Abstract
Cyclic dinucleotides are signaling molecules that modulate many processes, including immune response and virulence factor production. Their cellular levels in bacteria are fine-tuned by metal-dependent phosphodiesterases, namely, the EAL and HD-GYP proteins, with HD-GYPs belonging to the larger HD domain superfamily. In this study, we first focus on the catalytic properties and the range of metal ions and substrates of the HD-[HD-GYP] subfamily, consisting of two HD domains. We identified SO3491 as a homologue of VCA0681 and the second example of an HD-[HD-GYP]. Both proteins hydrolyze c-di-GMP and 3′3′ c-GAMP and coordinate various metal ions, but only Fe and to a lesser extent Co support hydrolysis. The proteins are active only in the diferrous form and not in the one-electron more oxidized FeIIFeIII state. Although the C-terminal HD-GYP domain is essential for activity, the role of the N-terminal HD domain remains unknown. We show that the N-terminal site is important for protein stability, influences the individual apparent kcat and KM (but not kcat/KM), and cannot bind c-di-GMP, thus precluding its involvement in cyclic dinucleotide sensing. We proceeded to perform phylogenetic analyses to examine the distribution and functional relationships of the HD-[HD-GYP]s to the rest of the HD-GYPs. The phylogeny provides a correlation map that draws a link between the evolutionary and functional diversification of HD-GYPs, serving as a template for predicting the chemical nature of the metallocofactor, level of activity, and reaction outcome.
Graphical Abstract
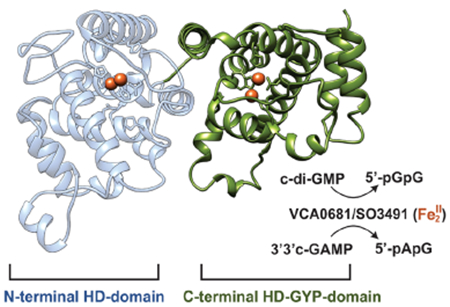
Cyclic dinucleotides (CDNs) are ubiquitous signaling molecules in bacterial and mammalian cells.1–3 They modulate many processes, including innate immune response,4 motility,5 virulence factor production,6 antibiotic production,7 phage resistance,8 and biofilm formation.9 The four known CDNs are the cyclic diguanosine monophosphate (c-di-GMP), cyclic diadenosine monophosphate (c-di-AMP), and the hybrid c-UAMP and c-GAMP, the latter of which exists as two isomers (the bacterial 3′3′ c-GAMP and the mammalian 2′3′ c-GAMP).6,10–12 Among these, c-di-GMP is the most intensively studied and a major regulator in virulence, biofilm formation, and cell motility.3,13 c-di-AMP is involved in cell growth, survival, and regulation of virulence.7,14,15 Although hybrid CDNs have only recently been identified, the evidence of their biological importance is growing. c-UAMP is a novel signaling nucleotide discovered in Escherichia coli that is thought to have a role in innate immune response.12 3′3′ c-GAMP is required for efficient intestinal colonization, chemotaxis regulation, and phage resistance in the bacterium Vibrio cholerae,6,8 whereas 2′3′ c-GAMP acts as a second messenger to activate innate immune response in eukaryotes.10,16
The intracellular levels of CDNs are tuned by the concerted (and opposing) action of two types of enzymes: (a) dinucleotide synthases (synthesis of CDN from two NTP molecules) and (b) phosphodiesterases (hydrolysis to either the linearized dinucleotide or two NMPs). These phosphodiesterases (PDEs) are metallohydrolases that include the EAL proteins (named after their conserved Glu-Ala-Leu motif) and HD-GYP proteins.3 HD-GYPs (IPR037522) count approximately 67000 members, while EALs (IPR001633) are more prevalent with ~245000 representatives. Although they were discovered almost at the same time,17,18 HD-GYPs are less well structurally and functionally characterized with respect to the chemical nature of their metallocofactor and catalytic outcomes.
HD-GYPs make up a subclass of the larger HD domain superfamily.17,19 HD domain proteins contain the characteristic tandem histidine-aspartate sequence motif, which is involved in the coordination of at least one transition metal ion.19 They are predominantly (phospho)hydrolases, but three known cases of oxygenases exist,20–22 demonstrating that this protein superfamily has a diverse functional repertoire. The hydrolases are further subdivided into PDEs and phosphatases (mono-, di-, or triphosphatases).23–25 Dinuclear HD domain cofactors are held to the protein polypeptide by six protein residues in a motif arranged as H···HD···H···H···D.17,19 HD-GYPs are PDEs that have (i) a seventh ligand added to the active site (i.e., an extra histidine adjacent to the last His of the motif) and (ii) a GYP residue triad in a loop close to the active site.17,26,27 Some HD-GYPs can have additional features such as (i) extra regulatory, sensory, or catalytic protein domains27–29 and (ii) an additional glutamate for coordination of a trimetal site.26 Assembly of a dinuclear or trinuclear cofactor is thought to be linked to different reaction outcomes, i.e., one-step or two-step hydrolysis.26,30,31
There is an increasing number of annotated HD-GYP proteins, highlighting their importance in the regulation of CDNs. Just the genome of the human pathogen V. cholerae encodes nine putative HD-GYPs.28,32 Among these, VCA0681 significantly decreases c-di-GMP levels and suppresses bacterial infectivity. It is also one of the three V. cholerae HD-GYPs that hydrolyze the hybrid 3′3′ c-GAMP,28 showing that some of them may be more specialized than others and not all HD-GYPs act on 3′3′ c-GAMP. VCA0681 consists of two metal-binding domains, an N-terminal HD domain and a C-terminal HD-GYP domain. Only the C-terminal domain is involved in CDN hydrolysis, whereas the N-terminal domain has an unknown functional role.30
The specific activities of HD-GYPs, and of VCA0681 in particular, are not known. The four available HD-GYP crystal structures,26,33–35 three of which are of holoproteins, reveal little about the substrate specificity, the chemical nature of the active cofactor, or the role of the GYP triad in catalysis. To bridge the existing gap in our knowledge of HD-GYPs, with emphasis on the HD-[HD-GYP] subfamily, we further biochemically characterized VCA0681 and its homologue SO3491 from Shewanella oneidensis. We show that HD-[HD-GYP]s can incorporate Fe-, Co-, Ni-, and Mn-based bimetallic cofactors, with the Fe form being the most active. This strong Fe dependence is more typical for HD domain oxygenases rather than HD domain hydrolases that typically utilize non-redox divalent metal ions.23–25,30,36,37 VCA0681 and SO3491 are active in their diferrous (Fe2II) form and not in the one-electron more oxidized FeIIFeIII form (which is in contrast to oxygenases). SO3491 and VCA0681 hydrolyze c-di-GMP and 3′3′ c-GAMP via one-step hydrolysis to the linearized products 5′-pGpG and 5′-pApG,38 respectively. The kinetic parameters for SO3491 and VCA0681 compare well to those of the EALs, demonstrating similar catalytic efficiencies. The hydrolytically inactive N-terminal HD site is important for protein stability, does not bind c-di-GMP, but influences the apparent kcat and KM for c-di-GMP hydrolysis. The C-terminal HD-GYP domain is critical for hydrolysis, as reported previously.30
In an attempt to bring together and better understand the diversity of HD-GYP proteins, their choice of metallocofactors, and the catalytic outcome, we performed bioinformatic analyses by collecting representatives of known biochemical function and/or structure. This allowed us to depict a correlation among substrate specificity, metallocofactor type, and different hydrolytic products among HD-GYPs. The overall outcome is a combined functional and evolutionary map that depicts the diversification within the HD-GYP protein superfamily and can serve as a molecular predictor for newly identified HD-GYP proteins.
MATERIALS AND METHODS
Materials.
All chemicals unless specified were obtained from Fisher Scientific and were of high purity grade. All monoand dinucleotides were purchased from Axxora, LLC (Farmingdale, NY).
Sequence Similarity Network (SSN).
The SSN was generated with the web-based Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) using 27488 sequences from InterPro family IRP037522, which includes proteins that have the HD-GYP domain.39 The network was further refined using protein sequences with an alignment score of 120.
Preparation of S. oneidensis pSUMO-SO3491 and MBP-SO3491.
Detailed procedures for overexpression, purification, and mutagenesis of the proteins are provided in the Supporting Information.
Metal Quantification.
Metal analysis of the as-purified proteins was achieved by inductively coupled atomic emission spectrometry (ICP-AES) at the Environmental Sustainability Laboratories (EESL) at The Pennsylvania State University (University Park, PA).
Steady-State Experiments.
