Abstract
The interferons (IFNs) are cytokines with important antineoplastic and immune modulatory effects. These cytokines have been conserved through evolution as important elements of the immune surveillance against cancer. Despite this, defining their precise and specific roles in the generation of antitumor responses remains challenging. Emerging evidence suggests the existence of previously unknown roles for IFNs in the control of the immune response against cancer that may redefine our understanding on how these cytokines function. Beyond the engagement of classical JAK-STAT signaling pathways that promote transcription and expression of gene products, the IFNs engage multiple other signaling cascades to generate products that mediate biological responses and outcomes. There is recent emerging evidence indicating that IFNs control the expression of both traditional immune checkpoints like the PD-L1/PD1 axis, but also less well understood “intracellular” immune checkpoints whose targeting may define new approaches for the treatment of malignancies.
Keywords: Interferon, signaling, cancer, immune response, immune checkpoint, non-canonical pathways
1. Introduction
The interferons (IFNs) were originally described as soluble factors with antiviral properties [1–3]. Years later it was understood that the biological functions of these cytokines extend well beyond their antiviral activities and that they have important immune-modulatory and antineoplastic effects [4–9]. It is now well established that IFNs are key elements of the immune surveillance against cancer and have important antineoplastic activities when administered to patients suffering from various types of cancers by exerting both direct effects on the malignant cells, as well as indirect effects on cells of the immune system [10].
There are three known types of IFNs: I, II and III. Type I (IFNα/β) and type III (IFNλ) IFNs are produced by many different types of mammalian cells, whereas the only type II IFN, IFNγ, is produced mainly by T cells and natural killer (NK) cells [11–13]. Once secreted, the different IFNs bind to specific cell surface receptors to initiate biochemical signals that mediate their biological effects. The different IFN receptors have each two binding subunits and include the Type I (IFNα/β) receptor (IFNAR1 and IFNAR2), the Type II (IFNγ) receptor (IFNGR1 and IFNGR2), and the Type III (IFNλ) receptor (IFNLR1 and IL10R), respectively [11–13]. The binding of the different IFNs to their corresponding receptors triggers activation of multiple intracellular signaling cascades that ultimately drive the expression of IFN-stimulated genes (ISGs) that control cell cycle progression, proliferation, apoptosis, differentiation, migration, and survival [12–14].
After the realization that IFNs have important antitumor activities in the late 70’s and early 80’s, there was high hope and expectation that the use of these cytokines as therapeutic agents was going to dramatically transform cancer treatment for many types of tumors and cure many patients [15–18]. With time it became clear that IFN-treatment will not meet this very high expectation. Nevertheless, over the years IFNs have had a major impact in the treatment of several malignancies and they improved the survival of hundreds of thousands of patients with specific types of cancer, including different hematologic malignancies, melanoma, renal cell carcinoma, and Kaposi’s sarcoma [14, 19]. However, a complicating factor of IFN-treatment has been the substantial toxicity of these cytokines when used at high doses, and this has been a limiting factor in a number of cases [14, 19].
The early clinical studies using IFNs underscored the need for a better understanding of the mechanisms by which these cytokines mediate direct antitumor effects and elicit immune responses. This is particularly important in the current medical era, with emerging new immune therapeutic approaches resulting in improved outcomes for patients with previously fatal malignancies [20]. Surprisingly, activation of IFN-signaling pathways and downstream expression of ISGs has been shown to correlate with either response or resistance to immune therapies [21–23]. The complex mechanisms by which IFNs impact anti-tumor immune responses are still far from being completely understood. Dissecting and defining these mechanisms may allow the development of novel immune therapeutic approaches that circumvent pro-tumorigenic IFN-activated resistance mechanisms, while at the same time promoting IFN-dependent antitumor immunity. New insights on the direct and indirect antitumor effects of IFNs resulting from research in the last few years have uncovered new treatment opportunities and potential therapeutic combinations that are currently being exploited or may be investigated in the future [24].
2. Overview of non-canonical signaling pathways in the regulation of ISG expression
The Type I and II IFN receptors are expressed widely in mammalian cells [13, 14, 25–29], while the Type III IFN receptor appears to show more tissue selectivity towards epithelial cells [3, 30]. Engagement of all three types of IFN receptors (IFNRs) results in activation of JAK/STAT signaling pathways [13, 26–32]. The engagement of JAK/STAT cascades appears to be a “universal” or so called “classical” [27, 28] or “canonical” [31] pathway, required for transcription and ultimate expression of specific ISGs with interferon stimulating responsive elements (ISRE) in their promoter region. This definition of Jak-Stats as “classical” or “canonical” is more reflective of historical events that led to the discovery of the pathway rather than proof that these pathways constitute the single or main cellular mechanism by which IFN-responses occur [32]. Notably, the initial discovery of Jaks and Stats was from scientists in the IFN-signaling field [reviewed in 13, 26–28] and subsequently impacted many other cytokine and growth factor research fields. However, IFN-responses involve activation of additional IFN- cellular cascades, called “non-canonical” signaling pathways, which are required for the optimal transcription and translation of IFN genes and their products (Fig. 1) [13, 28, 32]. Although the term “non-canonical” is used to describe these cascades, the function of all these signaling pathways appears to be essential and critical for the generation of the biological effects of IFNs on malignant cells and/or for induction of antiviral responses. These pathways include broadly MAP kinase (MAPK) pathways, mTOR pathways and guanine exchange factor (GEF) involving pathways (Fig. 1). Activation of MAP kinase or mTOR pathways leads to signals that control ISG transcription, ISG mRNA translation, or both, with specific elements or responses accounting for regulation of these processes. In addition, MAP kinase- or mTOR-generated signals directly or indirectly modify STAT function, underscoring the requirement for coordination of different IFN-generated signals for optimal induction of biological outcomes (Fig. 1) [13, 28, 32].
Figure 1:
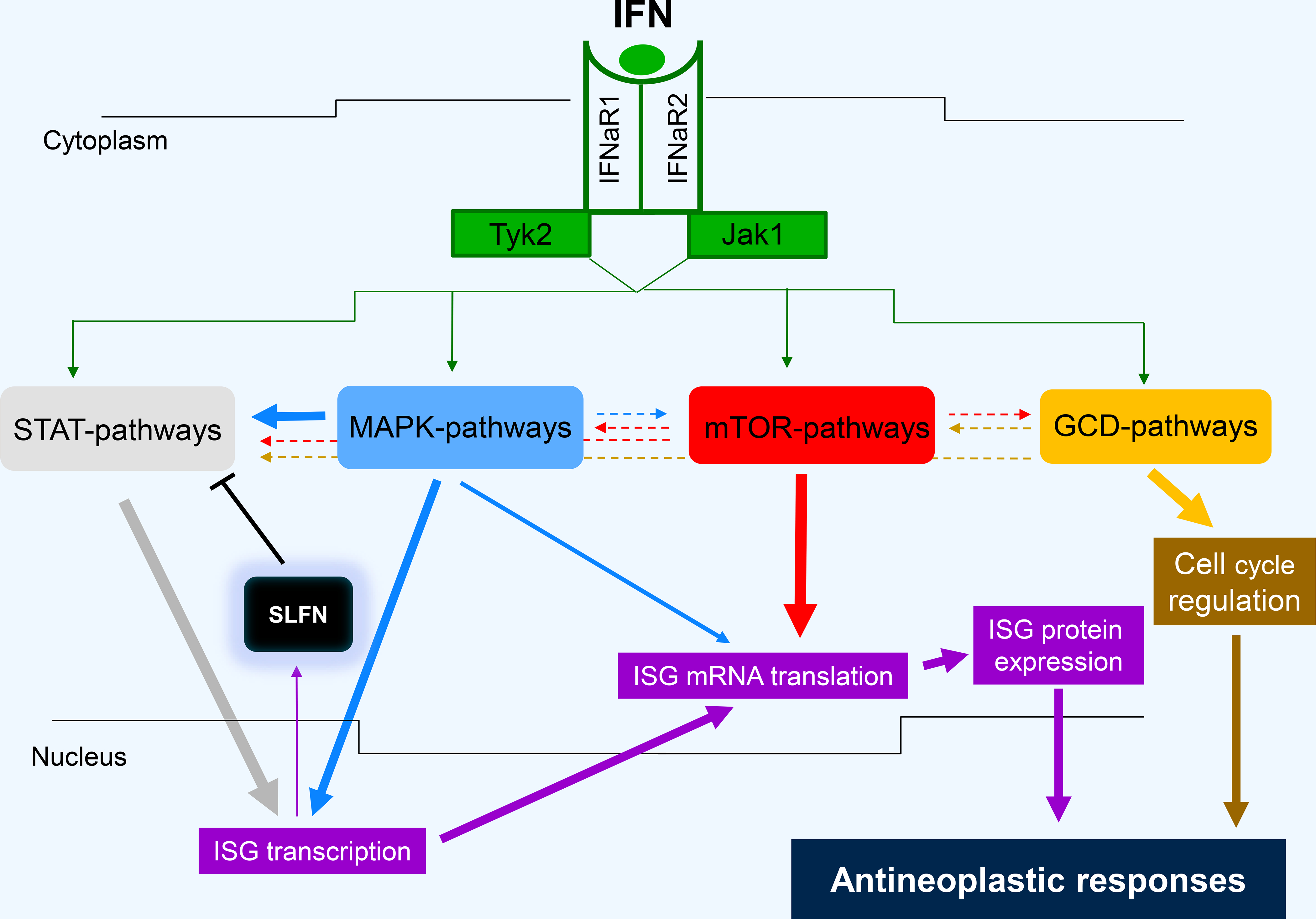
Non-canonical pathways and related interactions in the generation of Type I IFN antineoplastic responses. ISG - Interferon stimulated genes, STAT - signal transducers and activators of transcription, MAPK - Mitogen activated protein kinase, SLFN - Schlafen, mTOR - mammalian target of rapamycin, GCD - GTPases/Cyclin Dependent Kinases.
