Abstract
Background:
The discovery that short oligonucleotides, termed aptamers, can fold into three-dimensional structures that allow them to selectively bind and inhibit the activity of pathogenic proteins is now over 25 years old. The invention of the SELEX methodology heralded in an era in which such nucleic acid-based ligands could be generated against a wide variety of therapeutic targets.
Results:
A large number of aptamers have now been identified by combinatorial chemistry methods in the laboratory and moreover, an increasing number have been discovered in nature. The affinities and activities of such aptamers have often been compared to that of antibodies, yet only a few of these agents have made it into clinical studies compared to a large and increasing number of therapeutic antibodies. One therapeutic aptamer targeting VEGF has made it to market, while 3 others have advanced as far as phase III clinical trials.
Conclusion:
In this manuscript, we hope the reader appreciates that the success of aptamers becoming a class of drugs is less about nucleic acid biochemistry and more about target validation and overall drug chemistry.
Keywords: Aptamer, DNA, RNA, therapeutic, clinical trial, ligonucleotides
1. INTRODUCTION
Observations in the 1980s that viruses had evolved nucleic acid ligands, subsequently termed aptamers [1], to modulate the activities of viral and cellular proteins essential for their replication [2], or to interfere with cellular proteins involved in antiviral responses [3], led translational researchers to investigate whether such ligands could be employed for therapeutic applications. The first two studies demonstrating that such nucleic acid aptamers can inhibit the activities of therapeutically relevant proteins were published in 1990 [4, 5]. Sullenger and colleagues demonstrated that an RNA aptamer derived from the Human Immunodeficiency Virus (HIV) RNA genome, a stem-loop structured RNA ligand called Transactivation response element (TAR), Fig. (1) could act as a decoy and squelch the activity of the viral Trans-activator of transcription (tat) protein and potently inhibit HIV replication in CD4+ T-cells [4]. Bielinska et al. demonstrated that a DNA aptamer derived from the DNA binding site of the transcription factor NF-kappa B could limit NF-kappa B activation of gene expression from the Interleukin-2 (IL-2) and HIV promoters in B and T cells [5]. These results suggested that RNA and DNA aptamers derived from nature represent novel therapeutic agents to control the activities of clinically relevant nucleic acid-binding proteins. During the intervening 25 years, numerous naturally occurring aptamers have been discovered that selectively bind to many clinically relevant nucleic acid-binding proteins as well as cellular metabolites [6, 7]. A few of these naturally derived aptamers have been evaluated in clinical studies as potential treatments for maladies ranging from cardiovascular to infectious diseases.
Fig. (1).
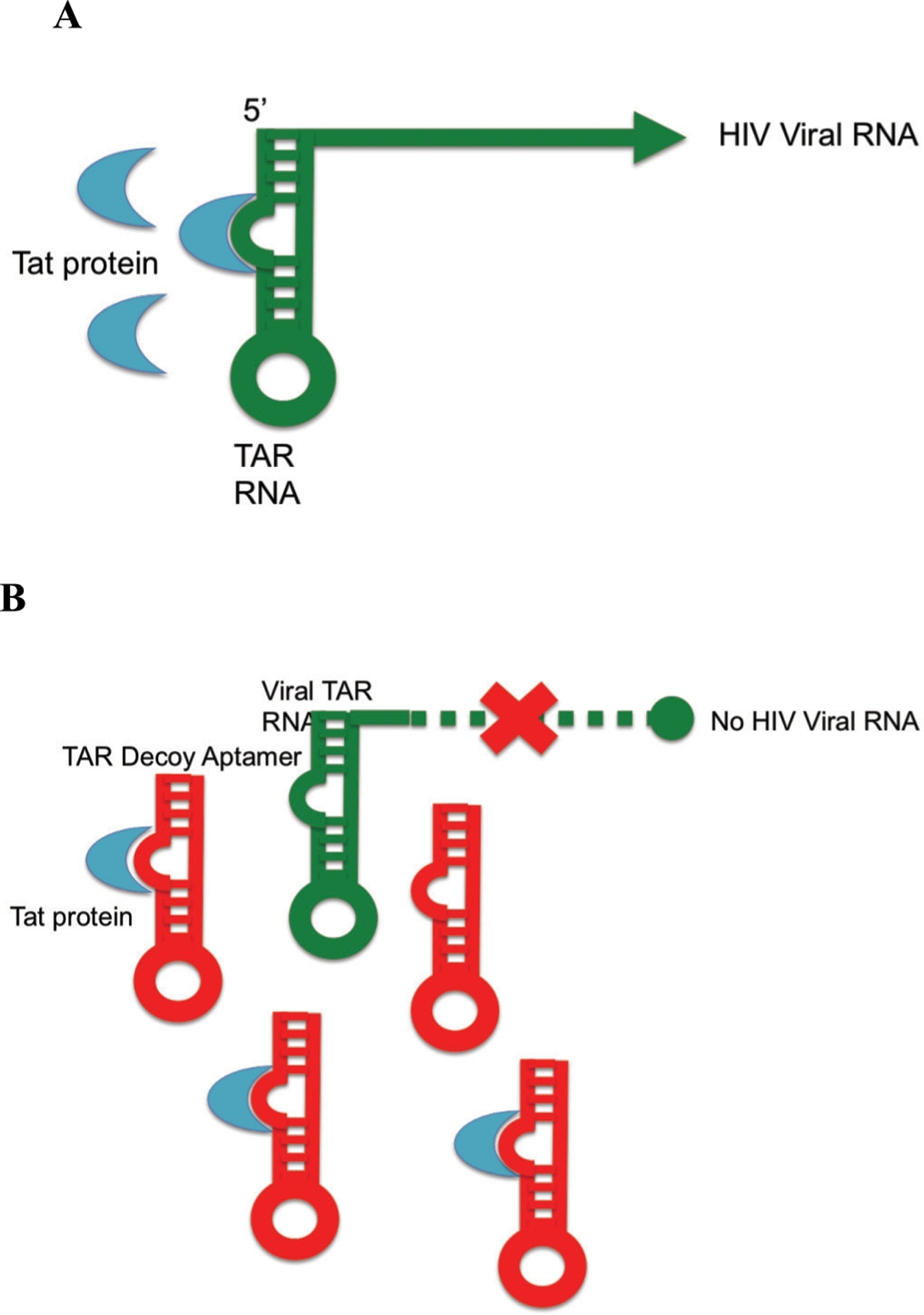
A: HIV evolution and use of an RNA aptamer as a decoy. A: HIV evolved an RNA aptamer termed Trans-Activator Response (TAR) element to control its gene expression and replication. The viral trans-activator of transcription (tat) protein binds to TAR at the 5’ end of all viral RNAs and together with cellular factors activates viral gene expression and replication. B: Inhibition of HIV replication by the first described therapeutic aptamer. TAR decoy RNA aptamers bind the tat protein, preventing them from binding the viral TAR sequence, thereby inhibiting tat-mediated activation of HIV gene expression and replication [4, 14].
In 1990, two additional seminal publications demonstrated that RNA aptamers could also be generated in the laboratory using combinatorial chemistry methods (Fig. 2) [1, 8]. In these studies, large libraries of artificially created randomized RNA molecules were screened in the test tube for those molecules in the library that could be ligands and bind T4 DNA polymerase [8] or an organic dye [1] with high affinity. The term aptamer, which has been adopted by the field to mean nucleic acid ligand, was coined by Ellington and Szostak [1] and the in vitro selection process to identify them in the laboratory was termed SELEX (systematic evolution of ligands by exponential enrichment) by Tuerk and Gold [8]. The invention of the SELEX process fundamentally changed the aptamer field because it offered the possibility of generating aptamers to target proteins, or other types molecules, that are not known to interact with nucleic acid ligands in nature. Moreover, since the SELEX process is performed in the test tube, one is not limited to using naturally occurring nucleotides in the RNA or DNA libraries, which allows for modifications of aptamers to make them more amenable to drug development. Since 1990, thousands of aptamers have been generated by the SELEX methodology or derivatives of it to a vast array of target proteins most of which do not have natural aptamers that bind them [9–11]. Thus, the invention of SELEX by Tuerk and Gold [8] and Ellington and Szostak [1] in 1990 suggested that the concept of therapeutic aptamers first described by Sullenger et al. [4] and Bielinska et al. [5] that same year might become more broadly useful than initially envisioned. As detailed below, this prediction has been proven correct. The FDA has approved one in vitro selected aptamer, while three others have made their way into large phase 3 clinical trials.
