Abstract
The lncRNA LINC01123 has been reported to act as an oncogene in many human cancers. Nevertheless, the function and underlying mechanism of LINC01123 in osteosarcoma (OS) remain unclear. This study aimed to explore the roles and mechanisms of LINC01123 in OS progression. In this study, the expression of LINC01123 was significantly upregulated in OS cell lines than in a human osteoblast cell line. Furthermore, in vitro and in vivo experiments confirmed that knockdown of LINC01123 suppressed cell progression. Mechanistically, LINC01123 acted as a competing endogenous RNA by sponging miR‐516b‐5p, thus, increasing Gli1 expression by directly targeting its 3ʹUTR. Taken together, LINC01123 enhances OS proliferation and metastasis via the miR‐516b‐5p/Gli1 axis. Therefore, LINC01123 may be a potential therapeutic target for OS treatment.
Keywords: Gli1, LINC01123, metastasis, osteosarcoma, proliferation
Our data showed that LINC01123 is an oncogene that promotes the progression of OS cells via competitively binding miR‐561b‐5p, thereby enhancing Gli1 expression. Our results suggest that LINC01123 may be an important possible target for OS therapy, thus potentially providing a new therapy direction for OS treatment.
1. INTRODUCTION
Osteosarcoma (OS) is a common malignant bone tumor that is mainly found in the limbs, but can also appear in the axial skeleton. 1 OS occurs primarily in children and adolescents 15‐25 years of age. 2 OS has high malignancy, and the main treatment is surgery combined with neoadjuvant radiotherapy and chemotherapy. 3 However, the 5 year survival rate has not substantially improved in the past few decades. Consequently, a greater understanding of the underlying molecular mechanism is needed to provide more effective clinical treatment methods.
Long non‐coding RNAs (lncRNAs) are transcripts greater than 200 bp in length with no protein‐coding ability. LncRNAs influence the transcription and translation of some genes through many mechanisms, such as chromatin modification and protein inhibition. 4 Moreover, lncRNAs also act as competing endogenous RNAs (ceRNAs) that sponge microRNAs (miRNAs) that target messenger RNA (mRNA) expression. 5 LncRNAs are critical in the tumorigenesis of OS. Ding et al have found that the lncRNA CRNDE promotes OS growth via the Wnt/β‐catenin pathway. 6 The lncRNA LINC00511 enhances OS cell proliferation and migration through decreasing microRNA‐765 expression. 7 The recently discovered lncRNA LINC01123 was initially identified as an oncogene in non‐small cell lung cancer (NSCLC). 8 Accumulating evidence indicates that LINC01123 promotes tumor growth in various cancers. 6 , 9 However, the function of LINC01123 in OS remains unknown.
The Hedgehog (Hh) pathway is crucial in embryonic development, through its effects on cell growth and differentiation. 10 Accumulating evidence indicates that the Hh pathway is re‐activated and becomes a key factor determining cell proliferation, metastasis, and stemness in various malignant tumors. 11 , 12 The Hh pathway primarily relies on the Gli transcription factor family (Gli1, Gli2, and Gli3), whose members are downstream effectors. Moreover, Gli1 is the main transcription effector controlling gene expression when the Hh signal is activated. 13 Previous studies have reported that Gli‐mediated signaling pathways are activated in advanced OS. 14 However, the mechanism of the Hh pathway in OS remains unknown.
In this study, we found that LINC01123 is upregulated in OS cell lines and regulates OS proliferation and metastasis. Mechanistically, LINC01123 competes with Gli1 for miR‐516b‐5p and relieves the inhibitory effect of miR‐516b‐5p on Gli1, thereby increasing Gli1 expression. Therefore, the LINC01123/miR‐516b‐5p /Gli1 axis may provide theoretical support for identifying new therapeutic targets for OS treatment.
2. MATERIALS AND METHODS
2.1. Cell culture and reagents
Two human osteosarcoma cell lines (MG63 and Saos‐2) and human osteoblast hFOB1.19 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human osteosarcoma U‐2OS and 143B cell lines were obtained from the American type culture collection (ATCC, Manassas, VA). All cell lines were maintained per standard protocols. Briefly, MG63, Saos‐2, U‐2OS, and 143B cells were maintained in a 5% CO2 atmosphere at 37°C; hFOB1.19 cells were maintained in a 5% CO2 atmosphere at 34.5°C. Antibodies against OCT4 (ab181557; 1:1000 dilution), Nanog (ab109250; 1:1000 dilution), SOX2 (ab92494; 1:1000 dilution), ALDH1A1 (ab52492; 1:1000 dilution), β‐catenin (ab32572; 1:1000 dilution), RBPJ (ab25949; 1:1000 dilution), Gli1 (ab49314; 1:1000 dilution), Gli2 (ab26056; 1:1000 dilution), Gli3 (ab181130; 1:1000 dilution), and GAPDH (bsm‐33033 M; 1:1000 dilution) were used.
2.2. RNA extraction and Real‐time quantitative PCR (RT‐qPCR)
Total RNA from OS tissues and cells was extracted using TRIzol reagent (Invitrogen), followed by the synthesis of cDNA using a reverse transcription kit (Takara, Dalian, China). RT‐qPCR was carried out as reported previously. 15 The relative mRNA expression was calculated with the 2−△△Ct method. GAPDH served as internal controls. The primers are shown in Table S1.
2.3. Cell transfection
The miR‐516b‐5p mimic and control inhibitor, obtained from Gene‐Pharma (Shanghai, China), were transfected into OS cells with Lipofectamine 3000 (Invitrogen), in accordance with the manufacturer's protocol.
The plasmid for LINC01123 knockdown (sh‐LINC01123) and the empty plasmid (sh‐Ctrl) were purchased from Gene Pharma (Shanghai, China). The Gli1 overexpression plasmid and empty vector were also obtained from OBiO Technology (Shanghai, China). U‐2OS and 143B cells were plated on six‐well plates for transfection with Lipofectamine 2000 (Invitrogen). After 48 h, cells were collected.
2.4. Western blotting
Western blotting was carried out with methods as previously described. 15 Briefly, total protein was extracted with RIPA buffer (Sigma, USA). Protein samples were electrophoresed on 5‐10% SDS‐PAGE gels, transferred to a membrane, and incubated with antibodies against GAPDH (Abcam) at 4°C overnight. The membrane was then incubated for 1 h with secondary antibody and detected with ECL substrate (Share‐bio, Shanghai, China).
