| Prostatectomy Gleason score | 6 | 3 |
| 3 +4 =7 | 45 | |
| 4 +3 =7 | 12 | |
| 8-10 | 10 | |
| Tumor stage | pT2 | 41 |
| pT3a | 23 | |
| pT3b | 7 | |
| Nodal stage | pN0 | 59 |
| pNx | 11 | |
| Surgical margin | Negative | 57 |
| Positive | 13 | |
| Collection site | Iliac crest | 27 |
| Pubis | 43 | |
| Metastatic prostate cancer patients | ||
| Total | … | 8 |
| Age, y | ≤59 | 3 |
| 60-69 | 3 | |
| ≥70 | 2 | |
| PSA (ng/mL) before collection | ≤6 | 4 |
| 6.01-10 | 1 | |
| 10.01-20 | 0 | |
| >20 | 3 | |
| Collection site | Iliac crest | 6 |
| Vertebrae | 1 | |
| Femur | 1 | |
| Patients with no known cancer | ||
| Total | … | 8 |
| Sex | Male | 5 |
| Female | 3 | |
| Age, y | ≤29 | 4 |
| ≥30 | 4 | |
| Collection site | Iliac crest | 6 |
| Vertebrae | 2 | |
FIGURE 1.
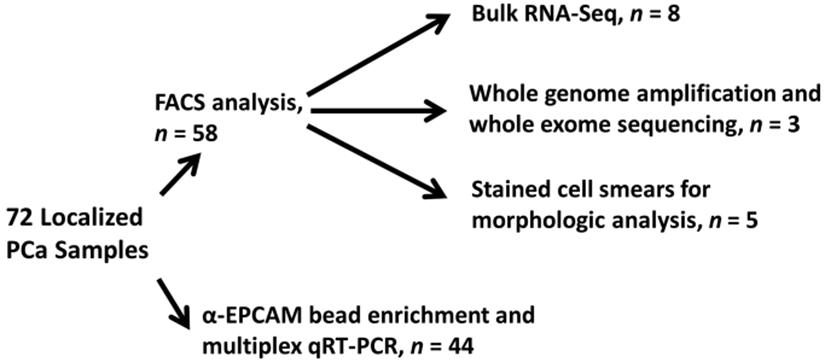
Use of localized PCa samples in complimentary analytic approaches. Fluorescence-activated cell sorting (FACS) with multiple markers followed by molecular or morphologic analysis of sorted cells (top), or enrichment with immunomagnetic beads for EPCAM positive cells and multiplex qRT-PCR (bottom). PCa, prostate cancer; qRT-PCR, quantitative reverse trancriptase polymerase chain reaction
2.2 ∣. Anti-EPCAM bead enrichment, mRNA isolation, and qRT-PCR
Enrichment of DTCs from 1mL of BM aspirate was performed as described for peripheral blood.27 Briefly, cells were bound to anti-EPCAM magnetic beads, washed, and directly lysed. mRNA was captured with Oligo(dT) 25 mRNA Dynabeads (Thermo Fisher Scientific) and reverse transcribed into complementary DNA (cDNA). Primer sequences were as previously described,27 and all pairs except KLK2 cross exon boundaries. Preamplification of up to 18 genes including controls was performed followed by qRT-PCR and relative quantification by the ΔΔCt method.
2.3 ∣. DTC enrichment and/or isolation by FACS
Marrow aspirates were mixed 1:1 with phosphate-buffered saline (PBS), layered onto Ficoll and centrifuged at 500g for 30 minutes to isolate the buffy coat/nucleated population. Subsequent steps were performed in cold flow cytometry buffer (PBS with 2% fetal calf serum and 1mM EDTA). The cells were washed and stained for 1 hour with the following antibodies: PE-Cy7 α-CD235a (1:20; #349112; BioLegend), Brilliant Violet 605 α-CD34 (1:20; #343529; BioLegend), APC α-EPCAM (1:20; #347200; BD), PerCP-Cy5.5 α-alkaline phosphatase (1:20; #561508; BD), and PE α-CD45 (1:5; #555483; BD). Cells were washed and resuspended in flow buffer containing 0.5 μg/mL DAPI and passed through a 40 μm filter to create a single cell suspension for FACS. All analyses were performed on a BD FACS-Aria IIu instrument with 405, 488, and 630 nm lasers. EPCAM positivity was defined as higher than unstained or isotype control stained cells. EPCAMhigh was defined as at least ×5 more intense than the center of the adjacent “dim” population. The putative DTC population was selected from single, viable cells as double negative on a plot of CD45 vs CD235a, then negative for alkaline phosphatase, then negative for CD34 and with high surface expression of EPCAM on a plot of CD34 vs EPCAM (Figure 2A). Sorted cells were collected in 10% FCS RPMI in a 0.2-mL tube, washed with cold PBS, leaving behind approximately 10 μL per tube. Ten thousand units of RNAse inhibitor (#55518-012; Invitrogen) was added, followed by freezing in liquid nitrogen.
FIGURE 2.
