ABSTRACT
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a virus that causes the infectious disease coronavirus disease-2019. Currently, there is no effective drug for the prevention and treatment of this virus. This study aimed to identify secondary metabolites that potentially inhibit the key proteins of SARS-CoV-2. This was an in silico molecular docking study of several secondary metabolites of Indonesian herbal plant compounds and other metabolites with antiviral testing history. Virtual screening using AutoDock Vina of 216 Lipinski rule-compliant plant metabolites was performed on 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), and spike glycoprotein. Ligand preparation was performed using JChem and Schrödinger's software, and virtual protein elucidation was performed using AutoDockTools version 1.5.6. Virtual screening identified several RdRp, spike, and 3CLpro inhibitors. Justicidin D had binding affinities of −8.7, −8.1, and −7.6 kcal mol−1 on RdRp, 3CLpro, and spike, respectively. 10-methoxycamptothecin had binding affinities of −8.5 and −8.2 kcal mol−1 on RdRp and spike, respectively. Inoxanthone had binding affinities of −8.3 and −8.1 kcal mol−1 on RdRp and spike, respectively, while binding affinities of caribine were −9.0 and −7.5 mol−1 on 3CLpro and spike, respectively. Secondary metabolites of compounds from several plants were identified as potential agents for SARS-CoV-2 therapy.
Keywords: AutoDock Vina, coronavirus disease-2019, Indonesian herbal medicine, molecular docking, infectious disease
INTRODUCTION
Coronavirus Disease-2019 (COVID-19) is a disease similar to pneumonia that caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Since the first case on December 12, 2019, the number of cases and deaths due to COVID-19 has continued to increase. As of December 27, 2020, there have been 79.2 million cases, and over 1.7 million deaths have been reported.[1]
SARS-CoV-2 is a positive-strand RNA virus. Patients infected with this virus experience symptoms such as fever, dry cough, and shortness of breath. However, many patients are carriers and show no symptoms. This is one of the reasons for the widespread transmission of this virus.[2] The treatment of COVID-19 patients currently uses drugs that have previously received approval from drug regulatory agencies, such as hydroxychloroquine, chloroquine, and remdesivir.[3]
The SARS-CoV-2 genome encodes several proteins that are important in the virulence process. These proteins include SARS-CoV-2 3C-like protease (3CLpro) and RNA-dependent RNA polymerase (RdRp), which are useful for viral replication and transcription, as well as spike glycoproteins that mediate virus-host binding and viral fusion.[2] Inhibition of these proteins in SARS-CoV-2 could lead to a potential therapy for COVID-19.
This study aimed to identify secondary metabolites that potentially inhibit the key proteins of SARS-CoV-2 by virtual screening of plant metabolites against the 3CLpro, RdRp proteins, and spike glycoprotein. These metabolites were derived from several species of Indonesian traditional medicine as well as other metabolites that had an antiviral testing history.
MATERIALS AND METHODS
Ligand and protein preparation
The ligands used were metabolites of Cymbopogon citratus, Kaempferia galangal, Curcuma longa, Curcuma xanthorrhiza, and Zingiber officinale. Plant metabolites that had a history of antiviral testing were also used as ligands. All compounds were obtained from http://www.knapsackfamily.com. Screening using SwissADME (http://www.swissadme.ch/index.php) according to the five Lipinski rules was performed on 240 metabolites. The ligand structure was formed using JChem software and optimized using the LigPrep module from Schrödinger's software (Schrödinger LLC, New York, NY, USA). Atomic protonation was adjusted to pH 7.0 with Epik software (Schrödinger LLC), and geometry optimization used OPLS_2005 Force Field software (Schrödinger LLC).
Protein structures targeted for docking were obtained from the Protein Data Bank (https://www.rcsb.org/). The proteins used were 3CLpro (PDB ID: 6M2N chain A), RdRp (PDB ID: 6M71 chain A), and spike glycoprotein (PDB ID: 6VXX chain B). Protein preparation was performed using AutoDockTools version 1.5.6. Protein validation for 3CLpro was performed by calculating the root mean square deviation (RMSD) of the protein with its co-crystalline ligand using PyMOL software version 2.3.4 (Schrödinger LLC).