Fig. (2).
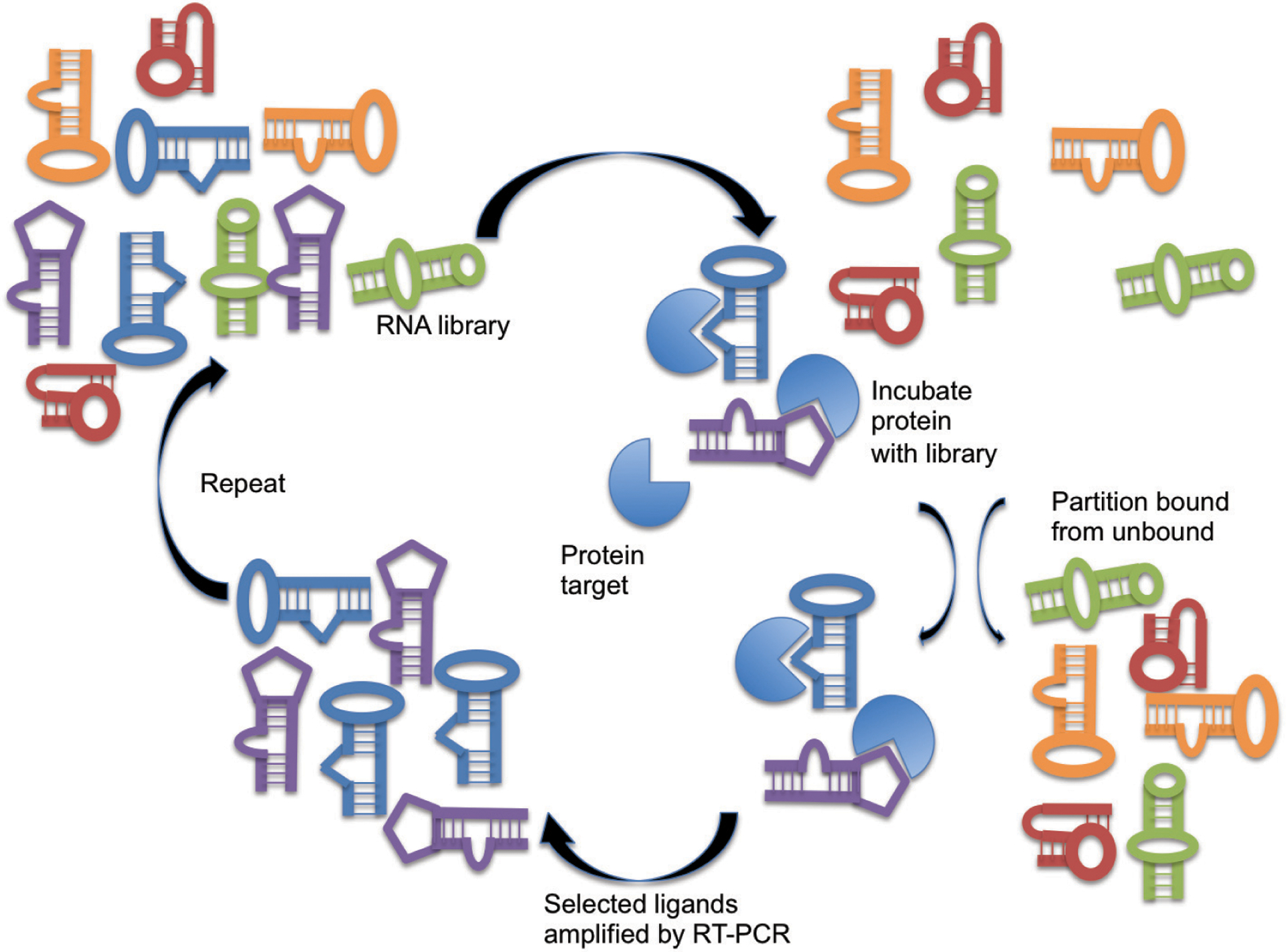
In Vitro Evolution of Aptamers via SELEX. Systematic Evolution of Ligands by EXponential enrichment (SELEX) is an iterative process that exposes a vast randomized library of RNA/DNA molecules of different structures to a target protein, partitions the RNA/DNA molecules that bind to the target protein from those that do not and amplifies those RNA/DNA molecules by RT-PCR [1].
2. TRANSLATION OF APTAMERS FOUND IN NATURE INTO THE CLINIC
To date, fourteen aptamers have been translated from the laboratory to the clinic (Table 1). Of these, five were evolved in nature. The first two aptamers to be evaluated in clinical trials were derived from nature: an RNA-based RRE (rev response element) decoy aptamer targeting the HIV rev protein [12] and a DNA decoy aptamer targeting the E2F transcription factor family [13]. Results from phase I clinical trials using both of these aptamers were published in 1999 by Kohn and colleagues (RRE decoy aptamer) [12] and Mann and colleagues (E2F decoy aptamer) [13]. Thus in nine years, the concept of inhibiting the activity of a therapeutically relevant protein with a nucleic acid ligand was translated into first in human studies. Results from clinical studies on these two aptamers as well as clinical studies using a second RNA decoy aptamer and two additional DNA decoy aptamers all of which target intracellular nucleic acid-binding proteins are briefly described below.
Table 1.
Aptamers that have been translated into clinical studies.
