ABSTRACT
α-mangostin is a xanthone compound isolated from mangosteen pericarp. It is known as an anticancer through induction of apoptotic process by inhibiting fatty acid synthase (FAS) receptor. α-mangostin is a potentially useful ligand for diagnostic purposes in the form of complexes with a radionuclide such as68Gallium (68Ga). Unfortunately, α-mangostin could not be directly labeled with radionuclides. In order to be labeled, a chelator such as 1,4,7-triazacyclononane-1,4,7-triacetic acid (NOTA), a derivative (NO2At), is required. The aim of this study was to find out the interaction of Fe-NO2At-α-mangostin complex compound against FAS receptor using molecular dynamics software. Both the metals have similar chemical characteristics. The results showed a strong interaction between Fe-NO2At-α-mangostin complex compound and FAS receptor. The molecular dynamic showed the complex compound Fe-NO2At-α-mangostin in FAS-KS which produced a bond-free energy values (ΔG) of − 96.7 kcal/mol, forming hydrogen bonds with amino acid residues Glu 115 and Ser 114. The model of molecular dynamic result could be used as a model for the production of 68Ga-α-mangostin in radiopharmaceutical.
Keywords: 68Ga, fatty acid synthase, Fe, molecular dynamics, NO2At, α-mangostin
INTRODUCTION
Positron-emission tomography/computed tomography (PET/CT) scan using 68Gallium (68Ga)-labeled radiopharmaceutical is a molecular imaging diagnostic modality to detect and localized tumor, as well as and monitoring treatment.[1]
Radiometal 68Ga-based radiopharmaceuticals usually require chelators that can conjugate with molecules as target receptors. The chelator protects the radiometal ion to avoid transchelation and hydrolysis reaction, so the radiopharmaceutical has specific biological benefits.[2] Chelators are covalently bonded to the target molecule, producing biologically active radiopharmaceuticals. The most chelator used for 68Ga3+ compound complex is a derivative of a macrocyclic chelator known as NOTA.[3]
Ga3+ has a small ionic radius (0.62Å)[3,4] and compatible with the hole size of the NOTA cavity that leads to the stability of compound complex of Ga3+-NOTA.[2,5] This phenomenon is known as a size-match selectivity.[4] NOTA and its derivatives are considered to be the gold standard chelator for 68Ga[3,5] since the kinetics of the formation of complex compounds are fast at room temperature[2] and excellent in vivo stability.[5]
Discovering a new drug for cancer is challenging, such as drugs that can induce apoptosis in cancer cells. In this regard, there is an anticancer compound through induction of apoptotic process by inhibiting fatty acid synthase (FAS) receptor[6] known as α-mangostin. It is a major isolated compound of Garcinia mangostana.[7] α-mangostin is a potential ligand for cancer theranogtic if it can be labeled with different radionuclides. Unfortunately, α-mangostin could not be directly labeled with radionuclide. In order to be labeled, a chelator NOTA derivates such as NO2At is required.
FAS is an enzyme required to regulate de novo biosynthesis of long-chain fatty acids and has seven domains. The catalytic domains include acyl carrier protein, dehydrogenase (DH), enoyl reductase (ER), β-ketoacyl reductase (KR), β-ketoacyl synthase (KS), malonyl/acetyltransferase (MAT), and thioesterase domain (TE).[8,9] The are two types of FAS. Human FAS and others mammals are type I, which consisted of two identical proteins (homodimerics) and the seven domains forming one binding. FAS in plants is a type II and the seven domains are independent.[8]
High expression of FAS in cancer cell is a poor indicator in patients with breast, ovary, and prostate cancer.[10] The inhibitory reaction of α-mangostin (IC50 value) at FAS was 5.54 mM. α-mangostin can inhibit FAS at the acetyl binding site of KS and stronger than MAT.[11] The study aim was to find out the interaction between Fe-NO2At-α-mangostin complex compound and FAS receptor using molecular dynamics software. In this study, Ga was substituted by Fe since the Ga parameter is not available in the system. Both the metals have similar chemical characteristics.