Steady-state parameters were examined in multiple-turnover assays in three independent experiments. The reaction conditions utilizing c-di-GMP as a substrate included 1 μM (diiron concentration) WT and variant (native, SUMO-tagged, and MBP-tagged) SO3491 and MBP-VCA0681, 100 μM MgCl2, and 2 mM sodium dithionite. The reactions were carried out in 100 mM HEPES buffer containing 150 mM NaCl and 10% glycerol (pH 8). The reaction conditions utilizing 3′3′ c-GAMP as a substrate were exactly the same apart from the enzyme concentration (5 μM). Time-dependent reactions to compare the PDE activities of native SO3491 and MBP-VCA0681 were carried out with 1 μM protein (diiron concentration), 100 μM MgCl2, 2 mM sodium dithionite, and 150 μM c-di-GMP or 3′3′ c-GAMP. End-point reactions to examine the substrate specificity of the HD-GYP proteins were carried out with 5 μM WT MBP-VCA0681 and MBP-SO3491 (diiron concentration), 100 μM MgCl2, 2 mM sodium dithionite, and 50 μM di- and mononucleotides. The reactions were carried out under O2-free conditions in an anaerobic glovebox, and the redox state of the cofactor was verified by 57Fe Mössbauer spectroscopy. All reactions were initiated with the addition of substrate or substrate analogues and quenched at the described time points by incubation at 95 °C for 10 min. The efficiency of heat quenching for rapid inactivation of the enzymes at the desired time points was confirmed by comparison to quenching in 2 M acetic acid. The initial rates for c-di-GMP and 3′3′ c-GAMP were fitted by linear regression to the Michaelis–Menten equation using the Kaleidagraph software.
| (1) |
High-Performance Liquid Chromatography (HPLC).
Denatured reaction samples were centrifuged at 21130g, and the supernatant was filtered with a 0.22 μm nylon Spin-X Centrifuge Filter (Corning Inc., Corning, NY). The filtered samples were analyzed on a model 1260 Infinity Liquid Chromatography system (Agilent) and a model 1260 Infinity Photodiode Array Detector WR by monitoring the absorbance at 254 nm. Samples were injected on an Agilent reverse-phase C18-A Polaris column (particle size of 5 μm, 150 mm × 4.6 mm). Educts and products were separated using a gradient method utilizing a water-based mobile phase [10 mM KH2PO4 and 10 mM tetrabutylammonium hydroxide (TBAH) (pH 6), solvent A] and an organic-based mobile phase (methanol with 10 mM TBAH, solvent B). Quantification was afforded by the relative integrated peak intensities of educts and products.
Mössbauer Spectroscopy.
All samples were prepared in storage buffer under O2-free conditions in an anaerobic glovebox (Coylab). Samples were reacted with a small excess of sodium ascorbate (3–5 equiv) for 20 min to obtain the FeIIFeIII redox state. Samples to obtain the Fe2II state were reacted with an excess of sodium dithionite (10–20 equiv) for 30 min at 22 °C prior to being frozen in liquid N2. Mössbauer spectra were recorded on a WEB Research (Edina, MN) instrument that has been described previously.40 The spectrometer used to acquire the weak-field spectra is equipped with a Janis SVT-400 variable-temperature cryostat. The external magnetic field was applied parallel to the γ beam. All isomer shifts are quoted relative to the centroid of the spectrum of α-iron metal at room temperature. Mössbauer spectra were fitted using the WMOSS spectral analysis software (www.wmoss.org, WEB Research).
Electron Paramagnetic Resonance (EPR) Spectroscopy.
All samples were prepared in storage buffer under O2-free conditions in an anaerobic glovebox (CoyLab). The samples were reacted with sodium ascorbate for 20 min at 22 °C prior to being frozen in liquid N2. EPR spectra were recorded on a Bruker E500 Elexsys continuous wave (CW) X-band spectrometer (operating at approximately 9.38 GHz) equipped with a rectangular resonator (TE102) and a continuous-flow cryostat (Oxford 910) with a temperature controller (Oxford ITC 503). The spectra were recorded at variable temperatures between 10 and 40 K at a microwave power of 0.2 mW, using a modulation amplitude of 1 mT and a microwave frequency of 9.38 GHz.
Thermal Shift Assays.
The apparent melting temperatures for the various forms of SO3491 were determined by performing fluorescence thermal shift assays on a StepOne Plus Real Time PCR System. Experiments were carried out in MicroAmp Fast Optical 96-Well plates, in 25 μL reaction mixtures including 10× SYPRO fluorescent dye and 30 μM protein in a buffer containing 50 mM HEPES (pH 8.0) and 200 mM NaCl. Reaction mixtures were heated from 25 to 95 °C, and the fluorescence intensity at 570 nm was recorded in 0.3 °C temperature increments.
Phylogenetic Analysis.
A BLAST search was performed using the sequence of the V. cholerae VCA0681 (NCBI Reference Sequence NP_233069.1) against proteins in the RefSeq database. We selected 109 unique and nonredundant sequences of HD domain-containing PDEs for which there is some in vitro or in vivo biochemical information available. The selected subgroups are denoted as VCA0681, VCA0210, VCA0931, VC1295, PA4781, Bd1817, PA4108, VC1348, VC2340, and TM0186, on the basis of their annotation. Ten to twelve sequences were selected from each of these groups of PDEs. Ten sequences of the HD domain of the CRISPR-associated helicase/endonuclease Cas3 were used as an outgroup. Sequences were aligned with the MAFFT software (https://www.ebi.ac.uk/Tools/msa/mafft/). The maximum-likelihood phylogenetic rooted tree was subsequently computed with the IQ-tree software employing the LG+R8 model.41 A second phylogenetic analysis included the recently characterized HD-GYP protein PmxA from Myxococcus xanthus29 together with the 121 unique and nonredundant sequences of HD-GYP proteins and used the c-di-AMP specific PDE PgpH42 as an outgroup (10 sequences). The maximum-likelihood phylogenetic rooted tree was computed with the LG +R5 model.
RESULTS
S. oneidensis SO3491 Is a Catalytically Active HD-[HD-GYP] Protein.
We constructed a sequence similarity network (SSN) to map the HD-GYP protein space and select the closest isofunctional ortholog of V. cholerae VCA0681 (Figure 1A), which is not stable after cleavage of the fusion partner.30 SO3491 from S. oneidensis was selected on the basis of its sequence similarity and predicted protein solubility. Similar to VCA0681 (Figure 1B), SO3491 consists of two metal-binding domains. Native (untagged) SO3491 or as a fusion with SUMO and MBP hydrolyzed c-di-GMP and 3′3′ c-GAMP in an Fe-dependent manner, similar to MBP-VCA0681 (Figure 1C,D).30 SO3491 hydrolyzed c-di-GMP and 3′3′ c-GAMP with observed rates that are 2–3-fold faster compared to those of VCA0681, demonstrating that both proteins are catalytically comparable.
Figure 1.
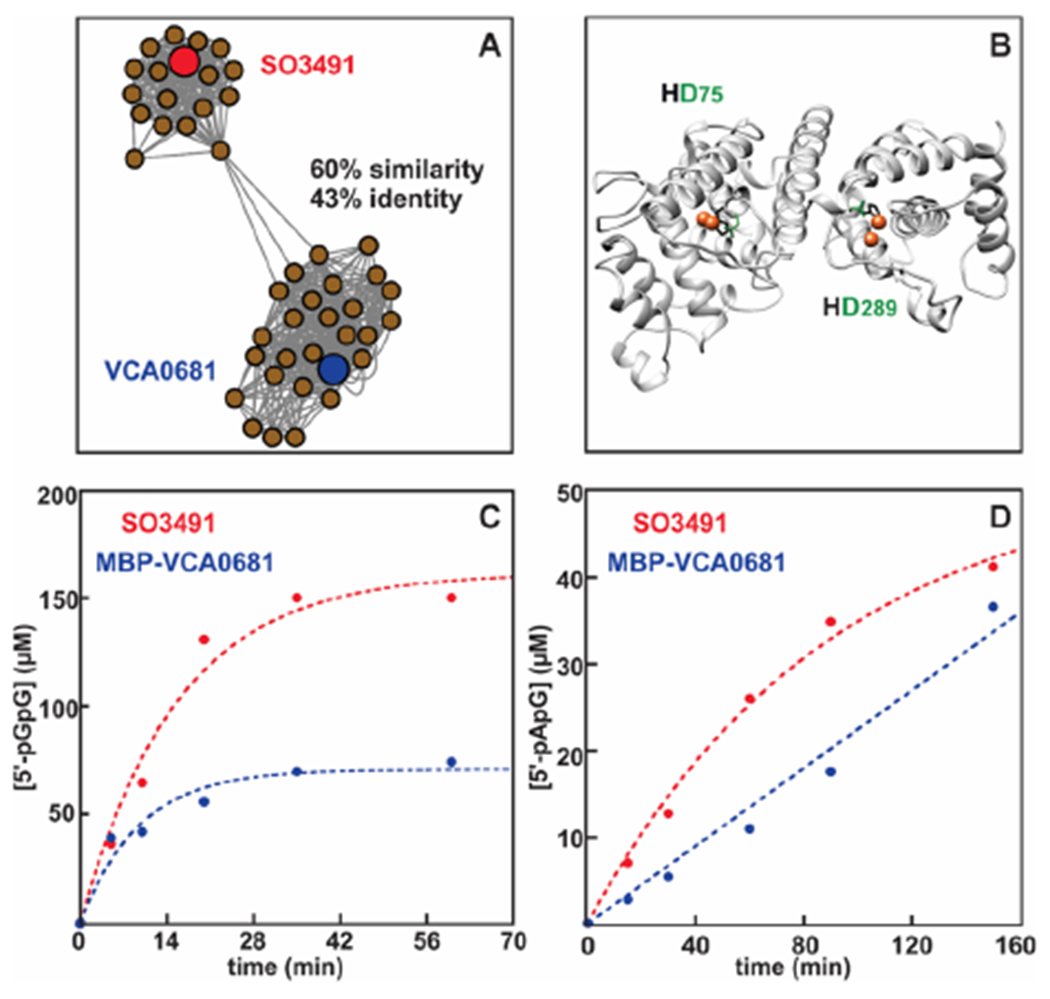
Sequence-based distribution of HD-GYP proteins, homology model of VCA0681, and hydrolytic activities of two HD-[HD-GYP] proteins with c-di-GMP and 3′3′ c-GAMP. (A) Cluster of the SSN containing only the HD-[HD-GYP] proteins (187 sequences). (B) Homology model of VCA0681. The two diiron cofactors have been modeled and are shown as orange spheres. The histidine and aspartate ligands coordinating the metal ions are colored black and green, respectively. (C) c-di-GMP hydrolysis by the diiron WT SO3491 and MBP-VCA0681 in the dithionite-reduced form. Experimental conditions: 1 μM diFe and 150 μM c-di-GMP. (D) 3′3′ c-GAMP hydrolysis by the diiron WT SO3491 and MBP-VCA0681 in the dithionite-reduced form. Experimental conditions: 5 μM diFe, 150 μM c-di-GMP, and 200 μM dithionite. Experiments were performed under O2-free conditions. Similar results were obtained for different protein preparations.