| Aptamer | Company | Target | Origin | DNA/RNA | Indication(s) | Phase(s) | Refs. |
|---|---|---|---|---|---|---|---|
| Edifoligide (E2F Decoy) |
Academic Institution Corgentech | E2F | Nature | DNA | Bypass graft surgery Restenosis | I, II and III | [13,18] |
| RRE Decoy | Academic Institution | rev | Nature | RNA | Human Immunodeficiency Virus-1 (HIV-1) | I/II | [12] |
| TAR Decoy | Benitec Biopharma | tat | Nature | RNA | Acquired immunodeficiency syndrome (AIDS)-related lymphoma | I/II | [14] |
| NF-kappaB Decoy | Academic Institution | Nuclear factor kappa B (NF-kB) | Nature | DNA | Atopic dermatitis Restenosis | I/II | [19] |
| STAT-3 Decoy | Ophthotech | Signal Transducer and Activator of Transcription (STAT-3) | Nature | DNA | Head and Neck Cancer | 0 | [20] |
| Macugen (Pegaptanib) |
Nexstar; Ophthotech | VEGF | SELEX | RNA | Age-related macular degeneration | I, II & III Approved | [27, 29, 30] |
| Fovista (El0030) |
Ophthotech | PDGF | SELEX | DNA/RNA | Age-related macular degeneration | I, II & III | [27] |
| Zimura | Ophthotech | Complement Factor C5 | SELEX | DNA/RNA | Dry age-related macular degeneration | I, II & III | [27] |
| RB006 (Pegnivacogin) |
Regado | Coagulation Factor IXa | SELEX | RNA | Acute Coronary Syndrome, Percutaneous Coronary Intervention | I, II & III | [36,40,41, 43] |
| ARC1779 | Archemix | von Willebrand factor | SELEX | DNA/RNA | Thrombotic microangiopathy; Thrombotic thrombocytopenic purpura (TTP)Acute coronary syndrome; vWD; CEA | I & II | [45,47–50] |
| NOX-A12 (olaptesed pegol) |
Noxxon | Stromal cell-derived factor 1 (SDF-1) | SELEX | RNA | Hematopoietic stem cell transplantation | I & Ha | [52] |
| NOX-E36 (emapticap pegol), |
Noxxon | CCL2 (Monocyte Chemoattractant Protein l.MCP-1) | SELEX | RNA | Chronic inflammatory diseases; type 2 diabetes mellitus; systemic lupus erythematosis Autologous stem cell transplantation |
I & IIa | [52, 53] |
| NOX-H94 (lexaptepid pegol) |
Noxxon | Hepcidin | SELEX | RNA | Anemia of chronic disease; inflammation | I & IIa | [52] |
| AS1411 | Antisoma | Nucleolin | In Vitro Cell Screen | DNA | Acute myeloid leukemia and Metastatic Renal Cell Carcinoma | I & II | [54, 55] |
Two RNA aptamers taken from nature [12, 14] have been translated into clinical studies and both target viral RNA binding proteins, rev and tat, essential for HIV replication (Fig. 1). Kohn and colleagues introduced an aptamer derived from the HIV rev Response Element (RRE) into hematopoietic cells of HIV infected children in an effort to inhibit the viral rev protein and virus replication [12]. Because this aptamer is comprised of normal RNA and the rev protein is an intracellular protein, a MoMLV-based retroviral vector was utilized to transfer an expression cassette for the RRE decoy aptamer into CD34+ bone marrow-derived stem cells in an effort to continuously produce the aptamer inside the treated cells. No evidence of adverse events was noted in the treated pediatric patients but the levels of aptamer containing cells in the peripheral blood of treated patients were found to be very modest one year following treatment [12]. The investigators concluded that gene therapy using RNA aptamers to inhibit HIV replication is possible but that improved gene transfer strategies need to be developed for the strategy to be effective. Since this initial study in 1999, significant advances have been made in the field of gene therapy including the development of lentivirus-based vectors. DiGiusto and colleagues utilized such improved gene transfer technology to deliver an expression cassette for an aptamer derived from HIV TAR RNA (Fig. 1), in conjunction with two other anti-HIV RNAs - a short hairpin RNA and a ribozyme, to stem cells from patients with AIDS-related lymphoma [14]. In four patients, persistent therapeutic RNA expression in multiple cell lineages has been observed for years following transplant with no unexpected toxicities. However, the level of gene transfer was observed to be too low to evaluate clinical efficacy. Therefore current efforts are focused upon developing optimal gene transfer and expression cassettes for the evaluation of such decoy RNA aptamers or other short RNA therapeutic agents [15]. Thus, though it took nearly 20 years, the concept of using a TAR decoy aptamer to inhibit HIV replication [4] has been successfully translated into human studies [14]. Evaluation of additional gene transfer strategies and longer-term follow up for the patients treated in this trial are now needed to better assess the utility of this therapeutic approach for treating HIV infected patients by providing a reservoir of HIV-resistant hematopoietic cells in the treated individuals. Three DNA decoy aptamers derived from normal human genomic DNA have also been evaluated in clinical studies. The first to be translated from the lab to the clinic was an aptamer that targets the E2F family of transcription factors. This aptamer was derived from the double-stranded DNA binding site for E2F1 and is bound by the entire family of E2F transcription factors [16]. Since the E2F family regulates cell cycle progression, Mann and colleagues evaluated the ability of the E2F decoy aptamer termed Edifoligide, to limit cell proliferation and vein graft intimal hyperplasia [13]. Vein grafts were pre-treated with Edifoligide ex vivo before being implanted during graft bypass surgery. Phase I and II clinical studies suggested that aptamer-treated grafts had a lower incidence of graft stenosis and occlusion [13]. However, in two large randomized phase III clinical trials (Table 1), no improvements were observed in primary or secondary outcomes related to graft patency, stenosis and occlusion in coronary or peripheral vein grafts with Edifoligide treatment [17, 18]. This limited clinical efficacy is believed to be a consequence of inefficient delivery of the DNA aptamer into cells in the various layers of the vein grafts.
Subsequently, clinical studies have been initiated to begin to evaluate a DNA decoy aptamer targeting nuclear factor kappa-B (NF-KB) for treatment of patients with atopic dermatitis and restenosis following percutaneous coronary intervention [19] while a DNA decoy aptamer to Signal Transducer and Activator of Transcription-3 (STAT3) for treatment of cancer patients (Table 1). In a phase 0 clinical trial, Sen and colleagues demonstrated that direct intratumoral delivery of the STAT3 DNA Decoy Aptamer into 30 patients with head and neck cancer led to a decrease in STAT3 target gene expression in the patients’ tumors [20]. Current studies are underway to enhance the systemic delivery and tumor uptake of such STAT3 DNA Decoy aptamers including generating cyclic decoys and appending the STAT3 aptamer to cytosine guanine dinucleotide (CpG) DNA motifs to facilitate cancer cell internalization, as efficient intracellular delivery of these DNA decoy aptamers will most likely be essential for therapeutic efficacy [21].
The major challenge with the development of aptamers derived from nature has been and continues to be that their protein targets are intracellular. Thus, as with si/miRNA, ribozyme, antisense oligonucleotide, and Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR)-based therapeutics, efficient delivery of such natural aptamers into cells is critical. If improvements in gene transfer technology continue to occur, the delivery of expression cassettes for RNA aptamers that target intracellular proteins will attract increasing therapeutic interest as is suggested by the TAR decoy aptamer clinical studies underway. Given the increasing appreciation for the regulatory functions that intracellular RNA binding proteins play in human biology and disease, RNA decoy aptamers may prove very useful for the treatment of a wide range of clinical applications once this delivery challenge is overcome. Similarly, for short double-stranded DNA decoy aptamers, improvements in the delivery of synthetic oligonucleotides into cells are required for such DNA aptamers to fulfill their potential to inhibit the activities of the numerous therapeutically-relevant nucleic acid-binding proteins inside the cells of patients. In this regard, aptamers that bind cell surface receptors may prove to be effective targeting and intracellular delivery agents for other aptamers that recognize intracellular proteins [22].
3. CLINICAL STUDIES WITH APTAMERS DERIVED IN THE LABORATORY
The 1990 discovery by Tuerk and Gold [8] and Ellington and Szostak [1] that aptamers could be isolated from large libraries of randomized RNA or DNA by SELEX in the test tube, immediately changed the aptamer field in two fundamental ways (Fig. 2). First, it allowed scientists to isolate aptamers against protein targets that do not naturally bind nucleic acids including extracellular proteins [9, 10]. Second, it allowed for unnatural nucleotide chemistries to be incorporated into aptamers to increase their stability in biological fluids [11, 23–26]. Because of these properties, taken together with the challenge of delivering oligonucleotides into cells, eight of the nine aptamers generated in the laboratory that have been evaluated in the clinic target extracellular factors and are highly modified oligonucleotides (Table 1). Thus, they do not have to enter cells to exert their therapeutic effects.
4. DELIVERY INTO THE EYE
Groundbreaking work from investigators at NeXstar Pharmaceuticals led to the first aptamer that was generated by SELEX, to enter clinical studies [27]. This aptamer, which targets Vascular Endothelial Growth Factor165 (VEGF165) [28] is called pegaptanib sodium or Macugen. It is currently the only aptamer approved by the FDA. Following intravitreal injection, pegaptanib inhibits VEGF165 activity and reduces neovascularization and slows vision loss in patients with age-related macular degeneration (AMD, Fig. 3) [29–31]. The initial phase I safety clinical trial for pegaptanib was published in 2002 [29], with phase II and definitive phase III clinical studies published in 2003 and 2004, respectively (Table 1) [30, 31]. Thus, in 12 years, in vitro generated aptamers were translated from concept to first in human studies and by 14 years, through definitive clinical studies. The phase III clinical results led the FDA to approve pegaptanib or Macugen for the treatment of AMD in 2004, which was a major milestone for the aptamer field. In 2006, however, the FDA approved an Fab derivative of the anti-VEGF antibody bevacizumab called ranibizumab or Lucentis, for the treatment of AMD as well. Clinical studies demonstrated that Lucentis not only slows the loss of vision in AMD patients but actually improves it following intravitreal injection [32]. It has been speculated that the improved clinical results with ranibizumab are due to pegaptanib being specific for the VEGF165 isoform, whereas ranibizumab binds to and inhibits VEGF165 and shorter isoforms of VEGF [27, 33]. Thus, the enhanced specificity of the VEGF aptamer may limit its clinical efficacy relative to a less specific VEGF inhibitor. Regardless, the clinical approval of Macugen demonstrated that an aptamer could address an unmet medical need and also validated neovascularization factors as important targets for the treatment of AMD.