MATERIALS AND METHODS
Hardware and software
Hardware: the hardware used is a personal computer with Intel Xeon 6 core, NVIDIA GTX 1080Ti, WD Green SSD, and 16GB RAM. Software: The softwares used were as follows: AutoDock 4 (The Scripps Research Institute, USA),[12] Biovia Discovery Studio 2019 (Dassault Systemes, Sandiego), Protein Data Bank (rcsb.org/pdb/), PubChem (NCBI, NIH, USA), VMD, MGL Tools 1.5, and ChemDraw Ultra 15.1 programs (PerkinElmer Inc., downloaded at http://www.cambridgesoft.com/).
Materials
The three-dimensional (3D) FAS-KS structure of PDB with PDB ID code 3HHD and 1EK4 was used as a target protein complex in this study. Meanwhile, the 3D of α-mangostin and its derivative structures was created using Biovia Discovery Studio 2019.
Methods
Recognizing structure of fatty acid synthase-ketoacyl synthase
This study was conducted by recognizing the structure of FAS-KS. The crystal structure of the FAS-KS used in this study was downloaded from the PDB site with PDB ID 3HHD from human and 1EK4 from of Escherichia coli. The next step was comparing 3HHD with 1EK4 structures to look for the binding site of FAS-KS.
α-Mangostin modeling
α-mangostin and the complexed compound of Fe-NO2At-α-mangostin were modeled usingBiovia Discovery Studio program and the results in the form of 3D form were stored in PDB format. The charge and atom types were added to those complex compounds using the MGL tools 1.5.6 program and saved in PDB format.
Molecular docking
Molecular docking simulation of α-mangostin on the active site of FAS-KS 3HHD was tested using Autodock program with the grid box parameter set at 60 × 60 × 60 with a grid spacing of 0.376 Å placed at the midpoint of the active site. The docking was done by 100 times searching conformation using the Lamarckian-Genetic Algorithm protocol.
Molecular dynamics simulation
The results of the docking were carried out on molecular dynamics simulations using Amber18 (University of California, San Francisco, USA). The input files for simulations were prepared using the LeaP module. The antechamber program was used for ligand parameterization by a semi-empirical calculation of AM1-BCC. The receptor was prepared with the PDB4amber module to change the name of amino acid residue to the Amber format and regulate the protonation state of those amino acids. The tleap program was used to connect the receptors to ligands and their parameters, such as ff14SB, gaff, water TIP3P. The next step was adding the water molecule box into the system with the TIP3P water model. The minimization of structure was done by using the steepest descent method and conjugating each gradient for 5000 times, followed by three steps of the increasing grade of heat. Sequentially, the steps were 0–100oK for 20 ps, 100–200oK for 20 ps, and 200–300oK for 20 ps and balanced gradually using the module pmemd for 1 ns. The production phase was carried out for 50 ns. Then, the trajectory of the simulation results was analyzed to obtain root mean square deviation (RMSD), root mean square fluctuation (RMSF), and ligand binding energy with receptors. The bond energy was obtained using the Molecular Mechanic-Generalized Boltzmann Surface Area (MMGBSA) method.
RESULTS
The KS domain of FAS enzyme is a homodimer protein where the active site of this enzyme is located between the two monomers [red circle in Figure 1].
Figure 1.
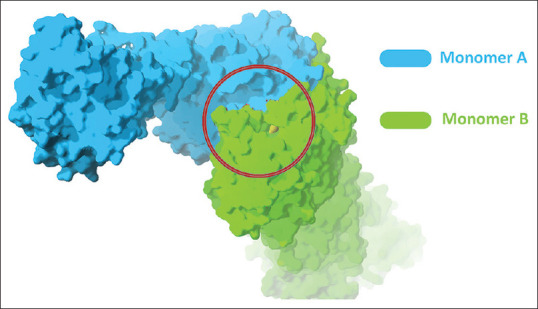
The active side of fatty acid synthase-ketoacyl synthaseenzyme
The minimization process of apo to be holo structure
Apo structure changes can be done by inserting dodecanoic acid into the active site of 3HHD, then relaxing it by minimizing the structure in the Amber program by 10,000 steps with 5000 steps steepest descends and 5000 conjugate gradient. There was a shift of amino acids on the active site of the KS-FAS apo structure after the minimization process is presented in Figure 2.
Figure 2.
