Abstract
Upon encountering an antigen, antibodies mature through various rounds of somatic mutations, resulting in higher affinities and specificities to the particular antigen. We review recent progress in four areas of antibody maturation studies. (1) Next-generation and single-cell sequencing have revolutionized the analysis of antibody repertoires by dramatically increasing the sequences available to study the state and evolution of the immune system. Computational methods, including machine learning tools, have been developed for reconstituting antibody clonal lineages and for general repertoire analysis. (2) The availability of X-ray structures, thermodynamic and kinetic data, and molecular dynamics simulations provide information on the biophysical mechanisms responsible for improved affinity. (3) In addition to improved binding to a specific antigen, providing affinity-independent diversity and self/nonself discrimination are fundamental functions of the immune system. Recent studies, including X-ray structures, yield improved understanding of both mechanisms. (4) Results from in vivo maturation help to develop methods of in vitro maturation to improve antibody properties for therapeutic applications, frequently combining computational and experimental approaches.
Introduction
After exposure to an antigen, antibodies specific to that antigen will be enriched through the process of antibody maturation, which involves clonal selection, expansion and somatic hypermutation [1]. Immunoglobulin (Ig) genes are mutated and any resulting B cell receptors (BCRs) which have acquired higher affinity are favored for survival; the humoral response will become dominated by these mutated receptors, which confer protection in subsequent antigen exposures. Such a rapid cycle of mutation and selection bolsters the host defense, with antibody affinity improving 10 to 5,000 fold during the immune response [2]. This complex process raises a number of interesting questions. Where do the mutations occur? What is the effect of the mutations on antibody structure and flexibility? How do the mutations change antibody properties, primarily affinity, on and off rates, specificity, and stability? How does the immune system provide self/nonself discrimination? How deterministic are the developmental pathways (lineages)? Are these similar in different individuals? While interpreting experimental data to answer these questions provides necessary insight, the major test of understanding is whether the changes associated with antibody maturation can be predicted with any reasonable accuracy, and whether there is sufficient information for developing therapeutic antibodies. As shown in this short review focusing on aspects of antibody maturation (Figure 1), during the last two or three years a number of important discoveries substantially improved our understanding of the immune system, and large scale collection of data and the development of novel methods predict further progress.
Figure 1.

Focus areas in antibody maturation. (a) Large collections of sequences representing entire antibody repertoires are now available to establish models of somatic hypermutation and to reconstruct antibody clonal lineages. (b) Enthalpy-driven improvement of affinity is frequently caused by mutations of contact residues in the CDRs that increase shape complementarity interface, improve electrostatic interactions, hydrogen bonding, and promote increased burial of hydrophobic regions in the interface. (c) Alternatively, the binding affinity can be improved by the rigidification of some interacting loops, in most cases H3, thereby reducing the entropy loss upon binding. Binding can be improved by mutations outside the loops directly contacting the antigen. (d) The panels represent mutations on the antibody (yellow spheres) to compensate for the naturally occurring mutations of the antigen (shown as red spheres) in frequently mutating viruses such as influenza.
Analysis of antibody repertoires
The collection, or repertoire, of antibodies within an organism convey its immune status, describes its innate ability to deal with invading or harmful substances, and acts as a history of how the organism has previously responded to similar challenges [1]. Recent advances in next-generation sequencing (NGS) have revolutionized strategies for antibody repertoire analysis by dramatically increasing sample depth compared to previous low-throughput methods [2]. New methods have also been developed for single-cell sequencing, which allow large-scale determination of paired light (L) and heavy (H) chains. In addition to computational tools for reconstituting antibody clonal lineages [3], these advances can provide valuable insights into how the immune system works, including how it is initially capable of protecting against diverse threats, but produces higher affinity antibodies after antigen exposure [1]. Researchers now have easy access to a vast number of sequences. For example, the Observed Antibody Space (OAS) database, contains over 1 billion sequences [4]. A number of specialized sequence analysis tools are also available [5], and have enabled accurate models of somatic hypermutation to be established [6], leading to the creation of software that simulates the repertoires [3,7]. In particular, the analyses were employed to study the effect of disease on the immune system [8] and to monitor the impact of organ transplant [9]. Machine learning was also used to predict vaccination status or the presence of disease [10], and in view of the availability of sequence data it is expected to become a major tool to study the repertoires.
