Abstract
The phosphatidylinositol 3‐kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway plays a vital role in cell proliferation, apoptosis, metabolism, and angiogenesis in various human cancers, including head and neck squamous cell carcinoma (HNSCC). In the present study, we aimed to clarify the role of AKT, which is a major downstream effector of the PI3K‐AKT‐mTOR pathway, in HNSCC. We first investigated the mRNA expression of AKT isoforms using RNA‐sequencing data from The Cancer Genome Atlas database. We observed a specific elevation of AKT3 expression in HNSCC tissues when compared with that in normal tissues. Furthermore, AKT3 expression correlated with genes related to the immunosuppressive microenvironment more than the other AKT isoforms and PIK3CA. Accordingly, we focused on AKT3 and performed a knockdown approach using an HNSCC cell line. AKT3 knockdown cells exhibited impaired proliferation, a shift in the cell cycle from G2/M to G1/G0 phase, an increase in apoptotic cells, and downregulation of gene expression related to immunosuppression, as well as the knockdown of its upstream regulator PIK3CA. We also performed immunohistochemistry for both AKT3 and PIK3CA using surgical specimens from 72 patients with HNSCC. AKT3 expression in tumor cells correlated with immune cell infiltration and unfavorable prognosis when compared with PIK3CA. These findings suggested that AKT3 expression is a potential biomarker for predicting the immunoreactivity and prognosis of HNSCC. Furthermore, the isoform‐specific inhibition of AKT3 could be developed as a novel cancer therapy that efficiently suppresses the PI3K‐AKT‐mTOR pathway.
Keywords: AKT, head and neck squamous cell carcinoma, immunosuppression, PIK3CA, tumor microenvironment
This research work demonstrated that AKT3 is a key regulator that modulates the proliferative and immunosuppressive functions of HNSCC. AKT3 is a potential candidate for a novel biomarker of the immunosuppressive microenvironment and shorter survival. The isoform‐specific inhibition of AKT3 could be developed as a novel cancer therapy that efficiently suppresses the PI3K‐AKT‐mTOR pathway.
1. INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, with a 5‐y survival rate of 66%; approximately 890 000 new cases and 450 000 deaths reportedly occur annually. 1 , 2 Exposure to tobacco‐derived carcinogens and excessive alcohol consumption are known key risk factors for HNSCC. 3 Furthermore, oropharyngeal cancers associated with prior infection with oncogenic strains of human papillomavirus (HPV) are reportedly increasing in the USA and Western Europe. 4
In HPV‐positive HNSCCs, viral proteins, including E6 and E7, facilitate oncogenic transformation by ubiquitylation and proteasomal degradation of p53 or by interacting with several cell cycle regulatory proteins such as E2F. 5 By contrast, in HPV‐negative HNSCCs, frequent alterations in TP53 and CDKN2A were reported by analyzing The Cancer Genome Atlas (TCGA). 6 Notably, PIK3CA, which encodes the catalytic subunit of phosphoinositide 3‐kinase (PI3K), has been recognized as a common HNSCC oncogene, which is frequently mutated in both HPV‐positive and HPV‐negative HNSCCs. 6 , 7 PIK3CA is a key component of the PI3K‐AKT‐mammalian target of rapamycin (mTOR) pathway, which plays a vital role in cell proliferation, apoptosis, metabolism, and angiogenesis in various human cancers, including HNSCC. 8 In addition to PIK3CA mutations, several mechanisms, including epidermal growth factor receptor activation, PIK3CA amplification, PI3K overexpression, and phosphatase and tensin homolog (PTEN) mutations, have been reported to activate the PI3K‐AKT‐mTOR pathway. In the PI3K‐AKT‐mTOR pathway, AKT, also known as protein kinase B (PKB), is the major downstream effector. 9 , 10 , 11 AKT is comprised of 3 mammalian isoforms: AKT1, AKT2, and AKT3. AKT1 is usually expressed at higher levels than other isoforms, indicating its importance, especially in cell survival. AKT2 plays a vital role in the insulin signal transduction pathway. AKT3 is primarily expressed in the brain and testis, however the specific role of AKT3 remains elusive. 12 Meanwhile, AKT overactivation has been reported in various malignancies, including HNSCC, 13 , 14 and the activation of AKT has been reported to negatively correlate with prognosis in patients with esophageal squamous cell carcinoma and non–small‐cell lung cancer. 15 , 16 However, the specific role of each AKT isoform needs to be comprehensively elucidated in malignant tumors, especially in HNSCC.
In the present study, we sought to clarify the role of AKT activation in HNSCC. We first investigated the mRNA expression of each AKT isoform using RNA‐sequencing data obtained from TCGA database. Based on this analysis, we focused on AKT3 and performed a knockdown approach using HNSCC cells. Moreover, we investigated the significance of AKT3 in surgical specimens derived from HNSCC patients.
2. MATERIALS AND METHODS
2.1. The Cancer Genome Atlas data analysis
RNA‐sequencing data (Illumina HiSeq RNA‐seq V2, RSEM normalized) and clinical information were obtained from TCGA Research Network (TCGA provisional version updated in 2016, http://cancergenome.nih.gov/). In total, 564 cases consisting of 97 HPV‐positive HNSCC, 423 HPV‐negative HNSCC, and 44 normal samples were available. The log10‐transformed value of mRNA expression levels was calculated.