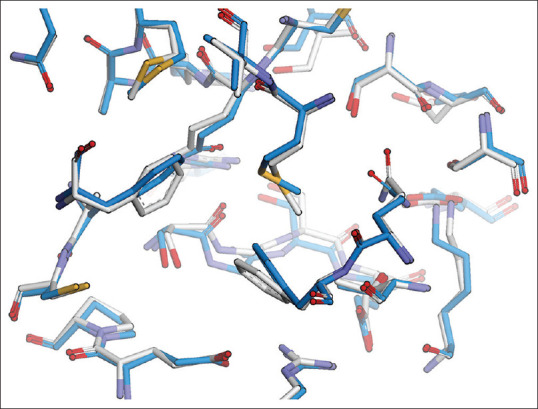
Amino acids on the active side of the fatty acid synthase-ketoacyl synthase apo structure after the minimization process with an root mean square deviation of 2.5 Ao (before relaxation silver and after relaxation blue)
α-mangostin modeling
The modeling of α-mangostin and Fe-NO2At-α-mangostin using the BIOVIA Discovery Studio program is presented in Figure 3.
Figure 3.
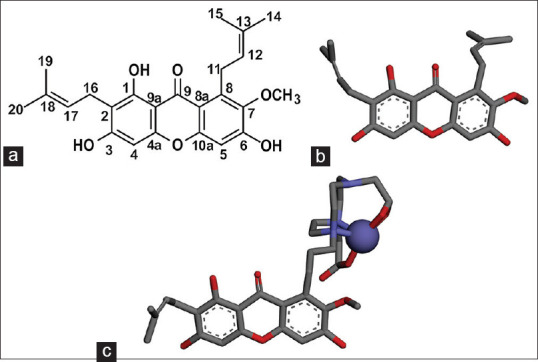
(a) Two-dimensional structure α-mangostin (b) three-dimensional structure α-mangostin, (c) The three-dimensional structure Fe-NO2At-α-mangostin
The molecular docking of α-mangostin to fatty acid synthase-ketoacyl synthase apo
The process of docking α-mangostin to the structure of the FAS-KS apo which was relaxed with the Amber 18 program is presented in Figure 4.
Figure 4.
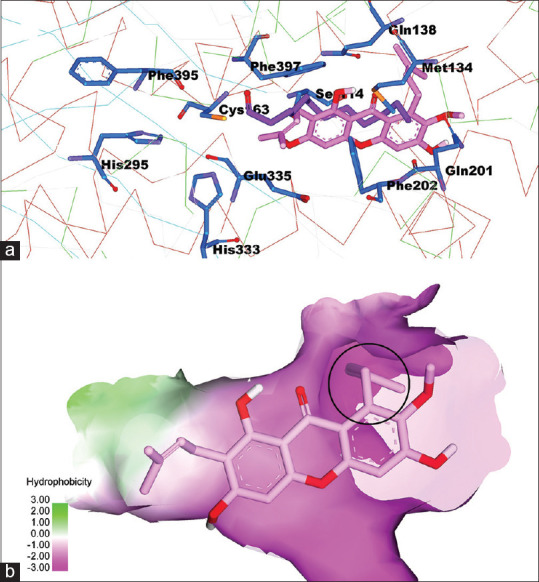
(a) α-mangostin interactions on the active site of fatty acid synthase-ketoacyl synthase, (b) the FAS-KS active surface based on hydrophobicity properties
The NO2At-Fe conjugation in α-mangostin structure
The NO2At-Fe conjugation in the α-mangostin structure is carried out in the blue-circled group in Figure 4b and conjugated at C number 13 of α-mangostin compound [Figure 3a]. The interaction of these substituents with the active site of FAS-KS occurs in the hydrophilic region. NO2 At-Fe is a polar compound; thus, the polar environment is appropriate for the interaction between these molecules. The docking was carried out for Fe-NO2At-alpha-mangostin in FAS-KS and pose was obtained is presented in Figure 5.
Figure 5.