Fig. (3).
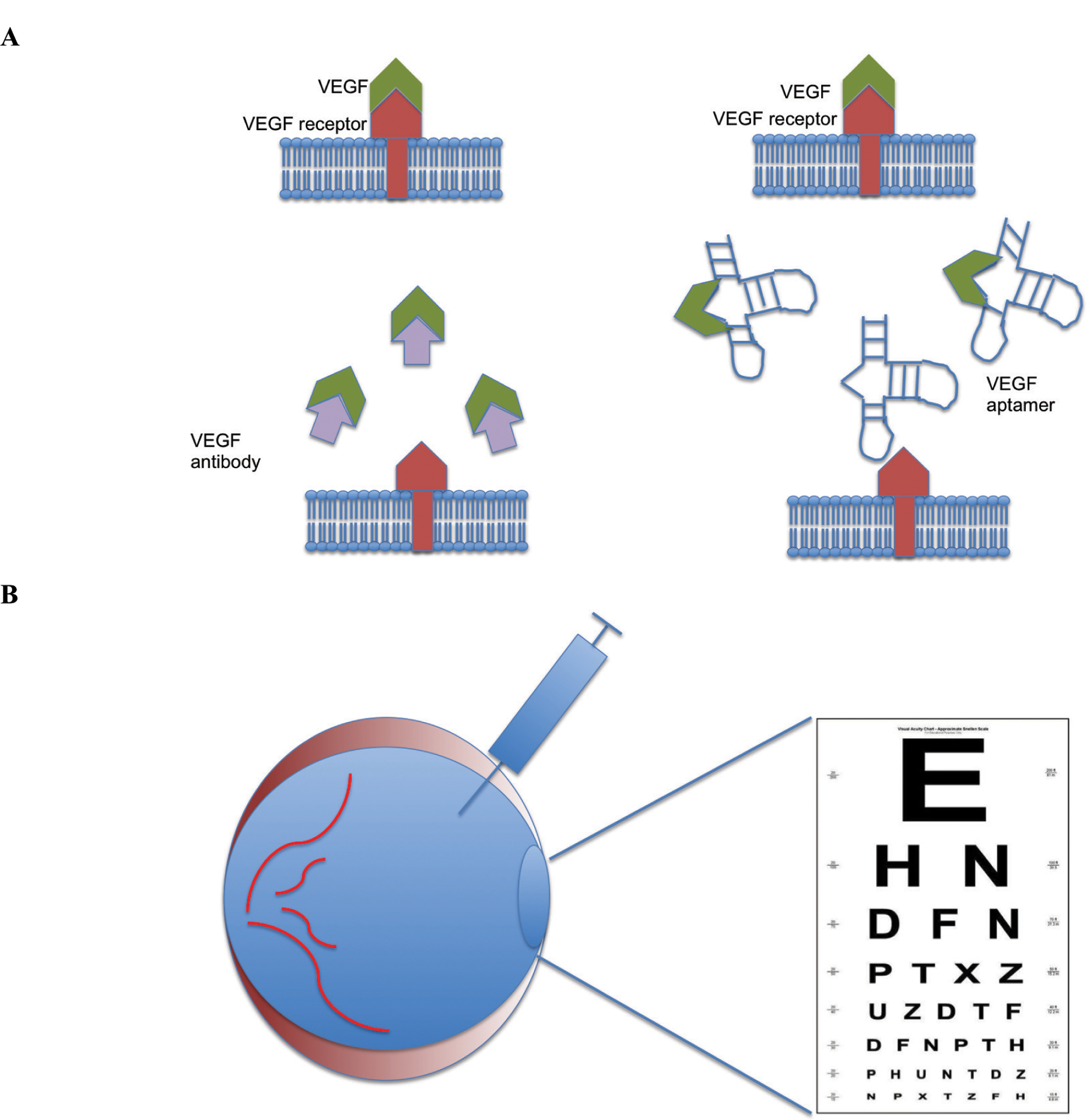
Treating Age-Related Macular Degeneration (AMD) with an aptamer. A: Macugen or VEGF antibodies target Vascular Endothelial Growth Factor (VEGF) that induces neovascularization and vision loss in AMD. B: Aptamer or antibody-based VEGF inhibitors are injected into the vitreous space to bind VEGF in the eye, prevent it from binding to its target receptor and thereby inhibit neovascularization, effectively treating AMD.
With the initial success of aptamer-based therapy to treat eye pathology, two other aptamers, E10030 (Fovista) and Zimura entered clinical trials for the treatment age-related macular degeneration (Table 1) [27]. Fovista targets Platelet-Derived Growth Factor (PDGF) while Zimura targets Complement Factor 5 (C5). Encouraging phase IIb clinical trial results using Fovista in combination with protein-based VEGF inhibitors to treat AMD have been reported that indicate that the combination may be superior to treatment with VEGF inhibitors alone [27]. Unfortunately, phase III clinical studies conducted by Ophthotech demonstrated no improved clinical benefit of combining Fovista with Ranibizumab (an anti-VEGF antibody) compared to Ranibizumab alone [34]. Ophthotech is conducting ongoing clinical trials to evaluate Zimura in a form of AMD called Geographic Atrophy (GA). It remains to be seen if aptamer-based therapeutics will once again become front line treatment for a common ocular disease such as AMD. The fact that direct delivery into the eye (Fig. 3) limits the amount of aptamer required for therapeutic efficacy and also eliminates any challenges associated with systemic delivery makes the development of additional therapeutic aptamers for ocular or other diseases associated with well-defined, self-contained, and easily accessible tissues particularly attractive.
5. SYSTEMIC DELIVERY
The first aptamer to be evaluated in humans following systemic administration is an aptamer that targets coagulation factor IXa. This aptamer, originally identified in our laboratory [35], was licensed to Regado Biosciences for clinical development. A fully optimized version of the aptamer, termed RB006 or pegnivacogin, is formulated with a 40 kDa polyethylene glycol (PEG) to increase the aptamer’s circulating half-life (Table 1). In a phase I clinical study, intravenous administration of RB006 was shown to be a rapid onset and dose-dependent anticoagulant in healthy volunteers with a median duration of effect of up to 30 hours [36]. Because we had observed that potent inhibition of factor IXa activity with the aptamer could result in increased bleeding in animals [37], we invented a strategy to rapidly reverse the activity of aptamers using antidote oligonucleotides [35, 37, 38]. Administration of such an antidote oligonucleotide termed RB007 to patients who had been given the RB006 aptamer resulted in rapid and durable neutralization of the pharmacodynamics effect of the aptamer within 1 to 5 minutes of antidote administration [36]. This result was particularly significant for the aptamer therapeutics field because it highlighted a distinguishing feature of aptamer-based medicines compared to antibody-based drugs; aptamer activity can be rapidly controlled with antidotes making them potentially safer therapeutic agents for clinical applications where target inhibition can result in severe and potentially life-threatening side effects such as bleeding in the case of anticoagulant therapy. A subsequent phase I study of pegnivacogin demonstrated that aptamers can also be administered by subcutaneous injection with a resulting calculated mean residence time of nearly 10 days [39].
To begin to evaluate the potential advantage of being able to rapidly reverse the activity of pegnivacogin, Povsic and colleagues performed a phase IIb clinical study comparing pegnivacogin and its antidote to heparin in patients with Acute Coronary Syndrome (ACS) undergoing cardiac catheterization (Fig. 4) [40]. Results from this study indicate that intravenous administration of the factor IXa aptamer was able to rapidly and adequately anticoagulate patients to allow for cardiac catheterization and reduce ischemic events compared to heparin treatment. Moreover, administration of the antidote rapidly restored normal hemostasis and resulted in numerically fewer patients experiencing significant bleeding compared to heparin treated patients [40]. Results from this phase IIb study indicated that an adequately powered phase III clinical trial of pegnivacogin and its antidote in patients who require rapid onset anticoagulation was warranted.