2.2. Cell culture
The squamous cell carcinoma of the head and neck (SCCHN) cell line HSC‐3 was purchased from the Japanese Collection of Research Bioresources (JCRB) Cell Bank. The cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin.
2.3. Transfection
shRNAs for luciferase (LUC), AKT3, and PIK3CA were transfected into HSC‐3 cells. The shRNA oligonucleotides were annealed at 90°C for 15 min, 70°C for 30 min, and 25°C for 30 min, and then cloned into the pLKO.1 puro lentiviral shRNA expression vector between the AgeI and EcoRI sites. The oligonucleotide target sequences used for shRNAs were as follows: AKT3#4, 5′‐GTAGTCCAACTTCACAAAT‐3′; AKT3#5, 5′‐GATGTGGATTTACCTTATC‐3′; PIK3CA#1, 5′‐CCAGATGTATTGCTTGGTA‐3′; and PIK3CA#4, 5′‐GCACAATCCATGAACAGC‐3′.
2.4. Reverse transcription‐polymerase chain reaction (RT‐PCR)
Total RNA was extracted from transduced HSC‐3 cells using an RNeasy Mini Kit (QIAGEN). Quantitative RT‐PCR was performed in triplicate using a Power SYBR Green RNA‐to‐CT 1‐Step Kit and an Applied Biosystems StepOne instrument (Thermo Fisher Scientific). A melting curve was recorded at the end of every run to assess the product specificity. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was used as an internal control. The relative expression level was evaluated using the 2−ΔΔCt method, in which Ct represents the threshold cycle. The PCR primers used are listed in Supporting Information Table S1.
2.5. Cell proliferation assay
Transfected HSC‐3 cells were harvested and plated in 96‐well plates (1 × 103 cells/well). After incubation, 20 µL of CellTiter 96® AQueous One Solution Reagent (Promega) was added to each well, followed by incubation for 2 h. The absorbance at 490 nm was measured using an iMark™ Microplate Absorbance Reader (Bio‐Rad) to measure cell proliferation, which was evaluated at 24, 48, and 72 h.
2.6. Cell cycle analysis
Transduced HSC‐3 cells were harvested at approximately 70% confluency. For cell fixation, 70% ice‐cold ethanol was added dropwise while vortexing and incubated at 4°C for 30 min. After washing the cells with PBS, the samples were treated with RNase A (1 μg/mL, Promega), followed by nuclear staining with propidium iodide (PI; 50 μg/mL, BioLegend). Cell cycle phases were analyzed by flow cytometry.
2.7. Apoptosis assay
Transduced HSC‐3 cells were harvested at approximately 70% confluency. After washing with PBS, cells were stained with A CaspACE™ fluorescein isothiocyanate (FITC)‐VAD‐FMK In Situ Marker (Promega) according to the manufacturer's instructions. The proportion of apoptotic cells was measured by flow cytometry. 7‐Amino‐actinomycin D (7‐AAD; BD Bioscience) was added 10 min before analysis. FITC‐VAD‐FMK‐positive and 7‐AAD‐negative cells were defined as apoptotic cells.
2.8. T cell proliferation assay
Peripheral blood mononuclear cells (PBMCs) obtained from healthy donors were labeled with 1 µmol/L carboxyfluorescein succinimidyl ester (CFSE; Thermo Fisher Scientific) according to the manufacturer's protocol. CFSE‐labeled PBMCs (1 × 105) were plated into 96‐well plates with transduced HSC‐3 cells (2 × 104) in RPMI medium supplemented with 10% FCS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Following the addition of anti‐CD3/anti‐CD28 stimulus (Treg Suppression Inspector human; Miltenyi Biotec), the cells were incubated for 96 h. After incubation, floating cells were harvested and stained with allophycocyanin (APC)‐CD3 antibody (BD Bioscience) at 4°C for 1 h. Then, 7‐AAD (BD Bioscience) was added 10 min before the analysis. The proliferation of CD3‐positive 7‐AAD‐negative cells was measured by dilution of CFSE staining using flow cytometry.
2.9. Flow cytometry
Flow cytometry was used to assess cell cycle, apoptosis, and T cell proliferation. Multicolor flow cytometry was performed using a BD FACSVerse Flow Cytometer (BD Bioscience). Acquired data were analyzed using FlowJo software (TreeStar).
2.10. Patient specimens for immunohistochemistry
In total, 72 HNSCC surgical specimens resected from primary tongue cancers were examined using immunohistochemistry (IHC). Patients who underwent surgery at Gunma University Hospital between November 2000 and January 2012 were included in the study; patients who received preoperative chemotherapy, radiotherapy, or immunotherapy were excluded. Clinical and pathological information, including age, sex, tumor‐node‐metastasis (TNM) stage, lymphatic/vascular invasion, overall survival (OS), and disease‐free survival (DFS), were obtained from electronic medical records. TNM classification was based on the 7th edition of the International System for Staging adopted by the American Joint Committee on Cancer and the Union for International Cancer Control. The present study was approved by the Institutional Review Board of Gunma University (No. 12‐12) and was performed in accordance with the Declaration of Helsinki of 1996. Written informed consent was obtained from all the patients.
