Abstract
Osimertinib is a third‐generation epidermal growth factor receptor tyrosine kinase inhibitor (EGFR‐TKI) that is effective in treating both naïve and T790M‐mutated EGFR‐TKI‐resistant non–small cell lung cancer patients. The EGFR C797S mutation is the major osimertinib resistance mechanism. The present study monitored the EGFR C797S mutation during osimertinib treatment in Japanese patients using droplet digital PCR (ddPCR). In our first cohort, C797S detection was validated with tumor specimens and/or plasma samples from 26 patients using ddPCR with custom‐designed probes detecting and discriminating T790M and C797S in cis and trans positions. In our second cohort, 18 patients with EGFR‐T790M who were going to start osimertinib were analyzed using ddPCR by collecting the plasma samples every month from the beginning of the course of osimertinib. In the first cohort, C797S was detected in 15.4% of patients. C797S and T790M in cis and trans positions were distinguished using ddPCR. In the second cohort, serial cfDNA evaluation revealed that the rate of EGFR mutation changes with disease state. Increases of EGFR mutation were detected, including C797S several months before the diagnosis of disease progression. As with the first cohort, C797S and T790M in cis and trans position were distinguished by ddPCR at disease progression. Coincidentally, in the first cohort, next generation sequencing detected NRAS Q61K mutation and the resistance with NRAS Q61K mutation was overcome by trametinib. In the second cohort, serial cfDNA analysis was useful for evaluating bone oligo‐progression and local radiation therapy.
Keywords: C797S, droplet digital PCR, EGFR, osimertinib, resistance, T790M
In this study, we monitored osimertinib resistance mechanisms mainly using cfDNA in two cohorts: (i) tumor specimens and/or plasma samples from 26 patients with progressive disease during the osimertinib treatment after T790M confirmation were examined; and (ii) 18 patients with non–small cell lung cancer who were positive for EGFR‐T790M and who started osimertinib were analyzed by collecting plasma samples every month from the beginning of osimertinib treatment to disease progression or to 2 years after the osimertinib treatment. We found that C797S and T790M in cis and in trans positions can be detected by ddPCR after the disease progression and confirmed the usefulness of serial evaluation of EGFR mutation in the plasma cfDNA for detecting the emergence of tumor resistance 1‐2 months earlier than the diagnosis of disease progression and identifying the response to the radiation therapy for oligo‐progression.

1. INTRODUCTION
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR‐TKI) inhibit the EGFR and its downstream signaling pathways by binding to the adenosine triphosphate (ATP)‐binding pocket of the EGFR‐tyrosine kinase domain, which strongly suppresses the growth of EGFR‐mutant non–small cell lung cancer (NSCLC) cells. 1 EGFR‐TKI have been shown to prolong the progression‐free survival (PFS) of patients with EGFR‐mutant NSCLC. 2 , 3 However, cancer cells acquire resistance to EGFR‐TKI. One of the resistance mechanisms is via the secondary point mutation Thr790Met (T790M), which decreases the relative binding affinity of TKI to the ATP‐binding site of EGFR. 5 , 6
Osimertinib, a third‐generation EGFR‐TKI, was developed to overcome this resistance by covalently binding to T790M‐mutated EGFR. 7 Its efficacy in T790M‐positive patients with advanced NSCLC has been evaluated in a phase 3 clinical trial. The results showed that the median PFS was significantly longer with osimertinib than that with platinum‐based chemotherapy. 8 However, a wide variety of osimertinib‐resistant mechanisms have been reported (eg, C797S, G796D, L718Q, MET amplification, BRAF V600E mutation, and small‐cell lung cancer [SCLC] transformation). 9 , 10 , 11 , 12 , 13 , 14 C797S is one of the major resistance mechanisms that impair the covalent binding between EGFR and osimertinib. Preclinical model studies suggested some strategies to overcome resistance due to C797S. 15 Therefore, detection of C797S and understanding osimertinib resistance mechanisms are important.
Cell‐free DNA (cfDNA) from plasma samples contains the DNA of malignant tumors. Thus, it is possible to detect EGFR mutations, not only activating mutations but also resistance mutations like T790M from plasma cfDNA. A clinical trial confirmed the efficacy of osimertinib treatment based on EGFR mutation status in plasma cfDNA 16 . Moreover, the amount of cfDNA with EGFR mutation has been reported to change with treatment response and disease progression. 17 , 18 , 19 For example, after EGFR‐TKI treatment, the EGFR mutation becomes negative in plasma cfDNA, whereas after the disease progression, the EGFR mutation becomes positive again. However, EGFR mutation in cfDNA has not been fully evaluated during osimertinib treatment. For example, the mechanism of each EGFR mutation status change (activating mutation, T790M, and C797S) under osimertinib treatment beyond progression or after local radiation therapy remains unknown.
Therefore, in our first cohort we validated the C797S detection from plasma cfDNA using digital droplet PCR (ddPCR) by evaluating the concordance between tissue and plasma clinical samples. In addition to a commercially available independent ddPCR probe for C797S and T790M, a novel original probe set that could detect and distinguish C797S and T790M mutations in cis and trans positions was used. In the second cohort, we checked the EGFR mutation profile (exon 19 deletion, L858R, T790M, and C797S) in the plasma cfDNA during and after the osimertinib treatment in the serial analysis. Serial EGFR mutation examination with cfDNA in the plasma enables early detection of resistance mutation emergence, including EGFR C797S, some months earlier than the radiological progression. Thus, serial monitoring of the EGFR mutation status, including C797S, would be useful in developing an appropriate treatment strategy involving the administration of osimertinib.