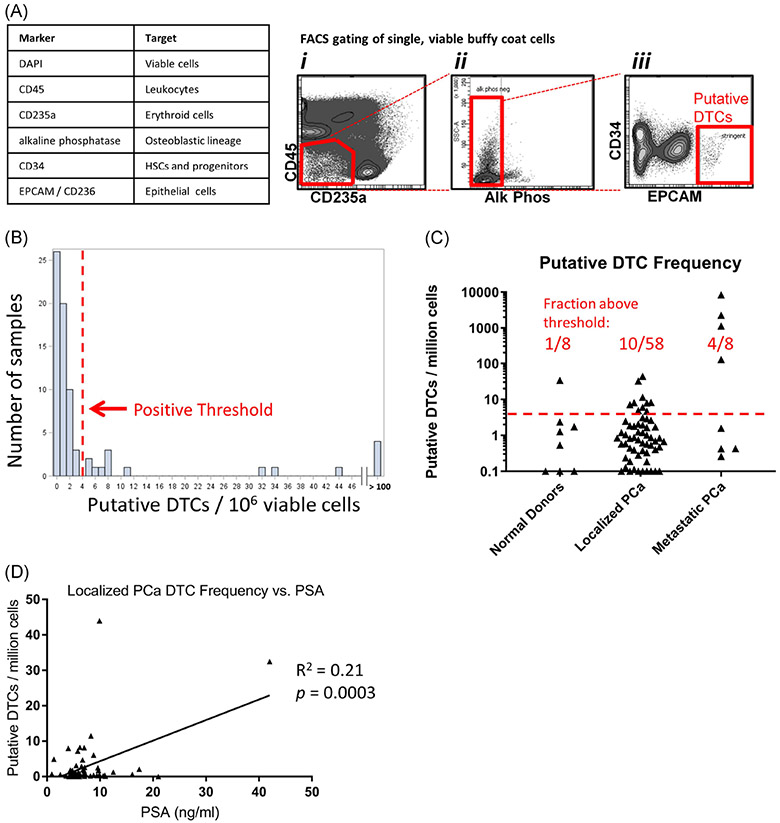
Analysis and isolation of putative DTCs using multi-parameter FACS. A, Flow cytometry markers and gating strategy. B, Histogram of the frequency of putative DTCs per 106single, viable, BM cells from all sample types: normal donors; n = 8, localized PCa; n = 58, and metastatic PCa; n = 8. A red threshold line is drawn at a local minimum at 4 cells per 106 where no data points were present. C, Frequency of the putative DTC population in normal donors, localized PCa patients and metastatic PCa patients. The red dotted line indicates the FACS positive threshold of four cells per 106. D, Correlation of localized PCa patient DTC frequency vs blood PSA concentration by linear regression analysis. BM, bone marrow; DTCs, disseminated tumor cells; FACS, fluorescence-activated cell sorting; PCa, prostate cancer
2.4 ∣. Bulk RNA-Seq
FACS-isolated marrow samples or 100 C42B cells as a positive control were processed without initial RNA purification using the Takara SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (#634889). cDNA was amplified between 8 and 15 cycles depending on cell number, followed by 16 cycles of library amplification with mRNA optimized methods. 40 million 50-cycle single end reads were obtained on an Illumina HiSeq-4000 instrument. FastQC was used to assess read quality. Data were processed using published methods.28 Briefly, RNA-Seq reads were aligned to the GRCh38 genome using HiSAT2 (v 2.1.0).29 Stringtie (v 1.3.4) was subsequently used to assemble and quantify transcripts from the alignment data.30 Individual data files were converted to transcripts per million (TPM).
2.5 ∣. Analysis of gene expression data
Morpheus software (Broad Institute) was used for visualization and hierarchical clustering of gene expression data using default parameters. Gene Ontology analysis was conducted with gene set enrichment analysis (GSEA) software (Broad Institute) of the c5.bp.v6.2 gene sets, parameter metric Diff_of_classes, using five permutations. The CIBERSORT algorithm31 (Stanford University) was used to estimate the identity of any immune cells present in RNA-Seq data. Data were uploaded as a text file of TPM values. Data were analyzed using the LM22 (default) signature gene file and the “absolute” mode with 100 permutations.
2.6 ∣. Whole genome amplification and whole-exome sequencing
Samples were processed directly after FACS with no intervening DNA purification. A total of 100 PC3 cells were used as a positive control. Whole genome amplification was performed with the Qiagen Single Cell REPLI-g Kit (#150343). Libraries were constructed using an Agilent SureSelectXT Human All Exon V6 Kit. Sequencing was performed to approximately 100× coverage on an Illumina HiSeq PE150 instrument with 40 million paired end reads per sample. Somatic single nucleotide polymorphisms (SNPs) were analyzed using MuTect2 software (Broad Institute) tumor vs normal approach with CD45+ cells from the same patient as the control for each putative DTC sample. The tumor-only approach was used for PC3 cells. Manta software (omicX, Rouen, France) was used for structural variant (SV) analysis again using the tumor vs normal approach except for PC3 cells. We queried a list of the following 75 genes known to be altered in PCa: PTEN, ZNF292, TP53, FOXA1, ERG, CDKN1B, NEAT1, PDE4D, ROBO2, PPAP2A, ETV3, MLL3, SPOP, MYST3, CDH12, KMT2C, PPP2R2A, ADAM28, IL6ST, UBTF, AR, APC, GPATCH8, ASH1L, DLC1, NCOR2, ZFHX3, TBL1XR1, SENP6, ANTXR2, ARID4B, ASXL2, LCE2B, DOCK10, NDST4, RPL11, RB1, USP28, ARID1A, CASZ1, CNOT3, ATM, PIK3R1, BRCA2, TBX3, ZMYM3, CDK12, KDM6A, NCOR1, CTNNB1, SMAD2, SMAD4, AKT1, BRAF, HRAS, IDH1, KMT2D, MLL2, MTUS1, PIK3CA, PIK3CB, RNF43, FOXP1, SHQ1, RYBP, CDH1, ROBO1, ZBTB16, NCOA7, MYC, MAP3K1, LRP1B, PPE4D, CSMD3, and NKX3-1.32 We reported those genes with an SNP or SV in the putative DTC sample but not in the internal control.
2.7 ∣. Cell smear morphologic analysis
Cells were dried on slides, fixed, and stained using the Hema-3 Kit (Fisher Scientific). Five hundred cell counts were performed by a blinded pathologist.
2.8 ∣. Hematopoietic colony formation
A total of 24-well plates were seeded with 1000 flow events per well and then combined with 500 μL of methylcellulose media with hematopoietic growth factors (#13M53790; H4434 Classic; Stem Cell Technologies). Cells were cultured under standard conditions for 7 days without disruption and then photomicrographed. CFU-E, CFU-GM, and CFU-GEMM colonies were counted by a blinded observer.
2.9 ∣. Statistical analyses
Comparison of two groups was performed in the GraphPad Prism software by the two samples unpaired Student t test. Linear regression was used to assess the correlation between putative DTC percentage and patient PSA. Histograms were generated using SAS software.
