Abstract
Reliable and noninvasive biomarkers for the early diagnosis of non‐small‐cell lung cancer (NSCLC) are an unmet need. This study aimed to screen and validate potential urinary biomarkers for the early diagnosis of NSCLC. Using protein mass spectrometry, urinary MDH2 was found to be abundant both in patients with lung cancer and lung cancer model mice compared with controls. Urine samples obtained as retrospective and prospective cohorts including 1091 NSCLC patients and 736 healthy controls were measured using ELISA. Patients with stage I NSCLC had higher urinary MDH2 compared with healthy controls. The area under the receiver‐operating characteristic curve (AUC) for the urinary MDH2 was 0.7679 and 0.7234 in retrospective and prospective cohorts to distinguish stage I cases from controls. Urinary MDH2 levels correlated with gender and smoking history. MDH2 expression levels were elevated in lung cancer tissues. MDH2 knockdown using shRNA inhibited the proliferation of lung cancer cells. Our study demonstrated that urinary MDH2 concentration was higher in early‐stage NSCLC patients compared with that in controls and that MDH2 could serve as a potential biomarker for early detection of NSCLC.
Keywords: diagnosis, early detection, malate dehydrogenase 2, non‐small‐cell lung cancer, urinary biomarker
Malate dehydrogenase 2 was significantly elevated both in urine and in cancer tissues of NSCLC patients. The level of MDH2 in urine could serve as an assistant biomarker for the early diagnosis of NSCLC.

Abbreviations
- AUC
area under the curve
- DIA
data‐independent acquisition
- FNR
false negative rate
- FPR
false‐positive rate
- GTEx
genotype‐tissue expression
- IAA
iodoacetamide
- LC
liquid chromatography
- LDCT
low‐dose computed tomography
- LUAD
lung adenocarcinoma
- LUSC
lung squamous carcinoma
- MDH2
malate dehydrogenase 2
- MS
mass spectrometry
- NSCLC
non‐small‐cell lung cancer
- PPV
positive predictive value
- ROC
receiver‐operating characteristic
1. INTRODUCTION
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer death in both more developed and less developed countries globally. 1 Although there has been much progress in the diagnosis and prognosis of lung cancer in previous years, most patients with lung cancer have advanced‐stage disease at diagnosis. 2 Lung cancer has a 5‐y survival rate of merely 19%, but the 5‐y survival rate of patients with stage I lung cancer who undergo surgical treatment can be as high as 77%‐92%. 3 , 4 Common methods of detecting lung cancer include computed tomography (CT), positron emission tomography, blood tumor biomarker analysis, sputum analysis, bronchoscopy with biopsies, endobronchial ultrasound, and CT‐guided transthoracic biopsy and mediastinoscopy. CT is the main screening method but has a high FPR. 5 , 6 Biopsies can confirm a diagnosis in most cases but present risks such as hemorrhage or pneumothorax, therefore the urgent need to find reliable and noninvasive biomarkers is intensified.
Urine has recently revealed its potential as a carrier of cancer biomarkers. Compared with other biofluids, urine has the following characteristics: (a) abundance; (b) easy and noninvasive sampling; (c) little change in content after collection if properly stored; and (d) richness in metabolites, reflecting the metabolic changes caused by diseases. 7 , 8 Studies using MS‐based methods have attempted to profile the metabolite changes of several cancers such as prostate cancer, bladder cancer, pancreatic cancer, and breast cancer. 9 , 10 , 11 , 12 , 13 For lung cancer, previous studies have discovered a range of urinary metabolites as an indication of lung cancer, 14 , 15 , 16 but only a few metabolites have been further validated for their specificity for lung cancer, 17 , 18 therefore urinary biomarkers for lung cancer remain unclear.
In this study, we used MS to determine urinary proteins that were abundant in patients with lung cancer and assessed the performance of urinary proteins as biomarkers for the early detection of non‐small‐cell lung cancer.
2. MATERIALS AND METHODS
2.1. Study subjects
Urine samples were collected from lung cancer patients and healthy controls at Tianjin Medical University Cancer Institute and Hospital between February 2017 and June 2019. Only patients with pathologically or histologically confirmed NSCLC, before any treatment, and who were not simultaneously diagnosed with other malignant diseases were included in this study. In total, 1091 lung cancer patients and 736 control subjects were analyzed. Among these subjects, 4 patients and 4 control subjects were used for MS assay, urine samples from 318 stage I NSCLC patients and 239 control subjects were selected from February 2017 to December 2017 to be included in the retrospective cohort, and 769 NSCLC patients and 493 control subjects were enrolled prospectively from April 2018 to June 2019. In addition, 13 patients pathologically confirmed with benign pulmonary nodules (BPNs) were also recruited for diagnostic evaluation of the biomarker. The Tianjin Medical University Cancer Institute and Hospital ethics committee approved this study (approval numbers: bc2016014, bc2018009, bc2019091). Human urine samples were collected after informed consent was signed. The study methodologies conformed to the standards set by the Declaration of Helsinki. The study schematic diagram is shown in Figure 1A.