Virtual screening and molecular docking
Virtual screening was performed using an Intel® Celeron® 2955U at 1.40 GHZ processor, 2.00 GB of RAM 64-bit operating system. Targeted docking was performed on the 3CLpro protein. The grid box was 40 Å × 40 Å × 40 Å centered at −32,981, −65,436, and 41.404. The validity of the docking protocol was assessed based on the RMSD. For the RdRp protein and spike glycoprotein, blind docking and targeted docking were performed. Blind docking was useful for finding receptor pockets that bind positive control compounds (remdesivir, hydroxychloroquine, and chloroquine). The chains targeted for blind docking were chains that had binding pockets with high (top five) drug score, as determined by DoGSiteScorer (https://proteinsplus.zbh.uni-hamburg.de/#dogsite), which were chain A of RdRp and chain B of spike. Blind docking was performed by making the grid box sufficient to cover all sides of the protein, and the docking was performed three times in a row. Remdesivir had the lowest binding affinity at both the receptors and was consistent in one binding pocket, so the remdesivir position after docking was used as a grid box for RdRp and spike glycoprotein. The grid box for RdRp was 40 Å × 40 Å × 40 Å centered at 132,754, 112,571, and 103,907, while spike glycoproteins was 40 Å × 40 Å × 40 Å centered at 175,288, 177,632, and 235,662. Molecular docking was performed using AutoDock Vina, and the ligand–receptor interaction was visualized using Discovery Studio Visualizer v17.2.016349 software (Dassault Systèmes, San Diego, California, USA).
RESULTS
Two hundred and sixteen compounds that complied with Lipinski's rules were subjected to virtual screening against 3CLpro, RdRp, and the spike glycoprotein [Figure 1]. The compounds that potentially become inhibitors of the proteins were compounds that had same or lower than each benchmark's binding affinities [Table 1]. Virtual screening showed that four compounds had the potential to become RdRp inhibitors [Figure 2 and Table 2]. On the other hand, there were 79 compounds which had the potential to become spike inhibitors. Of these 79 compounds, 11 were selected with the lowest binding affinities [Figure 3 and Table 3]. While for the 3CLpro receptor, they were 11 compounds that potentially inhibit the protein [Figure 4 and Table 4].
Figure 1.
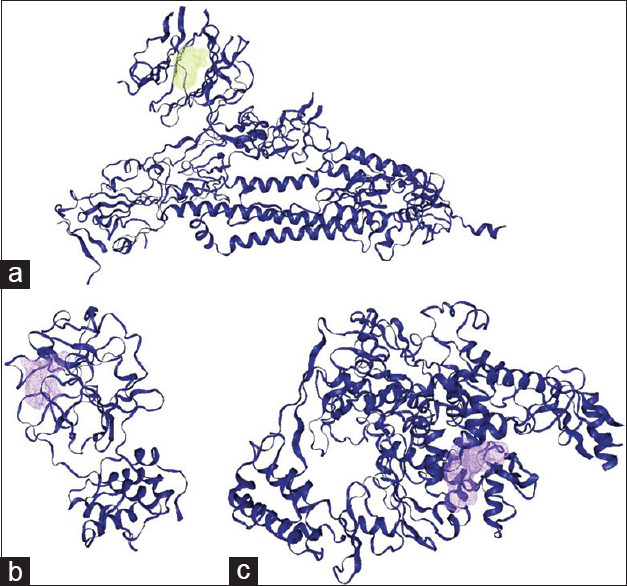
Cartoon representation of spike glycoprotein (a), 3C-like protease (b), and RNA-dependent RNA polymerase (c) of Severe acute respiratory syndrome coronavirus-2, with their binding sites and selected grid box as a targeted docking. The binding site were determined with DoGSiteScorer
Table 1.
