Abstract
Acetylation was initially discovered as a post-translational modification (PTM) on the unstructured, highly basic N-terminal tails of eukaryotic histones in the 1960s. Histone acetylation constitutes part of the “histone code”, which regulates chromosome compaction and various DNA processes such as gene expression, recombination, and DNA replication. In bacteria, nucleoid-associated proteins (NAPs) are responsible these functions in that they organize and compact the chromosome and regulate some DNA processes. The highly conserved DNABII family of proteins are considered functional homologues of eukaryotic histones despite having no sequence or structural conservation. Within the past decade, a growing interest in Nε-lysine acetylation led to the discovery that hundreds of bacterial proteins are acetylated with diverse cellular functions, in direct contrast to the original thought that this was a rare phenomenon. Similarly, other previously undiscovered bacterial PTMs, like serine, threonine, and tyrosine phosphorylation, have also been characterized. In this review, the various PTMs that were discovered among DNABII family proteins, specifically histone-like protein (HU) orthologues, from large-scale proteomic studies are discussed. The functional significance of these modifications and the enzymes involved are also addressed. The discovery of novel PTMs on these proteins begs this question: is there a histone-like code in bacteria?
Keywords: post-translational modification, HU, HupA, HupB, HBsu, acetylation, phosphorylation, succinylation, acetyltransferase, deacetylase
Graphical Abstract
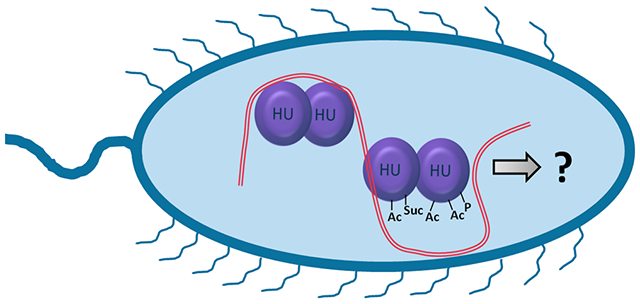
INTRODUCTION
Histones Regulate Chromosome Compaction and Gene Expression in Eukaryotes
In eukaryotes, meters-long linear stretches of DNA must be compacted in order to fit inside the ~6 μm nucleus of the cell. The DNA must be compacted in a way that allows for accurate replication, proper expression of genes, and correct repair, and therefore, it must remain accessible to various protein machines. This is achieved by wrapping the DNA molecule twice around a nucleosome, which is composed of two copies of each histone, H2A, H2B, H3, H4 (Figure 1A), or one of several histone variants.1 The degree of chromatin compaction is correlated with the level of gene expression and is regulated by numerous factors, including chromatin remodeling proteins and biochemical post-translational modifications (PTMs) on the N-terminal tails of histones.2–4 The first modifications discovered on histones were lysine acetylation and methylation and then phosphorylation in the 1960s.5,6 Since then, many more PTMs have been identified, including multiple lysine acylations; arginine methylation and citrullination; lysine sumoylation and ubiquitination; ADP-ribosylation; proline isomerization; glycosylation; biotinylation; and serine, threonine, and tyrosine phosphorylation.7,8
Figure 1.
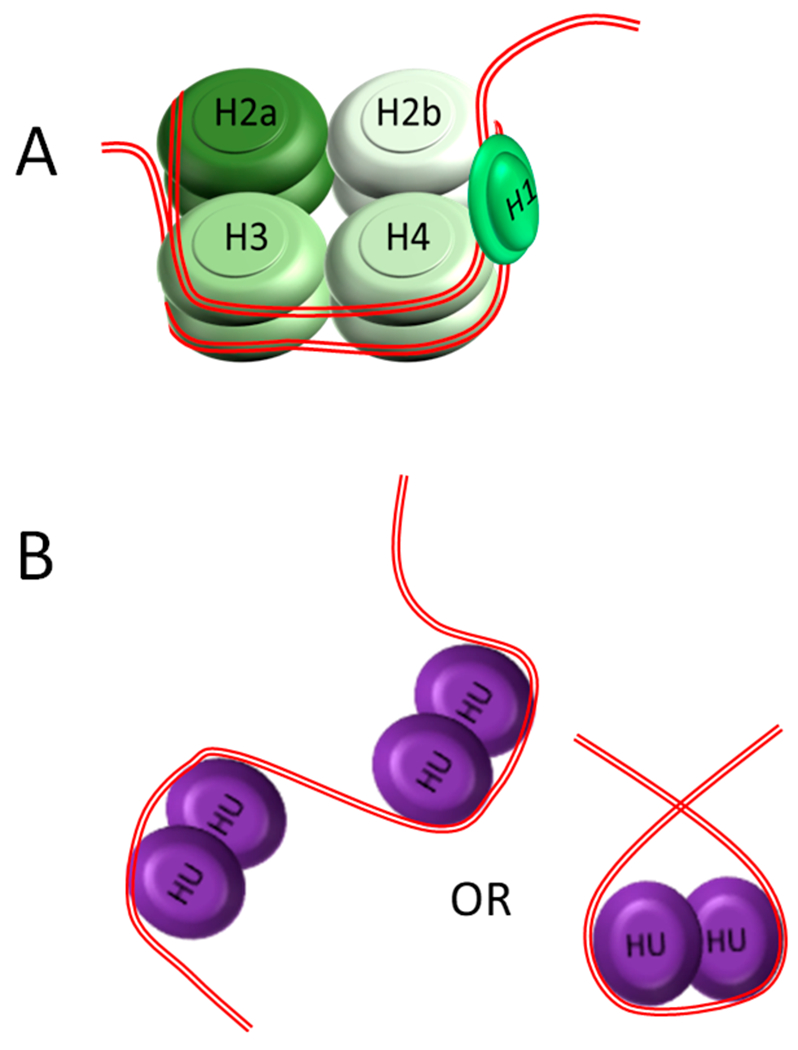
Comparison of DNA organization and condensation in histones and HU orthologues. (A) In the nucleus, the DNA is wrapped approximately two times around a core of histone proteins, composed of two copies of each histone (H2A, H2B, H3, and H4) or a histone variant. Histone H1 serves to hold the wrapped DNA in place and stabilize linker DNA between nucleosomes. The wrapping of DNA around histones at regular intervals condenses and further organizes the chromatin. (B) In bacteria, HU family proteins, either hetero- or homodimers, bind nonspecifically along the chromosome and introduce large bends (105° to > 180°). DNA can also wrap around an HU dimer in a right-handed orientation. These two actions help organize and condense the circular bacterial chromosome analogously to histones.
The presence and combination of these epigenetic modifications are regulatory in nature, and it has been hypothesized that these PTMs influence chromatin structure and transcription. This regulatory process is called the “histone code”. These modifications act as signaling hubs and regulate the interaction of the DNA with histones and other accessory proteins and machines.9 This concept was originally proposed on the basis of observations that specific histone modifications, like serine 10 phosphorylation of H3 (H3S10), played a role in both chromatin decompaction and the opposite function of condensation.10,11 One possible explanation of this phenomenon is that although the S10 phosphorylation mark alone is not sufficient to determine the chromatin state, perhaps it becomes important in the context of the overall PTM landscape. In other words, it may be the case that this mark works in combination with other PTMs on one or more histone tails. Possibly the PTMs have a direct effect on chromatin structure by influencing protein–DNA interactions. Additionally, various combinations of histone PTMs lead to the recruitment of chromatin remodeling or transcription factors, called “reader” proteins, which bind modified histones with specific functional consequences.9,12 For example, the mammalian protein Brd2, which contains two acetyllysine binding domains called bromodomains, binds to acetylated lysine residues (K5, K8, and K12) in histone H4. Binding of the Brd2 reader protein results in the recruitment of the general transcription factor TATA-binding protein (TBP) to the E2F complex, which activates a subset of cell cycle genes.13
Bacterial Nucleoid-Associated Proteins Are Functional Equivalents of Histones
While bacteria contain circular chromosomes that are much smaller than their linear eukaryotic counterparts, bacterial chromosomes must still be compacted at least 1000-fold to fit inside the nucleoid region.14 Negative supercoiling accounts for one level of chromosome compaction, and the supercoils are further organized into loop domains that are topologically isolated from one another.15 The domains are in part defined by DNA interactions with small, basic, nucleoid-associated proteins (NAPs),16 which are essential for chromosome compaction and the coordination of DNA transactions. Bacterial NAPs bind either specifically or nonspecifically at sites distributed throughout the chromosome. They condense and organize the nucleoid by introducing bends and bridging chromosomal loci (as reviewed in ref 17).
The most widely conserved NAPs that are present in all eubacteria are the members of the DNABII family, also called the HU family.18 The DNABII family of proteins consists of orthologues of histone-like protein (HU) and integration host factor (IHF). These proteins are structurally and functionally related, with the distinction that only IHF binds with sequence specificity.19 This review focuses on the nonspecific DNA binding proteins, the HU orthologues. This family of proteins has been extensively studied in Escherichia coli. E. coli HU consists of two subunits, α and β, and exists mostly as a heterodimeric species.20–24 It bends and condenses DNA into a fiber (Figure 1B)25,26 and binds preferentially to single-stranded (ss) forks and 3′ overhangs or to certain double-stranded (ds) DNA structures (e.g., forks, kinks, junctions, or bends) as well as to RNA.27–32 At least one function of HU is to form and constrain negative supercoils,33 which leads to wrapping of DNA in a right-handed orientation for compaction.26,33–35 HU has a similar amino acid content as eukaryotic histone H2B, and despite the lack of sequence or structural homology, this family of proteins are considered to be functional homologues of eukaryotic histones.36 Like histones, HU regulates several DNA transactions such as replication initiation,37–39 inversion,40 bacteriophage Mu transposition,41,42 and DNA repair.43,44 Similarly to eukaryotic histones, there is evidence that HU regulates gene expression. Deletion of both subunits of HU affects the transcription of ~1500 genes, including those involved in translation and stress responses, implicating this protein in global transcriptional control.45–47 However, E. coli HU mutants exhibit pleiotropic defects, leaving open the possibility that some of these transcriptional changes are not due to the absence of HU per se but are secondary consequences of the mutant phenotype, such as slow growth, prolonged lag phase, or defective cell division. Thus, the role of HU in gene expression is not completely understood.
