Multiple structures of full-length calcium-sensing receptors reveal the mechanism of activation and allosteric modulation.
Abstract
Calcium-sensing receptor (CaSR) is a class C G protein–coupled receptor (GPCR) that plays an important role in calcium homeostasis and parathyroid hormone secretion. Here, we present multiple cryo–electron microscopy structures of full-length CaSR in distinct ligand-bound states. Ligands (Ca2+ and l-tryptophan) bind to the extracellular domain of CaSR and induce large-scale conformational changes, leading to the closure of two heptahelical transmembrane domains (7TMDs) for activation. The positive modulator (evocalcet) and the negative allosteric modulator (NPS-2143) occupy the similar binding pocket in 7TMD. The binding of NPS-2143 causes a considerable rearrangement of two 7TMDs, forming an inactivated TM6/TM6 interface. Moreover, a total of 305 disease-causing missense mutations of CaSR have been mapped to the structure in the active state, creating hotspot maps of five clinical endocrine disorders. Our results provide a structural framework for understanding the activation, allosteric modulation mechanism, and disease therapy for class C GPCRs.
INTRODUCTION
Calcium signaling is necessary for various cellular functions. The calcium-sensing receptor (CaSR) is an essential component for the transduction of calcium signals and maintenance of calcium homeostasis (1, 2). In addition to acting as a second messenger in the process of signal transduction, calcium can also serve as a first messenger to be directly sensed by receptors on the plasma membrane (3). In general, subtle changes in the extracellular Ca2+ concentration can be sensed by CaSR, which is highly expressed in parathyroid glands, and then be precisely regulated by the secretion of parathyroid hormone (PTH) through the reabsorption of Ca2+ from bone, urinary, and intestine (4). Abnormal expression levels and receptor activities of CaSR will cause many diseases, such as familial hypocalciuric hypercalcemia type 1 (FHH1) (5), neonatal severe hyperparathyroidism (NSHPT) (6), autosomal dominant hypocalcemia type 1 (ADH1) (7), and asthma (8).
CaSR belongs to the class C G protein–coupled receptor (GPCR) family and functions as a homodimer tethered by disulfide bonds in the cytoplasmic membrane (9). The single-copy human CaSR gene that maps to 3q13.3-21 encodes 1078 amino acid residues (4, 10). The overall structure of CaSR is presumably similar to other class C GPCRs. In response to different stimuli, the CaSR homodimer can bind to various ligands (11–15) and allosteric modulators (16, 17), and then mediates downstream signaling through G proteins, including Gq, Gi, G12/13, and Gs (18). In addition to endogenous allosteric modulators such as l-aromatic amino acids, CaSR can also be regulated by synthetic allosteric modulators, including positive and negative allosteric modulators (19–21). Evocalcet (4-[(3S)-3-[[(1R)-1-(1-naphthalenyl)ethyl]amino]-1-pyrrolidinyl]) is a positive allosteric modulator widely used in the treatment of secondary hyperparathyroidism (22). Evocalcet was first approved in Japan in 2018 for its potent positive allosteric efficacy and little gastrointestinal-related side effect compared with cinacalcet (23). On the other hand, the synthetic molecule NPS-2143 (2-chloro-6-[(2R)-2-hydroxy-3-[(2-methyl-1-naphthalen-2-ylpropan-2-yl)amino]propoxy]benzonitrile) is a selective and potent CaSR antagonist that can correct the increased signaling pathway associated with gain-of-function CaSR and Gα11 mutations (24, 25). In vivo studies have shown that NPS-2143 notably increases the cytoplasmic concentrations of calcium and PTH in ADH-associated mouse models (16, 26).
To date, the crystal structure of the N-terminal human CaSR in the active and inactive states has been determined (27, 28). However, the lack of intact structural information on CaSR limits the understanding of receptor function, biased signal transduction, and clinical drug development against multiple diseases. In this research, we determined the structures of full-length CaSR from Gallus gallus (ggCaSR) by cryo–electron microscopy (cryo-EM) in several different conformational states, including ggCaSR in complex with l-tryptophan (l-Trp) and Ca2+, representing the active conformational state (ggCaSR-active); ggCaSR in complex with l-Trp, Ca2+, and NPS-2143, representing the negative allosteric modulating state (ggCaSR-NAM); ggCaSR in complex with l-Trp, Ca2+, and evocalcet, representing the positive allosteric modulating state (ggCaSR-PAM); and ggCaSR alone, representing the inactive state (ggCaSR-inactive). These structures provide the framework for understanding the mechanism of allosteric regulation and biased signal transduction for class C GPCRs.
RESULTS
Structure determination
The structural determination of full-length CaSR was conducted using cryo-EM. After screening different CaSR proteins from various species, ggCaSR was selected and purified. The amino acid sequence of ggCaSR is more than 80% identical to that of Homo sapiens CaSR (fig. S1A). We transfected the DNA encoding FLAG-tagged full-length ggCaSR into human embryonic kidney (HEK) 293T cells and carried out an inositol monophosphate (IP1) production assay. Compared with the control, evocalcet showed positive allosteric effects, while NPS-2143 showed negative allosteric effects (fig. S1B), indicating that ggCaSR has a similar biological function to its human counterpart in response to extracellular Ca2+ (fig. S1, B and C). The purified ggCaSR protein showed a molecular weight of approximately 150 kDa as a monomer by SDS–polyacrylamide gel electrophoresis, whereas due to glycosylation, detergent micelles, and molecular shape, a larger molecular weight was observed (approximately 670 kDa) from an analytical size exclusion column (fig. S2A).
Details of the cryo-EM sample preparation, data acquisition and processing, model building, and structure refinement can be found in Materials and Methods and in figs. S2 to S5. The two-dimensional (2D) class average of the particles for the four structures reveals that ggCaSR forms a dimer (figs. S2D, S3B, S4B, and S5B) with approximate C2 symmetry and consists of a transmembrane domain wrapped in detergent micelles and a large extracellular region. Because of the conformational differences of the transmembrane domains between the two protomers (figs. S3H, S4H, and S5H), only C1 symmetry was used in the structural reconstruction of all four structures. The breakdown of C2 symmetry in the ggCaSR homodimer was unexpected because another class C GPCR, the metabotropic glutamate receptor 5 (mGlu5), is twofold symmetric (29). The correlation of the asymmetric ggCaSR homodimer with its biological function may require further investigation.
To further improve the densities of the transmembrane helices of the ggCaSR-active, ggCaSR-NAM, and ggCaSR-PAM structures, detergent density subtraction, application of a mask around the transmembrane helices, and focused refinement were applied. The final cryo-EM maps for ggCaSR-active, ggCaSR-NAM, ggCaSR-PAM, and ggCaSR-inactive were determined at resolutions of 3.2, 3.2, 3.2, and 7.2 Å, respectively. Three structures (ggCaSR-active, ggCaSR-NAM, and ggCaSR-PAM) show well-resolvable density, and atomic models were built. The cryo-electron density map of ggCaSR-inactive did not provide sufficient quality for atomic model building due to the low resolution. We therefore fitted separated domains from the ggCaSR-active monomer structure into the density map of ggCaSR-inactive to obtain a low-resolution model. It has been noted that the intracellular C-terminal portion (amino acids 868 to 1059) of ggCaSR in the four conformational states is not included in models due to the intrinsic flexibility and will not be discussed here. This portion in the human counterpart was shown to bind with multiple intracellular partners and mediate various receptor-specific signaling pathways (30–32).
