Abstract
Hepatocellular carcinoma (HCC) is one of the most lethal cancers in humans. The inhibition of peptidyl‐prolyl cis/trans isomerase (Pin1) gene expression may have great potential in the treatment of HCC. N‐Acetylgalactosamine (GalNAc) was used to target the liver. Cholesterol‐modified antimicrobial peptide DP7 (DP7‐C) acts as a carrier, the GalNAc‐siRNA/DP7‐C complex increases the uptake of GalNAc‐siRNA and the escape of endosomes in hepatocytes. In addition, DP7‐C nanoparticles and hydrogel‐assisted GalNAc‐Pin1 siRNA delivery can effectively enhance the stability and prolong the silencing effects of Pin1 siRNA. In an orthotopic liver cancer model, the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel complex can potentially regulate Pin1 expression in hepatocellular carcinoma cells and effectively inhibit tumor progression. Our study proves that Pin1 siRNA is an efficient method for the treatment of HCC and provides a sustainable and effective drug delivery system for the suppression of liver cancer.
Keywords: DP7‐C, hepatocellular carcinoma, hydrogel, peptidyl‐prolyl cis/trans isomerase
Summary of the synthesis of the GalNAc‐siRNA/DP7‐C complex and the sustained release strategy of the thermosensitive hydrogel and the process of Pin1 siRNA entering the cells to induce Pin1 inhibition. Inhibition of Pin1 slows the process of tumor growth.
Abbreviations
- EEA1
early endosome antigen 1
- GalNAc
N‐acetylgalactosamine
- HCC
hepatocellular carcinoma
- LAMP1
lysosomal associated membrane protein 1
- Pin1
peptidyl‐prolyl cis/trans isomerase
- PLEL
PDLLA‐PEG‐PDLLA
- XPO5
Exportin5
1. INTRODUCTION
As the most common primary malignant tumor of the liver, hepatocellular carcinoma (HCC) is the second leading cause of cancer‐related death worldwide. 1 , 2 At present, only surgical resection and liver transplantation are considered to be effective treatments for HCC. 3 However, because of the high mortality rate, the overall survival rate of patients with HCC is still very low. 4 Therefore, more effective treatments need to be developed. Pin1, a key peptidyl‐prolyl cis/trans isomerase (PPIase) in cells, is strongly overexpressed in several tumors, including HCC. 5 It is well known that miRNAs are generally downregulated in many malignant tumors. 6 Exportin5 (XPO5) is one of the key proteins involved in miRNA maturation processes. The output of XPO5 is the rate‐limiting step in miRNA biogenesis. 7 Specifically, Pin1 inhibits the expression of miRNA and promotes the development of hepatocellular carcinoma (HCC). 8 In addition, Pin1 knockout can restore XPO5 function and miRNA biogenesis, therefore inhibiting HCC proliferation and migration. 9 These findings verify that Pin1 plays a central role in tumorigenesis and tumor development by activating and/or amplifying numerous cancer‐driving pathways. 5
RNA interference (RNAi) therapy has become a promising candidate therapy for many diseases since its discovery in 1998. 10 , 11 , 12 Small interfering RNA (siRNA) can inhibit gene expression in a highly specific way. Although siRNA is a powerful gene regulator, its rapid degradation in biological liquids and its inherent physical and chemical properties (high molecular weight, negative charge, and rigid structure) lead to poor cell uptake and low siRNA system delivery efficiency and intracellular transport, and it is difficult for siRNA to escape from the endoplasmic reticulum and be released into the cytoplasm. 13 , 14
To solve the problems and challenges above, we have successfully screened a cationic antibacterial peptide DP7 composed of 12 amino acids through computer simulation. Then cholesterol‐modified antimicrobial peptide DP7 (DP7‐C), 15 as a carrier, improved the transfection efficacy. The early methods of siRNA delivery mainly focused on lipid nanoparticles (LNPs) and synthetic nanoparticles. 16 , 17 The function of LNPs is to mask the charge of siRNA, prevent it from being degraded by RNase, and promote its endosomal escape into the cytoplasm.
In addition, using the biodegradable hydrogel, which is a three‐dimensional (3D) network composed of cross‐linked hydrophilic polymers, has great potential to support the transmission of biomolecules (including siRNA). 18 , 19 , 20 Recently, new injectable thermosensitive PDLLA‐PEG‐PDLLA (PLEL) copolymers were developed to achieve slow degradation over the course of 3‐4 weeks. 21 As a promising biomaterial, thermosensitive hydrogel has been extensively studied in sustained drug delivery, tissue regeneration, and post‐operative adhesion prevention. 22 In this study, PLEL solution is solution state at room temperature while spontaneously turning into the solid phase at physiological temperature. Its properties enable pharmaceutical agents to be easily incorporated into the hydrogel aqueous solutions through simply mixing. The corresponding formulations were then injected subcutaneously on the backs of mice to form a standing gel, which acted as a sustained drug delivery depot. At the same time, this chemical‐reaction free, and minimal invasiveness method is beneficial for medical application. The PLEL hydrogel can help to achieve a sustained release effect with a single dose. 23
In conclusion, our study provides a subcutaneous drug delivery system that can achieve a liver targeting effect for 21 d and effective cell transfection efficiency of siRNA, which opens up new possibilities for the treatment of liver‐related diseases.
2. MATERIALS AND METHODS
2.1. Material synthesis and characterization
DP7 was synthesized by the standard Fmoc solid‐phase peptide synthesis method on an automatic peptide synthesizer (CSBio 136XT); DP7‐C was obtained using the hydrophobic modification and purification of DP7 with cholesterol. PDLLA‐PEG‐PDLLA triblock copolymer hydrogel was provided by the State Key Laboratory of Biotherapy, West China Hospital, Sichuan University. 21 GalNAc‐Pin1 siRNA was synthesized by Bsyntech Co., Ltd., China.
The particle size and zeta potential of the DP7‐C diluent were measured using a Malvern laser particle size analyzer (Nano‐ZS 90, Malvern).
2.2. Complexation and characterization of GalNAc‐siRNA/DP7‐C
Briefly, GalNAc‐siRNA and DP7‐C were incubated at room temperature for 15 minutes for complexation at different mass ratios. The mass ratio between DP7‐C and GalNAc‐siRNA at which there was no uncomplexed GalNAc‐siRNA was determined by gel electrophoresis. 24 The size and zeta potential of the GalNAc‐siRNA/DP7‐C complex were characterized using a Malvern Zetasizer. The size of the complexes was examined by transmission electron microscopy (TEM; H‐6009IV Hitachi).