With the constant improvements of mass-spectrometry-based proteomics, many bacterial PTMs, with lysine acetylation the most extensively characterized,48–52 have now been cataloged on the global scale. Examination of these data sets led to the observation that many of the HU orthologues are post-translationally modified. In this review, the evolutionary conservation of Nε-lysine acetylation of HU orthologues is explored. Furthermore, the functional significance of these modifications and their possible enzymatic regulation are addressed. Finally, the discovery of other PTMs on HU proteins is discussed, leading to the exciting possibility of the existence of a histone-like code in bacteria.
Nε-LYSINE ACETYLATION OF HU PROTEINS IS AN EVOLUTIONARILY CONSERVED PHENOMENON
Prior to the year 2000, only a few examples of acetylated bacterial proteins were known, and Nε-lysine acetylation was thought to be rare in bacteria.48 Around 2009, the first large-scale mass-spectrometry-based proteomic characterizations of a bacterial acetylome were performed in E. coli.50,51With the rapid advances in mass spectrometry technology and improvements in reagents, such as anti-acetyllysine antibodies, the lists of acetylated proteins have expanded considerably. Indeed, the bacterial acetylomes of more than 30 different species are now catalogued.48,49 It is apparent that lysine acetylation is far more widespread and perhaps more important than previously thought. So then, what is the functional significance of these thousands of acetylation sites on hundreds of proteins? This open-ended question is ever-present in the field, as the significance of the vast majority of these modifications remains unclear.
With the daunting task ahead to understand the biological relevance of bacterial protein acetylation, specific examples to date have focused largely on proteins involved in important functions like virulence, transcription, translation, and central carbon metabolism.48,49,53 From acetylome studies in Bacillus subtilis,54 Mycobacterium tuberculosis,55 and Acinetobacter baumannii,56 it was realized that the histone-like HU family proteins are acetylated. This conserved feature of distant species prompted a deeper examination of the previously published acetylome data (Table 1). HU orthologues were found to contain at least one acetylation site in 21 different species. Despite the fact that some published bacterial acetylomes were performed in species where the genome was not completely annotated, HU acetylation nonetheless showed broad distribution in the phyla Proteobacteria, Deinococcales, Cyanobacteria, Firmicutes, and Actinobacteria. These acetylome analyses were performed using different growth conditions, lysis conditions, acetyllysine antibodies for enrichment, proteomics workflows, and mass spectrometry instrumentation. With this lack of uniformity across studies, it is possible that the acetylation of HU family proteins is actually even more widespread and universally conserved. From these 21 organisms, the most well conserved acetylation sites are K3, K18, and K86, which are also the three most highly conserved lysine residues (Figure 2). The high sequence conservation of the HU family, specifically the positively charged residues, implies that these modifications occur in important functional domains, likely those that make contact with the DNA backbone. There are three regions in which many acetylation sites lie. The first is within the N-terminal 20 amino acids, in which almost every species has at least one site (Figure 2). The next hot spot is in the middle of the protein, centered on amino acid 40, which is less conserved. Finally, the C-terminal 25 amino acids are also enriched for acetylation sites. If these acetylation sites occur in functional domains for DNA binding, acetylation of these residues would partially negate the positive charge of the lysine side chain and could drastically alter the HU–DNA binding properties. To date, the available evidence supports this idea, as discussed below.56–58
Table 1.
Lysine Acetylation Observed in HU Orthologues
| bacterial species and protein | identified acetylation site(s) |
|---|---|
| Acinetobacter baumannii122 | |
| HU-β | K3, K13, K23, K64, K72 |
| Acinetobacter baumannii SK1756 | |
| HU-β | K13 |
| Aeromonas hydrophila98 | |
| HU-α | K91 |
| HU-β | K3, K9, K18, K75, K80 |
| Bacillus amyloliquefaciens123 | |
| Hbs | K3, K37, K86 |
| Bacillus subtilis75 | |
| HBsu | K18 |
| Bacillus subtilis54 | |
| HBsu | K3, K18, K37, K41, K75, K80, K86 |
| Bacillus subtilis76 | |
| HBsu | K18, K19, K23, K80 |
| Clostridium acetobutylicum124 | |
| HU | K38 |
| Escherichia coli125 | |
| HU-α | K13 |
| Escherichia coli126 | |
| HU-β | K67 or K75, K83 or K86a |
| Escherichia coli127 | |
| HU-α | K22, K51, K86 |
| HU-β | K9, K18, K86 |
| Escherichia coli128 | |
| HU-α | K3, K13, K18, K22 |
| HU-β | K9, K18, K67 |
| Francisella novicida129 | |
| HU-β | K37, K53 |
| Geobacillus kaustophilus130 | |
| Hbs | K3, K18, K23, K41 |
| Mycobacterium tuberculosis55 | |
| HU-β | K13, K86 |
| Pseudomonas aeruginosa131 | |
| HU | K3, K18, K67 |
| Pseudomonas aeruginosa132 | |
| HU | K80 |
| Saccharopolyspora erythraea133 | |
| SACE_6142 (HU) | K51, K85 |
| Spiroplasma eriocheiris134 | |
| HupA | K18, K31, K38, K70, K75, K86 |
| Staphylococcus aureus135 | |
| HU | K13 |
| Streptomyces roseosporus136 | |
| HU-β | K13, K86 |
| Sulfurospirillum halorespirans137 | |
| HU-β | one siteb |
| Synechococcus sp. PCC 7002138 | |
| HU | K68, K86 |
| Synechocystis sp. PCC 139 | |
| HU | K3, K13, K15, K24 |
| Thermus thermophilus140 | |
| HU | K37 |
| Vibrio alginolyticus141 | |
| HU-α | K3, K18, K22, K37, K51, K64, K83 |
| HU-β | K3, K18, K64, K75 |
| Vibrio cholerae142 | |
| HU-α | K18, K64, K70, K83 |
| HU-β | K18, K70, K75, K80 |
| Vibrio parahemolyticus143 | |
| HU-β | K18, K64 |
The identified peptides contained two lysine residues, and the exact location of the acetylation site was not specified.
Specific site location information was not provided.
Figure 2.
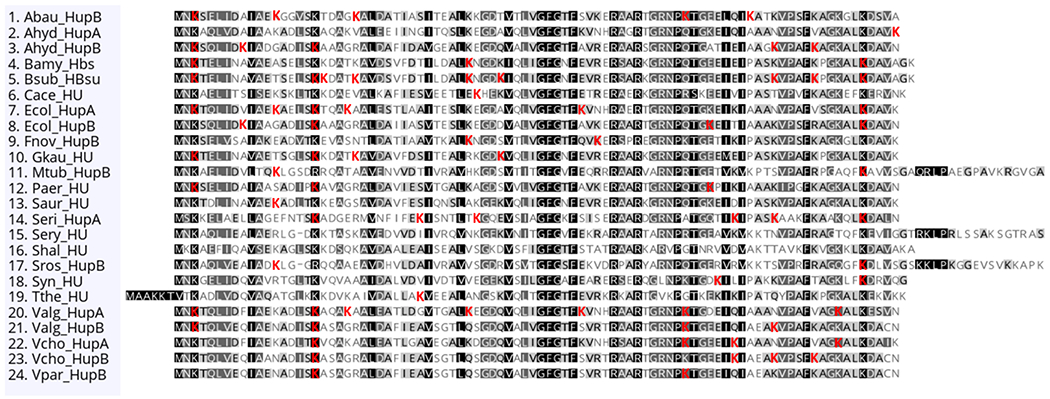
Evolutionary conservation of lysine acetylation of HU orthologues. HU orthologous sequences were obtained from the NCBI protein database. For some bacterial species, the HU orthologues exist as heterodimers, and the two subunits are denoted as HupA and HupB. The sequence alignment was performed using Geneious Prime, version 2020.1.1; the level of shading indicates the level of conservation, with black the best conserved. The identified acetylation sites from the acetylome studies listed in Table 1 are highlighted in red. The Actinobacteria (Mycobacterium tuberculosis, Saccharopolyspora erythraea, and Streptomyces roseosporus) contain a unique C-terminal extension that is rich in basic amino acids. The entire extension is about 110 amino acids and is not shown. Species included: Acinetobacter baumannii (Abau), Aeromonas hydrophila (Ahyd), Bacillus amyloliquefaciens (Bamy), Bacillus subtilis (Bsub), Clostridium acetobutylicum (Cace), Escherichia coli (Ecol), Francisella novicida (Fnov), Geobacillus kaustophilus (Gkau), M. tuberculosis (Mtub), Pseudomonas aeruginosa (Paer), Staphylococcus aureus (Saur), Spiroplasma eriocheiris (Seri), S. erythraea (Sery), Sulfurospirillum halorespirans (Shal), S. roseosporus (Sros), Synechococcus sp. (Syn), Thermus thermophilus (Tthe), Vibrio alginolyticus (Valg), Vibrio cholerae (Vcho), and Vibrio parahemolyticus (Vpar).
