Abstract
Efficient cellular uptake of nanoparticles (NPs) is necessary for the development of nanomedicine in biomedical applications. Recently, the coadministration of functionalized NPs (FNPs) was shown to stimulate the cellular uptake of nonfunctionalized NPs (termed bystander NPs, BNPs), which presents a new strategy to achieve synergistic delivery. However, a mechanistic understanding of the underlying mechanism is still lacking. In this work, the bystander uptake effect was investigated at the cell membrane level by combining the coarse-grained molecular dynamics, potential of mean force calculation and theoretical energy analysis methods. The membrane internalization efficiency of BNPs was enhanced by co-administered FNPs, and such activity depends on the affinity of both NPs to the membrane and the resultant membrane deformation. The membrane-curvature-mediated attraction and aggregation of NPs facilitated the membrane uptake of BNPs. Furthermore, quantitative suggestions were given to modulate the BNP internalization through controlling the FNP properties such as size, concentration and surface-ligand density. Our results provide insight into the molecular mechanism of the bystander uptake effect, and offer a practical guide to regulate the cellular internalization of NPs for targeted and efficient delivery to cells.
Graphical Abstract
A mechanistic understanding and efficient modulation of the bystander uptake effect are given for synergistic delivery of NPs to cells.

1. Introduction
Enormous efforts have been taken over the past decades to develop novel nanoparticle (NP)-based nanomedicines due to the huge potential of NPs in varying applications such as bioimaging,1-3 therapeutic drug/gene delivery4-7 and cell tracking.8, 9 For these biomedical applications, the entry of NPs into the target cells is a critical but challenging step.10, 11 NPs enter cells by several distinct pathways12-15 with endocytosis being dominant.16 The endocytic process of cells is extremely complex.17-21 It is usually initiated by chemo-targeting that relies on NP surface functional ligands, which specifically bind to the cell membrane-embedded receptors in a molecular “lock-and-key” form.22 The process is further modulated by the characteristic properties of NPs, such as type,22-27 shape,28-34 elasticity,35-37 and surface properties.38-42 However, due to the complexity of the corresponding biological processes, more promising strategies are still required to regulate the cell entry of NPs, which currently limits their use for many advanced and diverse clinical applications.43-45
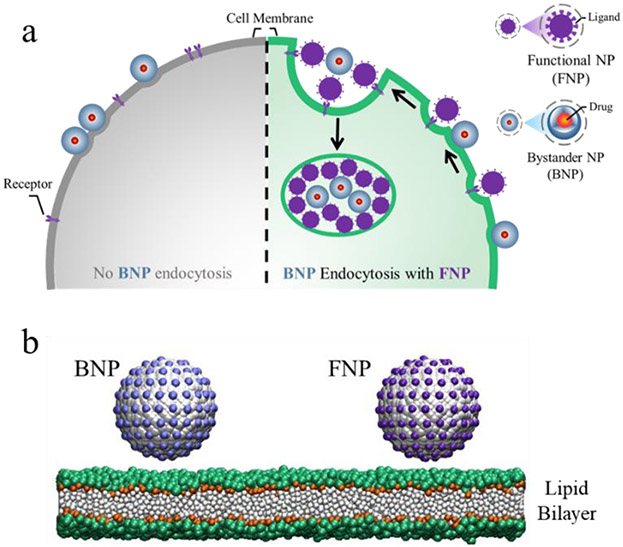
It was recently reported that the cellular internalization of NPs functionalized with transactivator of transcription (TAT) peptides (i.e., FNPs) triggers the cell entry of bystander NPs (BNPs), that cannot be taken up by the cell alone through a receptor-dependent micropinocytosis pathway (Fig. 1a).46 Additionally, Xiang et al reported that the co-administration of peptosomes can act like FNP to improve the delivery efficiency of BNPs to tumours.47 Such an enhanced cellular uptake activity of BNPs, described as the bystander uptake effect herein, promises an efficient manner to regulate the cellular uptake of cargo NPs or a new strategy to realize NP delivery to target cells. However, a mechanistic understanding of this cellular endocytosis behaviour, such as the physical origin behind the FNP-triggered BNP-uptake process, remains largely unknown.
Fig. 1.
Schematic illustration of the bystander uptake effect (a) and simulation models (b). Colour codes used in (b): the core of the NP in grey, ligands on FNP and BNP surface in purple and light blue, respectively, NC3 and PO4 groups of lipids in green, GL1 and GL2 groups of lipids in orange, lipid tails in white.
In this work, the bystander uptake effect was investigated using a coarse-grained (CG) molecular dynamics (MD) simulation and theoretical modelling. The FNPs (mimicking the TAT-coated NPs used in the experiments) and BNPs were constructed and their interactions with the lipid bilayer membrane was performed (Fig. 1). Simulations showed that the membrane internalization of BNPs is dependent on the membrane interaction with both NPs, and thus such an effect could be effectively modulated by controlling the FNP properties such as surficial ligand density and particle size and number. Moreover, calculations of the potential of mean force (PMF) and theoretical energy analysis revealed that the co-endocytosis of BNPs and FNPs can be ascribed to the membrane-curvature-mediated attraction and aggregation of the particles. These results provide insight into the molecular mechanism of the bystander uptake effect of NPs and can guide development of means to regulate the cellular uptake behaviour of NPs as well as novel strategies for targeted delivery of NPs to cells.