2.11. Immunohistochemistry
Formalin‐fixed (10% formaldehyde) paraffin tissue sections (5‐µm thick) were used for IHC. After deparaffinization and hydration, antigen retrieval was performed using Proteinase K (Dako) at room temperature for 5 min for CD68 or by autoclaving at 121°C for 20 min in citrate buffer (pH 6.0) for the other antigens. Endogenous peroxidase and non‐specific binding sites were blocked with 3% H2O2 and 1% BSA/5% normal horse serum, respectively. Primary antibody staining was performed at 4°C overnight with antibodies listed in Table S2, followed by secondary staining with Labeled Polymer‐HRP anti‐mouse/rabbit (Dako) at room temperature for 45 min. Detection was performed using 3.3′‐diaminobenzidine (DAB Dako), followed by counterstaining with Mayer's hematoxylin (FUJIFILM Wako Pure Chemical Corporation). After dehydration, the slides were mounted with a non‐aqueous mounting medium DPX (Merck). Stained sections were evaluated using an Axioscope (Carl Zeiss Microscopy GmbH) light microscope combined with AxioVision LE image acquisition software by 2 independent observers in a blinded manner. Immunostaining of AKT3 and PIK3CA was scored based on the intensity and heterogeneity. The staining intensity was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The proportion of stained tumor cells was scored as follows: 0 (<5%), 1 (5%‐25%), 2 (26%‐50%), and 3 (>50%). Immunostaining was scored by multiplying the intensity and proportion scores (final score: 0‐9). Low expression was defined as <2, whereas high expression was defined as ≥2. To enumerate CD68+ and CD163+ macrophages, more than 4 representative area fields adjacent to cancer cells were examined at a magnification of ×200. More than 5 areas of a representative field were assessed for CD1a+ DC and CD56+ NK cells at ×200 magnification. Infiltration of CD3+ T cells in more than 5 ×400 fields (HPF) was considered as grade 1 (<10 positive cells/HPF), grade 2 (10‐30/HPF), grade 3 (31‐100/HPF), and grade 4 (>101/HPF), followed by calculation of the average of multiple counted areas.
2.12. Statistical analysis
Data were analyzed using GraphPad Prism version 8 (GraphPad Software) and R (The R Foundation for Statistical Computing, Vienna, Austria). Student t test (for 2 groups), one‐way ANOVA with Tukey post‐hoc test for multiple pairwise testing (for more than 2 groups), chi‐square test for independence, and Fisher exact test were used to examine differences in continuous and categorical variables. Pearson correlation coefficient was used to evaluate the correlation between the 2 continuous variables. Two‐sided P‐values < .05 were considered statistically significant. Survival curves were calculated using the Kaplan‐Meier method and compared using the log‐rank test. Multivariate regression analyses were performed using the Cox proportional hazards model. Variables were included in subsequent multivariate analyses when P‐values were < .10 in univariate analyses.
3. RESULTS
3.1. mRNA expression of AKT isoforms/PIK3CA and immune‐related genes in HNSCC
To compare the mRNA expression of each AKT isoform in the normal epithelium of the head and neck region and HNSCC, we evaluated RNA‐sequencing data obtained from TCGA database. The expression of AKT1 and AKT3, especially AKT3, was higher in HNSCC than that in normal tissues, as well as PIK3CA expression. In contrast, AKT2 expression was lower in HNSCC tissues than in normal tissues (Figure 1A,B). We next analyzed the correlation between the expression of AKT1/AKT2/AKT3/PIK3CA and stromal cell markers in 520 HNSCC samples (Figure 1C, Table S3). Among AKT1/AKT2/AKT3/PIK3CA, AKT3 expression exhibited a strong positive correlation with genes related to the immunosuppressive microenvironment when compared with other genes. For instance, AKT3 expression was positively correlated with tumor‐associated macrophage (TAM)‐related genes, including FCGR1A, CD68, CD163, MRC1, CCL2, CXCL12, and CSF1. Regulatory T cell (Treg)‐related genes, including CD4, FOXP3, and IL10, were positively correlated with AKT3 expression. Furthermore, AKT3 expression positively correlated with cytotoxic T cell markers, NK cell marker, and T cell exhaustion markers, including CD3E, CD8A, NCAM1, PDCD1, and HAVCR2, and negatively correlated with T cell activation markers IFNG, and GZMB. Meanwhile, AKT1 and AKT3, especially AKT3 expression, positively correlated with marker genes for cancer‐associated fibroblasts (CAFs), including FAP, COL11A1, PDGFRB, and POSTN. Based on these results, we focused on AKT3 and investigated its role in the following experiments.
FIGURE 1.