Structural examination of the physical location of these modification sites reveals that many are found in regions predicted to directly contact the DNA. As shown in Figure 3, the top of the HU dimeric structure contains “arms” that create a channel where the DNA molecule rests. This family of proteins introduces large, flexible bends in the DNA molecule of ~105° to >180°, causing the DNA to bend downward and make contacts along the front and back faces of the HU dimer.59 For the E. coli (Figure 3A) and Vibrio alginolyticus (Figure 3B) HupA/HupB heterodimers, most of the modified lysine side chains (red) point toward the DNA backbone, which would be at the center of the HU structure. This implies that acetylation of these residues could affect DNA binding. The same is true of B. subtilis (Figure 3D). In each of these structures, some lysines are oriented to point away from where DNA binding likely occurs, so these sites may mediate other important features like protein–protein interactions. Interestingly, the lysine modifications of HupA from the distantly related, spiral shaped, cell-wall-lacking Spiroplasma eriocheiris (Figure 3C) mostly point away from where the DNA molecule lies. This suggests that although acetylation is broadly conserved, the main regulatory mechanisms by which they function probably vary from species to species.
Figure 3.
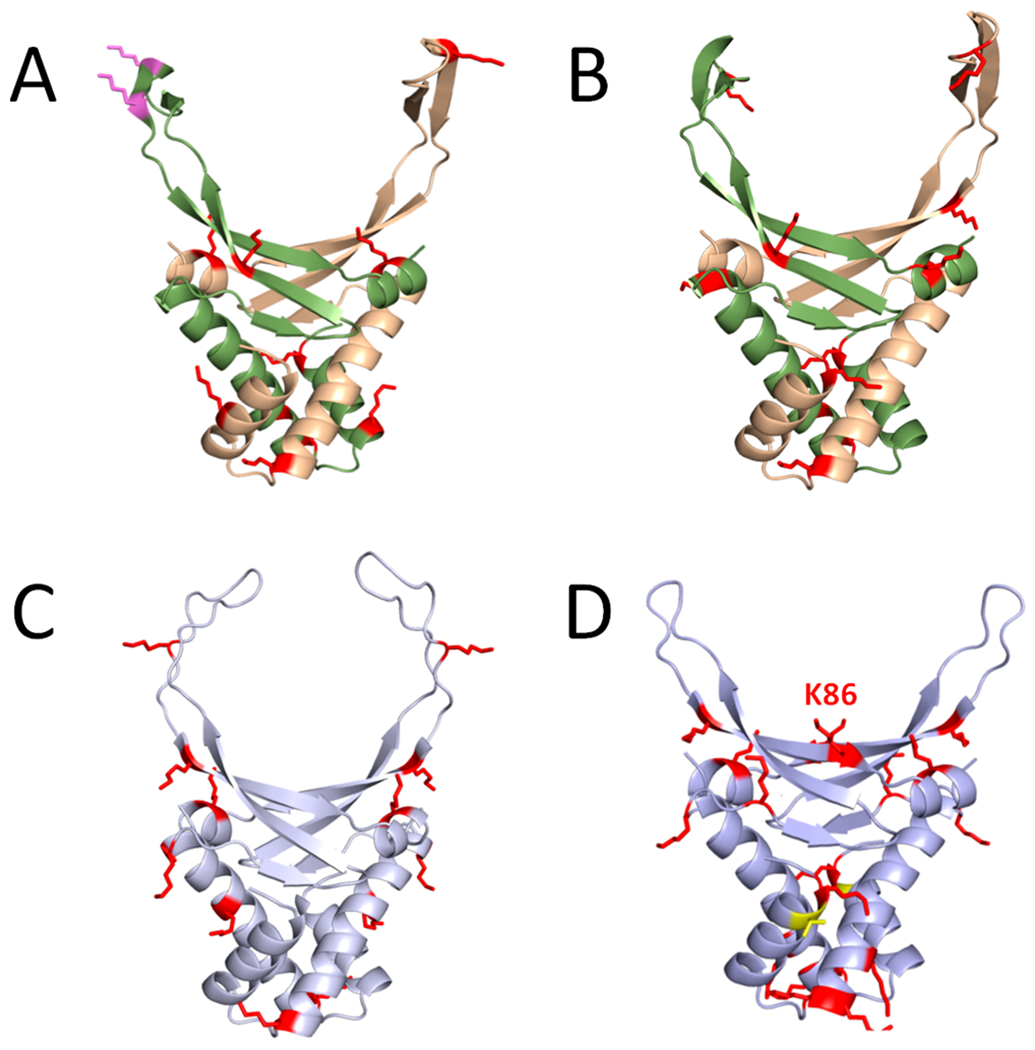
Structural location of PTMs on HU orthologues. Computational models of (A) E. coli HupA–HupB heterodimer, (B) V. alginolyticus HupA–HupB heterodimer, (C) S. eriocheiris HupA homodimer, and (D) B. subtilis HBsu homodimer are shown. For the heterodimers depicted in (A) and (B), the HupA subunit is colored green, and the HupB subunit is colored tan. Both monomers of the homodimers are depicted in (C) and (D). Acetylated residues are shown as red sticks, and phosphorylated residues are shown as yellow sticks; succinylated residues that do not overlap with an acetylation site are shown as pink sticks. The comparative models were generated using Phyre2.120 The Anabaena HU–DNA cocrystal structure (Protein Data Bank code 1P71) was used as the modeling template. Molecular graphics were produced with PyMOL.121
FUNCTIONAL SIGNIFICANCE OF Nε-LYSINE ACETYLATION OF HU PROTEINS
Within the past 5 years, several reports about the functional significance of the acetylation of HU orthologues have been published.56–58,60 Acetylome analysis of A. baumannii revealed that HU was acetylated at a single site, lysine 13.56 In contrast to E. coli, A baumannii HU is encoded by hupB and exists as a homodimer.61 This acetylation site is well-conserved in other bacterial species (Table 1 and Figure 2). HU protein that was homogeneously acetylated at K13 was generated through the use of genetic code expansion using a strain with an evolved tRNA synthetase/tRNA pair for protein purification.62 It was found that lysine acetylation does not alter the oligomeric state but does lower the thermal stability of the protein. Similarly, tetraacetylation of histone H4 lowers the thermal stability of the nucleosome particle.63 Therefore, one plausible explanation for this observation is that acetylation destabilizes the HU dimer. Additionally, the kinetics of HU–DNA binding was altered in that K13-acetylated protein had order of magnitude lower association (kon) and dissociation (koff) rate constants compared with unacetylated protein. The equilibrium constants (KD) of the acetylated and unacetylated forms were similar. It was proposed that lysine 13, which is not predicted to directly contact the DNA, may serve as an epigenetic regulator that controls the availability of the pool of free HU in the cell. In this model, unmodified HU associates with the DNA because of faster kinetics compared with the acetylated form. Furthermore, it is possible that acetylation is required to maintain the HU–DNA complexes because of a lower dissociation rate. While these data suggest a functional role for a specific acetylation site that likely does not contact the DNA, the in vivo significance of these observations was not determined.
The HU orthologues in the phylum Actinobacteria are encoded by a single hupB gene and are composed of two domains. The N-terminal domain is highly homologous to those of other HU orthologues,64 as depicted in Figure 2. The HU orthologues in this phylum also contain a unique C-terminal extension that is rich in basic amino acids.61,64 Large-scale acetylome analysis showed that the M. tuberculosis HU protein was acetylated at two sites, K13 and K86.55 Proteomic analysis of HU alone identified eight total sites, with three present in the conserved N-terminal domain.60 By the use of protein acetylated in vitro, it was discovered that acetylation reduced the DNA binding affinity 10-fold relative to the unacetylated protein. Moreover, microscopic analysis of HU–DNA complexes by TEM and AFM demonstrated that acetylation leads to less compacted DNA and other architectural changes.58,65 Again, the biological significance of these observations remains unknown. However, one previously identified site from both acetylome analysis55 and the previously discussed study58 (K86) has been further evaluated. Lysine 86 is predicted to fall in the region that directly contacts the DNA (Figure 3D). In support of this, in B. subtilis, alanine substitution at K86 resulted in a 70% reduction of the DNA binding activity of the orthologue HBsu.66 When mycobacteria are grown in the presence of a suboptimal concentration of a drug, drug-tolerant variants readily arise.60 Drug tolerance in bacterial populations is defined as the ability to survive a transient exposure to antibiotics at a lethal concentration.67 Clinically, these slower-growing, tolerant variants often lead to treatment failure and relapses. By the use of the model organism Mycobacterium smegmatis, lysine 86 was mutated into an arginine residue, which cannot be modified.60 The HU K86R mutant resulted in specific elimination of the isoniazid-tolerant small colony variant (SCV) subpopulation. These SCV populations have altered gene transcription and metabolic potential. Altogether, these data suggest that acetylation of HU proteins may globally alter transcription, and one possible mechanism of action is by alteration of DNA binding activity through PTMs, analogously to eukaryotic histone modifications.