2.5. CCK‐8 assay and colony formation assay
CCK‐8 assay and colony formation assay were carried out with methods as reported previously. 16
2.6. Migration and invasion assays
Cell migration and invasion were assessed with Transwell assays with 24 well Transwell chambers (8 μm pore size; Corning, Corning, NY) with or without Matrigel matrix (BD Biosciences, CA, USA). In brief, 2 × 104 treated U‐2OS and 143B cells were cultured in the upper chamber in 200 μl serum‐free medium. Medium with 10% fetal bovine serum was added to the lower chamber. After culturing for 48 h, the cells that had migrated or invaded across the membrane were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Finally, cells were counted under an inverted microscope (Olympus).
2.7. Sphere formation assay
A total of 1 × 105 transfected U‐2OS and 143B cells were cultured on ultra‐low attachment surface six‐well culture plates (Corning, NY, USA). The cells were resuspended in 2 ml serum‐free DMEM/F12 containing 20 mg/L EGF (Peprotech, USA), 20 mg/L hFGF (Peprotech, USA), and 4 U/L insulin (Sigma, USA). The number of spheroids was counted under a microscope after culturing for 7 days.
2.8. Flow cytometry
For flow‐cytometry analysis, cells (1 × 105 cells/100 mL buffer, 5% FBS in PBS) were placed on ice for 30 min and then incubated with a pure anti‐CD133 antibody (#130‐105‐225, Miltenyi Biotec) for 45 min. Mouse isotype control immunoglobulin G antibody (#130‐104‐580, Miltenyi Biotec) was used as negative control for fluorescence‐activated cell sorting (FACS) gating. After being washed, the cells were incubated with an Alexa 488‐conjugated secondary antibody (Invitrogen) for 30 min and washed again before analysis using a BD FACScalibur flow cytometer (BD Biosciences). The fluorescence intensity was analyzed with Flow Jo.
2.9. EdU incorporation assay
The EdU incorporation assay was performed as previously mentioned. 17
2.10. Mouse xenograft assay
Animal experiments were performed in strict accordance with the guidelines of the Research Ethics Committee of East China Normal University. First, transfected cells were subcutaneously injected into male BALB/C nude mice (5‐6 weeks old). Tumor volume was measured weekly. The mice were euthanized after 4 weeks. All animal protocols were approved by the Affiliated Hospital of Nanjing Medical University Animal Protection and Use Committee.
2.11. TUNEL assay
Apoptosis of transfected U‐2OS cells in the xenograft tumors was assessed with the TUNEL method, as previously described. 17
2.12. IHC staining
The IHC assay was implemented as previously described. 17 Ki67 was detected with primary antibodies (GB13030; Servicebio, Wuhan, China) at 1:200 dilution.
2.13. Fluorescence in situ hybridization (FISH)
A microarray containing tissue from 40 OS patients was obtained from Alena Biotechnology Co., Ltd. (Xi'an, China). OS tissue sections were hybridized with the LINC01123 probes (Servicebio, Wuhan, China). Fluorescence in situ hybridization was performed as described previously. 17
2.14. Dual‐luciferase reporter assay
The predicted and mutant binding sites of miR‐516b‐5p in the Gli1 sequence or the predicted and mutant binding sites of miR‐516b‐5p in the LINC01123 sequence were synthesized and cloned into the pmirGLO Vector. Then, miR‐516b‐5p mimic or negative control with luciferase reporters were co‐transfected into U‐2OS and 143B cells with Lipofectamine 2000 (Invitrogen). After 48 h, the relative luciferase activity was evaluated with the Dual‐luciferase Reporter assay system (Promega).
2.15. RNA immunoprecipitation (RIP) assay
The relationships among LINC01123, miR‐516b‐5p, and Gli1 were investigated via RIP assays. The experimental procedure was performed according to the manufacturer's instructions.
2.16. RNA pull‐down assay
Briefly, LINC01123 biotin probe was purchased from Genepharma (Shanghai, China) and transfected into U‐2OS and 143B cells for 48 h. Then, the cell lysate was incubated with Dynabeads M‐280 Streptavidin (Sigma, MO, USA). Finally, the expression levels were detected by RT‐qPCR.
2.17. Database analysis
Data from TargetScan (http://www.targetscan.org/) and StarBase (http://starbase.sysu.edu.cn/ind ex.php) were used to analyze potential genes interacting with miR‐516b‐5p or LINC01123.
2.18. Statistical analyses
The monitoring data were analyzed in GraphPad Prism 8.0 and SPSS 26.0. All experimental data are presented as the mean ±standard deviation. All experiments were performed in triplicate. P values <0.05 were considered statistically significant.
3. RESULTS
3.1. The LINC01123 was upregulated in OS cell lines and promoted cell proliferation and metastasis
The expression of LINC01123 in human osteosarcoma cell lines was first detected by RT‐qPCR. As shown in Figure 1A, all OS cell lines (U‐2OS, 143B, MG63, and Saos‐2) showed significantly higher expression of LINC01123 than the control human osteoblast cell line (hFOB1.19), especially in the U‐2OS and 143B cells. Furthermore, we detected differences in the expression of LINC01123 in OS (n = 40) tissue samples through fluorescence in situ hybridization (FISH). We found that the higher expression of LINC01123 was related to the advanced pathological staging in OS (Figure S1A and B). To further examine the role of LINC01123 in OS progression, we silenced LINC01123 in two OS cell lines (U‐2OS and 143B). The knockdown efficiency was evaluated with qRT‐PCR (Figure 1B). The cell counting kit‐8 (CCK‐8) assay (Figure 1C and D), colony formation assay (Figure 1E) and EdU assay (Figure 1F and G) showed that knockdown of LINC01123 partly inhibited the proliferation of U‐2OS and 143B cells. The results of Transwell assays confirmed that LINC01123 silencing inhibited cell migration (Figure 1H and I) and invasion (Figure 1J and K) in U‐2OS and 143B cells. Collectively, these results revealed that the LINC01123 was upregulated in OS and promoted OS cell proliferation and migration.
FIGURE 1.