Binding affinity (kcal/mol) of anti-severe acute respiratory syndrome-coronavirus-2 comparator on the targeted protein RNA-dependent RNA polymerase, spike glycoprotein, and 3C-like protease
| Compounds | Receptor | ||
|---|---|---|---|
| RdRp | Spike | 3CLpro | |
| 5,6,7-trihydroxy- 2-phenyl-4H-chromen - 4-one |
ND | ND | −7.8 |
| Remdesivir | −8.2 | −6.6 | −6.7 |
| Hydroxychloroquine | −6.7 | −5.7 | −6.5 |
| Chloroquine | −5.8 | −5.9 | −6.0 |
ND: Not determined, 3CLpro: 3C-like protease, RdRp: RNA-dependent RNA polymerase
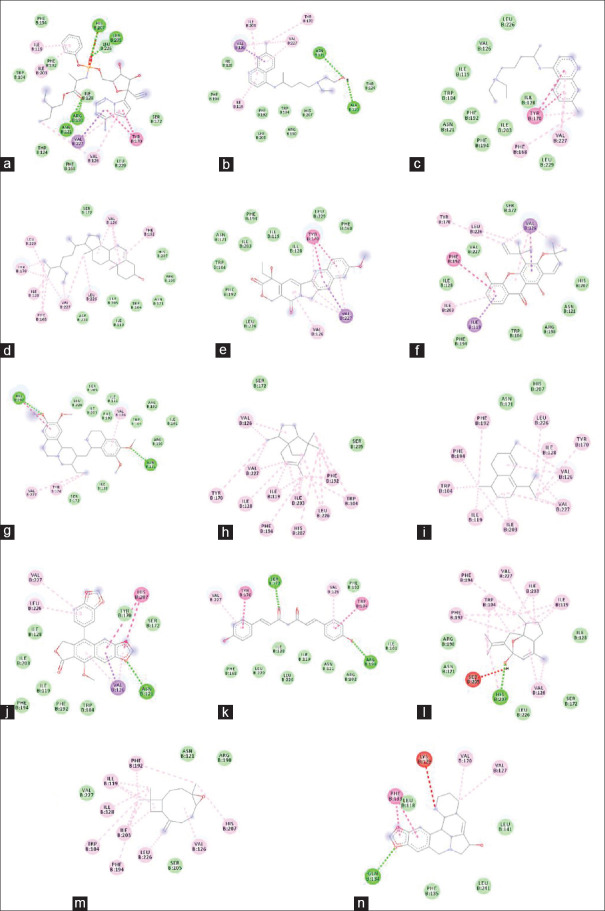
Figure 2.
Molecular interaction of selected ligands with RNA-dependent RNA polymerase of severe acute respiratory syndrome coronavirus-2. Remdesivir (a), hydroxychloroquine (b), chloroquine (c), justicidin D (d), 10-methoxycamptothecin (e), inoxanthone (f), 3-O-Caffeoylquinic acid (g)
Table 2.