2. Methods
2.1. Simulation system and details
The Dry Martini force field was used in the simulations considering the large spatio-temporal scale involved in the cellular endocytosis process. The force field includes four main types of beads (i.e., P: polar, C: apolar, N: nonpolar, Q: charged), and each type has four or five subtypes (such as Q0, Qd and Nda). More details of the Dry Martini force field can be found in the paper of Arnarez et al.48 All simulations were performed with a time step of 30 fs. The second-order stochastic dynamics (SD) integrator with the friction in the Langevin equation (τt = 4.0 ps) was used in the simulations. The temperature was set to 310 K. In the simulations, the Berendsen and MTK barostats were used to control the system pressure in the equilibrium and production processes, respectively. Specifically, the compressibility was set to 0 bar−1 in the direction normal to the bilayer surface and 3 × 10−4 bar−1 in the directions along the bilayer plane with a time constant of τp = 12.0 ps. Accordingly, the tensionless state of the membrane in the pure lipid bilayer system or the membrane system containing FNP and BNP could be well controlled in the simulations (Fig. S1). Each system experienced an equilibrium process of 1 μs, after which a production run of 12 μs was performed. All simulations were performed using GALAMOST (version 4.0) package for a full use of computational power of GPUs.49, 50
2.2. Simulation models
Lipid DLPC (1,2-dodecanol-sn-glycerol-3-phosphocholine) was applied to build the planar lipid bilayer membrane with insane.py script51 (Fig. 1b), and some lipids were chosen as receptors with a special affinity between their head-groups and the ligands coated on the NP surface. Initially, the bilayer had a size of 50 x 50 nm2 unless stated otherwise. The NP was constructed by arranging the Nda-type beads on a face-cantered cubic (FCC) lattice with the nearest distance of 0.37 nm. The spherical diameter ranged from 6 to 10 nm where DF and DB represent the diameters of FNP and BNP, respectively. The ligand beads (Qd1 for FNP and Qd2 for BNP) were evenly decorated on the NP surface at a density of ρ (ρF and ρB referring to FNP and BNP, respectively, Fig. 1b). In the simulations, the receptor-ligand interaction was demonstrated by a non-bonded interaction between the Qd1 or Qd2 bead and Q0 bead (i.e., the head-group of the receptor), as Ur−l = 4ε[(σ/r)12 − (σ/r)6]. Accordingly, the strength of Ur−l could be easily modulated by adjusting the parameter, ε. Noted that in our experiments, FNPs and BNPs differ mainly in their chemical properties or membrane affinities; as a result, unlike FNPs, BNPs are not taken up by cells alone.46 Therefore, to realize such behaviour in the simulations (Fig. S2), for FNP, Ur−l was set to have a larger ε = εF (e.g., εF = 3.5 kJ/mol) due to its stronger membrane affinity; while for BNP, its weaker affinity with receptors was achieved with a smaller ε = εB (e.g., εB = 1.5 kJ/mol). In addition, the time for membrane wrapping of NPs is on the order of a second,52 which is well much beyond the simulation timescale. To overcome this limitation, a high density (30% or 100%) was used as in previous simulations.23, 34, 53-55 Additionally, it was found that even with a low total receptor density of 30%, the local number density grew quickly upon FNP binding to more than 80% around the membrane bound particles (Fig. S3a). In the cases with varying receptor percentages, the membrane internalization degrees of NPs were still similar (Fig. S3b). Thus, to improve the computational efficiency, all lipids were considered as receptors in the simulations except where noted to mimic the situations with sufficient receptors in the system. However, this necessary compromise is due to the current limitation of computational ability. The actual membrane internalization rate of NPs, including the occurrence time, is still influenced by the receptor density as well as their diffusion behaviour in the membrane (e.g., Fig. S3b).
2.3. Potential of mean force calculation
The PMF as a function of the distance between FNP and BNP, L, was calculated based on the umbrella sampling method following the most-used protocol (see ESI for details).56-58 In short, the initial FNP-BNP distance was divided into the configuration windows spaced by 0.2 nm. Then in each window, the system was simulated for 400 ns with a spring force using a constant of 1000 J/mol/nm2 applied between BNP and FNP to restrain the special NPs’ configuration on the membrane. The last 300 ns of such trajectories was then used for the Weighted Histogram Analysis Method with the program g_wham in the GROMACS package.59 In each window for sampling, the membrane morphology remained stable without obvious change under the NPs’ interactions (Fig. S4).
2.4. Data analysis
The membrane internalization degree of a NP could be described by the extent to which the nanoparticle is wrapped by the membrane, denoted as η = n/N. Here, n is the number of the surface beads in the membrane deformation region, and N is the total number of the surface beads of NP. As such, a larger value of η corresponds to a higher membrane internalization degree. To quantitatively describe the bystander uptake effect, the relative membrane internalization degree of a BNP, ηR = ηB1/ηB0, was used in our simulations, where ηB0 is the membrane internalization degree of the BNP alone (without FNPs) and ηB1 is the internalization degree of the BNP with the help of FNPs. For each system, the last 1 μs trajectory was used for the calculation of η or ηR.
3. Results and discussion
3.1. FNP-triggered membrane internalization of BNPs
The affinity between ligands and receptors drives the receptor-mediated endocytosis of NPs. For the BNPs with a weak membrane affinity (i.e., εB = 1.5 kJ/mol), the particles only adsorb to the membrane surface without internalization throughout the simulations (Fig. S2a). However, for the FNPs with εF = 3.5 kJ/mol, a strong membrane deformation occurs due to FNP actions in the same simulation conditions (Fig. S2b). As a result, the FNPs are mostly wrapped by the membrane (η ≈ 70 %). These observations agree well with previous studies.34 Moreover, an effective membrane internalization of BNPs could be realized with the help of FNPs, when both of them were placed on the membrane surface (Figs. 2a and S5). The internalization was also dependent on εF and εB. When εB ≥ 1.5 kJ/mol, the BNP spontaneously moves towards the FNP (Fig. 2b), which energetically causes lipid membrane deformation (Fig. S5). Eventually, the BNP is wrapped by the membrane together with the FNP (Figs. 2a and S5). The simulation thus provides a plausible mechanism for the promotion of BNPs internalization by FNPs that is also corroborates experimental observations.46 Prior to associating with FNP, BNPs undergoes lateral surface diffusion with the duration dependent on the value of εB (Fig. S7). That is, large values promote FNP-BNP association, but if the BNP-membrane affinity is too weak (e.g., εB = 1.0 kJ/mol), the BNP remain distant (Fig. 2b) or even detach from the surface (Fig. S5). In these latter situations, little bystander uptake is observed. This is also consistent with experimental observation of TAT-NPs, where little or no bystander activity to solute tracers was seen (the tracer has no affinity to the cell membrane).39
Fig. 2.