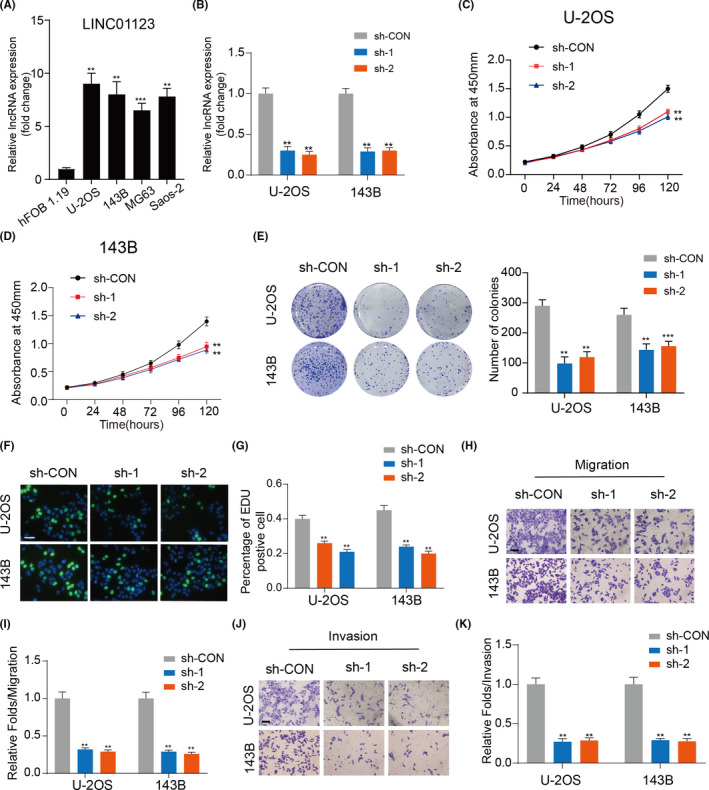
LINC01123 expression is significantly increased in OS cell lines and knockdown of LINC01123 inhibits OS cells growth. A, LINC01123 expression was detected in a normal osteoblast cell line (hFOB1.19) and OS cell lines (U‐2OS, 143B, MG63, and Saos‐2). Data were presented as the mean ±SD, **P <.01, ***P <.001. B, The knockdown efficiency of LINC01123 was detected in U2OS and 143B cells by qRT‐PCR. Data were presented as the mean ±SD, **P <.01. C and D, Knockdown of LINC01123 suppressed proliferation capability of U‐2OS and 143B cells using the CCK‐8 assay. Data were presented as the mean ±SD, **P <.01. E, Cell viability was measured in OS cells with LINC01123 knockdown or not using the colony formation assay. Data were presented as the mean ±SD, **P <.01. F and G, LINC01123 shRNA decreased the percentage of EdU‐positive OS cells, Scale bars =100 μm. Data were presented as the mean ±SD, **P <.01. H and I, Cell migration ability was detected in OS cells with LINC01123 knockdown or not. Scale bars =50 μm. Data were presented as the mean ±SD, **P <.01. J and K, Cell invasion ability was detected in OS cells with LINC01123 knockdown or not. Scale bars =50 μm. Data were presented as the mean ±SD, **P <.01
3.2. LINC01123 promoted OS cell growth by activating the Hh pathway
Cancer stem cells (CSCs) have critical roles in OS proliferation and metastasis. 18 , 19 As shown in Figure 2A, OS cells with knockdown of LINC01123 showed lower expression of key CSC marker genes (OCT4, Nanog and SOX2) than did control cells. As shown in Figure 2B and Figure S2, knockdown of LINC01123 reduced the ratio of CD133+ cells in the U‐2OS and 143B cells. The sphere formation ability of U‐2OS and 143B cells also was restrained by LINC01123 deficiency (Figure 2C and D). The Notch, Wnt, and Hh pathways are known to play significant roles in the differentiation of cancer stem‐like cells. 20 , 21 , 22 To explore whether LINC01123 might regulate one of these pathways, we detected the significant transcriptional regulators of these three pathways. As shown in Figure 2E, knockdown of LINC01123 inhibited Hh signaling, as reflected by the downregulated expression of Gli1. However, knockdown of LINC01123 did not inhibit Notch or Wnt signaling, as reflected by the lack of changes in the expression of their key effectors, RBPJ or β‐catenin. Gli1 is the main transcription effector of Hh pathway and control the downstream hedgehog target genes. Therefore, RT‐qPCR was used to detect the expression of Gli1 and main target genes of Hh pathway (PTCH1, HHIP and Snail). The results showed that knockdown of LINC01123 remarkably reduced the expression of Gli1 and these target genes (Figure 2F and G).
FIGURE 2.
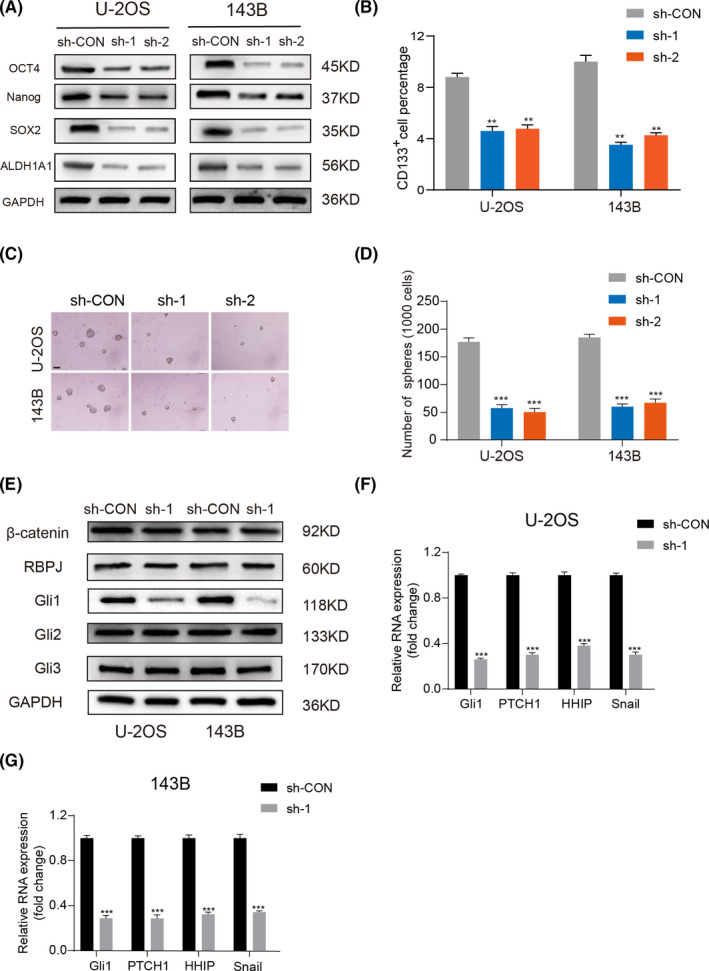
LINC01123 regulated cell stemness by activating the Hedgehog pathway. A, The protein level of CSC markers (OCT4, Nanog and SOX2) were examined in OS cells with LINC01123 knockdown or not. B, Fluorescence‐activated cell sorting (FACS) analysis of the CD133+ subpopulation of OS cells with LINC01123 knockdown or not. Data were presented as the mean ±SD, **P <.01. C and D, Self‐renewal ability was detected in OS cells with LINC01123 knockdown or not by sphere formation assays. Scale bars =50 μm. Data were presented as the mean ±SD, ***P <.001. E, LINC01123 activated the Hh signaling, but did not activate the Notch signaling pathway and the Wnt signaling pathway. F and G, The expression of target genes of Hh pathway were detected by qRT‐PCR. Data were presented as the mean ±SD, **P <.01, ***P <.001
Next, we determined whether LINC01123 activated the Hh pathway via Gli1 and modulated OS growth. First, we overexpressed Gli1 in LINC01123 knockdown OS cell lines (Figure 3A). As shown in Figure 3B‐E, overexpression of Gli1 partly reversed the inhibitory effect of LINC01123 knockdown on cell proliferation. Meanwhile, the inhibitory effect of LINC01123 knockdown on cell metastasis and stemness was reduced by overexpressing of Gli1 (Figure 3F‐K). Collectively, these data showed that LINC01123 activates the Hh pathway via Gli1, thus, regulating OS growth.