2.3. Dynamic rheological study of PDLLA‐PEG‐PDLLA solutions
The rheological properties of PDLLA‐PEG‐PDLLA copolymer solution at defined concentrations were measured using a HAAKE Rheostress 6000 rheometer (Thermo Scientific). During the temperature scanning experiment, the heating and cooling rates were set as 1°C/min. The change of storage modulus (G′) and loss modulus (G″) was measured.
2.4. Sustained release study in vitro
To investigate the release of GalNAc‐siRNA/DP7‐C in the PLEL hydrogel system, the release medium was collected and replaced with new PBS at a predetermined time. The standard curve was calculated according to the known concentration of FAM‐GalNAc‐siRNA/DP7‐C and the corresponding fluorescence intensity. The fluorescence intensity of FAM‐GalNAc‐siRNA in the medium was measured using a fluorescence microplate reader (Synergy H1), so the corresponding drug concentration was calculated.
2.5. Analysis of the cellular uptake of GalNAc‐Pin1 siRNA/DP7‐C
Hep3B cells were seeded in 6‐well plates at 2 × 105 cells per well 1 d prior to siRNA treatment. Hep3B cells were incubated with cell culture medium (untreated), free FAM‐labeled GalNAc‐siRNA (negative control), and FAM‐labeled GalNAc‐siRNA/Lipo 2000 (positive control). After 6 hours, the Hep3B cells were monitored for GalNAc‐siRNA delivery by fluorescence microscopy. In addition, Hep3B cells were harvested and analyzed on an Accuri C6 flow cytometer.
2.6. Cell viability assay
To verify the cytotoxicity of DP7‐C in normal hepatocytes, cell viability was measured with a cell counting kit‐8 (CCK‐8; Sigma, UK) assay. Briefly, LO2 cells were seeded in a 96‐well culture plate (8000 cells/well) and then treated with different concentrations of DP7‐C or Lipofectamine 2000 for 24 hours. Next, 10 µL of CCK‐8 solution (1 mg/mL) was added into each well. After a 1‐h incubation, the absorbance was measured at 450 nm.
2.7. Study of the mechanism of GalNAc‐siRNA/DP7‐C complex uptake in vitro
To study the pathway by which the GalNAc‐siRNA/DP7‐C complex is taken up by Hep3B cells, 3 × 104 Hep3B cells were seeded into 24‐well plates for 24 hours and then treated with 800 μmol/L amiloride, an inhibitor of the macropinocytosis pathway; 200 μmol/L genistein, an inhibitor of the caveolin‐mediated endocytosis pathway; or 10 μmol/L chlorpromazine, an inhibitor of the clathrin‐mediated endocytosis pathway, for 2 hours. Next, Cy3‐GalNAc‐siRNA/DP7‐C was added to the cells for 4 hours. The cells were then collected for flow cytometry analysis.
2.8. Confocal microscopy analysis
To further explore the process of GalNAc‐siRNA/DP7‐C complex transport through early endosomes, late endosomes, and lysosomes, the time‐dependent colocalization of different transport stages with specific markers, such as early endosome antigen 1 (EEA1) and lysosomal associated membrane protein 1 (LAMP1) for late endosomes, was evaluated. Hep3B cells were incubated with FAM‐GalNAc‐siRNA/DP7‐C or FAM‐GalNAc‐siRNA/Lipo 2000 for 4 hours or 22 hours, respectively. Then, the cells were fixed in 4% paraformaldehyde for at least 20 minutes at room temperature. After washing with PBS, the cells were permeabilized with 0.1% Triton for 15 minutes at room temperature. Then, the samples were blocked with 0.05% BSA/PBS blocking buffer for 20 minutes. The transfected cells were then incubated for 2 hours with anti‐EEA1 (1:100, Invitrogen) and anti‐LAMP‐1‐PE (1:100, Invitrogen) antibodies in PBS. Then, the cells were labeled with monkey anti‐rabbit PE‐conjugated secondary antibodies (1:100, for 1 hours, at room temperature). The coverslips were gently washed with distilled water and mounted onto slides using Antifade Mounting Medium with DAPI (Solarbio), and the samples were analyzed by laser confocal microscopy (Olympus).
LysoTracker Red (Beyotime Biotechnology) was used to detect lysosomes. After transfection 4 hours or 22 hours later, Hep3B cells were subjected to LysoTracker Red staining at 37°C for 2 hours. Then, the cells were fixed with 4% paraformaldehyde, sealed with Antifade Mounting Medium with DAPI (Solarbio) and imaged using laser confocal microscopy (Olympus).
2.9. qPCR and western blot
Hep3B cells were seeded in 6‐well plates at 2 × 105 cells/well. DP7‐C and GalNAc‐Pin1 siRNA complex was incubated with the Hep3B cells at doses of 1 μg GalNAc‐Pin1 siRNA. Hep3B cells were harvested 48 hours later for analysis. Total RNA was extracted with a kit (ForeGene). qPCR reactions were performed on a Bio‐Rad CFX96 system.
The western blot results better showed the silencing efficiency of the siRNA. The protein samples were probed with anti‐GAPDH (Cell Signaling Technology, CST), anti‐Pin1 (CST), anti‐Beta Tubulin (CST), anti‐Lamin B (CST), and anti‐Exportin 5 (CST) antibodies. The cells were incubated with primary antibodies at 4°C overnight. Then, the membranes were incubated with secondary antibodies (CST) for 60 minutes and developed using the ECL detection system (Vazyme).
2.10. Analysis of sustained silencing mediated by hydrogel‐released siRNA/DP7‐C in vitro
Similar to the gene silencing experiments, Hep3B cells were seeded into 6‐well plates at 2 × 105 cells/well 1 d before treatment. For the GalNAc‐Pin1 siRNA/DP7‐C‐treated groups, 4 µg DP7‐C complexed with 1 µg GalNAc‐Pin1 siRNA was used to treat cells. For the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel‐treated groups, GalNAc‐Pin1 siRNA (1 µg) was complexed with 4 µg DP7‐C and trapped in the hydrogel (100 µl, 10%). The hydrogels and complexes were placed in transwell inserts with 800 µl of cell culture medium, the medium was replaced, and the cells were collected every 3 d. RNA extraction, cDNA synthesis, and RT‐PCR were performed as previously described. The expression of Pin1 and some other mRNAs was quantified based on the reaction efficiency, normalized to the expression of β‐actin and compared with the expression in the untreated negative control cells. 25
2.11. In vivo sustained release of GalNAc‐siRNA/DP7‐C/hydrogel
BALB/c nude mice (4‐6 weeks old, female) were purchased from Beijing Vital River Laboratory Animal Co., Ltd. All animal experiments complied with the ARRIVE guidelines. Cy3‐labeled GalNAc‐siRNA complexed with DP7‐C was loaded onto the hydrogel. Here 300 μL of the drug delivery system, which included 150 µL of 20% hydrogel, 60 µg of siRNA (2 µg/µL) 26 and 240 µg of DP7‐C (2 µg/µL), was subcutaneously injected into the backs of the mice. In other groups, local injection of Cy3‐GalNAc‐siRNA, Cy3‐GalNAc‐siRNA/DP7‐C and Cy3‐GalNAc‐siRNA/hydrogel at the same site was used as a control. Fluorescence intensity was monitored longitudinally using an IVIS imaging system (PerkinElmer IVIS Lumina III), and IVIS analysis was performed on days 0, 1, 3, 7, 14, and 21 after injection.