Overall structure of the full-length CaSR
ggCaSR forms a disulfide-tethered homodimer and consists of a large N-terminal extracellular domain (ECD), which can be further divided into a Venus flytrap module (VFT) with bilobed architecture, a cysteine-rich module (CR), and a heptahelical transmembrane domain (7TMD). Two protomers exist side by side and face opposite directions (Fig. 1). The ggCaSR-active VFT dimer is highly similar to previous crystallographic studies (27, 28) with root mean square deviations (RMSDs) of 1.09 and 0.89 Å (fig. S6, A and B). The ggCaSR-active CR dimer is also similar to that of its human counterpart (27), giving an RMSD value of 2.13 Å. Moreover, we observed that in ggCaSR-active, the distance between the two CR protomers gradually increased from the VFT end to the 7TMD end compared with the crystallographic human counterpart (fig. S6B). Compared with another class C GPCR, the mGlu5 dimer (29), the overall structure of ggCaSR is similar. Superimposition of these two structures in their active conformations gives an RMSD value of 2.77 Å (fig. S6C), and two main structural differences were observed. One is located within the interaction interface of two VFTs (fig. S6D). These two loops in ggCaSR are significantly longer than those in mGlu5 and play important roles in dimerization and regulating the functions of CaSR (28). The other is located in the CR and covers residues 581 to 599 (fig. S6E). The apical region (residues 586 to 589) points toward the outside flank of the CR and is away from extracellular loop 2 (ECL2) of the 7TMD. In the mGlu5 protomer, this tip region interacts with ECL2 of the 7TMD (29).
Fig. 1. The overall structure of full-length CaSR.
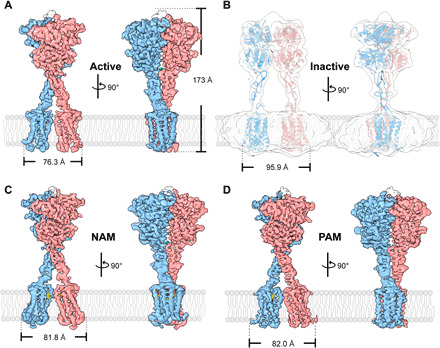
(A) Surface representation of the cryo-EM density map of the ggCaSR-active state bound to Ca2+, Cl−, and l-Trp. (B) Rough cryo-EM density map of the ggCaSR-inactive receptor. (C) Surface representation of the cryo-EM density map of the ggCaSR-NAM structure with Ca2+, Cl−, l-Trp, and NPS-2143. (D) Surface representation of the cryo-EM density map of the ggCaSR-PAM structure with Ca2+, Cl−, l-Trp, and evocalcet. Each structure is shown in two different side views that are rotated by 90° along the vertical axis. NPS-2143 and evocalcet are colored yellow, Ca2+ ions are colored cyan, l-Trp moieties are colored orange, and Cl− ions are colored green. Blue and pink represent the two different protomers in the cryo-EM density maps.
The entire ggCaSR height for the four conformational states is similar and was estimated to be approximately 173 Å. The ggCaSR widths on the cytosolic side for the active, NAM, PAM, and inactive states are approximately 76.3, 81.8, 82, and 95.9 Å, respectively. In the ggCaSR-active, ggCaSR-NAM, and ggCaSR-PAM structures, the ECD of the homodimeric receptor adopts a closed conformation, which presumably drives the closure of two 7TMDs (Fig. 1, A, C, and D). The NAM molecule NPS-2143 was bound to the ligand-binding pocket of the 7TMD of CaSR, further resulting in the drastic rearrangement of two of the 7TMDs (Fig. 1C). The PAM molecule evocalcet bound to a ligand-binding pocket similar to that of the 7TMD. Compared with ggCaSR-active, ggCaSR-PAM maintains a similar overall conformation (Fig. 1D). In sharp contrast, the ECD portion of the homodimeric receptor remained in an open state in the ggCaSR-inactive structure, leading to the good separation of these two 7TMDs (Fig. 1B).
Ligand recognition
In the ggCaSR-NAM structure, three Ca2+ binding sites were identified within each protomer (Fig. 2A). Site 1 is positioned around the side chain of residue T100, and Ca2+ is stabilized by the hydroxyl group (Fig. 2B). Site 2 is positioned around S302, and Ca2+ is coordinated by its backbone carbonyl oxygen atom (Fig. 2C). Site 3 is positioned at the edge of the interdomain cleft of the VFT and CR (Fig. 2D). Ca2+ is coordinated by the carboxylate group of D234 and backbone carbonyl oxygen atoms of E231 and G556 from the other protomer. This Ca2+ binding site is part of the homodimer interface formed upon agonist binding, indicating that this site is most likely associated with receptor activation (27). Mutations of residues in the Ca2+ binding site greatly affected the Ca2+-dependent ggCaSR receptor response (fig. S7, A and B). Moreover, many natural inactive mutants appear at the Ca2+ binding site, which can impair Ca2+-induced human CaSR receptor activity and cause FHH1 or hyperparathyroidism, such as the corresponding human CaSR mutations T100I (33), S302F (34), and G557E (35). These three Ca2+ binding sites are quite similar to those previously reported (27).
Fig. 2. Ligand recognition of the ggCaSR-NAM structure.
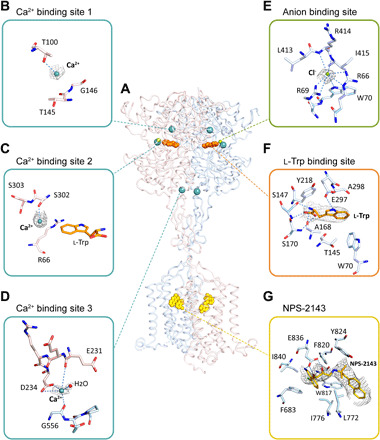
(A) Overall picture of ligands bound in the receptor structure. Ligands are shown as spheres with different colors. (B to D) Three Ca2+-binding sites in the ggCaSR-NAM structure. Ca2+ ions are drawn as cyan spheres. (E) Anion binding site. Cl− ions are drawn in green. (F) l-Trp binding site. l-Trp is shown in orange. (G) Binding pocket of NPS-2143 in the 7TMD. NPS-2143 is shown in yellow. For (B) to (G), the surrounding residues around different ligands are shown in stick models. The gray mesh represents the density map of the ligand, and the dotted line represents a hydrogen bond. Oxygen and nitrogen atoms are colored red and blue, respectively, and carbon atoms are colored according to the protomer color.
In our structure, we identified only one anion binding site and assigned it as Cl− (Fig. 2E). This binding site is located close to the binding pocket of l-Trp (Fig. 2A). This Cl− is stabilized by the side chains of R66, R69, and W70 as well as the main-chain nitrogen atoms of L413 and R414. This site is very similar to the previously reported PO43− binding site (27) or bicarbonate binding site (28). Mutations of residues in this site notably affected the Ca2+-dependent ggCaSR receptor response (fig. S7C). Moreover, previous studies have reported that four inactive mutations of the corresponding human CaSR—R66H (36), R66C (36, 37), R69C (38), and R69H (39)—may affect glycosylation or ligand binding of CaSR and lead to FHH1 or NSHPT.