FIGURE 3.
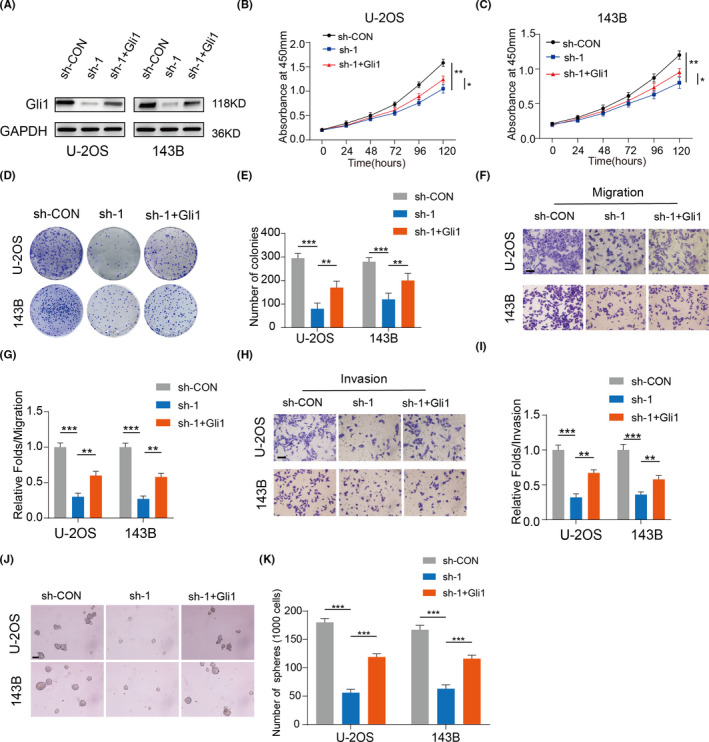
LINC01123 enhanced cell proliferation by modulating Gli1 expression. A, Overexpression efficacy of Gli1 in sh‐ LINC01123 OS cells (U‐2OS and 143B) was detected by western blotting. B and C, Overexpression of Gli1 partly reversed the suppressed effects of LINC01123‐knockdown on the cell viability of U‐2OS and 143B cells using the CCK‐8 assay. Data were presented as the mean ±SD, *P <.05, **P <.01. D and E, Overexpression of Gli1 partly reversed the suppressed effects of LINC01123‐knockdown on the cell viability of U‐2OS and 143B cells using the colony formation assay. Data were presented as the mean ±SD, **P <.01, ***P <.001. F‐I, Overexpression of Gli1 partly reversed the suppressed effects of LINC01123‐knockdown on the Cell migration (F and G) and invasion (H and I) ability of U‐2OS and 143B cells. Scale bars =50 μm. Data were presented as the mean ±SD, **P <.01, ***P <.001. J and K, Overexpression of Gli1 partly reversed the inhibitory effects of LINC01123‐knockdown on the self‐renewal ability of U‐2OS and 143B cells. Scale bars =50 μm. Data were presented as the mean ±SD, ***P <.001
3.3. MiR‐561b‐5p directly targeted Gli1 in OS cells
Various lncRNAs are increasingly being shown to sponge microRNAs, thus, regulating target gene expression. 23 Therefore, we identified 75 potential microRNAs that target Gli1, on the basis of the prediction website TargetScan, and 19 potential microRNAs regulated by LINC01123, on the basis of the prediction website StarBase. As shown in Figure 4A, we found the miR‐561b‐5p after screening. Next, we found that overexpression of miR‐561b‐5p inhibited the mRNA and protein expression of Gli1 in U‐2OS and 143B cells (Figure 4B‐C). The potential target site of miR‐561b‐5p on Gli1 is shown in Figure 4D. We then used dual luciferase reporter assays to validate the binding between miR‐561b‐5p and Gli1. The miR‐561b‐5p mimic markedly decreased the luciferase activity of the WT Gli1 3′ UTR (Gli1‐WT) reporter but not the reporter vector containing Gli1 with mutated (MUT) miR‐561b‐5p‐binding sites in U‐2OS and 143B cells (Figure 4E‐F). Moreover, RIP assays showed that overexpression of miR‐561b‐5p increased the enrichment of Gli1 in the Argonaute 2 (Ago2) group (Figure 4G‐H). Together, these results demonstrated that miR‐561b‐5p inhibits Gli1 expression by directly targeting its 3ʹ UTR in OS cells.
FIGURE 4.