Chemical Nature of the Active Diiron Cofactor.
We combined activity assays with EPR and Mössbauer spectroscopies to identify the redox state of the reduced cofactor that is hydrolytically competent. Figure 2 and Figure S1 show the Mössbauer spectra of WT MBP-SO3491 under different redox conditions. The 4.2 K, 78 mT Mössbauer spectrum of the aerobically isolated protein shows only quadrupole doublets, characteristic of antiferromagnetically coupled diferric sites, demonstrating that the Fe in the sample is assembled exclusively in bimetallic sites and not in monomeric species (Figure S1). The diferric form is hydrolytically inactive. To examine which of the two possible reduced cofactor states (FeIIFeIII or Fe2II) can hydrolyze c-di-GMP, we reduced the enzyme with ascorbate and dithionite under conditions that would afford a maximum yield of each reduced species.20,21
Figure 2.
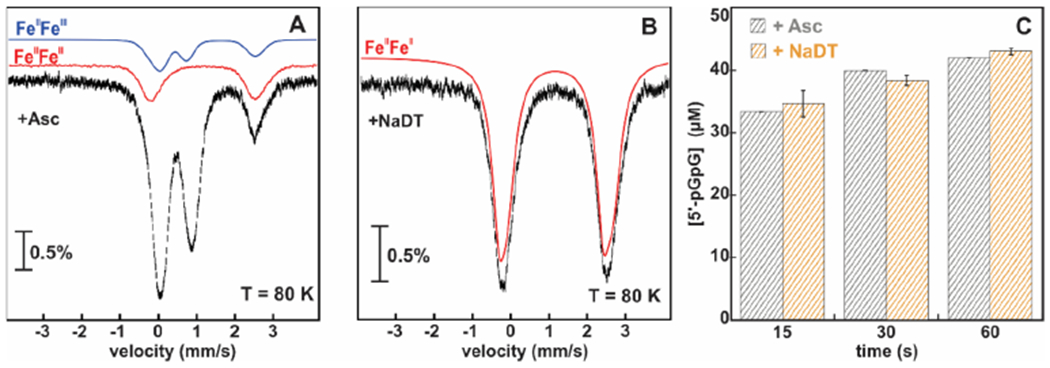
Mössbauer spectra and time-dependent c-di-GMP hydrolysis by the 57Fe-labeled WT MBP-SO3491 in different redox states. (A) 80 K Mössbauer spectrum of WT MBP-SO3491 reduced with 5 mM ascorbate. (B) 80 K Mössbauer spectrum of WT MBP-SO3491 reduced with 10 mM dithionite. The amount of Fe2II is shown with the red trace (experimental spectrum), and the amount of FeIIFeIII with the blue trace (simulation). (C) c-di-GMP hydrolysis by ascorbate- and dithionite-reduced 57Fe-labeled WT MBP-SO3491. The error bars represent the standard deviation.
At a high temperature (80 K), electronic relaxation is much faster than the nuclear Larmor frequency.43 As a result, both paramagnetic and diamagnetic states produce quadrupole doublets, allowing for their reliable quantification when combined with low-temperature EPR measurements. Combined analyses of the 80 K Mössbauer and 10 K EPR spectra (Figure 2A and Figure S2) show that ascorbate-reduced MBP-SO3491 contains the following speciation of diiron cofactors: 65% diamagnetic Fe2III, 15% paramagnetic , and 20% EPR-silent Fe2II forms. Under the same conditions, reduction with ascorbate of MBP-VCA0681 yields a similar amount of FeIIFeIII, but the majority species is now the Fe2II form (Figure S3). Reduction with dithionite yields homogeneously reduced samples to the all-ferrous state for both proteins (Figure 2B).
c-di-GMP hydrolysis was assayed on the 57Fe-labeled MBP-SO3491, for which concentrations of the reduced cofactor states were quantified by both EPR and Mössbauer spectroscopies (Figure 2A,B and Figure S2). The ascorbate-reduced sample contains 1 μM FeIIFeIII and 1.2 μM Fe2II (the rest of the sample is in the diferric form), while the dithionite-reduced sample contains only 1 μM Fe2II. Figure 2C shows the extent of c-di-GMP hydrolysis at different time points for the ascorbate- and dithionite-reduced samples. The production of 5′-pGpG is similar for both cases (Figure 2C), suggesting that the FeIIFeIII, present only in ascorbate-reduced MBP-SO3491, hardly contributes to the observed hydrolysis. Therefore, only the all-ferrous form can hydrolyze c-di-GMP, whereas the mixed-valent form is essentially inactive.
Substrate Scope and Specificity of the HD-[HD-GYP] Proteins.
We screened nine di- and mononucleotides as potential substrates28,30,42,44 for MBP-SO3491 and MBP-VCA0681 (Figure 3 and Figure S4). MBP-SO3491 and MBP-VCA0681 are very specific for c-di-GMP and 3′3′ c-GAMP and afford hydrolyses to 5′-pGpG and 5′-pApG, respectively (without any other detectable products). c-di-AMP cannot be hydrolyzed by either MBP-SO3491 or MBP-VCA0681. Mononucleotides are poor substrates, with only 2′3′-cAMP and 2′3′-cGMP producing a small amount of linear monophosphates (i.e., 3′-AMP, 2′-AMP, and 3′-GMP).
Figure 3.
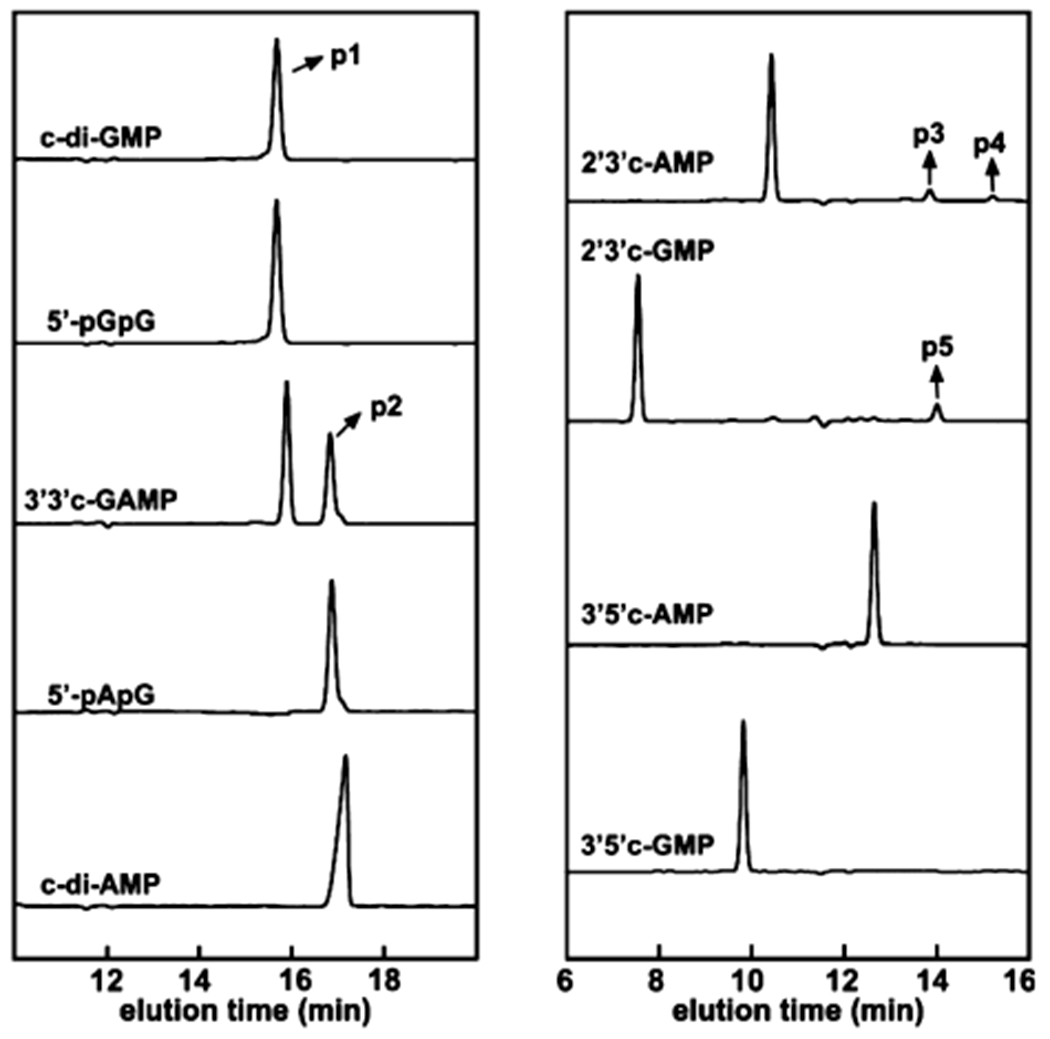
HPLC chromatograms monitoring the reaction of WT MBP-SO3491 with different nucleotides. The x-axis is the elution time, and the y-axis is the absorbance at 254 nm. All reactions were performed with 5 μM protein (diiron concentration) and 50 μM substrate analogues under O2-free conditions. The trace label corresponds to the substrate nucleotide. p1 corresponds to 5′-pGpG, p2 to 5′-pApG,38 p3 to 3′-AMP, p4 to 2′-AMP, and p5 to 3′-GMP.