3 ∣. RESULTS
We analyzed BM samples from 72 patients with localized PCa, 8 patients with metastatic PCa (6 living donors, and 2 rapid autopsy patients), and 8 patients with no known cancer as negative controls (6 iliac crest aspirates from paid donors and 2 vertebral marrow samples from noncancer spine surgeries; Table 1). There were incomplete clinical data for two of the localized PCa patients. The living donor metastatic PCa patients all had treated castration-resistant disease and had varying degrees of disease control at the time of donation as approximated by their plasma PSA level. Four were well controlled with PSA values of less than 6 ng/mL. As described in more detail below, the samples were analyzed using either multicolor FACS followed by downstream analyses on the isolated cells, or enrichment of EPCAM+ cells with immunomagnetic beads followed by qRT-PCR for PCa markers (Figure 1). To maximize the data collected from these valuable human samples, we split samples between immunomagnetic bead enrichment (1 mL of marrow aspirate) and FACS analyses (5-6 mL) as limited by personnel and equipment availability. Therefore, some patients have data for both the immunomagnetic bead and FACS-based approaches.
Our FACS-based method utilized negative markers for erythroid lineage cells (CD235a), hematopoietic stem and progenitor cells, and endothelial cells (CD34), osteoblastic lineage cells (alkaline phosphatase), and set a high threshold for EPCAM positivity (Figure 2A). To dichotomize the presence of putative DTCs, we set a threshold of greater than four cells per million single, viable BM cells. This threshold was selected at a local minimum of a histogram of the frequency of these cells in all samples (Figure 2B, red line and arrow). Based on the current data, this seemed to be the most reasonable threshold, but we acknowledge that this threshold line could shift slightly if data from more patients were acquired. Furthermore, because of the possibility that DTCs were less numerous than four per million, we continued to collect and subsequently analyze the putative DTC population in samples with lower cell frequencies if at least 20 cells could be sorted. Because of the rarity of these cells, we were limited to collect a maximum of 300 cells from a tube of marrow from localized PCa patients but were able to collect more from some metastatic patients. Using this threshold of more than 4 per million cells, we detected the presence of the putative DTC population in 4 of 8 metastatic PCa patients, 10 of 58 localized PCa patients, and 1 of 8 normal donors (Figure 2C). In support of a malignant identity for the putative DTC population, we also noted that the putative DTC frequency is significantly correlated with localized PCa patients’ plasma PSA concentration, a predictor of recurrence.33 Clinical recurrence data for this patient cohort will take years to mature.
In interpreting these results, we note that some of the metastatic PCa patients were under good disease control (PSA <6 ng/mL) at the time of sample collection (Table 1). The four metastatic patients with PSA more than 6 ng/mL all had a putative DTC population frequency of more than 4 cells per million, whereas the metastatic patients with PSA less than 6 ng/mL had DTC population frequencies of less than 4 cells per million. With regard to the one normal donor with this population present above threshold, we also note that these methods could theoretically detect most epithelial cancers and at present we are unable to exclude this possibility. Further, the marrow from this normal donor subject (AC_6297) demonstrated no molecular evidence of PCa marker gene expression by RNA-Seq (Figure 3A) or qRT-PCR (Figure 6A), thus suggesting a false positive result not due to PCa cells.
FIGURE 3.
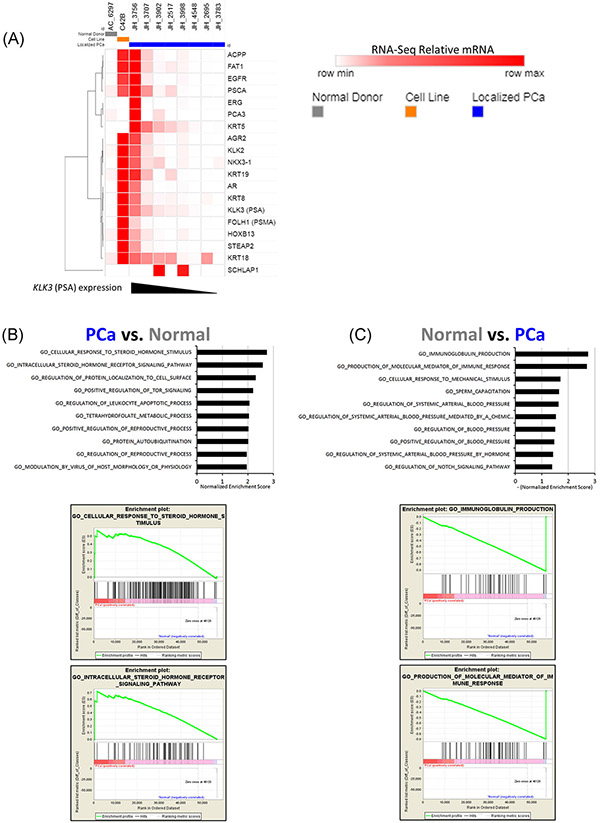
Examination of BM putative DTC expression profiles by RNA sequencing. A, Heat map of relative TPM values of proposed PCa marker genes from samples of the FACS population containing putative DTCs. Samples are color coded as follows: gray, the one control patient with this population; orange, C42B cell line as a positive control; and blue, 8 Localized PCa patients. The localized PCa samples are ordered from left to right in decreasing order of KLK3 (PSA) expression. B, Gene Ontology analysis using GSEA software comparing the four highest KLK3 expressing localized PCa samples relative to the normal donor sample. Top: gene sets with the 10 highest normalized enrichment scores are listed. Bottom: Enrichment plots for the gene sets with the two highest normalized enrichment scores. C, Results from the gene ontology analysis shown in (B), for gene sets enriched in the normal donor relative to the localized PCa patient samples. BM, bone marrow; DTCs, disseminated tumor cells; FACS, fluorescence-activated cell sorting; GSEA, gene set enrichment analysis; PCa, prostate cancer; PSA, prostate-specific antigen; TPM, transcripts per million
FIGURE 6.