2. MATERIALS AND METHODS
2.1. Patients and samples
The first cohort included 60 Japanese patients with NSCLC with EGFR T790M mutation who started osimertinib treatment after the first or second generation EGFR‐TKI treatment in the Department of Thoracic Medical Oncology at the Cancer Institute Hospital of the Japanese Foundation for Cancer Research (JFCR) from June 2014 to January 2018. Among these patients, 50.0% had EGFR exon 19 deletion, 48.3% had EGFR L858R, and 1.7% had EGFR L861Q. A total of 26 patients agreed to undergo a tumor specimen assay and/or plasma sample analysis after the disease progression.
In the second cohort, 18 Japanese patients with NSCLC with EGFR T790M mutation started osimertinib treatment after the first‐generation or second‐generation EGFR‐TKI treatment at the Cancer Institute Hospital of the JFCR from December 2017 to November 2018. Among them, 83.3% had EGFR exon 19 deletion, and 16.7% had EGFR L858R. Plasma samples were collected almost every month from the start to 6 months after the osimertinib treatment.
The systemic response to osimertinib was evaluated using RECIST v1.1. All patients underwent computed tomography every 2‐3 months, with a few exceptions. All patients provided written informed consent, and this study was approved by the Institutional Review Board of the Cancer Institute Hospital in the JFCR.
2.2. Nucleic acid extraction
DNA was extracted from cytology or histology samples using the cobas EGFR Mutation Test kit (Roche Molecular Systems), the RNeasy Mini Kit (Qiagen), or the DNeasy Blood & Tissue Kit (Qiagen) as per the manufacturer’s instructions. cfDNA was extracted from plasma samples using the Maxwell RSC ccfDNA Plasma Kit (Promega) or the QIAamp MinElute ccfDNA Kit (Qiagen) following the manufacturer’s instructions. DNA concentration was measured using the Qubit dsDNA HS Assay Kit (Invitrogen, Life Technologies) on a Qubit 2.0 Fluorometer (Invitrogen, Life Technologies).
2.3. Digital droplet PCR for detecting C797S
The method used in ddPCR for detecting EGFR exon 19 deletion, L858R, and T790M is described in our previously published report. 20 In brief, a probe was obtained from Riken Genesis to detect C797S. Cycling conditions for PCR were determined according to the manual. Assays were considered positive if ≥10 copies were observed per reaction (20 µL) based on false‐positive droplet counts from human reference genomic DNA (average C797S copies, 2.36). For the detection of cis/trans‐C797S with T790M, novel probes were designed. Forward primer: CATCTGCCTCACCTCCAC, reverse primer: CGTATCTCCCTTCCCTGATTAC. T790M mutation probe: FAM/IBFQ, CA+TC+A+T+GC+A+GC. C797S mutation probe‐1 (c.2390): FAM/IBFQ, CG+GC+T+C+CCTC. C797S mutation probe‐2 (c.2389): FAM/IBFQ, TCGG+C+A+GCCT+C. WT probe: HEX/IBFQ, TCA+TC+A+C+GC+A+GC.
2.4. Deep sequencing
A targeted amplicon sequencing library was prepared by amplifying exon 20 of EGFR by PCR. Paired‐end sequencing (2 × 150 bp) was performed using the MiSeq platform. To test whether the two mutations, T790M and C797S, were located in the same allele (cis) or different alleles (trans), four reference FASTA files with or without these mutations were prepared. The obtained sequence reads were then mapped to these four reference sequences using BLAT and the reads with lowest mismatches were counted.
For the target sequencing analysis, the library was prepared using a Haloplex custom panel (Agilent), which was designed to detect well‐known cancer‐associated somatic mutations. 21 Paired‐end sequencing (2 × 150 bp) was performed on the MiSeq platform. Sequence reads were aligned to the UCSC hg19 reference genome using the Burrows‐Wheeler Aligner (BWA) (version 0.7.10). Read pairs with a mapping quality of <30 and with mismatches of more than 5% of the read length were excluded. Somatic variants were called by in‐house pipeline.
2.5. Establishment of osimertinib‐resistant PC9 cells with NRAS Q61K
PC9 cells, harboring the EGFR del19 mutation, were treated with increasing concentrations of osimertinib for 6‐12 months and several osimertinib‐resistant PC9 cell lines were established. From the in‐house NGS analysis detecting cancer‐related 108 genes, 21 NRAS Q61K mutation was found in osimertinib‐resistant PC9 cells.
2.6. Cell viability assays
We carried out 3‐day cell viability assays by plating 2000 cells per well into black transparent‐bottom 96‐well plates. On the following day, the cells were treated with the indicated TKI across a 10‐dose range from 0.3 nmol/L to 10 μmol/L. After 72 hours of drug treatment, cell viability was measured using the CellTiter‐Glo assay (Promega).
2.7. Statistical analysis
Fisher’s exact and χ2 tests were used for categorical comparison of data, based on group number, and the Mann‐Whitney U‐test was used to compare differences in continuous data. Paired t tests were used to compare differences in continuous and paired data. All P‐values were based on a two‐sided hypothesis. We performed all statistical analyses using the JMP statistical software package 13 (SAS Institute).