Fig. (4).
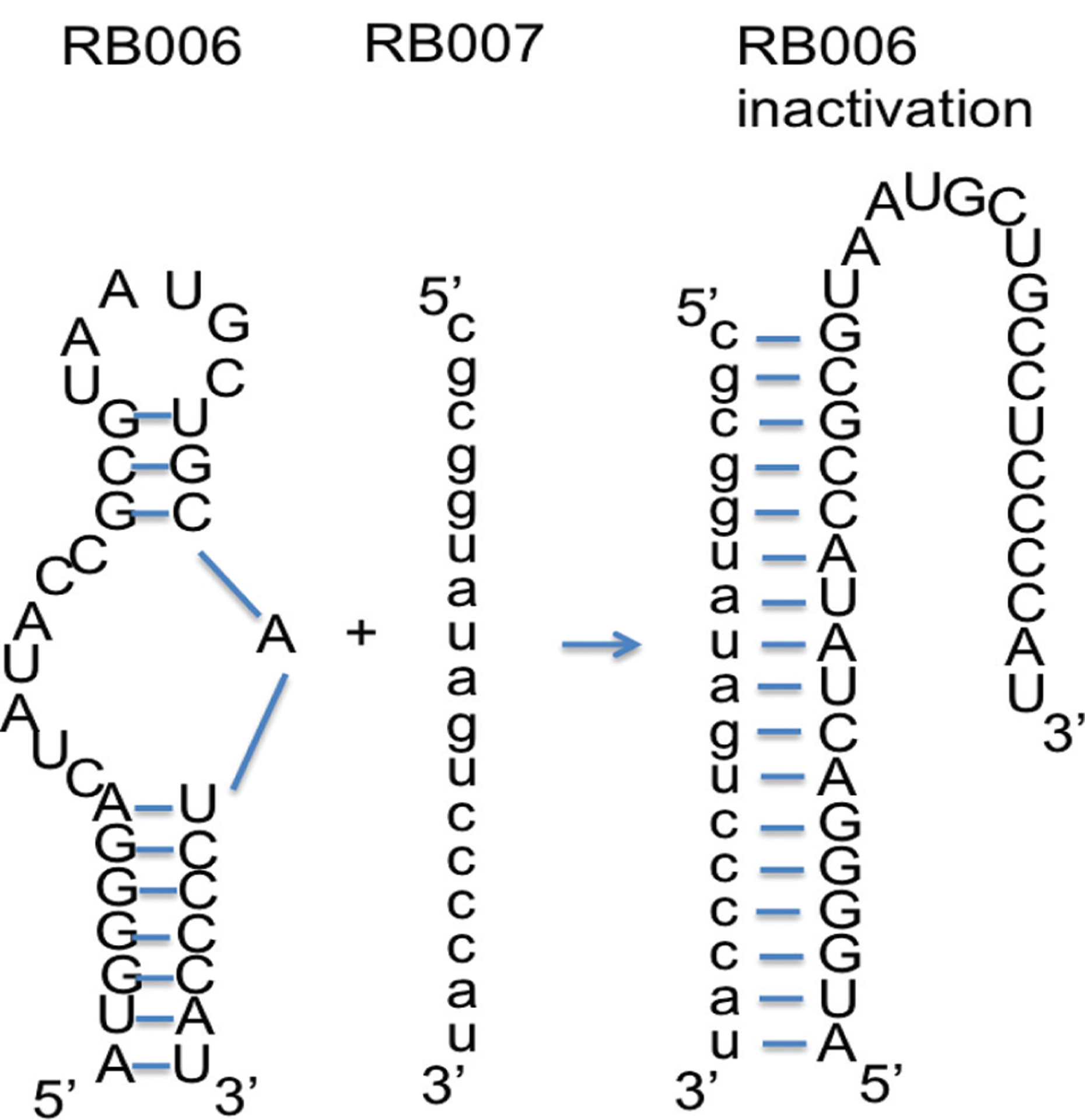
Aptamer-mediated anticoagulation during percutaneous coronary intervention and antidote-mediated control of factor IXa aptamer function. Utilizing Watson-Crick base-pairing rules, an antidote oligonucleotide (RB007) can be designed to interact with a portion of the factor IXa aptamer (RB006). Upon binding and unwinding the aptamer, the antidote converts the aptamer’s structure from a functional one to an inactive conformation thereby reversing the aptamer’s ability to inhibit factor IXa activity.
A phase III study designed to enroll 13,200 patients undergoing Percutaneous Coronary Intervention (PCI) was initiated to compare pegnivacogin to bivalarudin for anticoagulation in this common clinical setting. After enrolling 25% of the intended patients (3,232 patients randomized 1:1 between bivalarudin and RB006), the trial was halted because 10 patients experienced severe allergic reactions immediately after receiving the pegylated aptamer [41]. Subsequent analyses demonstrated that all of these patients had high levels of pre-existing antibodies against PEG [42, 43] but did not have any antibodies that recognized the RNA aptamer portion of the drug. Clinical results from this truncated study did demonstrate that the factor IXa aptamer was able to rapidly and adequately anticoagulate over 1,600 patients to perform PCI and limit ischemic events as well as the standard of care. However, because the trial was stopped early, it was not possible to determine if the aptamer could limit thrombotic events more effectively than bivalarudin. Nevertheless, the study demonstrated that RNA aptamer-antidote pairs represent rapid-onset and rapidly reversible clinical agents that may prove useful for a number of clinical settings [43, 44]. In addition the observed allergic reactions indicate that alternative, non-PEG, formulations should be considered for improving aptamer PK properties, at least for indications requiring large amounts of rapidly administered drug, or alternatively, the presence of anti-PEG antibodies should be determined prior to treatment and patients with such pre-existing antibodies be excluded to avoid rare allergic events.
A second aptamer, originally developed by Archemix Corporation, has been studied for the treatment of thrombosis. ARC1779, a DNA/RNA aptamer that targets von Willebrand factor has been studied as an anti-thrombotic agent and to treat thrombocytopenia in patients with Thrombotic Thrombocytopenic Purpura (TTP). It is formulated with a 20 kDa PEG. In evaluating its effect in preventing thrombosis, ARC1779 inhibited von Willebrand-dependent platelet activity and in patients undergoing carotid endarterectomy. Aptamer-treated patients had fewer embolic events post-operatively compared to placebo controls [45, 46]. In treating patients with TTP, ARC1779 revealed a dose-dependent relationship in platelet function inhibition as well as recovery of platelet count in phase II trials [47–50]. Currently, the future clinical development of this interesting vWF aptamer is unclear. However, it was reported that one patient out of 47 experienced a moderately severe hypersensitivity reaction at the injection site following treatment with the pegylated vWF aptamer. It is unclear if this reaction was directed toward the PEG portion of ARC1779 and if this hypersensitivity has hindered its clinical development [45].
Four additional aptamers have been evaluated in the clinic. Three are spiegelmers or L-RNA enantiomers of naturally occurring D-RNA. The chiral configuration of spiegelmer, which means “mirror image” in German, renders them highly resistant to exonuclease degradation [51]. NOXXON Pharma has pioneered the highly innovative development of spiegelmers and moved three of these into clinical trials [52]. The NOX-A12 (olaptesed pegol) spiegelmer targets Stromal Cell-Derived Factor 1 (SDF-1) and is being developed for use in stem cell transplantation. NOX-E36 (emapticap pegol), which targets cytokine Monocyte Chemoattractant Protein 1 (MCP-1), is being evaluated for the treatment of chronic inflammatory disease, type-2 diabetes mellitus, Systemic Lupus Erythmatosis (SLE) and in stem cell transplantation. A recent phase 2 clinical trial with NOX-E36 demonstrated that the Spiegelmer was able to significantly reduce albuminuria in type 2 diabetic patients and reduce HbA1c levels as well [53]. Spiegelmer NOX-H94 (lexaptepid pegol) targets hepcidin and is currently under evaluation for treatment of anemia. All three of these Spiegelmers appear to be well tolerated and are prime candidates for additional clinical studies [52]. Finally, AS1411 is a DNA G-quartet aptamer targeting nucleolin that is being clinically evaluated for the treatment of a number of cancers including acute myeloid leukemia (AML) [54] and metastatic Renal Cell Carcinoma (RCC) [55]. Results from a phase II trial in RCC patients demonstrated that AS1411 has minimal effect on most RCC patients but that rare individuals can experience dramatic and durable responses following therapy [55]. Thus personalize medicine efforts appear to be warranted to determine which subset of RCC patients may significantly benefit from AS1411 therapy.