Dependence of the membrane internalization of BNPs on εB and εF. (a) Representative NPs-membrane interaction states (side-view snapshots) under the various εB and εF (kJ/mol). All the snapshots in the table are obtained at 12 μs. (b) Evolution of BNP-FNP distance during the membrane interaction process. εF = 3.5 kJ/mol. (c) Change of the relative membrane internalization degree (ηR) of BNPs with εB and εF. In all cases, ρF = ρB = 1.6 nm−2, DF = DB = 8 nm.
The relative membrane internalization degree of the BNPs, ηR, provides a quantitative measure of the bystander uptake effect the value of ηR is determined by both εB and εF. For most values, ηR > 100% suggesting an enhanced membrane internalization (i.e., cellular uptake) of BNPs with the help of FNPs. With a certain εB, ηR increases with the increase of εF (Fig. 2c), indicating that a stronger ligand-receptor affinity of the FNPs would further improve the membrane internalization efficiency of BNPs. The influence of εB on the BNP internalization behaviour is more complicated. On the one hand, ηR decreases with an increase in εB, and when εB is sufficiently strong (e.g., εB = 2.7 kJ/mol), individual BNPs without FNPs can be enveloped by the membrane to some extent (e.g., η > 50%, Fig. S5 and S6). In this case, the enhancing role of FNPs is effectively reduced. On the other hand, when εB is rather weak, ηR is less than 100% (e.g., ηR < 80% with εB = 1.0 kJ/mol, dashed columns in Fig. 2c and Fig. S5). It appears that the ensuing large perturbation of membrane due to FNP adsorption interferes with BNP membrane association and/or binding to FNP, which results in a small ηR value (Fig. S8). Overall, our simulations indicate that the bystander uptake effect occurs only when the BNP-membrane affinity exceeds a threshold value (e.g., εB ≥ 1.5 kJ/mol) and a larger discrepancy in membrane affinity between BNPs and FNPs would further enhance such effect.
3.2. Influence of FNP properties on the bystander uptake effect
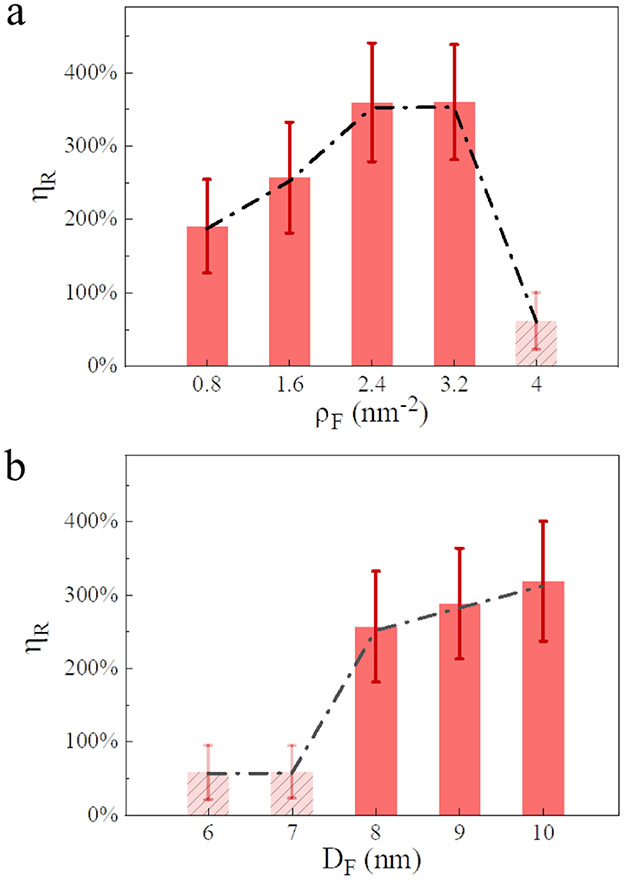
The density of ligands on the FNP surface, ρF, is an important factor determining the FNP-membrane interaction process and thereby the bystander activity. Fig. 3a shows that ηR increases monotonically (from ~200% to ~360%) with an increase of ρF when ρF ≤ 2.4 nm−2, and then reaches a plateau value with 2.4 ≤ ρF ≤ 3.2 nm−2. However, when ρF is further increased (e.g., ρF = 4.0 nm−2), ηR drops significantly (to ~60%). That is, there appears to be an optimal ligand density for the bystander uptake effect. It may be that with the increase of ligand density, the membrane deformation under FNP actions becomes more dramatic, which significantly facilitates the membrane internalization of FNPs and enhances the cellular uptake efficiency of BNPs. However, for large ρF values, the FNP would be rapidly internalized by the membrane (e.g., <0.2 μs), and the BNP is thus left on the membrane without internalization (Fig. S9).
Fig. 3.
Influences of ligand density (a) and particle size (b) of FNPs on the relative membrane internalization degree of BNPs (ηR). εF = 3.5 kJ/mol, εB = 1.5 kJ/mol. In (a), ρB = 1.6 nm−2 while ρF varies, DF = DB = 8 nm; In (b), ρF = ρB = 1.6 nm−2, DB = 8 nm while DF varies. Dashed lines are used to guide eyes.
