Abstract
Antiprotozoal drug nitazoxanide (NTZ) has shown diverse pharmacological properties and has appeared in several clinical trials. Herein we present the synthesis, characterization, in vitro biological investigation, and in silico study of four hetero aryl amide analogs of NTZ. Among the synthesized molecules, compound 2 and compound 4 exhibited promising antibacterial activity against Escherichia coli (E. coli), superior to that displayed by the parent drug nitazoxanide as revealed from the in vitro antibacterial assay. Compound 2 displayed zone of inhibition of 20 mm, twice as large as the parent drug NTZ (10 mm) in their least concentration (12.5 µg/ml). Compound 1 also showed antibacterial effect similar to that of nitazoxanide. The analogs were also tested for in vitro cytotoxic activity by employing cell counting kit‐8 (CCK‐8) assay technique in HeLa cell line, and compound 2 was identified as a potential anticancer agent having IC50 value of 172 µg which proves it to be more potent than nitazoxanide (IC50 = 428 µg). Furthermore, the compounds were subjected to molecular docking study against various bacterial and cancer signaling proteins. The in vitro test results corroborated with the in silico docking study as compound 2 and compound 4 had comparatively stronger binding affinity against the proteins and showed a higher docking score than nitazoxanide toward human mitogen‐activated protein kinase (MAPK9) and fatty acid biosynthesis enzyme (FabH) of E. coli. Moreover, the docking study demonstrated dihydrofolate reductase (DHFR) and thymidylate synthase (TS) as probable new targets for nitazoxanide and its synthetic analogs. Overall, the study suggests that nitazoxanide and its analogs can be a potential lead compound in the drug development.
Keywords: cytotoxicity assay, DHFR, E. coli, MAPK9, nitazoxanide, TS
Four heteroaryl nitazoxanide analogues were synthesized efficiently. Among the synthesized analogues, two compounds showed prominent antibacterial and anticancer potentials through in vitro and in silico studies.
![]()
Abbreviations
- CCK‐8
cell counting kit‐8
- DHFR
demonstrated dihydrofolate reductase
- MAPK9
mitogen‐activated protein kinase
- NTZ
nitazoxanide
- TS
thymidylate synthase
1. INTRODUCTION
Nitazoxanide (NTZ) or 2‐acetyloxy‐N‐(5‐nitro‐2‐thiazolyl) benzamide (Figure 1) belongs to 5‐nitrothiazole group of molecules which has already received ample attention in the field of drug discovery and drug development. 1 , 2 NTZ possessed broad spectrum of activity against protozoa, 3 , 4 helminthes, 5 , 6 and numerous Gram‐positive and Gram‐negative anaerobic bacteria. 6 , 7 , 8 NTZ also exhibited potential antiviral properties 9 , 10 and, recently, it has been found to be effective against SARS‐CoV‐2 11 and a number of clinical trials are underway. 12 , 13
FIGURE 1.
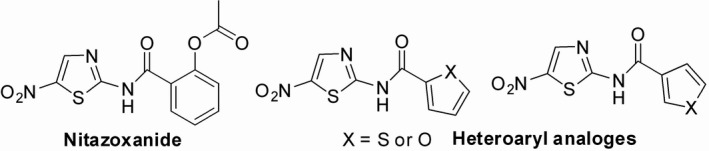
Structure of NTZ and heteroaryl analogs
Because of the potential biological properties, structurally modified NTZ‐based analogs or structurally related molecules were synthesized and investigated for their biological activity. Some recently synthesized analogs of NTZ reportedly possess prominent antiprotozoal, 14 , 15 , 16 , 17 antibacterial, and antimycobacterial 18 , 19 and antiviral 20 activities even in some cases better than the parent drug NTZ itself. Hence, NTZ became the drug of interest for our research where we focused on synthesis of hetero‐aryl amide analogs.
Although, NTZ and some of its analogs were investigated for a wide range of biological activities, heteroaryl amide analogs were not screened for several bioactivities. Therefore, we planned to synthesize some heteroaryl amide analogs (Figure 1) to evaluate their diverse biological activities such as antibacterial, antiviral, anticancer activity, in order to prove them to be a potential therapeutic choice. Based on the in vitro biological activities, we also conducted in silico molecular docking study to understand a mechanistic insight regarding the activity of NTZ and its synthetic analogs. So far, we have investigated in vitro antibacterial, cytotoxic and in vivo analgesic and anti‐inflammatory effects of NTZ and four synthesized analogs. Although, the analogs did not produce significant analgesic and anti‐inflammatory effects, we found some interesting results in antibacterial and cytotoxic screening which we are going to disclose in this paper.
2. MATERIALS AND METHODS
2.1. General experimental procedures
All the synthetic procedures were conducted in Chemical Biology and DNA Synthesis Laboratory, Faculty of Pharmacy, University of Dhaka. The solvents were dried and properly distilled. Progress of the reactions was monitored by Thin Layer Chromatography (TLC), and visualization was accomplished by using UV light at 254 nm. For column chromatography, silica gel 60 (0.06–0.2 mm, ROTH) was employed. 1H NMR (400 MHz) was recorded on Ultra Shield Bruker 400 NMR instrument, using DMS0‐d6, and the chemical shifts are reported as δ (ppm) with respect to tetramethylsilane (TMS) (ppm) as internal standard. Fourier‐transform infrared (FT‐IR) Spectra were recorded with FT‐IR 8400S Shimadzu spectrophotometer in the range of 4000–400 cm−1 using KBr pressed pellet technique. All the reagents used in synthetic procedure were purchased from Sigma‐Aldrich, Germany. Nitazoxanide, metronidazole, and nalidixic acid were procured from Incepta Pharmaceuticals Ltd. as dried powder.
2.2. Synthesis
Triethylamine (TEA) (1.2 equiv.) was added to the solution of 2‐amino‐5‐nitrothiazole (0.00689 mol) and dichloromethane (DCM). After the mixture was stirred for 15 min at 5℃, a solution of hetero aryl acid chloride (1.1 equiv.) in dichloromethane was added dropwise. The reaction mixture was stirred at room temperature for 24 h and after the completion of reaction indicated by TLC, the resulting residue was neutralized by saturated NaHCO3 solution. The mixture was then extracted with ethyl acetate. The organic layer was washed with brine solution. The solvent was removed under reduced pressure and crude product was then subjected to column chromatography for purification.