CONCLUSION
We predict that the next two to three years will determine if in vitro generated aptamer-based therapeutics will re-emerge as a valuable therapeutic approach. In particular, C5 aptamer is currently being evaluated in phase II and III clinical studies in a therapeutic niche that seems particularly well suited for aptamer-based drugs, both in intra-ocular indications where local delivery is a major advantage.
However, we also believe that additional advances in the aptamer field will uncover additional clinical niches where aptamers may prove particularly useful. As discussed above, the invention of SELEX [1, 8] and the ability to incorporate modified nucleotides into aptamers were major advances for the therapeutic aptamer field [10, 23–25]. Recent advances in both the SELEX process as well as the chemical repertoire that can be incorporated into aptamers promises to herald a new era for the therapeutic aptamer field. Work by scientists at SomaLogic has recently led to the development of aptamers with additional chemical functionalities such as the addition of a benzyl or other groups at the 5-position of uridine. Selection utilizing such novel libraries has allowed for the generation of aptamers that bind proteins in previously unattainable and potentially very therapeutically useful ways [9, 11]. Hirota and colleagues recently reported that such a modified aptamer against interleukin-6 (IL-6) can inhibit the induction of arthritis in primates [56]. Thus we anticipate that these new classes of therapeutic aptamers will soon be evaluated in first-in-man studies.
Work by several groups including our own has demonstrated that in vitro selection methods can be performed to simultaneously identify aptamers against entire proteomes [57, 58] or cell surface targets [59, 60]. Moreover, selection for functions other than binding has now been described such as the identification of modified RNA or DNA molecules that efficiently internalize into cells [61] or even home to tumors in vivo [62, 63]. The ability to isolate such RNA or DNA molecules has led to much speculation that they may prove useful for the delivery of therapeutic agents [64, 65]. Initial results from these studies are encouraging in that they clearly demonstrate that these nucleic acid-based delivery agents can enter into cells and localize to the endosomal compartment efficiently. However, additional work remains to be performed to enhance the escape of the delivered therapeutic agents from this intracellular compartment. In addition to delivering cargoes to cells, aptamers have also been described that can deliver signals to cells by recognizing and cross-linking cell surface receptors [66–68]. Aptamers can also be conjugated to antineoplastic drugs that have significant systemic toxicity. These aptamer-conjugated cytotoxic drugs inhibited prostate cancer in vivo without systemic toxicity [69]. While beyond the scope of this manuscript, an excellent review on this has recently been published [70].
Thus, aptamer-based agonists can now be envisioned that can function as signaling molecules such as growth factors or cytokines. We anticipate that the flexibility of the in vitro selection process, taken together with the vastness of the nucleic acid libraries that can be explored and high through-put sequencing methods, will lead to additional innovative advances to further broaden the horizon of the therapeutic aptamer field in the coming years. Finally, as clinically amenable gene transfer technologies reach the market, we anticipate that a renaissance in the field of therapeutic RNA aptamers derived from nature will emerge. If any of the aptamers currently being evaluated clinically prove to be useful therapeutics, it would obviously facilitate such innovation by encouraging increased investment and interest in the therapeutic aptamer field in general.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- [1].Ellington AD; Szostak JW In vitro selection of RNA molecules that bind specific ligands. Nature, 1990, 346(6287), 818–822. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- [2].Cullen BR; Greene WC Regulatory pathways governing HIV-1 replication. Cell, 1989, 58(3), 423–426. 10.1016/0092-8674(89)90420-0 [DOI] [PubMed] [Google Scholar]
- [3].Burgert HG; Ruzsics Z; Obermeier S; Hilgendorf A; Windheim M; Elsing A Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol, 2002, 269, 273–318. 10.1007/978-3-642-59421-2_16 [DOI] [PubMed] [Google Scholar]
- [4].Sullenger BA; Gallardo HF; Ungers GE; Gilboa E Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell, 1990, 63(3), 601–608. 10.1016/0092-8674(90)90455-N [DOI] [PubMed] [Google Scholar]
- [5].Bielinska A; Shivdasani RA; Zhang LQ; Nabel GJ Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science, 1990, 250(4983), 997–1000. 10.1126/science.2237444 [DOI] [PubMed] [Google Scholar]
- [6].Mandal M; Breaker RR Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol, 2004, 5(6), 451–463. 10.1038/nrm1403 [DOI] [PubMed] [Google Scholar]
- [7].Cech TR; Steitz JA The noncoding RNA revolution-trashing old rules to forge new ones. Cell, 2014, 157(1), 77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- [8].Tuerk C; Gold L Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 1990, 249(4968), 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- [9].Gold L; Polisky B; Uhlenbeck O; Yarus M Diversity of oligonucleotide functions. Annu. Rev. Biochem, 1995, 64, 763–797. 10.1146/annurev.bi.64.070195.003555 [DOI] [PubMed] [Google Scholar]
- [10].Nimjee SM; Rusconi CP; Sullenger BA Aptamers: an emerging class of therapeutics. Annu. Rev. Med, 2005, 56, 555–583. 10.1146/annurev.med.56.062904.144915 [DOI] [PubMed] [Google Scholar]
- [11].Mehan MR; Ostroff R; Wilcox SK; Steele F; Schneider D; Jarvis TC; Baird GS; Gold L; Janjic N Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Adv. Exp. Med. Biol, 2013, 735, 283–300. 10.1007/978-1-4614-4118-2_20 [DOI] [PubMed] [Google Scholar]
- [12].Kohn DB; Bauer G; Rice CR; Rothschild JC; Carbonaro DA; Valdez P; Hao Ql.; Zhou C; Bahner I; Kearns K; Brody K; Fox S; Haden E; Wilson K; Salata C; Dolan C; Wetter C; Aguilar-Cordova E; Church J A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood, 1999, 94(1), 368–371. 10.1182/blood.V94.1.368.413a47_368_371 [DOI] [PubMed] [Google Scholar]
- [13].Mann MJ; Whittemore AD; Donaldson MC; Belkin M; Conte MS; Polak JF; Orav EJ; Ehsan A; Dell’Acqua G; Dzau VJ Ex-vivo gene therapy of human vascular bypass grafts with E2F decoy: the PREVENT single-centre, randomised, controlled trial. Lancet, 1999, 354(9189), 1493–1498. 10.1016/S0140-6736(99)09405-2 [DOI] [PubMed] [Google Scholar]