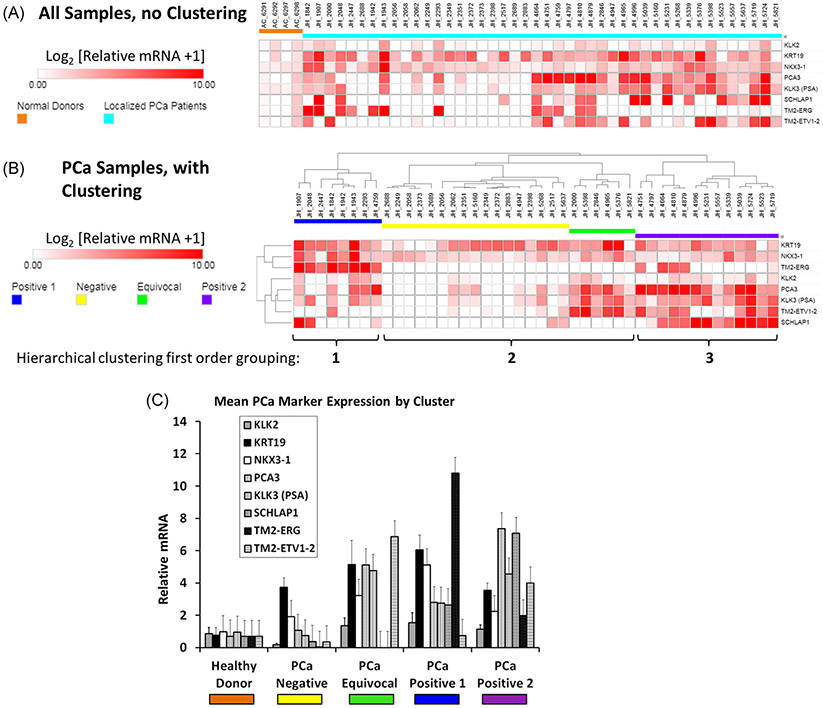
Evaluation of localized PCa DTCs by anti-EPCAM bead enrichment and qRT-PCR. A, Expression of 8 potential PCa marker genes in bone marrow from 44 patients with localized PCa (light blue) relative to four bone marrow donors with no known cancer (orange). B, Hierarchical clustering of marker gene expression in the PCa patients. Of the three main clusters, the outside two clusters are hypothesized to contain DTCs (“positive 1” [blue] and “positive 2” [purple]). The middle cluster is hypothesized to be “negative” (yellow) or “equivocal” (green) for the presence of DTCs. The three main clusters are indicated by the tree above, and brackets below the heat map. C, Mean ± SEM expression of the marker genes grouped by healthy donor or the four PCa patient expression clusters. DTCs, disseminated tumor cells; PCa, prostate cancer; qRT-PCR, quantitative reverse trancriptase polymerase chain reaction
To evaluate the tumor identity of the putative DTC population, we analyzed sorted cells from eight localized PCa patients by bulk RNA-Seq. Three of these patients (JH3998, JH2695, and JH2517) had a putative DTC frequency above the threshold of four cells per million, and five patients (JH3707, JH3756, JH3783, JH3902, and JH4548) were below this FACS threshold. As a comparator for these analyses, we included the putative DTC population from the one normal donor patient that had the putative DTC population present. We also used the C42B cell line as a positive control (Figure 3A). The localized PCa samples are ordered for presentation by decreasing KLK3 (PSA) expression from left to right. In keeping with the well-established role of PSA as a PCa marker, several other potential PCa marker genes follow the same pattern as PSA. None of the right-most samples (lowest PSA expression) have expression of the PCa marker genes higher than the normal donor sample, with the exception of one sample with KRT18 expression (patient JH_2695). We interpret this data that four of the eight localized PCa samples had expression of PCa marker genes consistent with the presence of PCa cells. Importantly, the one normal donor sample had no evidence of expression of PCa marker genes. We also analyzed this data set with Gene Ontology analyses using GSEA methods. Here, we compared the gene expression of the four localized PCa samples with the highest PSA values vs the one normal donor patient with this population present. Again, in keeping with the presence of PCa cells in the putative DTC population, we noted the highest level of enrichment for the “Cellular Response to Steroid Hormone Stimulus” and “Intracellular Steroid Hormone Receptor Signaling Pathway” gene sets (Figure 3B). We found this to be notable given the importance of testosterone and other steroid hormones in PCa. Conversely, when we compared putative DTC gene expression of the one normal donor to the PCa patients we observed the highest level of enrichment for two immune-related gene groups; “Immunoglobulin Production” and “Production of Molecular Mediator of Immune Response,” consistent with the presence of marrow cells in these samples.