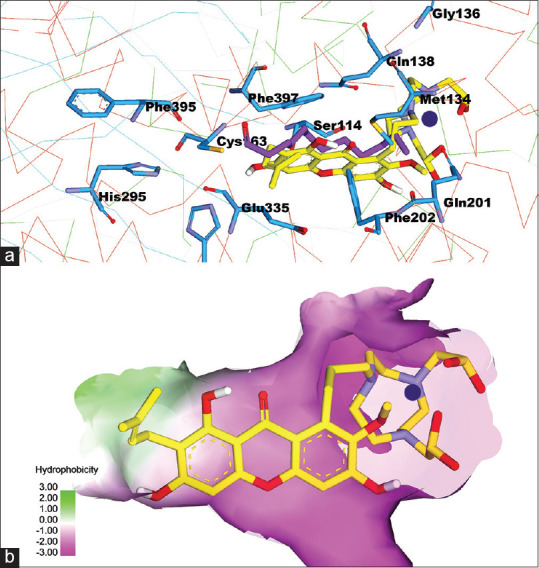
(a) complex compound Fe-NO2At-α-mangostin interactions on the active site of fatty acid synthase-ketoacyl synthase, (b) the fatty acid synthase-ketoacyl synthaseactive surface based on hydrophobicity properties
Molecular dynamic simulation
RSMF showed that the fluctuation of amino acid residues that made up receptors during simulation process could represent residual flexibility.[13] The area of 400–500 Ao on the RMSF chart at Figure 6a, FAS-KS bound to the ligand showed lower fluctuations compared to the apo structure. FAS-KS amino acid residues that bound to the ligands in the region 100–300 Ao showed a little bit higher fluctuation than apo structure (monomer 1) due to the position of monomer 1 which is the location, the binding site, of the FAS-KS bound to the ligand. The result of monomer-2 of RMSF in the area 850 Ao as a binding site between amino acid located in KSb portion and the ligand showed that the fluctuation was lower than the apo structure because the NO2At-Fe group was interacting with KSb portion, as presented in Figure 6b.
Figure 6.
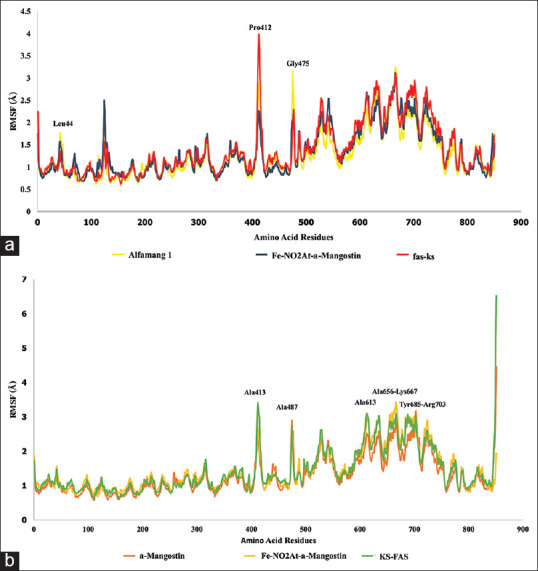
(a) RSMF of fatty acid synthase-ketoacyl synthasemonomer 1, (b) RSMF of fatty acid synthase-ketoacyl synthasemonomer 2
RMSD is an atomic displacement that occurs during a simulation compared to a native ligand structure.[13] RMSD results showed that the condition of binding between FAS-KS and the ligand tends to be more stable than the apo structure [Figures 7 and 8]. The RSMD ligand graphic showed a spike of 1 Ao on frame 228–240 for α-mangostin system [Figure 9].
Figure 7.
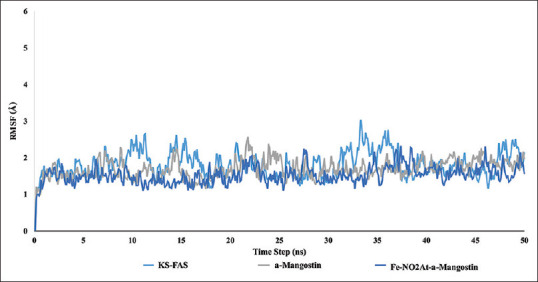
Root mean square deviation fatty acid synthase-ketoacyl synthasemonomer 1
Figure 8.
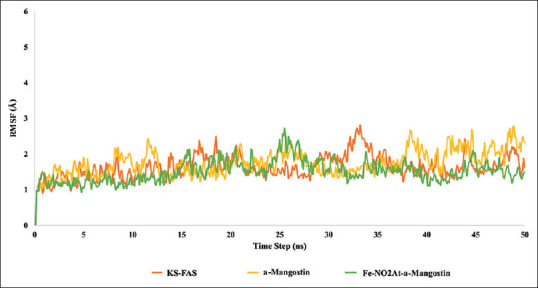
Root mean square deviation fatty acid synthase-ketoacyl synthasemonomer 2
Figure 9.