The acetylation of HBsu from the Gram-positive organism B. subtilis has also been studied. Similar to HU, HBsu binds nonspecifically to curved DNA,68 associates with the nucleoid region, and condenses DNA.69 HBsu is encoded by a single gene and assembles as a homodimer.70 Unlike E. coli HU, HBsu is essential for viability.70,71 To date, HBsu has been implicated in recombination72–74 and DNA repair.73 HBsu was identified as acetylated in three different acetylome analyses.54,75,76 The functional significance of seven of those sites was assessed with regard to nucleoid compaction.57 In mutants that could not be acetylated (arginine substitutions), the nucleoids were more compacted compared with the wild type. Additionally, the phenotype of acetyl mimic mutations (glutamine substitutions), specifically HBsuK3Q, -K41Q, and -K86Q, had decreased DNA content, suggesting that acetylation of HBsu regulates DNA replication. Interestingly, one of these acetyl site mutants, HBsuK41Q, was shown to have decreased DNA binding affinity. More work is required to determine the specific functions of HBsu in DNA compaction and replication and how they are modulated by acetylation. Taken together, however, these data suggest that acetylation of HBsu at specific lysine residues regulates both DNA compaction and replication, and reduced DNA binding affinity may be one mechanism by which acetylation induces these cellular responses.
ENZYMATIC CONTROL OF HU ACETYLATION
If this possible histone-like code is a regulatory element, there must be a means to add and remove such modifications. In bacteria, acetylation occurs by two mechanisms, nonenzymatic and enzymatic (as reviewed in refs 48, 49 and 53). Non-enzymatic acetylation is prevalent in bacteria and may be responsible for broad, low-level global acetylation.48,49 The local environment surrounding a lysine residue influences its nucleophilic properties, which provides a mechanism for autocatalysis. The acetyl donor for nonenzymatic acetylation is predominantly the secondary metabolite acetylphosphate. Enzymatic acetylation occurs via the action of lysine acetyltransferases (KATs), whereby acetyl-CoA serves as the acetyl donor. In eukaryotes, there are five classes of KATs, but only Gcn5-N-acetyltransferase (GNAT) is broadly conserved in all domains of life. The evidence to date suggests that HU family proteins are acetylated enzymatically.57,58 A mass-spectrometry-based interactome analysis performed with M. tuberculosis HU protein identified an interaction with a protein called enhanced intracellular survival (Eis).58 Eis is a GNAT that was known to be involved in acetylation of aminoglycoside antibiotics.77 By the use of purified proteins, it was established that Eis directly acetylates HU in vitro78 and is capable of acetylating 32 sites when present in excess. Moreover, the Michaelis constant (Km) was ~65 times lower for HU than for kanamycin. Finally, confocal microscopy analysis of strains overproducing Eis showed nucleoid decompaction, consistent with a role for acetylation in the regulation of nucleoid compaction.57 These data strongly support a role for Eis as a bona fide KAT for HU.
In B. subtilis, about 50 deletion mutants of putative acetyltransferases were screened for nucleoid compaction phenotypes.57 Five KATs were identified whose deletion resulted in a compacted nucleoid, similar to the arginine-substitution phenotype observed for HBsu. Through genetic bypass experiments, it was determined that two KATs acetylate HBsu at specific sites. One enzyme, YfmK, was shown to directly acetylate HBsu in vitro using purified proteins. Moreover, acetylation at K80 was shown to be significantly reduced in a yfmK mutant compared with the wild type using targeted mass spectrometry, which allows selective and specific quantification. Thus, as with Eis in M. tuberculosis, a KAT directly acetylates a B. subtilis HU family protein, and it is likely that this holds true in other bacteria. In eukaryotes, the reversal of lysine acetylation is catalyzed by the NAD+-dependent sirtuins and the Zn+-dependent lysine deacetylases (KDACs).79 The sirtuins are widely distributed in an evolutionary context and are prevalent in bacteria.48 KDAC orthologues are known to exist in only a few species.80,81 Recently, in M. tuberculosis, the sirtuin orthologue Rv1151c was demonstrated to interact with HU in vivo and deacetylate HU in vitro.65,78 The enzymes responsible for deacetylation of HU in other bacteria have not yet been identified. It is of course entirely possible that other, novel classes of enzymes exist as well. Future studies will be required to address this possibility and determine the complete mechanism of deacetylation of HU family proteins.
OTHER PTMS DISCOVERED ON HU PROTEINS
As noted above, many other modifications beside acetylation have been discovered that constitute part of the code.4,8 While the biological significance of many of these modifications is unclear, at least seven amino acids have been observed to undergo modification in histones. For example, in addition to acetylation, lysine residues can be modified by methylation (mono-, di-, and tri-), ubiquitination, sumolyation, biotinylation, ADP-ribosylation, and various additional short-chain acylations like succinylation, malonylation, and crotonylation.82 Arginine residues are modified by various methylation states, ADP-ribosylation, and citrullination. Serine, threonine, tyrosine, and histidine can be phosphorylated.4 These modifications are mostly involved in gene regulation, chromatin formation, and DNA replication and repair.
Over the past decade, numerous PTMs were discovered and characterized in bacteria,83 the most common of which include protein phosphorylation (Arg, Ser, Thr, Tyr), lysine acetylation and succinylation, glycosylation, and lipidation. Arginine phosphorylation is a relatively newly discovered PTM in some Gram-positive bacteria84,85 that may regulate important components of stress response systems.86,87 Arginine phosphorylation is believed to target proteins for degradation to the Clp protease, possibly serving as a protein quality control mechanism.88 Another PTM that signals for degradation is the prokaryotic ubiquitin-like protein (Pup).89 This modification is known to exist mostly in Actinobacteria, where it is a functional homologue of eukaryotic ubiquitin, targeting proteins to the proteasomal degradation machinery.
Phosphorylation and lysine acylation are the most common and best understood regulatory modifications in bacteria. As with lysine acetylation, large-scale mass-spectrometry-based “omics” studies in various bacteria have recently been published.90,91 If a “histone-like” code exists, it is probable that other modifications may be present among HU orthologues and constitute part of this code. HBsu in B. subtilis contains a single arginine phosphorylation site (R61).84 The significance of this modification is unclear, but two possibilities are likely. First, phosphorylation of R61 may regulate protein stability or turnover and serve in protein quality control. However, R61 does lie within the arms of the structure66 and thus may directly contact the DNA. In fact, mutation of this residue to leucine resulted in a 70% loss of DNA binding activity,66 suggesting that it is important for DNA binding. The effects of a charge reversal mutant were not investigated, as modified protein may retain even less DNA binding activity because of repulsive electrostatic interactions of the phosphoryl group with the DNA backbone.92,93
Examination of the phosphoproteomes from 24 bacterial species revealed that this PTM occurs in ~40% of HU orthologues (Table 2).91 The distribution and evolutionary conservation do not appear to be as widespread as lysine acetylation, but these studies are limited by the same caveats as described before, including lack of uniformity in study design and incomplete genome annotations. Additionally, TiO2 affinity chromatography, which is widely used for enrichment of phosphopeptides in eukaryotes,94 has not been optimized for use with bacterial samples. In bacteria there are far fewer STY phosphorylations than observed in eukaryotes,91 so this optimization is essential in order to broaden and expand the identification of these modifications. Nonetheless, in the diverse bacteria where STY phosphorylation of HU has been observed, there is typically a single phosphorylation site, and it occurs within the first 20 amino acids of the protein (Table 2). Structural examination of the T4 location in HBsu in B. subtilis (Figure 3D, yellow) shows that this modification likely occurs in a region of the protein that directly contacts the DNA. This suggests that modifications located near this region may be important for relaxation of compacted DNA, as phosphorylation introduces a large structural disturbance. As there is a 20 amino acid region where these modifications tend to occur, some of them may point in the other direction, away from bound DNA. In these cases, phosphorylation may be important for mediating protein–protein interactions and may recruit other transcription factors or modifying enzymes, analogously to histone modifications in eukaryotes. Further functional studies involving protein phosphorylation of HU orthologues must be performed to test these predictions.
Table 2.
Phosphorylation and Succinylation of HU Orthologues
| STY Phosphorylation | |
|---|---|
| Acinetobacter baumannii144 | S4 |
| Bacillus subtilis145 | T4 |
| Listeria monocytogenes146,147 | T5, S18 |
| Rhodopseudomonas palustris148 | S13 |
| Sinorhizobium meliloti149 | S8, S10 |
| Staphylococcus aureus85 | T4 |
| Streptomyces coelicolor150 | S4, T44 |
| Synechococcus sp.151 | T6, T74 |
| Synechocystis sp.152 | S21 |
| R Phosphorylation | |
| Bacillus subtilis84 | R61 |
| K Succinylation | |
| Aeromonas hydrophila153 | K3, K75, K80, K83 |
| Escherichia coli154 | |
| HU-α | K3, K13, K18, K22, K67, K70, K86 |
| HU-β | K9, K18, K67 |
| Porphyromonas gingivalis155 | K3 |
| Pseudomonas aeruginosa132 | K18, K61 |
| Mycobacterium tuberculosis156 | K70, K72, K103 |
| Synechococcus sp.157 | K3, K68, K86 |
| Vibrio parahemolyticus97 | K3, K13, K22, K64, K83 |
It has become appreciated that lysine acyl modifications are widespread and prevalent in bacteria.95 Numerous large-scale studies have recently been published, with lysine succinylation being the most well studied and understood modification after acetylation. Succinylation utilizes the metabolite succinyl-CoA as a donor, and like acetylation, it is a reversible process.96 Succinylation is drastically different than acetylation, as it is bulkier (100 Da vs 42 Da for acetylation) and thus has a larger impact on protein structure and function. Acetylation neutralizes the positive charge of the lysine residue, while succinylation results in a charge reversal (+1 to −1) due to the presence of a carboxylate group. Again, this may have large functional consequences for the modified protein. To date, HU succinylation has been observed in seven different bacterial species (Table 2), although the biological significance of these modifications has not been assessed. It is interesting to note that many of the succinylation sites overlap with the acetylation sites (Table 1), which has been previously noted.95,97,98 In fact, for the E. coli heterodimer (Figure 3A), all but two lysine residues (pink) were sites for both acetylation and succinylation. This interesting observation suggests that there may be crosstalk between these PTMs. It remains to be determined whether different acylations on the same lysine residue have different functional consequences.