2.3. Spectrometric characterization of synthesized compounds
The structure of the synthetic analogs of nitazoxanide was elucidated spectroscopically by using IR and 1H NMR and by comparing the data with that of the reported data. 17 , 21 IR and 1H‐NMR data are recorded as follows:
2.3.1. N‐(5‐nitrothiazol‐2‐yl) thiophene‐2‐carboxamide (compound 1)
Yield: 91%; light yellow solid; Rf: 0.77 (ethyl acetate: n‐hexane = 1:2); IR (KBr, cm−1): 1542 (Aryl C=C str), 3077 (Aryl C–H str), 1265 (C–N str thiazole), 1351, 1542 (N–O str), 1662 (C=O str amide), 3350 (N–H str amide); 1H NMR (400 MHz, DMSO‐d6): δ 8.69 (s, 1H), 8.29 (d, J = 3.2 Hz, 1H), 8.05 (d, J = 4.8 Hz, 1H), 7.27 (dd, J = 3.2, 4.8 Hz, 1H).
2.3.2. N‐(5‐nitrothiazol‐2‐yl) thiophene‐3‐carboxamide (compound 2)
Yield: 89%; yellow solid; Rf: 0.54 (ethyl acetate: n‐hexane = 1:2); IR (KBr, cm−1): 1548 (Aryl C=C str), 3103 (Aryl C–H str), 1317 (C–N str thiazole), 1354, 1548 (N–O str), 1674 (C=O str), 3120 (N–H str amide); 1H NMR (400 MHz, DMSO‐d6): 8.70 (s, 1H), 8.69 (s, 1H), 7.74 (d, J = 10 Hz, 1H), 7.72 (d, J = 10 Hz, 1H).
2.3.3. N‐(5‐nitrothiazol‐2‐yl) furan‐2‐carboxamide (compound 3)
Yield: 70%; beige solid; Rf: 0.90 (ethyl acetate: n‐hexane = 1:2); IR (KBr, cm−1): 1411 (Aryl C=C str), 3143 (Aryl C–H str), 1315 (C–N str thiazole), 1356, 1482 (N–O str), 1667(C=O stretching of amide functional group), 3350 (N–H stretching of amide group); 1H NMR (400 MHz, DMSO‐d6): δ 7.77 (s, 1H), 7.72 (br s, 1H), 7.32 (br s, 1H), 7.06 (s, 1H), 6.57 (d, J = 1.6 Hz, 1H).
2.3.4. N‐(5‐nitrothiazol‐2‐yl) furan‐3‐carboxamide (compound 4)
Yield: 53%; yellow solid; Rf: 0.96 (ethyl acetate: n‐hexane = 1:2); IR (KBr, cm−1): 1313 (Aryl C=C str), 3139 (Aryl C–H str), 1313 (C–N str thiazole) 1355, 1507 (N–O str), 1653 (C=O stretching of amide group), 3446 (N–H stretching of amide group); 1H NMR (400 MHz, DMSO‐d6): δ 8.67 (s,1H), 8.65 (s,1H), 7.86 (s,1H), 7.10 (s,1H).
2.4. Antibacterial activity assay against S. aureus and E. coli
NTZ and its four analogs were assayed for antibacterial activity against a gram‐positive Staphylococcus aureus (S. aureus – coagulase (+) ve ATCC: 20121107‐4) and a gram‐negative (E. coli ATCC: 0157‐CR3) strain by the standardized disc diffusion method. 22 All the test samples were dissolved in DMSO (0.1% v/v) and diluted to prepare four concentrations (100, 50, 25, and 12.5 µg/ml) for each test sample. Nalidixic acid was used as the standard drug. The zone of inhibition was compared with standard drug after 24 h of incubation at 37°C for antibacterial activity.
2.5. Cytotoxicity assay
HeLa, a human cervical carcinoma cell line, was used for the cytotoxicity assay. Here the assay was designed in two phases. Firstly, the quantity of cell viability was determined by Trypan blue dye exclusion technique. 23 Then MTT method was performed to determine the IC50 value (50% growth inhibitory concentration) from the calculation of percent growth inhibition.
2.5.1. Trypan blue dye exclusion method
A cell suspension was made with a fixed volume of cells (e.g. 1 ml) and 50 µl of cell suspension was taken in a cryo vial. Equal parts of 0.4% trypan blue dye were added to the cell suspension and mixed them well by pipetting up and down. The cell suspension was then transferred to a hemocytometer to count the live cell and dead cell. Viable cell will resist the dye to enter as it possesses intact cell membrane whereas dead cell will take up the dye and turn into blue staining. Typically, each side of the hemocytometer will take 10–20 μl of the solution. After that the hemocytometer was placed on the stage of an inverted microscope. All the cells (clear and blue) in each large square in each corner of the hemacytometer were counted. Blue cells were the non‐viable cells and clear cells were the viable cells. Calculation of the percentage of viable cells was done by using the following formula: Percentage (%) cell viability = (Live cell count/Total cell count) × 100.
2.5.2. MTT assay
Hela cell line was cultured in DMEM (Dulbecco's Modified Eagles’ medium) containing 1% penicillin–streptomycin (1:1), 0.2% gentamycin, and 10% fetal bovine serum (FBS) and was incubated at 37°C with an atmosphere of 5% CO2. Cell Counting Kit‐8 (CCK‐8), a non‐radioactive colorimetric cell proliferation and cytotoxic assay kit (Sigma‐Aldrich), was employed for the in vitro cytotoxicity test. 24 , 25 Cells were seeded onto 96‐well plates at a concentration of (2 × 104/100 µl) and incubated at 37°C with an atmosphere of 5% CO2 for 24 h. Each sample measuring 25 µl (filtered) was added into each well in duplicate. Positive control (NTZ) and compounds 2 and 4 (500 and 100 μg/ml) were dissolved in DMSO (0.1% v/v). Cells were periodically checked for granularity, shrinkage, and swelling using trinocular microscope with camera (Optika) during the incubation period of 48 h. After incubation, 10 μl of CCK‐8 (5 mg/ml) solution was added to each well followed by incubation at 37°C for 4 h for cytotoxicity. The viable cells were visualized by the presence of purple color formazan dye. As the amount of produced formazan dye is directly proportional to the number of living cells, the measurement of absorbance value will give the number of viable cells. 23 , 24 The absorbance values were measured using a microplate reader at 570 nm wavelength where DMSO was used as blank. The percentage growth inhibition was calculated using the following formula. 26
where, At = Mean absorbance value of test compound, Ab = Mean absorbance value of blank, Ac = Mean absorbance value of control.