2.12. Animal orthotopic liver tumor models
All the mice were administered 1% pentobarbital at a dose of 60 mg/kg by intraperitoneal injection. 27 The Hep3B‐luciferase cells (1 × 106 cells in 10 μL serum‐free DMEM) were implanted into the liver using a microinjector. At 2 weeks after tumor cell implantation, all the mice were randomly assigned to different groups (6 per group) and administered GalNAc‐Pin1 siRNA at a dose of 60 µg per mouse. 26 The day of injection was recorded as day 0.
2.13. Healing efficiency of orthotopic liver tumors treated with GalNAc‐Pin1 siRNA/DP7‐C/hydrogel in vivo
Tumor models were established as described in the previous sections. GalNAc‐Pin1 siRNA complexed with DP7‐C (60 µg GalNAc‐Pin1 siRNA 26 ) was loaded into 300 µL of hydrogel (10%) and implanted by subcutaneous injection into the backs of mice, as previously described. Untreated tumors were used as negative controls. Additional controls included groups that contained those treated with DP7‐C alone, GalNAc‐scramble siRNA/DP7‐C, GalNAc‐Pin1 siRNA/DP7‐C, GalNAc‐scramble siRNA/DP7‐C/hydrogel and GalNAc‐Pin1 siRNA/DP7‐C/hydrogel.
2.13.1. Xenogen IVIS imaging
The mice received 200 µL of d‐luciferin by intraperitoneal injection and, 10 minutes later, the mice were anesthetized by isoflurane (RuiWD). The total counts of the emitted light were measured and reflected the relative luciferase expression of the cells in the tumor. The data were analyzed by Living Imaging software.
2.13.2. Histological analysis
The mice were sacrificed 3 weeks after treatment. Tissues were fixed in 4% formaldehyde for at least 48 hours. Tissues were subsequently cut into small pieces, embedded in paraffin, and cut into 5‐µm sections. HE and TUNEL staining were used to observe the pathological changes of the liver tissues in the different groups.
For frozen sections, parts of liver were embedded in optimal cutting temperature (OCT) medium, and cut into 5‐μm sections. The sections were fixed in 4% formaldehyde for 20 minutes and mounted onto slides using Antifade Mounting Medium with DAPI.
2.14. Statistical analysis
All the data were statistically analyzed to express the mean value ± standard deviation (SD) of the mean. Statistical significance was evaluated at a level of P < .05 using T test analysis with *, ** and *** indicating statistical significance P < .05, P < .001 and P < .0001, respectively.
3. RESULTS
3.1. The characterization of DP7‐C and hydrogel
Gel retardation experiments showed that the best mass ratio of DP7‐C and GalNAc‐siRNA was 4:1 (Figure 1B). The particle size distribution of DP7‐C was approximately 36.08 ± 2.792 nm, the zeta potential of DP7‐C was approximately 37.7 ± 6.68 mV, and the polydispersity index was 0.131. After incubating DP7‐C with GalNAc‐siRNA, the particle size was larger than that of DP7‐C alone, while the zeta potential was lower than that of DP7‐C alone, indicating that DP7‐C can form complexes with GalNAc‐siRNA (Figure 1C,D). The shape of GalNAc‐siRNA/DP7‐C was spherical, and the size was similar to its particle size, as shown by TEM, Figure 1E). The particle size distribution of the complex was detected on the first day, the seventh day, and the twenty‐first day. The particle size did not change significantly (Figure 1F). The temperature of gelation was proved by testing the rheology of G′ and G″, and the hydrogel changes from liquid to solid state at approximately 36.37°C (Figure 1F). As Figure 1H shows, the sustained release effect of drugs in hydrogels can be up to 21 d or longer.
FIGURE 1.
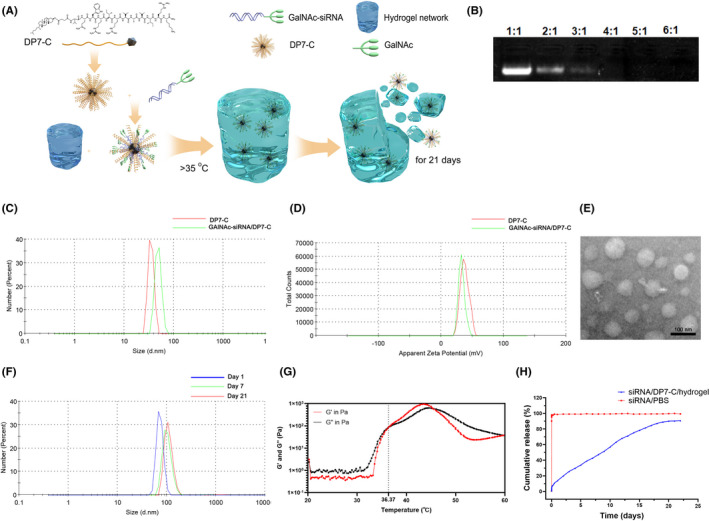
The characterization of DP7‐C and hydrogel. A, Schematic diagram. B, Gel retardation analysis. C, D, Particle size and zeta potential distributions of DP7‐C and the GalNAc‐siRNA/DP7‐C complexes. E, The morphology of GalNAc‐siRNA/DP7‐C complexes was evaluated by TEM. Scale bar, 50 nm. F, The stability of GAlNAc‐siRNA/DP7‐C. G, Temperature dependence curves of storage modulus (G′) and loss modulus (G″) of copolymer aqueous solution (20%). H, Study on drug release properties of hydrogel
3.2. The transfection efficiency of DP7‐C and its cytotoxicity in liver cells
The fluorescence microscopy and flow cytometry results showed that, compared with cells treated with free GalNAc‐siRNA and GalNAc‐siRNA/Lipo 2000, the GalNAc‐siRNA/DP7‐C group exhibited significantly higher median fluorescence intensities (Figure 2A‐D). According to the results of the flow cytometry, DP7‐C had higher transfection efficiency and stronger fluorescence intensity than Lipo (Figure S1).