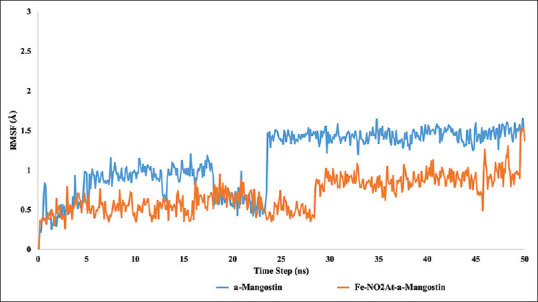
Root mean square deviation ligand
The calculation of ligand binding energy with energy calculation receptor was done using MMGBSA in Amber 18. The results of molecular dynamic of α-mangostin in FAS-KS produced a free binding energy with the value of ΔG – 57.4 kcal/mol and the hydrogen bonds that were formed with amino acid residues Glu 333 and Glu 115, as shown in Figure 10. The molecular dynamic results of the complex compound of Fe-NO2At-α-mangostin in FAS-KS produced bond energy with values of (ΔG) – 96.7 kcal/mol and the hydrogen bonds that were formed with amino acid residues Glu 115 and Ser 114, as shown in Figure 11. The interactions of Fe-NO2At-α-mangostin compounds with FAS-KS were similar to the interactions with α-mangostin in the presence of hydrogen bonding at the residues of Glu 115 and Phe 395.
Figure 10.
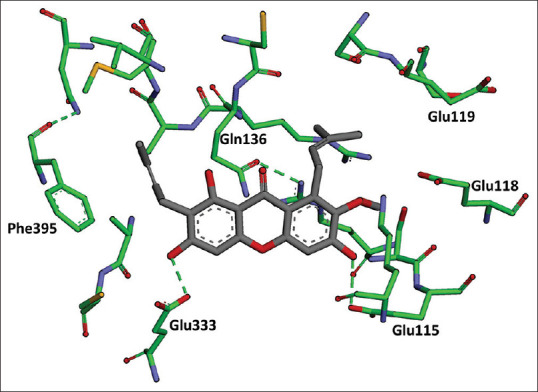
Results of molecular dynamic α-mangostin in fatty acid synthase-ketoacyl synthase
Figure 11.
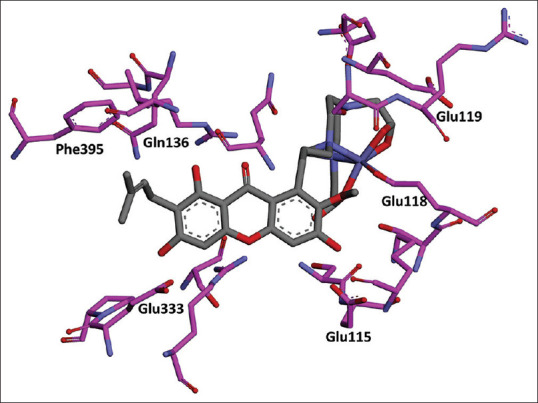
Results of molecular dynamic the complex compound Fe-NO2At-α-mangostin in fatty acid synthase-ketoacyl synthase
Moreover, the NO2At consists of two carboxylic groups. Both the oxygen atoms from the carboxylic group must be bound to Fe metal, but from the simulation molecular dynamic, only one oxygen atom was bound to Fe, while one Fe was bound to Glu 118, as the oxygen atoms were released from Fe and stabilized by the amino acid residue of Ser 114 [Figure 12].
Figure 12.
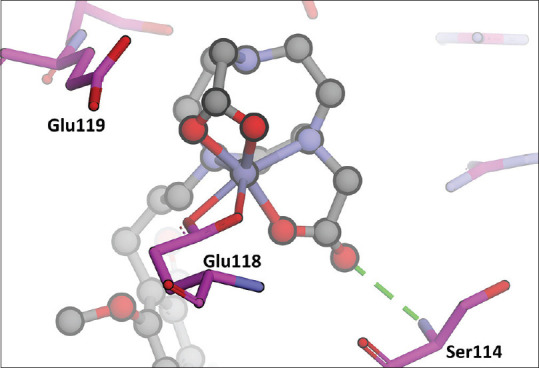
The interaction of complex compounds with the amino acid Glu118 in fatty acid synthase-ketoacyl synthase
Figure 12 shows the interaction of complex compounds with the amino acid Glu 118 in FAS-KS.