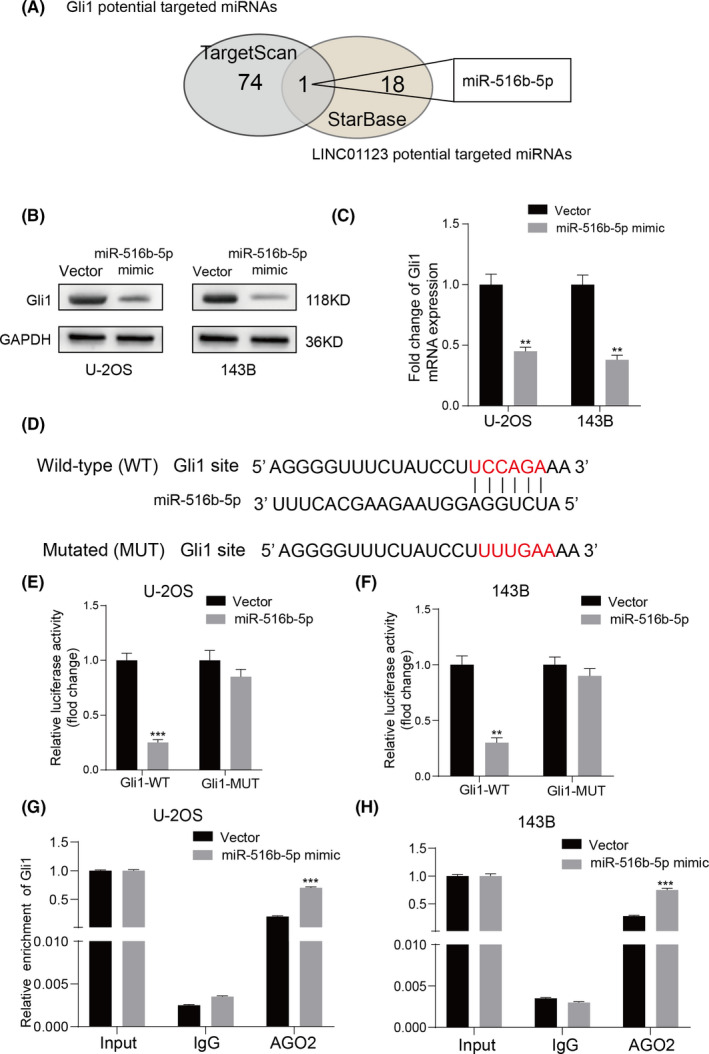
Gli1 was a direct target of miR‐561b‐5p. A, Venn diagram showing the predicted target genes of Gli1 and LINC01123 from databases (TargetScan and StarBase). B and C, Gli1 expression was detected in the U‐2OS and 143B cells transfected with miR‐561b‐5p mimics by qRT‐PCR and western blotting. D, The wild‐type and the mutated sequences of the Gli1 mRNA 3’‐UTR (mutation site: red). E and F, The relative luciferase activity of luciferase reporters containing Gli1 wild‐type (WT) or mutated (MUT) co‐transfected with miR‐561b‐5p or its negative control in U‐2OS and 143B cells was assessed. Data were presented as the mean ±SD, **P <.01, ***P <.001. G and H, RIP assays using antibodies against AGO2 or IgG were performed in cellular lysates from U‐2OS and 143B cells. The relative enrichment of Gli1 was measured in cells transfected with miR‐561b‐5p or NC mimics by qRT‐PCR, Data were presented as the mean ±SD, **P <.01, ***P <.001
3.4. LINC01123 acted as a competing endogenous RNA by sponging miR‐561b‐5p
To further investigate whether LINC01123 functioned as a competing endogenous RNA sponge for miR‐561b‐5p, we measured the expression of miR‐561b‐5p in U‐2OS and 143B cells transfected with sh‐LINC01123. Knockdown of LINC01123 markedly increased the expression of miR‐561b‐5p in OS cells (Figure 5A). The potential target site of LINC01123 on miR‐561b‐5p was shown in Figure 5B. Furthermore, MS2‐based RIP assays showed that miR‐561b‐5p was significantly more enriched in RNAs retrieved from the WT MS2bs‐LINC01123 group than the MUT MS2bs‐LINC01123 group (Figure 5C). As shown in Figure 5D, RNA pull‐down assays revealed that miR‐561b‐5p bound biotin‐labeled WT LINC01123, but not MUT LINC01123. Next, we demonstrated that miR‐561b‐5p mimics significantly decreased the luciferase activity of WT LINC01123 but not MUT LINC01123 in U‐2OS and 143B cells (Figure 5E and F). miRNAs have been found to bind their targets in an Ago2‐dependent manner. 24 To confirm whether LINC01123 binds miR‐561b‐5p in a similar manner, we performed anti‐Ago2 RIP in U‐2OS and 143B cells transiently overexpressing miR‐561b‐5p. Endogenous LINC01123 enrichment was greater in the miR‐561b‐5p mimic group than the vector group, as measured by RT‐qPCR (Figure 5G and H). These data indicated that LINC01123 acts as a ceRNA by sponging miR‐561b‐5p.
FIGURE 5.
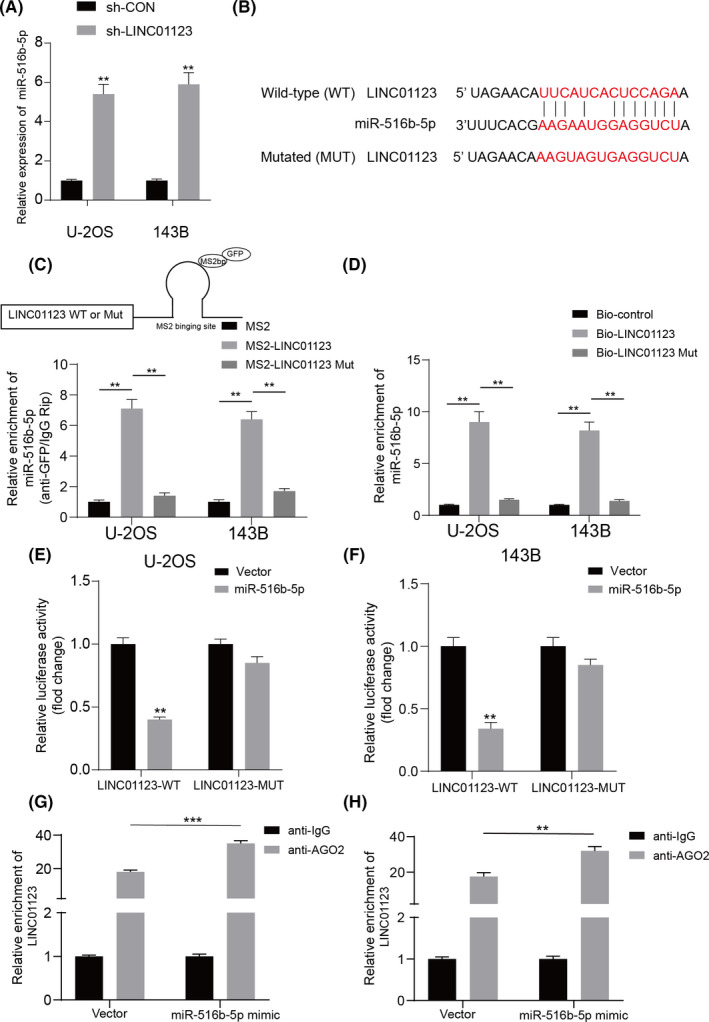
LINC01123 bound to miR‐561b‐5p and decreased its expression in the OS cells. A, miR‐561b‐5p expression was detected in OS cells with LINC01123 knockdown or not. B, The wild‐type and the mutated sequences of the LINC01123 mRNA 3’‐UTR (mutation site: red). C, Top panel shows a schematic image of a construction containing LINC01123 wild type combined with MS2 binding sequence. MS2‐RIP followed by miR‐561b‐5p qRT‐PCR to measure miR‐561b‐5p endogenously associated with LINC01123. Data were presented as the mean ±SD, **P <.01. D, U‐2OS and 143B cells lysate were incubated with biotin‐labeled LINC01123, qRT‐PCR measured miR‐561b‐5p expression in the products of pulldown by biotin, Data were presented as the mean ±SD, **P <.01. E and F, The relative luciferase activity of luciferase reporters containing LINC01123 wild‐type (WT) or mutated (MUT) co‐transfected with miR‐561b‐5p or its negative control in U‐2OS and 143B cells was assessed. Data were presented as the mean ±SD, **P <.01. G and H, AGO2‐RIP followed by qRT‐PCR to evaluate LINC01123 level after miR‐561b‐5p overexpression. **P <.01, ***P <.001
3.5. MiR‐561b‐5p inhibition reversed the inhibitory effect in cell growth caused by LINC01123 knockdown in OS cells