3. RESULTS
3.1. Concordance of C797S, T790M mutation detection between tumor cell DNA and plasma cfDNA
Osimertinib‐resistant tumor cell DNA and/or plasma cfDNA of 26 patients was analyzed in the first cohort. Patient characteristics, a flow chart representing the availability of each examination, a summary of treatment history, and the resistance mechanism of 26 patients are shown in Table S1, Figures S1‐S3. C797S was detected by ddPCR in 15.4% (4/26) of patients, and T790M loss was detected in 65.4% (17/26) (Figure 1A). The concordance rate between tumor cell DNA and plasma cell‐free DNA was 88.9% (8/9) for detecting C797S and 100% (9/9) for detecting T790M (Figure 1B).
FIGURE 1.
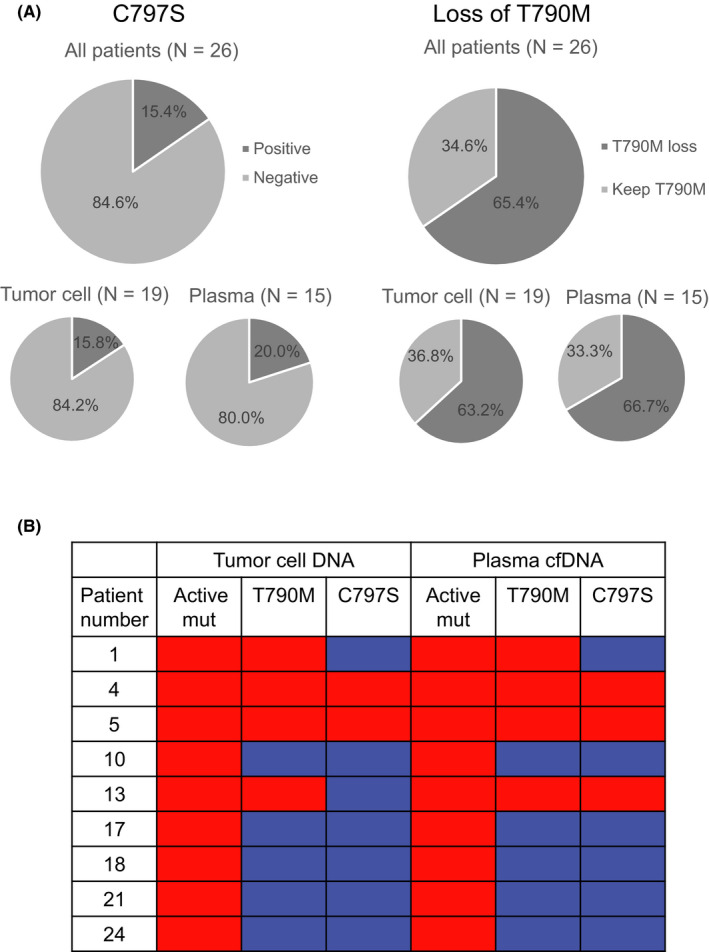
A, Frequency of C797S and T790M loss detected by droplet digital PCR (ddPCR). B, The concordance of T790M and C797S detection with ddPCR between tumor cell DNA and plasma cfDNA. Red: positive. Blue: negative
3.2. Validation to detect and discriminate EGFR‐T790M/C797S in cis and trans positions with droplet digital PCR
We validated the detection of C797S by ddPCR. JFCR163 is a cell line from the specimen of osimertinib‐resistant patient No. 20. The EGFR of this cell line was sequenced, and C797S was detected (Figure 2A). Using this sample, ddPCR was performed and showed “a positive” result in the cis position (Figure 2B). Next, we assayed the sample from patient No. 4 (JFCR‐129) using a custom probe that can distinguish between C797S in the cis and trans positions with T790M (Figure 2C). The results showed that C797S was in the trans position, which was validated by NGS (Figures 2D, 3E and Figure S4).
FIGURE 2.