Percentage of inhibition is plotted against the drug concentration, and IC50 value is determined.
2.6. Molecular docking study
The current protocol followed ‘rigid ligand–rigid receptor’ docking. The crystallographic structures of the selected target proteins were downloaded from RCSB Protein Data Bank (RCSB‐PDB) 27 in PDB format. The ligands were drawn in ChemBioDraw Ultra 12.0 and then collected in SDF format from the PubChem 28 database. Nitazoxanide's accession no. in Drug Bank database, DB00507, 29 CASTp, 30 and DoGSiteScorer 31 were used to predict the active sites of the target proteins. The structures of the target proteins were converted into PDBQT format after performing necessary modifications in AutoDoc Tools (ADT) (version 1.5.6). 32 The ligands were also converted into PDBQT format with Open Bable tool. 33 Finally, the docking of the ligands with the target proteins was done by using the Autodock Vina (version 1.1.2). 34
2.7. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY, 35 and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20. 36
3. RESULTS AND DISCUSSION
3.1. Synthesis of heteroaryl amide derivatives
In the present work, 2‐amino‐5‐nitrothiazole has been used as the starting material which was conjugated with various hetero arylchloride using well known Schotten Baumann 37 , 38 reaction in the presence of triethylamine (TEA) and dichloromethane (DCM) to produce the hetero aryl amide analogs of NTZ (Table 1). During synthesis of different analogs, 5‐nitro thiazole group was kept constant as drug activation largely depends on the redox potential of the 5‐nitro group, because reduction of 5‐nitro group by nitroreductases, including pyruvate ferredoxin oxidoreductase (PFOR), is responsible for NTZ’s mechanistic activation and, likewise, spectrum of activity. 39 Moreover, heterocyclic rings (e.g. thiophene and furan) were incorporated replacing the benzene ring in the synthetic analogs to understand the role of hetero aryl residues.
TABLE 1.
Synthesis of heteroaryl amide derivatives as NTZ analogues
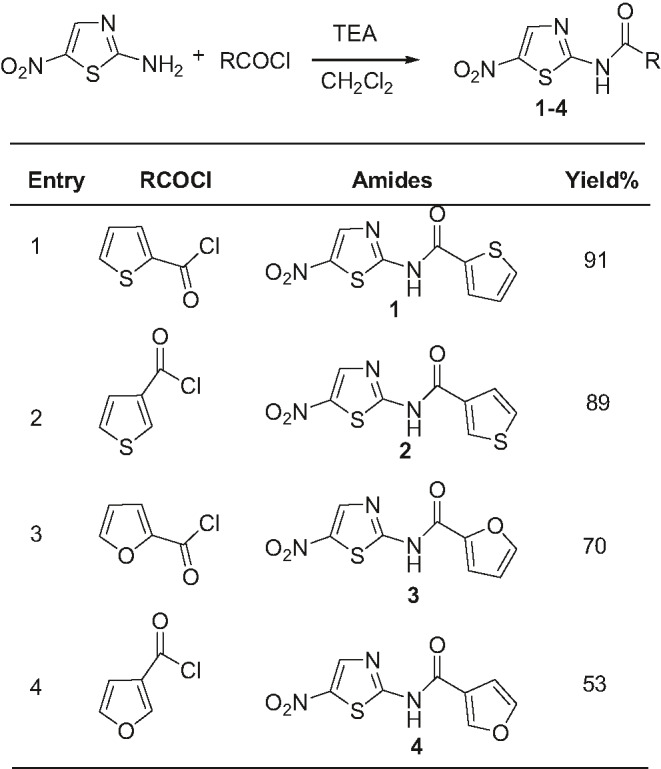
According to the synthetic method described above, four nitazoxanide analogs have been synthesized (Table 1). The analogs designated as compound 1, 2, 3 and 4 were produced having a good yield 91%, 89%, 70%, and 53%, respectively. Structures of the synthesized compounds were elucidated by analyses of their high resolution 1H‐NMR and FT‐IR spectroscopic data and were further confirmed by comparing their spectral data to that of the published values. 17 , 21 The spectral features are in close agreement with the published data.
3.2. Antibacterial activity assay against S. aureus and E. coli
In vitro antibacterial effect of the four NTZ analogs on E. coli and S. aureus was observed on the basis of differential concentration of the test samples. E. coli was found to be more sensitive toward the synthetic compounds as most of the samples exhibited zone of inhibition in the range of 10–22 mm (Table 2). By contrast, S. aureus was rather resistant toward NTZ and the synthetic compounds as most of them did not show notable zone of inhibition (6 mm). Noticeably, only compound 1 and compound 3, both at a concentration of 100 µg/ml, showed moderate zone of inhibition.
TABLE 2.
Antibacterial activity assay against Escherichia coli and Staphylococcus aureus in differential concentration
| Test samples | Zone of inhibition (mm) a | |||||||
|---|---|---|---|---|---|---|---|---|
| Concentration (μg/ml) | ||||||||
| 100 | 50 | 25 | 12.5 | 100 | 50 | 25 | 12.5 | |
| E. coli | S. aureus | |||||||
| 1 | 15 | 14 | 14 | 6 | 10 | 6 | 6 | 6 |
| 2 | 22 | 21 | 20 | 20 | 6 | 6 | 6 | 6 |
| 3 | 14 | 10 | 6 | 6 | 12 | 9 | 6 | 6 |
| 4 | 16 | 13 | 13 | 13 | 6 | 6 | 6 | 6 |
| NTZ | 15 | 12 | 12 | 10 | 6 | 6 | 6 | 6 |
Nalidixic acid (30 μg/disc): 20 mm in E. coli and 19 mm in S. aureu.
Compound 2 exhibited greatest activity among all the synthetic compounds including NTZ in all concentrations. This compound displayed zone of inhibition 20 mm which was twice as large as the parent drug NTZ (10 mm). The observed antibacterial effect against E. Coli was similar or superior to the standard Nalidixic acid. Compound 4 also showed remarkable activity by displaying larger zone of inhibition than the parent drug NTZ in all concentrations. The activity of Compound 1 and 3 was also moderate and comparable to that of NTZ. The present results indicate that hetero aryl NTZ analogs might be more potent antimicrobial agent.