The binding site of l-Trp was identified at the interdomain cleft of the VFT domain (Fig. 2F). The l-Trp molecule is stabilized by residues S147, A168, S170, and E297 through multiple hydrogen bonds and by residues W70 and Y218 through hydrophobic interactions. E297 serves as a disease-associated switch residue that is located in the region that undergoes large conformational changes upon ligand binding (40). Consequently, the E297K inactive mutation of the corresponding human CaSR and E297D active mutation cause FHH1 (or NSHPT) and ADH1, respectively (37, 41). This binding site is identical to the previously reported l-Trp binding site in the crystal structure of the human CaSR ECD (27) and the l-1,2,3,4-tetrahydronorharman-3-carboxylic acid binding site in the crystal structure of the human CaSR VFT (28).
The binding pocket of allosteric modulators
The NAM molecule NPS-2143 was found to bind in the 7TMD of the ggCaSR-NAM structure (Fig. 2G). This binding site is similar to the previously predicted site (20, 42). NPS-2143 binds at the upper half of the 7TMD and mainly contacts TM3, TM5, TM6, and TM7. NPS-2143 is packed by a hydrophobic pocket composed of residues I8407.37 [numbered according to Ballesteros-Weinstein (43)], F6833.36, I7765.44, W8176.50, L7725.40, Y8246.57, and F8206.53 and is coordinated by three hydrogen bonds with E8367.33 of TM7. The corresponding human CaSR mutations E837A (or I or Q), L773R, I841A, and F684A reduced the affinity and potency of NPS-2143 (16, 24, 42, 44).
The PAM molecule evocalcet was bound in the 7TMD of the ggCaSR-PAM structure (Fig. 3, A and B) and mainly contacted TM3, TM5, TM6, and TM7. In detail, the naphthalenyl group was buried inside the pocket and packed by hydrophobic interactions with the bulky side chains of residues F6833.36, F6873.40, W8176.50, and I8407.37 (Fig. 3B). The pyrrolidinyl group extends toward the open space between TM5 and TM6 and is stabilized by residue Y8246.57. Residues E8367.33 and Q6803.33 make two hydrogen bonds with the nitrogen atom of evocalcet in the center position (Fig. 3B). Compared with the wild type, mutations Y824A and W817A significantly reduced the affinity of evocalcet, and mutations F683A, E836A, Q680A, and F687A lost almost all evocalcet binding (Fig. 3, C and D).
Fig. 3. Evocalcet binding in the ggCaSR-PAM structure.
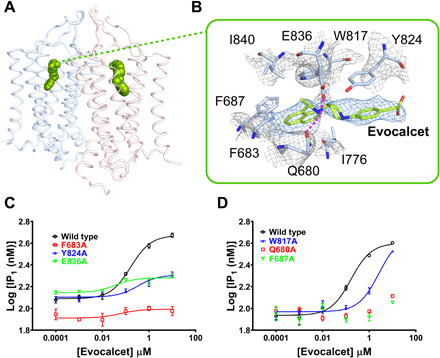
(A) Overall view of evocalcet bound in two 7TMDs of the receptor. Evocalcet is shown as green spheres. Two 7TMDs are represented as cartoon model and colored differently. (B) Binding pocket of evocalcet in the 7TMD. Evocalcet and the surrounding residues are shown in stick models. The mesh colored gray and blue represents the density map, and the magenta dotted lines represent hydrogen bonds. Oxygen, nitrogen, and carbon atoms in CaSR are colored red, blue, and pale blue, respectively. Carbon atoms in evocalcet are colored green. (C) Concentration-response binding curves of evocalcet for the ggCaSR wild type (black), the mutant F683A (red), the mutant Y824A (blue), and the mutant E836A (green) by IP1 production assay. (D) Concentration-response binding curves of evocalcet for the ggCaSR wild type (black), the mutant W817A, the mutant Q680A (red), and the mutant F687A (green) by IP1 production assay.
Although the PAM molecule evocalcet and the NAM molecule NPS-2143 have opposite allosteric effects, their binding pockets are located at a position similar to that of the 7TMD (Fig. 4A). A feature of such a binding pocket in one CaSR protomer is that one end is widely exposed to the phospholipid environment inside the membrane, and the other end is deeply buried inside the middle of the 7TMD (Fig. 4, B and D). Previous studies have revealed a large number of structures of class A, class B, class C (7TMD alone), and class F GPCRs in complex with allosteric modulators that showed that the binding site and pose of these modulators are diverse (45, 46). Compared with these published binding pockets for allosteric modulators, the binding pocket in CaSR partially overlaps with the 4-fluoro-N-(4-(6-(isopropylamino)pyrimidin-4-yl)thiazol-2-yl)-N-methylbenzamide (FITM) molecule binding pocket of the 7TMD of mGlu1 (47) and the tiotropium binding pocket of the human M5 muscarinic acetylcholine receptor (Fig. 4A) (48). Moreover, we identified that the NPS-2143 molecule in one protomer directly faces TM6 from the other protomer reciprocally (Fig. 4C), suggesting that a cooperative binding mode may exist between two NPS-2143 molecules. This is consistent with previous results that obtaining full inhibition by a negative allosteric modulator requires binding to both protomers of the full-length CaSR (49, 50). In contrast, two evocalcet molecules in the dimer were well separated by two TM6s and two TM7s (Fig. 4E).
Fig. 4. Binding pocket of allosteric modulators in CaSR.
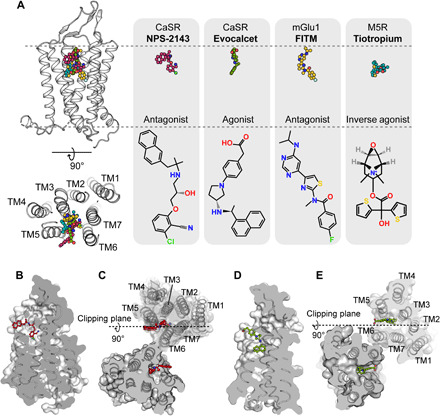
(A) Comparison of the similar binding pockets of the allosteric modulators in different GPCRs from a side view and an extracellular view. FITM in the mGlu1 structure (PDB code 4OR2) and tiotropium in the M5R structure (PDB code 6OL9) partially overlapped with the binding pocket of NPS-2143 and evocalcet in CaSR. Carbon atoms in the NPS-2143, evocalcet, FITM, and tiotropium models (upper row) are colored red, green, dark yellow, and cyan, respectively. Carbon atoms in the schematic drawing of NPS-2143, evocalcet, FITM, and tiotropium (bottom row) are shown in black. Nitrogen, oxygen, chloride, fluoride, and sulfide atoms are colored blue, red, light green, green, and yellow, respectively. (B and C) Cut-through surface side view of the NPS-2143 binding cavity in the protomer (B) and in the dimer (C) of the ggCaSR-NAM structure. NPS-2143 is represented as a ball-and-stick model. (D and E) Cut-through surface side view of the evocalcet binding cavity in the protomer (D) and in the dimer (E) of the ggCaSR-PAM structure. Evocalcet is represented as a ball-and-stick model.