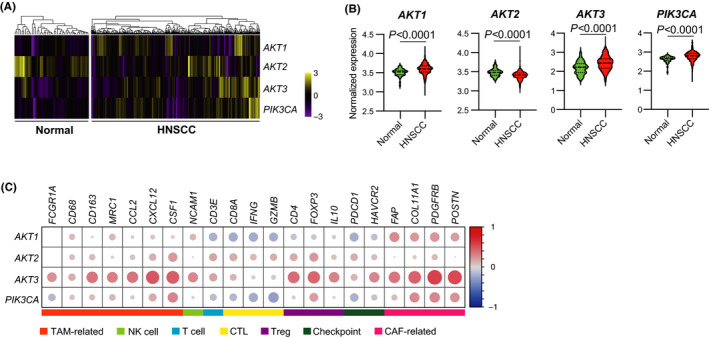
mRNA expression of AKT3 correlates with the immunosuppressive tumor microenvironment in HNSCC. A‐C, mRNA expression data and clinical information were obtained from TCGA. A, Heat map of AKT1/AKT2/AKT3/PIK3CA expression in 520 HNSCC and 44 normal tissues. Samples were clustered using hierarchical clustering based on z‐scores. B, Log10‐transformed expression of genes displayed in A. C, Correlation matrix displaying R‐values for assessing the correlation between expression of AKT1/AKT2/AKT3/PIK3CA and that of stromal cell markers across 520 HNSCC tissues. Each circle size represents the absolute value of the R‐value. CAF, cancer‐associated fibroblast; CTL, cytotoxic T lymphocyte; HNSCC, head and neck squamous cell carcinoma; TAM, tumor‐associated macrophage; TCGA, The Cancer Genome Atlas; Treg, regulatory T cell
3.2. AKT3 knockdown downregulated the activity of HSC‐3 cells as well as PIK3CA
We performed a knockdown approach to investigate the role of AKT3 in HNSCC. Two independent shRNAs against AKT3 (shAKT3#4 and shAKT#5) were transfected into HSC‐3 cells. Two independent shRNAs against PIK3CA (shPIK3CA3#1 and shPIK3CA#4) were also transfected into HSC‐3 cells for comparison with AKT3 knockdown cells. shRNA against luciferase (shLUC) was used as a control. As expected, quantitative RT‐PCR revealed that AKT3 or PIK3CA knockdown cells expressed less AKT3 or PIK3CA, respectively, than controls (Figure 2A). Moreover, AKT3/PIK3CA knockdown cells exhibited lower expression of several immune‐related genes, including IL1B, IL6, CXCL8, TGFB1, CD274, and PDCD1LG2 (Figure 2B). Furthermore, we analyzed the proliferation of knockdown cells. The proliferation of AKT3/PIK3CA knockdown cells was marginally suppressed. In contrast, cell cycle analysis revealed that a higher number of AKT3/PIK3CA knockdown cells were present in the G1/G0 phase than control cells, whereas lower numbers of knockdown cells were in the G2/M phase compared with control cells (Figure 2D). Moreover, AKT3/PIK3CA knockdown cells were more apoptotic than control cells (Figure 2E). To further characterize AKT3/PIK3CA knockdown cells, we examined their suppressive activity on T cell proliferation. T cells co‐cultured with AKT3/PIK3CA knockdown cells demonstrated greater proliferation than those co‐cultured with control cells, indicating the downregulation of the suppressive activity of AKT3/PIK3CA knockdown cells on T cell immunity (Figure 2F).
FIGURE 2.
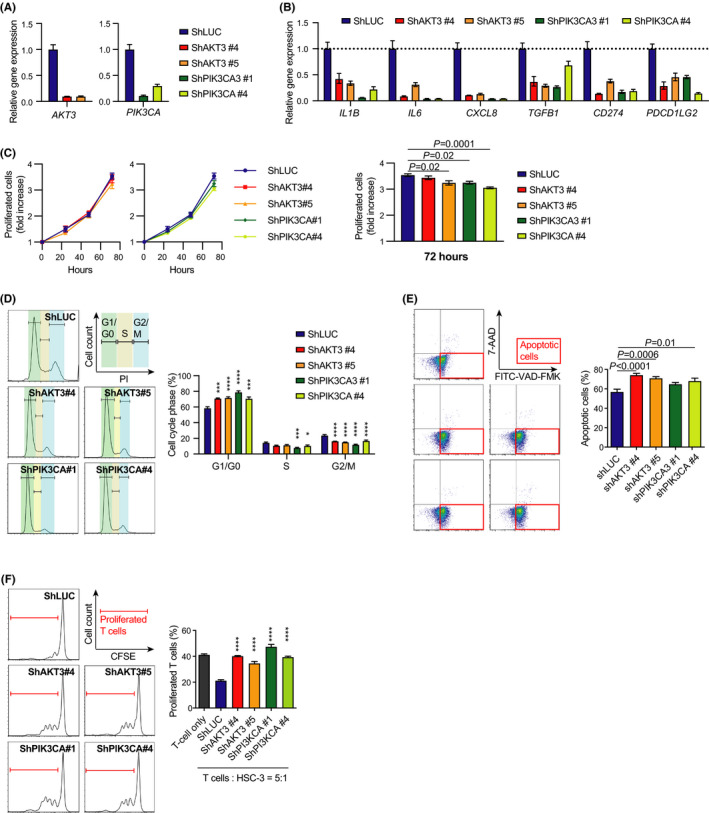
AKT3 is crucial to maintain the activities of HNSCC cells. A, Relative AKT3/PIK3CA expression in transduced HSC‐3 cells as measured by RT‐PCR. B, Relative mRNA expression of immune‐related genes in transduced HSC‐3 cells as measured by RT‐PCR. C, Proliferation of transduced HSC‐3 cells. A fold increase compared with that at 0 h is shown. D, Cell cycle analysis of transduced HSC‐3 cells. Histograms represent the cell cycle phases. E, Proportion of apoptotic cells in transduced HSC‐3 cells. F, Proliferation of T cells co‐cultured with transduced HSC‐3 cells. *P < .05; **P < .01; ***P < .001; ****P < .0001. HNSCC, head and neck squamous cell carcinoma; RT‐PCR, reverse transcription‐polymerase chain reaction
3.3. AKT3 expression in tumor cells correlates with stromal cell infiltration and shorter overall survival
We investigated the clinical significance of AKT3/PIK3CA expression in 72 HNSCC surgical specimens. The staining score of AKT3/PIK3CA was classified as low or high (mentioned in Materials and Methods). AKT3/PIK3CA staining exhibited no correlation with clinical parameters, including age, sex, differentiation, lymphatic invasion, vascular invasion, lymph node involvement, T factor, and TNM stage (Table 1). Meanwhile, AKT3 staining positively correlated with stromal cell infiltration, including TAMs (CD68), NK cells (CD56), T cells (CD3), and CAFs (αSMA) (Figure 3C). Moreover, high expression of AKT3 correlated with shorter OS (Figure 3B). PIK3CA staining was positively correlated with stromal cell infiltration, including M2‐like TAMs (CD163) and CAFs (αSMA) (Figure 3F). No correlation between PIK3CA staining and survival was observed (Figure 3E). We also performed multivariate regression analyses with both clinical parameters and AKT3/PIK3CA staining using Cox proportional hazards model (Table 2). Both age and AKT3 staining were independent prognostic factors for OS, whereas no independent prognostic factor for DFS was determined.