The size of FNPs also affects the bystander uptake effect, although to a lesser extent than that of ρF. As shown in Fig. 3b, for BNPs with DB = 8 nm, FNPs with a size similar to or larger undergo enhanced membrane internalization. Specifically, ηR increases from 50% (DF = 6 or 7 nm) to more than 260% (DF = 8~10 nm). In contrast, FNPs smaller than the BNP undergo little or no enhancement effect. As opposed to the large FNPs, the smaller FNP do not induce significant membrane perturbation (Fig. S10) most likely due to the higher energy required for membrane wrapping of the small NP. Additionally, it is shown by our simulations that the initial FNP-BNP distance only has a minor influence on the bystander uptake effect (Fig. S11). In general, it is obviously demonstrated that the bystander uptake effect can be effectively regulated by modulating the properties of FNPs.
3.3. Association between the bystander uptake effect and membrane curvature change
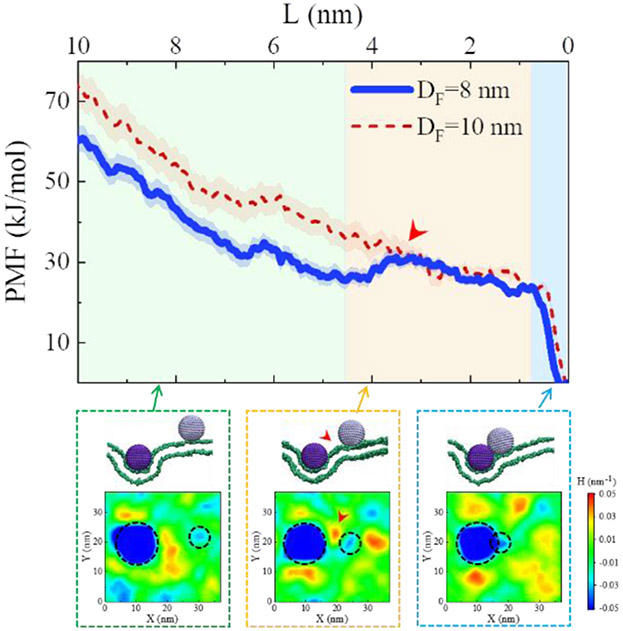
The potential mean force was calculated using the distance between FNP and BNP, L, as the reaction coordinate. Using the case of DF = 8 nm as an example, the PMF exhibited a complicated, stage-like decline with L (Fig. 4). The PMF displays a rapid decrease when the BNP and FNP are far away from each other (i.e., L > ~4.5 nm); and then the decrease becomes slower, even with a small increase in the middle stage; after that, a dramatic drop in PMF appears at L~0.8 nm, reflecting the aggregation of the two NPs in membrane (Fig. 4). Insofar as no specific affinity is assigned between the BNP and FNP (see Fig. S12c for the detailed interaction energy analysis), the decline of the calculated PMF profile with L suggests a membrane-mediated attraction between NPs. Moreover, at different FNP-BNP distances, the varying membrane deformation or membrane wrapping degrees of NPs occur under the NP-membrane interactions (Fig. S13). In a closer examination, the PMF was noted to be closely correlated to the change membrane curvature. As shown in Fig. 4, the decreasing of PMF exactly corresponds to the merging process of two membrane regions with negative curvatures induced by the membrane-bound FNP and BNP, respectively (the blue parts highlighted by dashed circles, Fig. 4). Even, a small membrane protrusion with positive curvature (indicated by red arrowheads in Fig. 4) appears during the approaching of the NPs, referring to the increase of the PMF profile observed in the middle stage. In addition, in the case of DF = 10 nm, the L-dependent decrease of PMF is even steeper and more smoothly, corresponding to the stronger membrane deformation and higher NP internalization efficiency. Similar membrane curvature changes around the NPs were also observed in the unbiased simulations with spontaneous movement of BNP towards FNP (Fig. S12a, b). In addition, the systems containing FNPs with varying ρF values also demonstrated a similar tendency in the PMF profile (Fig. S14). All these results suggest that the bystander uptake effect is a result of the membrane-curvature-mediated attraction and aggregation of the NPs.53, 55, 60-62
Fig. 4.
Change of the PMF profile with the BNP-FNP distance (L). Dashed boxes show the representative snapshots during the NP-membrane interactions with different L and the corresponding membrane curvature patterns (bottom). For clarity, only lipid heads (green beads) are shown in the snapshots. εF = 3.5 kJ/mol, εB = 1.5 kJ/mol, ρF = ρB = 1.6 nm−2, DB = 8 nm, DF = 8 or 10 nm.