2.1. Regulation of STAT phosphorylation by non-canonical signaling pathways
There is extensive evidence that IFN-dependent serine phosphorylation of Stats plays a key and essential role for optimal transcription of ISGs [reviewed in 13, 28]. Several elements/components of non-canonical pathways have been implicated in serine phosphorylation of Stat1 in response to either Type I and/or Type II IFN signaling, including protein kinase C-delta (PKCδ) [33, 34], phosphoinositide 3-kinase (PI3K) [35], TNK1 [36], calmodulin-dependent kinase II (CamKII) [37], mammalian target of rapamycin (mTOR) [38, 39], cyclin-dependent kinase 8 (CDK8) [40], and CDK9 downstream of sirtuin 2 (SIRT2) deacetylation [41]. Other recent studies have implicated Type I IFN-dependent phosphorylation at serine 354 in the transactivation domain of Stat3 via the TANK-binding kinase 1 (TBK1) in the negative regulation of Stat3 activity [42], while phosphorylation of Stat2 on serines 287 and 734 negatively regulates Stat2 transcriptional activity [43, 44]. Importantly, another recent study demonstrated that in addition to serine phosphorylation, threonine phosphorylation at specific residues also exerts important regulatory effects. Specifically, threonine 387 in Stat2 lies on a CDK consensus and appears to have negative regulatory effects on Stat2-mediated transcriptional activation [45].
It should be noted that some of the non-canonical pathways that modify the function of the canonical (STAT) pathway, also exhibit important regulatory effects on the induction of IFN-responses, unrelated to modification of STAT pathways. For instance, as outlined below, the PI3K pathway plays a key role in IFN-signaling, by controlling mTOR activation and ultimately mRNA translation and production of key ISG proteins that mediate IFN-responses [reviewed in 13, 26–28, 46]. Beyond the phosphorylation of Stat1 by PKCδ and other PKC isoforms [28, 47], there is also evidence that IFN-activated PKC pathways mediate a variety of other functions, such as ion channel modulations in neurocortical pyramidal neurons [48].
2.2. MAP kinase pathways in IFN-signaling
The p38 MAPK pathway plays key and essential roles in IFN-signaling and is activated by all different IFN types (I, II, III) [reviewed in 13, 28]. This signaling cascade is required for optimal transcription of ISGs in the Type I IFN system, via STAT-unrelated transcriptional mechanisms [13, 28]. Similarly, there has been previous extensive evidence for important regulatory roles of the ERK pathway in the control of IFN-responses [reviewed in 13, 28]. Importantly, the activation of IFN-induced MAP kinase pathways results in downstream activation of MNK kinases [reviewed in 28, 49] that play key and essential roles for mRNA translation of ISGs in malignant cells [50, 51] and cells of the immune system [52]. Importantly, the function of MNK kinases appears to be essential for the generation of the suppressive effects of IFNs on both normal and malignant hematopoietic cells [50, 51, 53], underscoring the critical role of these signals in the control of IFN-responses. Other key effectors downstream of the IFN-activated p38 MAPK pathway appear to be Sprouty (Spry) proteins 1, 2 and 4, which control IFN-responses downstream of the IFN-activated MAP kinase pathway, acting as negative regulators of downstream signals and generation of IFN-biological effects [54]. Altogether, there has been extensive evidence over the years that MAPK pathways are critical elements of IFN-signaling and that engagement of these cascades results in signaling events that are essential for mediating important cellular responses, such as transcription, mRNA translation and protein expression of ISGs. With the increasingly recognized roles of IFNs in immune-mediated effects and cancer immunotherapy, these pathways are potential target modulators for immune responses in cancer, as discussed below.
2.3. mTOR pathways in IFN-signaling
The mTOR kinase mediates signals critical for mRNA translation, cell metabolism, and cell proliferation of normal cells [46, 55]. At the same time, dysregulation of mTOR pathways during tumorigenesis plays key roles in malignant cell transformation and survival, mRNA translation of mitogenic genes and aberrant cell proliferation [56–58]. Although the pathway is generally discussed in the context of proliferation or mitogenic signals, observations in the early 2000s demonstrated that Type I IFN treatment activates mTOR effectors in different types of sensitive malignant hematopoietic cells [59–61]. Subsequent work established that the activation of this pathway occurs downstream of the PI3K/AKT pathway and is important for mRNA translation of IFN-sensitive genes, including genes whose transcription is controlled by JAK-STAT pathways [62–64]. These discoveries addressed an important outstanding issue in the IFN-signaling field, as up to that time the mechanisms of mRNA translation of ISGs had not been elucidated. They also established that the AKT/mTOR pathway is not an exclusively mitogenic pathway, but it is also engaged and utilized by growth suppressive cytokines [reviewed in 65, 66].
The accumulating evidence implicating the mTOR kinase in the IFN-response led to further studies aimed to explore whether different mTOR complexes are involved in the generation of IFN-responses. The mTOR kinase is present, as the common catalytic subunit, in two distinct complexes, mTORC1 and mTORC2 [55–58, 65–67]. mTORC1 components include Raptor and mLST8; while mTORC2 includes Rictor, mLST8, and SIN1 [55–58, 65–67]. Our group provided the first evidence for activation of mTORC2 complexes by the Type I IFN receptor [68]. The initial studies demonstrated surprising important functions of these complexes in the control of both ISG transcription and mRNA translation [68]. Further studies established that transient or stable disruption of Rictor or Sin1 expression results in defects in the formation of STAT-DNA binding complexes, providing a mechanism by which mTORC2 complexes control Type I IFN-dependent transcription [69]. Similarly, the Type II IFN receptor was also found to activate mTORC2 mRNA translation of type II IFN-stimulated genes and Rictor was shown to be essential for the generation of type II IFN-dependent antiviral and antiproliferative responses [70]. Surprisingly, disruption of Sin1 results in decreased Type II IFN-dependent tyrosine phosphorylation of Stat1 and this appears to be the mechanism by which it controls Type II ISG-transcription [71]. The same studies demonstrated that SIN1 interacts with the Type II IFNR, suggesting that the effects of SIN1 on Stat phosphorylation and gene transcription reflect a possible upstream regulatory role for SIN1 on IFNγ-signaling, at the Type II IFN receptor level [71].
The importance of mTOR pathways in both Type I and II IFN-biological responses is further underscored by more recent studies that demonstrated that modulation of mTOR activity controls IFNγ-dependent, Stat1-mediated, human and mouse mesenchymal stem cell immunomodulation [72] and that IFNβ enhances mesenchymal stromal stem cell immunomodulatory function through both Stat activation and mTOR-regulation of glucose metabolism [73]. Other recent studies have established that inflammatory interferon activates HIF-1α-mediated epithelial-to-mesenchymal transition via engagement of the mTOR pathway [74]. Altogether there is strong accumulating evidence suggesting an important role for mTOR complexes in the induction and regulation of the biological effects of IFNs. The recent discovery and description of CDK9 mTOR-like complexes (CTORC) in leukemia cells [75], raises the possibility of involvement of these complexes in the IFN-pathway as well, and this is something that should be examined in future studies.
2.4. ULK1 pathways in IFN signaling
ULK1, the mammalian homolog of the UNC-51 kinase of Caenorhabditis elegans [76], is a highly conserved serine/threonine kinase ubiquitously expressed in human adult tissues and exhibits important regulatory effects on autophagy [77–79]. Previous studies have shown that AMPK phosphorylates ULK1 on serine (Ser) 556, promoting its engagement into autophagy-related pathways, while activated mTORC1 phosphorylates ULK1 on Ser758 and prevents autophagy by disrupting the interaction between ULK1 and AMPK [80].