TABLE 1.
Relationship between AKT3/PIK3CA expression and clinical parameters in 72 HNSCC patients
| Variable | AKT3 in tumors | PIK3CA in tumors | ||||
|---|---|---|---|---|---|---|
| Negative | Positive | P‐value | Negative | Positive | P‐value | |
| Age (y) | ||||||
| <71 | 25 | 16 | .64 | 10 | 31 | .60 |
| ≥71 | 17 | 14 | 10 | 21 | ||
| Gender | ||||||
| Male | 27 | 18 | .81 | 13 | 32 | 1.00 |
| Female | 15 | 12 | 7 | 20 | ||
| Differentiation | ||||||
| Well/moderate | 35 | 27 | .81 | 16 | 46 | .93 |
| Poorly | 7 | 3 | 4 | 6 | ||
| Lymphatic invasion | ||||||
| Negative | 25 | 14 | 0.28 | 13 | 26 | .25 |
| Positive | 17 | 16 | 7 | 26 | ||
| Vascular invasion | ||||||
| Negative | 31 | 18 | .22 | 14 | 35 | .83 |
| Positive | 11 | 12 | 6 | 17 | ||
| Lymph node involvement | ||||||
| Negative | 28 | 20 | 1.00 | 13 | 35 | .85 |
| Positive | 14 | 10 | 7 | 17 | ||
| T factor | ||||||
| T1‐2 | 38 | 25 | .48 | 18 | 45 | 1.00 |
| T3‐4 | 4 | 5 | 2 | 7 | ||
| TNM stage | ||||||
| I‐II | 29 | 18 | .43 | 14 | 33 | .60 |
| III‐IV | 13 | 12 | 6 | 19 | ||
Abbreviations: HNSCC, head and neck squamous cell carcinoma; TNM, tumor‐node‐metastasis.
FIGURE 3.
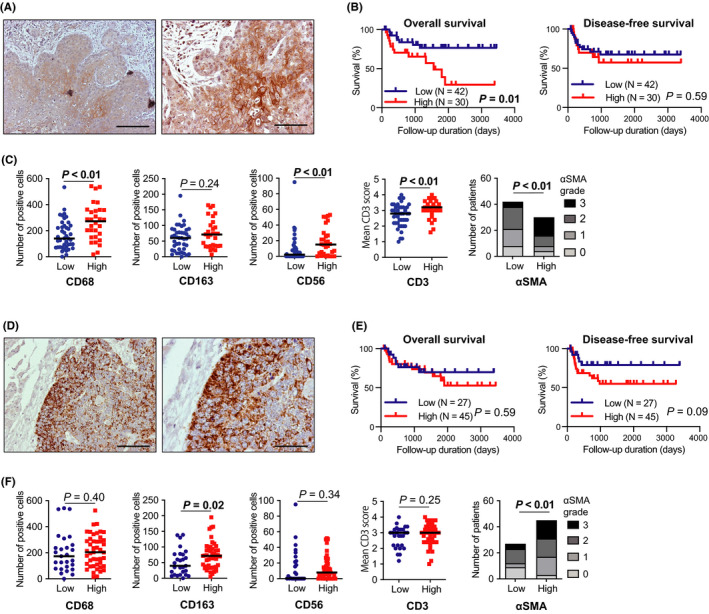
AKT3 expression in tumor cells correlates with immune cell infiltration and poor prognosis in HNSCC. Immunohistochemistry (IHC) staining of 72 surgical HNSCC specimens was performed. A, IHC staining positive for AKT3 in tumor cells; left, ×100 magnification, scale bar 200 μm; right, ×200 magnification, scale bar 100 μm. B, Kaplan‐Meier survival curves based on AKT3 staining scores in tumor cells. C, Relationship between IHC staining of stromal cells and AKT3 score in tumor cells. The αSMA grades represent the infiltration of the CAFs. D, IHC staining positive for PIK3CA in tumor cells; ×100 magnification, scale bar 400 μm; right, ×200 magnification, scale bar 200 μm. E, Kaplan‐Meier survival curves based on PIK3CA staining scores in tumor cells. F, Relationship between IHC staining of stromal cells and PIK3CA score in tumor cells. αSMA, α‐smooth muscle actin; CAFs, cancer‐associated fibroblasts; HNSCC, head and neck squamous cell carcinoma
TABLE 2.