Binding affinity of anti-severe acute respiratory syndrome-coronavirus-2 RNA-dependent RNA polymerase candidates
| C_ID | Compounds | Plant | Binding affinity (kcal/mol) |
|---|---|---|---|
| C00000720 | Justicidin D | J. procumbens[5] | −8.7 |
| C00029372 | 10-Methoxycamptothecin | C. acuminata[6] | −8.5 |
| C00035841 | Inoxanthone | C. inophyllum[7] | −8.3 |
| C00002724 | 3-O-Caffeoylquinic acid | Schefflera heptaphylla. L.[4] | −8.2 |
J. procumbens: Justicia procumbens, C. acuminata: Camptotheca acuminata, C. inophyllum: Calophyllum inophyllum
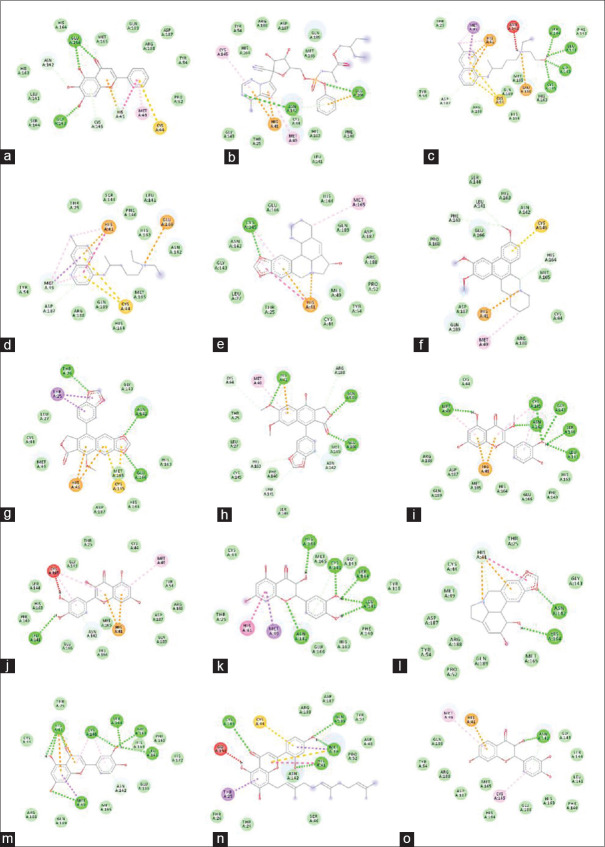
Figure 3.
Molecular interaction of selected ligands with spike glycoprotein of severe acute respiratory syndrome coronavirus-2. Remdesivir (a), hydroxychloroquine (b), chloroquine (c), (−)-beta-Sitosterol (d), 10-methoxycamptothecin (e), inoxanthone (f), emetine (g), alpha-cedrene (h), zonarene (i), justicidin D (j), bis-demethoxycurcumin (k), curcumenol (l), beta-caryophyllene epoxide (m), caribine (n)
Table 3.
Binding affinity of anti-Severe acute respiratory syndrome-coronavirus-2 spike glycoprotein candidates
| C_ID | Compounds | Plant | Binding affinity (kcal/mol) |
|---|---|---|---|
| C00003672 | (−)-beta-sitosterol | Z. officinale, T. capitatus[8,9] | −8.4 |
| C00029372 | 10-Methoxycamptothecin | C. acuminata[6] | −8.2 |
| C00035841 | Inoxanthone | C. inophyllum[7] | −8.1 |
| C00001849 | Emetine | C. ipecacuanha[10] | −7.7 |
| C00003111 | Alpha-cedrene | Z. officinale[11] | −7.6 |
| C00032554 | Zonarene | Z. officinale[12] | −7.6 |
| C00000720 | Justicidin D | J. procumbens[4] | −7.6 |
| C00032769 | Bis-demethoxycurcumin | C. longa[13] | −7.5 |
| C00020351 | Curcumenol | C. longa[14] | −7.5 |
| C00012483 | Beta-caryophyllene epoxide | C. zanthorrhiza[15] | −7.5 |
| C00001566 | Caribine | H. arenicola[16] | −7.5 |
Z. officinale: Zingiber officinale, T. capitatus: Thymus capitatus, C. acuminata: Camptotheca acuminata, C. inophyllum: Calophyllum inophyllum, J. procumbens: Justicia procumbens, C. longa: Curcuma longa, C. zanthorrhiza: Curcuma zanthorrhiza, H. arenicola: Hymenocallis arenicola, C. ipecacuanha: Carapichea ipecacuanha
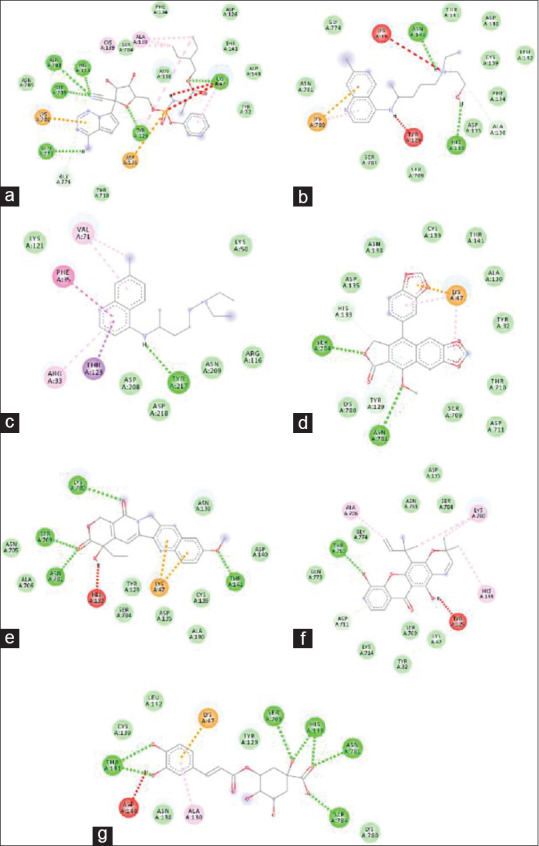
Figure 4.
Molecular interaction of selected ligands with 3C-like protease of severe acute respiratory syndrome coronavirus-2. 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one (a), remdesivir (b), hydroxychloroquine (c), chloroquine (d), caribine (e), cryptopleurine (f), justicidin D (g), diphyllin (h), quercetin 3-O-methyl ether (i), quercetin (j), dihydroquercetin (k), lycorine (l), luteolin (m), moralbanone (n), (2R,3R)-fustin (o)
Table 4.
Binding affinity of anti-severe acute respiratory syndrome-coronavirus-2 3C-like protease candidates
| C_ID | Compounds | Plant | Binding affinity (kcal mol−1) |