The impact of mutations on antibody structure, flexibility, and binding affinity
While sequences alone provide valuable information regarding the immune response, 3D structures are the best to determine how an antibody governs its binding properties and interacts with an antigen [1,11]. One of the mechanisms to achieve increased affinity in mature antibodies has been shown to be mutations to the residues in the complementarity-determining regions (CDRs) of the variable chains. The mutations in CDRs may drive affinity maturation through two main mechanisms and their combinations. On one extreme, mutations that increase shape complementarity of the interface, improve electrostatic interactions, hydrogen bonding, and promote increased burial of hydrophobic regions in the interface all improve binding by enthalpic means. The alternative and even better studied mechanism involves decreasing entropic penalties associated with complex formation due to the rigidification of some CDRs. The CDR H3 loop has proven to be of particular importance in both mechanism, as it has been shown to form the most contacts on average with the antigen, while also demonstrating highest structural variation even without direct mutations [12]. A well-studied example of the entropy-driven increase of affinity is a B-cell lineage expressing broadly neutralizing influenza virus antibodies, as discussed in Schmidt et al. [13]. The lineage was derived from a subject immunized with a trivalent vaccine and was comprised of three mature antibodies, the unmutated common ancestor, and a common intermediate (Figure 2), all with the CDR H3 inserting into the conserved receptor-binding pocket of influenza hemagglutinin. Mutations that almost exclusively occur in non-H3 CDR and framework regions rigidify the conformation of the H3 loop very close to its bound conformation, as demonstrated by the analysis of structures and binding kinetics. Long time-scale molecular dynamics simulations revealed that the maturation increases the probability of the H3 loop being close to its conformation in the antigen-bound structure [13]. Rigidification of the H3 loop by remote mutations was also reported for an anti-HIV neutralizing antibody [14]. In another recent study, Fernández-Quintero et al. [15,16] analyzed pairs of antibody fragments which differed in specificity and stage of affinity maturation. Using a combination of metadynamics and molecular dynamics (MD) simulations, they observed substantial rigidification in flexibility and plasticity as reflected by a decrease of conformational diversity. However, a large scale study by Jeliazkov et al. [17] focusing on CDR H3 loops did not find substantial differences in the flexibility of naïve and antigen-experienced antibodies. Molecular dynamics simulations revealed a spectrum of changes in flexibility, indicating that while rigidification may be important, it is not the only biophysical mechanism leading to improved affinity.
Figure 2.
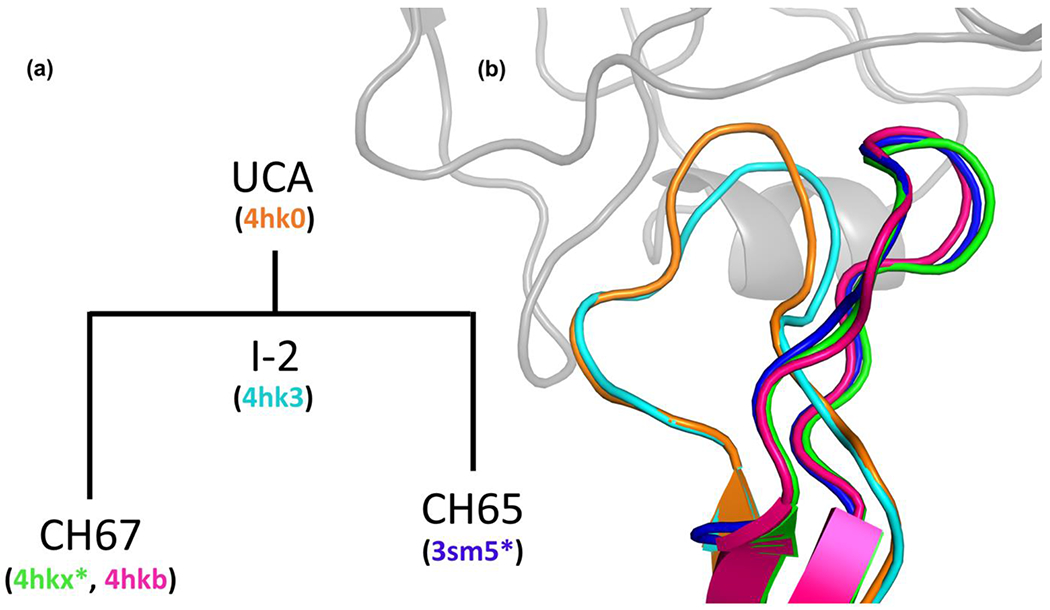
Example of preconfigured H3 loop in entropy-driven improvement of binding affinity. (a) B-cell lineage expressing broadly neutralizing influenza virus antibodies that bind to the sialic acid receptor region of the hemagglutinin (HA), including the UCA (PDB ID 4hk0), a common intermediate I-2 (4hk3) and two of the mature antibodies. X-ray structures are available for both the unbound and HA-bound variant CH67 (4hkb and 4hkx, respectively). Only the HA-bound structure is solved for the variant CH65. (b) The conformations of the H3 loop in the five structures shown on the left reveal that the unbound structure in the mature antibody CH67 (4hkb, purple) comes very close to the conformations of the loop in the HA-bound structures (4hkx and 3sm5, colored green and blue, respectively). The star indicates HA-bound structures.