Multivariate survival analyses on OS and DFS in 72 HNSCC patients
| Variables | Overall survival | Disease‐free survival | ||
|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | |
| Age (y) | ||||
| <71 | 1 | .020 | ||
| ≥71 | 3.285 (1.203‐8.965) | |||
| Differentiation | ||||
| Well/moderate | 1 | .052 | 1 | .072 |
| Poorly | 3.117 (0.989‐9.820) | 2.781 (0.914‐8.084) | ||
| Lymphatic invasion | ||||
| Negative | 1 | .205 | 1 | .734 |
| Positive | 2.575 (0.597‐11.113) | 1.265 (0.326‐4.907) | ||
| Vascular invasion | ||||
| Negative | 1 | .587 | 1 | .909 |
| Positive | 1.341 (0.465‐3.868) | 0.938 (0.310‐2.839) | ||
| N factor | ||||
| N0 | 1 | .682 | 1 | .071 |
| N1‐3 | 1.310 (0.359‐4.786) | 3.682 (0.893‐15.190) | ||
| TNM stage | ||||
| I‐II | 1 | .969 | 1 | .855 |
| III‐IV | 1.030 (0.229‐4.630) | 0.879 (0.220‐3.508) | ||
| AKT3 in tumor | ||||
| Low | 1 | .009 | ||
| High | 3.912 (1.399‐10.935) | |||
| PIK3CA in tumor | ||||
| Low | 1 | .076 | ||
| High | 2.883 (0.895‐9.287) | |||
Bold indicates significant values.
Abbreviations: CI, confidence interval; DFS, disease‐free survival; HNSCC, head and neck squamous cell carcinoma; HR, hazard ratio; OS, overall survival; TNM, tumor‐node‐metastasis.
4. DISCUSSION
A detailed analysis of 279 HNSCC tumors in TCGA database has previously elucidated several gene alterations specific to HNSCC. 6 The list of frequently mutated genes is comprised of several tumor suppressor genes, including TP53, CDKN2A, FAT1, NOTCH1, KMT2D, NSD1, and TGFBR2, however PIK3CA was the only oncogene that was mutated (~14%) in HNSCC. In the present study, we focused on AKT, the major downstream effector of the PI3K‐AKT‐mTOR pathway. We observed a specific elevation of AKT3 expression in HNSCC samples obtained from TCGA database. Furthermore, AKT3 expression was highly associated with genes related to the immunosuppressive microenvironment when compared with the other isoforms. The knockdown approach revealed that AKT3 promotes the proliferative and immunosuppressive functions of HNSCC cells. Moreover, AKT3 expression in HNSCC samples correlated with immune cell infiltration and unfavorable survival when compared with PIK3CA. These findings suggested that AKT3 expression is a potential biomarker for predicting the immunoreactivity and prognosis of HNSCC. Furthermore, AKT3 is a potential candidate for molecular targeted therapy of HNSCC.
In the present study, we focused on the role of AKT3 among the known AKT isoforms. Although the specific role of each AKT isoform in malignant tumors remains to be comprehensively elucidated, there have been a few studies in the field of breast cancer. AKT3, but not AKT1 and AKT2, is reported as a critical regulator of the growth of triple‐negative breast cancers. 17 In contrast, several studies have reported the opposing functions of AKT1 and AKT2 in cell migration and invasion. 18 , 19 , 20 , 21 Furthermore, AKT3 reportedly promotes the proliferation of human prostate cancer cells through the upregulation of both total AKT and B‐Raf, and the downregulation of tuberous sclerosis complex (TSC) 1 and TSC2. 22 The overexpression of AKT3 in prostate cancer cells promoted the shift of the cell cycle from the G1 to the S phase and stimulated tumor growth in vivo. In line with these studies observed in breast and prostate cancers, we hypothesized that the elevated mRNA expression of AKT3 in TCGA HNSCC cohort might be related to the upregulated proliferation and growth activity of HNSCC. Accordingly, we knocked down AKT3/PIK3CA in HSC‐3 cells and revealed their impaired proliferation, cell cycle shift from the G2/M to G1/G0 phase, and increased apoptotic cells in not only PIK3CA knockdown cells, but also AKT3 knockdown cells. These results are consistent with previous reports on breast and prostate cancers, highlighting the implications of AKT3‐targeted therapy. Several phase I and phase II clinical trials targeting AKT are currently ongoing for numerous types of cancers; however, no AKT3 isoform‐specific inhibitors are clinically available. 13 Our TCGA analyses indicated the increased expression of AKT1 and AKT3, especially AKT3, but not AKT2, in HNSCC. This finding suggests that AKT3‐specific therapy may suppress AKT function more efficiently than the other isoforms. Clinical trials have mainly been performed with pan‐AKT inhibitors. 13 These pan‐AKT inhibitors can suppress all AKT isoforms, however, as mentioned above, opposing functions of AKT1 and AKT2 have been reported in breast cancers, indicating the unexpected effects of AKT1/AKT2 suppression, including the promotion of cell migration or invasion. Moreover, Stottrup C and colleagues reported that AKT3 is a key regulator of resistance to the AKT inhibitor, MK2206. 23 They established an MK2206‐resistant breast cancer cell line that markedly upregulated AKT3 expression. Knockdown of AKT3 in these cells restored sensitivity to MK2206. These findings suggest that isoform‐specific inhibition of AKT3 is more efficient than pan‐AKT inhibition or PIK3CA inhibition. Further characterization of AKT3‐specific roles and anti‐tumor effects of AKT3 inhibition is warranted.