To explore whether LINC01123 might affect OS progression through miR‐561b‐5p, we performed a rescue experiment by silencing LINC01123 and transfecting miR‐561b‐5p inhibitors into U‐2OS and 143B cells. As shown in Figure 6A, the inhibition of miR‐561b‐5p partially rescued the decrease in Gli1 expression inducing by downregulation of LINC01123 in U‐2OS and 143B cells. Moreover, silencing miR‐561b‐5p alleviated the inhibitory effect of LINC01123 knockdown on the proliferation and metastasis of OS cells (Figure 6B‐F and Figure S3A‐C). Similarly, miR‐561b‐5p inhibition also partly restored stemness in U‐2OS and 143B cells (Figure 6G and Figure S3D). These results demonstrated that LINC01123 promotes OS cell growth via acting as a ceRNA that sponges miR‐561b‐5p.
FIGURE 6.
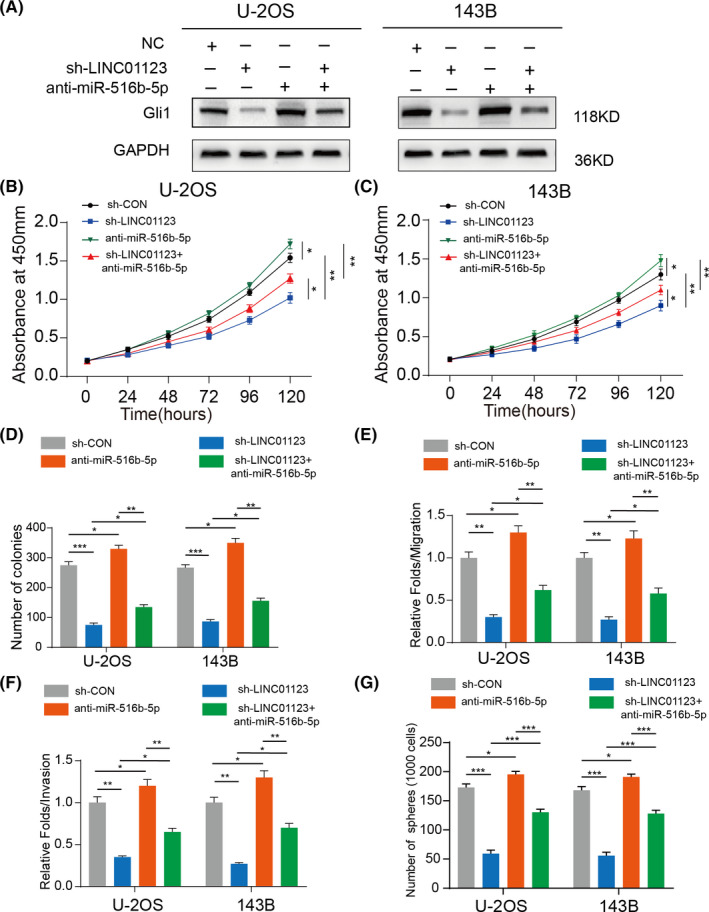
MiR‐561b‐5p inhibition partly rescued the LINC01123 knockdown effect in OS cells. A, Western blot showed the Gli1 expression in U‐2OS and 143B cells transfected with miR‐561b‐5p inhibitor, sh‐LINC01123, or negative control. B and C, miR‐561b‐5p‐knockdown partly reversed the inhibitory effects of LINC01123‐knockdown on the cell viability of U‐2OS and 143B cells using the CCK‐8 assay and silenced miR‐561b‐5p in wide type OS cells (U‐2OS and 143B) also promoted their proliferation. Data were presented as the mean ±SD, *P <.05, **P <.01. D, miR‐561b‐5p‐knockdown partly reversed the inhibitory effects of LINC01123‐knockdown on the colony formation properties of U‐2OS and 143B cells and silenced miR‐561b‐5p in wide type OS cells (U‐2OS and 143B) also promoted their proliferation. Data were presented as the mean ±SD, *P <.05, **P <.01, ***P <.001. E, miR‐561b‐5p‐knockdown partly reversed the suppressed effects of LINC01123‐knockdown on the cell migration ability of U‐2OS and 143B cells. Silenced miR‐561b‐5p in wide type OS cells (U‐2OS and 143B) also promoted their cell migration ability. Data were presented as the mean ±SD, *P <.05, **P <.01. F, miR‐561b‐5p‐knockdown partly reversed the suppressed effects of LINC01123‐knockdown on the cell invasion ability of U‐2OS and 143B cells. Silenced miR‐561b‐5p in wide type OS cells (U‐2OS and 143B) also promoted their cell invasion ability. Data were presented as the mean ±SD, *P <.05, **P <.01, ***P <.001. G, miR‐561b‐5p‐knockdown partly reversed the suppressed effects of LINC01123‐knockdown on the cell stemness ability of U‐2OS and 143B cells. Silenced miR‐561b‐5p in wide type OS cells (U‐2OS and 143B) also promoted their cell stemness ability. Data were presented as the mean ±SD, *P <.05, **P <.01, ***P <.001
3.6. LINC01123 promoted OS cell growth by sponging miR‐561b‐5p in vivo
To investigate the role of LINC01123 in tumor growth in vivo, we established a xenograft model by stably silencing LINC01123 and transfecting miR‐561b‐5p inhibitors into U‐2OS cells. As shown in Figure 7A‐D, the stable LINC01123‐ knockdown group exhibited significantly decreased the xenografted tumor growth and a decreased tumor burden compared to the control group. Moreover, the compromised tumorigenic potential in the LINC01123‐knockdown group was partly offset by silencing of miR‐561b‐5p. TUNEL and immunohistochemical staining assays showed that revealed declining expression of Ki67 and a rising rate of apoptosis in the xenografted tumors of LINC01123‐knockdown group (Figure 7E). In addition, the knockdown efficiency of LINC01123 was evaluated with qRT‐PCR (Figure 7F). As shown in Figure 7G and H, the inhibition of miR‐561b‐5p partially rescued the decrease in Gli1 expression inducing by downregulation of LINC01123 in the xenografted tumors. These data further indicate that LINC01123 promoted OS cell growth by sponging miR‐561b‐5p in vivo.