3.3. Trypan blue dye exclusion test for cytotoxicity
Trypan blue dye exclusion test is a widely used technique for determining the number of viable cells present in a cell suspension. It is based on the principle that viable cells possess intact cell membranes that is impermeable to the dyes such as trypan blue. On the other hand, dead cells will easily take up the dye and get blue staining. Thus, the viable cells can be easily distinguished from the dead cells as the viable cell will have a clear cytoplasm, whereas a nonviable cell will have a blue cytoplasm under a microscope. 23 According to the procedure stated above, percentage of the viable cells was determined and the results are summarized in Table 3. It is clear that the % cell viability decreased with increasing concentration of test compounds. Among all of the synthetic compounds, 2 and 4 exhibited the highest growth inhibition (30%‐40% and 45%–55%, respectively at 500 μg/ml concentration) which were similar or superior to that obtained by NTZ (40%–50% inhibition at the same concentration).
TABLE 3.
Trypan blue dye exclusion test to determine the percentage of cell viability
| Compound | Concentration μg/ml | Live cell count | % cell viability |
|---|---|---|---|
| Control | – | 2 × 104 | 100% |
| DMSO | – | 19 × 103 | >95% |
| 1 | 20 | 19 × 103 | >95% |
| 100 | 19 × 103 | >95% | |
| 500 | 12 × 103–14 × 103 | 60%–70% | |
| 2 | 20 | 19 × 103 | >95% |
| 100 | 11 × 103–12 × 103 | 50%–60% | |
| 500 | 6 × 103–8 × 103 | 30%–40% | |
| 3 | 20 | 19 × 103 | >95% |
| 100 | 19 × 103 | >95% | |
| 500 | 19 × 103 | >95%% | |
| 4 | 20 | 19 × 103 | >95% |
| 100 | 19 × 103 | >95% | |
| 500 | 9 × 103–11 × 103 | 45%–55% | |
| NTZ | 20 | 19 × 103 | >95% |
| 100 | 18 × 103 | 92% | |
| 500 | 8 × 103–10 × 103 | 40%–50% |
3.4. MTT assay for cytotoxicity
Based on the result of the trypan blue dye exclusion test, compound 2 and compound 4 were further subjected to MTT assay for determination of their cytotoxic potentials as these two derivatives along with the parent NTZ demonstrated less than 50% cell viability. In the MTT assay, compound 2 demonstrated the highest growth inhibition with an IC50 value 172 µg which is superior to that exhibited by. NTZ (IC50 = 498 µg) (Table 4). This result indicates that compound 2 may act as a potential anticancer agent and proved it to be more potent than nitazoxanide. Compound 4 manifested moderate cytotoxic activity (IC50 = 490 μg) Recently, some studies have proposed that NTZ might exhibit anticancer activity in ovarian 40 and colorectal cancer 40 , 41 , 42 by means of interfering various crucial metabolic and pro‐death signaling mechanisms such as drug detoxification, unfolded protein response (UPR), autophagy, anti‐cytokines activities, and c‐Myc inhibition. 43 These reports justify our in vitro cytotoxic test results. It is the very first report of in vitro cytotoxic activity of the synthesized NTZ analogs.
TABLE 4.
Cytotoxicity of the synthesized compounds
| Name of the sample | Concentration (µg/ml) | Mean absorbance at 570 nm | % cell viability | % growth inhibition | IC50 μg |
|---|---|---|---|---|---|
| Blank (DMSO and Media) | — | 0.0515 | — | — | — |
| Control (Cells with DMSO) | 0 | 0.4445 | 100 | 0 | — |
| Compound 2 | 500 | 0.1860 | 34.23 | 65.77 | 172 |
| 100 | 0.2615 | 53.44 | 46.56 | ||
| Compound 4 | 500 | 0.2460 | 49.50 | 50.50 | 490 |
| 100 | 0.3275 | 70.23 | 29.77 | ||
| NTZ | 500 | 0.2126 | 41.01 | 58.99 | 428 |
| 100 | 0.4087 | 90.89 | 9.11 |
3.5. Molecular docking
For better understanding the impact of NTZ and its heteroaryl analogs on the above biological activities, the molecular docking study was performed. The probable target proteins chosen for the current docking simulation study were those which are somewhat involved in the biochemical pathways regarding their respective biological activities. For understanding anticancer activity, we targeted some cancer signaling proteins, for example thymidylate synthase (TS), dihydrofolate reductase (DHFR), mechanistic target of Rapamycin complex 1 (mTORC1), mitogen activated kinase 8, 9 and 10 (MAPK 8, MAPK9, MAPK10), etc., and for investigating antibacterial activity, some bacterial proteins for example pyruvate ferredoxin oxidoreductase (PFOR), pyruvate dehydrogenase (PDH), β‐ketoacyl‐acyl carrier protein synthase III (FabH), and glucosamine‐6‐phosphate synthase (G6PS) were chosen. All the interactions having RMSD (Root Mean Square Deviation) value 0 indicates a good docking prediction. 44
In case of interaction with the bacterial enzymes for example PFOR, PDH, G6PS, and FabH, the compounds showed a remarkable binding affinity (−4.7 to −8.2 kcal/mol) toward them (Table 5). For instance, in case of PFOR, all the compounds along with NTZ showed an excellent interaction (−7 to −9 kcal/mol) with Thr31, Glu64, Arg114, and Asp996, the active site of PFOR. NTZ demonstrates its bioactivity by blocking PFOR, an essential enzyme for energy metabolism in anaerobes, leading to bacterial and protozoal death. 45 , 46 Among the synthetic compounds, 2 and 4 showed better interaction than the other compounds (Figure 2). This computational finding is in consistence with a previous study by Scior, 15 where a notable binding affinity was reported by the author between their synthetic NTZ analogs and PFOR. Like PFOR, pyruvate dehydrogenase (PDH) also plays the same role in glucose consumption and energy metabolism via oxidation of pyruvate in aerobic organism (e.g. E. coli) and mammals. 17 NTZ had been reported to inhibit 35 to 80% growth of E. coli in a dose‐dependent manner by inhibiting PDH. 46 This fact is well supported by our excellent docking score ranging from −7 to −9.1 kcal/mol, for NTZ and the analogs toward PDH (Table 5, Figure 3A and B). Another enzyme glucosamine‐6‐phosphate synthase (G6PS) is required for building cell wall macromolecules like chitin and mannoproteins in fungi and peptidoglycans in bacteria, via the production of a molecule named uridine diphosphate N‐acetylglucosamine (UDP‐GlcNAc). 47 Recently G6PS has been targeted for in silico study involving substituted thiazoles that showed significant binding affinity toward the enzyme 47 as is evident in current docking experiment where all the compounds including NTZ presented strong interaction (−7.2 to −8.3 kcal/mol) with G6PS in its active site Cys2 and Lys604. NTZ and its analogs showed moderate (−4.7 to −6.3 kcal/mol) affinity with FabH, a key enzyme in the fatty acid biosynthesis pathway (FAB) of prokaryotes which gets inhibited by metronidazole‐thiazole derivative in its active site. 48 Here, Compound 2 (−5.7 kcal/mol) and compound 4 (−6.3 kcal/mol) had greater docking score than NTZ (−5.2 kcal/mol) against FabH.