In the present study, we also focused on the relationship between AKT3 and the immune microenvironment. TCGA analysis revealed that AKT3 expression was positively correlated with cytotoxic T cell and NK cell markers, including CD3E, CD8, and NCAM1. The AKT3 staining score in 72 HNSCC samples positively correlated with the infiltration of CD3+ T cells and CD56+ NK cells. The infiltration of both T cells and NK cells into the tumor stroma has been recognized as a favorable prognostic factor in HNSCC 24 , 25 , however the AKT3 staining score was an unfavorable prognostic factor for OS in 72 HNSCC samples. Accordingly, AKT3 in tumor cells may facilitate the dysfunction of tumor‐infiltrated cytotoxic cells or recruit an immunosuppressive population of T cells. The increased proliferation of T cells co‐cultured with AKT3 knockdown HNSCC cells, compared with control cells, highlights the suppressive effects of tumor AKT3 on T cell immunity. The decreased gene expression of PD‐1 ligands, CD274 and PDCD1LG2, which are negative regulators of activated T cells, 26 also indicates that AKT3 may suppress T cell functions by inducing these immune checkpoint molecules. Furthermore, the downregulation of proinflammatory and immunosuppressive cytokine genes, including IL1B, IL6, CXCL8, and TGFB1, in AKT3 knockdown cells clarifies the relationship between tumor AKT3 and T cell dyfunction. 27 , 28 , 29 , 30 Infiltration of other stromal cell types, such as TAMs and CAFs, may provide an alternate rationale, facilitating the dysfunction of effector cells. In TCGA analysis, AKT3 expression was positively correlated with both TAM‐related and CAF‐related genes. Moreover, the AKT3 staining score was positively correlated with the infiltration of both TAMs and CAFs in HNSCC samples. TAM has been recognized as a key stromal cell type that facilitates tumor progression through various mechanisms, including immunosuppression, angiogenesis, and tumor invasion. 31 , 32 , 33 CAF is also a known key stromal player that promotes tumor progression by contributing to epithelial‐to‐mesenchymal transition, immunosuppression, and angiogenesis. 34 , 35 , 36 We previously demonstrated that CAFs suppress T cell immunity and induce M2‐like TAMs. 37 , 38 In the tumor microenvironment of AKT3‐upregulated HNSCCs, an abundance of TAMs and CAFs may suppress the infiltration of cytotoxic T cells, resulting in dysfunction and exhaustion of T cells. The positive correlation between AKT3 expression and T cell exhaustion markers in TCGA database supports this prediction. Furthermore, a positive correlation between AKT3 expression and the expression of Treg‐related genes was observed in TCGA database. Tregs are an immunosuppressive subset of CD4+ T cells that suppresses anticancer immunity. 39 In the 72 HNSCC samples investigated in the present study, the highly infiltrated CD3+ T cells might consist of numerous Tregs. In line with these results, AKT3 may be a key regulator of the immunosuppressive functions of HNSCCs, resulting in shorter survival of patients with HNSCC. AKT3 expression may be a novel biomarker, reflecting both the complex immunosuppressive microenvironment and poor prognosis.
In conclusion, we demonstrated that AKT3 is a key regulator that modulates the proliferative and immunosuppressive functions of HNSCC. AKT3 is a potential candidate for a novel biomarker of the immunosuppressive microenvironment and shorter survival. The isoform‐specific inhibition of AKT3 could be developed as a novel cancer therapy that efficiently suppresses the PI3K‐AKT‐mTOR pathway.
DISCLOSURE
The authors declare no potential conflicts of interest.