3.4. Cooperation of FNPs regulates the bystander activity
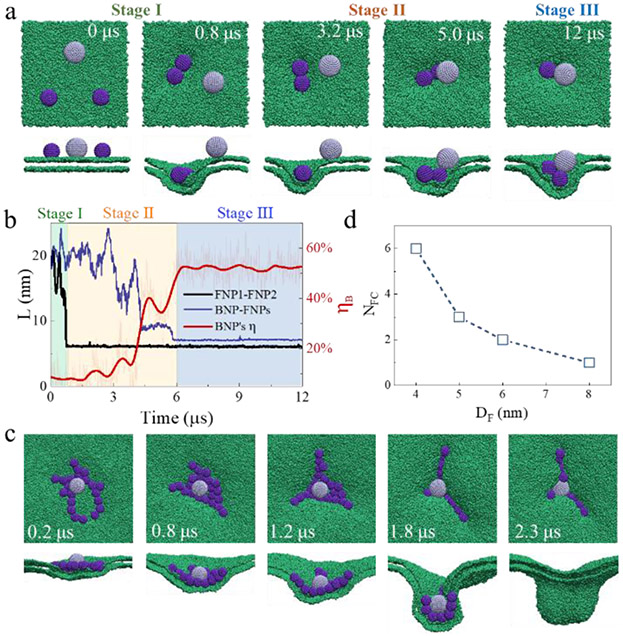
In the simulations above, the simplest situation consisting of a single FNP and a BNP was considered. However, the FNP number (NF), i.e., the relative concentration of FNPs to BNPs, also plays a role in determining the NP-membrane interactions. For example, as shown in Fig. 3b, it is difficult for a small FNP with DF = 6 nm to activate the membrane internalization of a BNP with DB = 8 nm due to the weak membrane actions of the FNP. However, at a higher FNP number, e.g., from one to two, the bystander uptake became obvious (Fig. 5a). In this case, the occurrence of the BNP internalization is followed by the aggregation of the two small FNPs (Fig. 5a). This point is also reflected by the close association between the changes of ηR and that of the BNP-FNP distance (Fig. 5b): the two small FNPs aggregate together rapidly (Stage I), which triggers the gradual approaching of the BNP (Stage II), and the final FNP-BNP aggregation is accompanied by a much-enhanced membrane internalization degree of the particles (Stage III).
Fig. 5.
FNP number and size dependence. (a) Representative simulation snapshots showing the membrane internalization of a BNP with the help of two FNPs. (b) Time-evolutions of the NP-NP distance and the membrane internalization degree of a BNP. Background colours refer to the three stages. (c) Representative simulation snapshots showing the membrane internalization of a BNP with the help of 21 FNPs. In this case, the membrane size is 70 nm × 70 nm. (d) Correlation between the critical number of FNPs required for the internalization of a BNP and the FNP size. In all cases, εF = 3.5 kJ/mol, εB = 1.5 kJ/mol, ρF = ρB = 1.6 nm−2, DB =8 nm; in (a-c), DF = 6 nm.
An increased FNP number in the system would also further strengthen the degree of membrane deformation and facilitate the bystander uptake effect. Fig. 5c shows the events associated with a FNP number of NF = 21, in which a large-scale membrane deformation occurs (even like the vesiculation observed in experiments),46 along with a strong membrane wrapping of the BNP closely surrounded by the FNPs.
In addition, the effect of FNP number is dependent on FNP size. We calculated the minimum number of FNPs (NFc) needed to activate the bystander uptake of a BNP with DB = 8 nm (i.e., ηR ≥ 200%). As shown in Fig. 5d, with the decrease of DF, more FNPs are required, but the initial locations of FNPs relative to the BNP only have a minor effect on ηR of the BNP (Fig. S15). Generally, all these results suggest the complicated cooperative effect of FNPs, which is influenced by the particle size and number (i.e., concentration), in activating the membrane uptake of BNPs.
3.5. Theoretical energy analysis of the NP-membrane interactions
A theoretical energy analysis of the interaction between the membrane and particles (i.e., FNPs and BNP) and the possible membrane wrapping situations were examined based on the Helfrich model (Fig. 6a).63 For such a multiple-NP-membrane system, the total energy E could be taken as:
| (1) |
Fig. 6.
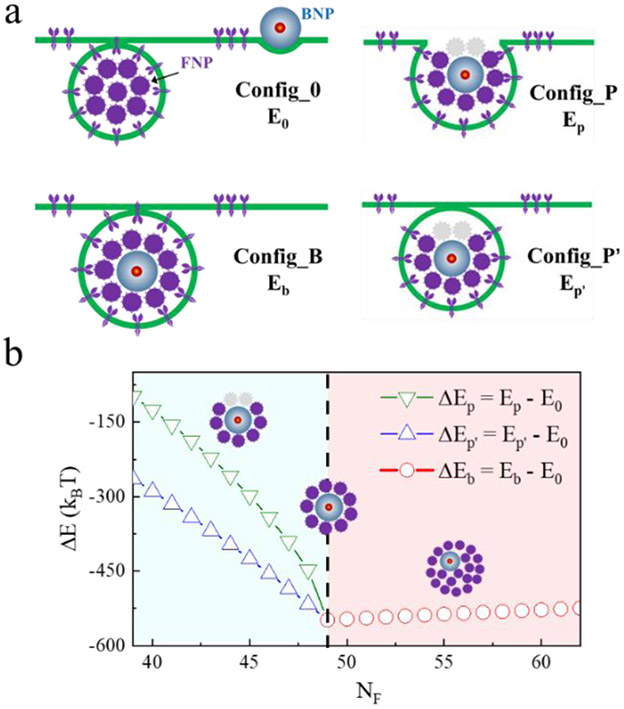
Energy analysis of the bystander uptake effect. (a) Sketches showing some typical interaction states between the NPs (i.e., FNPs and BNPs) and the membrane. (b) Changes of energy differences between the different states with the FNP number NF. Insets show the possible covering situations of FNPs around BNPs. NB = 1, rF = 10 nm, rB = 25 nm.