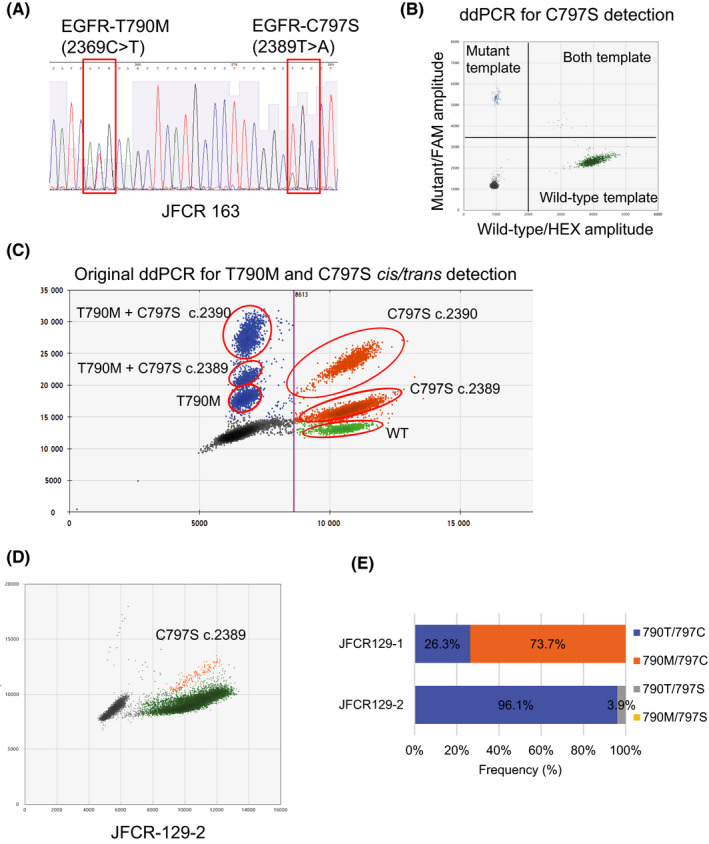
Detection of epidermal growth factor receptor (EGFR) T790M and C797S by droplet digital PCR (ddPCR). Detection of T790M and C797S in the trans position was possible using ddPCR. A, Detection of EGFR T790M and C797S with Sanger sequencing from JFCR163. B, Detection of EGFR C797S mutation with ddPCR from JFCR 163. Mutant positive template is depicted with blue dots. C, Example of our original ddPCR probe to distinguish cis or trans EGFR‐T790M and C797S mutations. T790M mutation was detected as FAM positive with low amplitude and depicted with blue dots. C797S and T790M in cis position was detected as FAM positive with high amplitude and depicted with blue dots. C797S in trans position was detected as both FAM and HEX positive and depicted with yellow dots. D, Sample from patient No. 4 (JFCR‐129‐2) showed yellow dots positive with ddPCR, which indicated the detection of T790M and C797S in the trans position. E, T790M and C797S in the trans position was also confirmed by NGS using the PCR amplified exon 20 of EGFR, the results of JFCR‐129‐2. (JFCR‐129‐1 as a comparison)
FIGURE 3.
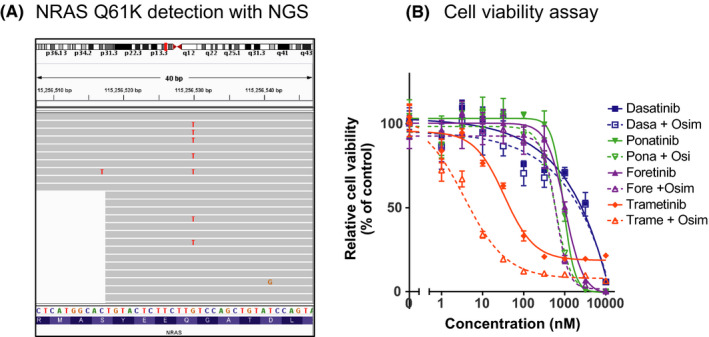
Detection of NRAS‐Q61K mutation in osimertinib‐resistant case. A, NRAS Q61K was detected from the clinical samples from patient No. 24 with NGS. B, Cell viability assay using osimertinib‐resistant PC9 with NRAS Q61K showed that trametinib and osimertinib combination can overcome the resistance with NRAS Q61K mutation
3.3. NRAS Q61K mutation is an osimertinib‐resistant mechanism, which is overcome with trametinib and osimertinib treatment
While we were analyzing the osimertinib resistance mechanism including C797S in the first cohort, coincidentally NGS sequencing detected the NRAS Q61K mutation in 1 patient (Figure 3A). As an independent experiment, we established osimertinib‐resistant PC9 cells that harbored the NRAS‐Q61K mutation; resistance was overcome with trametinib treatment combined with osimertinib (Figure 3B).
3.4. Fractional abundance of epidermal growth factor receptor mutation including C797S in cfDNA changes with osimertinib treatment response and disease progression
In the second cohort, 18 patients started osimertinib treatment after T790M detection. In 44.4% (8/18) of patients, EGFR‐activating mutation in the plasma cfDNA was positive with ddPCR at least once during the follow‐up period. All patients’ clinical backgrounds are shown in Table S2. The EGFR‐activating mutation tended to be negative, especially when disease was limited to the intrathoracic region and the number of metastatic organs was small.
The fractional abundance chart of the EGFR mutation in 8 patients is depicted in Figures 4 and 5. Pretreatment, EGFR‐activating mutation in the plasma cfDNA was positive in 7 patients, whereas posttreatment, the fractional abundance of EGFR‐activating mutation in plasma cfDNA decreased to almost zero in all 7 patients. Disease progression was confirmed in 6 patients, and 5 of them had EGFR‐activating mutations. EGFR C797S mutation was detected in cfDNA before disease progression in 3 cases. EGFR T790M/C797S in cis and trans positions was also discriminated with ddPCR using our original probes (both case A7 and case A19 harbored EGFR T790M/C797S in cis). Fractional abundance changes of EGFR‐activating mutation are significantly more correlated with the treatment response and disease progression than carcinoembryonic antigen (CEA), a major clinical tumor marker (Figure 6).
FIGURE 4.