Supporting information
Table S1‐S3
Table S1‐S3
ACKNOWLEDGMENTS
This work was supported in part by a Grant‐in‐Aid for Scientific Research (B) 20H03834 (KC) and Grant‐in‐Aid for Young Scientists 20K18243 (HT) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Takahashi H, Rokudai S, Kawabata‐Iwakawa R, et al. AKT3 is a key regulator of head and neck squamous cell carcinoma. Cancer Sci. 2021;112:2325–2334. 10.1111/cas.14911
REFERENCES
- 1. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941‐1953. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777‐789. [DOI] [PubMed] [Google Scholar]
- 4. Mehanna H, Beech T, Nicholson T, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta‐analysis of trends by time and region. Head Neck. 2013;35:747‐755. [DOI] [PubMed] [Google Scholar]
- 5. Faraji F, Zaidi M, Fakhry C, Gaykalova DA. Molecular mechanisms of human papillomavirus‐related carcinogenesis in head and neck cancer. Microbes Infect. 2017;19:464‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lui VW, Hedberg ML, Li H, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761‐769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marquard FE, Jücker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem Pharmacol. 2020;172:113729. [DOI] [PubMed] [Google Scholar]
- 9. Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473‐6488. [DOI] [PubMed] [Google Scholar]
- 10. Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372‐383. [DOI] [PubMed] [Google Scholar]
- 11. Hoxhaj G, Manning BD. The PI3K‐AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wadhwa B, Makhdoomi U, Vishwakarma R, Malik F. Protein kinase B: emerging mechanisms of isoform‐specific regulation of cellular signaling in cancer. Anticancer Drugs. 2017;28:569‐580. [DOI] [PubMed] [Google Scholar]
- 13. Song M, Bode AM, Dong Z, Lee MH. AKT as a therapeutic target for cancer. Cancer Res. 2019;79:1019‐1031. [DOI] [PubMed] [Google Scholar]
- 14. Amornphimoltham P, Sriuranpong V, Patel V, et al. Persistent activation of the Akt pathway in head and neck squamous cell carcinoma: a potential target for UCN‐01. Clin Cancer Res. 2004;10:4029‐4037. [DOI] [PubMed] [Google Scholar]
- 15. Shan ZZ, Chen PN, Wang F, Wang J, Fan QX. Expression of P‐EGFR and P‐Akt protein in esophageal squamous cell carcinoma and its prognosis. Oncol Lett. 2017;14:2859‐2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobsen K, Bertran‐Alamillo J, Molina MA, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun. 2017;8:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chin YR, Yoshida T, Marusyk A, Beck AH, Polyak K, Toker A. Targeting Akt3 signaling in triple‐negative breast cancer. Cancer Res. 2014;74:964‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chin YR, Toker A. The actin‐bundling protein palladin is an Akt1‐specific substrate that regulates breast cancer cell migration. Mol Cell. 2010;38:333‐344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu H, Radisky DC, Nelson CM, et al. Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc Natl Acad Sci USA. 2006;103:4134‐4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoeli‐Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539‐550. [DOI] [PubMed] [Google Scholar]
- 21. Arboleda MJ, Lyons JF, Kabbinavar FF, et al. Overexpression of AKT2/protein kinase Bbeta leads to up‐regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196‐206. [PubMed] [Google Scholar]
- 22. Lin HP, Lin CY, Huo C, et al. AKT3 promotes prostate cancer proliferation cells through regulation of Akt, B‐Raf, and TSC1/TSC2. Oncotarget. 2015;6:27097‐27112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stottrup C, Tsang T, Chin YR. Upregulation of AKT3 confers resistance to the AKT Inhibitor MK2206 in breast cancer. Mol Cancer Ther. 2016;15:1964‐1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bisheshar SK, De Ruiter EJ, Devriese LA, Willems SM. The prognostic role of NK cells and their ligands in squamous cell carcinoma of the head and neck: a systematic review and meta‐analysis. Oncoimmunology. 2020;9:1747345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T‐lymphocytes in squamous cell carcinoma of the head and neck: a systematic review and meta‐analysis. Oncoimmunology. 2017;6:e1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang Y, Chen M, Nie H, Yuan Y. PD‐1 and PD‐L1 in cancer immunotherapy: clinical implications and future considerations. Hum Vaccin Immunother. 2019;15:1111‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaplanov I, Carmi Y, Kornetsky R, et al. Blocking IL‐1β reverses the immunosuppression in mouse breast cancer and synergizes with anti‐PD‐1 for tumor abrogation. Proc Natl Acad Sci USA. 2019;116:1361‐1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsukamoto H, Fujieda K, Senju S, Ikeda T, Oshiumi H, Nishimura Y. Immune‐suppressive effects of interleukin‐6 on T‐cell‐mediated anti‐tumor immunity. Cancer Sci. 2018;109:523‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. David JM, Dominguez C, Hamilton DH, Palena C. The IL‐8/IL‐8R axis: a double agent in tumor immune resistance. Vaccines (Basel). 2016;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh SA, Li MO. TGF‐β: guardian of T cell function. J Immunol. 2013;191:3973‐3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noy R, Pollard JW. Tumor‐associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou J, Tang Z, Gao S, Li C, Feng Y, Zhou X. Tumor‐associated macrophages: recent insights and therapies. Front Oncol. 2020;10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang L, Zhang Y. Tumor‐associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puram SV, Tirosh I, Parikh AS, et al. Single‐cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611‐1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer‐associated fibroblasts. Nat Rev Cancer. 2020;20:174‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monteran L, Erez N. The dark side of fibroblasts: cancer‐associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front Immunol. 2019;10:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi H, Sakakura K, Kawabata‐Iwakawa R, et al. Immunosuppressive activity of cancer‐associated fibroblasts in head and neck squamous cell carcinoma. Cancer Immunol Immunother. 2015;64:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi H, Sakakura K, Kudo T, et al. Cancer‐associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget. 2017;8:8633‐8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression ‐ implications for anticancer therapy. Nat Rev Clin Oncol. 2019;16:356‐371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S3
Table S1‐S3