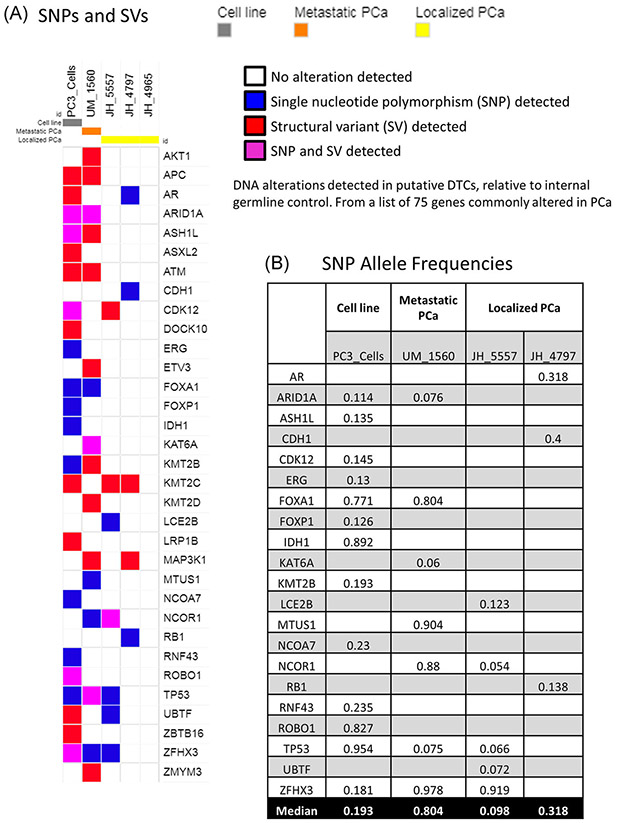
For further validation of the tumor origin of cells in the putative DTC population, we performed whole genome amplification and whole-exome sequencing to approximately 100× coverage of leukocytes and between 100 and 250 putative DTCs from three localized PCa patients, one metastatic patient as a FACS control, and the PC3 cell line as a sequencing technical control. The metastatic patient (UM_1560) and two of the localized patients (JH_4797 and JH_4905) had more than four putative DTCs per 106 BM cells, and localized patient JH_5557 had three putative DTCs per 106 BM cells. The leukocyte (CD45+) population was used as an internal germline and technical control for each patient. To determine if these populations contained small alterations characteristic of PCa, we focused our analysis on a list of 75 genes previously reported to be altered in PCa (gene list in methods).32 Figure 4A reports genes from this list which had either SNPs or gene level SVs in the putative DTC population but not in the corresponding internal leukocyte control. We observed alterations of genes characteristic of PCa in the metastatic sample and two of the three localized PCa putative DTC samples. Unfortunately, the data across the genome was not sufficiently uniform to visualize chromosomal level alterations (deletions, amplifications, and translocations), likely due to unequal amplification, which has been reported previously with the Repli-G whole genome amplification system.34 Finally, to gain an appreciation for the approximate fraction of cancer cells in the putative DTC population, we examined the allele frequency for SNPs. The allele frequency was lower for the localized PCa samples than the metastatic sample, suggesting the continued presence of a normal marrow population at a higher fraction in the localized samples than the metastatic sample (Figure 4B). We did not analyze the allele frequency for SVs because it is more difficult to estimate with the bioinformatics methods used.
FIGURE 4.
Whole genome amplification and whole-exome sequencing of the putative DTC population. A, Map showing somatic single nucleotide polymorphisms (SNPs) or small structural variants (SVs), detected in each experimental sample but not CD45+ cells from the same patient as an internal control. Sample types are annotated as follows: gray, PC3 PCa cell line; orange, metastatic PCa patient; yellow, three localized PCa patients. Detected alterations are indicated by: white, no alteration detected; blue, SNP detected; red, SV detected; purple, SNP and SV both detected. All genes were selected from a list of 75 genes previously reported to be altered in prostate cancer. B, Allele frequencies for SNPs detected in the PCa putative DTC samples and PC3 cells. The median allele frequency for each sample is listed in the bottom row. DTC, disseminated tumor cell; PCa, prostate cancer
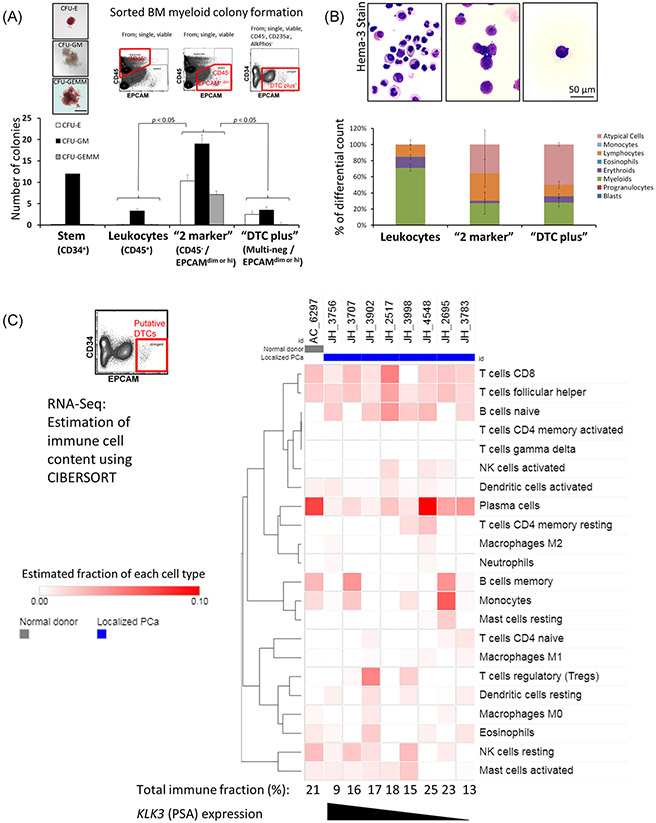
To improve isolation methods and to guide selection of any additional negative markers for future protocol refinement, we further examined the contaminating marrow cell types in sorted populations (Figure 5). We first performed myeloid colony formation assays to assess the cell types present (Figure 5A). The putative DTC population from a localized PCa patient does not yield enough cells for most in vitro assays. Therefore, to achieve a usable number of cells, we decreased the stringency of the EPCAM gate to yield a population we refer to as “DTC plus,” which is like putative DTCs but also includes EPCAM dim cells in addition to EPCAM high expressing cells. For comparator cell populations, we also isolated CD34+ cells, CD45+ cells, and “2 marker” cells which were CD45–/EPCAM dim or hi but did not include the other negative markers. The “DTC plus” population showed lower numbers of myeloid colonies than the “2 marker” population—consistent with contaminating nonmyeloid cells in the putative DTC population. We also analyzed the same populations by morphology on cell smears (Figure 5B). We observed low numbers of granulocytes in the “DTC plus” population but a higher number of “atypical cells” with an appearance consistent with plasma cells, erythroid precursors or tumor cells. Then, using the RNA-Seq data set first described in Figure 3, we used the CIBERSORT algorithm to estimate the immune cell composition in these samples.31 The samples are again arranged in decreasing order of PSA expression for the localized PCa samples (Figure 5C). In agreement with the morphological analyses, this highlighted the presence of B-cell subsets, especially of plasma cells. We noted a higher estimated content of plasma cells in the samples on the right side of the heat map, which also has lower expression of PCa markers, thus consistent with an increased percentage of plasma cells in samples where the DTC content appears lower. Importantly, the patient with the highest apparent PCa content in Figure 3A (JH_3756) also had the lowest estimated immune cell fraction. Overall, these analyses further support the PCa marker data from Figure 3 and highlight nonmyeloid cells, especially plasma cells, as potential contaminating cell types of our putative DTC population.