Transition Metal Ions That Support Cofactor Assembly and Activity.
Guided by the range of transition metal ions that confer activity to HD domain hydrolases,23–25,31,42,45 we interrogated which metallocofactor type can afford hydrolysis in HD-[HD-GYP]s. We expressed and purified the Fe-, Mn-, Co-, and Ni-containing forms of WT MBP-SO3491 and proceeded to perform end-point assays monitoring c-di-GMP hydrolysis (Figure 4). On the basis of dimetal cofactor concentration (Table 1), the most active form of the enzyme is the diferrous form, followed by the Co-containing form. The Ni and Mn forms are essentially inactive. Inactivity in the case of the Mn-containing form does not stem from incomplete assembly of a dimetal site, because EPR experiments confirm the presence of a dimanganous state (Figure S2).
Figure 4.
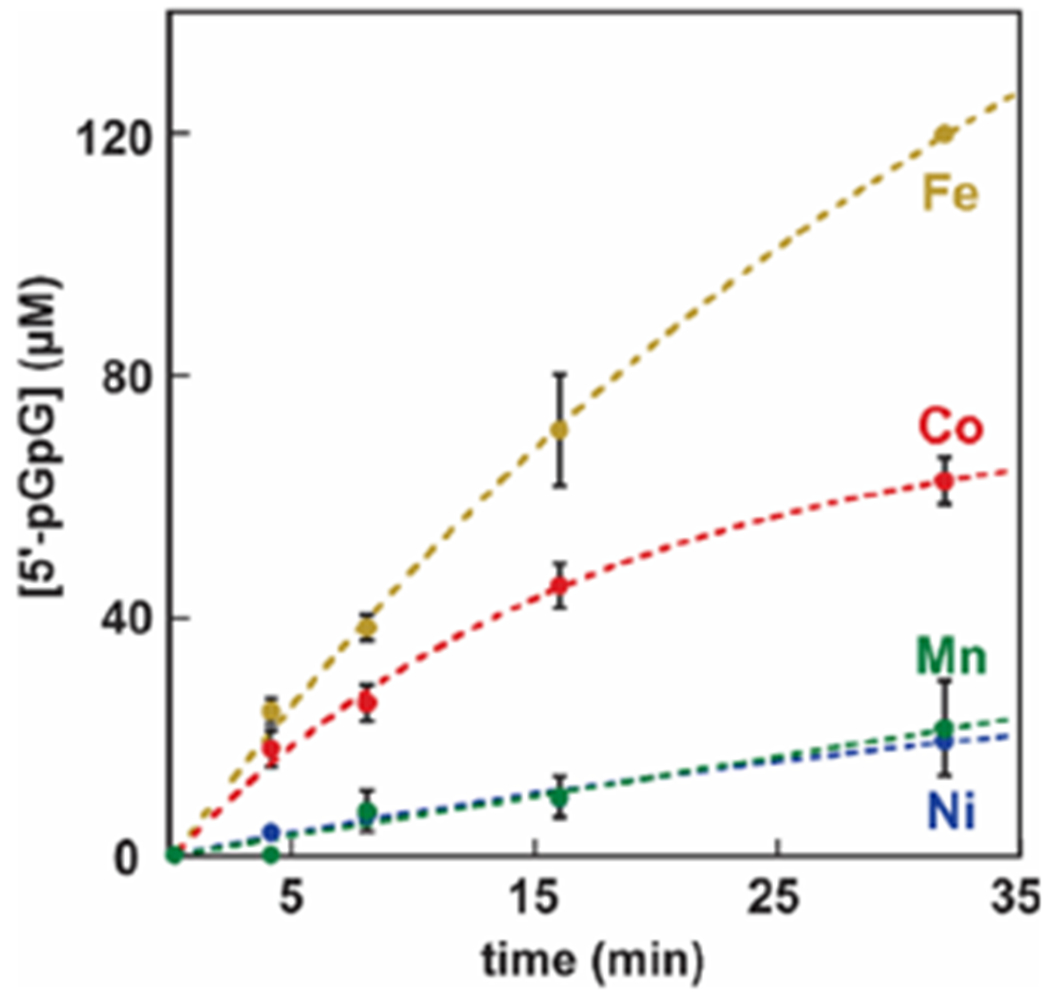
c-di-GMP hydrolysis by Co, Fe, Ni, and Mn forms of WT MBP-SO3491. The dithionite-reduced Fe form of MBP-SO3491 (1 μM diiron) was assayed under O2-free conditions, while the Co, Ni, and Mn forms of MBP-SO3491 (1 μM dimetal) were assayed under aerobic conditions. The c-di-GMP concentration was 120 μM.
Table 1.
Metal Ion Content of WT MBP-SO3491 Determined by ICP-AES
| Fe (per protein) | Mn (per protein) | Ni (per protein) | Co (per protein) | |
|---|---|---|---|---|
| M9-Fe | 2.68 | <0.001 | 0.07 | <0.001 |
| M9-Mn | 1.04 | 0.98 | 0.10 | <0.001 |
| M9-Ni | 0.21 | <0.001 | 3.51 | <0.001 |
| M9-Co | 0.11 | <0.001 | 0.10 | 1.80 |
Catalytic Proficiencies of HD-[HD-GYP]s and Comparison to Other PDEs.
We determined the catalytic proficiencies of the HD-[HD-GYP]s for c-di-GMP and 3′3′-cGAMP that are currently unknown. To obviate the possibility that a solubility tag affects activity, we first examined the activity of native SO3491. The Fe2II form hydrolyzes c-di-GMP with a catalytic constant (kcat) of 0.16 ± 0.01 s−1 and an apparent Michaelis constant (KM) of 3.2 ± 2 μM (Table 2) and a kcat/KM of 5 × 104 M−1 s−1 (Table 2 and Figure S5).46 3′3′ c-GAMP hydrolysis is slower and occurs with a kcat of 0.007 ± 0.002 s−1 and an apparent KM of 119 ± 60 μM. Interestingly, the catalytic proficiency of SO3491 for 3′3′ c-GAMP is ~6 M−1 s−1, almost 3 orders of magnitude lower than that for c-di-GMP. We also examined MBP-SO3491 to determine whether the presence of MBP affects the steady-state properties and to compare the kinetics of SO3491 to those of VCA0681 (insoluble without MBP). The presence of MBP in SO3491 does not influence the kcat for c-di-GMP but leads to an overestimation of the apparent KM, which is increased to 16 ± 1.8 μM (Table 2 and Figure 5B). MBP-VCA0681 exhibits a 5-fold smaller kcat for c-di-GMP (0.029 ± 0.003 s−1) compared to that of MBP-SO3491 but a similar apparent KM (Table 2 and Figure 5B). VCA0681 hydrolyzes 3′3′ c-GAMP slower than c-di-GMP (data not shown), similar to SO3491, showing that the two HD-[HD-GYP]s are catalytically equivalent for both substrates.
Table 2.
Steady-State Parameters of HD-GYP and EAL PDEs
| PDE | catalytic domain | metal ion | nuclearity | CDN | KM (μM) | kcat (s−1) | kcat/KM (M−1 s−1) |
|---|---|---|---|---|---|---|---|
| MBP-VCA0681 (V. cholerae) | HD-GYP | Fe2+ | dinuclear | c-di-GMP | 18 ± 8.1 | 0.029 ± 0.003 | 1.6 × 103 |
| MBP-SO3491 (S. oneidensis) | HD-GYP | Fe2+ | dinuclear | c-di-GMP | 16 ± 1.8 | 0.15 ± 0.005 | 9.5 × 103 |
| SO3491 (S. oneidensis) | HD-GYP | Fe2+ | dinuclear | c-di-GMP | 3.2 ± 2 | 0.16 ± 0.01 | 5.0 × 104 |
| PA410835 (Psudomonas aeruginosa) | HD-GYP | Mn2+ | dinuclear | c-di-GMP | 20 ± 5 | 1.5 × 10−4 | 7.5 |
| PA478135 (P. aeruginosa) | HD-GYP | Mn2+ | dinuclear | c-di-GMP | 119 ± 30 | 2.0 × 10−4 | 2.7 |
| PdeB47 (Borrelia burgdorferi) | HD-GYP | Mn2+ | nd | c-di-GMP | 0.0029 | nd | nd |
| CC339648 (Caulobacter crescentus) | EAL | Mg2+ | nd | c-di-GMP | >100 | nd | nd |
| RocR49 (P. aeruginosa) | EAL | Mg2+ | mononuclear | c-di-GMP | 3.2 | 0.67 | 2.1 × 105 |
| PA256750 (P. aeruginosa) | EAL | Mg2+ | nd | c-di-GMP | 5.2 | 0.39 | 7.5 × 104 |
| VieA51 (V. cholerae) | EAL | Mn2+ | nd | c-di-GMP | 0.06 | nd | nd |
| BlrP152 (Klebsiella pneumoniae) | EAL | Mn2+ | dinuclear | c-di-GMP | nd | 0.13 | nd |
| VcEAL (V. cholerae)53 | EAL | Ca2+ | dinuclear | c-di-GMP | 23.7 ± 2.3 | 0.16 | 6.6 × 103 |
| VcEAL (V. cholera)53 | EAL | Ca2+ | dinuclear | 3′3′ c-GAMP | 26.5 ± 2.7 | 0.15 | 5.8 × 103 |
| SO3491 (S. oneidensis) | HD-GYP | Fe2+ | dinuclear | 3′3′ c-GAMP | 119 ± 60 | 0.007 ± 0.002 | 5.9 |
Figure 5.