PERSPECTIVES
Eukaryotic histones contain long, unstructured N-terminal and C-terminal tails, which are largely absent from bacterial NAPs, with the exception of the C-terminal extensions seen in the Actinobacteria. While the majority of histone PTMs tend to be located among these tails, a substantial number are found within the histone fold domain; these regulate histone–DNA and histone–histone associations and constitute part of the code.99,100 It seems possible that the widespread conservation of bacterial HU PTMs (Figure 2 and Tables 1 and 2) may represent the evolutionary origins of the eukaryotic histone code. While this is an exciting possibility, significant questions still remain.
The first important question is the following: what are the functions of these identified modifications? HU family proteins are known to play roles in the regulation of various DNA transactions in addition to nucleoid compaction. Determining how these modifications impact those other roles is essential to our understanding of the language of the code. Another important issue concerns the extent to which modifications are conserved. In other words, is the potential code universal, or do different phyla or species have different languages? It is becoming clearer that other PTMs are waiting to be characterized in bacteria. More recently, large-scale proteomics analyses of other PTMs have become available, such as propionylation101,102 and malonylation.103 The Thermus thermophilus HU is propionylated at K23, and the E. coli HupA and HupB are malonylated at K18 and K70 and at K6, respectively. As enrichment strategies and mass spectrometry workflows continue to improve, the landscape of PTMs on HU family proteins will undoubtedly continue to expand. With new modifications continually being discovered, it will be interesting to move away from characterizing single PTMs and determine how combinations of these modifications function together to regulate protein activity. Mass-spectrometry-based techniques are currently available to identify and quantify multiple different PTMs on histones following nuclei isolation and acid extraction.104 For this reason, the dynamic interplay of crosstalk between these modifications on eukaryotic histones is already being examined.105,106 As HU proteins are highly abundant, methods to extract native HU proteins without affinity enrichment have been described,107,108 but mass spectrometry workflows need to be developed and optimized in order to study multiple PTMs.
In eukaryotes, enzymes that add histone PTMs are called “writers”, and those that remove them are called “erasers”. As we learn more about the complete landscape of HU PTMs, the enzymatic mechanisms by which these modifications are regulated will need to be addressed. It is also important to determine which proteins “read” or decipher the code within the bacterial cell. In eukaryotes, there are specific reader proteins that have domains that bind to specific modified amino acids. For example, proteins containing the bromodomain bind to acetylated lysine residues.109 This binding typically enables recruitment of other important transcription factors or chromatin remodeling factors and thus regulates gene expression.110 To date, no orthologues or functional homologues of the bromodomain have been discovered in bacteria, which remains an important task going forward toward understanding how this histone-like code affects cellular physiology.
The role of protein acetylation in bacterial virulence and drug resistance is beginning to be elucidated.111–115 With proper study, targeting the enzymes involved in regulation (i.e., KATs, sirtuins, and KDACs) or the acetylated form of a key protein (i.e., virulence factors, essential genes, etc.) may allow for the design of new antimicrobials. Indeed, altering acetylation (acetyltransferase and deacetylase inhibitors) has been a promising therapy for the treatment of some cancers116,117 and latent viral infections.118 Additionally, it was recently determined that treatment of mycobacteria with the antibiotic fusidic acid followed by treatment with the Eis inhibitor 1a* (a KAT inhibitor) was more efficient at killing the bacteria than with antibiotics alone.119 This suggests that targeting protein acetylation of HU family proteins may be a therapeutic strategy worth further exploration. Our knowledge of this histone-like code is far from complete, and additional PTMs that constitute it are continually being revealed. More PTM writers, erasers, and readers are waiting to be discovered, and similar to the enzymes involved in acetylation, all of them could represent promising new drug targets.
ACKNOWLEDGMENTS
This work was supported by Grant GM138303. I thank A. Khataokar for generating the HU structural models and D. Dubnau, T. Greco, and A. Khataokar for helpful discussions and critical reading of the manuscript.
ABBREVIATIONS
- AFM
atomic force microscopy
- GNAT
Gcn5-N-acetyltransferase
- KAT
lysine acetyltransferase
- KDAC
lysine deacetylase
- NAP
nucleoid-associated protein
- PTM
post-translational modification
- SCV
small colony variant
- TEM
transmission electron microscopy
Footnotes
The author declares no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.0c00442
REFERENCES
- (1).Kornberg RD; Lorch Y Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98 (3), 285–94. [DOI] [PubMed] [Google Scholar]
- (2).Narlikar GJ; Sundaramoorthy R; Owen-Hughes T Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154 (3), 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bannister AJ; Kouzarides T Regulation of chromatin by histone modifications. Cell Res. 2011, 21 (3), 381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rothbart SB; Strahl BD Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta, Gene Regul. Mech 2014, 1839 (8), 627–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Allfrey VG; Faulkner R; Mirsky AE Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A 1964, 51 (5), 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gutierrez RM; Hnilica LS Tissue specificity of histone phosphorylation. Science 1967, 157 (3794), 1324–5. [DOI] [PubMed] [Google Scholar]
- (7).Janssen KA; Sidoli S; Garcia BA Recent Achievements in characterizing the histone code and approaches to integrating epigenomics and systems biology. Methods Enzymol 2017, 586, 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sadakierska-Chudy A; Filip M A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotoxic. Res 2015, 27 (2), 172–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Strahl BD; Allis CD The language of covalent histone modifications. Nature 2000, 403 (6765), 41–5. [DOI] [PubMed] [Google Scholar]
- (10).Koshland D; Strunnikov A Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol 1996, 12, 305–33. [DOI] [PubMed] [Google Scholar]
- (11).Prigent C; Dimitrov S Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci 2003, 116 (18), 3677–3685. [DOI] [PubMed] [Google Scholar]
- (12).Patel DJ; Wang Z Readout of epigenetic modifications. Annu. Rev. Biochem 2013, 82, 81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Josling GA; Selvarajah SA; Petter M; Duffy MF The role of bromodomain proteins in regulating gene expression. Genes 2012, 3 (2), 320–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Trun NJ; Marko JF Architecture of a bacterial chromosome. ASM News 1998, 64 (5), 276–283. [Google Scholar]
- (15).Worcel A; Burgi E On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol 1972, 71, 127–147. [DOI] [PubMed] [Google Scholar]
- (16).Dillon SC; Dorman CJ Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol 2010, 8 (3), 185–195. [DOI] [PubMed] [Google Scholar]
- (17).Wang XD; Llopis PM; Rudner DZ Organization and segregation of bacterial chromosomes. Nat. Rev. Genet 2013, 14 (3), 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ohniwa RL; Ushijima Y; Saito S; Morikawa K Proteomic Analyses of Nucleoid-Associated Proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One 2011, 6 (4), No. e19172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Craig NL; Nash HA E. coli integration host factor binds to specific sites in DNA. Cell 1984, 39 (3), 707–716. [DOI] [PubMed] [Google Scholar]
- (20).Losso MA; Pawlik RT; Canonaco MA; Gualerzi CO Proteins from the Prokaryotic Nucleoid - a Protein-Protein Cross-Linking Study on the Quaternary Structure of Escherichia coli DNA-Binding Protein Ns (Hu). Eur. J. Biochem 1986, 155 (1), 27–32. [DOI] [PubMed] [Google Scholar]
- (21).Kano Y; Yoshino S; Wada M; Yokoyama K; Nobuhara M; Imamoto F Molecular cloning and nucleotide-sequence of the HU-1 gene of Eschericia coli. Mol. Gen. Genet 1985, 201 (2), 360–362. [DOI] [PubMed] [Google Scholar]
- (22).Claret L; RouviereYaniv J Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J. Mol. Biol 1997, 273 (1), 93–104. [DOI] [PubMed] [Google Scholar]
- (23).Rouviereyaniv J; Kjeldgaard NO Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979, 106 (2), 297–300. [DOI] [PubMed] [Google Scholar]
- (24).Kano Y; Yoshino S; Wada M; Yokoyama K; Nobuhara M; Imamoto F Molecular cloning and nucleotide sequence of the HU-1 gene of Escherichia coli. Mol. Gen. Genet 1985, 201 (2), 360–2. [DOI] [PubMed] [Google Scholar]