Allosteric modulation mechanism
To uncover the mechanism of the allosteric modulation mechanism of CaSR by different allosteric effectors, we superimposed the ggCaSR-active structure onto the ggCaSR-NAM structure and the ggCaSR-PAM structure. The ECD of CaSR is largely similar between the two structures, indicating that the binding of allosteric effectors did not cause noticeable conformational changes in the ECD (fig. S8A). However, upon binding of the NAM molecule NPS-2143, the 7TMD rearranges the dimeric formation (fig. S8B). In the ggCaSR-active structure, TM6 from one protomer faces two helices, TM6 and TM7, from the other protomer (Fig. 5A). In the ggCaSR-NAM structure, TM6 from one protomer faces two helices, TM6 and TM5, as well as NPS-2143 from the other protomer (Fig. 5B). In the ggCaSR-PAM structure, the 7TMD dimer conformation is similar to that in the ggCaSR-active structure (Fig. 5C and fig. S8C). From the extracellular view, we observed that the binding of NPS-2143 to the single 7TMD pushes away TM5 by approximately 1.9 Å, TM6 by approximately 3.4 Å, and TM7 by approximately 2.2 Å, which induces the movement (approximately 10.1 Å) of the other entire 7TMD (Fig. 5D). From the cytosolic view, the movement distance (approximately 16.8 Å) of the other entire 7TMD was observed (Fig. 5E). Therefore, the intersubunit interfaces within the two 7TMDs are quite different between the ggCaSR-active and ggCaSR-NAM structures (fig. S8, D and E). The binding of evocalcet to the single 7TMD pushes away TM5, TM6, and TM7 by minor distances (Fig. 5F), resulting in a closer distance between the extracellular ends of the two TM6s (Fig. 5F) and a greater distance between the intracellular ends of the two TM6s (Fig. 5G). To further distinguish the two different kinds of intersubunit interfaces, we introduced cysteine-mediated cross-links at TM6. As expected, mutant ggCaSR S826C-T827C was constitutively active (Fig. 5H), while mutant ggCaSR-P822C showed attenuated activity (Fig. 5I). Surface expression levels of the S826C-T827C and P822C mutants are comparable to those of the wild type (fig. S8F).
Fig. 5. Allosteric regulation mechanism of CaSR.
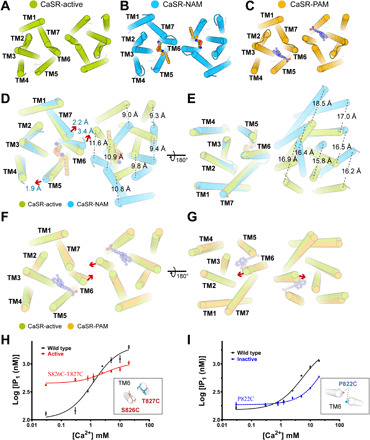
(A) Overall view of the dimeric 7TMD in ggCaSR-active from the extracellular view. (B) Overall view of the dimeric 7TMD in ggCaSR-NAM from the extracellular view. NPS-2143 is shown as a ball-and-stick model. (C) Overall view of the dimeric 7TMD in ggCaSR-PAM from the extracellular view. Evocalcet is shown as a ball-and-stick model. (D and E) Structural superimposition of the dimeric 7TMD upon NPS-2143 binding from the extracellular view (D) and the intracellular view (E). The moving distance of each TM is labeled. Red arrows denote the moving direction. (F and G) Structural superimposition of the dimeric 7TMD upon evocalcet binding from the extracellular view (F) and the intracellular view (G). Red arrows denote the moving direction of TM6s. (H) Dose-dependent Ca2+-stimulated IP production in cells transiently expressing the wild-type or constitutively active mutant (S826C-T827C). Inset indicates the position of S826 and T827 in the ggCaSR-active state. For clarity, only two TM6s are shown. (I) Dose-dependent Ca2+-simulated IP production in cells transiently expressing wild-type or inactive mutant (P822C). Inset indicates the position of P822 in the ggCaSR-NPS state. For clarity, only two TM6s were shown.
DISCUSSION
In this research, we present the structures of full-length ggCaSR in four different conformational states. The overall structure of ggCaSR is a disulfide bond–tethered homodimer similar to other reported class C GPCRs, such as mGlu5 (29). ggCaSR-inactive is in an open state. Upon the binding of the ligands Ca2+ and l-Trp to the VFT domains, the closed conformation of the VFT domains then induces the CR domains to move into close proximity to each other and brings the 7TMDs together, leading to the activation of CaSR. The orientation of the two 7TMDs in the ggCaSR-active structure established one kind of TM interface (named activated TM6/TM6 hereafter), which is quite similar to that of the activated mGlu5 structure (fig. S8, G and H). Such an activated TM6/TM6 interface was observed in our ggCaSR-PAM structure (Fig. 5C) and was also recently reported for metabotropic GABAB (γ-aminobutyric acid type B) receptor structures (51, 52). These results suggest that class C GPCRs share a universal activation mechanism (27, 29, 53, 54). Strikingly, NPS-2143 can further bind to the activated receptor. The ECD of the receptor undergoes hardly noticeable conformational changes, while drastic conformational changes occur in the 7TMDs, leading to the formation of another kind of TM interface (named inactivated TM6/TM6 hereafter). The unconventional dimeric interface of the 7TMDs has not been observed in previously published GPCR structures, presumably representing the negatively modulated allosteric state that is unable to couple with G proteins. Together, allosteric modulators bind to the 7TMDs, inducing different TM interface arrangements (activated TM6/TM6 versus inactivated TM6/TM6) to cause various allosteric effects.
CaSR is critical in regulating calcium hemostasis and maintains the extracellular Ca2+ concentration within a tight range (1, 2, 17, 55). Consequently, more than 400 different germline gain- and loss-of-function CaSR mutations have been found (32, 56). Using the high-resolution structure of full-length ggCaSR-active, we were able to map a total of 305 missense mutations into the structure for five kinds of clinical endocrine disorders, including 70 gain-of-function missense mutations (62 and 8 mutations for ADH1 and hypoparathyroidism, respectively) and 235 loss-of-function missense mutations (206, 19, and 10 mutations for FHH1, NSHPT, and hyperparathyroidism, respectively) (Fig. 6 and table S1). We found that more than one-third (25 of 70) of the gain-of-function missense mutations related to ADH1 and hypoparathyroidism are located in the homodimer interface of the ligand-bound CaSR-active structure (Fig. 6, A and B), suggesting that CaSR may exhibit a superactive state induced by a positive allosteric modulator or intracellular inducers (45, 57). However, loss-of-function missense mutations related to FHH1, NSHPT, and hyperparathyroidism are universally distributed over the whole CaSR receptor (Fig. 6, C to E) and prefer to cluster in the ligand-binding regions that directly affect the affinity of ligands or gather in the regions that undergo a large conformational change upon ligand binding (32). In addition, 11 human CaSR mutations (Q27E, V104I, L273F, N178Y, P221L, E297D, S657C, R680G, N802I, S820F, and T828N) have been found to cause opposite phenotypes, which may require special attention for these mutations associated with disease therapy. Overall, our hotspot map of CaSR mutations related to five clinical endocrine disorders will be beneficial for identifying the potential disease risk for patients and providing insights for related disease therapy.
Fig. 6. Hotspot map of CaSR missense mutations.
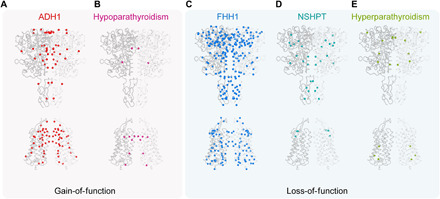
(A to E) Missense mutations associated with five endocrine disorders—ADH1 (A), hypoparathyroidism (B), FHH1 (C), NSHPT (D), and hyperparathyroidism (E)—were mapped onto the ggCaSR-active structure. Each Ca atom of the mutated residues is represented by a sphere and colored red, magenta, blue, cyan, and green, respectively. ADH1 and hypoparathyroidism are caused by the gain-of-function CaSR mutations, while FHH1, NSHPT, and hyperparathyroidism are caused by the loss-of-function CaSR mutations.