|---|---|---|---|
| C00001566 | Caribine | H. arenicola[16] | −9.0 |
| C00002331 | Cryptopleurine | T. indica[17] | −8.2 |
| C00000720 | Justicidin D | J. procumbens[7] | −8.1 |
| C00002600 | Diphyllin | J. gendarussa[18] | −8.0 |
| C00004632 | Quercetin 3-O-methyl ether | Widespread in medicinal plants | −7.9 |
| C00004631 | Quercetin | Widespread in medicinal plants | −7.9 |
| C00000677 | Dihydroquercetin | Widespread in medicinal plants | −7.9 |
| C00001576 | Lycorine | L. radiata[19] | −7.9 |
| C00000674 | Luteolin | Widespread in medicinal plants | −7.9 |
| C00013411 | Moralbanone | Morus alba L.[20] | −7.8 |
| C00000963 | (2R,3R)-fustin | R. verniciflua[21] | −7.8 |
H. arenicola: Hymenocallis arenicola, T. indica: Tylophora indica, Justicia procumbens: J. procumbens, J. gendarussa: Justicia gendarussa, L. radiata: Lycoris radiata, R. verniciflua: Rhus verniciflua
DISCUSSION
RNA-dependent RNA polymerase
As with other positive-strand RNA viruses, genome replication and transcription of SARS-CoV-2 are dependent on RdRp.[2] In this study, the potential SARS-CoV-2 RdRp inhibitors were 3-O-caffeoylquinic acid, justicidin D, 10-methoxycamptothecin, and inoxanthone. 3-O-caffeoylquinic acid is a caffeoylquinic acid derivative with a history of antiviral activity testing against respiratory syncytial virus; however, its antiviral activity is not as potent as the other two derivatives of caffeoylquinic acid.[4] Justicidin D is a metabolite isolated from Justicia procumbens var. leucantha and is known to have antiviral activity in vesicular stomatitis virus.[5] However, there have been no recent reports of these compounds. Moreover, methoxycamptothecin is a camptothecin derivative found in Camptotheca acuminata and has been reported to have antiviral activity.[6] In addition, inoxanthone is a xanthone derivative found in Calophyllum inophyllum and has known cytotoxicity. However, reports of antiviral activity of inoxanthone compound have not been found.[7,8]
Spike glycoprotein
Spike (S) glycoprotein is the protein that mediated the entry of SARS-CoV-2 into the host.[3] Based on the present study, some of the compounds that potentially inhibit the spike protein of SARS-CoV-2 were emetine and carbine. Emetine is an alkaloid found in ipecacuanha (ipecac) root and recently was reported to show activity against SARS-CoV-2 in vitro.[10] Carbine is a metabolite isolated from Hymenocallis arenicola, but no recent reports on this compound have been found.[16]