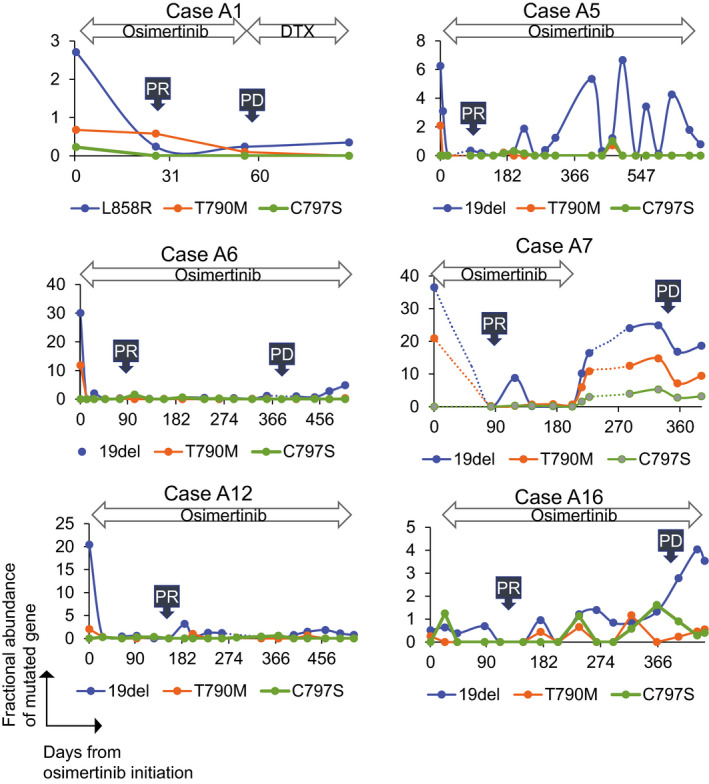
Fractional abundance chart of epidermal growth factor receptor (EGFR) mutation in 6 patients. Fractional abundance of EGFR mutation is shown on the vertical axis, whereas the days from osimertinib initiation is shown on the horizontal axis. C797S was detected in 1 patient (Case A7) at least once during the follow‐up period. RT, radiation therapy
FIGURE 5.
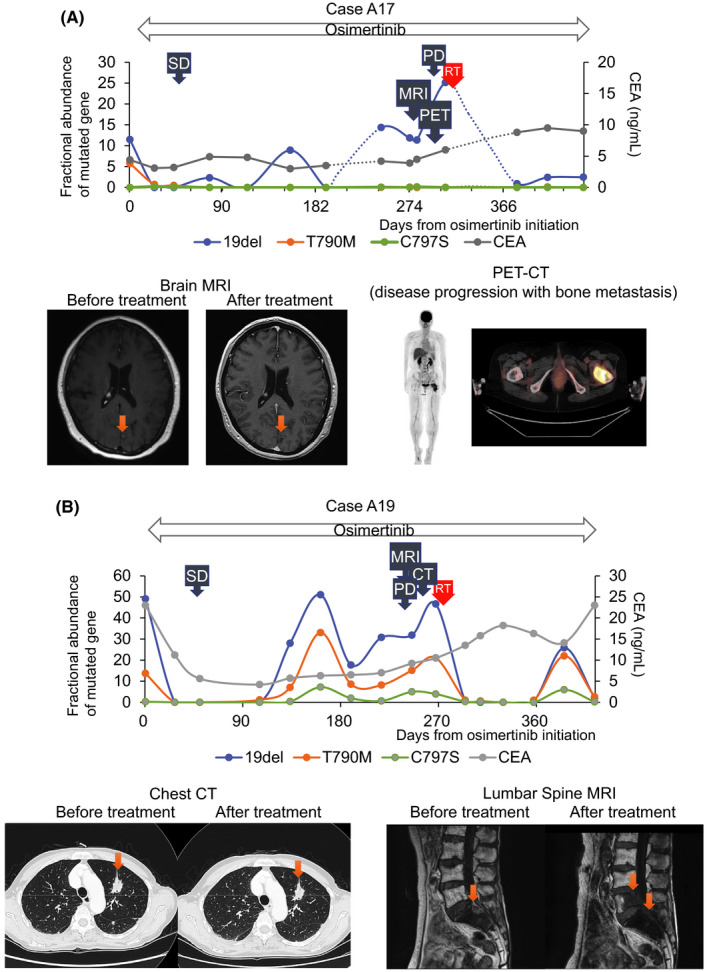
A, Change of fractional abundance of epidermal growth factor receptor (EGFR) mutation in Case A17 is shown. After osimertinib treatment, multiple brain metastasis remained in remission. PET‐CT examination revealed disease progression of left femur bone metastasis. B, Change of fractional abundance of EGFR mutation in Case A19 is shown. After osimertinib treatment, primary lung tumor and other metastasis remained stable with CT examination. MRI examination revealed L5 vertebrae metastasis progression
FIGURE 6.