Here, Ebind = −Npair μ, in which Npair is the number of ligand-receptor pairs and μ is the energy released upon the binding of a ligand-receptor pair, whose value is on the order of 20kBT based on experimental measurements.64 Usually, the cross-sectional area of a receptor is about a0 ≈ 192 nm2.64 For a NP with the radius r partially wrapped by the membrane, its contact area with membrane is 4πr2η, where η is the membrane wrapping degree of the NP. Thus, η = 1 refers to the fully membrane-wrapping state of the NP, η = 0 is the slight membrane-adhesion state of the NP, and 0 < η < 1 corresponds to the partially membrane-wrapping state of the NP. Accordingly, Npair could be taken as Npair = 4πr2η/a0. The bending energy, Ebend, of the membrane was calculated as Ebend = 1/2κA(H1 + H2)2 where the bending rigidity κ is typically on the order of 20 kBT,65 H1 and H2 are the local principal curvatures of the deformed membrane region, and A is the area of the deformed membrane. For a membrane-wrapped spherical NP, , and A = 4πr2η. Erim = γL is the line energy of the rim of the deformed membrane region, in which γ is the line tension(γ~ kB T/nm) and L is the length of the rim.66 Similarly, L was taken as . Thus, for a simple NP-membrane interaction system, the associated energy could be written as:
| (2) |
Due to the complexity of the membrane wrapping process in the bystander uptake activity, the energy differences between some typical states (ΔE) associated with the bystander uptake effect was the focus. That is, the bystander uptake or non-bystander uptake states shown in Fig. 6a were examined to determine the energetically favourable state, instead of the wrapping pathway. The system contained multiple FNPs (note that NF is a variable here) with a radius of rF = 10 nm and one BNP with the radius of rB = 25 nm. As shown in Fig. 6a, the Config_0 state refers to the state without the bystander effect, that is, all FNPs are wrapped by the membrane while the BNP remains at the membrane surface. For simplification, the membrane-wrapped FNPs are regarded approximately as a “big NP” with the radius of R0, which has the same volume as that of the membrane-wrapped FNPs (i.e.,). The membrane wrapping degree of the BNP was assumed to be ηB0. Thus,
| (3) |
The first term on the right of (3) stems from the energy contribution of the FNP-membrane interactions and the second term is for the BNP. It is noted that there is also a ligand-receptor binding energy for BNP, although μB ≪ μF. Here, we assume μF = 20 kBT, μB = 4 kBT, and γB0 = 20%.
In contrast, the Config_B state could be considered as an ideal bystander uptake state (Fig. 6a). Based on the simulation (e.g., the snapshots in Fig. 5c) and experimental observations,46 we assume that, in this state, the BNP is always surrounded by the FNPs, and then they are wrapped by the membrane completely. Also, all these wrapped NPs (including both FNPs and BNP) are considered as a “big NP” with the same volume. Thus,
| (4) |
In (4), R1 the radius of the equivalent “NP”, whose volume .
The partial bystander uptake state probably exists, if there are not enough FNPs or the BNP is too large. Under this situation, only part of the BNP surface could be covered by the FNPs, and the coverage ratio is approximately . Two possibilities were taken into consideration (Fig. 6a). One, the membrane is only able to wrap the part of the BNP which is covered by the FNPs (i.e., Config_P). In this case, the system energy is about:
| (5) |
The other, the BNP and the surrounding FNPs are still able to be internalized by the membrane completely, if the binding between the BNP and the membrane is considered and ηc is large enough (Config_P’, Fig. 6a). Accordingly, the energy is:
| (6) |
Also, R2 or R2, is the radius of the equivalent “NP”. For simplification, we only consider the situation with a large ηc (ηc > 80%), namely a large part of the BNP surface is covered by the FNPs. Thus, R2 or R2, is approximately equal to R1. Based on these considerations, the energy difference between the states associated with the bystander activity could be calculated as ΔEb = Eb – E0, ΔEp = Ep – E0, and ΔEp′ = Ep′ – E0.
As shown in Fig. 6b, the calculated values of all three ΔE are less than zero. This indicates that the co-endocytosis of BNP with FNPs is more energy favourable compared with the non-bystander uptake state (i.e., Config_0 state). Moreover, with the increase of NF, both ΔEp and ΔEp′ decrease rapidly, suggesting that an increased FNP number would effectively facilitate the bystander uptake behaviour, being consistent with the simulation observations (Fig. 5). With a further increased in NF (e.g., NF ≥ 49 here, with which the FNPs are more than enough to fully cover the BNP surface), ΔEb increases slowly with NF, although is still less than zero. This is mainly ascribed to the decrease in the surface area gain of the membrane-wrapped NPs with NF (ΔS, see Fig. S16). These results demonstrate that an optimal FNP number (i.e., relative concentration of FNPs to BNPs) is suggested in improving the bystander uptake efficiency.
For simplification, the shape of the vesicle containing wrapped NPs is assumed to be spherical in the energy analysis above. However, based on previous theoretical models and especially experimental observations,46, 67-70 both spherical and tube-like endocytic vesicles are possibly formed. Therefore, energy analysis of the energy differences between some typical states in the bystander uptake activity with tube-like vesicle forms was also performed (see ESI for details). Similarly, it is found that, compared with the non-bystander uptake state, the bystander uptake states are more energetically favourable (Fig. S17) although the membrane is deformed in a tube-like form.
4. Conclusion
The bystander uptake effect is one of the newly found pathways for NPs entrance into cells.46 We investigated the underlying molecular mechanism by combining the MD simulations with theoretical model analysis. The simulations revealed that the membrane wrapping of FNPs will enhance the membrane internalization of BNPs, consistent with experimental observations. From PMF calculations and membrane curvature analysis, the bystander uptake effect was closely correlated with the FNP/BNP-membrane interactions and the resultant membrane-curvature-mediated attraction and aggregation of the NPs. The FNP properties of size, number (i.e., relative concentration of FNPs to BNPs), and the number density of ligands coated on the FNP surface, play complicated roles in regulating the bystander uptake effect. Note that the cooperative wrapping behaviours of NPs by the membrane have been reported before, which, however, were mostly concentrated on the NPs with the same or similar properties.53, 55, 60, 69-71 It was clearly demonstrated by our simulations and energy analysis that membrane-mediated cooperation of NPs could also occur between NPs with distinct properties. Furthermore, the strong dependence of the bystander uptake effect on the properties of BNPs and especially FNPs, such as the cargo selectivity effect or the quantitative modulating ability of the membrane internalization degree of BNPs by the FNP number and size, provides a larger parametric space to control the cooperation or cellular uptake of NPs. Moreover, the stage-like internalization process of BNP with multiple FNPs also provides a deeper insight into the complexity of the membrane-mediated cooperation of NPs. In all, our results provide a mechanistic insight into the bystander uptake effect, and suggest the practical ways to regulate such a new cellular uptake behaviour for applications.