There has been recent evidence by our group and others implicating ULK1 in the control of the innate immune response [81–85]. Previous studies demonstrated that cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) synthase (cGAS) binds cytosolic DNA leading to the synthesis of cGAMP, which activates the stimulator of interferon genes (STING) and consequent IRF3 activation, leading to Type I IFN production [86, 87]. In a subsequent study, cGAS-mediated synthesis of cGAMP was found to trigger dephosphorylation of ULK1 on Ser556, igniting downstream phosphorylation of STING on Ser366, an event resulting in STING degradation and suppression of type I IFN production [81]. This novel function of ULK1 appears to be a negative-feedback control mechanism, aimed to regulate the extent of innate immune responses, and could have important implications in the effectiveness of cancer immunotherapies [81,88]. Interestingly, although de-phosphorylation of ULK1 on Ser556 is involved in inhibition of STING-dependent Type I IFN expression [81], IFNα/β-treatment induces phosphorylation of ULK1 at the mTORC1 phosphorylation site, Ser758, but not at Ser556 [82]. Our studies have shown that engagement of ULK1 downstream of the IFNAR is required for full phosphorylation/activation of p38 MAPK and transcription of ISGs involved in the generation of Type I IFN-dependent anti-neoplastic effects and responses [82] (Fig. 2). Thus, ULK1 plays a central role in Type I IFN signaling and complements the function of the canonical type I IFN-activated JAK/STAT1 pathways [82]. Notably, in that study it was also definitively established that the effect of ULK1 on selective ISG transcription is independent of its regulatory effects on autophagy [82]. As IFN-dependent AKT/mTORC1 activation is only required for mRNA translation of ISGs [64], it is possible that AKT/mTORC1 induces phosphorylation of ULK1 on Ser758 blocking its pro-autophagic functions, and another type I IFN-activated upstream ULK1 kinase (UULKK) induces phosphorylation of ULK1 on other amino acid residues triggering its transcriptional-related activity. Future studies should aim to identify and characterize these putative UULKKs (Fig. 2).
Figure 2:
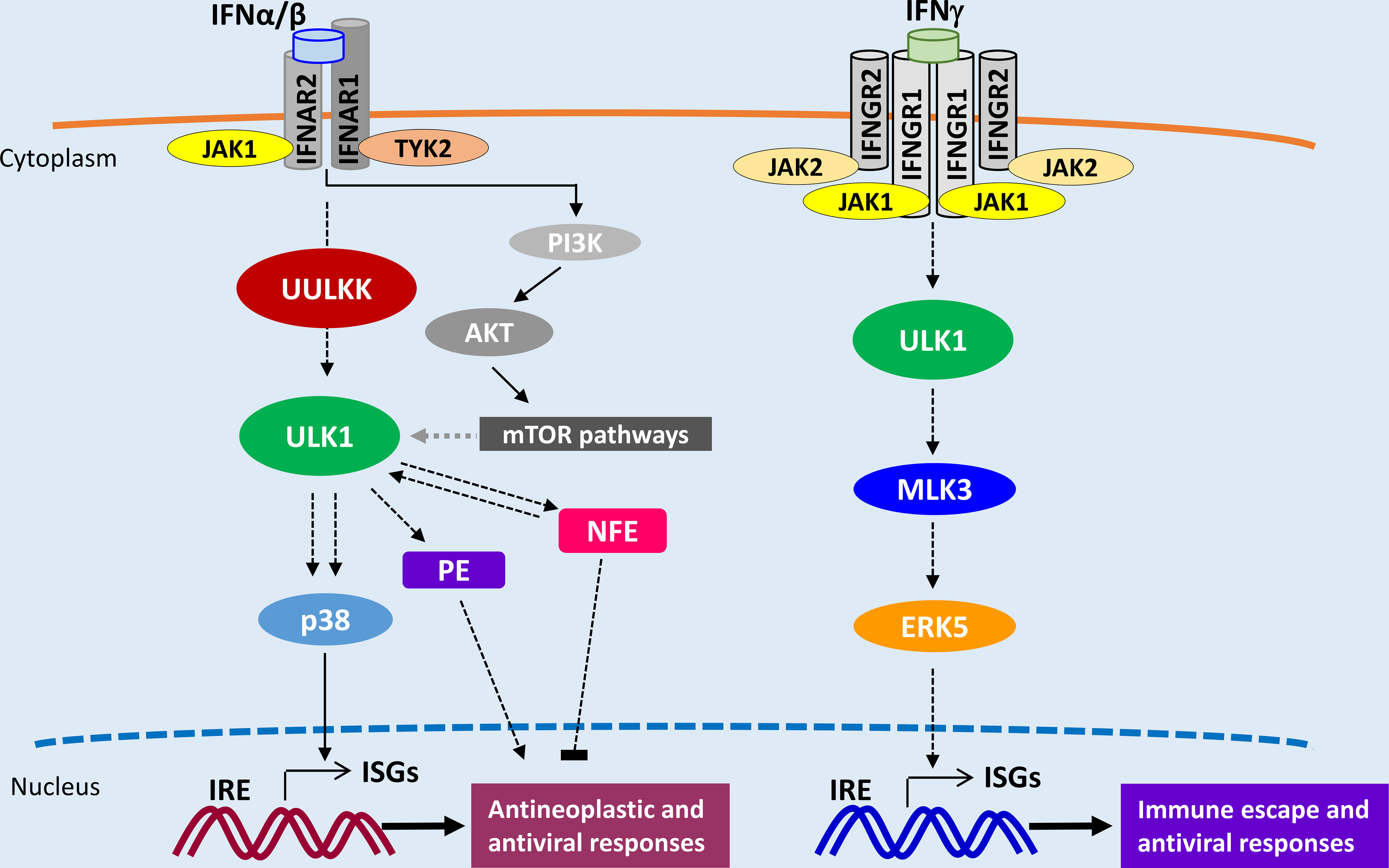
ULK1 pathways in Type I and II IFN signaling. UULKK – Upstream ULK1 kinase; ISGs – IFN-stimulated genes; IRE - IFN responsive elements; PE – Positive Effectors; NFE - Negative Feedback effectors
Beyond its involvement in the Type I IFN system, ULK1 is also activated during IFNγ- engagement of the Type II IFN receptor. We recently demonstrated that ULK1 binds and phosphorylates the MAPKKK MLK3 upon IFNγ stimulation, which in turn is required for activation of the MAPK ERK5 and transcriptional regulation of specific ISGs, in an autophagy-independent manner [83] (Fig. 2). Notably, RNA sequencing analysis revealed decreased expression of IFNγ-dependent genes involved in viral defense response and regulation of inflammatory responses, and increased IFNγ-mediated induction of genes involved in T cell activation in the absence of ULK1 expression [83]. Thus, engagement of ULK1 pathways downstream of the IFNGR seems to be essential for the tight regulation of immune anti-tumor and responses (Fig. 2). Further studies are warranted to fully elucidate the role of ULK1 in IFN signaling and whether its effects are cell type-specific.
2.5. SLFN proteins and regulatory effects on IFN signaling
The schlafen (SLFN) proteins are members of a family of IFN-stimulated genes that includes several human and mouse members with important regulatory roles on key cellular functions [89–91]. Extensive work from us and others in recent years has established that distinct mouse and human SLFNs are implicated in many important cellular activities, including proliferation, growth and cell cycle progression, translational control, cell migration and differentiation, as well as regulation of antiviral and immune responses [92–104]. There has been also evidence implicating certain human SLFNs in the pathophysiology of human diseases. SLFN5 expression has been shown to correlate with gastric carcinoma development on background of metaplasia [105], while SLFN14 mutations have been implicated in the development of a syndrome involving severe thrombocytopenia and platelet secretion defects, associated with excessive bleeding [106]. Moreover, heterozygous deletion of the human SLFN gene loci region on chromosome 17q12 has been associated with an immunodeficiency syndrome [107] reminiscent of the elektra phenotype in mice [108]. Remarkably, recent studies have provided evidence that human SLFN11 plays a key role in mediating the antitumor effects of some types of chemotherapeutic agents and, in some cases, its expression may correlate with response to treatment [109–115]. Similarly, mouse Slfns were also shown to mediate the effects of bleomycin in mouse models of idiopathic pulmonary fibrosis [116].
There is now a significant amount of evidence that has established that SLFN proteins have been implicated in the generation of IFN-responses, both in terms of antiproliferative/ antineoplastic effects, as well as generation of antiviral responses [89, 92–95]. These responses may depend on the cellular context and other, yet to be identified, co-existing factors. Human (SLFN5) [92, 95] and mouse (Slfn2, 3, 5) [93, 94] schlafens have been shown to suppress tumor growth and/or motility or invasiveness in renal cell carcinoma and malignant melanoma. Moreover, there appears to be a correlation between high levels of SLFN11 expression and an “IFN-gene signature” in breast cancer [96] and small cell lung cancer [115], as well as a correlation of high SLFN11 expression with PD-L1 expression in small cell lung cancer [115]. SLFN11 is also both IFN-inducible and has anti-HIV1 properties [104], suggesting that it may be part of the Type I IFN response to HIV infection [90]. On the other hand, under different contexts SLFN proteins may unexpectedly mediate opposing effects and suppress antitumor and antirviral responses. It was recently shown that in glioblastoma (GBM) SLFN5 promotes motility and invasion of GBM cells, while high expression of the SLFN5 gene is associated with worse clinical outcomes in GBM patients [117]. The mechanisms underlying the tumorigenic effects of SLFN5 in GBM appear to involve interaction of SLFN5 with Stat1 and repression of Stat1 transcriptional activity and consequent suppression of ISG expression [117]. These findings from our group have suggested the existence of a negative-feedback loop activated during engagement of the Type I IFN receptor that may account for suppression of antitumor immune responses in GBM and possibly other selected tumors [117]. Notably, we recently observed that mouse Slfn2 also suppresses mouse IFN-dependent transcription of genes and that targeted disruption of the mouse Slfn2 gene results in enhanced IFN-inducible antiviral responses [101]. In that case the mechanism appears to be related to the interaction of Slfn2 with the protein phosphatase 6 regulatory subunit 1 (PPP6R1), ultimately resulting in decreased activation of nuclear factor kappa B (NF-κB) signaling [101]. These findings have established that SLFNs can in certain cases suppress the immune response by interacting with elements of the IFN-signaling machinery. We therefore propose the term “intracellular immune checkpoints” to describe this SLFN-involving cellular processes. Future studies should determine if other SLFNs, beyond SLFN5, may be playing similar roles and whether specific SLFN targeting may provide an approach to enhance responses to cancer immunotherapy treatments.