Changes in conformation and flexibility also determine the kinetics of antibody-antigen binding. The already mentioned study of influenza antibody maturation by Schmidt et al. [13] reported two orders of magnitude increase in the on-rate and one order of magnitude decrease in the off-rate values, in good agreement with the observation that the major change is the preorganization of the CDR H3 region. In contrast, Rosenfeld et al. [18] found that improved antibody-based ricin neutralization by affinity maturation was correlated with slower off-rate values. We think that this variation is due to the difference in the shape of the antibody epitopes. The H3 loop of neutralizing antibodies targeting influenza HA must find the fairly narrow sialic acid binding site [13], which is the major binding energy hot spot [19,20]. This suggests that appropriate preorganization and rigidity of the H3 loop increases the kon values, whereas the koff values are less affected due the scarcity of mutations in H3. In contrast, modeling of the ricin-binding antibody suggests that the mutations may increase this variant’s conformational flexibility, which may improve its ability to bind ricin [18].
Maturation for improved specificity
The selection of antibody variants and somatic hypermutation play at least two main roles in generating a robust B cell immune response [21]. The first is the classical process of affinity maturation, in which the antibody adapts to fit more perfectly to the antigen structure. The second role is the generation of affinity-independent diversity and the ability to adapt to changes in the antigen. The latter outcome may be particularly important for protection against pathogens such as influenza virus that mutate rapidly enough to reinfect a previously exposed individual. McCarthy et al. [21] described an extensive structural and biophysical analysis of a lineage of B cell antigen receptors (BCRs) directed against the receptor binding site of subtype H1 influenza virus hemagglutinin (HA). The antibodies were obtained from a donor who was born in 1989 and in 2008 received a trivalent influenza vaccine. The lineage included 8 antibodies, three in one principal branch and five in the other. As described previously [13], the CDR H3 was found to fit with an invariant pose into the small sialic acid binding site of HA, but in each of the two branches the rest of the Fab reoriented specifically from its position in the unmutated common ancestor (UCA). The reorientation generated new contacts, which compensated for contacts lost as the HA itself mutated during the time between the donor’s initial exposure and his vaccination. The presence of cells producing antibodies from divergent branches like these thus offers broader protection when compared to cells from only a single, linear evolutionary trajectory [21]. In a large scale study, Shehata et al. [22] analyzed biophysical properties of human antibodies derived from multiple B cell subsets, and found that somatic hypermutation was associated with increased antibody specificity. However, they observed that maturation reduced both hydrophobicity and thermal stability compared with naive B cell-derived mAbs. In agreement with this finding, Julian et al. [23] reported that co-selection of compensatory mutations to maintain thermodynamic stability was required for efficient affinity maturation of antibody variable domains.