FIGURE 2.
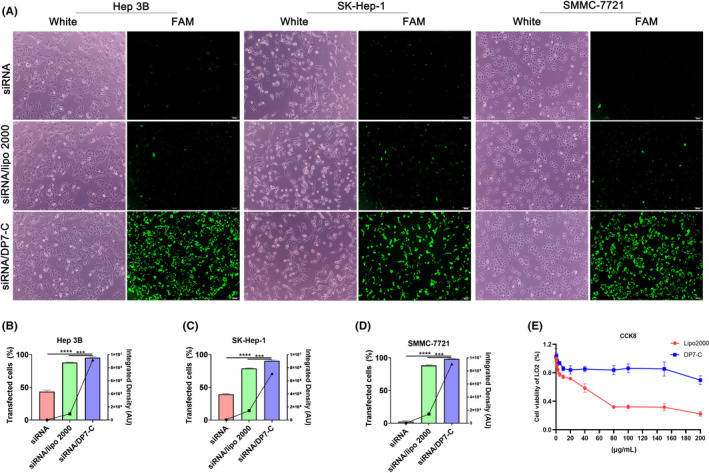
The transfection results of DP7‐C in different cells and its cytotoxicity in LO2 cells. A, Fluorescence images of Hep3B cells, SK‐Hep‐1 cells, and SMMC‐7721 cells transfected with FAM‐labeled GalNAc‐siRNA. B‐D, Cells successfully transfected with FAM‐labeled GalNAc‐siRNA were analyzed by flow cytometry, and the data were converted into digital form for statistical analysis (bar graph, corresponding to the left Y‐axis). The fluorescence intensity value was calculated and is expressed as a line graph, corresponding to the Y‐axis on the right. E, CCK‐8 assay of DP7‐C and Lipo‐treated LO2 cells. Data are expressed as the mean ± SEM. *** P < .001, **** P < .0001
Subsequently, we evaluated the cytotoxicity of DP7‐C in the liver cell line LO2. However, the cell survival rate gradually decreased after applying the same concentration of Lipofectamine 2000, the survival rate decreased by approximately 70% after applying 200 μg/mL Lipofectamine 2000, and the survival rate decreased by approximately 30% after applying 200 μg/mL DP7‐C (Figure 2E). The results indicated that DP7‐C has low toxicity in normal cells.
3.3. The intracellular fate of GalNAc‐siRNA/DP7‐C‐containing compartments
The pathways for extracellular substance uptake are mainly divided into macropinocytosis, caveolin‐mediated, and clathrin‐mediated endocytosis 28 (Figure 3A). Our results indicated that the uptake efficiency was significantly reduced after treatment with chlorpromazine, an inhibitor of the clathrin pathway (Figure 3B). However, the transfection efficiency of Lipo2000 was most significant with the inhibitor of clathrin (Figure 3D). After internalization, GalNAc‐siRNA/DP7‐C composite nanoparticles were transported in turn through early endosomes, late endosomes, and lysosomes. 29 At 6 h after GalNAc‐siRNA transfection by DP7‐C, FAM‐labeled GalNAc‐siRNA (green) colocalized with early endosomes (red) and late endosomes (red), as well as with lysosomes, which exhibited an obvious yellow signal (Figure 3C). However, only a few points of colocalization could be detected 24 hours after transfection. For gene‐based drugs, GalNAc‐siRNA released from lysosomes into the cytoplasm can play an effective role in the silencing of target genes (Figure 3D).
FIGURE 3.
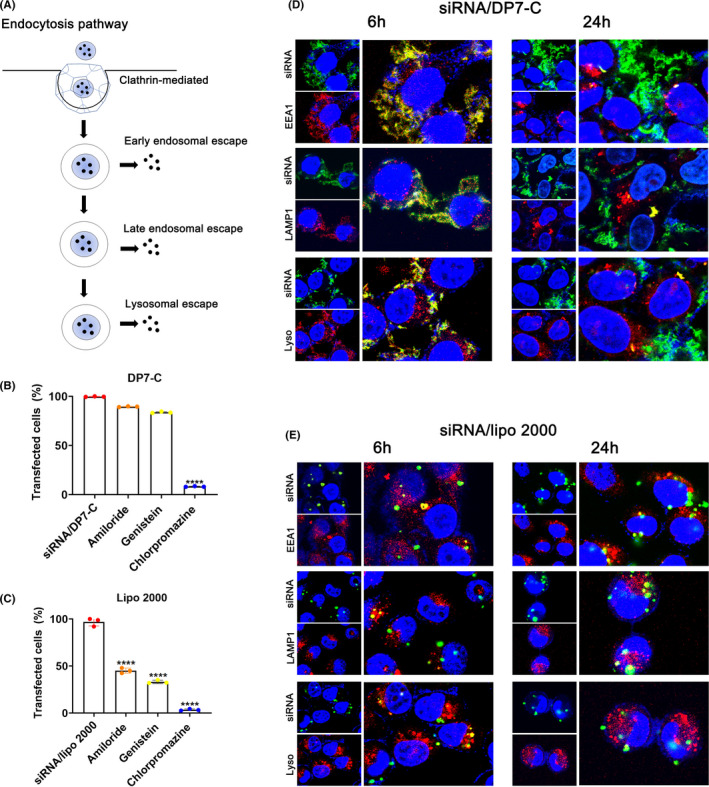
The pathway of GalNAc‐siRNA/DP7‐C uptake and the escape process after internalization of GalNAc‐siRNA/DP7‐C in Hep3B cells. A, Schematic diagram of cellular uptake and endosomal escape. B, Uptake pathway inhibitor experiments of DP7‐C. C, Uptake pathway inhibitor experiments of Lipo2000. D, E, Hep3B cells were stained with EEA1 (red, left panels), LAMP1 (red) and lysosome (red, left panels). Blue represents the localization of the DAPI‐stained nuclei, and yellow indicates the colocalization of siRNA and endosomes. Data are expressed as the mean ± SEM. **** P < .0001
3.4. Gene silencing of Pin1 in vitro
According to previous reports, the isomerization of Pin1 isomerase results in the retention of XPO5 in the nucleus and the downregulation of key tumor suppressive miRNAs 9 , 30 (Figure 3A). GalNAc‐Pin1 siRNA/DP7‐C exposure resulted in a 52% reduction in mRNA abundance (Figure 4B) and a certain degree of inhibition of Pin1 at the protein level (Figure 4C).