3. Antitumor Immune responses and non-canonical versus canonical IFN- pathways
The possibility that IFNs promote host’s anti-tumor responses was first raised from studies in which IFNα treatment increased survival [118] and inhibited the growth and formation of lung metastases [119] in mice inoculated with IFN-resistant cancer cells. Moreover, injection of immunocompetent mice with a neutralizing antibody to IFNα/β, which did not affect the growth of malignant cells in vitro, increased tumor growth and decreased survival of the mice, indicating that endogenous Type I IFNs play an inhibitory role against tumor growth by acting on host cells [120]. Similarly, IFNγ was shown to promote anti-tumor responses by acting on both host immune cells and directly on tumor cells [121–123]. Consistent with the important activities of IFNs as mediators of immune antitumor responses, subsequent work demonstrated that defects in IFN-signaling are commonly found in cells of the immune system or in the tumor cells themselves in different types of malignancies [122, 124, 125]. Moreover, mutations of genes that encode for protein elements of IFN-signaling pathways or defective expression of such elements frequently accounts for resistance to cancer immunotherapy [126–129]. It is worth noting that immune checkpoint blockade (ICB) therapies are highly effective, but only in a fraction of patients, due to primary and acquired resistance mechanisms occurring in the majority of cancer patients [126, 130, 131].
ICB therapies are designed to overcome tumor immune escape mechanisms by enhancing T-cell-mediated anti-tumor activities [132]. The FDA has approved four ICB treatments in the United States: ipilimumab, an anti-CTLA-4 monoclonal antibody (mAb) for melanoma, two anti-PD-1 mAbs, pembrolizumab (for melanoma and lung cancer) and nivolumab (for melanoma, Hodgkin’s lymphoma, and lung, kidney, bladder, and head and neck cancers), and an anti-PD-L1 mAb, atezolizumab, for the treatment of bladder, lung, and triple-negative breast cancer [20, 133, 134]. PD-1 and CTLA-4 are cell surface receptors that play complementary roles in the regulation of adaptive immune responses. PD-1 is expressed on activated T cells, B lymphocytes, and NK cells and controls T-cell exhaustion in peripheral tissues, whereas CTLA-4 inhibits T-cell activation at early time points in the lymph nodes [20, 135]. PD-L1, often expressed by tumor cells, is one of the ligands for the PD-1 receptor, blocking T-cell mediated recognition and tumor killing. The expression of the two known PD-1 receptor ligands, PD-L1 and PD-L2, is induced by both Type I and II IFNs [123,136–140]. The IFNγ-JAK-STAT-IRF1 pathway has been shown to primarily induce expression of PD-L1, with IRF1 binding to its promoter, whereas both IFNβ and IFNγ equally induce expression of PD-L2 in melanoma cells, with IRF1 and STAT3 binding to its promoter [136]. Other studies have shown that IFNγ induces expression of PD-L1 in lung cancer cells through activation of both JAK/STAT3 and PI3K/AKT pathways [137]. Similarly, IFNβ regulates PD-L1 expression in lung cancer cells through JAK/STAT/IRF9- and PI3K-dependent signaling pathways [138]. Also, IFNα induces PD-L1 expression on dendritic cells, via engagement of both STAT3 and p38 MAPK pathways [139], while JAK/STAT1 signaling is required for IFNγ-induced expression of PD-L1 in hematopoietic malignant cells and resistance to natural killer cell lysis [140]. Taken together, these studies suggest that cooperation of canonical and non-canonical signaling pathways is essential for IFN-dependent PD-L1/PD-L2 expression. In a recent study, Benci et al. [141] demonstrated that sustained IFNγ signaling promotes PD-L1-dependent and -independent resistance to ICB and that both IFNγ and IFNα/β pathways are required to maintain this resistance. In another study, dual ICB combination therapy with both anti-CTLA-4 and anti-PD-1 antibodies was associated with reduced anti-tumor immune responses in both animal models and melanoma patients due to high levels of IFNγ production [142]. Moreover, using breast cancer, lymphoma and fibrosarcoma mouse tumor models, Takeda et al. [143], showed that in vivo activation of cytotoxic T cells results in copy-number alterations in the tumor cells leading to the emergence of immune-resistant cancer cell clones. These effects were dependent on the presence of IFNγ within the tumor microenvironment, suggesting that IFNγ-induced genetic instability in the tumor cells could be one of the acquired resistance mechanisms against ICB [143]. Based on these studies [141–143], one can conclude that targeting IFNγ signaling could revert resistance to ICB. However, it could also negatively affect the positive effects of IFNγ signaling required for response to ICB [144]. This dual and complex role of IFN signaling in malignancies supports the need to identify means to selectively and specifically modulate its effects to support anti-tumor immune responses without promoting immunosuppression. In line with this thought, Gao et al. [145] showed that JAK2/STAT1 signaling activation is essential for the antiproliferative effects of IFNγ against lung cancer cells, whereas inhibition of PI3K activation promotes the anti-proliferative effects of IFNγ and decreases PD-L1 expression in these cells.
4. Conclusions and Future Directions
There has been a lot of progress in recent years in understanding the mechanisms by which IFNs mediate biological responses. The need to fully understand these mechanisms appears to be more urgent and impactful than ever because of the rapid advances in the cancer immunotherapy field and the central role that IFNs play in the control of immune checkpoint regulation in cells of the immune system. The better understanding of IFN-signaling mechanisms, especially as it relates to the roles of non-canonical pathways, creates new opportunities for targeted immune modulation approaches for the treatment of malignancies. For example, based on very recently acquired knowledge, ULK1 is a central pathway in IFN-signaling and its selective targeting might enhance IFNγ-dependent expression of immunostimulatory genes and reduce expression of immunosuppressive ones [83], while potentiating STING-dependent expression of type I IFNs [81]. Furthermore, the possibility that some of these non-canonical pathways, such SLFNs, may act as “intracellular immune checkpoints” raise the possibility for the development of a completely new class of antitumor drugs that target these IICs (Fig. 3).
Figure 3:
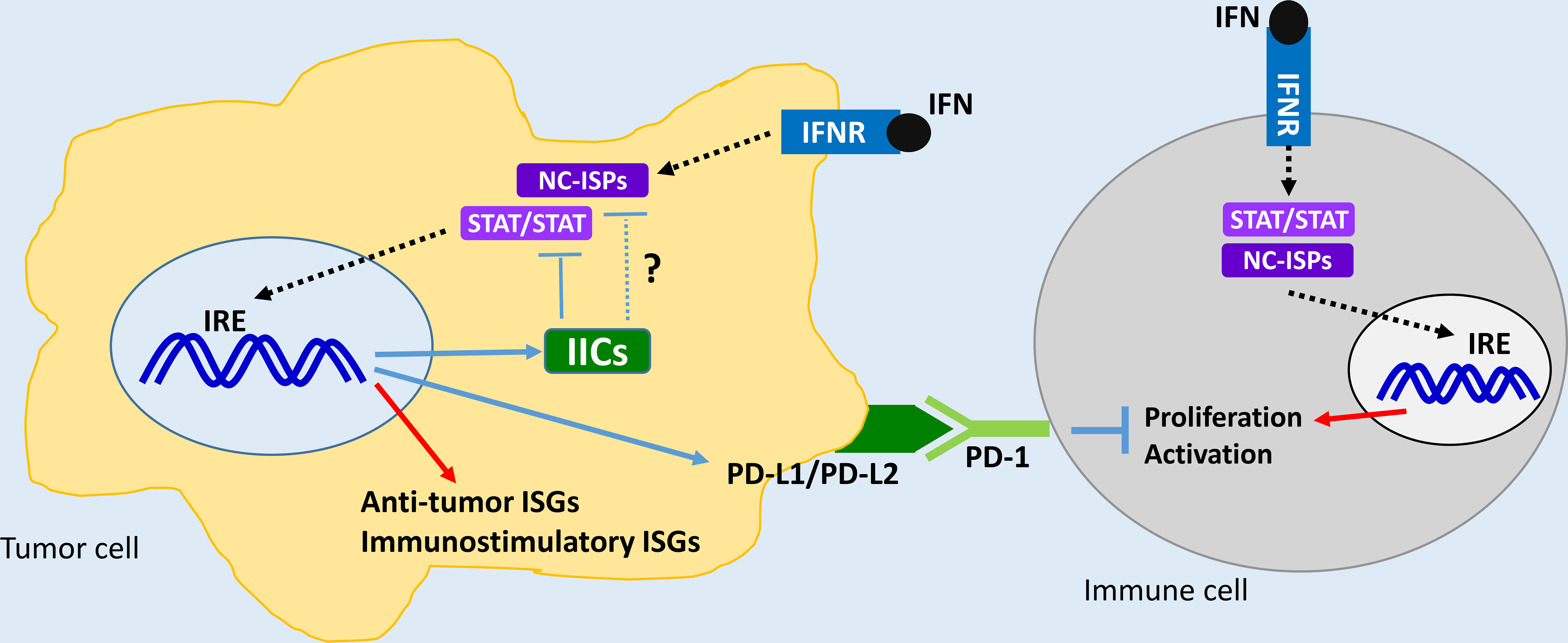
Proposed model on the roles of classical immune checkpoints and putative “intracellular immune checkpoints” related to IFN-signaling pathways. IICs - Intracellular Immune Checkpoints, NC-ISPs - Non-Canonical IFN Signaling Pathways
Highlights.