Three metabolites of Z. officinale are also potential spike protein inhibitors: (−)-beta-sitosterol, alpha-cedrene, and zonarene.[11,12] (−)-beta-sitosterol is also found in Thymus capitatus and known to have activity against herpes simplex virus type 2 (HSV-2).[9] Alpha-cedrene is a compound that improves and prevents liver steatosis in vivo.[22] On the other hand, zonarene is also found in essential oils and is known to have antifungal, mosquito larvicidal, and biting deterrent activities.[23] Based on the present study, Z. officinale may be useful as an herbal treatment for SARS-CoV-2. Nonetheless, further research for this is needed
Furthermore, two metabolites of C. longa, bis-demethoxycurcumin and curcumenol, are also potential inhibitors of spike glycoproteins. These compounds are known to have anticancer activity. However, activity against SARS-CoV-2 has been more widely reported for its analog, curcumin.[13,14] Moreover, one of the metabolites of C. xanthorrhiza, beta-caryophyllene epoxide, is also a potential inhibitor of the spike glycoprotein. This compound was previously reported to have activity against HSV type 1 in vitro.[15,24]
10-methoxycamptothecin, inoxanthone, and justicidin D were in the 11 compounds that had the lowest binding affinities for the spike. These compounds also had good binding affinity to the RdRp, as has been previously stated. Therefore, broad antiviral activity of these three compounds was possible in SARS-CoV-2.
3C-like protease
3CLpro is a protease that cuts polyproteins. Based on the present study, quercetin, quercetin 3-O-methyl ether and dihydroquercetin, has the potential to be 3CLpro SARS-CoV-2 inhibitors. These compounds are known to have various activities, including antiviral against SARS-CoV.[25] Lycorine and luteolin also have the potential to inhibit this protein. Lycorine is an alkaloid derived from Lycoris radiata, whereas luteolin is a metabolite that is widespread in medicinal plants.[19] Lycorine has antiviral activity against SARS-CoV-2 in vitro, with an EC50 of 0.31 μM.[19,26] On the other hand, luteolin has antiviral activity against SARS-CoV.[27]
In this study, cryptopleurine, diphyllin, fustin, and moralbanone potentially inhibit 3CLpro. Cryptopleurine is an alkaloid isolated from Tylophora indica and has been shown to have antiviral activity.[17] Diphyllin is a metabolite of Justicia gendarussa, a ATPase blocker that has activity against viruses.[18,28] Fustin is a metabolite of Rhus verniciflua and was reported to have antiviral activity in vitro.[21,29] Furthermore, moralbanone, a compound isolated from Morus alba L, also has the potential to inhibit 3CLpro.[20] However, no recent reports of moralbanone have been found.
Furthermore, virtual screening suggested that caribine and justicidin D had the potential to inhibit the 3CLpro receptor. As previously explained, this compound also may inhibit both the RdRp and spike receptors, so these two compounds may have broad-spectrum antiviral activity by targeting those three receptors.