An important question is how antibodies develop the specificity to differentiate foreign antigens that mimic self-antigens. Burnett et al. [24] generated B cells in a mouse model displaying an antibody that cross-reacted with two related protein antigens expressed on self versus foreign cells. They found that the concentration of B cells remained low until challenged with a high-density foreign antigen, which initiated germinal center recruitment and antibody gene hypermutation. The mutations primarily decreased self-affinity, and increased foreign affinity at a slower rate. Crystal structures revealed that these mutations exploit subtle structural differences in order to achieve 5000-fold preferential binding to foreign over self-epitopes. The interesting conclusion was that antibody mutation away from self-reactivity deferred the need to acquire stringent self-tolerance until after an infection. However, retaining self-reactive clones in the naïve antibody repertoire as substrates for protective antibody responses was required to retain the ability to detect all foreign antigens.
Watanabe et al. [25] used single-cell cultures to determine the repertoires of human B cell antigen receptors before and after the second B cell tolerance checkpoint in both healthy donors and in patients with systemic lupus erythematosus (SLE). Among healthy donors, roughly 70% of transitional B cells before the second checkpoint recognizing foreign antigens also bound human self-antigens, but peripheral tolerance halved the frequency of the self-reactive mature B cells. However, in SLE patients who are defective in the second tolerance checkpoint, frequencies self-reactive B cells remained unchanged during maturation. The authors concluded that cross-reactivity between foreign and self-epitopes may be more common than previously believed [25]. This agrees with the observations of Burnett et al. [24] that such cell are needed in the native repertoire, but their concentration is low and are increasingly eliminated upon mutations to respond to an infection.
In vitro maturation of therapeutic antibodies
Antibodies have become very important therapeutics, as evidenced by an increasing number of FDA-approved monoclonal antibodies [26–28]. Antibody drugs have many advantages over small-molecule drugs, including superior specificity, prolonged serum half-life, and high druggability [26,29]. Antibody discovery platforms use either a display-based library approach (phage, yeast, ribosome, mammalian, or other systems) or an immunization and hybridoma screening strategy for antibody isolation [30]. In vitro affinity is needed when the affinity of antibodies generated by these methods does not meet the requirement for drug development. Moreover, to reduce their antigenicity, humanization of antibodies generated from non-humanized animals frequently results in reduction of antibody affinity, which has to be restored [31]. The display methods mentioned above can be used for in vitro affinity maturation, and successful applications have been reported [32]. Other tools are random mutagenesis by error-prone PCR, and combinatorial mutagenesis limited to the CDRs [33]. Reprogramming the antigen specificity of B cells using CRISPR-Cas9 genome-editing technologies is a more recent and very innovative approach [34]. However, these methods of in vitro affinity maturation can be laborious and time consuming, and hence a variety of computational approaches have been developed [28,30,35]. Although the methods of in silico antibody maturation and methods of de novo antibody design partially overlap, here we focus only on the first application, and refer to recent reviews [11,36] for the design tools.
Computational antibody maturation generally requires a high-quality antibody-antigen co-crystal structure as the starting point, and an algorithm which calculates the energy change ocurring upon mutation. As an example, Purisima and co-workers developed the ADAPT (Assisted Design of Antibody and Protein Therapeutics) platform for improving and modulating antibody affinity [37,38]. The method uses a combination of three scoring functions, and tests the impact of mutating residues one-by-one without changing the initial conformation of the backbone. In spite of these simplifying assumptions, the platform provided triple mutants that exhibited over 30-fold improvements in binding affinity. Kuroda and Tsumoto [35] and Cannon et al. [30] also provided examples of successful application, although in the latter the computational method was guided by experimental alanine scanning.