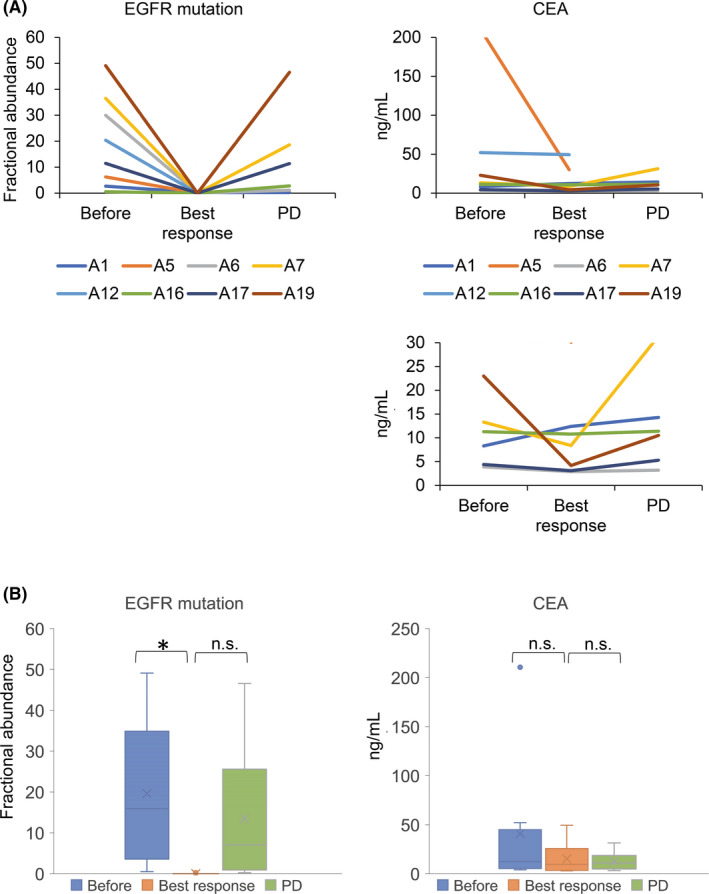
A, Fractional abundance of epidermal growth factor receptor (EGFR)‐activating mutation and carcinoembryonic antigen (CEA) before the osimertinib treatment, best osimertinib response, and disease progression are described in the graph. The two types of vertical axis were shown to estimate the variation precisely. B, Fractional abundance of EGFR‐activating mutation was significantly different between pretreatment and at the time of best response
3.5. Fractional abundance changes of epidermal growth factor receptor mutation in cases of bone oligo‐progression and after a local radiation therapy
We focused on 2 patients. In both cases, EGFR mutation in the plasma cfDNA increased a few months before the radiological progression. It became negative after the local radiation therapy for bone oligo‐progression.
Case A17 was a 56‐year‐old woman with NSCLC with EGFR exon 19 deletion. She was treated with erlotinib; however, the disease progressed with multiple brain metastases. EGFR T790M mutation was confirmed in the plasma cfDNA, and she started osimertinib treatment. The EGFR mutation in the plasma cfDNA became negative and she continued osimertinib treatment. The EGFR mutation in the plasma cfDNA started to increase after 8 months of continuous treatment; however, her routine CT and brain MRI examination did not reveal disease progression. After that, she complained of left leg pain and PET‐CT examination revealed disease progression of left femur bone metastasis; however, the other metastases remained in remission. Local radiation therapy (30 Gy/10 Fr) was performed on the left femur bone for pain control. Consequently, the EGFR mutation in the plasma cfDNA became negative again. The patient continued osimertinib treatment beyond progression, and the disease remained stable for several months without disease progression (Figure 5A).
Case A19 was a 76‐year‐old woman with NSCLC with EGFR exon 19 deletion. Disease progression was confirmed as an S1 vertebrae metastasis under afatinib treatment. The patient started osimertinib after detection of T790M mutation in the plasma cfDNA. The EGFR mutation in the plasma cfDNA became negative but soon increased again. Routine CT scans did not detect disease progression at around 1‐year posttreatment. However, back pain occurred after 8 months of treatment, and repeat MRI examination revealed L5 vertebrae metastasis progression. Local radiation therapy (39 Gy/13 Fr) was performed on the L5 vertebrae for pain control. Consequently, EGFR mutation in the plasma cfDNA decreased to zero, and the patient continued osimertinib treatment beyond progression for 6 months without disease progression (Figure 5B).
4. DISCUSSION
We first validated the C797S detection from plasma cfDNA with ddPCR using clinical samples. The concordance rate between tumor cell DNA and plasma cfDNA was high at disease progression after osimertinib treatment. Then, ddPCR was used to distinguish cis and trans C797S from T790M using novel specific probes. Past reports also showed the efficacy of ddPCR in detecting C797S; 22 , 23 however, the efficacy of ddPCR in distinguishing EGFR‐T790M/C797S in cis and trans, validated with clinical samples, has not been reported. A difference in treatment strategies was observed between T790M and C797S in the cis and trans positions: while brigatinib with an anti–EGFR antibody has been a promising treatment option for tumors with cis‐C797S/T790M, osimertinib combined with gefitinib has been found to be effective against trans‐C797S and T790M. 15 , 24 , 25 Therefore, detection of C797S in the cis and trans positions using ddPCR would be useful in clinical practice. At present, treatment targeting C797S has not been approved in Japan; therefore, we are trying to include these patients in clinical trials. NGS has been widely used to detect multiple gene mutations. Although NGS can also distinguish EGFR T790M/C797S in cis and trans positions, ddPCR is cheaper to use and has a faster turnaround time than NGS. 26 , 27 Therefore, ddPCR would be useful, especially in serially monitoring EGFR mutations during osimertinib treatment.
Coincidentally, we found the NRAS Q61K mutation through the NGS in analyzing the osimertinib‐resistant tumor cells. Trametinib with osimertinib was effective against tumors with NRAS Q61K mutation in the cell line model. Therefore, a clinical trial to evaluate the effect of trametinib combination treatment for NSCLC patients resistant to osimertinib is expected.