CONCLUSION
Virtual screenings targeting the RdRp, spike glycoprotein, and 3CLpro receptors showed that several plant metabolites had the same or lower binding affinities of compounds currently used as SARS-CoV-2 therapeutic agents. Several compounds such as emetine, lycorine, luteolin and quercetin are widely known to have antiviral activity. On the other hand, other compounds such as caribine and justicidin D that are also predicted to have broad antiviral SARS-CoV-2 activity have not much pharmacological activities reported. These metabolites could potentially be used in the management of COVID-19. However, further studies are needed.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We thank Schrödinger Inc. and ChemAxon Ltd. for granting an academic licence.
REFERENCES
- 1.World Health Organization. Weekly Epidemiological Update – 29 December 2020. World Health Organization. 2020. [Last accessed on 2021 Jan 17]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update--- 29-december-2020 .
- 2.Goyal B, Goyal D. Targeting the dimerization of the main protease of coronaviruses: A potential broad-spectrum therapeutic strategy. ACS Comb Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 3.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, et al. Remdesivir: A review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6:672–83. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, But PP, Ooi VE. Antiviral activity and mode of action of caffeoylquinic acids from Schefflera heptaphylla (L.) Frodin. Antiviral Res. 2005;68:1–9. doi: 10.1016/j.antiviral.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Asano J, Chiba K, Tada M, Yoshii T. Antiviral activity of lignans and their glycosides from Justicia procumbens. Phytochemistry. 1996;42:713–7. doi: 10.1016/0031-9422(96)00024-6. [DOI] [PubMed] [Google Scholar]
- 6.Salim V, Jones AD, DellaPenna D. Camptotheca acuminata 10-hydroxycamptothecin O-methyltransferase: An alkaloid biosynthetic enzyme co-opted from flavonoid metabolism. Plant J. 2018;95:112–25. doi: 10.1111/tpj.13936. [DOI] [PubMed] [Google Scholar]
- 7.Yimdjo MC, Azebaze AG, Nkengfack AE, Meyer AM, Bodo B, Fomum ZT. Antimicrobial and cytotoxic agents from Calophyllum inophyllum. Phytochemistry. 2004;65:2789–95. doi: 10.1016/j.phytochem.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Mah SH, Lian Ee GC, Teh SS, Sukari MA. Antiproliferative xanthone derivatives from Calophyllum inophyllum and Calophyllum soulattri. Pak J Pharm Sci. 2015;28:425–9. [PubMed] [Google Scholar]
- 9.Toujani MM, Rittà M, Civra A, Genovese S, Epifano F, Ghram A, et al. Inhibition of HSV-2 infection by pure compounds from Thymus capitatus extract in vitro. Phytotherapy Res. 2018;32:1555–63. doi: 10.1002/ptr.6084. [DOI] [PubMed] [Google Scholar]
- 10.Bleasel MD, Peterson GM. Emetine, ipecac, ipecac alkaloids and analogues as potential antiviral agents for coronaviruses. Pharmaceuticals (Basel) 2020;13:51. doi: 10.3390/ph13030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H, Xie Z, Koo HJ, McLaughlin SP, Timmermann BN, Gang DR. Metabolic profiling and phylogenetic analysis of medicinal Zingiber species: Tools for authentication of ginger (Zingiber officinale Rosc) Phytochemistry. 2006;67:1673–85. doi: 10.1016/j.phytochem.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Hui YH, Sherkat F. Handbook of Food Science, Technology, and Engineering. Florida: CRC Press; 2005. [Google Scholar]