- (25).van Noort J; Verbrugge S; Goosen N; Dekker C; Dame RT Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (18), 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kar S; Choi EJ; Guo F; Dimitriadis EK; Kotova SL; Adhya S Right-handed DNA supercoiling by an octameric form of histone-like protein HU - Modulation of cellular transcription. J. Biol. Chem 2006, 281 (52), 40144–40153. [DOI] [PubMed] [Google Scholar]
- (27).Kamashev D; Balandina A; Mazur AK; Arimondo PB; Rouviere-Yaniv J HU binds and folds single-stranded DNA. Nucleic Acids Res 2007, 36 (3), 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Pontiggia A; Negri A; Beltrame M; Bianchi ME Protein HU Binds Specifically to Kinked DNA. Mol. Microbiol 1993, 7 (3), 343–350. [DOI] [PubMed] [Google Scholar]
- (29).Balandina A; Claret L; Hengge-Aronis R; Rouviere-Yaniv J The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol 2001, 39 (4), 1069–1079. [DOI] [PubMed] [Google Scholar]
- (30).Balandina A; Kamashev D; Rouviere-Yaniv J The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids - DNA, RNA, and their hybrids. J. Biol. Chem 2002, 277 (31), 27622–27628. [DOI] [PubMed] [Google Scholar]
- (31).Kamashev D; Rouviere-Yaniv J The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000, 19 (23), 6527–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kamashev D; Balandina A; Rouviere-Yaniv J The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999, 18 (19), 5434–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tanaka H; Yasuzawa K; Kohno K; Goshima N; Kano Y; Saiki T; Imamoto F Role of HU proteins in forming and constraining supercoils of chromosomal DNA in Escherichia coli. Mol. Gen. Genet 1995, 248 (5), 518–526. [DOI] [PubMed] [Google Scholar]
- (34).Malik M; Bensaid A; RouviereYaniv J; Drlica K Histone-like protein HU and bacterial DNA topology: Suppression of an HU deficiency by gyrase mutations. J. Mol. Biol 1996, 256 (1), 66–76. [DOI] [PubMed] [Google Scholar]
- (35).Broyles SS; Pettijohn DE Interaction of the Escherichia coli HU protein with DNA - Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol 1986, 187 (1), 47–60. [DOI] [PubMed] [Google Scholar]
- (36).Rouviere-Yaniv J; Gros F Characterization of a novel, low molecular weight DNA-binding protein from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1975, 72, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jaffe A; Vinella D; D’Ari R The Escherichia coli histone-like protein HU affects DNA initiation, chromosome partitioning via MukB, and cell division via MinCDE. J. Bacteriol 1997, 179 (11), 3494–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Dixon NE; Kornberg A Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1984, 81 (2), 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hwang DS; Kornberg A Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem 1992, 267 (32), 23083–23086. [PubMed] [Google Scholar]
- (40).Wada M; Kutsukake K; Komano T; Imamoto F; Kano Y Participation of the Hup Gene-Product in Site-Specific DNA Inversion in Escherichia coli. Gene 1989, 76 (2), 345–352. [DOI] [PubMed] [Google Scholar]
- (41).Kano Y; Goshima N; Wada M; Imamoto F Participation of Hup Gene Product in Replicative Transposition of Mu Phage in Escherichia coli. Gene 1989, 76 (2), 353–358. [DOI] [PubMed] [Google Scholar]
- (42).Huisman O; Faelen M; Girard D; Jaffe A; Toussaint A; Rouviereyaniv J Multiple Defects in Escherichia coli Mutants Lacking HU Protein. J. Bacteriol 1989, 171 (7), 3704–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Castaing B; Zelwer C; Laval J; Boiteux S HU protein of Escherichia coli binds specifically to DNA that contains single strand breaks or gaps. J. Biol. Chem 1995, 270 (17), 10291–10296. [DOI] [PubMed] [Google Scholar]
- (44).Miyabe I; Zhang QM; Kano Y; Yonei S Histone-like protein HU is required for recA gene-dependent DNA repair and SOS induction pathways in UV-irradiated Escherichia coli. Int. J. Radiat. Biol 2000, 76 (1), 43–49. [DOI] [PubMed] [Google Scholar]
- (45).Prieto AI; Kahramanoglou C; Ali RM; Fraser GM; Seshasayee ASN; Luscombe NM Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012, 40 (8), 3524–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Oberto J; Nabti S; Jooste V; Mignot H; Rouviere-Yaniv J The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 2009, 4 (2), No. e4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Berger M; Farcas A; Geertz M; Zhelyazkova P; Brix K; Travers A; Muskhelishvili G Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010, 11 (1), 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Carabetta VJ; Cristea IM Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol 2017, 199 (16), No. e00107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Christensen DG; Baumgartner JT; Xie X; Jew KM; Basisty N; Schilling B; Kuhn ML; Wolfe AJ Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 2019, 10 (2), No. e02708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhang J; Sprung R; Pei J; Tan X; Kim S; Zhu H; Liu CF; Grishin NV; Zhao Y Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 2009, 8 (2), 215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yu BJ; Kim JA; Moon JH; Ryu SE; Pan J-G The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol 2008, 18 (9), 1529–36. [PubMed] [Google Scholar]
- (52).Kim GW; Yang XJ Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci 2011, 36 (4), 211–20. [DOI] [PubMed] [Google Scholar]
- (53).VanDrisse CM; Escalante-Semerena JC Protein acetylation in bacteria. Annu. Rev. Microbiol 2019, 73, 111–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Carabetta VJ; Greco TM; Tanner AW; Cristea IM; Dubnau D Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems 2016, 1 (3), No. e00005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Xie L; Wang X; Zeng J; Zhou M; Duan X; Li Q; Zhang Z; Luo H; Pang L; Li W; Liao G; Yu X; Li Y; Huang H; Xie J Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol 2015, 59, 193–202. [DOI] [PubMed] [Google Scholar]
- (56).Liao J-H; Tsai C-H; Patel SG; Yang J-T; Tu I-F; Lo Cicero M; Lipka-Lloyd M; Wu W-L; Shen W-J; Ho M-R; Chou C-C; Sharma GR; Okanishi H; Luk LYP; Tsai Y-H; Wu S-H Acetylome of Acinetobacter baumannii SK17 reveals a highly-conserved modification of histone-like protein HU. Front. Mol. Biosci 2017, 4, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Carabetta VJ; Greco TM; Cristea IM; Dubnau D YfmK is an N(epsilon)-lysine acetyltransferase that directly acetylates the histone-like protein HBsu in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (9), 3752–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ghosh S; Padmanabhan B; Anand C; Nagaraja V Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol. Microbiol 2016, 100 (4), 577–88. [DOI] [PubMed] [Google Scholar]
- (59).Swinger KK; Rice PA Structure-based analysis of HU–DNA binding. J. Mol. Biol 2007, 365 (4), 1005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Sakatos A; Babunovic GH; Chase MR; Dills A; Leszyk J; Rosebrock T; Bryson B; Fortune SM Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci. Adv 2018, 4 (5), No. eaao1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Grove A Functional evolution of bacterial histone-like HU proteins. Curr. Issues Mol. Biol 2011, 13 (1), 1–12. [PubMed] [Google Scholar]
- (62).Neumann H; Hancock SM; Buning R; Routh A; Chapman L; Somers J; Owen-Hughes T; van Noort J; Rhodes D; Chin JW A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 2009, 36 (1), 153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Siino JS; Yau PM; Imai BS; Gatewood JM; Morton Bradbury E Effect of DNA length and H4 acetylation on the thermal stability of reconstituted nucleosome particles. Biochem. Biophys. Res. Commun 2003, 302 (4), 885–891. [DOI] [PubMed] [Google Scholar]
- (64).Bhowmick T; Ghosh S; Dixit K; Ganesan V; Ramagopal UA; Dey D; Sarma SP; Ramakumar S; Nagaraja V Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors. Nat. Commun 2014, 5, 4124. [DOI] [PubMed] [Google Scholar]
- (65).Anand C; Garg R; Ghosh S; Nagaraja V A Sir2 family protein Rv1151c deacetylates HU to alter its DNA binding mode in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun 2017, 493 (3), 1204–1209. [DOI] [PubMed] [Google Scholar]
- (66).Köhler P; Marahiel MA Mutational analysis of the nucleoid-associated protein HBsu of Bacillus subtilis. Mol. Gen. Genet 1998, 260 (5), 487–91. [DOI] [PubMed] [Google Scholar]
- (67).Brauner A; Fridman O; Gefen O; Balaban NQ Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol 2016, 14 (5), 320–30. [DOI] [PubMed] [Google Scholar]
- (68).Kohler P; Marahiel MA Mutational analysis of the nucleoid-associated protein HBsu of Bacillus subtilis. Mol. Gen. Genet 1998, 260 (5), 487–491. [DOI] [PubMed] [Google Scholar]
- (69).Kohler P; Marahiel MA Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J. Bacteriol 1997, 179 (6), 2060–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Micka B; Marahiel MA The DNA-binding protein HBsu is essential for normal growth and development in Bacillus subtilis. Biochimie 1992, 74 (7–8), 641–650. [DOI] [PubMed] [Google Scholar]
- (71).Micka B; Groch N; Heinemann U; Marahiel MA Molecular-Cloning, Nucleotide-Sequence, and Characterization of the Bacillus subtilis Gene Encoding the DNA-Binding Protein HBsu. J. Bacteriol 1991, 173 (10), 3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Alonso JC; Gutierrez C; Rojo F The role of the chromatin-associated protein HBsu in beta-mediated DNA recombination is to facilitate the joining of distant recombination sites. Mol. Microbiol 1995, 18 (3), 471–478. [DOI] [PubMed] [Google Scholar]