In vitro maturation generally attempts to optimize several properties, including affinity, specificity, stability, and solubility. A common challenge is that an improvement in one property (e.g., affinity) can lead to a deficits in another (e.g., stability). Rabia et al. [29] studied potential trade-offs and the possibility of co-optimizing multiple antibody properties [29]. An additional but very important goal of antibody maturation is avoiding “developability issues” such as poor stability or high levels of aggregation. Raybould et al. [27] provided guideline values for five metrics implicated in poor developability, including the total length of CDRs, the extent and magnitude of surface hydrophobicity, positive charge and negative charge in the CDRs, and asymmetry in the net heavy- and light-chain surface charges. The guideline cutoffs for each property were derived from the values seen in clinical-stage antibody therapeutics.
Acknowledgements
This work was supported in part by the Division of General Medical Sciences of the National Institute Health (NIH grant R35GM118078) and by the the National Science Foundation (NSF grants DBI 1759277 and AF 1645512).
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Marks C, Deane CM: How repertoire data are changing antibody science. J Biol Chem 2020, 295:9823–9837. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper is an excellent review of the opportunities provided by the availability of large collections of antibody repertoires from next-generation and single-cell sequencing efforts.
- 2.Mishra AK, Mariuzza RA: Insights into the structural basis of antibody affinity maturation from next-generation sequencing. Front Immunol 2018, 9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kepler TB, Wiehe K: Genetic and structural analyses of affinity maturation in the humoral response to HIV-1. Immunol Rev 2017, 275:129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovaltsuk A, Leem J, Kelm S, Snowden J, Deane CM, Krawczyk K: Observed antibody space: a resource for data mining next-generation sequencing of antibody repertoires. J Immunol 2018, 201:2502–2509. [DOI] [PubMed] [Google Scholar]
- 5.Teraguchi S, Saputri DS, Llamas-Covarrubias MA, Davila A, Diez D, Nazlica SA, Rozewicki J, Ismanto HS, Wilamowski J, Xie J, et al. : Methods for sequence and structural analysis of B and T cell receptor repertoires. Comput Struct Biotechnol J 2020, 18:2000–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper lists the sequence analysis and 3D modeling software tools currently available to study B-cell and T-cell receptor repertoirs.
- 6.Horns F, Vollmers C, Dekker CL, Quake SR: Signatures of selection in the human antibody repertoire: Selective sweeps, competing subclones, and neutral drift. Proc Natl Acad Sci U S A 2019, 116:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yermanos A, Greiff V, Krautler NJ, Menzel U, Dounas A, Miho E, Oxenius A, Stadler T, Reddy ST: Comparison of methods for phylogenetic B-cell lineage inference using time-resolved antibody repertoire simulations (AbSim). Bioinformatics 2017, 33:3938–3946. [DOI] [PubMed] [Google Scholar]
- 8.Bashford-Rogers RJM, Bergamaschi L, McKinney EF, Pombal DC, Mescia F, Lee JC, Thomas DC, Flint SM, Kellam P, Jayne DRW, et al. : Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature 2019, 574:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai L, Zhou X, Chen H, Luo Y, Sui W, Zhang J, Tang D, Yan Q, Dai Y: Composition and diversity analysis of the B-cell receptor immunoglobulin heavy chain complementarity-determining region 3 repertoire in patients with acute rejection after kidney transplantation using high-throughput sequencing. Exp Ther Med 2019, 17:2206–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora R, Kaplinsky J, Li A, Arnaout R: Repertoire-based diagnostics using statistical biophysics. bioRxiv 2019:519108. [Google Scholar]
- 11.Raybould MI, Wong WK, Deane CM: Antibody–antigen complex modelling in the era of immunoglobulin repertoire sequencing. Molecular Systems Design & Engineering 2019, 4:679–688. [Google Scholar]
- 12.Regep C, Georges G, Shi J, Popovic B, Deane CM: The H3 loop of antibodies shows unique structural characteristics. Proteins 2017, 85:1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. : Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 2013, 110:264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes the analysis of a lineage of broadly neutralizing influenza virus antibodies binding to the head region of the hemagglutinin (HA) protein. The lineage includes three mature antibodies, the unmutated common ancestor (UCA), and a common intermediate (I-2). X-ray structures are available for UCA, I-2, one unbound mature antibody, and two mature antibodies bound to HA. Their CDR H3 inserts into HA’s conserved receptor-binding pocket. The structures show that during maturation the H3 loop becomes preconfigured in its HA-bound conformation. Long time-scale molecular dynamics simulations reveal that the maturation increases the probability of the H3 loop being in such conformation, and thus the maturation is primarily entropy driven.