Supplementary Material
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Nos. 21774092, U1532108, and U1932121), the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, National Institute of Health of USA (R01CA214550, R01GM133885), and the State of Minnesota (MNP#19.08).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Wolfbeis OS, Chem. Soc. Rev, 2015, 44, 4743–4768. [DOI] [PubMed] [Google Scholar]
- 2.Rodríguez-Fajardo V, Sanz V, de Miguel I, Berthelot J, Aćimović SS, Porcar-Guezenec R and Quidant R, Nanoscale, 2018, 10, 4019–4027. [DOI] [PubMed] [Google Scholar]
- 3.Gao X, Cui Y, Levenson RM, Chung LW and Nie S, Nat. Biotechnol, 2004, 22, 969–976. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA and Siegwart DJ, Nat. Nanotechnol, 2020, 15, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnstone TC, Suntharalingam K and Lippard SJ, Chem. Rev, 2016, 116, 3436–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T, Zhang YS, Pang B, Hyun DC, Yang M and Xia Y, Angew. Chem. Int. Ed, 2014, 53, 12320–12364. [DOI] [PubMed] [Google Scholar]
- 7.Lung P, Yang J and Li Q, Nanoscale, 2020, 12, 5746–5763. [DOI] [PubMed] [Google Scholar]
- 8.Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT and Weissleder R, Nat. Biotechnol, 2000, 18, 410–414. [DOI] [PubMed] [Google Scholar]
- 9.Jin D, Xi P, Wang B, Zhang L, Enderlein J and van Oijen AM, Nat. Meth, 2018, 15, 415–423. [DOI] [PubMed] [Google Scholar]
- 10.Barua S and Mitragotri S, Nano Today, 2014, 9, 223–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Gao H and Bao G, ACS Nano, 2015, 9, 8655–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong-Ekkabut J, Baoukina S, Triampo W, Tang IM, Tieleman DP and Monticelli L, Nat. Nanotechnol, 2008, 3, 363–368. [DOI] [PubMed] [Google Scholar]
- 13.Ding HM, Tian WD and Ma YQ, ACS Nano, 2012, 6, 1230–1238. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, Terazzi E, Seemann R, Fleury JB and Baulin VA, Sci. Adv, 2016, 2, e1600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Yang R, Zhang F, Yuan B, Yang K and Ma YQ, Nanoscale, 2018, 10, 11670–11678. [DOI] [PubMed] [Google Scholar]
- 16.Sahay G, Alakhova DY and Kabanov AV, J. Control. Release, 2010, 145, 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson JA, Nat. Rev. Mol. Cell Biol, 2008, 9, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA and Thompson CB, Nature, 2013, 497, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaksonen M and Roux A, Nat. Rev. Mol. Cell Biol, 2018, 19, 313. [DOI] [PubMed] [Google Scholar]
- 20.Nabi IR and Le PU, J. Cell Biol, 2003, 161, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayor S and Pagano RE, Nat. Rev. Mol. Cell Biol, 2007, 8, 603–612. [DOI] [PubMed] [Google Scholar]
- 22.Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V and Thompson M, Nat. Mater, 2009, 8, 543–557. [DOI] [PubMed] [Google Scholar]
- 23.Shi X, von Dem Bussche A, Hurt RH, Kane AB and Gao H, Nat. Nanotechnol, 2011, 6, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P, Xu Z, Zhu G, Dai X and Yan LT, Phys. Rev. Lett, 2020, 124, 198102. [DOI] [PubMed] [Google Scholar]
- 25.Ji QJ, Yuan B, Lu XM, Yang K and Ma YQ, Small, 2016, 12, 1140–1146. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Li J, Lykotrafitis G, Bao G and Suresh S, Adv. Mater, 2009, 21, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Ding HM, Xu Y, Zeng D, Zhu H, Zang DM, Bao SS, Ma YQ and Zheng LM, Nat. Commun, 2017, 8, 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang K and Ma YQ, Nat. Nanotechnol, 2010, 5, 579–583. [DOI] [PubMed] [Google Scholar]
- 29.Chithrani BD, Ghazani AA and Chan WC, Nano Lett., 2006, 6, 662–668. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Yue T, Yang K and Zhang X, Biomaterials, 2012, 33, 4965–4973. [DOI] [PubMed] [Google Scholar]
- 31.Huang C, Butler PJ, Tong S, Muddana HS, Bao G and Zhang S, Nano Lett., 2013, 13, 1611–1615. [DOI] [PubMed] [Google Scholar]
- 32.Yang K, Yuan B and Ma YQ, Nanoscale, 2013, 5, 7998–8006. [DOI] [PubMed] [Google Scholar]
- 33.Ding H, Li J, Chen N, Hu X, Yang X, Guo L, Li Q, Zuo X, Wang L and Ma Y, ACS Cent. Sci, 2018, 4, 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vácha R, Martinez-Veracoechea FJ and Frenkel D, Nano Lett., 2011, 11, 5391–5395. [DOI] [PubMed] [Google Scholar]
- 35.Yi X, Shi X and Gao H, Phys. Rev. Lett, 2011, 107, 098101. [DOI] [PubMed] [Google Scholar]