FIGURE 5.
Identity of contaminating cell types. A, Estimation of the number of contaminating myeloid precursors by colony formation. Upper left; example images of each colony type. Upper right: Example FACS plots describing the cell populations examined. Data are a representative experiment mean ± SD of the six rows of a 24-well plate. Means were compared by the Student t test. B, Estimation of contaminating cell types by morphology. Top: Hema3 (Wright-Giemsa) stained smears prepared after sorting the indicated cell populations; bottom: 500 cell marrow differential cell counts were performed by a blinded observer of smears from five patients. Data represent mean ± SEM. C, Estimation of immune cell content in the putative DTC population from the RNA-Seq data (as in Figure 3) using the CIBERSORT algorithm. FACS, fluorescence-activated cell sorting
Finally, to achieve another estimate of the prevalence of BM DTCs in localized PCa, we used α-EPCAM magnetic bead enrichment and multiplex qRT-PCR for PCa markers to assess the presence of DTCs in 44 localized PCa patients and 4 normal marrow donors (Figure 6). We previously developed these techniques for peripheral blood and reliably detected ≥10 PCa cells.27 In the current work, we observed upregulation of one or more PCa-related genes in approximately half of the localized PCa samples as compared to the normal donor controls (Figure 6A). Importantly, the one normal donor with the putative DTC population present on FACS (AC_6297) was one of these normal controls and did not show increased PCa marker gene expression. To objectively separate putative DTC positive vs DTC negative patients, we analyzed the localized PCa patient data with hierarchical clustering and observed segregation into three main trunks (Figure 6B). After clustering analysis, we interpreted the outer two trunks as positive—labeled “positive 1” or “positive 2” (collectively 21 of 44 samples or 47%) and the center trunk as less likely to contain DTCs—labeled “negative” or “equivocal.” The six patients labeled “equivocal” cluster with the “negative” patients but also share characteristics with “positive 2.” Thus a less conservative interpretation of the data would estimate DTC prevalence at 27 of 44 (61%). We noted that the “positive 1” trunk was characterized by expression of the pathognomonic PCa fusion gene TMPRSS2-ERG and that the “positive 2” trunk was characterized by expression of PCA3 and SCHLAP1. When the same data were analyzed and presented as the mean gene expression for all patients in each gene expression cluster, we observed increased expression relative to normal donor marrow of 6 of the 8 PCa genes in “positive 1” and/or “positive 2” but not in the “negative” gene expression group (Figure 6C).
Utilization and results from all of the PCa patient and normal donor samples is summarized in Table 2. Because the EPCAM bead enrichment and qRT-PCR panel utilized only 1 ml of sample, some patients have both PCR data and either RNA-Seq or DNA sequencing data. Not all patients have molecular data available.
TABLE 2.
Results summary. Left column “negative” and “positive” denote either less than four putative DTCs per million BM cells or more than four putative DTCs per million BM cells respectively. “RNA-Seq positive” denotes increased expression of PCa marker genes on RNA sequencing of putative DTCs. “WGA/WES positive” denotes the presence of mutations in PCa genes after whole genome amplification and whole-exome sequencing
| Clinical/FACS category | Total | DTCs collected | RNA-Seq positive | RNA-Seq negative | WGA/WES positive | WGA/WES negative | qRT-PCR positive | qRT-PCR negative or equivocal |
|---|---|---|---|---|---|---|---|---|
| Normal donor DTC negative | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Normal donor DTC positive | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| Localized PCa DTC negative | 48 | 8 | 3 | 2 | 1 | 1 | 11 | 13 |
| Localized PCa DTC positive | 10 | 9 | 1 | 2 | 1 | 0 | 1 | 5 |
| Localized PCa no FACS data | 14 | 0 | 0 | 0 | 0 | 0 | 10 | 4 |
| Metastatic PCa DTC negative | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Metastatic PCa DTC positive | 4 | 4 | 0 | 0 | 1 | 0 | 0 | 0 |
Abbreviations: BM, bone marrow; DTCs, disseminated tumor cells; FACS, fluorescence-activated cell sorting; PCa, prostate cancer; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
4 ∣. DISCUSSION
Here we present multiple lines of evidence indicating BM DTCs are detectable in many men with localized PCa and describe a novel method for their enrichment and collection by FACS—a widely available technique. This study provides important information on the anatomic distribution of PCa cells in clinically localized disease. At a threshold of four putative DTCs per 106 BM cells, we conservatively estimate the prevalence of DTCs at 10 of 58 patients (17%). However, as shown by our RNA-Seq data, some patients with a detectable putative DTC population below this threshold are likely to contain DTCs as well, although their isolation becomes increasingly difficult with current methods. Using qRT-PCR for a panel of PCa marker genes, after EPCAM bead enrichment, we estimate the prevalence of DTCs at 21/44 (47%). Perhaps the real prevalence lies somewhere in the middle. Nevertheless, we feel given the current data it is reasonable to posit that at least some PCa patients do have BM DTCs at the time or radical prostatectomy. We think that these results lend additional insight into the field of PCa dormancy and recurrence, and provide an invaluable tool for investigators studying these processes. These essential techniques could be used in the current form for additional downstream analyses in subsequent work or could serve as the basis for further refinements in DTC isolation.