FIGURE 4.
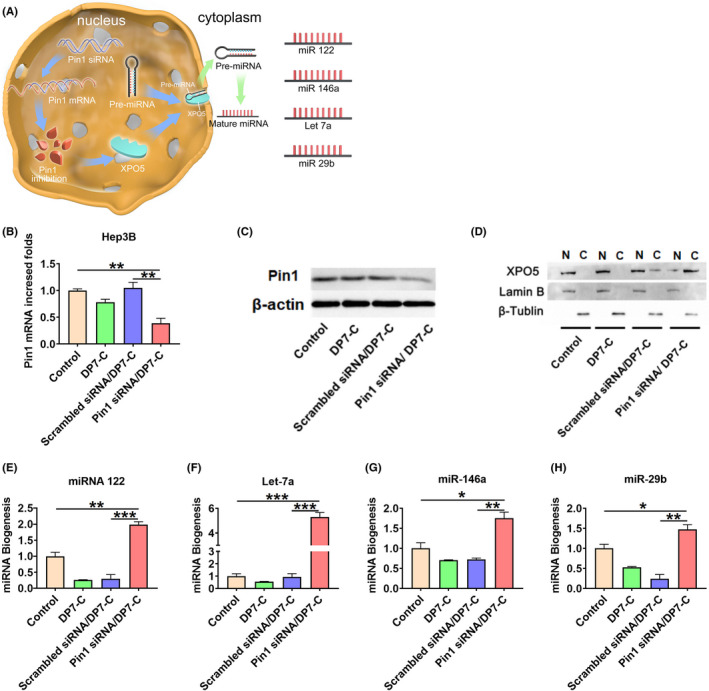
Effective suppression of Pin1 in Hep3B cells by GalNAc‐Pin1 siRNA/DP7‐C transfection in vitro. A, Schematic diagram. B, Pin1 mRNA expression in Hep3B cells quantified by qPCR. C, Pin1 protein expression in Hep3B cells quantified by western blot. D, Western blot assay of XPO5 subcellular distribution in Hep3B cells. Expression of related mature miRNAs, including (E) miR‐122, (F) miRNA Let‐7a, (G) miR‐146a and (H) miR‐29b, was analyzed using qPCR. Data are expressed as the mean ± SEM. * P < .05, ** P < .01, *** P < .001
The western blotting results showed that Pin1 siRNA could restore the nucleus‐to‐cytoplasm export ability of XPO5 (Figure 4D). Considering that XPO5 plays an important role in the cytoplasmic transport of pre‐miRNAs, we investigated whether Pin1 siRNA could affect the biogenesis of mature miRNAs. Based on the abundance and important biological activities of miR‐122, let‐7a, mir‐29b, and miR‐146a, the biogenesis of these miRNAs was examined. miR‐122 is the most abundant miRNA in the adult liver and plays a significant role. 31 , 32 The let‐7 family inhibits the proliferation of HCC cells by downregulating c‐myc and upregulating p16 (INK4a). 33 , 34 In addition, miR‐29b inhibits the expression of tumor suppressor genes 35 and activates the expression of tumor suppressor genes. qPCR results showed that Pin1 siRNA significantly increased the expression of these miRNAs in tumor cells compared with the control treatment (Figure 4E‐H). Therefore, Pin1 siRNA has the advantage of inhibiting the growth of HCC cells by repairing miRNA biogenesis.
3.5. Sustained gene silencing ability of DP7‐C/Pin1 siRNA released from hydrogel in vitro
To show that the DP7‐C/hydrogel system is capable of sustained siRNA‐mediated gene silencing, Hep3B cells were treated in vitro with hydrogel‐released DP7‐C/Pin1 siRNA in Transwell 6‐well plates. qPCR analysis showed that GalNAc‐Pin1 siRNA/DP7‐C provided a transient Pin1 knockdown of 26% at day 1 (Figure 5A). By day 7, Pin1 expression was reduced to 15%. For the cells treated with the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel, at day 3, Pin1 expression was reduced to 43% in the control Hep3B cells. At day 7, the siRNA/DP7‐C/hydrogel‐treated cells maintained a low level of Pin1 expression (2.59%), which was significantly different from the expression in the negative control cells. Sustained knockdown was consistent with the time course of siRNA/DP7‐C release.
FIGURE 5.
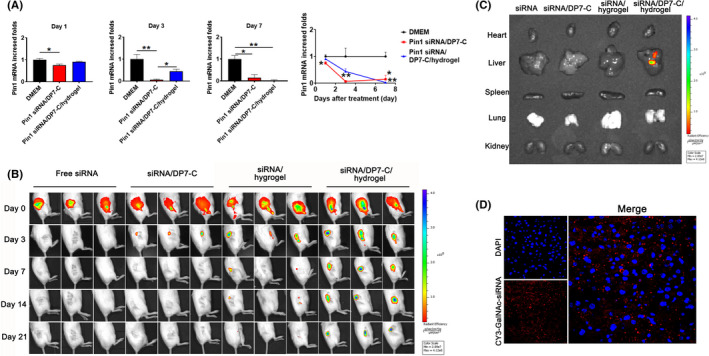
Sustained gene silencing ability of GalNAc‐Pin1 siRNA DP7‐C released from hydrogels in vitro and the sustained release effect of hydrogels in vivo. A, Sustained knockdown of Pin1 in the Hep3B cells treated with GalNAc‐Pin1‐siRNA/DP7‐C, n = 3. B, Cy3‐GalNAc‐siRNA/DP7‐C and hydrogels were colocalized. C, The viscous tissue was observed through in vivo fluorescence imaging on day 21. D, The infiltration of siRNA into liver cells. Data are expressed as the mean ± SEM. * P < .05, ** P < .01
3.6. Localization and sustained release of GalNAc‐siRNA/DP7‐C/hydrogel in vivo
The controlled release behavior was further investigated in vivo. Injections of GalNAc‐siRNA led to rapid clearance from the injection site before day 3, GalNAc‐siRNA/DP7‐C persisted at the injection site for more than 3 d, and GalNAc‐siRNA released from the hydrogel without DP7‐C showed a shorter persistence time of 14 d, siRNA/DP7‐C released from the hydrogel persisted at the injection site for 21 d (Figure 5B). We sacrificed the mice on the twenty‐first day and immediately took the viscera of the mice to perform IVIS imaging. The results showed that fluorescence could only be seen in the livers of the GalNAc‐siRNA/DP7‐C/hydrogel group (Figure 5C). Subsequently, the liver was immediately frozen and sectioned. After DAPI staining, under a confocal microscope GalNAc‐siRNA (red) was observed in the sections (Figure 5D). This finding not only proved the targeting of our GalNAc‐siRNA to the liver but also indicated the effective sustained release of the drug delivery system.