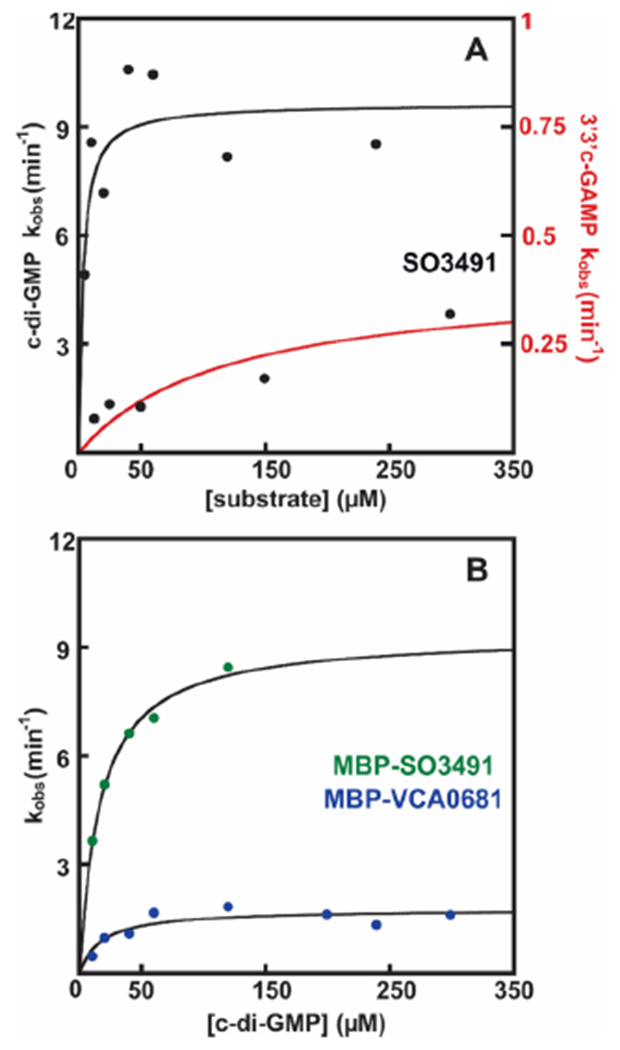
Steady-state kinetics of SO3491 and VCA0681. (A) Steady-state kinetics of WT native SO3491 with c-di-GMP (black) and 3′3′ c-GAMP (red). (B) Steady-state kinetics of WT MBP-SO3491 (green circles) and MBP-VCA0681 (blue circles) with c-di-GMP. The data are an average of values from at least three different experiments. Standard errors are listed in Table 2 and in the text.
Role of the N-Terminal HD Domain in HD-[HD-GYP]s.
The N-terminal HD domain is hydrolytically inactive, and little to no information exists about its possible roles in structure and function. We introduced the D75A and D69A substitutions in VCA0681 and SO3491, respectively, which disrupts putative carboxylate coordination to the metal centers.30 The Fe2II D75A MBP-VCA0681 hydrolyzes c-di-GMP with a kcat of 0.14 ± 0.02 s−1 and an apparent KM of 147 ± 59 μM, resulting in a kcat/KM of 1000 M−1 s−1 (Figure 6B). The catalytic efficiency of the D75A variant for c-di-GMP hydrolysis is thus on par with that of WT MBP-VCA0681, suggesting that the N-terminal metallosite does not have a significant effect on PDE activity. Removal of the C-terminal metallocofactor, introduced by the D289A and D283A substitutions in VCA0681 and SO3491, respectively, leads to completely inactive enzymes (Figure S6), confirming previous reports.30
Figure 6.
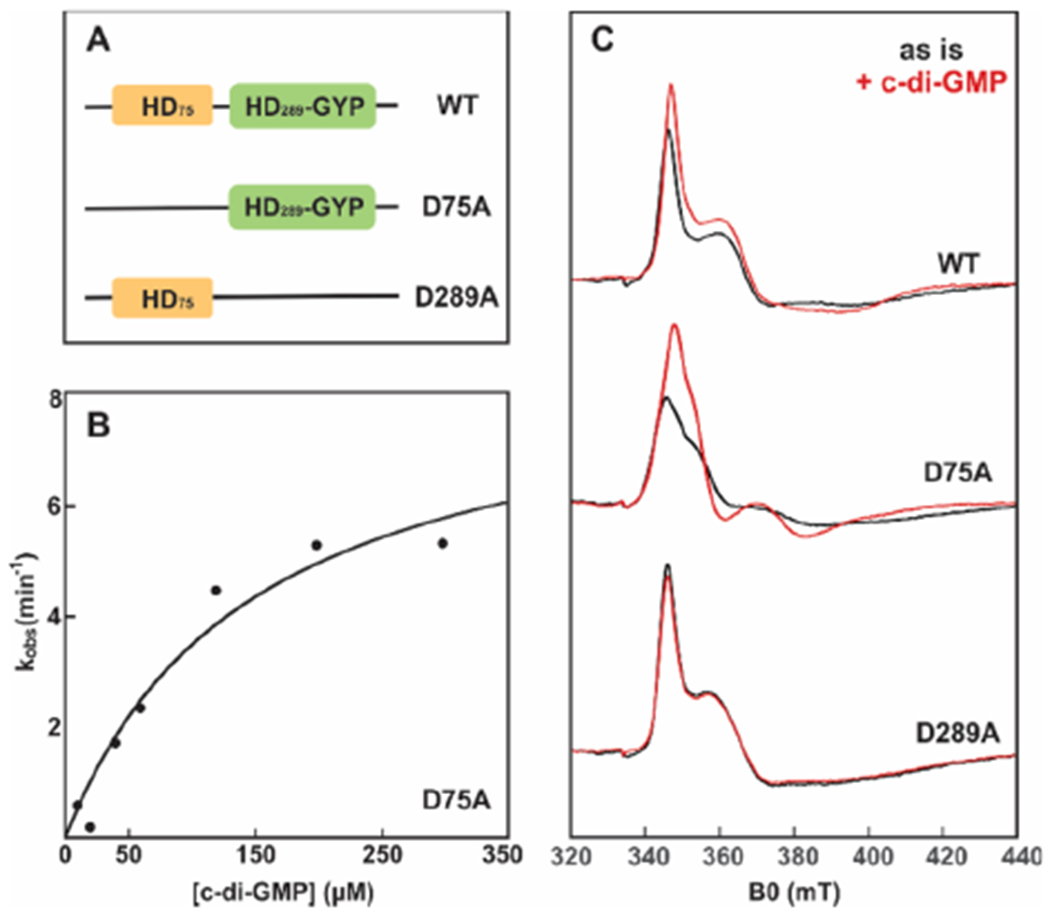
Steady-state kinetics for D75A MBP-VCA0681 and CW EPR spectra monitoring binding of c-di-GMP to WT and variant MBP-VCA0681. (A) Cartoon illustrations of the domain architectures for WT and variant VCA0681 referring to their two respective metallosites (HD). (B) Steady-state kinetics of D75A MBP-VCA0681 with c-di-GMP. The data are an average of at least three different experiments. (C) CW EPR spectra of ascorbate-reduced WT (top), D75A (middle), and D289A (bottom) MBP-VCA0681 in the absence (black) and presence (red) of 1.5 mM c-di-GMP. All samples were reduced with 5 mM sodium ascorbate, and spectra were recorded at 10 K. Experimental conditions: 100 μM diFe, microwave frequency of 9.38 GHz, microwave power of 0.2 mW, and modulation amplitude of 1 mT.
We then asked whether the N-terminal site is involved in substrate binding and thus c-di-GMP sensing. Figure 6C shows the 10 K CW EPR spectra of ascorbate-reduced WT, D75A, and D289A MBP-VCA0681, all of which exhibit signals characteristic of antiferromagnetically coupled FeIIFeIII centers (black traces). The EPR spectra of the two variants are different and can thus serve for spectroscopically distinguishing each of the two sites. To examine whether c-di-GMP can associate with either or both diiron centers, the dinucleotide was added in excess and samples were quickly frozen to avoid hydrolysis due to the diferrous form present. The EPR spectrum of the active D75A variant shows large differences upon addition of c-di-GMP (Figure 6C, red trace), consistent with coordination of c-di-GMP to the C-terminal HD-GYP site. Addition of c-di-GMP to the inactive D289A variant, however, has no observable effects on its EPR spectrum, demonstrating that the N-terminal HD site does not bind c-di-GMP. Addition of c-di-GMP to the WT protein that harbors both HD and HD-GYP centers results in intermediary spectral changes, which can be deconvoluted to the individual mixed-valent spectra of c-di-GMP-bound D75A and substrate-free D289A.