TABLE 5.
Result of molecular docking of the compounds against 14 target proteins
| Protein | PDB ID | Organism | UniProt ID | Active site | Affinity (kcal/mol) with | ||||
|---|---|---|---|---|---|---|---|---|---|
| NTZ | 1 | 2 | 3 | 4 | |||||
| PDH | 2IEA | E. coli | — | Pocket prediction (whole) | −9.1 | −7.3 | −7.5 | −8.1 | −7.4 |
| PFOR | 2C42 | D. africanus | P94692 | 31T, 64E, 114R, 996N | −9 | −7 | −8.1 | −7.6 | −8.2 |
| G6PS | 2J6H | E. coli | P17169 | 2C, 604K | −8.3 | −7.7 | −7.2 | −7.8 | −7.2 |
| MAPK8 | 3O17 | H. sapiens | P45983 | 151D | −7.9 | −7.2 | −6.5 | −7.4 | −6.8 |
| MAPK9 | 3E7O | H. sapiens | P45984 | 151D | −7 | −5.3 | −7.9 | −5.6 | −8.8 |
| MAPK10 | 4KKE | H. sapiens | P53779 | 189D | −7.9 | −6.3 | −6.4 | −6.2 | −6.7 |
| DHFR | 4M6J | H. sapiens | P00374 | NADPH binding site | −7.7 | −6.4 | −7.2 | −6.4 | −7.5 |
| GSTP1 | 17GS | H. sapiens | P09211 | 8Y, 39W, 45K | −7.3 | −5.3 | −6.1 | −5.8 | −6.2 |
| TS | 4UP1 | H. sapiens | P04818 | 195C | −7.1 | −5.9 | −5.9 | −6 | −6.3 |
| mTORC1 | 5H64 | H. sapiens | P42345 | 2340H | −6.7 | −5.9 | −6 | −6 | −6.1 |
| FabH | 3IL9 | E. coli | P0A6R0 | Pocket prediction (DogSiteScorer) | −5.2 | −4.7 | −5.7 | −5.2 | −6.3 |
FIGURE 2.
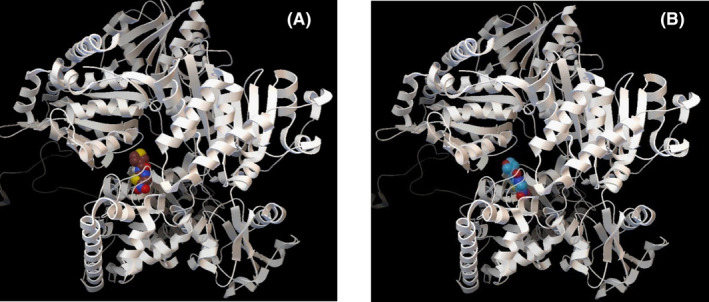
Binding Interaction of Synthesized analogs with PFOR. (A) Compound 2 and PFOR, (B) Compound 4 and PFOR
FIGURE 3.
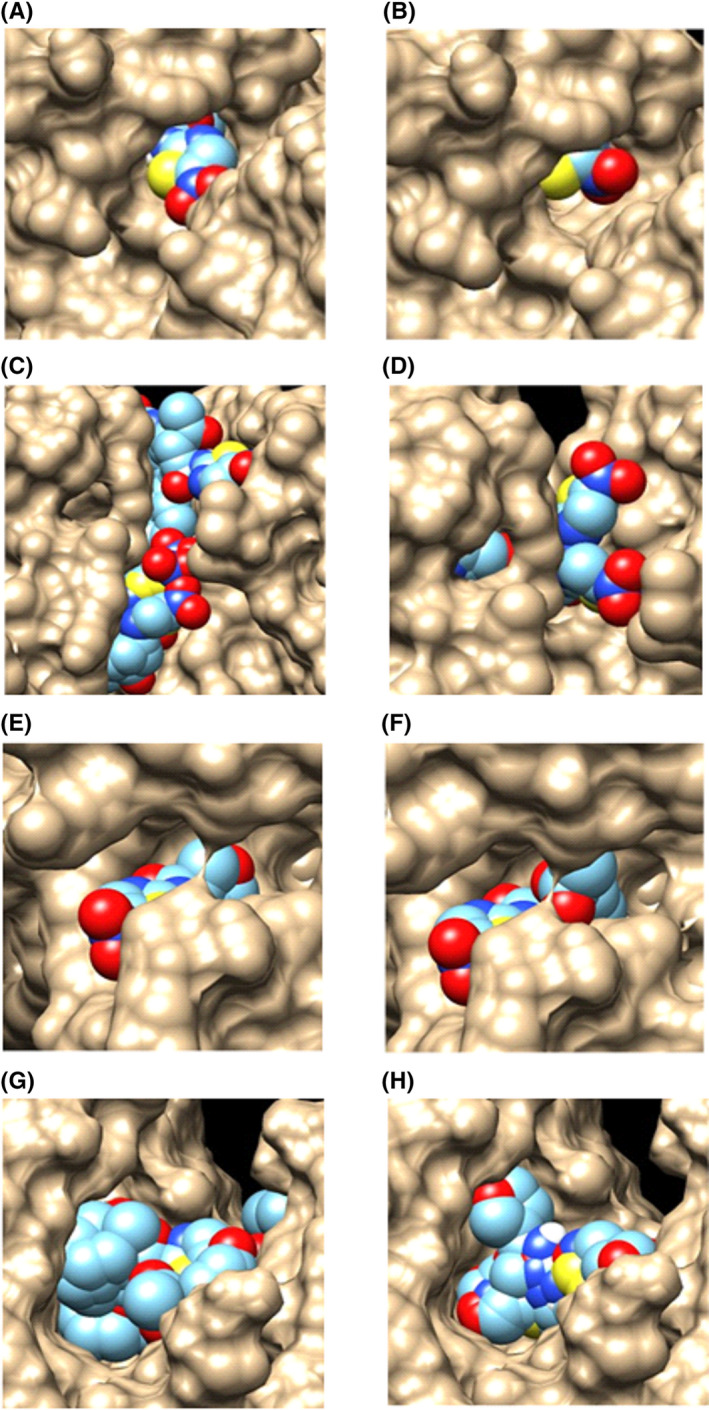
Graphical representation of the interaction of NTZ (multicolor) and the synthesized compounds (multicolor) with the respective proteins (brown) obtained by docking study. (A) Binding mode of NTZ with active site of PDH. (B) Binding mode of Compound 3 with active site of PDH. (C) Binding mode of NTZ with active site of MAPK9. (D) Binding mode of Compound 4 with active site of MAPK9. (E) Binding mode of NTZ with active site of DHFR. (F) Binding mode of Compound 4 with active site of DHFR. (G) Binding mode of NTZ with active site of TS. (H) Binding mode of Compound 4 with active site of TS. Abbreviations: DHFR, Dihydrofolate reductase; MAPK9, Mitogen activated kinase 9; NTZ, Nitazoxanide; PDH, Pyruvate dehydrogenase; TS, Thymidylate synthase
Furthermore, NTZ and all the synthesized compounds were examined for their interaction with various cancer signaling proteins including MAPK9 (Figure 3C and D), DHFR (Figure 3E and F), and TS (Figure 3G and H). All the synthesized compounds were found to have prominent binding affinity (−5.3 to −8.8 kcal/mol) in case of interaction with various cancer signaling proteins for example DHFR, TS, mTORC1, GSTP1, MAPK8, MAPK9, and MAPK10 (Table 5). For instance, MAPK 8, MAPK 9, and MAPK10 were identified as potential cellular targets for NTZ in a recent in silico study. 49 All compounds exhibited a favorable interaction (−5.3 to −8.8 kcal/mol) at their active sites; Asp151 in MAPK 8, 9 and with Asp189 in MAPK10. Among the synthetic analogs, compound 2 and compound 4 (Figure 3) showed stronger affinity than NTZ toward MAPK9.
Another two cancer signaling proteins, glutathione‐S‐transferase of the Pi class (GSTPI) 50 and mechanistic target of rapamycin complex 1 (mTORC1) 40 , 43 have been reported to be a promising mammalian target for NTZ. NTZ inhibits these proteins and causes cancer cell death in colorectal and ovarian cancer. 40 , 41 , 50 These reports support our docking finding where the compounds seemed to have considerable affinity toward GSTPI (−5.3 to −7.3 kcal/mol) in its active site Tyr8, Trp39, Lys45, and mTORC1 (−5.9 to −6.7 kcal/mol) in its active site His2350.