MATERIALS AND METHODS
Plasmid construct
The synthetic gene encoding full-length wild-type ggCaSR (accession no. XP_416491.5) followed by a C-terminal enhanced green fluorescent protein (eGFP) cleavable by the protease rhinovirus 3C was cloned into the pEG-BacMam vector and used to generate baculovirus using the BacMam Baculovirus system (Thermo Fisher Scientific).
Expression and purification of ggCaSR
Baculovirus bacmid containing the CaSR expression cassette was generated by the transformation of DH10Bac Escherichia coli cells with the pEG-BacMam-CaSR-3C-eGFP plasmid. Spodoptera frugiperda (Sf9) cells grown in a six-well plate at 27°C were transfected with the bacmid using X-tremeGENE HP DNA transfection reagent (Roche). The first-generation virus (P1) was collected 72 hours after transfection, and the P1 virus was added (1%, v/v) to Sf9 cells to produce the second-generation virus (P2). P2 was used to infect HEK293S-GnTi− cells at a cell density of 2.0 × 106 cells/ml in 5% CO2 at 37°C. After 24 hours, sodium butyrate was added to the cell culture at a final concentration of 10 mM, and the culture temperature was reduced to 30°C to facilitate protein expression. Transfected cells were further cultured at 30°C for 60 hours.
For protein purification, two liters of infected HEK293S-GnTi− cells were harvested by centrifugation at 800g. The collected cells were resuspended and lysed in buffer A [20 mM tris-HCl (pH 7.5) and 200 mM NaCl] supplemented with 1% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace), 0.2% cholesteryl hemisuccinate (CHS; Sigma-Aldrich), and 1× protease inhibitor cocktail (TargetMol) and solubilized for 2 hours at 4°C. The supernatant was collected by centrifugation at 120,000g for 60 min at 4°C and then incubated with anti-GFP nanobody affinity gel (BioLegend) at 4°C for 2.5 hours. After washing five times with buffer B containing 20 mM tris-HCl (pH 7.5), 200 mM NaCl, 0.025% DDM, 0.005% CHS, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), the column was washed with high-salt wash buffer composed of buffer B supplemented with 0.5 and 1 M NaCl. Afterward, the beads were washed twice with buffer B supplemented with 10 mM adenosine triphosphate (ATP) and 10 mM Mg2+ and incubated at 4°C for 30 min. Then, the resin was resuspended in buffer C containing 20 mM tris-HCl (pH 7.5), 200 mM NaCl, 0.01% glycol-diosgenin (GDN; Anatrace), and 0.1 mM PMSF and incubated for 1 hour at 4°C for detergent exchange. Last, the beads were mixed with buffer C plus rhinovirus 3C protease overnight (1:30, v/v) to cleave the eGFP tag.
After digestion, the protein was eluted with buffer C, and the eluate was concentrated using a 100-kDa cutoff concentrator (Millipore). For the preparation of the ggCaSR-inactive sample, the protein was injected into Superose 6 Increase 10/300 GL (GE Healthcare) equilibrated with 20 mM tris (pH 7.5), 200 mM NaCl, 2 mM Ca2+, and 0.0063% GDN. For the preparation of the ggCaSR-active sample, the protein was further purified by the Superose 6 Increase column, which was equilibrated with 20 mM tris (pH 7.5), 200 mM NaCl, 10 mM Ca2+, 10 mM l-Trp, and 0.0063% GDN. In addition, we used buffer containing 20 mM tris (pH 7.5), 200 mM NaCl, 10 mM Ca2+, 10 mM l-Trp, 10 μM NPS-2143, and 0.0063% GDN to induce the ggCaSR-NAM state, which has an active ECD and inactive TMD. For the preparation of the ggCaSR-PAM sample, we used buffer containing 20 mM tris (pH 7.5), 200 mM NaCl, 10 mM Ca2+, 10 mM l-Trp, 20 μM evocalcet, and 0.0063% GDN. The peak fractions were pooled and concentrated to ~6 to 9 mg/ml for the preparation of the cryo-EM grid.
Cryo-EM sample preparation and imaging
For all ggCaSR samples, an aliquot of 2.5 μl of purified ggCaSR sample was applied onto a glow-discharged holey carbon film grid (200 mesh, R2/1, Quantifoil). The grid was blotted and plunge-frozen in liquid ethane with FEI Vitrobot Mark IV. The grids were loaded onto an FEI Titan Krios electron microscope operated at an accelerating voltage of 300 kV. Image stacks were recorded on a K2 Summit direct electron counting detector (Gatan) set in counting mode using SerialEM (http://bio3d.colorado.edu/SerialEM/) with a defocus range of −1.5 and −2.5 μm at a pixel size of 1.014 Å per pixel. A total of 40 frames were acquired in 8 s for each image stack, giving a total electron dose of 50 to 53 e−/A2. Last, 4465, 2143, 2504, and 2320 image stacks were acquired for ggCaSR-active, ggCaSR-NAM, ggCaSR-PAM, and ggCaSR-inactive sample, respectively.
Image processing
The recorded image stacks were processed by MotionCor2 (58) for a 5 × 5 patch drift correction with dose weighting. The non–dose-weighted images were used for contrast transfer function (CTF) estimation by CTFFIND 4 (59). Images of poor quality were removed before particle picking. Particles were semiautomatically picked from dose-weighted images by Gautomatch (www.mrc-lmb.cam.ac.uk/kzhang/) and extracted by RELION-3 (60) in a box size of 280 pixels.
For the ggCaSR-active dataset, two rounds of 2D classification were performed in RELION-3 to remove contaminations, ice, and bad particles, yielding 486,941 good particles. The selected particles were then used to generate the initial model with an ab initio method with C1 symmetry in CryoSPARC-2 (https://cryosparc.com/). The initial model was pass-filtered to 30-Å resolution and applied to the first round of 3D classification in RELION-3. The particles were divided into four subsets, two of the four classes showed structure feathers, and their particles were selected for 3D autorefine with C1 symmetry. Particle subtraction was performed to remove the contribution of the detergent density from alignments and subsequent 3D refinement. The subtracted particles were further selected by two rounds of 3D classification. The selected particles were subjected for another round of 3D autorefine. The subsequent 3D classification was carried out by applying a mask around the transmembrane domain and without particle rotation and translation alignment using the 3D-autorefined particles. After this round of 3D classification, one of the three classes containing 100,488 particles with better density in the TM domain was selected for focused refinement of the TM region and then refined to 4.3-Å resolution in RELION-3. The selected 100,488 particles were re-extracted and transferred to CryoSPARC-2 for 3D classification using heterogeneous refinement. A total of 86,921 particles were selected for nonuniform refinement and then refined to 3.2-Å resolution. These particles were duplicated by symmetry expansion according to C2 symmetry and through another round of local refinement with nonuniform adaptive regulation in CryoSPARC-3 focused on the ECD region, resulting in map of ECD region at 2.9-Å resolution (C1 symmetry). The two individual maps of TM and ECD were aligned on ggCaSR-active whole map and merged in Chimera software (www.rbvi.ucsf.edu/chimera). Image process protocols of the ggCaSR-NAM dataset and the ggCaSR-PAM dataset are similar to those described for the ggCaSR-active dataset.
For the ggCaSR-inactive dataset, two rounds of 2D classification were performed in RELION-3 to remove contaminations, ice, and bad particles, yielding 312,631 food particles. The selected particles were transferred to CryoSPARC-2 to generate the three initial models with an ab initio method with C1 symmetry. The initial models were applied to a further round of 3D classification using heterogeneous refinement, and one of the three classes with better structure feathers was selected for a further round of 3D heterogeneous refinement. The final selected 80,184 particles were refined to 7.2-Å resolution using nonuniform refinement.