- (73).Fernandez S; Rojo F; Alonso JC The Bacillus subtilis chromatin-associated protein HBsu is involved in DNA repair and recombination. Mol. Microbiol 1997, 23 (6), 1169–1179. [DOI] [PubMed] [Google Scholar]
- (74).Alonso JC; Weise F; Rojo F The Bacillus-Subtilis Histone-Like Protein Hbsu Is Required for DNA Resolution and DNA Inversion Mediated by the Beta-Recombinase of Plasmid Psm19035. J. Biol. Chem 1995, 270 (7), 2938–2945. [DOI] [PubMed] [Google Scholar]
- (75).Gramlich WM; Kim IL; Burdick JA Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiolnorbornene chemistry. Biomaterials 2013, 34 (38), 9803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Kosono S; Tamura M; Suzuki S; Kawamura Y; Yoshida A; Nishiyama M; Yoshida M Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 2015,10 (6), No. e0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Houghton JL; Biswas T; Chen W; Tsodikov OV; Garneau-Tsodikova S Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase Eis. ChemBioChem 2013, 14 (16), 2127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Green KD; Biswas T; Pang AH; Willby MJ; Reed MS; Stuchlik O; Pohl J; Posey JE; Tsodikov OV; Garneau-Tsodikova S Acetylation by Eis and deacetylation by Rv1151c of Mycobacterium tuberculosis HupB: Biochemical and structural insight. Biochemistry 2018, 57 (5), 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Grozinger CM; Schreiber SL Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol 2002, 9 (1), 3–16. [DOI] [PubMed] [Google Scholar]
- (80).Crosby HA; Heiniger EK; Harwood CS; Escalante-Semerena JC Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol 2010, 76 (4), 874–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Gardner JG; Escalante-Semerena JC In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol 2009, 191 (6), 1749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Sabari BR; Zhang D; Allis CD; Zhao Y Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol 2017, 18 (2), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Macek B; Forchhammer K; Hardouin J; Weber-Ban E; Grangeasse C; Mijakovic I Protein post-translational modifications in bacteria. Nat. Rev. Microbiol 2019, 17 (11), 651–664. [DOI] [PubMed] [Google Scholar]
- (84).Elsholz AKW; Turgay K; Michalik S; Hessling B; Gronau K; Oertel D; Mäder U; Bernhardt J; Becher D; Hecker M; Gerth U Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (19), 7451–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Junker S; Maaβ S; Otto A; Michalik S; Morgenroth F; Gerth U; Hecker M; Becher D Spectral library based analysis of arginine phosphorylations in Staphylococcus aureus. Mol. Cell. Proteomics 2018, 17 (2), 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Fuhrmann J; Subramanian V; Kojetin DJ; Thompson PR Activity-based profiling reveals a regulatory link between oxidative stress and protein arginine phosphorylation. Cell Chem. Biol 2016, 23 (8), 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Schmidt A; Trentini DB; Spiess S; Fuhrmann J; Ammerer G; Mechtler K; Clausen T Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response. Mol. Cell. Proteomics 2014, 13 (2), 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Trentini DB; Suskiewicz MJ; Heuck A; Kurzbauer R; Deszcz L; Mechtler K; Clausen T Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature 2016, 539 (7627), 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Pearce MJ; Mintseris J; Ferreyra J; Gygi SP; Darwin KH Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 2008, 322 (5904), 1104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Ge R; Shan W Bacterial phosphoproteomic analysis reveals the correlation between protein phosphorylation and bacterial pathogenicity. Genomics, Proteomics Bioinf. 2011, 9 (4–5), 119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Yagüe P; Gonzalez-Quiñonez N; Fernánez-García G; Alonso-Fernández S; Manteca A Goals and challenges in bacterial phosphoproteomics. Int. J. Mol. Sci 2019, 20 (22), 5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Brehove M; Wang T; North J; Luo Y; Dreher SJ; Shimko JC; Ottesen JJ; Luger K; Poirier MG Histone core phosphorylation regulates DNA accessibility. J. Biol. Chem 2015, 290 (37), 22612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Bannister AJ; Kouzarides T Regulation of chromatin by histone modifications. Cell Res. 2011, 21 (3), 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Thingholm TE; Larsen MR Phosphopeptide enrichment by immobilized metal affinity chromatography. Methods Mol. Biol 2016, 1355, 123–33. [DOI] [PubMed] [Google Scholar]
- (95).Weinert BT; Schölz C; Wagner SA; Iesmantavicius V; Su D; Daniel JA; Choudhary C Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4 (4), 842–851. [DOI] [PubMed] [Google Scholar]
- (96).Alleyn M; Breitzig M; Lockey R; Kolliputi N The dawn of succinylation: a posttranslational modification. Am. J. Physiol.: Cell Physiol 2018, 314 (2), C228–C232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Pan J; Chen R; Li C; Li W; Ye Z Global analysis of protein lysine succinylation profiles and their overlap with lysine acetylation in the marine bacterium Vibrio parahemolyticus. J. Proteome Res 2015, 14 (10), 4309–18. [DOI] [PubMed] [Google Scholar]
- (98).Sun L; Yao Z; Guo Z; Zhang L; Wang Y; Mao R; Lin Y; Fu Y; Lin X Comprehensive analysis of the lysine acetylome in Aeromonas hydrophila reveals cross-talk between lysine acetylation and succinylation in LuxS. Emerging Microbes Infect. 2019, 8 (1), 1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Arnaudo AM; Garcia BA Proteomic characterization of novel histone post-translational modifications. Epigenet. Chromatin 2013, 6 (1), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Tropberger P; Schneider R Going global: novel histone modifications in the globular domain of H3. Epigenetics 2010, 5 (2), 112–7. [DOI] [PubMed] [Google Scholar]
- (101).Okanishi H; Kim K; Masui R; Kuramitsu S Lysine propionylation is a prevalent post-translational modification in Thermus thermophilus. Mol. Cell. Proteomics 2014, 13 (9), 2382–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Okanishi H; Kim K; Masui R; Kuramitsu S Proteome-wide identification of lysine propionylation in thermophilic and mesophilic bacteria: Geobacillus kaustophilus, Thermus thermophilus, Escherichia coli, Bacillus subtilis, and Rhodothermus marinus. Extremophiles 2017, 21 (2), 283–296. [DOI] [PubMed] [Google Scholar]
- (103).Qian L; Nie L; Chen M; Liu P; Zhu J; Zhai L; Tao SC; Cheng Z; Zhao Y; Tan M Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism. J. Proteome Res 2016, 15 (6), 2060–71. [DOI] [PubMed] [Google Scholar]
- (104).Karch KR; Sidoli S; Garcia BA Identification and quantification of histone PTMs using high-resolution mass spectrometry. Methods Enzymol. 2016, 574, 3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Zhang C; Gao S; Molascon AJ; Wang Z; Gorovsky MA; Liu Y; Andrews PC Bioinformatic and proteomic analysis of bulk histones reveals PTM crosstalk and chromatin features. J. Proteome Res 2014, 13 (7), 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Wojcik F; Dann GP; Beh LY; Debelouchina GT; Hofmann R; Muir TW Functional crosstalk between histone H2B ubiquitylation and H2A modifications and variants. Nat. Commun 2018, 9 (1), 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Ghadam P; Samadi R Rapid purification of HU protein from Halobacillus karajensis. Mol. Biol. Res. Commun 2014, 3 (1), 1–8. [PMC free article] [PubMed] [Google Scholar]
- (108).Ghadam P; Mollasalehi M One-step purification of histone-like protein (HU) from Halobacillus litoralis. Acta Biol. (Szeged) 2015, 59 (1), 19–23. [Google Scholar]
- (109).Marmorstein R; Zhou M-M Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor Perspect. Biol 2014, 6 (7), No. a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Musselman CA; Lalonde M-E; Côté J; Kutateladze TG Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol 2012, 19 (12), 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Sakatos A; Babunovic GH; Chase MR; Dills A; Leszyk J; Rosebrock T; Bryson B; Fortune SM Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci. Adv 2018, 4 (5), No. eaao1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Gaviard C; Cosette P; Jouenne T; Hardouin J LasB and CbpD Virulence Factors of Pseudomonas aeruginosa Carry Multiple Post-Translational Modifications on Their Lysine Residues. J. Proteome Res 2019, 18 (3), 923–933. [DOI] [PubMed] [Google Scholar]
- (113).Ren J; Sang Y; Lu J; Yao YF Protein acetylation and its role in bacterial virulence. Trends Microbiol. 2017, 25 (9), 768–779. [DOI] [PubMed] [Google Scholar]
- (114).Ren J; Sang Y; Tan Y; Tao J; Ni J; Liu S; Fan X; Zhao W; Lu J; Wu W; Yao YF Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 2016, 12 (3), No. e1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Sang Y; Ren J; Ni J; Tao J; Lu J; Yao YF Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J. Infect. Dis 2016, 213 (11), 1836–45. [DOI] [PubMed] [Google Scholar]