IFNs activate many signaling pathways in cancer cells
Non-canonical, non-STAT, pathways are critical for the IFN-antitumor response
IFN-engaged mTORC1 and/or mTORC2 pathways control ISG mRNA translation and/or transcription
SLFNs control IFN-induced STAT responses in some malignant cells and may act as intracellular immune checkpoints
Acknowledgments
Dr. Platanias’s research is supported by the National Institutes of Health grants R01-CA121192 and R01-CA77816, and grant I01-CX000916 from the Department of Veterans Affairs. This article is dedicated to the memory of Barbara Kroczynska who over the years made significant contributions to the IFN-signaling field.
Footnotes
Conflict of interest statement
Declarations of interest: none.
References
- [1].Isaacs A, Lindenmann J, Virus interference. I. The interferon, Proc. R. Soc. Lond. B Biol. Sci, 147 (1957) 258–267. [PubMed] [Google Scholar]
- [2].Wheelock EF, Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin, Science, 149 (1965) 310–311. [PubMed] [Google Scholar]
- [3].Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP, IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex, Nat. Immunol, 4 (2003) 69–77. [DOI] [PubMed] [Google Scholar]
- [4].Falcoff R, Some properties of virus and immune-induced human lymphocyte interferons, J. Gen. Virol, 16 (1972) 251–253. [DOI] [PubMed] [Google Scholar]
- [5].Youngner JS, Salvin SB, Production and properties of migration inhibitory factor and interferon in the circulation of mice with delayed hypersensitivity, J. Immunol, 111 (1973) 1914–1922. [PubMed] [Google Scholar]
- [6].Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM, IL-28, IL-29 and their class II cytokine receptor IL-28R, Nat. Immunol, 4 (2003) 63–68. [DOI] [PubMed] [Google Scholar]
- [7].Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, Reuhl K, Dickensheets H, Sheikh F, Donnelly RP, Raveche E, Kotenko SV, Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma, Cancer Res. 66 (2006) 4468–4477. [DOI] [PubMed] [Google Scholar]
- [8].Sato A, Ohtsuki M, Hata M, Kobayashi E, Murakami T, Antitumor activity of IFN-lambda in murine tumor models, J. Immunol, 176 (2006) 7686–7694. [DOI] [PubMed] [Google Scholar]
- [9].George PM, Badiger R, Alazawi W, Foster GR, Mitchell JA, Pharmacology and therapeutic potential of interferons, Pharmacol Ther, 135 (2012) 44–45. [DOI] [PubMed] [Google Scholar]
- [10].Platanias LC, Interferons and their antitumor properties, J Interferon Cytokine Res, 33 (2013) 143–1434. [DOI] [PubMed] [Google Scholar]
- [11].Pestka S, Kraus CD, Walter MR, Interferons, interferon-like cytokines, and their receptors, Immunol Rev, 202 (2004) 8–32. [DOI] [PubMed] [Google Scholar]
- [12].Lazear HM, Nice TJ, Diamond MS MS, Interferon-λ: Immune functions at barrier surfaces and beyond, Immunity, 43 (2015) 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Platanias LC, Mechanisms of type-I- and type-II-interferon-mediated signaling, Nat Rev Immunol, 5 (2005) 375–386. [DOI] [PubMed] [Google Scholar]
- [14].Parker BS, Rautela J, Hertzog PJ, Antitumour actions of interferons: implications for cancer therapy, Nat Rev Cancer, 16 (2016) 131–144. [DOI] [PubMed] [Google Scholar]
- [15].Strander H, Interferons. Antineoplastic drugs? Blut, 35 (1977) 288–289. [DOI] [PubMed] [Google Scholar]
- [16].Sikora K, Does interferon cure cancer? Br Med J, 281 (1980) 855–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Merigan TC, Sikora K, Breeden JH, Levy R, Rosenberg SA, Preliminary observations on the effect of human leukocyte interferon in non-Hodgkin’s lymphoma, N Engl J Med, 299 (1978) 1449–1453. [DOI] [PubMed] [Google Scholar]
- [18].Horning SJ, Levine JF, Meyer M, Merigan TC, Rosenberg SA, Phase I study of human leukocyte interferon in patients with advanced cancer, J Biol Response Mod., 2 (1983) 47–56. [PubMed] [Google Scholar]
- [19].Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR, Interferons at age 50: past, current and future impact on biomedicine, Nat Rev Drug Discov., 6 (2007) 975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sharma P, Allison JP, The future of immune checkpoint therapy, Science, 348 (2015) 56–61. [DOI] [PubMed] [Google Scholar]
- [21].Minn AJ, Wherry EJ, Combination cancer therapies with immune checkpoint blockade: Convergence on interferon signaling, Cell, 165 (2016) 272–275. [DOI] [PubMed] [Google Scholar]
- [22].Fragale A, Romagnoli G, Licursi V, Buoncervello M, Del Vecchio G, Giuliani C, Parlato S, Leone C, De Angelis M, Canini I, Toschi E, Belardelli F, Negri R, Capone I, Presutti C, Gabriele L, Antitumor effects of Epidrug/IFNα combination driven by modulated gene signatures in both colorectal cancer and dendritic cells, Cancer Immunol Res., 5 (2017) 604–616. [DOI] [PubMed] [Google Scholar]
- [23].Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, Zitvogel L, Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors, Immunity, 44 (2016) 1255–1269. [DOI] [PubMed] [Google Scholar]
- [24].Borden EC, Interferons α and β in cancer: therapeutic opportunities from new insights, Nat Rev Drug Discov., 3 (2019) 219–234. [DOI] [PubMed] [Google Scholar]
- [25].Pestka S, Langer JA, Zoon KC, Samuel CE, Interferons and their actions, Annu Rev Biochem, 56 (1987) 727–777. [DOI] [PubMed] [Google Scholar]
- [26].Stark GR, Kerr IM, Williams BRG, Silverman RH, Screiber RD, How cells respond to interferons, Annu Rev Biochem, 67 (1998) 227–264. [DOI] [PubMed] [Google Scholar]
- [27].Platanias LC, Fish EN, Signaling pathways activated by interferons, Exp Hematol, 27 (1999) 1583–1592. [DOI] [PubMed] [Google Scholar]
- [28].Fish EN, Platanias LC, Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes, Mol. Cancer Res, 12 (2014) 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ivashkiv LB, IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy, Nat. Rev. Immunol, 18 (2018) 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zanoni I, Granucci F, Broggi A, Interferon (IFN)-λ takes the helm: Immunomodulatory roles of Type III IFNs, Front Immunol, 28 (2017) 1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Majoros A, Platanitis F, Kernbauer-Hölzl E, Rosebrock F, Müller M, Decker T, 2017. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol, 8, 29. 10.3389/fimmu.2017.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Platanias LC, Introduction: interferon signals: what is classical and what is non-classical? J Interferon Cytokine Res, 25 (2005) 732. [DOI] [PubMed] [Google Scholar]
- [33].Uddin S, Sassano A, Deb DK, Verma A, Majchrzak B, Rahman A, Malik AB, Fish EN, Platanias LC, Protein kinase C-delta (PKC-delta) is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727, J Biol Chem, 277 (2002) 14408–16. [DOI] [PubMed] [Google Scholar]
- [34].Deb DK, Sassano A, Lekmine F, Majchrzak B, Verma A, Kambhampati S, Uddin S, Rahman A, Fish EN, Platanias LC, Activation of protein kinase C delta by IFN-gamma, J Immunol, 171 (2003) 267–73. [DOI] [PubMed] [Google Scholar]
- [35].Nguyen H, Ramana CV, Bayes J, Stark GR, Roles of phosphatidylinositol 3-kinase in interferon-gamma-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression, J Biol Chem, 276 (2001) 33361–33368. [DOI] [PubMed] [Google Scholar]
- [36].Ooi EL, Chan ST, Cho NE, Wilkins C, Woodward J, Li M, Kikkawa U, Tellinghuisen T, Gale M Jr, Saito T, Novel antiviral host factor, TNK1, regulates IFN signaling through serine phosphorylation of STAT1, Proc Natl Acad Sci USA, 111 (2014) 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nair JS, DaFonseca CJ, Tjernberg A, Sun W, Darnell JE Jr, Chait BT, Zhang JJ, Requirement of Ca2þ and CaMKII for Stat1 Ser-727 phosphorylation in response to IFN-0γ, Proc Natl Acad Sci USA, 99 (2002) 5971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kristof AS, Marks-Konczalik J, Billings E, Moss J, Stimulation of signal transducer and activator of transcription-1 (STAT1)-dependent gene transcription by lipopolysaccharide and interferon-gamma is regulated by mammalian target of rapamycin, J Biol Chem, 278 (2003) 33637–44. [DOI] [PubMed] [Google Scholar]