3.7. GalNAc‐Pin1 siRNA/DP7‐C/hydrogel suppresses tumor growth in mice
In vivo imaging was performed every 3 d to monitor tumor growth (Figure 6A), before day 9 the group treated with GalNAc‐Pin1 siRNA/DP7‐C showed more effective inhibition, which was similar to the in vitro data. However, throughout the rest of the experiment, the hydrogel group displayed the best inhibitory effect (Figure 6B). By contrast, the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel‐treated tumors exhibited approximately 8.2‐fold decreased counts compared with the control‐treated tumors (Figure 6C). H&E staining was performed on liver tissues to observe the pathological changes in the liver after treatment (Figure 6D). Taken together, these data indicated that the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel suppressed liver tumor growth in a hypodermic way and that the hydrogel prolonged the inhibitory effect.
FIGURE 6.
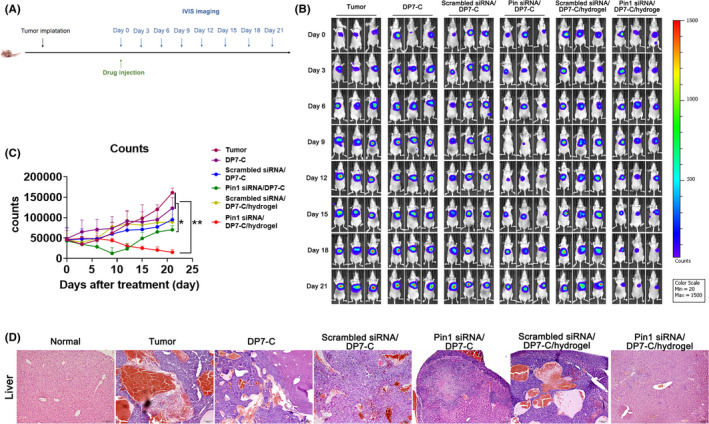
Administration of siRNA/DP7‐C/hydrogel extended antitumour effects. A, Experimental setup. B, Live images were taken every 3 d to observe the growth of the tumors. C, Quantification of tumor growth based on IVIS imaging. Data show the total radiant efficiency of the drawn region of interest (ROI) in the IVIS images normalized to day 0. Mean ± SD, n = 6. D, Pathological changes in the livers of the mice were observed by H&E staining. Data are expressed as the mean ± SEM. * P < .05, ** P < .01
An additional experiment was conducted to evaluate the function of DP7‐C in vivo, as shown in Figure S4A. IVIS imaging was performed every 3 d to monitor tumor growth (Figure S4B), siRNA/hydrogel group and siRNA/DP7‐C/hydrogel treatment group showed the same tumor inhibition effect in the first 9 d. By contrast, the siRNA/DP7‐C/hydrogel group showed a sustained and effective treatment effect in the whole treatment process. We explored the effect of hydrogel sustained release in vivo initially (Figure 5B), on day 14, only a small amount of siRNA was found in the siRNA/hydrogel group, which may be the reason for the poor therapeutic effect of siRNA/hydrogel group in the later stage. From the statistical results of photon counts (Figure S4C) and H&E staining (Figure S4D) results, it was obvious that the therapeutic effect of Pin1 siRNA/DP7‐C/hydrogel group was significantly better than that of the group without DP7‐C (siRNA/hydrogel), which showed the superiority of DP7‐C in the treatment of liver cancer in situ model mice.
3.8. Pin1 siRNA promotes miRNA expression to suppress HCC development
Consistent with the mechanism revealed in vitro, Pin1 mRNA expression was downregulated in the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel group, and tumor suppressor miRNAs were obviously increased (Figure 7A). We also evaluated the apoptosis of Hep3B carcinoma cells in an in situ model. The apoptotic cells in the GalNAc‐Pin1 siRNA/DP7‐C/hydrogel group were significantly increased (Figure 7B,C).
FIGURE 7.
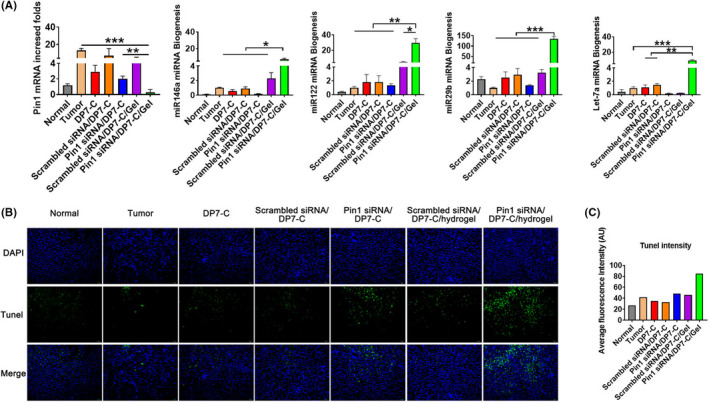
Pin1 siRNA promotes miRNA expression to suppress HCC development. A, The expression levels of Pin1, miR‐122, miR‐146a, miR‐29b, and let 7a were detected in Hep3B xenografts in nude mice. B, Tumor‐bearing mouse liver sections were observed by TUNEL staining; scale bar, 100 mm. C, Fluorescence intensity analysis of TUNEL staining (middle panel in Figure 7B). Data are expressed as the mean ± SEM. * P < .05, ** P < .01, *** P < .001
3.9. Preliminary evaluation of the safety
The weight of the tumor‐bearing mice in the control groups significantly decreased, while there was no significant weight change in other control groups (Figure S2). H&E staining showed that there was no abnormal or necrotic tissue except for some metastases in the lung (Figure S3). In addition, no adverse reactions, such as vomiting or diarrhea (data not shown).
4. DISCUSSION
In this study, we developed a gene regulator based on siRNA, namely, the GalNAc‐siRNA/DP7‐C/hydrogel system, to effectively inhibit HCC progression by depleting Pin1.