- [14].DiGiusto DL; Krishnan A; Li L; Li H; Li S; Rao A; Mi S; Yam P; Stinson S; Kalos M; Alvarnas J; Lacey SF; Yee JK; Li M; Couture L; Hsu D; Forman SJ; Rossi JJ; Zaia JA RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med, 2010, 2(36), 36ra43. 10.1126/scitranslmed.3000931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chung J; Scherer LJ; Gu A; Gardner AM; Torres-Coronado M; Epps EW; Digiusto DL; Rossi JJ Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Mol. Ther, 2014, 22(5), 952–963. 10.1038/mt.2014.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Morishita R; Gibbons GH; Horiuchi M; Ellison KE; Nakama M; Zhang L; Kaneda Y; Ogihara T; Dzau VJ A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl. Acad. Sci. USA, 1995, 92(13), 5855–5859. 10.1073/pnas.92.13.5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Alexander JH; Hafley G; Harrington RA; Peterson ED; Ferguson TB Jr; Lorenz TJ; Goyal A; Gibson M; Mack MJ; Gennevois D; Califf RM; Kouchoukos NT PREVENT IV Investigators. Efficacy and safety of edifoligide, an E2F transcription factor decoy, for prevention of vein graft failure following coronary artery bypass graft surgery: PREVENT IV: a randomized controlled trial. JAMA, 2005, 294(19), 2446–2454. 10.1001/jama.294.19.2446 [DOI] [PubMed] [Google Scholar]
- [18].Conte MS; Bandyk DF; Clowes AW; Moneta GL; Seely L; Lorenz TJ; Namini H; Hamdan AD; Roddy SP; Belkin M; Berceli SA; DeMasi RJ; Samson RH; Berman SS PREVENT III Investigators. Results of PREVENT III: a multicenter, randomized trial of edifoligide for the prevention of vein graft failure in lower extremity bypass surgery. J. Vasc. Surg, 2006, 43(4), 742–751. 10.1016/j.jvs.2005.12.058 [DOI] [PubMed] [Google Scholar]
- [19].Suzuki J; Tezuka D; Morishita R; Isobe M An initial case of suppressed restenosis with nuclear factor-kappa B decoy transfection after percutaneous coronary intervention. J. Gene Med, 2009, 11(1), 89–91. 10.1002/jgm.1266 [DOI] [PubMed] [Google Scholar]
- [20].Sen M; Thomas SM; Kim S; Yeh JI; Ferris RL; Johnson JT; Duvvuri U; Lee J; Sahu N; Joyce S; Freilino ML; Shi H; Li C; Ly D; Rapireddy S; Etter JP; Li PK; Wang L; Chiosea S; Seethala RR; Gooding WE; Chen X; Kaminski N; Pandit K; Johnson DE; Grandis JR First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov, 2012, 2(8), 694–705. 10.1158/2159-8290.CD-12-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang Q; Hossain DM; Duttagupta P; Moreira D; Zhao X; Won H; Buettner R; Nechaev S; Majka M; Zhang B; Cai Q; Swiderski P; Kuo YH; Forman S; Marcucci G; Kortylewski M Serum-resistant CpG-STAT3 decoy for targeting survival and immune checkpoint signaling in acute myeloid leukemia. Blood, 2016, 127(13), 1687–1700. 10.1182/blood-2015-08-665604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kotula JW; Sun J; Li M; Pratico ED; Fereshteh MP; Ahrens DP; Sullenger BA; Kovacs JJ Targeted disruption of β-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS One, 2014, 9(4), e9344. 10.1371/journal.pone.0093441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beigelman L; McSwiggen JA; Draper KG; Gonzalez C; Jensen K; Karpeisky AM; Modak AS; Matulic-Adamic J; DiRenzo AB; Haeberli P Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J. Biol. Chem, 1995, 270(43), 25702–25708. 10.1074/jbc.270.43.25702 [DOI] [PubMed] [Google Scholar]
- [24].Jellinek D; Green LS; Bell C; Lynott CK; Gill N; Vargeese C; Kirschenheuter G; McGee DP; Abesinghe P; Pieken WA Potent 2’-amino-2’-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry, 1995, 34(36), 11363–11372. 10.1021/bi00036a009 [DOI] [PubMed] [Google Scholar]
- [25].Tucker CE; Chen LS; Judkins MB; Farmer JA; Gill SC; Drolet DW Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J. Chromatogr. B Biomed. Sci. Appl, 1999, 732(1), 203–212. 10.1016/S0378-4347(99)00285-6 [DOI] [PubMed] [Google Scholar]
- [26].Willis MC; Collins BD; Zhang T; Green LS; Sebesta DP; Bell C; Kellogg E; Gill SC; Magallanez A; Knauer S; Bendele RA; Gill PS; Janjić N; Collins B Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug. Chem, 1998, 9(5), 573–582. 10.1021/bc980002x [DOI] [PubMed] [Google Scholar]
- [27].Drolet DW; Green LS; Gold L; Janjic N Fit for the eye: aptamers in ocular disorders. Nucleic Acid Ther, 2016, 26(3), 127–146. 10.1089/nat.2015.0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ruckman J; Green LS; Beeson J; Waugh S; Gillette WL; Henninger DD; Claesson-Welsh L; Janjić N 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem, 1998, 273(32), 20556–20567. 10.1074/jbc.273.32.20556 [DOI] [PubMed] [Google Scholar]
- [29].Eyetech Study Group. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina, 2002, 22(2), 143–152. 10.1097/00006982-200204000-00002 [DOI] [PubMed] [Google Scholar]
- [30].Eyetech Study Group. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology, 2003, 110(5), 979–986. 10.1016/S0161-6420(03)00085-X [DOI] [PubMed] [Google Scholar]
- [31].Gragoudas ES; Adamis AP; Cunningham ET Jr; Feinsod M; Guyer DR VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med, 2004, 351(27), 2805–2816. 10.1056/NEJMoa042760 [DOI] [PubMed] [Google Scholar]
- [32].Rosenfeld PJ; Brown DM; Heier JS; Boyer DS; Kaiser PK; Chung CY; Kim RY MARINA Study Group. Ranibizumab for neovascular age-related macular degeneration. N. Engl. J. Med, 2006, 355(14), 1419–1431. 10.1056/NEJMoa054481 [DOI] [PubMed] [Google Scholar]
- [33].Ferrara N; Gerber H-P; LeCouter J The biology of VEGF and its receptors. Nat. Med, 2003, 9(6), 669–676. 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- [34].Dunn EN; Hariprasad SM; Sheth VS An Overview of the fovista and rinucumab trials and the fate of anti-pdgf medications. Ophthalmic Surg. Lasers Imaging Retina, 2017, 48(2), 100–104. 10.3928/23258160-20170130-02 [DOI] [PubMed] [Google Scholar]
- [35].Rusconi CP; Scardino E; Layzer J; Pitoc GA; Ortel TL; Monroe D; Sullenger BA RNA aptamers as reversible antagonists of coagulation factor IXa. Nature, 2002, 419(6902), 90–94. 10.1038/nature00963 [DOI] [PubMed] [Google Scholar]
- [36].Dyke CK; Steinhubl SR; Kleiman NS; Cannon RO; Aberle LG; Lin M; Myles SK; Melloni C; Harrington RA; Alexander JH; Becker RC; Rusconi CP First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation, 2006, 114(23), 2490–2497. 10.1161/CIRCULATIONAHA.106.668434 [DOI] [PubMed] [Google Scholar]
- [37].Rusconi CP; Roberts JD; Pitoc GA; Nimjee SM; White RR; Quick G Jr; Scardino E; Fay WP; Sullenger BA Antidote-mediated control of an anticoagulant aptamer in vivo. Nat. Biotechnol, 2004, 22(11), 1423–1428. 10.1038/nbt1023 [DOI] [PubMed] [Google Scholar]
- [38].Nimjee SM; Keys JR; Pitoc GA; Quick G; Rusconi CP; Sullenger BA A novel antidote-controlled anticoagulant reduces thrombin generation and inflammation and improves cardiac function in cardiopulmonary bypass surgery. Mol. Ther, 2006, 14(3), 408–415. 10.1016/j.ymthe.2006.04.006 [DOI] [PubMed] [Google Scholar]
- [39].Vavalle JP; Rusconi CP; Zelenkofske S; Wargin WA; Alexander JH; Becker RC A phase 1 ascending dose study of a subcutaneously administered factor IXa inhibitor and its active control agent. J. Thromb. Haemost, 2012, 10(7), 1303–1311. 10.1111/j.1538-7836.2012.04742.x [DOI] [PubMed] [Google Scholar]
- [40].Povsic TJ; Vavalle JP; Aberle LH; Kasprzak JD; Cohen MG; Mehran R; Bode C; Buller CE; Montale-scot G; Cornel JH; Rynkiewicz A; Ring ME; Zeymer U; Natarajan M; Delarche N; Zelenkofske SL; Becker RC; Alexander JH A Phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur. Heart J, 2012. 10.1093/eurheartj/ehs232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lincoff AM; Mehran R; Povsic TJ; Zelenkofske SL; Huang Z; Armstrong PW; Steg PG; Bode C; Cohen MG; Buller C; Laanmets P; Valgimigli M; Marandi T; Fridrich V; Cantor WJ; Merkely B; Lopez-Sendon J; Cornel JH; Kasprzak JD; Aschermann M; Guetta V; Morais J; Sinnaeve PR; Huber K; Stables R; Sellers MA; Borgman M; Glenn L; Levinson AI; Lopes RD; Hasselblad V; Becker RC; Alexander JH REGULATE-PCI Investigators. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. Lancet, 2016, 387(10016), 349–356. 10.1016/S0140-6736(15)00515-2 [DOI] [PubMed] [Google Scholar]