Interestingly, the synthesized analogs and NTZ also displayed a notable interaction (−6.4 to −7.7 kcal/mol) with DHFR in its NADPH binding site and with TS (−5.9 to −7.1 kcal/mol) in its active site Cys195 (Figure 3). These two enzymes are essential for cell proliferation and cell growth, 51 and inhibition of them causes disruption of DNA synthesis and cell death consequently. 52 , 53 Therefore, the mentioned molecular affinity of the compounds toward DHFR and TS makes them a potential novel target for NTZ and its synthetic analogs in anticancer therapy.
From the docking scores provided in Table 5, some important inferences can be made. For instance, NTZ and the synthesized analogs showed excellent binding affinity with all the proteins except FaBH. Additionally, NTZ and the synthesized analogs revealed potential binding affinity with TS and DHFR. To the best of our knowledge, till date NTZ and analogs have never been subjected to molecular docking study against TS and DHFR, which postulates them as novel targets for NTZ. Moreover, the synthesized compounds have never encountered any molecular docking study against the mentioned proteins previously, and the docking score reveals their potential of having new pharmacological activities.
In most of the cases, compound 2 and 4 accounted for stronger binding affinity with the target proteins than do the other two compounds (Table 5). In fact, compound 2 and compound 4 showed better affinity than the parent drug NTZ against MAPK9 and FabH. These observations further reinforce both the in vitro antibacterial and cytotoxicity test results, where compound 2 and 4 exhibited greater inhibitory activity than the other compounds. Also compound 2 displayed larger zone of inhibition and proved to be more cytotoxic in cancer cell line, compared with NTZ. Thus, an important correlation can be made between the docking prediction and biological activity which is well supported by the previous studies that elucidated the role of NTZ as antibacterial and anticancer agent by blocking its essential metabolic process and interfering in multiple cancer signaling pathways, respectively.
4. CONCLUSION
We have synthesized four heteroaryl analogs of NTZ by condensing 2‐amino‐5‐nitrothiazole with some acid chlorides. Among the four synthesized analogs of NTZ, compound 2 and compound 4 displayed better antibacterial activity than NTZ against E. coli. Additionally, cytotoxicity assay in HeLa cell line demonstrated greater cell growth inhibition for compound 2, which was proved to be more potent than NTZ. This is the very first report of cytotoxic as well as antibacterial activities of these synthesized compounds. The results of in vitro bioassays further corroborated with the molecular docking study that helped exploring probable mechanistic insights underlying their bioactivity. Moreover, the computational investigation identified DHFR and TS as potential novel targets for NTZ and its synthesized analogs. Although further detail experiments are warranted, these findings are unique and hence should draw attentions of structural, as well as chemical biologists, for future drug development considering nitazoxanide along with its analogs for repurposing it as a potential anticancer agent besides its conventional antibacterial and antiprotozoal uses. In addition, the analogs might be effective as antiviral agent against SARS‐CoV‐2 and, therefore, synthesis of new series of molecules followed by antibacterial, antiviral, and cytotoxicity assays will be focused in the future work. In addition, our future study will also focus on the in vivo and biochemical investigation of the synthesized analogs.