- 14.Kondo HX, Kiribayashi R, Kuroda D, Kohda J, Kugimiya A, Nakano Y, Tsumoto K, Takano Y: Effects of a remote mutation from the contact paratope on the structure of CDR-H3 in the anti-HIV neutralizing antibody PG16. Sci Rep 2019, 9:19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Quintero ML, Loeffler JR, Bacher LM, Waibl F, Seidler CA, Liedl KR: Local and global rigidification upon antibody affinity maturation. Front Mol Biosci 2020, 7:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Quintero ML, Loeffler JR, Kraml J, Kahler U, Kamenik AS, Liedl KR: Characterizing the diversity of the CDR-H3 loop conformational ensembles in relationship to antibody binding properties. Front Immunol 2018, 9:3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeliazkov JR, Sljoka A, Kuroda D, Tsuchimura N, Katoh N, Tsumoto K, Gray JJ: Repertoire analysis of antibody CDR-H3 loops suggests affinity maturation does not typically result in rigidification. Front Immunol 2018, 9:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeld R, Alcalay R, Mechaly A, Lapidoth G, Epstein E, Kronman C, S JF, Mazor O: Improved antibody-based ricin neutralization by affinity maturation is correlated with slower off-rate values. Protein Eng Des Sel 2017, 30:611–617. [DOI] [PubMed] [Google Scholar]
- 19.Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S: The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc 2015, 10:733–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran PR, Radoux CJ, Smilova MD, Sykes RA, Higueruelo AP, Bradley AR, Marsden BD, Spring DR, Blundell TL, Leach AR, et al. : Hotspots API: A Python package for the detection of small molecule binding hotspots and application to structure-based drug design. J Chem Inf Model 2020, 60:1911–1916. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy KR, Raymond DD, Do KT, Schmidt AG, Harrison SC: Affinity maturation in a human humoral response to influenza hemagglutinin. Proc Natl Acad Sci U S A 2019, 10.1073/pnas.1915620116. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper shows how the loss of affinity due to mutations in the highly variable influenza hemagglutinin protein are compensated by mutations of the antibodies outside the largely conserved H3 region.
- 22.Shehata L, Maurer DP, Wec AZ, Lilov A, Champney E, Sun T, Archambault K, Burnina I, Lynaugh H, Zhi X, et al. : Affinity maturation enhances antibody specificity but compromises conformational stability. Cell Rep 2019, 28:3300–3308 e3304. [DOI] [PubMed] [Google Scholar]
- 23.Julian MC, Li L, Garde S, Wilen R, Tessier PM: Efficient affinity maturation of antibody variable domains requires co-selection of compensatory mutations to maintain thermodynamic stability. Sci Rep 2017, 7:45259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burnett DL, Langley DB, Schofield P, Hermes JR, Chan TD, Jackson J, Bourne K, Reed JH, Patterson K, Porebski BT, et al. : Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science 2018, 360:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper is a very interesting analysis of self/non-self discrimination. It describes the observation of rapid selection for mutations that decrease self affinity and slower selection for epistatic mutations that specifically increase foreign affinity. The conclusion is that the presence of cross-reactive antibodies is necessary to avoid holes in the naïve repertoir, but the mutation away from self-reactivity in germinal center reactions defers the need to acquire stringent self-tolerance until after an infection.