DISCUSSION
In this study, the FAS-KS-MAT domain, a human FAS, as the target protein of complex compound Fe-NO2At-α-mangostin is based on Pappenberger et al. study. They showed that the human FAS with high-resolution crystal structure is KS-MAT domain which consists of a tandem domain KS which is linked by the linker domain (LD) to the malonyltransferase MAT.[14] Mammalian FAS can also be used as a target, but there are some limitations in size and flexibility and its crystal structure resolution is medium.
The KS domain consists of a sequence of amino acid residues (1–852), and the final position of the KS domain is a series of 28 amino acid residues from the sequence 825–852 (KSb). KSb locations are far from the previous residual sequence because of an insertion of MAT and LD in the KS domain.[14] The KS domain of FAS enzyme is a homodimer protein where the active site of this enzyme is located between two monomers. A previous study by Quan et al. 2012 showed that the α-mangostin can inhibit FAS by competing with the acetyl group in the KS domain.[11] Since the conjugation between crystalline structure of FAS-KS and α-mangostin is not available, an α-mangostin docking simulation must be performed on the FAS-KS.
The 3D structure of 3HHD comes from human FAS-MAT-KS in the form of apo that has no ligand with a resolution of 2.15 Ao. The 3D structure of EEK4 derived from E. coli FAS-KS binds with dodecanoic acid as a ligand with a resolution of 1.85 Ao (holo). In this case, the dodecanoic acid acts as an inhibitor in the acetyl domain. Both FAS-KS domains derived from E. coli and human have a high amino acid sequence identity on the active site. This shows that the FAS-KS domain of E. coli can act as a surrogate to design human FAS inhibitors.[14] The difference of these two structures is shown in Figure 13. α-mangostin interactions on the active site of FAS-KS can be seen more apparently in Figure 4. It shows that the appearance of the FAS-KS active site surface is based on hydrophobes. The results of the α-mangostin docking were parallel to the dedocanoic acid. It means that the conformation results of α-mangostin molecule docking were almost the same as the dodecanoic acid conformation. Figure 6b shows the active surface of FAS-KS based on hydrophobicity properties, while the alkyl substituents on α-mangostin occupy the hydrophilic region (pink surface).
Figure 13.
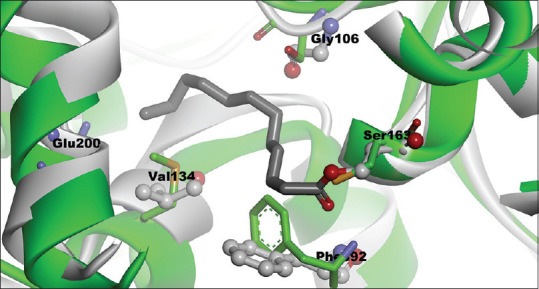
The difference of 3HDD and 1EK4 structures after superpose, 1EK4 (gray), 3HHD (green)
In this study, iron was used instead of gallium metal since the parameter of gallium metal is not available in the Amber program. Fe and Ga have similar characteristics. The ionic radius characteristics of gallium (III) are similar to iron (III). The octahedral radii of Ga3 + and Fe3 + are 0.620 Ao and 0.645 Ao, respectively. The tetrahedral ion radii of Ga3 + and Fe3 + are 0.47 Ao for Ga3 + and 0.49 Ao, respectively.[15] In this regard, Cusnir et al. conducted a study to develop the hydroxypyridinone ligand into Fe3 + chelation and how the design of chelators could be suitable for replacing68Ga as a radiopharmaceutical for PET.[16]
CONCLUSION
The NO2At-Fe compound could be conjugated in C Number 13 of α-mangostin compounds. The Fe-NO2At-α-mangostin forms a binding hydrogen with amino acid residues of Glu 115 and Ser 114, with a bond strength (ΔG) of − 96.7 kcal/mol. Based on the dynamic molecular computation, α-mangostin and Fe complex compounds with NO2At chelator were the models for the development of PET radiopharmaceutical according to 68Ga3+ toradionuclide since Fe3+ and Ga3+ have the same coordination number.