ETHICS APPROVAL STATEMENT
No ethical permission required for this in vitro and in silico studies.
DISCLOSURE
None.
AUTHOR CONTRIBUTIONS
Rahman, S. M. A. and Chowdhury, A. K. A. conceptualized, designed, and supervised the research work. Ahmed, T. performed the synthesis and biological work. Asaduzzaman, M and Islam A. B. M. K performed molecular docking studies. Asaduzzaman M and Ahmed T did the data analysis. Rahman, S. M. A., Ahmed, T. and Asaduzzaman, M. wrote the manuscript. All authors reviewed the manuscript and approved it.
ACKNOWLEDGEMENTS
Authors wish to thank Bangladesh Council of Scientific and Industrial Research (BCSIR) for providing facilities for 1H NMR and Centre of Advanced Research in Sciences (CARS), University of Dhaka, for their support in antibacterial and cytotoxic assays.
Ahmed T, Rahman SMA, Asaduzzaman M, Islam ABMMK, Chowdhury AKA. Synthesis, in vitro bioassays, and computational study of heteroaryl nitazoxanide analogs. Pharmacol Res Perspect. 2021;9:e00800. 10.1002/prp2.800
Contributor Information
S. M. Abdur Rahman, Email: smarahman@du.ac.bd.
A. K. Azad Chowdhury, Email: akchowdhury2003@yahoo.com.
DATA AVAILABILITY STATEMENT
The data that support these results are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Shakya A, Bhat HR, Ghosh SK. Update on nitazoxanide: a multifunctional chemotherapeutic agent. Curr Drug Discov Technol. 2018;15(3):201‐213. [DOI] [PubMed] [Google Scholar]
- 2. Hemphill A, Muller N, Muller J. Structure‐function relationship of thiazolides, a novel class of anti‐parasitic drugs, investigated in intracellular and extracellular protozoan parasites and larval‐stage cestodes. Anti‐Infect Agents Med Chem. 2007;6(4):273‐282. [Google Scholar]
- 3. Ortiz JJ, Ayoub A, Gargala G, Chegne NL, Favennec L. Randomized clinical study of nitazoxanide compared to metronidazole in the treatment of symptomatic giardiasis in children from Northern Peru. Aliment Pharmacol Ther. 2001;15(9):1409‐1415. [DOI] [PubMed] [Google Scholar]
- 4. Moron‐Soto M, Gutierrez L, Sumano H, Tapia G, Alcala‐canto Y. Efficacy of nitazoxanide to treat natural Giardia infections in dog. Parasit Vectors. 2017;10:52. 10.1186/s13071-017-1998-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossignol JFA, Ayoub A, Ayers MS. Treatment of diarrhea caused by Cryptosporidium parvum: a prospective randomized, double‐blind, placebo‐controlled study of nitazoxanide. J Infect Dis. 2001;184(1):103‐106. [DOI] [PubMed] [Google Scholar]
- 6. Mégraud F, Occhialini A, Rossignol JF. Nitazoxanide, a potential drug for eradication of Helicobacter pylori with no cross‐resistance to metronidazole. Antimicrob Agents Chemother. 1998;42(11):2836‐2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McVay CS, Rolfe RD. In vitro and in vivo activities of nitazoxanide against Clostridium difficile . Antimicrob Agents Chemother. 2000;44(9):2254‐2258. 10.1128/aac.44.9.2254-2258.2000. PMID: 10952564; PMCID: PMC90054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bailey MA, Na H, Duthie MS, Gillis TP, Lahiri R. Parish T nitazoxanide is active against mycobacterium leprae. PLoS One. 2017;12(8):e0184107. 10.1371/journal.pone.0184107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rossignol JF, Abu‐Zekry M, Hussein A, Santoro MG. Effect of nitazoxanide for treatment of severe rotavirus diarrhoea: randomised double‐blind placebo‐controlled trial. Lancet. 2006;368(9530):124‐129. [DOI] [PubMed] [Google Scholar]
- 10. Rossignol JF. Nitazoxanide: a first‐in‐class broad‐sectrum antiviral agent. Antiviral Res. 2014;110:94‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30(3):269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelleni MT. Nitazoxanide/azithromycin combination for COVID‐19: a suggested new protocol for early management. Pharmacol Res. 2020;157:104874. 10.1016/j.phrs.2020.104874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hossain MJ, Rahman SMA. Repurposing therapeutic agents against SARS‐CoV‐2 infection: most promising and neoteric progress. Expert Rev Anti‐Infect Ther. 2020;23:1‐19. 10.1080/14787210.2021.1864327 [DOI] [PubMed] [Google Scholar]
- 14. Navarrete‐Vazquez G, Chávez‐Silva F, Argotte‐Ramos R, et al. Synthesis of benzologues of nitazoxanide and tizoxanide: a comparative study of their in vitro broad‐spectrum antiprotozoal activity. Bioorg Med Chem Lett. 2011;21(10):3168‐3171. [DOI] [PubMed] [Google Scholar]
- 15. Scior T, Lozano‐Aponte J, Ajmani S, et al. Antiprotozoal nitazoxanide derivatives: synthesis, bioassays and QSAR study combined with docking for mechanistic insight. Curr Comput Aided Drug Des. 2015;11(1):21‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soria‐Arteche O, Hernández‐Campos A, Yépez‐Mulia L, et al. Synthesis and antiprotozoal activity of nitazoxanide–N‐methylbenzimidazole hybrids. Bioorg Med Chem Lett. 2013;23(24):6838‐6841. [DOI] [PubMed] [Google Scholar]
- 17. Ballard TE, Wang X, Olekhnovich I, et al. Biological activity of modified and exchanged 2‐amino‐5‐nitrothiazole amide analogues of nitazoxanide. Bioorg Med Chem Lett. 2010;20(12):3537‐3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gau JS, Lin WP, Kuo LC, Hu MK. Nitazoxanide analogues as antimicrobial agents against nosocomial pathogens. Med Chem. 2016;12(6):544‐552. 10.2174/1573406412666160129105719. PMID: 26825066. [DOI] [PubMed] [Google Scholar]
- 19. Jeankumar VU, Chandran M, Samala G, et al. Development of 5‐nitrothiazole derivatives: identification of leads against both replicative and latent Mycobacterium tuberculosis. Bioorg Med Chem Lett. 2012;22(24):7414‐7417. [DOI] [PubMed] [Google Scholar]
- 20. Stachulski AV, Santoro MG, Piacentini S, et al. Second‐generation nitazoxanide derivatives: thiazolides are effective inhibitors of the influenza A virus. Future Med Chem. 2018;10(8):851‐862. [DOI] [PubMed] [Google Scholar]