The stated resolutions were evaluated using the “gold-standard” FSC (Fourier shell correlation) = 0.143 criterion. The local resolution was calculated by ResMap (61). Data collection and reconstruction statistics are presented in table S2.
Model building and refinement
Model building of the VFT and CR-TM region of ggCaSR-active took advantage of the crystal structure of H. sapiens CaSR extracellular domain [Protein Data Bank (PDB): 5K5S] and cryo-EM structure of mGlu5 (PDB: 6N52), respectively. Model of H. sapiens CaSR VFT domain and CRD-TMD of H. sapiens mGlu5 were fitted into ggCaSR-active map in Chimera, and the prefitted model was refined by PHENIX (62) in real space with secondary structure and geometry restraints. The residue assignment and side-chain orientation were further refined in COOT software (63) according to the density map. Sequence assignment was mainly guided by secondary structure prediction results from PSIPRED (64) and visible densities of residues with bulky side chains (Trp, Phe, Tyr, and Arg). The model was refined by PHENIX real_space_refinement with secondary structure and geometry restraints. The ggCaSR-NAM structure and the ggCaSR-PAM structure were built and modified from the refined ggCaSR-active model. The quality of the models was assessed with MolProbity (65). Refinement statistics and validation statistics for the ggCaSR-active, ggCaSR-NAM, and ggCaSR-PAM models are shown in table S2.
Inositol phosphate measurement
Inositol phosphate accumulation was quantified with the homogeneous time-resolved fluorescence (HTRF) IP-One Tb kit (Cisbio Bioassays, Codolet, France), which measures the accumulation of IP1, a metabolite of inositol 1,4,5-trisphosphate (IP3). HEK293T cells were transfected with plasmids encoding FLAG-ggCaSR. One day after transfection, the cells were resuspended in the stimulation buffer [10 mM Hepes-NaOH, 0.5 mM MgCl2, 4.2 mM KCl, 146 mM NaCl, 5.5 mM glucose, 10 mM l-Trp, and 50 mM LiCl (pH 7.4)] containing either dimethyl sulfoxide (DMSO), 30 μM NPS-2143, or 30 μM evocalcet and an increasing concentration of extracellular calcium. The cells were seeded in HTRF 96-well low-volume plate (Cisbio Bioassays, Codolet, France) and incubated at 37°C for 1 hour. The stimulated cells were lysed, and the native IP1 that had been produced was incubated with a d2 fluorophore–labeled IP1 analog (the acceptor) to compete for binding to an Tb2+ cryptate–coupled anti-IP1 monoclonal antibody (the donor). Fluorescence data were collected at 620 and 665 nm with the Varioskan LUX Microplate Reader (Thermo Fisher Scientific, Waltham, USA) after laser excitation at 340 nm. The fluorescence resonance energy transfer (FRET) signal was calculated as the fluorescence ratio (665/620 nm) and is inversely proportional to the concentration of native IP1 produced. Data analysis was performed using the nonlinear regression algorithm in Prism (GraphPad Software, San Diego, CA, USA). Data points represent averages ± SEM of multiple experiments, each consisting of triplicate measurements.
Cell surface receptor expression
Cell surface expression was measured by flow cytometry. HEK293T cells cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum were seeded onto a six-well plate 12 hours before transfection. Cells were transfected with 3 μg of DNA and 9 μg of polyethyleneimine per well. Twenty-four hours after transfection, cells were washed and detached in 1 ml of Dulbecco’s phosphate-buffered saline (DPBS) supplemented with 3% bovine serum albumin (BSA). Cells (1 × 106) were then resuspended in DPBS containing 3% BSA and anti-FLAG M2 antibody (Sigma-Aldrich, F1804). After incubation for 60 min at room temperature, the cells were washed five times by centrifugation at 200g for 3 min and resuspended in ice-cold DPBS containing 3% BSA and Alexa Fluor 488–conjugated secondary antibody (IgG H&L, Abcam, ab150113). After incubation at 4°C for 60 min in the dark, the cells were washed with DPBS, passed through a cell strainer, and analyzed by flow cytometry on BD LSR Fortessa (BD Biosciences) with an excitation wavelength of 488 nm and emission at 530 nm. Expression levels as measured by mean fluorescence were normalized to the level of wild-type ggCaSR expression.
Acknowledgments
We thank the Center of Cryo-Electron Microscopy, Zhejiang University for data acquisition and A. Li for the support of electron microscopy at Nankai University. Funding: This study was supported by the National Key Research and Development Program of China (2017YFA0504800 to Y.S. and X.Z.), the National Key Research and Development Program of China (2018YFA0507700 to X.Z.), the National Natural Science Foundation of China (91954119 to X.Y., 31870736 to X.Y., 31870834 to Y.S., and 32071231 to Y.S.), the Fundamental Research Funds for the Central Universities (035-63201110 to X.Y.), and the Fundamental Research Funds for the Central Universities (035-63201109 to Y.S.). Author contributions: Conceptualization: Y.S. and X.Y.; methodology: T.W.; investigation: T.W., Z.W., X.C., Y.R., X.L., Y.X., J.L., S.C., X.Z., and X.Y.; supervision: Y.S., X.Z., and X.Y.; writing (original draft): T.W., Y.S., and X.Y.; writing (review and editing): T.W., Y.S., and X.Y. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Atomic coordinates have been deposited in the Protein Data Bank under accession number 7DD6 for ggCaSR-active, 7DD5 for ggCaSR-NAM, and 7DD7 for ggCaSR-PAM. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-30646, EMD-30645, EMD-30644, and EMD-30647 for ggCaSR-inactive, ggCaSR-active, ggCaSR-NAM, and ggCaSR-PAM, respectively. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg1483/DC1
REFERENCES AND NOTES
- 1.Brown E. M., Gamba G., Riccardi D., Lombardi M., Butters R., Kifor O., Sun A., Hediger M. A., Lytton J., Hebert S. C., Cloning and characterization of an extracellular Ca2+-sensing receptor from bovine parathyroid. Nature 366, 575–580 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Hannan F. M., Babinsky V. N., Thakker R. V., Disorders of the calcium-sensing receptor and partner proteins: Insights into the molecular basis of calcium homeostasis. J. Mol. Endocrinol. 57, R127–R142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown E. M., Extracellular Ca2+ sensing, regulation of parathyroid cell function, and role of Ca2+ and other ions as extracellular (first) messengers. Physiol. Rev. 71, 371–411 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Hannan F. M., Kallay E., Chang W., Brandi M. L., Thakker R. V., The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat. Rev. Endocrinol. 15, 33–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearce S. H., Williamson C., Kifor O., Bai M., Coulthard M. G., Davies M., Lewis-Barned N., McCredie D., Powell H., Kendall-Taylor P., Brown E. M., Thakker R. V., A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N. Engl. J. Med. 335, 1115–1122 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Pollak M. R., Brown E. M., Chou Y. H., Hebert S. C., Marx S. J., Steinmann B., Levi T., Seidman C. E., Seidman J. G., Mutations in the human Ca2+-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell 75, 1297–1303 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Pollak M. R., Brown E. M., Estep H. L., McLaine P. N., Kifor O., Park J., Hebert S. C., Seidman C. E., Seidman J. G., Autosomal dominant hypocalcaemia caused by a Ca2+-sensing receptor gene mutation. Nat. Genet. 