The incidence of HCC has risen in recent decades. Unfortunately, most patients with HCC are diagnosed at an advanced stage. Therefore, there is a great demand for an effective treatment. miRNAs are closely related to oncogenes and tumor suppressor genes, however they are downregulated in many tumors, including HCC. For example, miR‐122 deletion increased the tumor incidence rate, 36 , 37 injection of miR‐122 mimics resulted in suppression of xenograft growth, 38 and c‐myc transcription in the liver was negatively regulated by miR‐122. 39 miR‐29b reduces the Warburg effect of ovarian cancer by downregulating Akt expression 40 and induces apoptosis of myeloma cells by targeting MCL‐1. Previous studies have shown that Pin1 may activate oncogenes or growth‐promoting factors and promote HCC development by inhibiting miRNA expression by limiting XPO5 transport out of the nucleus, 9 which provides another mechanism for Pin1‐mediated signal transmission in cancer. This mechanism indicates that inhibiting Pin1 to enhance miRNA biogenesis is a promising strategy for HCC treatment. 41 Pin1 is involved in a variety of cellular processes, including cell cycle, 42 differentiation, senescence, and apoptosis. 43 We provide possible experimental support for the inhibition of the occurrence and development of cancer by the inhibition of the effect of Pin1 on miRNA.
siRNA has the advantages of good targeting and minimal side effects on normal tissues and cells, 44 , 45 while the rapid degradation of siRNA by serum nucleases and low cellular uptake reduce its effectiveness in vivo. 46 , 47 Therefore, related research has focused on the development of vectors to enhance its cellular uptake and endosomal escape. 48 GalNAc binding to ASGPR occurs at the hepatocyte surface, 49 and then, the rapid, local aggregation of the ligand binding to its receptor leads to its greater aggregation in clathrin‐coated pits and its entry by endocytosis. 50 Subsequent studies showed that acidification during endosomal maturation led to dissociation of the GalNAc ligand from ASGPR, and then GalNAc was degraded in the lysosome. 51 We have developed a novel nanoparticle system for the systemic delivery of siRNA, namely, DP7‐C, an amphiphilic and cationic lipid, with the assistance of biocompatible and biodegradable hydrogels. As described in our schematic diagram, on the one hand, DP7‐C can deliver GalNAc‐siRNA into cells with high transfection efficiency. On the other hand, DP7‐C promotes the rapid escape of GalNAc‐siRNA, therefore allowing siRNA to play a better role in gene silencing.
To prolong the efficacy of siRNA, siRNA‐complexed nanoparticles (GalNAc‐Pin1 siRNA/DP7‐C) were entrapped within the PLEL polymer hydrogel, the release of embedded GalNAc‐Pin1 siRNA/DP7‐C as the mesh size increases. As these hydrogels hydrolytically degrade, the secondary retrieval of hydrogel materials is unnecessary. For therapeutic schedules that require local, liver‐targeted, and sustained delivery of siRNA, our system can adequately achieve the continuous delivery of siRNA to the liver. However, this method did not show significant advantages in the first 9 d after treatment, which may reduce the efficacy of siRNA in the early stage of the treatment process. It may be possible to achieve better control by changing the weight percentage of the hydrogel.
In conclusion, we designed a DP7‐C/hydrogel as a siRNA delivery platform to target tumor suppressor miRNA interactions through Pin1 depletion to treat liver cancer. The GalNAc‐Pin1 siRNA/DP7‐C/hydrogel system supplies Pin1 siRNA with not only a protective structure against poor biological conditions but also an opportunity for passively targeting tumor tissue. This system can effectively inhibit the development of tumors without obvious adverse reactions or toxicity, and the delivery of Pin1 siRNA by DP7‐C/hydrogel is a promising treatment for HCC.
DISCLOSURE
This manuscript, or any part of it, has not been published and will not be submitted elsewhere for publication while being considered. The authors declare that there are no competing financial interests.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Science and Technology Major Project (No. 2018ZX09201018‐013 and No. 2018ZX09733001‐001‐007), “Leading Academic of Ten‐thousand Talents Program” and 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University. All the funding from these sources were used for data collection and analysis.
Zhao B, Zhou B, Shi K, et al. Sustained and targeted delivery of siRNA/DP7‐C nanoparticles from injectable thermosensitive hydrogel for hepatocellular carcinoma therapy. Cancer Sci. 2021;112:2481–2492. 10.1111/cas.14903
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2. Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Discovery. 2019;18:13‐14. 10.1038/nrd.2018.146 [DOI] [PubMed] [Google Scholar]
- 3. Kim DW, Talati C, Kim R. Hepatocellular carcinoma (HCC): beyond sorafenib‐chemotherapy. J Gastrointest Oncol. 2017;8:256‐265. 10.21037/jgo.2016.09.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang G, Zhang GY. Upregulation of FoxP4 in HCC promotes migration and invasion through regulation of EMT. Oncol Lett. 2019;17:3944‐3951. 10.3892/ol.2019.10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer‐driving pathways and is a unique drug target. Nat Rev Cancer. 2016;16:463‐478. 10.1038/nrc.2016.49 [DOI] [PubMed] [Google Scholar]
- 6. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834‐838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 7. Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220‐226. 10.1261/rna.7233305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun HL, Cui R, Zhou JK, et al. ERK activation globally downregulates miRNAs through phosphorylating exportin‐5. Cancer Cell. 2016;30:723‐736. 10.1016/j.ccell.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Pu W, Sun H‐L, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018;25:1612‐1624. 10.1038/s41418-018-0065-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis ME, Zuckerman JE, Choi CHJ, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067‐1070. 10.1038/nature08956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar P, Ban H‐S, Kim S‐S, et al. T cell‐specific siRNA delivery suppresses HIV‐1 infection in humanized mice. Cell. 2008;134:577‐586. 10.1016/j.cell.2008.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte‐directed siRNA delivery revealing cyclin D1 as an anti‐inflammatory target. Science. 2008;319:627‐630. 10.1126/science.1149859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li JM, Zhao M‐X, Su H, et al. Multifunctional quantum‐dot‐based siRNA delivery for HPV18 E6 gene silence and intracellular imaging. Biomaterials. 2011;32:7978‐7987. 10.1016/j.biomaterials.2011.07.011 [DOI] [PubMed] [Google Scholar]
- 14. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129‐138. 10.1038/nrd2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang R, Tang L, Tian Y, et al. Cholesterol‐modified DP7 enhances the effect of individualized cancer immunotherapy based on neoantigens. Biomaterials. 2020;241:119852. 10.1016/j.biomaterials.2020.119852 [DOI] [PubMed] [Google Scholar]