- 21. Ballard TE, Wang X, Olekhnovich I, et al. Synthesis and antimicrobial evaluation of nitazoxanide‐based analogues: identification of selective and broad spectrum activity. ChemMedChem. 2011;6(2):362‐377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Path. 1966;45(4_ts):493‐496. [PubMed] [Google Scholar]
- 23. Strober W. Trypan blue dye exclusion test of cell viability. Curr Protoc Immunol. 2001. May, Appendix 3(Appendix 3B). 10.1002/0471142735.ima03bs21. PMID: 18432654. [DOI] [PubMed] [Google Scholar]
- 24. Ishiyama M, Miyazono Y, Sasamoto K, Ohkura Y, Ueno K. A highly water‐soluble disulfonated tetrazolium salt as a chromogenic indicator for NADH as well as cell viability. Talanta. 1997;44(7):1299‐1305. [DOI] [PubMed] [Google Scholar]
- 25. Tominaga H, Ishiyama M, Ohseto F, et al. A water‐soluble tetrazolium salt useful for colorimetric cell viability assay. Anal Commun. 1999;36(2):47‐50. [Google Scholar]
- 26. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1‐2):55‐63. [DOI] [PubMed] [Google Scholar]
- 27. Berman HM, Westbrook J, Feng Z. The protein data bank, 1999. In: Rossmann MG, Arnold E, eds. International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules. Netherlands: Springer; 2006:675‐684. [Google Scholar]
- 28. Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res. 2015;44(D1):D1202‐D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knox C, Law V, Jewison T, et al. Drug Bank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2010;39(suppl_1):D1035‐D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(suppl_2):W116‐W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Volkamer A, Kuhn D, Rippmann F, Rarey M. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28(15):2074‐2075. [DOI] [PubMed] [Google Scholar]
- 32. Morris GM, Goodsell DS, Halliday RS. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639‐1662. [Google Scholar]
- 33. O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminformatics. 2011;3(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harding SD, Sharman J, Faccenda E, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2019: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091‐D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alexander SPH, Kelly E, Mathie A, et al. The Concise guide to pharmacology 2019/20: introduction and other protein targets. Br J Pharmacol. 2019;176(S1):S1‐S20. 10.1111/bph.14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schotten C. Ueber die oxydation des piperidins. Ber Dtsch Chem Ges. 1884;17(2):2544‐2547. 10.1002/cber.188401702178 [DOI] [Google Scholar]
- 38. Baumann E. Ueber eine einfache Methode der Darstellung von Benzoësäureäthern. Ber Dtsch Chem Ges. 1986;19(2):3218‐3222. 10.1002/cber.188601902348 [DOI] [Google Scholar]
- 39. Sison G, Goodwin A, Raudonikiene A, et al. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori . Antimicrob Agents Chemother. 2002;46(7):2116‐2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Di Santo N, Ehrisman J. Research perspective: potential role of nitazoxanide in ovarian cancer treatment. Old drug, new purpose? Cancers. 2013;5(3):1163‐1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Senkowski W, Zhang X, Olofsson MH, et al. Three‐dimensional cell culture‐based screening identifies the anthelmintic drug nitazoxanide as a candidate for treatment of colorectal cancer. Mol Cancer Ther. 2015;14(6):1504‐1516. [DOI] [PubMed] [Google Scholar]
- 42. Yu J, Yang K, Zheng J, et al. Synergistic tumor inhibition of colon cancer cells by nitazoxanide and obeticholic acid, a farnesoid X receptor ligand. Cancer Gene Ther. 2020. 10.1038/s41417-020-00239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Di Santo N, Ehrisman J. A functional perspective of nitazoxanide as a potential anticancer drug. Mutat Res. 2014;768:16‐21. [DOI] [PubMed] [Google Scholar]
- 44. Cole JC, Murray CW, Nissink JWM, Taylor RD, Taylor R. Comparing protein–ligand docking programs is difficult. Proteins: Struct Funct Bioinf. 2005;60(3):325‐332. [DOI] [PubMed] [Google Scholar]
- 45. Cremades N, Bueno M, Toja M, Sancho J. Towards a new therapeutic target: Helicobacter pylori flavodoxin. Biophys Chem. 2005;115(2–3):267‐276. [DOI] [PubMed] [Google Scholar]
- 46. Hoffman PS, Sisson G, Croxen MA, et al. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni . Antimicrob Agents Chemother. 2007;51(3):868‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arora P, Narang R, Bhatia S, Nayak SK, Singh SK, Narasimhan B. Synthesis, molecular docking and QSAR studies of 2, 4‐disubstituted thiazoles as antimicrobial agents. J Appl Pharm Sci. 2015;5(2):28‐42. [Google Scholar]
- 48. Qin YJ, Wang PF, Makawana JA, et al. Design, synthesis and biological evaluation of metronidazole–thiazole derivatives as antibacterial inhibitors. Bioorg Med Chem Lett. 2014;24(22):5279‐5283. [PubMed] [Google Scholar]
- 49. Zidan AM. Computational prediction of the protein kinase that mediates the anti‐HCV effect of nitazoxanide. Doctoral dissertation, University of Sadat City. 2014.
- 50. Sidler D, Brockmann A, Mueller J, et al. Thiazolide‐induced apoptosis in colorectal cancer cells is mediated via the Jun kinase–Bim axis and reveals glutathione‐S‐transferase P1 as Achilles’ heel. Oncogene. 2012;31(37):4095‐4106. [DOI] [PubMed] [Google Scholar]
- 51. Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119‐140. [DOI] [PubMed] [Google Scholar]
- 52. Cowman AF, Lew AM. Antifolate drug selection results in duplication and rearrangement of chromosome 7 in Plasmodium chabaudi. Mol Cell Biol. 1989;9(11):5182‐5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nicolini A, Conte M, Rossi G, et al. Additional 5‐FU‐LV significantly increases survival in gastrointestinal cancer. Front Biosci. 2011;2011(3):1475‐1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support these results are available from the corresponding author upon reasonable request.