- 13.Basile V, Ferrari E, Lazzari S, Belluti S, Pignedoli F, Imbriano C. Curcumin derivatives: molecular basis of their anti-cancer activity. Biochem Pharmacol. 2009;78:1305–15. doi: 10.1016/j.bcp.2009.06.105. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Long L, Zhang F, Chen Q, Chen C, Yu X, et al. Antifungal activity, main active components and mechanism of Curcuma longa extract against Fusarium graminearum. PloS one. 2018;13:e0194284. doi: 10.1371/journal.pone.0194284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechtenberg M, Quandt B, Nahrstedt A. Quantitative determination of curcuminoids in Curcuma rhizomes and rapid differentiation of Curcuma domestica Val. and Curcuma xanthorrhiza Roxb. by capillary electrophoresis. Phytochem Anal. 2004;15:152–8. doi: 10.1002/pca.759. [DOI] [PubMed] [Google Scholar]
- 16.Harborne JB, Baxter H. The Handbook of Natural Flavonoids. New York: Wiley & Sons; 1999. [Google Scholar]
- 17.Wang Y, Lee S, Ha Y, Lam W, Chen SR, Dutschman GE, et al. Tylophorine analogs allosterically regulates heat shock cognate protein 70 and inhibits hepatitis C virus replication. Sci Rep. 2017;7:10037. doi: 10.1038/s41598-017-08815-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Lopez A, Persaud M, Chavez MP, Zhang H, Rong L, Liu S, et al. Glycosylated diphyllin as a broad-spectrum antiviral agent against Zika virus. EBioMedicine. 2019;47:269–83. doi: 10.1016/j.ebiom.2019.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evidente A, Cicala MR, Giudicianni I, Randazzo G, Riccio R. 1H and 13C NMR analysis of lycorine and α-dihydrolycorine. Phytochemistry. 1983;22:581–4. [Google Scholar]
- 20.Du J, He ZD, Jiang RW, Ye WC, Xu HX, But PP. Antiviral flavonoids from the root bark of Morus alba L. Phytochemistry. 2003;62:1235–8. doi: 10.1016/s0031-9422(02)00753-7. [DOI] [PubMed] [Google Scholar]
- 21.Park KY, Jung GO, Lee KT, Choi J, Choi MY, Kim GT, et al. Antimutagenic activity of flavonoids from the heartwood of Rhus verniciflua. J Ethnopharmacol. 2004;90:73–9. doi: 10.1016/j.jep.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 22.Tong T, Yu R, Park T. α-Cedrene protects rodents from high-fat diet-induced adiposity via adenylyl cyclase 3. Int J Obes. 2019;43:202–16. doi: 10.1038/s41366-018-0176-0. [DOI] [PubMed] [Google Scholar]
- 23.Ali A, Tabanca N, Demirci B, Baser KHC, Ellis J, Gray S, et al. Composition, mosquito larvicidal, biting deterrent and antifungal activity of essential oils of different plant parts of Cupressus arizonica var.glabra (’Carolina Sapphire’) Nat Prod Commun. 2013;8:257–60. [PubMed] [Google Scholar]
- 24.Astani A, Reichling J, Schnitzler P. Screening for antiviral activities of isolated compounds from essential oils. Evid Based Complement Alternat Med 2011. 2011:253643. doi: 10.1093/ecam/nep187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park HR, Yoon H, Kim MK, Lee SD, Chong Y. Synthesis and antiviral evaluation of 7-O-arylmethylquercetin derivatives against SARS-associated coronavirus (SCV) and hepatitis C virus (HCV) Arch Pharm Res. 2012;35:77–85. doi: 10.1007/s12272-012-0108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YN, Zhang QY, Li XD, Xiong J, Xiao SQ, Wang Z, et al. Gemcitabine, lycorine and oxysophoridine inhibit novel coronavirus (SARS-CoV-2) in cell culture. Emerg Microbes Infec. 2020;9:1170–3. doi: 10.1080/22221751.2020.1772676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi L, Li Z, Yuan K, Qu X, Chen J, Wang G, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78:11334–9. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu CM, Chang WS, Fang ZS, Chen YT, Wang WL, Tsai HH, et al. Nanoparticulate vacuolar ATPase blocker exhibits potent host-targeted antiviral activity against feline coronavirus. Sci Rep. 2017;7:13043. doi: 10.1038/s41598-017-13316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang SY, Kang JY, Oh MJ. Antiviral activities of flavonoids isolated from the bark of Rhus verniciflua stokes against fish pathogenic viruses in vitro. J Microbiol. 2012;50:293–300. doi: 10.1007/s12275-012-2068-7. [DOI] [PubMed] [Google Scholar]