- 36.Hui Y, Yi X, Wibowo D, Yang G, Middelberg AP, Gao H and Zhao CX, Sci. Adv, 2020, 6, eaaz4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi X and Gao H, Nanoscale, 2017, 9, 454–463. [DOI] [PubMed] [Google Scholar]
- 38.Correa S, Boehnke N, Barberio AE, Deiss-Yehiely E, Shi A, Oberlton B, Smith SG, Zervantonakis I, Dreaden EC and Hammond PT, ACS Nano, 2020, 14, 2224–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X, Xu P, Ding HM, Yu YS, Huo D and Ma YQ, Nat. Commun, 2019, 10, 4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song E, Gaudin A, King AR, Seo YE, Suh HW, Deng Y, Cui J, Tietjen GT, Huttner A and Saltzman WM, Nat. Commun, 2017, 8, 15322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walkey CD, Olsen JB, Guo H, Emili A and Chan WC, J. Am. Chem. Soc, 2012, 134, 2139–2147. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Ding H, Lin M, Yin F, Song L, Mao X, Li F, Ge Z, Wang L, Zuo X, Ma Y and Fan C, J. Am. Chem. Soc, 2019, 141, 18910–18915. [DOI] [PubMed] [Google Scholar]
- 43.Wei Q, Huang C, Zhang Y, Zhao T, Zhao P, Butler P and Zhang S, Adv. Mater, 2018, 30, 1707464. [DOI] [PubMed] [Google Scholar]
- 44.Forster JC, Krausser J, Vuyyuru MR, Baum B and Šarić A, Phys. Rev. Lett, 2020, 125, 228101. [DOI] [PubMed] [Google Scholar]
- 45.Blanco E, Shen H and Ferrari M, Nat. Biotechnol, 2015, 33, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y, Tang T and Pang HB, Nat. Commun, 2019, 10, 3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang Z, Jiang G, Yang X, Fan D, Nan X, Li D, Hu Z and Fang Q, Biomolecules, 2019, 9, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnarez C, Uusitalo JJ, Masman MF, Ingólfsson HI, de Jong DH, Melo MN, Periole X, de Vries AH and Marrink SJ, J. Chem. Theory Comput, 2014, 11, 260–275. [DOI] [PubMed] [Google Scholar]
- 49.Zhu YL, Liu H, Li ZW, Qian HJ, Milano G and Lu ZY, J. Comput. Chem, 2013, 34, 2197–2211. [DOI] [PubMed] [Google Scholar]
- 50.Zhu YL, Pan D, Li ZW, Liu H, Qian HJ, Zhao Y, Lu ZY and Sun ZY, Mol. Phys, 2018, 116, 1065–1077. [Google Scholar]
- 51.Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP and Marrink SJ, J. Chem. Theory Comput, 2015, 11, 2144–2155. [DOI] [PubMed] [Google Scholar]
- 52.Gao H, Shi W and Freund LB, Proc. Natl. Acad. Sci. U. S. A, 2005, 102, 9469–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K and Deserno M, Nature, 2007, 447, 461–464. [DOI] [PubMed] [Google Scholar]
- 54.Liang J, Chen P, Dong B, Huang Z, Zhao K and Yan LT, Biomacromolecules, 2016, 17, 1834–1844. [DOI] [PubMed] [Google Scholar]
- 55.Yan Z, Wu Z, Li S, Zhang X, Yi X and Yue T, Nanoscale, 2019, 11, 19751–19762. [DOI] [PubMed] [Google Scholar]
- 56.Angelikopoulos P, Sarkisov L, Cournia Z and Gkeka P, Nanoscale, 2017, 9, 1040–1048. [DOI] [PubMed] [Google Scholar]
- 57.Lavagna E, Barnoud J, Rossi G and Monticelli L, Nanoscale, 2020, 12, 9452–9461. [DOI] [PubMed] [Google Scholar]
- 58.Šarić A and Cacciuto A, Phys. Rev. Lett, 2012, 108, 118101. [DOI] [PubMed] [Google Scholar]
- 59.Hub JS, De Groot BL and Van Der Spoel D, J. Chem. Theory Comput, 2010, 6, 3713–3720. [Google Scholar]
- 60.Yue T and Zhang X, ACS Nano, 2012, 6, 3196–3205. [DOI] [PubMed] [Google Scholar]
- 61.Yue T, Wang X, Huang F and Zhang X, Nanoscale, 2013, 5, 9888–9896. [DOI] [PubMed] [Google Scholar]
- 62.Zhu G, Xu Z and Yan LT, Nano Lett., 2020, 20, 5616–5624. [DOI] [PubMed] [Google Scholar]
- 63.Ou Yang ZC and Helfrich W, Phys. Rev. Lett, 1987, 59, 2486–2488. [DOI] [PubMed] [Google Scholar]
- 64.Tzlil S, Deserno M, Gelbart WM and Ben-Shaul A, Biophys. J, 2004, 86, 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helfrich W, Z. Naturforsch. C, 1973, 28, 693–703. [DOI] [PubMed] [Google Scholar]
- 66.Baumgart T, Hess ST and Webb WW, Nature, 2003, 425, 821–824. [DOI] [PubMed] [Google Scholar]
- 67.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR and McMahon HT, Nature, 2002, 419, 361–366. [DOI] [PubMed] [Google Scholar]
- 68.Messerschmidt C, Hofmann D, Kroeger A, Landfester K, Mailänder V and Lieberwirth I, Beilstein J. Nanotechnol, 2016, 7, 1296–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Yao H and Shi X, J. Mech. Phys. Solids, 2014, 73, 151–165. [Google Scholar]
- 70.Raatz M, Lipowsky R and Weikl TR, Soft Matter, 2014, 10, 3570–3577. [DOI] [PubMed] [Google Scholar]
- 71.Bahrami AH, Lipowsky R and Weikl TR, Phys. Rev. Lett, 2012, 109, 188102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.