After validating the detection of the EGFR C797S mutation in plasma cfDNA, serial evaluation of EGFR mutation (active mutation, T790M and C797S) was performed in the plasma cfDNA during and after osimertinib treatment in the second cohort. EGFR mutation in the plasma cfDNA has been reported to decrease with EGFR‐TKI treatment and to increase with disease progression, 17 , 18 , 19 but a serial evaluation of EGFR T790M and C797S status during the osimertinib treatment has not been fully reported. We found that C797S in cfDNA also changes with treatment response, and confirmed that the EGFR T790M and C797S in cis or trans position were detectable in plasma cfDNA some months before clinical disease progression. We further found that the fractional abundance of EGFR mutation in the plasma cfDNA might be a more reliable plasma biomarker of osimertinib response than CEA, a widely used tumor maker. While we were preparing this manuscript, Romero et al 28 reported on the EGFR mutation, including C797S monitoring during osimertinib treatment. The present study has some significance because we validated the detection of EGFR C797S mutation with clinical samples, showed C797S monitoring including cis/trans analysis, conducted closed and periodic monitoring, and illustrated the relationship between EGFR mutation and CEA in a Japanese cohort.
We focused on 2 patients who had disease progression with bone oligo‐progression, who were treated with local radiation therapy and continued osimertinib treatment. Several reports showed the usefulness of cfDNA for early detection of disease progression, and local radiation therapy decreases the amount of mutant EGFR DNA. 28 , 29 , 30 In some cases, early detection of bone metastasis is difficult with CT scans. 31 Indeed, not routine CT scans but PET‐CT and MRI successfully detected the bone metastasis in 2 patients. Therefore, the usefulness of cfDNA in detecting bone oligo progression should be emphasized. Second, the treatment strategy for oligo‐progression is still debatable. 32 There is no reliable biomarker to continue osimertinib treatment after local therapy. We clearly showed that EGFR mutation in the plasma cfDNA became negative after the local radiation therapy, and patients continued the osimertinib treatment over several months without disease progression. During the osimertinib treatment beyond progression, EGFR mutation in the plasma cfDNA was maintained at low levels, which might indicate that it could be a reliable biomarker to continue osimertinib beyond progression.
This study had certain limitations. First, the number of patients in this study is small, which decreases the importance of our results. Increasing the cohort size would be necessary in a future study. Second, the detection rate of EGFR mutation in the plasma cfDNA was not high, and the negative result in liquid biopsy can be a sensitivity issue and not a real negative. The technical improvement would be desirable to introduce it into clinical practice. A past report showed that the sensitivity of EGFR mutation detection in cfDNA is low when diseases are limited to the intrathoracic region. 33 Therefore, we also evaluated the disease state in the second cohort. We found that EGFR mutation in cfDNA tended not to be detected when the disease was in intrathoracic areas. Therefore, analysis of the cfDNA from patients with intrathoracic disease requires careful interpretation.
In conclusion, we found that EGFR‐T790M/C797S mutations in cis and trans positions are discriminately detectable, distinguished by ddPCR from plasma cfDNA. Serial evaluation of EGFR mutation in plasma cfDNA showed the emergence of EGFR mutations, including C797S, during osimertinib treatment before disease progression.
DISCLOSURE
M. Nishio received research funding from Novartis, ONO Pharmaceutical, Chugai Pharmaceutical, Bristol‐Myers Squibb, TAIHO Pharmaceutical, Eli Lilly, Pfizer, Astellas Pharma, and AstraZeneca and received honorarium from Pfizer, Bristol‐Myers Squibb, ONO Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, TAIHO Pharmaceutical, and AstraZeneca. N. Yanagitani is a consultant of Chugai Pharmaceutical. R. Katayama received research funding from Chugai, TAKEDA, Toppan Printing, and Daiichi‐Sankyo. T. Sasaki received research funding from Novartis, Pfizer, and Boehringer Ingelheim and received honorarium from AstraZeneca. All other authors declare no conflict of interest.
Supporting information
Supplementary Material
Appendix S1
Ariyasu R, Uchibori K, Sasaki T, et al. Monitoring epidermal growth factor receptor C797S mutation in Japanese non–small cell lung cancer patients with serial cell‐free DNA evaluation using digital droplet PCR. Cancer Sci. 2021;112:2371–2380. 10.1111/cas.14879
Funding information
This study was supported in part by MEXT/JSPS KAKENHI grant number JP17H06327 (to NF), JP19H03524 (to R. Katayama), JP19H04715 (to R. Katayama), JP15K14716 (to YO), and JP17K09472 (to YM), as well as a grant from the AMED, grant number JP20cm0106203h0005 and JP20ck0106472h0002 (to R. Katayama), and grants from the Uehara Memorial Foundation (to R. Katayama) and Nippon Foundation (to NF).
Contributor Information
Makoto Nishio, Email: mnishio@jfcr.or.jp.
Ryohei Katayama, Email: ryohei.katayama@jfcr.or.jp.