- 25.Watanabe A, Su KY, Kuraoka M, Yang G, Reynolds AE, Schmidt AG, Harrison SC, Haynes BF, St Clair EW, Kelsoe G: Self-tolerance curtails the B cell repertoire to microbial epitopes. JCI Insight 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, An L, Zhao Y, Zhang C, Li S, Ye C, Jing S, Hang H: In vitro affinity maturation of antibody against membrane-bound GPCR molecules. Appl Microbiol Biotechnol 2019, 103:7703–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raybould MIJ, Marks C, Krawczyk K, Taddese B, Nowak J, Lewis AP, Bujotzek A, Shi J, Deane CM: Five computational developability guidelines for therapeutic antibody profiling. Proc Natl Acad Sci U S A 2019, 116:4025–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabasinezhad M, Talebkhan Y, Wenzel W, Rahimi H, Omidinia E, Mahboudi F: Trends in therapeutic antibody affinity maturation: From in-vitro towards next-generation sequencing approaches. Immunol Lett 2019, 212:106–113. [DOI] [PubMed] [Google Scholar]
- 29.Rabia LA, Desai AA, Jhajj HS, Tessier PM: Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem Eng J 2018, 137:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannon DA, Shan L, Du Q, Shirinian L, Rickert KW, Rosenthal KL, Korade M 3rd, van Vlerken-Ysla LE, Buchanan A, Vaughan TJ, et al. : Experimentally guided computational antibody affinity maturation with de novo docking, modelling and rational design. PLoS Comput Biol 2019, 15:e1006980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Xia L, Zhang T, You M, Huang Y, He M, Su R, Tang J, Zhang J, Li S, et al. : Structure guided maturation of a novel humanized anti-HBV antibody and its preclinical development. Antiviral Res 2020, 180:104757. [DOI] [PubMed] [Google Scholar]
- 32.Colley CS, Popovic B, Sridharan S, Debreczeni JE, Hargeaves D, Fung M, An LL, Edwards B, Arnold J, England E, et al. : Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors. MAbs 2018, 10:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons JF, Lim YW, Carter KP, Wagner EK, Wayham N, Adler AS, Johnson DS: Affinity maturation of antibodies by combinatorial codon mutagenesis versus error-prone PCR. MAbs 2020, 12:1803646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss JE, Gonzalez-Martin A, Andrabi R, Fuller RP, Murrell B, McCoy LE, Porter K, Huang D, Li W, Sok D, et al. : Reprogramming the antigen specificity of B cells using genome-editing technologies. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper describes a novel approach to in vitro antibody maturation. The immunoglobulin genes of mature B cells are directly edited using CRISPR-Cas9 in a homology directed repair strategy, to replace the heavy chain variable region with that from an HIV broadly neutralizing antibody. Endogenous activation-induced cytidine deaminase in engineered cells generated B cell receptor variants with improved HIV neutralizing activity.
- 35.Kuroda D, Tsumoto K: Antibody affinity maturation by computational design. Methods Mol Biol 2018, 1827:15–34. [DOI] [PubMed] [Google Scholar]
- 36.Norman RA, Ambrosetti F, Bonvin A, Colwell LJ, Kelm S, Kumar S, Krawczyk K: Computational approaches to therapeutic antibody design: established methods and emerging trends. Brief Bioinform 2020, 21:1549–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper is an excellent review of the design tools that can be used for de novo therapeutic antibody design beyond the scope of in silico antibody maturation.
- 37.Sulea T, Hussack G, Ryan S, Tanha J, Purisima EO: Application of Assisted Design of Antibody and Protein Therapeutics (ADAPT) improves efficacy of a Clostridium difficile toxin A single-domain antibody. Sci Rep 2018, 8:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivcharuk V, Baardsnes J, Deprez C, Sulea T, Jaramillo M, Corbeil CR, Mullick A, Magoon J, Marcil A, Durocher Y, et al. : Assisted Design of Antibody and Protein Therapeutics (ADAPT). PLoS One 2017, 12:e0181490. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper describes a remarkably simple approach to in vitro antibody maturation. Amino acid mutations are introduced and tested one-by-one using a consensus z-score from three well established scoring functions. In spite of this simplicity, triple mutants exhibited at least 30-fold improvements in binding affinity, and over 90% of all the intermediate single and double mutants showed higher affinities than the parent sequence. Results suggest that current scoring functions are accurate enough to perform in vitro antibody maturation.