- [39].Kroczynska B, Blyth GT, Rafidi RL, Majchrzak-Kita B, Xu L, Saleiro D, Kosciuczuk EM, Jemielity J, Su B, Altman JK, Eklund EA, Fish EN, Platanias LC, Central Regulatory Role for SIN1 in Interferon γ (IFNγ) Signaling and Generation of Biological Responses, J. Biol. Chem, 292 (2017) 4743–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P, CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response, Immunity, 38 (2013) 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kosciuczuk EM, Mehrotra S, Saleiro D, Kroczynska B, Majchrzak-Kita B, Lisowski P, Driehaus C, Rogalska A, Turner A, Lienhoop T, Gius D, Fish EN, Vassilopoulos A, Platanias LC, Sirtuin 2-mediated deacetylation of cyclin-dependent kinase 9 promotes STAT1 signaling in type I interferon responses, J. Biol. Chem, 294 (2019) 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hsia HC, Hutti JE, Baldwin AS, Cytosolic DNA Promotes Signal Transducer and Activator of Transcription 3 (STAT3) Phosphorylation by TANK-binding Kinase 1 (TBK1) to Restrain STAT3 Activity, J. Biol. Chem, 292 (2017) 5405–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Steen HC, Kotredes KP, Nogusa S, Harris MY, Balachandran S, Gamero AM, Phosphorylation of STAT2 on serine-734 negatively regulates the IFN-α-induced antiviral response, J. Cell. Sci, 129 (2016) 4190–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Steen HC, Nogusa S, Thapa RJ, Basagoudanavar SH, Gill AL, Merali S, Barrero CA, Balachandran S, Gamero AM, Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses. J. Biol. Chem, 288 (2013) 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wang Y, Nan J, Willard B, Wang X, Yang J, Stark GR, Negative regulation of type I IFN signaling by phosphorylation of STAT2 on T387, EMBO J, 36 (2017) 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Saleiro D, Platanias LC, Intersection of mTOR and STAT signaling in immunity, Trends Immunol, 36 (2015) 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Choudhury GG, A linear signal transduction pathway involving phosphatidylinositol 3-kinase, protein kinase C-ε, and MAPK in mesangial cells regulates interferon-γ-induced STAT1α transcriptional activation, J. Biol. Chem, 279 (2004) 27399–27409. [DOI] [PubMed] [Google Scholar]
- [48].Reetz O, Stadler K, Strauss U, 2014. Protein kinase C activation mediates interferon-β-induced neuronal excitability changes in neocortical pyramidal neurons, J. Neuroinflammation, 11, 185. 10.1186/s12974-014-0185-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kroczynska B, Mehrotra S, Arslan AD, Kaur S, Platanias LC Regulation of interferon-dependent mRNA translation of target genes, J. Interfer. Cytok. Res, 34 (2014) 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Joshi S, Kaur S, Redig AJ, Goldsborough K, David K, Ueda T, Watanabe-Fukunaga R, Baker DP, Fish EN, Fukunaga R, Platanias LC, Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses, Proc. Natl. Acad. Sci. USA, 106 (2009) 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Joshi S, Sharma B, Kaur S, Majchrzak B, Ueda T, Fukunaga R, Verma AK, Fish EN, Platanias LC, Essential role for Mnk kinases in type II interferon (IFNγ) signaling and its suppressive effects on normal hematopoiesis, J. Biol. Chem, 286 (2011) 6017–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Rätsch G, Rice CM, Ivashkiv LB, Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation, Nat. Immunol, 16 (2015) 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mehrotra S, Sharma B, Joshi S, Kroczynska B, Majchrzak B, Stein BL, McMahon B, Altman JK, Licht JD, Baker DP, Eklund EA, Wickrema A, Verma A, Fish EN, Platanias LC, Essential role for the Mnk pathway in the inhibitory effects of type I interferons on myeloproliferative neoplasm (MPN) precursors, J. Biol. Chem, 288 (2013) 23814–23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sharma B, Joshi S, Sassano A, Majchrzak B, Kaur S, Aggarwal P, Nabet B, Bulic M, Stein BL, McMahon B, Baker DP, Fukunaga R, Altman JK, Licht JD, Fish EN, Platanias LC, Sprouty proteins are negative regulators of interferon (IFN) signaling and IFN-inducible biological responses, J. Biol. Chem, 287 (2012) 42352–42360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saxton RA, Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease, Cell, 168 (2017) 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Beauchamp EM, Platanias LC, The evolution of the TOR pathway and its role in cancer, Oncogene, 32 (2013) 3923–3932. 10.1038/onc.2012.567 [DOI] [PubMed] [Google Scholar]
- [57].Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N, Translation deregulation in human disease, Nat. Rev. Mol. Cell Biol, 19 (2018) 791–807. [DOI] [PubMed] [Google Scholar]
- [58].Mossmann D, Park S, Hall MN, mTOR signalling and cellular metabolism are mutual determinants in cancer, Nat. Rev. Cancer, 18 (2018) 744–757. [DOI] [PubMed] [Google Scholar]
- [59].Lekmine F, Uddin S, Sassano A, Parmar S, Brachmann SM, Majchrzak B, Sonenberg N, Hay N, Fish EN, Platanias LC, Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J. Biol. Chem, 278 (2003) 27772–27780. 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- [60].Lekmine F, Sassano A, Uddin S, Smith J, Majchrzak B, Brachmann SM, Hay N, Fish EN, Platanias LC, Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein, Exp. Cell Res, 295 (2004) 173–182. [DOI] [PubMed] [Google Scholar]
- [61].Parmar S, Smith J, Sassano A, Uddin S, Katsoulidis E, Majchrzak B, Kambhampati S, Eklund EA, Tallman MS, Fish EN, Platanias LC Differential regulation of the p70 S6 kinase pathway by interferon alpha (IFNalpha) and imatinib mesylate (STI571) in chronic myelogenous leukemia cells. Blood, 106 (2005) 2436–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kaur S, Lal L, Sassano A, Majchrzak-Kita B, Srikanth M, Baker DP, Petroulakis E, Hay N, Sonenberg N, Fish EN, Platanias LC, Regulatory effects of mammalian target of rapamycin-activated pathways in type I and II interferon signaling, J. Biol. Chem, 282 (2007) 1757–1768. [DOI] [PubMed] [Google Scholar]
- [63].Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, Brachmann SM, Fish EN, Platanias LC, Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling, J. Immunol, 181 (2008) 7316–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC, Role of the Akt pathway in mRNA translation of interferon-stimulated genes, Proc. Natl. Acad. Sci., USA, 105 (2008) 4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kroczynska B, Kaur S, Platanias LC, Growth suppressive cytokines and the AKT/mTOR pathway, Cytokine, 48 (2009) 138–143. [DOI] [PubMed] [Google Scholar]
- [66].Kroczynska B, Mehrotra S, Arslan AD, Kaur S, Platanias LC, Regulation of interferon-dependent mRNA translation of target genes. J. Interferon. Cytokine Res, 34 (2014) 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Laplante M, Sabatini DM, mTOR signaling in growth control and disease, Cell 149 (2012) 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kaur S, Sassano A, Majchrzak-Kita B, Baker DP, Su B, Fish EN, Platanias LC, Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses, Proc. Natl. Acad. Sci. USA, 109 (2012) 7723–7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kaur S, Kroczynska B, Sharma B, Sassano A, Arslan AD, Majchrzak-Kita B, Stein BL, McMahon B, Altman JK, Su B, Calogero RA, Fish EN, Platanias LC, Critical roles for Rictor/Sin1 complexes in interferon-dependent gene transcription and generation of antiproliferative responses, J. Biol. Chem, 289 (2014) 6581–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kroczynska B, Rafidi RL, Majchrzak-Kita B, Kosciuczuk EM, Blyth GT, Jemielity J, Warminska Z, Saleiro D, Mehrotra S, Arslan AD, Fish EN, Platanias LC, Interferon γ (IFNγ) signaling via mechanistic Ttarget of rapamycin complex 2 (mTORC2) and regulatory effects in the generation of Type II Interferon biological responses, J. Biol. Chem, 291 (2016) 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kroczynska B, Blyth GT, Rafidi RL, Majchrzak-Kita B, Xu L, Saleiro D, Kosciuczuk EM, Jemielity J, Su B, Altman JK, Eklund EA, Fish EN, Platanias LC, Central Regulatory Role for SIN1 in Interferon γ (IFNγ) signaling and generation of biological responses, J. Biol. Chem, 292 (2017) 4743–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Vigo T, Procaccini C, Ferrara G, Baranzini S, Oksenberg JR, Matarese G, Diaspro A, Kerlero de Rosbo N, Uccelli A, IFN-γ orchestrates mesenchymal stem cell plasticity through the signal transducer and activator of transcription 1 and 3 and mammalian target of rapamycin pathways, J. Allergy Clin. Immunol, 139 (2017) 1667–1676. [DOI] [PubMed] [Google Scholar]