We further explored the effect of the N-terminal HD domain in protein solubility and thermostability (and therefore activity). Depending on the presence and type of the tag, D69A SO3491 shows very different activities for c-di-GMP. The SUMO-tagged protein is largely insoluble and completely inactive, but substitution of SUMO with MBP restores solubility and protein activity (Figure S7). Thermal shift assays of WT and variant SUMO-SO3491 and MBP-SO3491 showed a weak correlation of protein thermostability and the size of the tag (Figure S7).
Phylogenetic Distribution of HD-GYP Proteins.
We carried out bioinformatic analyses with the aim of portraying the phylogenetic relationships of HD-[HD-GYP]s within the HD-GYP superfamily and draw any possible connections to HD-GYP type and their exact chemistry (cofactor and outcome). Figure 7 shows the maximum-likelihood rooted tree that contains 109 sequences of HD-GYPs, for which some biochemical information is available. The HD-GYPs are segregated in three major clades. The first clade contains SO3491 and VCA0681 (encoding two tandem HD motifs), as well as the less well-studied V. cholerae VCA0931. All are active HD-GYPs that hydrolyze c-di-GMP and 3′3′ c-GAMP.28,30
Figure 7.
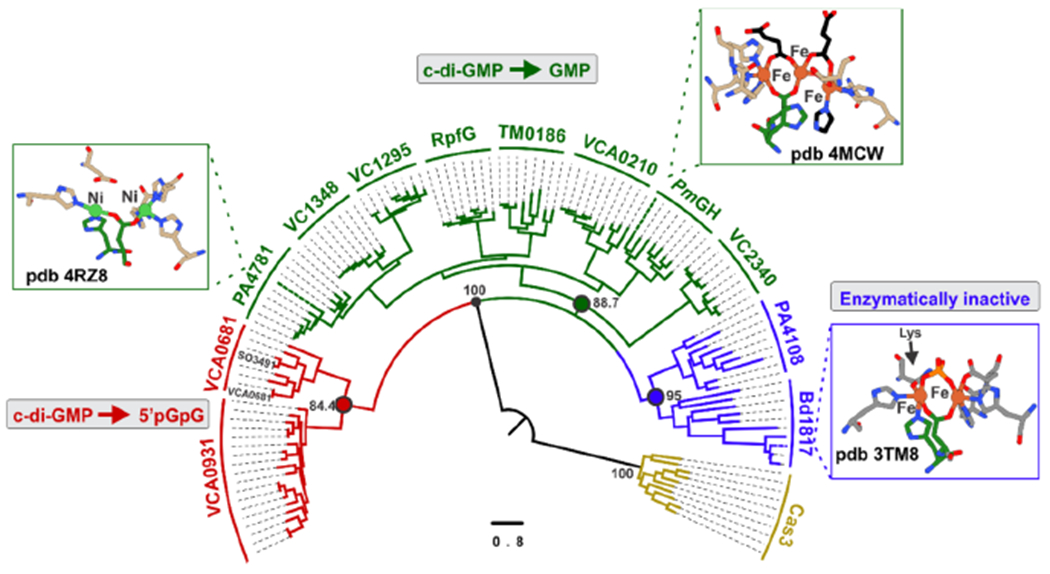
Phylogenetic distribution of HD-GYPs. The maximum-likelihood phylogenetic tree of HD-GYPs contains 109 sequences of HD-GYPs and was computed with the IQ-tree software using the LG+R8 model. The scale bar represents the number of substitutions per site. Cas3 was used as an outgroup (gold).
The second and largest clade includes the HD-GYPs from Persephonella marina [PmGH, Protein Data Bank (PDB) entry 4MCW] and PA4781 from Pseudomonas aeruginosa (PDB entry 4R8Z), for which crystal structures exist. PmGH hydrolyzes c-di-GMP completely to GMP and assembles a trinuclear center, with the third metal ion being coordinated by an extra glutamate.26,54 PA4781 also forms GMP, but the site has two nickel ions that are too far apart to form a bimetallic center.35,55 TM0186, also part of this clade (Figure 7), shows GMP formation when coordinating (at least transiently) a trinuclear site.31 The less well-studied V. cholerae VC1295, VC2340, and VCA0210 and the Xanthomonas campestris RpfG are also reported in some cases to yield GMP.27,32 Similar to PmGH, all HD-GYPs in this clade contain an extra glutamate in their sequence (Figure S8) and form GMP, suggesting that they all assemble trimetal centers and perform a two-step hydrolysis. The recently characterized 3′3′ c-GAMP specific M. xanthus PmxA, when included in a secondary analysis (Figure S9), co-localizes with the putative trinuclear proteins. PmxA yields mononucleotides similar to the trinuclear HD-GYPs but lacks the extra glutamate, which serves as a signature residue for trinuclear assembly.29
The third clade includes the crystallographically characterized Bdellovibrio bacteriovorus Bd1817 and the biochemically studied P. aeruginosa PA4108. Bd1817 is completely inactive with c-di-GMP,33 whereas PA4108 does exhibit some activity for c-di-GMP, albeit very low.35 It appears that this branch contains proteins that have lost completely or to a significant extent their ability to hydrolyze c-di-GMP (or perhaps they are missing a prior activating step or factor).
DISCUSSION
HD-GYP proteins regulate degradation of CDNs that are important secondary messengers.3,6,15 We have identified and biochemically characterized SO3491 from S. oneidensis MR-1 as the second protein from the HD-[HD-GYP] subclass. The CDN-related pathways in S. oneidensis MR-1 are less well-known, but these are of appreciable interest for applications in bioremediation and/or microbial fuel cells.56 SO3491 is a structural and functional homologue of VCA0681 with the same substrate preference and chemical nature of the active cofactor. In the study presented here, the Fe form of MBP-VCA0681 (and MBP-SO3491) exhibited the highest hydrolysis rates. The Mn and Ni forms were largely inactive, which comes in contrast to EALs and other HD-GYPs.31,35,47,49,51 Interestingly, Co also supported significant activity. However, whether Co-dependent hydrolysis represents a general property of the HD domain scaffold or is biologically relevant for HD-[HD-GYP]s remains to be elucidated.24,36
The choice of iron as a cofactor in HD-[HD-GYP]s may be linked to the environmental niches of these bacteria (i.e., V. cholera and S. oneidensis) and/or their targeted “activation” among other HD-GYPs within the cell that are Mn-dependent. In addition, the choice of iron may be related to the function of one or both metallocenters in small molecule sensing (such as O2 or NO). Binding of O2 (or NO) to HD domain sites can affect virulence by altering c-di-GMP hydrolysis. There are cases of other PDEs that have incorporated a secondary Fe-dependent component to regulate their activities. Examples include the heme domain of the E. coli direct oxygen sensor (DOS) involved in nucleotide-dependent signaling and the recently identified hemerythrin domain in the Ferrovum sp. PN-J185 suggested to act as an O2-sensing and -binding unit to regulate activity under oxidative stress.57,58 In the case of the HD-[HD-GYP]> proteins, the N-terminal (and/or the C-terminal) diiron center may serve a role similar to that of a redox sensor to modulate cellular activity.
Previous studies demonstrated that VCA0681 hydrolyzes c-di-GMP in its reduced form, but it was unclear whether the mixed-valent (FeIIFeIII) or diferrous (Fe2II) form is catalytically active.30 In the study presented here, we show that the Fe2II form in HD-[HD-GYP]s accumulates under mild reducing conditions and is the only cofactor form competent to hydrolyze c-di-GMP. In contrast, in HD domain oxygenases under the same reducing conditions, FeIIFeIII is the major species and the only active cofactor form.21 The observed stabilization of the Fe2II state at more positive potentials (when comparing the extent of accumulation of the diferrous state under the exact same redox conditions)21,22 suggests that the Fe2III/FeIIFeIII and FeIIFeIII/Fe2II redox couples are “closer” to each other in hydrolases than in oxygenases. The different redox properties allow oxygenases (active in the FeIIFeIII state) and HD domain PDEs (active in the FeIIFeII form) to be constitutively active within the cell.
HD-[HD-GYP]s and EALs are shown to be catalytically similar and exhibit comparable kcat/KM values for c-di-GMP.47–50,53 The apparent KMs of SO3491 (and VCA0681) can be well matched to the cellular levels of c-di-GMP in V. cholerae, which are reported to be 0.9–20 μM.59 However, SO3491 (and VCA0681)60 exhibits an almost 3 order of magnitude lower kcat/KM for 3′3′ c-GAMP than for c-di-GMP. This finding is to a certain degree unexpected, because (i) VCA0681 is purported to be the primary regulator of 3′3′ c-GAMP V. cholerae28 and (ii) VcEAL, a 3′3′ c-GAMP specific EAL domain PDE, exhibits the same catalytic efficiency for both c-di-GMP and 3′3′ c-GAMP.53 At present, it is unclear whether the differential activity for the two substrates is specific to HD-[HD-GYP]s. The recently characterized HD-GYP, PmxA, shows a strict preference for 3′3′ c-GAMP (over c-di-GMP), which has been attributed to a glutamine in the vicinity of the active site, proposed to act as a substrate specificity residue.29 SO3491 and VCA0681 have an arginine instead of a glutamine at the same position, which lacks the side-chain oxygen that would allow hydrogen bonding to the amine of the adenosine base of 3′3′ c-GAMP. Although PmxA is essentially inactive with c-di-GMP, both SO3491 and VCA0681 can hydrolyze 3′3′ c-GAMP. These observations suggest that substrate recognition mechanisms do not simply rely on single amino acid residues and can vary between different HD-GYPs.