This study must be placed in the context of prior research on PCa DTCs, as rates of detection have ranged from near zero to 72% of localized patients (reviewed in25). Before 2018, investigators have reported the presence of DTCs using independent methodologies including; RT-PCR for KLK3 (PSA), immunocytochemistry for PSA or pan-cytokeratin, and EPCAM-based immune-magnetic enrichment and single-cell isolation coupled with negative depletion of leukocytes and megakaryocytes. At least one work using each of these techniques has also shown correlation of DTC detection with patient data on recurrence or risk of recurrence.25 Additionally, our data should be considered in the context of a recent study in which PCa DTCs were very rarely detected at the time of radical prostatectomy.26 It is challenging to prove the absence of a cell population, as suggested in the study by Chalfin and colleagues, and is also difficult to know the sensitivity of their methodologies. This is particularly true given the use of semi-automated platforms which had been previously validated for the detection of CTCs (from peripheral blood) rather than for DTCs, and with minimal assessment of positive controls for DTC detection. The unexpected findings of the Chalfin study highlights the need for a further confirmatory investigation, such as ours, in part to provide balance. The bulk of the literature supports that BM DTCs are present in some patients with localized PCa—as we have recently reviewed.25 However, very little is known of the phenotype and behavior of these cells, which will be greatly aided by the techniques for viable cell isolation that we present here. With a few exceptions from groups at the University of Regensburg and University of Washington,4,7,8,18,24 prior investigations of PCa DTCs did not isolate viable cells.6,9-11,13-17,19-23
The key limitations of the current study include the relatively small sample size and the difficulty isolating sufficient numbers of cells for genetic analyses. Single cell rather than bulk next generation sequencing (NGS) analyses would also be useful but are a significant technical challenge also because of the rarity of the cells. The commonly used single-cell platforms from Fluidigm and 10x Genomics require many input cells; on the order of 104 or 105—a hundred or thousand fold more DTCs than are likely to exist in a tube of marrow from a localized PCa patient.
An additional limitation of this study is the continued use of EPCAM to help identify DTCs, as EPCAM can lead to both false positives and false negatives. Chery et al4 used single cell microarray analyses on DTCs isolated using EPCAM-based methods and observed significant EPCAM expression by normal BM cells.4 Furthermore, DTCs in the BM may downregulate EPCAM as they assume a mesenchymal or stem-like phenotype.35,36 We acknowledge that some DTCs with a stem or mesenchymal phenotype might not be captured by our current FACS methods. However, we did not think it possible to empirically choose a marker for mesenchymal phenotype DTCs at the onset of this work and hoped to refine our techniques based on data. We think that the data presented here provide an invaluable framework, with which to add additional positive or negative markers in future studies. Our RNA-Seq data are useful in this regard. Of the cell surface genes, prostate stem cell antigen appears to be highly expressed in DTCs, though analysis of additional patients will be useful. Our work examining the potential contaminating cell types highlights the presence of plasma cells and therefore suggests potential use of plasma cell negative markers. Our data are not inconsistent with the presence of contaminating erythroid precursors as well, as another group previously reported.4 Together, these data suggest that while normal marrow residents may express EPCAM, higher levels of expression can nevertheless provide valuable information on the presence of DTCs and allow for continued improvement in understanding their biology and how best to isolate them.
5 ∣. CONCLUSION
We present multiple lines of evidence demonstrating the presence of BM DTCs in localized PCa patients at the time of radical prostatectomy. Although the fraction of positive patients is perhaps lower than previously reported, DTCs are present in at least some patients at the time of prostatectomy, which provides a clear rationale for further investigation of the BM as a reservoir for PCa recurrence and for the eventual development of therapies targeted to this site. Lastly, we provide an invaluable tool for viable DTC isolation to better understand the biology of this process, which we hope will lead to fewer cases of deadly recurrent PCa.
ACKNOWLEDGMENTS
The authors graciously thank Emily Caruso, Megan Kuczler, and Amy Kasputis for management of sample collection. We also thank Dr Arul Chinnaiyan, Dr David Smith, and Dr Zachary Reichert for helpful discussions. We thank Melissa Coon and Dr Robert Lyons of the University of Michigan DNA Sequencing Core for sample preparation and generation of raw RNA-Seq data. Most importantly, we thank all the patients who donated samples to make this study possible. Direct funding was provided by the NIH/NCI P01-CA093900, Department of Defense W81XWH-14-1-0403 and The Prostate Cancer Foundation Challenge award 16CHAL05. RT received support as the Major McKinley Ash Colligate Professor. FC receives support from a Career Enhancement Award from the NIH/NCI Prostate Cancer Specialized Program in Research Excellence (SPORE) at the University of Michigan F048931, sub-award F036250, and Prostate Cancer Foundation Young Investigator Award 18YOUN04.