- [73].Vigo T, La Rocca C, Faicchia D, Procaccini C, Ruggieri M, Salvetti M, Centonze D, Matarese G, Uccelli A, IFNβ enhances mesenchymal stromal (stem) cells immunomodulatory function through STAT1–3 activation and mTOR-associated promotion of glucose metabolism, Cell Death Dis., 10 (2019) 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yeh YH, Hsiao HF, Yeh YC, Chen TW, Li TK, Inflammatory interferon activates HIF-1α-mediated epithelial-to-mesenchymal transition via PI3K/AKT/mTOR pathway, J. Exp. Clin. Cancer Res, 37 (2018) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Beauchamp EM, Abedin SM, Radecki SG, Fischietti M, Arslan AD, Blyth GT, ang A, Lantz C, Nelson A, Goo YA, Akpan I, Eklund EA, Frankfurt O, Fish EN, Thomas PM, Altman JK, Platanias LC, Identification and targeting of novel CDK9 complexes in acute myeloid leukemia, Blood. 133 (2019) 1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T, Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment, Genomics, 51 (1998) 76–85. [DOI] [PubMed] [Google Scholar]
- [77].Alers S, Löffler AS, Wesselborg S, Stork B The incredible ULKs. Cell Commun. Signal, 10 (2012) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chan EY, Kir S, Tooze SA, siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy, J. Biol. Chem, 282 (2007) 25464–25474. [DOI] [PubMed] [Google Scholar]
- [79].Lee EJ, Tournier C, The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy, Autophagy, 7 (2011) 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1, Nat. Cell Biol, 13 (2011) 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Konno H, Konno K, Barber GN, Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling, Cell, 155 (2013) 688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Saleiro D, Mehrotra S, Kroczynska B, Beauchamp EM, Lisowski P, Majchrzak-Kita B, Bhagat TD, Stein BL, McMahon B, Altman JK, Kosciuczuk EM, Baker DP, Jie C, Jafari N, Thompson CB, Levine RL, Fish EN, Verma AK, Platanias LC, Central role of ULK1 in type I interferon signaling, Cell Rep. 11 (2015) 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Saleiro D, Blyth GT, Kosciuczuk EM, Ozark PA, Majchrzak-Kita B, Arslan AD, Fischietti M, Reddy NK, Horvath CM, Davis RJ, Fish EN, Platanias LC, 2018. IFN-γ-inducible antiviral responses require ULK1-mediated activation of MLK3 and ERK5 . Sci. Signal. 11, eaap9921. 10.1126/scisignal.aap9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Saleiro D, Kosciuczuk EM, Platanias LC, Beyond autophagy: New roles for ULK1 in immune signaling and interferon responses, Cytokine Growth Factor Rev, 29 (2016) 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang B, Kundu M, Canonical and noncanonical functions of ULK/Atg1, Curr. Opin. Cell Biol, 45 (2017) 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway, Science, 339 (2013) 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ, Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA, Science, 339 (2013) 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kemp MG, Lindsey-Boltz LA, Sancar A, UV light potentiates STING (stimulator of interferon genes)-dependent innate immune signaling through deregulation of ULK1 (Unc51-like kinase 1), J. Biol. Chem, 290 (2015) 12184–12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mavrommatis E, Fish EN, Platanias LC, The schlafen family of proteins and their regulation by interferons, J. Interferon Cytokine Res, 33 (2013) 206–210. 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Soper A, Kimura I, Nagaoka S, Konno Y, Yamamoto K, Koyanagi Y, Sato K, 2018. Type I interferon responses by HIV-1 Infection: Association with disease progression and control. Front. Immunol. 8, 1823. 10.3389/fimmu.2017.01823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu F, Zhou P, Wang Q, Zhang M, Li D, The Schlafen family: complex roles in different cell types and virus replication. Cell Biol. Int. 42 (2018) 2–8. 10.1002/cbin.10778. [DOI] [PubMed] [Google Scholar]
- [92].Sassano A, Mavrommatis E, Arslan AD, Kroczynska B, Beauchamp EM, Khuon S, Chew TL, Green KJ, Munshi HG, Verma AK, Platanias LC, Human Schlafen 5 (SLFN5) Is a Regulator of Motility and Invasiveness of Renal Cell Carcinoma Cells, Mol. Cell Biol, 35 (2015) 2684–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mavrommatis E, Arslan AD, Sassano A, Hua Y, Kroczynska B, Platanias LC, Expression and regulatory effects of murine Schlafen (Slfn) genes in malignant melanoma and renal cell carcinoma, J. Biol. Chem. 288 (2013) 33006–33015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Katsoulidis E, Carayol N, Woodard J, Konieczna I, Majchrzak-Kita B, Jordan J, Sassano A, Eklund EA, Fish EN, Platanias LC, Role of Schlafen 2 (SLFN2) in the generation of IFNα-induced growth inhibitory responses, J. Biol. Chem. 284 (2009) 25051–25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N, Sawicki K, Munshi H, Platanias LC, Role of Interferon α (IFNα)-inducible Schlafen-5 (SLFN5) in regulation of anchorage - independent growth and invasion of malignant melanoma cells, J. Biol. Chem. 285 (2010) 40333–40341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Isnaldi E, Ferraioli D, Ferrando L, Brohée S, Ferrando F, Fregatti P, Bedognetti D, Ballestrero A, Zoppoli G, Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer, Breast Cancer Res. Treat. 177 (2019) 335–343. [DOI] [PubMed] [Google Scholar]
- [97].Chaturvedi LS, Wang Q, More SK, Vomhof-DeKrey EE, Basson MD, Schlafen 12 mediates the effects of butyrate and repetitive mechanical deformation on intestinal epithelial differentiation in human Caco-2 intestinal epithelial cells, Hum. Cell. 32 (2019) 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, Brummelkamp TR, Schumacher TN, 2019. SLFN11 can sensitize tumor cells towards IFN-γ-mediated T cell killing. PLoS One. 14, e0212053 10.1371/journal.pone.0212053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wan G, Liu Y, Zhu J, Guo L, Li C, Yang Y, Gu X, Deng LL, Lu C, SLFN5 suppresses cancer cell migration and invasion by inhibiting MT1-MMP expression via AKT/GSK-3β/β-catenin pathway, Cell Signal. 59 (2019) 1–12. [DOI] [PubMed] [Google Scholar]
- [100].Basson MD, Wang Q, Chaturvedi LS, More S, Vomhof-DeKrey EE, Al-Marsoummi S, Sun K, Kuhn LA, Kovalenko P, Kiupel M, Schlafen 12 Interaction with SerpinB12 and deubiquitylases drives human enterocyte differentiation, Cell Physiol. Biochem, 48 (2018) 1274–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fischietti M, Arslan AD, Sassano A, Saleiro D, Majchrzak-Kita B, Ebine K, Munshi HG, Fish EN, Platanias LC, (2018). Slfn2 regulates Type I Interferon responses by modulating the NF-κB pathway. Mol. Cell. Biol, 38, e00053–18 10.1128/MCB.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yang JY, Deng XY, Li YS, Ma XC, Feng JX, Yu B, Chen Y, Luo YL, Wang X, Chen ML, Fang ZX, Zheng FX, Li YP, Zhong Q, Kang TB, Song LB, Xu RH, Zeng MS, Chen W, Zhang H, Xie W, Gao S, Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control, Nat. Commun., 9 (2018) 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Seong RK, Seo SW, Kim JA, Fletcher SJ, Morgan NV, Kumar M, Choi YK, Shin OS, Schlafen 14 (SLFN14) is a novel antiviral factor involved in the control of viral replication, Immunobiology. 222 (2017) 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M, Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11, Nature, 491 (2012) 125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Companioni Nápoles O, Tsao AC, Sanz-Anquela JM, Sala N, Bonet C, Pardo ML, Ding L, Simo O, Saqui-Salces M, Blanco VP, Gonzalez CA, Merchant JL, SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer, J. Gastroenterol. 52 (2017) 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Fletcher SJ, Johnson B, Lowe GC, Bem D, Drake S, Lordkipanidzé M, Guiú IS, Dawood B, Rivera J, Simpson MA, Daly ME, Motwani J, Collins PW, Watson SP, Morgan NV and UK Genotyping and Phenotyping of Platelets study group, SLFN14 mutations underlie thrombocytopenia with excessive bleeding and platelet secretion defects. J. Clin. Invest. 125 (2015) 3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Recher M, Karjalainen-Lindsberg ML, Lindlöf M, Söderlund-Venermo M, Lanzi G, Väisänen E, Kumar A, Sadeghi M, Berger CT, Alitalo T, Anttila P, Kolehmainen M, Franssila R, Chen T, Siitonen S, Delmonte OM, Walter JE, Pessach I, Hess C, Simpson MA, Navarini AA, Giliani S, Hedman K, Seppänen M, Notarangelo LD, Genetic variation in schlafen genes in a patient with a recapitulation of the murine Elektra phenotype, J. Allergy Clin. Immunol. 133 (2014) 1462–1465. [DOI] [PubMed] [Google Scholar]
- [108].Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, Xia Y, Khovananth K, Moresco EM, Satoh T, Takeuchi O, Akira S, Beutler B, An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence, Nat. Immunol, 11 (2010) 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Iwasaki J, Komori T, Nakagawa F, Nagase H, Uchida J, Matsuo K, Uto Y, Schlafen11 expression Is associated with the antitumor activity of trabectedin in human sarcoma cell lines, Anticancer Res, 39 (2019) 3553–3563. [DOI] [PubMed] [Google Scholar]