Financial support and sponsorship
This study was financially supported the Directorate General of Higher Education of The Ministry of Research and Technology of Indonesia through Seeds Basic Research of Higher Education (PDUPT) Grants no. 1123ak/UN6.O/LT/2019.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This study was supported by Academic Leadership Grant no. 1427/UN6.3.1/LT/2020 and Competitive Grant no. and Doctoral Dissertation Research of Universitas Padjadjaran. We are grateful to Ade Rizqi Ridwan for his guidance of computation work.
REFERENCES
- 1.Singh AN, Liu W, Hao G, Kumar A, Gupta A, Öz OK, et al. Multivalent bifunctional chelator scaffolds for gallium-68 based positron emission tomography imaging probe design: Signal amplification via multivalency. Bioconjug Chem. 2011;22:1650–62. doi: 10.1021/bc200227d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prata MI. Gallium-68: A new trend in PET radiopharmacy. Curr Radiopharm. 2012;5:142–9. doi: 10.2174/1874471011205020142. [DOI] [PubMed] [Google Scholar]
- 3.Price EW, Orvig C. Matching chelators to radiometals for radiopharmaceuticals. Chem Soc Rev. 2014;43:260–90. doi: 10.1039/c3cs60304k. [DOI] [PubMed] [Google Scholar]
- 4.Saw MM. Medicinal radioparmaceutical chemistry of metal radiopharmacuticals. COSMOS. 2012;8:11–81. [Google Scholar]
- 5.Wei W, Rosenkrans ZT, Liu J, Huang G, Luo QY, Cai W. ImmunoPET: Concept, design, and applications. Chem Rev. 2020;120:3787–851. doi: 10.1021/acs.chemrev.9b00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, Tian W, Ma X. Alpha-mangostin inhibits intracellular fatty acid synthase and induces apoptosis in breast cancer cells. Mol Cancer. 2014;13:138. doi: 10.1186/1476-4598-13-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muchtaridi M, Suryani D, Qosim W, Saptarini NM. Quantitative analysis of A-mangostin in mangosteen (Garcinia mangostana L.) pericarp extract from four district of West Java by HPLC method. Inter J Pharm Pharm Sci. 2016;8:232–6. [Google Scholar]
- 8.Cheng C, Wang Z, Chen J. Targeting FASN in breast cancer and the discovery of promising inhibitors from natural products derived from traditional Chinese medicine. Evid Based Complement Alternat Med 2014. 2014:232946. doi: 10.1155/2014/232946. doi: 10.1155/2014/232946. Epub 2014 Mar 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panman W, Nutho B, Chamni S, Dokmaisrijan S, Kungwan N, Rungrotmongkol T. Computational screening of fatty acid synthase inhibitors against thioesterase domain. J Biomol Struct Dyn. 2018;36:4114–25. doi: 10.1080/07391102.2017.1408496. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci U S A. 2004;101:15567–72. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan X, Wang Y, Ma X, Liang Y, Tian W, Ma Q, et al. α-mangostin induces apoptosis and suppresses differentiation of 3T3-L1 cells via inhibiting fatty acid synthase. PLoS One. 2012;7:1–16. doi: 10.1371/journal.pone.0033376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong YW, Liao ML, Meng XL, Somero GN. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc Natl Acad Sci U S A. 2018;115:1274–9. doi: 10.1073/pnas.1718910115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappenberger G, Benz J, Gsell B, Hennig M, Ruf A, Stihle M, et al. Structure of the human fatty acid synthase KS–MAT didomain as a framework for inhibitor design. J Mol Biol. 2010;397:508–19. doi: 10.1016/j.jmb.2010.01.066. [DOI] [PubMed] [Google Scholar]
- 15.Chitambar CR. Gallium-containing anticancer compunds. Fut Med Chem. 2012;4:1257–72. doi: 10.4155/fmc.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cusnir R, Imberti C, Hilder RC, Blower PJ, Ma MT. Hydroxypyridinone chelators: From iron scavenging to radiopharmaceuticals for PET imaging with gallium-68. Int J Mol Sci. 2017;18:116. doi: 10.3390/ijms18010116. [DOI] [PMC free article] [PubMed] [Google Scholar]