Funding information
Prostate Cancer Foundation, Grant/Award Numbers: 16CHAL05, 18YOUN04; National Cancer Institute, Grant/Award Numbers: F048931, P01-CA093900, sub-award F036250; Congressionally Directed Medical Research Programs, Grant/Award Number: W81XWH-14-1-0403
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
REFERENCES
- 1.SEER Cancer Statistics Factsheets: Prostate Cancer. National Cancer Institute. Bethesda, MD. https://seer.cancer.gov/statfacts/html/prost.html. Accessed 9 December, 2016. [Google Scholar]
- 2.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281(17):1591–1597. [DOI] [PubMed] [Google Scholar]
- 3.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14(9):611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chéry L, Lam HM, Coleman I, et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget. 2014;5(20):9939–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubendorf L, Schöpfer A, Wagner U, et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31(5):578–583. [DOI] [PubMed] [Google Scholar]
- 6.Berg A, Bruland øS, Fosså SD, et al. Disseminated tumor cells in bone marrow following definitive radiotherapy for intermediate or high-risk prostate cancer. Prostate. 2008;68(15):1607–1614. [DOI] [PubMed] [Google Scholar]
- 7.Ellis WJ, Pfitzenmaier J, Colli J, Arfman E, Lange PH, Vessella RL. Detection and isolation of prostate cancer cells from peripheral blood and bone marrow. Urology. 2003;61(2):277–281. [DOI] [PubMed] [Google Scholar]
- 8.Holcomb IN, Grove DI, Kinnunen M, et al. Genomic alterations indicate tumor origin and varied metastatic potential of disseminated cells from prostate cancer patients. Cancer Res. 2008;68(14):5599–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köllermann J, Weikert S, Schostak M, et al. Prognostic significance of disseminated tumor cells in the bone marrow of prostate cancer patients treated with neoadjuvant hormone treatment. J Clin Oncol. 2008;26(30):4928–4933. [DOI] [PubMed] [Google Scholar]
- 10.Mansi JL, Berger U, Wilson P, Shearer R, Coombes RC. Detection of tumor cells in bone marrow of patients with prostatic carcinoma by immunocytochemical techniques. J Urol. 1988;139(3):545–548. [DOI] [PubMed] [Google Scholar]
- 11.Melchior SW, Corey E, Ellis WJ, et al. Early tumor cell dissemination in patients with clinically localized carcinoma of the prostate. Clin Cancer Res. 1997;3(2):249–256. [PubMed] [Google Scholar]
- 12.Morgan TM, Lange PH, Porter MP, et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clin Cancer Res. 2009;15(2):677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray NP, Aedo S, Fuentealba C, Reyes E, Salazar A. Minimum residual disease in patients post radical prostatectomy for prostate cancer: theoretical considerations, clinical implications and treatment outcome. Asian Pac J Cancer Prev. 2018;19(1):229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray NP, Reyes E, Orellana N, et al. Secondary circulating prostate cells predict biochemical failure in prostate cancer patients after radical prostatectomy and without evidence of disease. Scientific- WorldJournal. 2013;2013:762064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray NP, Reyes E, Orellana N, Fuentealba C, Jacob O. Comparison of the Walz Nomogram and presence of secondary circulating prostate cells for predicting early biochemical failure after radical prostatectomy for prostate cancer in Chilean men. Asian Pac J Cancer Prev. 2015;16(16):7123–7127. [DOI] [PubMed] [Google Scholar]
- 16.Murray NP, Reyes E, Tapia P, Badínez L, Orellana N. Differential expression of matrix metalloproteinase-2 expression in disseminated tumor cells and micrometastasis in bone marrow of patients with nonmetastatic and metastatic prostate cancer: theoretical considerations and clinical implications—an immunocytochemical study. Bone Marrow Res. 2012;2012:259351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantel K, Aignherr C, Köllermann J, Caprano J, Riethmüller G, Köllermann MW. Immunocytochemical detection of isolated tumour cells in bone marrow of patients with untreated stage C prostatic cancer. Eur J Cancer. 1995;31A(10):1627–1632. [DOI] [PubMed] [Google Scholar]
- 18.Pfitzenmaier J, Ellis WJ, Hawley S, et al. The detection and isolation of viable prostate-specific antigen positive epithelial cells by enrichment: a comparison to standard prostate-specific antigen reverse transcriptase polymerase chain reaction and its clinical relevance in prostate cancer. Urol Oncol. 2007;25(3):214–220. [DOI] [PubMed] [Google Scholar]
- 19.Schlimok G, Funke I, Holzmann B, et al. Micrometastatic cancer cells in bone marrow: in vitro detection with anti-cytokeratin and in vivo labeling with anti-17-1A monoclonal antibodies. Proc Natl Acad Sci USA. 1987;84(23):8672–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todenhöfer T, Hennenlotter J, Faber F, et al. Significance of apoptotic and non-apoptotic disseminated tumor cells in the bone marrow of patients with clinically localized prostate cancer. Prostate. 2015;75(6):637–645. [DOI] [PubMed] [Google Scholar]
- 21.Weckermann D, Müller P, Wawroschek F, Harzmann R, Riethmüller G, Schlimok G. Disseminated cytokeratin positive tumor cells in the bone marrow of patients with prostate cancer: detection and prognostic value. J Urol. 2001;166(2):699–703. [PubMed] [Google Scholar]
- 22.Wood DP Jr, Banerjee M. Presence of circulating prostate cells in the bone marrow of patients undergoing radical prostatectomy is predictive of disease-free survival. J Clin Oncol. 1997;15(12):3451–3457. [DOI] [PubMed] [Google Scholar]
- 23.Wood DP Jr, Banks ER, Humphreys S, McRoberts JW, Rangnekar VM. Identification of bone marrow micrometastases in patients with prostate cancer. Cancer. 1994;74(9):2533–2540. [DOI] [PubMed] [Google Scholar]
- 24.Gužvić M, Braun B, Ganzer R, et al. Combined genome and transcriptome analysis of single disseminated cancer cells from bone marrow of prostate cancer patients reveals unexpected transcriptomes. Cancer Res. 2014;74(24):7383–7394. [DOI] [PubMed] [Google Scholar]
- 25.Cackowski FC, Taichman RS. Advances in experimental medicine and biology. Adv Exp Med Biol. 2018;1100:47–53. [DOI] [PubMed] [Google Scholar]
- 26.Chalfin HJ, Glavaris SA, Malihi PD, et al. Prostate cancer disseminated tumor cells are rarely detected in the bone marrow of patients with localized disease undergoing radical prostatectomy across multiple rare cell detection platforms. J Urol. 2018;199:1494–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singhal U, Wang Y, Henderson J, et al. Multigene profiling of CTCs in mCRPC identifies a clinically relevant prognostic signature. Mol Cancer Res. 2018;16(4):643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wedge DC, Gundem G, Mitchell T, et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet. 2018;50(5):682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117(22):5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borgström E, Paterlini M, Mold JE, Frisen J, Lundeberg J. Comparison of whole genome amplification techniques for human single cell exome sequencing. PLoS One. 2017;12(2):e0171566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiozawa Y, Berry JE, Eber MR, et al. The marrow niche controls the cancer stem cell phenotype of disseminated prostate cancer. Oncotarget. 2016;7:41217–41232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller L, Werner S, Pantel K. Biology and clinical relevance of EpCAM. Cell Stress. 2019;3(6):165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]