- [110].Murai J, Thomas A, Miettinen M, Pommier Y, 2019. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol Ther. pii: S0163–7258(19)30091–9 10.1016/j.pharmthera.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Luan J, Gao X, Hu F, Zhang Y, Gou X, SLFN11 is a general target for enhancing the sensitivity of cancer to chemotherapy (DNA-damaging agents), J Drug Target. (2019) 1–8. 10.1080/1061186X.2019.1616746. [DOI] [PubMed] [Google Scholar]
- [112].Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, Owonikoko TK, Woo KM, Cardnell RJ, Fujimoto J, Long L, Diao L, Wang J, Bensman Y, Hurtado B, de Groot P, Sulman EP, Wistuba II II, Chen A, Fleisher M, Heymach JV, Kris MG, Rudin CM, Byers LA, Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer, J Clin Oncol. 36 (2018) 2386–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Inno A, Stagno A, Gori S, Schlafen-11 (SLFN11): a step forward towards personalized medicine in small-cell lung cancer? Transl. Lung Cancer Res., 7 (2018) S341–S345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tang SW, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, Pommier Y, Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors, Clin Cancer Res. 24 (2018) 1944–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Allison Stewart C, Tong P, Cardnell RJ, et al. , Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 17 (2017) 28575–28587 10.18632/oncotarget.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Jang S, Ryu SM, Lee J, Lee H, Hong SH, Ha KS, Park WS, Han ET, Yang SR, Bleomycin inhibits proliferation via Schlafen-mediated cell cycle arrest in mouse alveolar epithelial cells, Tuberc. Respir. Dis. (Seoul). 82 (2019) 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Arslan AD, Sassano A, Saleiro D, Lisowski P, Kosciuczuk EM, Fischietti M, Eckerdt F, Fish EN, Platanias LC, Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma, Oncogene. 36 (2017) 6006–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Gresser I, Maury C, Brouty-Boye D, Mechanism of the antitumour effect of interferon in mice, Nature, 239 (1972) 167–168. [DOI] [PubMed] [Google Scholar]
- [119].Nishimura J, Mitsui K, Ishikawa T, Tanaka Y, Yamamoto R, Suhara Y, Ishitsuka H, Antitumor and antimetastatic activities of human recombinant interferon alpha A/D, Clin. Exp. Metastasis, 3 (1985) 295–304. [DOI] [PubMed] [Google Scholar]
- [120].Gresser I, Belardelli F, Maury C, Maunoury MT, Tovey MG, Injection of mice with antibody to interferon enhances the growth of transplantable murine tumors, J. Exp. Med, 158 (1983) 2095–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dighe AS, Richards E, Old LJ, Schreiber RD Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors, Immunity, 1 (1994) 447–456. [DOI] [PubMed] [Google Scholar]
- [122].Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD, Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice, Proc. Natl. Acad. Sci. USA. 95 (1998) 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zaidi MR, Merlino G The two faces of interferon-γ in cancer, Clin. Cancer Res, 17 (2011) 6118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Critchley-Thorne RJ, Simons DL, Yan N, Miyahira AK, Dirbas FM, Johnson DL, Swetter SM, Carlson RW, Fisher GA, Koong A, Holmes S, Lee PP, Impaired interferon signaling is a common immune defect in human cancer, Pro. Natl. Acad. Sci. USA 106 (2009) 9010–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Colamonici OR, Domanski P, Platanias LC, O Diaz M, Correlation between interferon (IFN) alpha resistance and deletion of the IFN alpha/beta genes in acute leukemia cell lines suggests selection against the IFN system, Blood. 80 (1992) 744–749. [PubMed] [Google Scholar]
- [126].Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A, Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy, Cell, 168 (2017) 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Budczies J, Bockmayr M, Klauschen F, Endris V, Fröhling S, Schirmacher P, Denkert C, Stenzinger A, Mutation patterns in genes encoding interferon signaling and antigen presentation: A pan-cancer survey with implications for the use of immune checkpoint inhibitors, Genes Chromosomes Cancer, 56 (2017) 651–659. [DOI] [PubMed] [Google Scholar]
- [128].Respa A, Bukur J, Ferrone S, Pawelec G, Zhao Y, Wang E, Marincola FM, Seliger B, Association of IFN-gamma signal transduction defects with impaired HLA class I antigen processing in melanoma cell lines, Clin. Cancer Res, 17 (2011) 2668–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Meissner M, Whiteside TL, van Kuik-Romein P, Valesky EM, van den Elsen PJ, Kaufmann R, Seliger B, Loss of interferon-gamma inducibility of the MHC class II antigen processing pathway in head and neck cancer: evidence for post-transcriptional as well as epigenetic regulation, Br. J. Dermatol, 158 (2008) 930–940. [DOI] [PubMed] [Google Scholar]
- [130].Liu K, Caldwell SA, Abrams SI, Immune selection and emergence of aggressive tumor variants as negative consequences of Fas-mediated cytotoxicity and altered IFN-gamma-regulated gene expression, Cancer Res, 65 (2005) 4376–4388. [DOI] [PubMed] [Google Scholar]
- [131].Syn NL, Teng MWL, Mok TSK, Soo R, De-novo and acquired resistance to immune checkpoint targeting, Lancet Oncol, 18 (2017) e731–e741. [DOI] [PubMed] [Google Scholar]
- [132].Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy, Nat. Rev. Cancer, 12 (2012) 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cameron F, Whiteside G, Perry C, Ipilimumab: first global approval, Drugs, 71 (2011) 1093–1104. [DOI] [PubMed] [Google Scholar]
- [134].Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA; IMpassion130 Trial Investigators, Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med, 379 (2018) 2108–2121. [DOI] [PubMed] [Google Scholar]
- [135].Topalian SL, Drake CG, Pardoll DM, Immune checkpoint blockade: a common denominator approach to cancer therapy, Cancer Cell, 27 (2015) 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression, Cell Rep., 19 (2017) 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning W, Zeng H, Zhang N, Du W, Chen C, Huang JA, PD-L1 induced by IFN-γ from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer, Int. J. Clin. Oncol., 22 (2017) 1026–1033. [DOI] [PubMed] [Google Scholar]
- [138].Morimoto Y, Kishida T, Kotani SI, Takayama K, Mazda O, Interferon-β signal may up-regulate PD-L1 expression through IRF9-dependent and independent pathways in lung cancer cells, Biochem. Biophys. Res. Commun, 507 (2018) 330–336. [DOI] [PubMed] [Google Scholar]
- [139].Bazhin AV, von Ahn K, Fritz J, Werner J, Karakhanova S, Interferon-α Up-Regulates the Expression of PD-L1 Molecules on Immune Cells Through STAT3 and p38 Signaling, Front. Immunol., 9 (2018) 2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Bellucci R, Martin A, Bommarito D, Wang K, Hansen SH, Freeman GJ, Ritz J, 2015. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology. 4, e1008824 10.1080/2162402X.2015.1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DSM, Pauken KE, Huang AC, Gangadhar TC, Amaravadi RK, Schuchter LM, Feldman MD, Ishwaran H, Vonderheide RH, Maity A, Wherry EJ, Minn AJ, Tumor Interferon Signaling regulates a multigenic resistance program to immune checkpoint blockade, Cell, 167 (2016) 1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Pai CS, Huang JT, Lu X, Simons DM, Park C, Chang A, Tamaki W, Liu E, Roybal KT, Seagal J, Chen M, Hagihara K, Wei XX, DuPage M, Kwek SS, Oh DY, Daud A, Tsai KK, Wu C, Zhang L, Fasso M, Sachidanandam R, Jayaprakash A, Lin I, Casbon AJ, Kinsbury GA, Fong L, Clonal deletion of tumor-specific T cells by interferon-γ confers therapeutic resistance to combination immune checkpoint blockade, Immunity, 50 (2019) 477–492.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Takeda K, Nakayama M, Hayakawa Y, Kojima Y, Ikeda H, Imai N, Ogasawara K, Okumura K, Thomas DM, Smyth MJ, IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting, Nat. Commun., 8 (2017) 14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ, Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion, Front. Immunol., 9 (2018) 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L, IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling, Int. J. Cancer. 143 (2018) 931–943. [DOI] [PubMed] [Google Scholar]