- [42].Ganson NJ; Povsic TJ; Sullenger BA; Alexander JH; Zelenkofske SL; Sailstad JM; Rusconi CP; Hershfield MS Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGy-lated RNA aptamer. J. Allergy Clin. Immunol, 2015, 137(5), 1610–1613. 10.1016/j.jaci.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nimjee SM; Povsic TJ; Sullenger BA; Becker RC Translation and clinical development of antithrombotic aptamers. translation and clinical development of antithrombotic aptamers. Nucleic Acid Ther, 2016, 26(3), 147–155. 10.1089/nat.2015.0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nimjee SM; White RR; Becker RC; Sullenger BA Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol, 2017, 57, 61–79. 10.1146/annurev-pharmtox-010716-104558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gilbert JC; DeFeo-Fraulini T; Hutabarat RM; Horvath CJ; Merlino PG; Marsh HN; Healy JM; Boufakhreddine S; Holohan TV; Schaub RG First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation, 2007, 116(23), 2678–2686. 10.1161/CIRCULATIONAHA.107.724864 [DOI] [PubMed] [Google Scholar]
- [46].Markus HS; McCollum C; Imray C; Goulder MA; Gilbert J; King A The von Willebrand inhibitor ARC1779 reduces cerebral embolization after carotid endarterectomy: a randomized trial. Stroke, 2011, 42(8), 2149–2153. 10.1161/STROKEAHA.111.616649 [DOI] [PubMed] [Google Scholar]
- [47].Jilma B; Paulinska P; Jilma-Stohlawetz P; Gilbert JC; Hutabarat R; Knöbl P A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb. Haemost 2010, 104(3), 563–570. 10.1160/TH10-01-0027 [DOI] [PubMed] [Google Scholar]
- [48].Jilma-Stohlawetz P; Gilbert JC; Gorczyca ME; Knöbl P; Jilma B A dose ranging phase I/II trial of the von Willebrand factor inhibiting aptamer ARC1779 in patients with congenital thrombotic thrombocytopenic purpura. Thromb. Haemost 2011, 106(3), 539–547. 10.1160/TH11-02-0069 [DOI] [PubMed] [Google Scholar]
- [49].Jilma-Stohlawetz P; Gorczyca ME; Jilma B; Siller-Matula J; Gilbert JC; Knöbl P Inhibition of von Willebrand factor by ARC1779 in patients with acute thrombotic thrombocytopenic purpura. Thromb. Haemost, 2011, 105(3), 545–552. 10.1160/TH10-08-0520 [DOI] [PubMed] [Google Scholar]
- [50].Jilma-Stohlawetz P; Knöbl P; Gilbert JC; Jilma B The anti-von Willebrand factor aptamer ARC1779 increases von Willebrand factor levels and platelet counts in patients with type 2B von Willebrand disease. Thromb. Haemost 2012, 108(2), 284–290. 10.1160/TH11-12-0889 [DOI] [PubMed] [Google Scholar]
- [51].Eulberg D; Klussmann S Spiegelmers: biostable aptamers. ChemBioChem, 2003, 4(10), 979–983. 10.1002/cbic.200300663 [DOI] [PubMed] [Google Scholar]
- [52].Vater A; Klussmann S Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer® therapeutics. Drug Discov. Today, 2015, 20(1), 147–155. 10.1016/j.drudis.2014.09.004 [DOI] [PubMed] [Google Scholar]
- [53].Menne J; Eulberg D; Beyer D; Baumann M; Saudek F; Valkusz Z; Wiecek A; Haller H Emapticap Study, G., C-C motif-ligand 2 inhibition with emapticap pegol (NOX-E36) in type 2 diabetic patients with albuminuria. Nephrol. Dial. Transplant, 2016. 10.1093/ndt/gfv459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bates PJ; Laber DA; Miller DM; Thomas SD; Trent JO Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol, 2009, 86(3), 151–164. 10.1016/j.yexmp.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rosenberg JE; Bambury RM; Van Allen EM; Drabkin HA; Lara PN Jr; Harzstark AL; Wagle N; Figlin RA; Smith GW; Garraway LA; Choueiri T; Erlandsson F; Laber DA A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs, 2014, 32(1), 178–187. 10.1007/s10637-013-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hirota M; Murakami I; Ishikawa Y; Suzuki T; Sumida S; Ibaragi S; Kasai H; Horai N; Drolet DW; Gupta S; Janjic N; Schneider DJ Chemically modified interleukin-6 aptamer inhibits development of collagen-induced arthritis in Cynomolgus monkeys. Nucleic Acid Ther, 2016, 26(1), 10–19. 10.1089/nat.2015.0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Layzer JM; Sullenger BA Simultaneous generation of aptamers to multiple gamma-carboxyglutamic acid proteins from a focused aptamer library using DeSELEX and convergent selection. Oligonucleotides, 2007, 17(1), 1–11. 10.1089/oli.2006.0059 [DOI] [PubMed] [Google Scholar]
- [58].Ray P; Rialon-Guevara KL; Veras E; Sullenger BA; White RR Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J. Clin. Invest, 2012, 122(5), 1734–1741. 10.1172/JCI62385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dua P; Kim S; Lee DK Nucleic acid aptamers targeting cell-surface proteins. Methods, 2011, 54(2), 215–225. 10.1016/j.ymeth.2011.02.002 [DOI] [PubMed] [Google Scholar]
- [60].Wang J; Li G Aptamers against cell surface receptors: selection, modification and application. Curr. Med. Chem, 2011, 18(27), 4107–4116. 10.2174/092986711797189628 [DOI] [PubMed] [Google Scholar]
- [61].Magalhães ML; Byrom M; Yan A; Kelly L; Li N; Furtado R; Palliser D; Ellington AD; Levy M A general RNA motif for cellular transfection. Mol. Ther, 2012, 20(3), 616–624. 10.1038/mt.2011.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mi J; Liu Y; Rabbani ZN; Yang Z; Urban JH; Sullenger BA; Clary BM In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol, 2010, 6(1), 22–24. 10.1038/nchembio.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mi J; Ray P; Liu J; Kuan CT; Xu J; Hsu D; Sullenger BA; White RR; Clary BM In vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Mol. Ther. Nucleic Acids, 2016, 5e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].McNamara JO II; Andrechek ER; Wang Y; Viles KD; Rempel RE; Gilboa E; Sullenger BA; Giangrande PH Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol, 2006, 24(8), 1005–1015. 10.1038/nbt1223 [DOI] [PubMed] [Google Scholar]
- [65].Zhou J; Rossi JJ Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides, 2011, 21(1), 1–10. 10.1089/oli.2010.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dollins CM; Nair S; Boczkowski D; Lee J; Layzer JM; Gilboa E; Sullenger BA Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol, 2008, 15(7), 675–682. 10.1016/j.chembiol.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McNamara JO; Kolonias D; Pastor F; Mittler RS; Chen L; Giangrande PH; Sullenger B; Gilboa E Multi-valent 4–1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest, 2008, 118(1), 376–386. 10.1172/JCI33365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pastor F; Kolonias D; McNamara JO II; Gilboa E Targeting 4–1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther, 2011, 19(10), 1878–1886. 10.1038/mt.2011.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Powell Gray B; Kelly L; Ahrens DP; Barry AP; Kratschmer C; Levy M; Sullenger BA Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. USA, 2018, 115(18), 4761–4766. 10.1073/pnas.1717705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yan AC; Levy M Aptamer-mediated delivery and cell-targeting aptamers: room for improvement. Nucleic Acid Ther, 2018, 28(3), 194–199. 10.1089/nat.2018.0732 [DOI] [PMC free article] [PubMed] [Google Scholar]