- (116).West AC; Johnstone RW New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest 2014, 124 (1), 30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Gajer JM; Furdas SD; Grunder A; Gothwal M; Heinicke U; Keller K; Colland F; Fulda S; Pahl HL; Fichtner I; Sippl W; Jung M Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis 2015, 4, No. e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Herbein G; Wendling D Histone deacetylases in viral infections. Clin. Epigenet 2010, 1 (1–2), 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Scutigliani EM; Scholl ER; Grootemaat AE; Khanal S; Kochan JA; Krawczyk PM; Reits EA; Garzan A; Ngo HX; Green KD; Garneau-Tsodikova S; Ruijter JM; van Veen HA; van der Wel NN Interfering with DNA decondensation as a strategy against Mycobacteria. Front. Microbiol 2018, 9, 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Kelley LA; Mezulis S; Yates CM; Wass MN; Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 2015, 10 (6), 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).PyMOL Molecular Graphics System, ver. 1.8; Schrödinger, LLC: New York, 2015. [Google Scholar]
- (122).Kentache T; Jouenne T; De E; Hardouin J Proteomic characterization of Nα-and Nε-acetylation in Acinetobacter baumannii. J. Proteomics 2016, 144, 148–58. [DOI] [PubMed] [Google Scholar]
- (123).Liu L; Wang G; Song L; Lv B; Liang W Acetylome analysis reveals the involvement of lysine acetylation in biosynthesis of antibiotics in Bacillus amyloliquefaciens. Sci. Rep 2016, 6, 20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Xu JY; Xu Z; Liu X; Tan M; Ye BC Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol. Cell. Proteomics 2018, 17 (6), 1156–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Zhang K; Zheng S; Yang JS; Chen Y; Cheng Z Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res 2013, 12 (2), 844–51. [DOI] [PubMed] [Google Scholar]
- (126).Baeza J; Dowell JA; Smallegan MJ; Fan J; Amador-Noguez D; Khan Z; Denu JM Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem 2014, 289 (31), 21326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Meyer JG; D’Souza AK; Sorensen DJ; Rardin MJ; Wolfe AJ; Gibson BW; Schilling B Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J. Am. Soc. Mass Spectrom 2016, 27 (11), 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Schilling B; Basisty N; Christensen DG; Sorensen D; Orr JS; Wolfe AJ; Rao CV Global lysine acetylation in Escherichia coli results from growth conditions that favor acetate fermentation. J. Bacteriol 2019, 201 (9), No. e00768–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Marakasova E; Ii A; Nelson KT; van Hoek ML Proteome wide profiling of N-epsilon-lysine acetylation reveals a novel mechanism of regulation of the Chitinase activity in Francisella novicida. J. Proteome Res 2020, 19 (4), 1409–1422. [DOI] [PubMed] [Google Scholar]
- (130).Lee DW; Kim D; Lee YJ; Kim JA; Choi JY; Kang S; Pan JG Proteomic analysis of acetylation in thermophilic Geobacillus kaustophilus. Proteomics 2013, 13 (15), 2278–82. [DOI] [PubMed] [Google Scholar]
- (131).Ouidir T; Cosette P; Jouenne T; Hardouin J Proteomic profiling of lysine acetylation in Pseudomonas aeruginosa reveals the diversity of acetylated proteins. Proteomics 2015, 15 (13), 2152–7. [DOI] [PubMed] [Google Scholar]
- (132).Gaviard C; Broutin I; Cosette P; Dé E; Jouenne T; Hardouin J Lysine succinylation and acetylation in Pseudomonas aeruginosa. J. Proteome Res 2018, 17 (7), 2449–2459. [DOI] [PubMed] [Google Scholar]
- (133).Huang D; Li ZH; You D; Zhou Y; Ye BC Lysine acetylproteome analysis suggests its roles in primary and secondary metabolism in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol 2015, 99 (3), 1399–413. [DOI] [PubMed] [Google Scholar]
- (134).Meng Q; Liu P; Wang J; Wang Y; Hou L; Gu W; Wang W Systematic analysis of the lysine acetylome of the pathogenic bacterium Spiroplasma eriocheiris reveals acetylated proteins related to metabolism and helical structure. J. Proteomics 2016, 148, 159–69. [DOI] [PubMed] [Google Scholar]
- (135).Zhang Y; Wu Z; Wan X; Liu P; Zhang J; Ye Y; Zhao Y; Tan M Comprehensive profiling of lysine acetylome in Staphylococcus aureus. Sci. China: Chem 2014, 57 (5), 732–738. [Google Scholar]
- (136).Liao G; Xie L; Li X; Cheng Z; Xie J Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteomics 2014, 106, 260–9. [DOI] [PubMed] [Google Scholar]
- (137).Turkowsky D; Esken J; Goris T; Schubert T; Diekert G; Jehmlich N; von Bergen M A retentive memory of tetrachloroethene respiration in Sulfurospirillum halorespirans - involved proteins and a possible link to acetylation of a two-component regulatory system. J. Proteomics 2018, 181, 36–46. [DOI] [PubMed] [Google Scholar]
- (138).Chen Z; Zhang G; Yang M; Li T; Ge F; Zhao J Lysine acetylome analysis reveals Photosystem II Manganese-stabilizing protein acetylation is involved in negative regulation of oxygen evolution in model Cyanobacterium Synechococcus sp. PCC 7002. Mol. Cell. Proteomics 2017, 16 (7), 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Mo R; Yang M; Chen Z; Cheng Z; Yi X; Li C; He C; Xiong Q; Chen H; Wang Q; Ge F Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J. Proteome Res 2015, 14 (2), 1275–86. [DOI] [PubMed] [Google Scholar]
- (140).Okanishi H; Kim K; Masui R; Kuramitsu S Acetylome with structural mapping reveals the significance of lysine acetylation in Thermus thermophilus. J. Proteome Res 2013, 12 (9), 3952–68. [DOI] [PubMed] [Google Scholar]
- (141).Pang H; Li W; Zhang W; Zhou S; Hoare R; Monaghan SJ; Jian J; Lin X Acetylome profiling of Vibrio alginolyticus reveals its role in bacterial virulence. J. Proteomics 2020, 211, 103543. [DOI] [PubMed] [Google Scholar]
- (142).Jers C; Ravikumar V; Lezyk M; Sultan A; Sjoling A; Wai SN; Mijakovic I The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front. Cell. Infect. Microbiol 2018, 7, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Pan J; Ye Z; Cheng Z; Peng X; Wen L; Zhao F Systematic analysis of the lysine acetylome in Vibrio parahemolyticus. J. Proteome Res 2014, 13 (7), 3294–302. [DOI] [PubMed] [Google Scholar]
- (144).Lai JH; Yang JT; Chern J; Chen TL; Wu WL; Liao JH; Tsai SF; Liang SY; Chou CC; Wu SH Comparative phosphoproteomics reveals the role of AmpC β-lactamase phosphorylation in the clinical imipenem-resistant strain Acinetobacter baumannii SK17. Mol. Cell. Proteomics 2016, 15 (1), 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Macek B; Mijakovic I; Olsen JV; Gnad F; Kumar C; Jensen PR; Mann M The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 2007, 6 (4), 697–707. [DOI] [PubMed] [Google Scholar]
- (146).Misra SK; Moussan Désirée Aké F; Wu Z; Milohanic E; Cao TN; Cossart P; Deutscher J; Monnet V; Archambaud C; Henry C Quantitative proteome analyses identify PrfA-responsive proteins and phosphoproteins in Listeria monocytogenes. J. Proteome Res 2014, 13 (12), 6046–57. [DOI] [PubMed] [Google Scholar]
- (147).Misra SK; Milohanic E; Aké F; Mijakovic I; Deutscher J; Monnet V; Henry C Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 2011, 11 (21), 4155–65. [DOI] [PubMed] [Google Scholar]
- (148).Hu CW; Lin MH; Huang HC; Ku WC; Yi TH; Tsai CF; Chen YJ; Sugiyama N; Ishihama Y; Juan HF; Wu SH Phosphoproteomic analysis of Rhodopseudomonas palustris reveals the role of pyruvate phosphate dikinase phosphorylation in lipid production. J. Proteome Res 2012, 11 (11), 5362–75. [DOI] [PubMed] [Google Scholar]
- (149).Liu T; Tian CF; Chen WX Site-specific Ser/Thr/Tyr phosphoproteome of Sinorhizobium meliloti at stationary phase. PLoS One 2015, 10 (9), No. e0139143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Manteca A; Ye J; Sánchez J;Jensen ON Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res 2011, 10 (12), 5481–5492. [DOI] [PubMed] [Google Scholar]
- (151).Yang MK; Qiao ZX; Zhang WY; Xiong Q; Zhang J; Li T; Ge F; Zhao JD Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res 2013, 12 (4), 1909–23. [DOI] [PubMed] [Google Scholar]
- (152).Spät P; Maček B; Forchhammer K Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol 2015, 6, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Yao Z; Guo Z; Wang Y; Li W; Fu Y; Lin Y; Lin W; Lin X Integrated succinylome and metabolome profiling reveals crucial role of S-ribosylhomocysteine lyase in quorum sensing and metabolism of Aeromonas hydrophila. Mol. Cell. Proteomics 2019, 18 (2), 200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Colak G; Xie Z; Zhu AY; Dai L; Lu Z; Zhang Y; Wan X; Chen Y; Cha YH; Lin H; Zhao Y; Tan M Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics 2013, 12 (12), 3509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Wu L; Gong T; Zhou X; Zeng J; Huang R; Wu Y; Li Y Global analysis of lysine succinylome in the periodontal pathogen Porphyromonas gingivalis. Mol. Oral Microbiol 2019, 34 (2), 74–83. [DOI] [PubMed] [Google Scholar]
- (156).Yang M; Wang Y; Chen Y; Cheng Z; Gu J; Deng J; Bi L; Chen C; Mo R; Wang X; Ge F Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis. Mol. Cell. Proteomics 2015, 14 (4), 796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Liu X; Yang M; Wang Y; Chen Z; Zhang J; Lin X; Ge F; Zhao J Effects of PSII manganese-stabilizing protein succinylation on photosynthesis in the model cyanobacterium Synechococcus sp. PCC 7002. Plant Cell Physiol. 2018, 59 (7), 1466–1482. [DOI] [PubMed] [Google Scholar]