FIGURE 7.
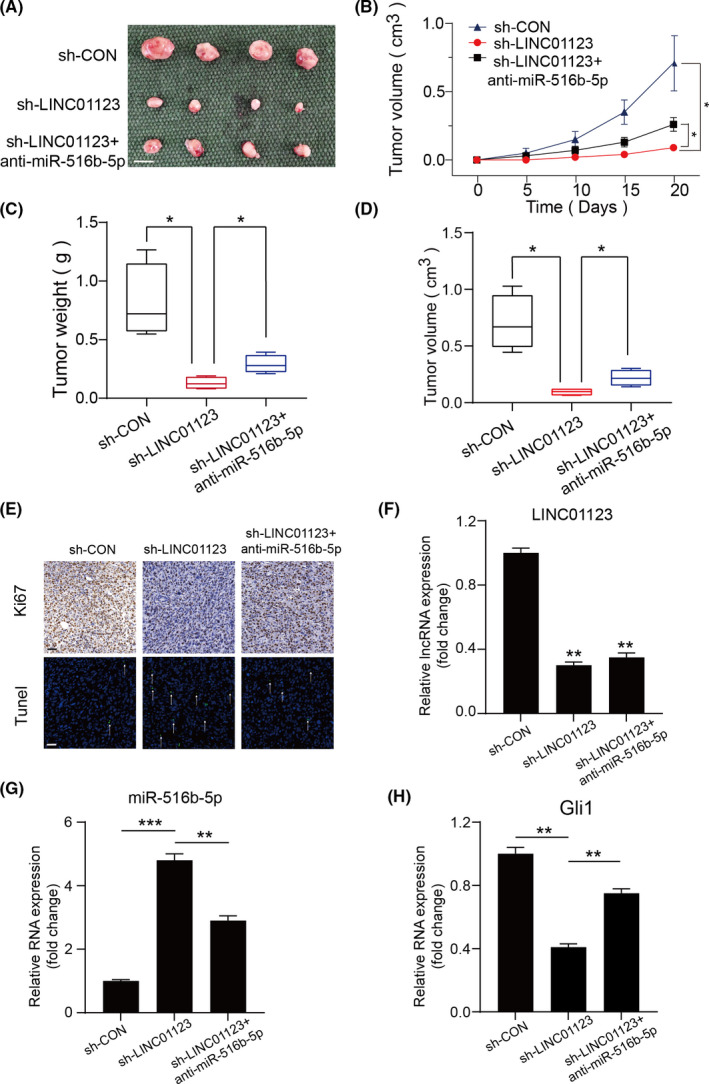
LINC01123 promoted cell growth by sponging miR‐561b‐5p in vivo. A, Morphologic characteristics of xenograft tumors from U‐2OS/sh‐Control group, U‐2OS/sh‐LINC01123 group and U‐2OS/sh‐LINC01123 + anti‐miR‐561b‐5p group (n = 4). Scale bars =1 cm. B, The tumor volumes were measured with calipers every 5 days. Data were presented as the mean ±SD, *P <.05. C, Tumor weights at 20 days were measured in each group. The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range of values that were outside of the intra‐quartile range. n = 4, *P <.05. D, Tumor volumes at 20 days were measured in each group. The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra‐quartile range. n = 4, *P <.05. E, Representative images of Ki67 and TUNEL staining in the xenograft tumors from the sh‐Control, sh‐ LINC01123 and sh‐LINC01123 + anti‐miR‐561b‐5p mice. A TUNEL positive cell is indicated (arrow). Scale bars =50μm. F‐H, The expression of LINC01123, miR‐561b‐5p and Gli1 in xenografts was examined by RT‐qPCR. Data were presented as the mean ±SD, **P <.01, ***P <.001
4. DISCUSSION
The lncRNA LINC01123 is involved in cell proliferation and invasion in various cancers. In NSCLC, LINC01123 is highly upregulated and serves as a ceRNA that promotes NSCLC proliferation and aerobic glycolysis through the miR‐199a‐5p/c‐Myc axis. 8 Previous work has demonstrated that knockdown of LINC01123 inhibits cell growth and invasion in colorectal cancer via the miR‐625‐5p/LASP1 axis. 25 In triple negative breast cancer, LINC01123, activated by FOXC1, regulates cell growth through miR‐663a/CMIP signaling. 6 In this study, we observed that the expression of LINC01123 was clearly upregulated in OS cell lines. Moreover, LINC01123 promoted cell proliferation, metastasis, and stemness in vitro. Our findings revealed that LINC01123 acts as an oncogene with a critical role in the OS cell progression.
Meanwhile, increasing reports indicate the existence of a comprehensive network of interactions involving ceRNAs in the biological function of non‐coding RNAs, in which lncRNA affect gene expression through competitively sponging miRNAs. 26 , 27 For instance, lncRNA CDC6 promotes breast cancer growth by acting as a ceRNA for miR‐215 7 ; lncRNA RMRP enhances bladder cancer progression via sponging miR‐206 8 ; and the lncRNA CDKN2BAS promotes metastasis in hepatocellular carcinoma by acting as a ceRNA for miR‐153‐5p. 28 In this study, we first confirmed that LINC01123 is primarily localized in the cytoplasm, thus suggesting that LINC01123 my serve as an endogenous miRNA sponge. Then, bioinformatics analysis and luciferase reporter analysis revealed that miR‐561b‐5p might be a target for LINC01123. The expression of miR‐561b‐5p was low in OS cell lines, and decreased expression of miR‐561b‐5p inhibited OS cell growth.