8, 303–307 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Yarova P. L., Stewart A. L., Sathish V., Britt R. D. Jr., Thompson M. A., Lowe A. P. P., Freeman M., Aravamudan B., Kita H., Brennan S. C., Schepelmann M., Davies T., Yung S., Cholisoh Z., Kidd E. J., Ford W. R., Broadley K. J., Rietdorf K., Chang W., Khayat M. E. B., Ward D. T., Corrigan C. J., Ward J. P. T., Kemp P. J., Pabelick C. M., Prakash Y. S., Riccardi D., Calcium-sensing receptor antagonists abrogate airway hyperresponsiveness and inflammation in allergic asthma. Sci. Transl. Med. 7, 284ra60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward D. T., Brown E. M., Harris H. W., Disulfide bonds in the extracellular calcium-polyvalent cation-sensing receptor correlate with dimer formation and its response to divalent cations in vitro. J. Biol. Chem. 273, 14476–14483 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Garrett J. E., Capuano I. V., Hammerland L. G., Hung B. C., Brown E. M., Hebert S. C., Nemeth E. F., Fuller F., Molecular cloning and functional expression of human parathyroid calcium receptor cDNAs. J. Biol. Chem. 270, 12919–12925 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Huang Y., Zhou Y., Yang W., Butters R., Lee H. W., Li S., Castiblanco A., Brown E. M., Yang J. J., Identification and dissection of Ca2+-binding sites in the extracellular domain of Ca2+-sensing receptor. J. Biol. Chem. 282, 19000–19010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Zhou Y., Castiblanco A., Yang W., Brown E. M., Yang J. J., Multiple Ca2+-binding sites in the extracellular domain of the Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 48, 388–398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Zhuo Y., Moniz H. A., Wang S., Moremen K. W., Prestegard J. H., Brown E. M., Yang J. J., Direct determination of multiple ligand interactions with the extracellular domain of the calcium-sensing receptor. J. Biol. Chem. 289, 33529–33542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng X., Xin Y., Miller C. L., Hamelberg D., Kirberger M., Moremen K. W., Hu J., Yang J. J., Structural mechanism of cooperative regulation of calcium-sensing receptor-mediated cellular signaling. Curr. Opin. Physiol. 17, 269–277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H., Yi P., Zhao W., Wu Y., Acher F., Pin J. P., Liu J., Rondard P., Illuminating the allosteric modulation of the calcium-sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 117, 21711–21722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leach K., Gregory K. J., Kufareva I., Khajehali E., Cook A. E., Abagyan R., Conigrave A. D., Sexton P. M., Christopoulos A., Towards a structural understanding of allosteric drugs at the human calcium-sensing receptor. Cell Res. 26, 574–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavez-Abiega S., Mos I., Centeno P. P., Elajnaf T., Schlattl W., Ward D. T., Goedhart J., Kallay E., Sensing extracellular calcium—An insight into the structure and function of the calcium-sensing receptor (CaSR). Adv. Exp. Med. Biol. 1131, 1031–1063 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Hofer A. M., Brown E. M., Extracellular calcium sensing and signalling. Nat. Rev. Mol. Cell Biol. 4, 530–538 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Davey A. E., Leach K., Valant C., Conigrave A. D., Sexton P. M., Christopoulos A., Positive and negative allosteric modulators promote biased signaling at the calcium-sensing receptor. Endocrinology 153, 1232–1241 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Petrel C., Kessler A., Dauban P., Dodd R., Rognan D., Ruat M., Positive and negative allosteric modulators of the Ca2+-sensing receptor interact within overlapping but not identical binding sites in the transmembrane domain. J. Biol. Chem. 279, 18990–18997 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Josephs T. M., Keller A. N., Khajehali E., DeBono A., Langmead C. J., Conigrave A. D., Capuano B., Kufareva I., Gregory K. J., Leach K., Negative allosteric modulators of the human calcium-sensing receptor bind to overlapping and distinct sites within the 7-transmembrane domain. Br. J. Pharmacol. 177, 1917–1930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki H., Ikeda Y., Sakurai O., Miyake T., Tsubota R., Okabe J., Kuroda M., Hisada Y., Yanagida T., Yoneda H., Tsukumo Y., Tokunaga S., Kawata T., Ohashi R., Fukuda H., Kojima K., Kannami A., Kifuji T., Sato N., Idei A., Iguchi T., Sakairi T., Moritani Y., Discovery of evocalcet, a next-generation calcium-sensing receptor agonist for the treatment of hyperparathyroidism. Bioorg. Med. Chem. Lett. 28, 2055–2060 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Leach K., Hannan F. M., Josephs T. M., Keller A. N., Moller T. C., Ward D. T., Kallay E., Mason R. S., Thakker R. V., Riccardi D., Conigrave A. D., Brauner-Osborne H., International union of basic and clinical pharmacology. CVIII. Calcium-sensing receptor nomenclature, pharmacology, and function. Pharmacol. Rev. 72, 558–604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leach K., Wen A., Cook A. E., Sexton P. M., Conigrave A. D., Christopoulos A., Impact of clinically relevant mutations on the pharmacoregulation and signaling bias of the calcium-sensing receptor by positive and negative allosteric modulators. Endocrinology 154, 1105–1116 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Gorvin C. M., Stokes V. J., Boon H., Cranston T., Glück A. K., Bahl S., Homfray T., Aung T., Shine B., Lines K. E., Hannan F. M., Thakker R. V., Activating mutations of the G-protein subunit α 11 interdomain interface cause autosomal dominant hypocalcemia type 2. J. Clin. Endocrinol. Metab. 105, 952–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannan F. M., Walls G. V., Babinsky V. N., Nesbit M. A., Kallay E., Hough T. A., Fraser W. D., Cox R. D., Hu J., Spiegel A. M., Thakker R. V., The calcilytic agent NPS 2143 rectifies hypocalcemia in a mouse model with an activating calcium-sensing receptor (CaSR) mutation: Relevance to autosomal dominant hypocalcemia type 1 (ADH1). Endocrinology 156, 3114–3121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geng Y., Mosyak L., Kurinov I., Zuo H., Sturchler E., Cheng T. C., Subramanyam P., Brown A. P., Brennan S. C., Mun H.-C., Bush M., Chen Y., Nguyen T. X., Cao B., Chang D. D., Quick M., Conigrave A. D., Colecraft H. M., McDonald P., Fan Q. R., Structural mechanism of ligand activation in human calcium-sensing receptor. eLife 5, e13662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C., Zhang T., Zou J., Miller C. L., Gorkhali R., Yang J.-Y., Schilmiller A., Wang S., Huang K., Brown E. M., Moremen K. W., Hu J., Yang J. J., Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci. Adv. 2, e1600241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koehl A., Hu H., Feng D., Sun B., Zhang Y., Robertson M. J., Chu M., Kobilka T. S., Laeremans T., Steyaert J., Tarrasch J., Dutta S., Fonseca R., Weis W. I., Mathiesen J. M., Skiniotis G., Kobilka B. K., Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y., Zhou Y., Wong H. C., Castiblanco A., Chen Y., Brown E. M., Yang J. J., Calmodulin regulates Ca2+-sensing receptor–mediated Ca2+ signaling and its cell surface expression. J. Biol. Chem. 285, 35919–35931 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuang X., Northup J. K., Ray K., Large putative PEST-like sequence motif at the carboxyl tail of human calcium receptor directs lysosomal degradation and regulates cell surface receptor level. J. Biol. Chem. 287, 4165–4176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorvin C. M., Molecular and clinical insights from studies of calcium-sensing receptor mutations. J. Mol. Endocrinol. 