SO3491 and VCA0681 are strictly tuned for c-di-GMP and 3′3′ c-GAMP degradation, but not for c-di-AMP. This result agrees well with previous studies28,30 and extends to the larger HD-GYP superfamily. Both proteins are inactive against 5′-pGpG or 5′-pApG, demonstrating that HD-[HD-GYP]s can neither hydrolyze their products to the individual nucleotides (via a second hydrolysis step) nor act as 5′-nucleotidases forming 5′-ApG, as previously stated.28 Although they act on 2′3′-cyclic mononucleotides, the catalytic efficiency is very poor. Overall, HD-[HD-GYP]s seem to be tailored for their specific substrates.
The role of the additional N-terminal HD domain in structure and function remains poorly understood. We show that the N-terminal bimetallic site (i) does affect the individual catalytic parameters, but under cellular c-di-GMP concentrations the overall catalytic efficiency remains the same, (ii) influences protein solubility, thermostability, and most likely structure and folding, and (iii) is not involved in substrate binding (and thus CDN sensing). The inability of the N-terminal HD domain to bind c-di-GMP may rationalize its lack of hydrolytic activity and may stem from changes in its local protein environment and/or the absence of the GYP residue triad. It also provides confidence that the obtained apparent KMs are not overestimated.
HD-GYPs are quite diverse with respect to their active metal cofactor, auxiliary domains, and possible catalytic outcomes. We thus analyzed their phylogenetic relationships to understand their diversification and specific functions. The phylogenetic tree consists of three main branches. The first branch contains the putative dinuclear HD-GYPs. VCA0681, SO3491, and VCA0931 hydrolyze c-di-GMP and 3′3′ c-GAMP to their linearized dinucleotides, suggesting common mechanisms.28,30 The chemical nature and specific activity of the metallocofactor of VCA0931 remain unknown, but on the basis of its sequence motif, we predict it coordinates a single bimetallic site.28,34
The HD-GYPs that occupy the second branch include the crystallographically characterized PmGH and PA478126,55 and biochemically studied TM0186.31 PmGH has an extra glutamate ligand that allows for formation of a trinuclear cofactor. All HD-GYPs of this clade have this extra glutamate residue conserved, suggesting that they can all coordinate a trimetal site. PmGH and TM0186 employ a mixed Fe/Mn or solely Fe- and Mn-containing cofactor for c-di-GMP hydrolysis. Both proteins are highly active and can break c-di-GMP completely to GMP via 5′pGpG, but the second hydrolysis occurs with a much slower rate. This observation may question the functional importance of hydrolysis to GMP under cellular conditions,29,31 especially because there are dedicated 5′-pGpG nucleases.61 PA4781 is unselective in its metal ion incorporation, has limited activity, and exhibits a preference for 5′-pGpG over c-di-GMP.35,55 The elongated distance between the two Ni ions in the crystal structure suggests that it can potentially assemble a trinuclear cofactor. Other proteins in this clade are VCA0210, VC1348, VC1295, VC2340, and RpfG, most of which are shown to yield GMP as the final product. VCA0210 can act on both c-di-GMP and 3′3′ c-GAMP, but there is some discrepancy about the hydrolytic outcome being mainly 5′pGpG or GMP. We hypothesize that VCA0210 can form GMP, only under conditions that favor assembly of a trinuclear cofactor. Thus, the differences reported for the VCA0210 activity may stem from protein expression conditions or a prior allosteric activation step (i.e., VCA0210 contains a REC domain that can be regulated by phosphorylation).
Overall, the functional (GMP formation) and sequence (extra glutamate) similarities of all of the HD-GYPs of this clade to PmGH suggest that (i) they can all incorporate trinuclear cofactors and (ii) there is a tight link between cofactor type and hydrolytic outcome. PmxA when included in the phylogenetic analysis is segregated together with putative trinuclear HD-GYPs. PmxA does not contain an extra glutamate residue to coordinate a third metal ion.29 Its co-localization with the putative trinuclear PDEs, however, may be related to its ability to produce GMP and AMP in a Mn-dependent manner. Therefore, either PmxA incorporates other ligands for stabilization of an extra metal ion, or the cofactor nuclearity of HD-GYPs may not be the sole determinant of their reaction outcome.
The third major clade comprises well-characterized Bd1817 and PA4108. Both proteins have very low or no activity toward c-di-GMP and are thus suggested to have other functions within the cell (c-di-GMP sensors).33,35 The observed loss of function in dinuclear Bd1817 may stem from the presence of an asparagine in place of the last coordinating aspartate or a degraded GYP motif. For PA4108, the molecular origins of the reduced hydrolytic activity cannot be easily inferred by the primary sequence, because all of the necessary residues are conserved. It may thus be inferred that this clade contains dinuclear HD-GYPs that can bind c-di-GMP (and/or 5′-pGpG) but are not competent toward hydrolysis.
Collectively, HD-GYPs are segregated in three main classes: (i) HD-GYPs employing a two-metal ion-based hydrolysis that may be preferentially Fe-dependent, (ii) HD-GYPs affording coordination of a trinuclear Mn- and Fe-dependent cofactor to allow for slow hydrolysis of the 5′-pGpG to GMP, and (iii) HD-GYPs coordinating a dinuclear site, which is hardly active. The diversity of HD-GYPs may reflect the tinkering of nature with this specific protein scaffold to expand its catalytic and structural repertoire and afford CDN hydrolysis under different stimuli and cellular cues.
CONCLUSION
HD-GYP proteins appear to be much more diverse than that originally thought, and we are at the beginning of uncovering their full functional potential. SO3491 is an Fe-dependent HD-[HD-GYP] and a functional ortholog of VCA0681. It can hydrolyze c-di-GMP and 3′3′ c-GAMP with rates that are on par with those of EALs. In contrast to HD domain oxygenases, HD-[HD-GYP]s employ the Fe2II form for hydrolysis and not the one-electron more oxidized FeIIFeIII state. The catalytically inactive N-terminal metal domain of HD-[HD-GYP]s neither influences the overall catalytic efficiency nor binds substrates. Our phylogenetic analysis provides insight into the distribution of HD-GYPs and assigns them a functional and evolutionary context with respect to their metallocofactors and specific activities.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Dr. Donald Kurtz (University of Texas at San Antonio, San Antonio, TX) for kindly providing us the plasmids of WT, D75A, and D289A MBP-VCA0681. The authors thank Paul S. Ralifo for providing us access to the EPR spectrometer at the Chemical Instrumentation Center of Boston University (Boston, MA). The authors also thank Laura J. Liermann for the ICP-AES analysis at the Laboratory for Isotopes and metals in Environment (The Pennsylvania State University, University Park, PA). The authors also thank Ben Pomerantz for initial protein characterization.
Funding
This work was supported by National Institutes of Health Grants GM111978 and GM126303 to M.-E.P. and the Brandeis University Innovation SPROUT program.
ABBREVIATIONS
- PDE
phosphodiesterase
- CDN
cyclic dinucleotide
- EAL
proteins that contain the conserved Glu-Ala-Leu motif
- c-di-GMP
bis(3′,5′)-cyclic dimeric guanosine monophosphate
- c-di-AMP
bis(3′,5′)-cyclic dimeric adenosine monophosphate
- NTP
nucleoside 5′-triphosphate
- NMP
nucleoside 5′-monophosphate
- 5′-pApG
5′-phosphoadenylyl-(3′→5′)-guanosine
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00257
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00257.
Procedures for protein purification, primers employed in this study (Table S1), and Mössbauer spectra, EPR spectra, HPLC assays, sequence alignment, and additional phylogenetic analysis (Figures S1–S9) (PDF)
Contributor Information
Sining Sun, Department of Biochemistry, Brandeis University, Waltham, Massachusetts 02453, United States.
Maria-Eirini Pandelia, Department of Biochemistry, Brandeis University, Waltham, Massachusetts 02453, United States.