Gli1 is a critical transcription factor in the Hh pathway. In various cancers, Gli1 is overexpressed and activated, and it subsequently regulates many cellular processes, including proliferation, metastasis, and stemness. Liao et al have reported that Gli1 is upregulated in tissues and promotes metastasis in glioma. 29 In addition, Gli1 has been found to promote cancer stemness through the PI3K/Akt/NFκB pathway in colorectal adenocarcinoma. 7 Furthermore, Gli1 is upregulated and controls gene expression in OS. 30 Yi et al have demonstrated that miR‐212 promotes the growth of OS cells via increasing the expression of Gli1. 31 Zhao et al have confirmed that degalactotigonin, extracted from Solanum nigrum L, inhibits the proliferation and metastasis of OS by regulating the Hh/Gli1 pathway. 15 In this study, we identified a new mechanism of Gli1 regulation in OS. Our results revealed that overexpression of Gli1 partly reversed the inhibitory effect of LINC01123 knockdown on OS cell progression. Simultaneously, LINC01123 activated the Hh pathway in OS by sponging miR‐561b‐5p, which directly targets the 3ʹUTR of Gli1.
In conclusion, our data showed that LINC01123 is an oncogene that promotes the progression of OS cells via competitively binding miR‐561b‐5p, thereby enhancing Gli1 expression. Our results suggest that LINC01123 may be an important possible target for OS therapy, thus potentially providing a new therapy direction for OS treatment.
CONFLICTS OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Table S1
Pan X, Tan J, Tao T, et al. LINC01123 enhances osteosarcoma cell growth by activating the Hedgehog pathway via the miR‐516b‐5p/Gli1 axis. Cancer Sci. 2021;112:2260–2271. 10.1111/cas.14913
Xiaohui Pan, Jingxue Tan, Tao Tao and Xiuwen Zhang are contributed equally to this work and shared the first authorship.
Contributor Information
Dong Zhou, Email: zhoudong1012@163.com.
Yifei Shen, Email: tjshenyifei@tongji.edu.cn.
REFERENCE
- 1. Ritter J, Bielack Ss. Osteosarcoma. Ann Oncol. 2010;7:vii320–vii325. [DOI] [PubMed] [Google Scholar]
- 2. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029‐3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16:543‐556. [DOI] [PubMed] [Google Scholar]
- 4. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47‐62. [DOI] [PubMed] [Google Scholar]
- 5. Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding Q, Mo F, Cai X, et al. LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial‐mesenchymal transition via Wnt/beta‐catenin signaling pathway. J Cell Biochem. 2020;121:3358‐3371. [DOI] [PubMed] [Google Scholar]
- 7. Kong X, Duan Y, Sang Y, et al. LncRNA‐CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA‐215. J Cell Physiol. 2019;234:9105‐9117. [DOI] [PubMed] [Google Scholar]
- 8. Cao HL, Liu ZJ, Huang PL, Yue YL, Xi JN. lncRNA‐RMRP promotes proliferation, migration and invasion of bladder cancer via miR‐206. Eur Rev Med Pharmacol Sci. 2019;23:1012‐1021. [DOI] [PubMed] [Google Scholar]
- 9. Pan X, Li H, Tan J, et al. miR‐1297 suppresses osteosarcoma proliferation and aerobic glycolysis by regulating PFKFB2. Onco Targets Ther. 2020;13:11265‐11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367‐372. [DOI] [PubMed] [Google Scholar]
- 11. Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416‐429. [DOI] [PubMed] [Google Scholar]
- 12. Kumar RMR, Fuchs B. Hedgehog signaling inhibitors as anti‐cancer agents in osteosarcoma. Cancers. 2015;7:784‐794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bangs F, Anderson KV. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9:a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohseny AB, Cai Y, Kuijjer M, et al. The activities of Smad and Gli mediated signalling pathways in high‐grade conventional osteosarcoma. Eur J Cancer. 2012;48:3429‐3438. [DOI] [PubMed] [Google Scholar]
- 15. Qu Y, Zheng S, Kang M, et al. Knockdown of long non‐coding RNA HOXD‐AS1 inhibits the progression of osteosarcoma. Biomed Pharmacother. 2018;98:899‐906. [DOI] [PubMed] [Google Scholar]
- 16. Weng Y, Shen Y, He Y, et al. The miR‐15b‐5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect. Biochem Biophys Res Commun. 2018;503:2749‐2757. [DOI] [PubMed] [Google Scholar]
- 17. Shen Y, Xu J, Pan X, et al. LncRNA KCNQ1OT1 sponges miR‐34c‐5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai X, Liu Y, Yang W, et al. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2016;34:932‐941. [DOI] [PubMed] [Google Scholar]
- 19. Schiavone K, Garnier D, Heymann M‐F, Heymann D. The heterogeneity of osteosarcoma: the role played by cancer stem cells. Stem Cells Heterogeneity in Cancer: Springer. 2019;187‐200. [DOI] [PubMed] [Google Scholar]
- 20. Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 2019;24:25‐40. [DOI] [PubMed] [Google Scholar]
- 21. Teeuwssen M, Fodde R. Wnt signaling in ovarian cancer stemness, EMT, and therapy resistance. J Clin Med. 2019;8:1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99:494‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bose M, Barman B, Goswami A. Spatiotemporal uncoupling of microRNA‐mediated translational repression and target RNA degradation controls microRNP recycling in mammalian cells. Mol Cell Biol. 2017;37:e00464‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shang T, Zhou X, Chen W. LINC01123 promotes progression of colorectal cancer via miR‐625‐5p/LASP1 axis. Cancer Biother Radiopharm. 2020. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 26. He X, Gao Z, Xu H, Zhang Z, Fu P. A meta‐analysis of randomized control trials of surgical methods with osteosarcoma outcomes. J Orthop Surg Res. 2017;12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965‐3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR‐153‐5p/ARHGAP18 signaling axis. Aging. 2018;10:3371‐3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao ZQ, Ye M, Yu PG, Xiao C, Lin FY. Glioma‐Associated Oncogene Homolog1 (Gli1)‐Aquaporin1 pathway promotes glioma cell metastasis. BMB Rep. 2016;49:394‐399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shahi MH, Holt R, Rebhun RB. Blocking signaling at the level of GLI regulates downstream gene expression and inhibits proliferation of canine osteosarcoma cells. PLoS One. 2014;9:e96593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhosle VK, Mukherjee T, Huang Y‐W, et al. SLIT2/ROBO1‐signaling inhibits macropinocytosis by opposing cortical cytoskeletal remodeling. Nat Commun. 2020;11:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Table S1