63, R1–R16 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Warner J., Epstein M., Sweet A., Singh D., Burgess J., Stranks S., Hill P., Perry-Keene D., Learoyd D., Robinson B., Birdsey P., Mackenzie E., Teh B. T., Prins J. B., Cardinal J., Genetic testing in familial isolated hyperparathyroidism: Unexpected results and their implications. J. Med. Genet. 41, 155–160 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vargas-Poussou R., Mansour-Hendili L., Baron S., Bertocchio J. P., Travers C., Simian C., Treard C., Baudouin V., Beltran S., Broux F., Camard O., Cloarec S., Cormier C., Debussche X., Dubosclard E., Eid C., Haymann J. P., Kiando S. R., Kuhn J. M., Lefort G., Linglart A., Lucas-Pouliquen B., Macher M. A., Maruani G., Ouzounian S., Polak M., Requeda E., Robier D., Silve C., Souberbielle J. C., Tack I., Vezzosi D., Jeunemaitre X., Houillier P., Familial hypocalciuric hypercalcemia types 1 and 3 and primary hyperparathyroidism: Similarities and differences. J. Clin. Endocrinol. Metab. 101, 2185–2195 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Nakayama T., Minato M., Nakagawa M., Soma M., Tobe H., Aoi N., Kosuge K., Sato M., Ozawa Y., Kanmatsuse K., Kokubun S., A novel mutation in Ca2+-sensing receptor gene in familial hypocalciuric hypercalcemia. Endocrine 15, 277–282 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Pidasheva S., Grant M., Canaff L., Ercan O., Kumar U., Hendy G. N., Calcium-sensing receptor dimerizes in the endoplasmic reticulum: Biochemical and biophysical characterization of CASR mutants retained intracellularly. Hum. Mol. Genet. 15, 2200–2209 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Bai M., Quinn S., Trivedi S., Kifor O., Pearce S. H., Pollak M. R., Krapcho K., Hebert S. C., Brown E. M., Expression and characterization of inactivating and activating mutations in the human Ca2+−sensing receptor. J. Biol. Chem. 271, 19537–19545 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Hannan F. M., Nesbit M. A., Zhang C., Cranston T., Curley A. J., Harding B., Fratter C., Rust N., Christie P. T., Turner J. J., Lemos M. C., Bowl M. R., Bouillon R., Brain C., Bridges N., Burren C., Connell J. M., Jung H., Marks E., McCredie D., Mughal Z., Rodda C., Tollefsen S., Brown E. M., Yang J. J., Thakker R. V., Identification of 70 calcium-sensing receptor mutations in hyper- and hypo-calcaemic patients: Evidence for clustering of extracellular domain mutations at calcium-binding sites. Hum. Mol. Genet. 21, 2768–2778 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Wilhelm-Bals A., Parvex P., Magdelaine C., Girardin E., Successful use of bisphosphonate and calcimimetic in neonatal severe primary hyperparathyroidism. Pediatrics 129, e812–e816 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Gorvin C. M., Frost M., Malinauskas T., Cranston T., Boon H., Siebold C., Jones E. Y., Hannan F. M., Thakker R. V., Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias. Hum. Mol. Genet. 27, 3720–3733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silve C., Petrel C., Leroy C., Bruel H., Mallet E., Rognan D., Ruat M., Delineating a Ca2+ binding pocket within the venus flytrap module of the human calcium-sensing receptor. J. Biol. Chem. 280, 37917–37923 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Miedlich S. U., Gama L., Seuwen K., Wolf R. M., Breitwieser G. E., Homology modeling of the transmembrane domain of the human calcium sensing receptor and localization of an allosteric binding site. J. Biol. Chem. 279, 7254–7263 (2004). [DOI] [PubMed] [Google Scholar]
- 43.J. A. Ballesteros, H. Weinstein, Integrated methods for the construction of threedimensional models and computational probing of structure-function relations in G protein–coupled receptors, in Methods in Neurosciences, S. C. Sealfon, Ed. (Academic Press, 1995), vol. 25, chap. 19, pp. 366–428. [Google Scholar]
- 44.Ma J. N., Owens M., Gustafsson M., Jensen J., Tabatabaei A., Schmelzer K., Olsson R., Burstein E. S., Characterization of highly efficacious allosteric agonists of the human calcium-sensing receptor. J. Pharmacol. Exp. Ther. 337, 275–284 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Thal D. M., Glukhova A., Sexton P. M., Christopoulos A., Structural insights into G protein–coupled receptor allostery. Nature 559, 45–53 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Wang C., Wu H., Evron T., Vardy E., Han G. W., Huang X. P., Hufeisen S. J., Mangano T. J., Urban D. J., Katritch V., Cherezov V., Caron M. G., Roth B. L., Stevens R. C., Structural basis for smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H., Wang C., Gregory K. J., Han G. W., Cho H. P., Xia Y., Niswender C. M., Katritch V., Meiler J., Cherezov V., Conn P. J., Stevens R. C., Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuckovic Z., Gentry P. R., Berizzi A. E., Hirata K., Varghese S., Thompson G., van der Westhuizen E. T., Burger W. A. C., Rahmani R., Valant C., Langmead C. J., Lindsley C. W., Baell J. B., Tobin A. B., Sexton P. M., Christopoulos A., Thal D. M., Crystal structure of the M5muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 116, 26001–26007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobsen S. E., Gether U., Brauner-Osborne H., Investigating the molecular mechanism of positive and negative allosteric modulators in the calcium-sensing receptor dimer. Sci. Rep. 7, 46355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregory K. J., Kufareva I., Keller A. N., Khajehali E., Mun H. C., Goolam M. A., Mason R. S., Capuano B., Conigrave A. D., Christopoulos A., Leach K., Dual action calcium-sensing receptor modulator unmasks novel mode-switching mechanism. ACS Pharmacol. Transl. Sci. 1, 96–109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaye H., Ishchenko A., Lam J. H., Han G. W., Xue L., Rondard P., Pin J. P., Katritch V., Gati C., Cherezov V., Structural basis of the activation of a metabotropic GABA receptor. Nature 584, 298–303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao C., Shen C., Li C., Shen D. D., Xu C., Zhang S., Zhou R., Shen Q., Chen L. N., Jiang Z., Liu J., Zhang Y., Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 30, 564–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geng Y., Bush M., Mosyak L., Wang F., Fan Q. R., Structural mechanism of ligand activation in human GABAB receptor. Nature 504, 254–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muto T., Tsuchiya D., Morikawa K., Jingami H., Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 104, 3759–3764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel B. S., Ravix J., Pabelick C., Prakash Y. S., Class C GPCRs in the airway. Curr. Opin. Pharmacol. 51, 19–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dershem R., Gorvin C. M., Metpally R. P. R., Krishnamurthy S., Smelser D. T., Hannan F. M., Carey D. J., Thakker R. V., Breitwieser G. E.; Regeneron Genetics Center , Familial hypocalciuric hypercalcemia type 1 and autosomal-dominant hypocalcemia type 1: Prevalence in a large healthcare population. Am. J. Hum. Genet. 106, 734–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wacker D., Stevens R. C., Roth B. L., How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr. Sect. D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swint-Kruse L., Brown C. S., Resmap: Automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Buchan D. W. A., Jones D. T., The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg1483/DC1