- 16. Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518‐6548. 10.1093/nar/gkw236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zimmermann TS, Lee ACH, Akinc A, et al. RNAi‐mediated gene silencing in non‐human primates. Nature. 2006;441:111‐114. 10.1038/nature04688 [DOI] [PubMed] [Google Scholar]
- 18. Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Hydrogels in regenerative medicine. Adv Mater. 2009;21:3307‐3329. 10.1002/adma.200802106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shapira K, Dikovsky D, Habib M, Gepstein L, Seliktar D. Hydrogels for cardiac tissue regeneration. Biomed Mater Eng. 2008;18:309‐314. [PubMed] [Google Scholar]
- 20. Krebs MD, Alsberg E. Localized, targeted, and sustained siRNA delivery. Chemistry. 2011;17:3054‐3062. 10.1002/chem.201003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi K, Wang Y‐L, Qu Y, et al. Synthesis, characterization, and application of reversible PDLLA‐PEG‐PDLLA copolymer thermogels in vitro and in vivo. Sci Rep. 2016;6:19077. 10.1038/srep19077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ko DY, Shinde UP, Yeon B, Jeong B. Recent progress of in situ formed gels for biomedical applications. Prog Polym Sci. 2013;38:672‐701. 10.1016/j.progpolymsci.2012.08.002 [DOI] [Google Scholar]
- 23. Shi K, Liu Z, Yang C, et al. Novel biocompatible thermoresponsive poly(N‐vinyl caprolactam)/clay nanocomposite hydrogels with macroporous structure and improved mechanical characteristics. ACS Appl Mater Interfaces. 2017;9:21979‐21990. 10.1021/acsami.7b04552 [DOI] [PubMed] [Google Scholar]
- 24. Malcolm DW, Sorrells JE, Van Twisk D, Thakar J, Benoit DS. Evaluating side effects of nanoparticle‐mediated siRNA delivery to mesenchymal stem cells using next generation sequencing and enrichment analysis. Bioeng Transl Med. 2016;1:193‐206. 10.1002/btm2.10035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu W, Saint DA. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal Biochem. 2002;302:52‐59. 10.1006/abio.2001.5530 [DOI] [PubMed] [Google Scholar]
- 26. Foster DJ, Brown CR, Shaikh S, et al. Advanced siRNA designs further improve in vivo performance of GaINAc‐siRNA conjugates. Mol Ther. 2018;26:708‐717. 10.1016/j.ymthe.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang XH, Ren L‐S, Wang G‐P, et al. A new method of establishing orthotopic bladder transplantable tumor in mice. Cancer Biol Med. 2012;9:261‐265. 10.7497/j.issn.2095-3941.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gestin M, Dowaidar M, Langel U. Uptake mechanism of cell‐penetrating peptides. Adv Exp Med Biol. 2017;1030:255‐264. 10.1007/978-3-319-66095-0_11 [DOI] [PubMed] [Google Scholar]
- 29. Gilleron J, Querbes W, Zeigerer A, et al. Image‐based analysis of lipid nanoparticle‐mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638‐646. 10.1038/nbt.2612 [DOI] [PubMed] [Google Scholar]
- 30. Pu W, Li J, Zheng Y, et al. Targeting Pin1 by inhibitor API‐1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology. 2018;68:547‐560. 10.1002/hep.29819 [DOI] [PubMed] [Google Scholar]
- 31. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR‐122–a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448‐457. 10.1016/j.jhep.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 32. Fornari F, Gramantieri L, Giovannini C, et al. MiR‐122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761‐5767. 10.1158/0008-5472.CAN-08-4797 [DOI] [PubMed] [Google Scholar]
- 33. Ji J, Zhao L, Budhu A, et al. Let‐7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690‐697. 10.1016/j.jhep.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lan FF, Wang H, Chen Y‐C, et al. Hsa‐let‐7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c‐Myc and upregulation of p16(INK4A). Int J Cancer. 2011;128:319‐331. 10.1002/ijc.25336 [DOI] [PubMed] [Google Scholar]
- 35. Qin L, Li R, Zhang J, Li A, Luo R. Special suppressive role of miR‐29b in HER2‐positive breast cancer cells by targeting Stat3. Am J Transl Res. 2015;7:878‐890. [PMC free article] [PubMed] [Google Scholar]
- 36. Filipowicz W, Grosshans H. The liver‐specific microRNA miR‐122: biology and therapeutic potential. Prog Drug Res. 2011;67:221‐238. [PubMed] [Google Scholar]
- 37. Hsu SH, Ghoshal K. MicroRNAs in liver health and disease. Curr Pathobiol Rep. 2013;1:53‐62. 10.1007/s40139-012-0005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai WC, Hsu S‐D, Hsu C‐S, et al. MicroRNA‐122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884‐2897. 10.1172/JCI63455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang B, Hsu S‐H, Wang X, et al. Reciprocal regulation of microRNA‐122 and c‐Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59:555‐566. 10.1002/hep.26712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teng Y, Zhang Y, Qu K, et al. MicroRNA‐29B (mir‐29b) regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3. Oncotarget. 2015;6:40799‐40814. 10.18632/oncotarget.5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu SH, Yu B, Wang X, et al. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine. 2013;9:1169‐1180. 10.1016/j.nano.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rippmann JF, Hobbie S, Daiber C, et al. Phosphorylation‐dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 2000;11:409‐416. [PubMed] [Google Scholar]
- 43. Toko H, Hariharan N, Konstandin MH, et al. Differential regulation of cellular senescence and differentiation by prolyl isomerase Pin1 in cardiac progenitor cells. J Biol Chem. 2014;289:5348‐5356. 10.1074/jbc.M113.526442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilead S, Gazit E. Self‐organization of short peptide fragments: from amyloid fibrils to nanoscale supramolecular assemblies. Supramol Chem. 2005;17:87‐92. 10.1080/10610270412331328943 [DOI] [Google Scholar]
- 45. Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543‐552. 10.1038/nrg3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492‐503. 10.1208/s12248-010-9210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Malcolm DW, Benoit DSW. Controlled and sustained delivery of siRNA/NPs from hydrogels expedites bone fracture healing. Biomaterials. 2017;139:127‐138. 10.1016/j.biomaterials.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castanotto D, Rossi JJ. The promises and pitfalls of RNA‐interference‐based therapeutics. Nature. 2009;457:426‐433. 10.1038/nature07758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D'Souza AA, Devarajan PV. Asialoglycoprotein receptor mediated hepatocyte targeting ‐ strategies and applications. J Control Release. 2015;203:126‐139. 10.1016/j.jconrel.2015.02.022 [DOI] [PubMed] [Google Scholar]
- 50. Springer AD, Dowdy SF. GalNAc‐siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 2018;28:109‐118. 10.1089/nat.2018.0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bridges K, Harford J, Ashwell G, Klausner RD. Fate of receptor and ligand during endocytosis of asialoglycoproteins by isolated hepatocytes. Proc Natl Acad Sci USA. 1982;79:350‐354. 10.1073/pnas.79.2.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