REFERENCES
- (1).Römling U, Gomelsky M, and Galperin MY (2005) C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol 57, 629–639. [DOI] [PubMed] [Google Scholar]
- (2).Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol 7, 263–273. [DOI] [PubMed] [Google Scholar]
- (3).Romling U, Galperin MY, and Gomelsky M (2013) Cyclic di-GMP: the First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Simm R, Morr M, Kader A, Nimtz M, and Römling U (2004) GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessibility to motility. Mol. Microbiol 53, 1123–1134. [DOI] [PubMed] [Google Scholar]
- (6).Davies BW, Bogard RW, Young TS, and Mekalanos JJ (2012) Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Luo Y, and Helmann JD (2012) Analysis of the role of Bacillus subtilis σ M in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol 83, 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, and Sorek R (2019) Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695. [DOI] [PubMed] [Google Scholar]
- (9).Tischler AD, and Camilli A (2004) Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol 53, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wu J, Sun L, Chen X, Du F, Shi H, Chen C, and Chen ZJ (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science (Washington, DC, U. S.) 339, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, and Hornung V (2013) CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, and Kranzusch PJ (2019) Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GCL, and Yildiz FH (2015) C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in Vibrio cholerae. PLoS Pathog. 11, e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mehne FMP, Gunka K, Eilers H, Herzberg C, Kaever V, and Stülke J (2013) Cyclic Di-AMP homeostasis in bacillus subtilis: Both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem 288, 2004–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, and Woodward JJ (2013) Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4, n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, and Patel DJ (2013) Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Galperin MY, Natale DA, Aravind L, and Koonin EV (1999) A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol 1, 303–305. [PMC free article] [PubMed] [Google Scholar]
- (18).Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, and Benziman M (1998) Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: Genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol 180, 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Aravind L, and Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci 23, 469–472. [DOI] [PubMed] [Google Scholar]
- (20).Xing G, Diao Y, Hoffart LM, Barr EW, Prabhu KS, Arner RJ, Reddy CC, Krebs C, and Bollinger JM (2006) Evidence for C-H cleavage by an iron-superoxide complex in the glycol cleavage reaction catalyzed by myo-inositol oxygenase. Proc. Natl. Acad. Sci. U. S. A 103, 6130–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wörsdörfer B, Lingaraju M, Yennawar NH, Boal AK, Krebs C, Bollinger JM, and Pandelia ME (2013) Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Proc. Natl. Acad. Sci. U. S. A 110, 18874–18879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Rajakovich LJ, Pandelia ME, Mitchell AJ, Chang WC, Zhang B, Boal AK, Krebs C, and Bollinger JM (2019) A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases. Biochemistry 58, 1627–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HCT, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, De Carvalho LPS, Stoye JP, Crow YJ, Taylor IA, and Webb M (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382. [DOI] [PubMed] [Google Scholar]
- (24).Bridwell-Rabb J, Kang G, Zhong A, Liu HW, and Drennan CL (2016) An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis. Proc. Natl. Acad. Sci. U. S. A 113, 13750–13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zimmerman MD, Proudfoot M, Yakunin A, and Minor W (2008) Structural Insight into the Mechanism of Substrate Specificity and Catalytic Activity of an HD-Domain Phosphohydrolase: The 5′-Deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol 378, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bellini D, Caly DL, Mccarthy Y, Bumann M, An SQ, Dow JM, Ryan RP, and Walsh MA (2014) Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol. Microbiol 91, 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, and Dow JM (2010) Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. U. S. A 107, 5989–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gao J, Tao J, Liang W, Zhao M, Du X, Cui S, Duan H, Kan B, Su X, and Jiang Z (2015) Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell Res. 25, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wright TA, Jiang L, Park JJ, Anderson WA, Chen G, Hallberg ZF, Nan B, and Hammond MC (2020) Second messengers and divergent HD-GYP phosphodiesterases regulate 3′,3′-cGAMP signaling. Mol. Microbiol 113, 222–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Miner KD, Klose KE, and Kurtz DM (2013) An HD-GYP cyclic di-guanosine monophosphate phosphodiesterase with a non-heme diiron-carboxylate active site. Biochemistry 52, 5329–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Miner KD, and Kurtz DM (2016) Active Site Metal Occupancy and Cyclic Di-GMP Phosphodiesterase Activity of Thermotoga maritima HD-GYP. Biochemistry 55, 970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).McKee RW, Kariisa A, Mudrak B, Whitaker C, and Tamayo R (2014) A systematic analysis of the in vitro and in vivo functions of the HD-GYP domain proteins of Vibrio cholerae. BMC Microbiol. 14, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lovering AL, Capeness MJ, Lambert C, Hobley L, and Sockett RE (2011) The structure of an unconventional HD-GYP protein from bdellovibrio reveals the roles of conserved residues in this class of Cyclic-di-GMP phosphodiesterases. mBio 2, n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Deng M.-j., Tao J, Chao E, Ye Z.-y., Jiang Z, Yu J, and Su X.-d. (2018) Novel Mechanism for Cyclic Dinucleotide Degradation Revealed by Structural Studies of Vibrio Phosphodiesterase V-cGAP3. J. Mol. Biol 430, 5080–5093. [DOI] [PubMed] [Google Scholar]
- (35).Stelitano V, Giardina G, Paiardini A, Castiglione N, Cutruzzolà F, and Rinaldo S (2013) C-di-GMP Hydrolysis by Pseudomonas aeruginosa HD-GYP Phosphodiesterases: Analysis of the Reaction Mechanism and Novel Roles for pGpG. PLoS One 8, e74920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD, Zhou S, Rajashankar K, Kurinov I, Zhang R, and Ke A (2014) Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol 21, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mashhadi Z, Xu H, and White RH (2009) An Fe2+-dependent cyclic phosphodiesterase catalyzes the hydrolysis of 7,8-dihydro-D-neopterin 2′,3′-cyclic phosphate in methanopterin biosynthesis. Biochemistry 48, 9384–9392. [DOI] [PubMed] [Google Scholar]
- (38).In our experiments, the identity of the product nucleotide as 5′-pApG or 5′-pGpA was concluded by comparison to a commercially available standard of 5′-pApG. However, we accept that there may be some ambiguity in its exact assignment (and it could be 5′-pGpA), if the HPLC separation was insufficient to resolve any shifts in elution times. [Google Scholar]
- (39).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Price JC, Barr EW, Tirupati B, Bollinger JM, and Krebs C (2003) The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: A high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry 42, 7497–7508. [DOI] [PubMed] [Google Scholar]
- (41).Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, and Vinh LS (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol 35, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Huynh TAN, Luo S, Pensinger D, Sauer JD, Tong L, and Woodward JJ (2015) An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U. S. A 112, E747–E756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gütlich P, Bill E, and Trautwein AX (2011) Mössbauer spectroscopy and transition metal chemistry: Fundamentals and applications. Mössbauer Spectrosc. Transit. Met. Chem. Fundam. Appl, DOI: 10.1007/978-3-540-88428-6. [DOI] [Google Scholar]
- (44).Nagata M, Kaito C, and Sekimizu K (2008) Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem 283, 2176–2184. [DOI] [PubMed] [Google Scholar]
- (45).Mulepati S, and Bailey S (2011) Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J. Biol. Chem 286, 31896–31903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Alternatively considering substrate/product inhibition at higher c-di-GMP concentrations, an apparent KM of 6 ± 3.7 μM and a kcat of 0.2 ± 0.04 s−1 and an apparent dissociation constant Ki of 553 ± 529 μM could be obtained. [Google Scholar]
- (47).Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, and Motaleb MA (2011) Analysis of the HD-GYP domain cyclic dimeric gmp phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect. Immun 79, 3273–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Christen M, Christen B, Folcher M, Schauerte A, and Jenal U (2005) Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem 280, 30829–30837. [DOI] [PubMed] [Google Scholar]
- (49).Rao F, Yang Y, Qi Y, and Liang ZX (2008) Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: A study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol 190, 3622–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, and Liang ZX (2009) The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol 191, 4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Tamayo R, Tischler AD, and Camilli A (2005) The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem 280, 33324–33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, and Schlichting I (2009) Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459, 1015–1018. [DOI] [PubMed] [Google Scholar]
- (53).Yadav M, Pal K, and Sen U (2019) Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: Identification of a novel conformational switch and its implication. Biochem. J 476, 3333–3353. [DOI] [PubMed] [Google Scholar]
- (54).Including three molecules from the crystallization solution (one imidazole and two succinates). [Google Scholar]
- (55).Rinaldo S, Paiardini A, Stelitano V, Brunotti P, Cervoni L, Fernicola S, Protano C, Vitali M, Cutruzzolà F, and Giardina G (2015) Structural basis of functional diversification of the HD-GYP domain revealed by the Pseudomonas aeruginosa PA4781 protein, which displays an unselective bimetallic binding site. J. Bacteriol 197, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Chao L, Rakshe S, Leff M, and Spormann AM (2013) PdeB, a cyclic di-GMP-specific phosphodiesterase that regulates Shewanella oneidensis MR-1 motility and biofilm formation. J. Bacteriol 195, 3827–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Tuckerman JR, Gonzalez G, Sousa EHS, Wan X, Saito JA, Alam M, and Gilles-Gonzalez MA (2009) An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48, 9764–9774. [DOI] [PubMed] [Google Scholar]
- (58).Kitanishi K, Igarashi J, Matsuoka A, and Unno M (2020) Identification and Characterization of a Redox Sensor Phosphodiesterase from Ferrovum sp. PN-J185 Containing Bacterial Hemerythrin and HD-GYP Domains. Biochemistry 59, 983–991. [DOI] [PubMed] [Google Scholar]
- (59).Koestler BJ, and Waters CM (2013) Exploring environmental control of cyclic di-gmp signaling in vibrio cholerae by using the ex vivo lysate cyclic di-GMP assay (TELCA). Appl. Environ. Microbiol 79, 5233–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).The observed rates of VCA0681 for 3′3′ c-GAMP hydrolysis are slower than those of SO3491 reported in this study. [Google Scholar]
- (61).Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, and Lee VT (2015) Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A 112, E5048–E5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