REFERENCES
- 1. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small‐cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129‐2139. [DOI] [PubMed] [Google Scholar]
- 2. Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non–small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121‐128. [DOI] [PubMed] [Google Scholar]
- 3. Zhou C, Wu Y‐L, Chen G, et al. Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non–small‐cell lung cancer (OPTIMAL, CTONG‐0802): a multicentre, open‐label, randomised, phase 3 study. Lancet Oncol. 2011;12:735‐742. [DOI] [PubMed] [Google Scholar]
- 4. Wu Y‐L, Zhou C, Hu C‐P, et al. Afatinib versus cisplatin plus gemcitabine for first‐line treatment of Asian patients with advanced non–small‐cell lung cancer harbouring EGFR mutations (LUX‐Lung 6): an open‐label, randomised phase 3 trial. Lancet Oncol. 2014;15:213‐222. [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non–small‐cell lung cancer to gefitinib. N Engl J Med. 2005;352:786‐792. [DOI] [PubMed] [Google Scholar]
- 6. Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid‐based assay. Clin Cancer Res. 2011;17:1169‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cross DAE, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M‐mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mok TS, Wu Y‐L, Ahn M‐J, et al. Osimertinib or platinum‐pemetrexed in EGFR T790M‐positive lung cancer. N Engl J Med. 2017;376:629‐640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin C‐C, Shih J‐Y, Yu C‐J, et al. Outcomes in patients with non–small‐cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med. 2018;6:107‐116. [DOI] [PubMed] [Google Scholar]
- 10. Zheng DI, Hu M, Bai YU, et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget. 2017;8:49671‐49679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third‐generation EGFR tyrosine kinase inhibitor osimertinib in non–small cell lung cancer patients. Clin Cancer Res. 2018;24:3097‐3107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 12. Planchard D, Loriot Y, André F, et al. EGFR‐independent mechanisms of acquired resistance to AZD9291 in EGFR T790M‐positive NSCLC patients. Ann Oncol. 2015;26:2073‐2078. [DOI] [PubMed] [Google Scholar]
- 13. Ho C‐C, Liao W‐Y, Lin C‐A, et al. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J Thorac Oncol. 2017;12:567‐572. [DOI] [PubMed] [Google Scholar]
- 14. Minari R, Bordi P, Del Re M, et al. Primary resistance to osimertinib due to SCLC transformation: issue of T790M determination on liquid re‐biopsy. Lung Cancer. 2018;115:21‐27. [DOI] [PubMed] [Google Scholar]
- 15. Uchibori K, Inase N, Araki M, et al. Brigatinib combined with anti–EGFR antibody overcomes osimertinib resistance in EGFR‐mutated non–small‐cell lung cancer. Nat Commun. 2017;8:14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non–small‐cell lung cancer. J Clin Oncol. 2016;34:3375‐3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR‐mutant lung cancer using quantitative next‐generation genotyping of cell‐free plasma DNA. Clin Cancer Res. 2014;20:1698‐1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JY, Qing XU, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC‐12‐02). Oncotarget. 2016;7:6984‐6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell‐free DNA is a marker to monitor osimertinib response. Br J Cancer. 2018;119:1252‐1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ariyasu R, Nishikawa S, Uchibori K, et al. High ratio of T790M to EGFR activating mutations correlate with the osimertinib response in non–small‐cell lung cancer. Lung Cancer. 2018;117:1‐6. [DOI] [PubMed] [Google Scholar]
- 21. Katayama R, Gong BO, Togashi N, et al. The new‐generation selective ROS1/NTRK inhibitor DS‐6051b overcomes crizotinib resistant ROS1‐G2032R mutation in preclinical models. Nat Commun. 2019;10:3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edwards V, Jackson L, Reese J, et al. Abstract 411: Rapid and sensitive detection of EGFR C797S mutations using a blood‐based droplet digital PCR assay. Cancer Res. 2019;79(13 Supplement):411. [Google Scholar]
- 23. Song X, Gong J, Zhang X, et al. Plasma‐based early screening and monitoring of EGFR mutations in NSCLC patients by a 3‐color digital PCR assay. Br J Cancer. 2020;123:1437‐1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niederst MJ, Hu H, Mulvey HE, et al. The allelic context of the C797S mutation acquired upon treatment with third‐generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res. 2015;21:3924‐3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arulananda S, Do H, Musafer A, et al. Combination osimertinib and gefitinib in C797S and T790M EGFR‐mutated non–small cell lung cancer. J Thorac Oncol. 2017;12:1728‐1732. [DOI] [PubMed] [Google Scholar]
- 26. Salto‐Tellez M, de Castro DG. Next‐generation sequencing: a change of paradigm in molecular diagnostic validation. J Pathol. 2014;234:5‐10. [DOI] [PubMed] [Google Scholar]
- 27. Takamatsu H. Comparison of minimal residual disease detection by multiparameter flow cytometry, ASO‐qPCR, droplet digital PCR, and deep sequencing in patients with multiple myeloma who underwent autologous stem cell transplantation. J Clin Med. 2017;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romero A, Serna‐Blasco R, Alfaro C, et al. ctDNA analysis reveals different molecular patterns upon disease progression in patients treated with osimertinib. Transl Lung Cancer Res. 2020;9:532‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep. 2016;6:33505. 10.1038/srep33505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ni J, Weng L, Liu YI, et al. Dynamic monitoring of EGFR mutations in circulating cell‐free DNA for EGFR‐mutant metastatic patients with lung cancer: early detection of drug resistance and prognostic significance. Oncol Lett. 2017;13:4549‐4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Sullivan GJ, Carty FL, Cronin CG. Imaging of bone metastasis: an update. World J Radiol. 2015;7:202‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tumati V, Iyengar P. The current state of oligometastatic and oligoprogressive non–small cell lung cancer. J Thorac Dis. 2018;10(Suppl 21):S2537‐S2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karlovich C, Goldman JW, Sun J‐M, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO‐1686). Clin Cancer Res. 2016;22:2386‐2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Appendix S1