FIGURE 1.
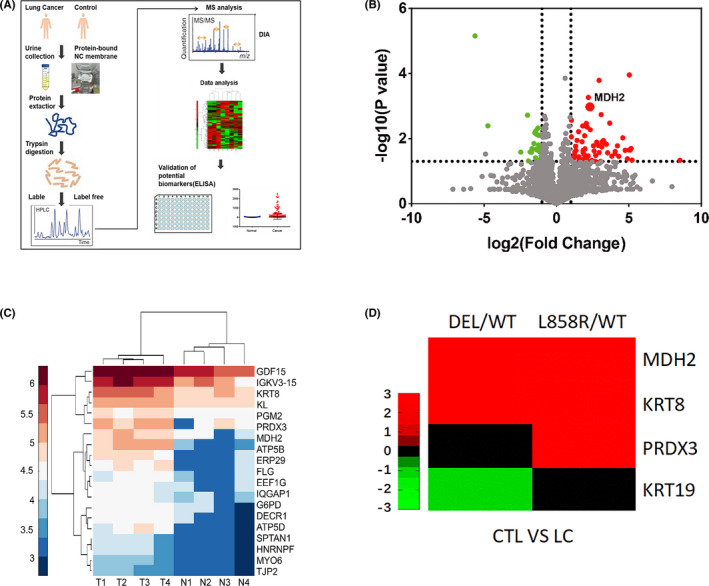
High urinary MDH2 levels were determined using DIA MS both in patients with NSCLC and lung cancer model mice. A, Schematic of discovery and assessment of lung cancer urinary biomarkers. B, Volcano plot of urine quantitative proteome results when comparing patients with lung cancer and healthy controls. Red dots show upregulated expression and green dots show downregulated expression in cancer. C, Heat map shows the top 20 most significantly upregulated urine proteins (fold change >2, P <.05) between patients with lung cancer (represented by T1‐T4) and healthy controls (represented by N1‐N4). D, Heat map shows the top 4 highly regulated urine proteins between EGFRL858R‐driven and EGFR19DEL‐driven lung cancer model mice and wild‐type mice, which were consistent with those in human (fold change >2, P <.05). CTL, controls; WT, wild‐type mice
2.2. Cell culture and reagents
A549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines were purchased from the ATCC. All cell lines were cultured at 37°C in the presence of 5% CO2 in RPMI 1640 medium containing 10% FBS (Gibco, Life Technologies) and 100 IU/mL penicillin/streptomycin (Gibco, Life Technologies). All human cell lines had been authenticated using short tandem repeat (STR) profiling within the last 3 y.
2.3. Plasmids and lentivirus packaging
For MDH2 knockdown experiments, shRNA fragments were cloned into the Tet‐pLKO‐zeo vector. The shRNAs sequence of MDH2 was based on the Sigma Mission shRNA Library: shMDH2 (TRCN0000028485). To generate cell lines for Dox‐inducible MDH2 knockdown, A‐549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines were infected with lentivirus packaged from the pLKO‐tet‐zeo vector harboring shRNA targeting MDH2 mRNA. Cells were selected with zeomycin (300 μg/mL) for 1 wk in appropriate cases.
2.4. Preparation of urine samples
Samples of 20‐30 mL of the first‐void urine prior to treatment were collected. Urine samples were centrifuged at 5000 g for 40 min at 4°C. The supernatants underwent a short storage period at −80°C. For longer storage, the urinary supernatants were diluted with phosphate buffer, pH 7.5 and then the urinary protein was fixed on 22‐μm nitrocellulose membranes (Millipore) using vacuum filtration and stored at −80°C. 19
For human urinary protein extraction, the nitrocellulose membranes on which sample urinary protein was fixed were cut into pieces and dissolved in acetone containing 0.5% ammonium bicarbonate with vigorously shaking. Samples were incubated at 55°C for 60 min with vigorous shaking every 20 min. Urinary protein was precipitated at 4°C followed by centrifugation at 16 000 g for 15 min. The protein precipitate was dried at room temperature. For protein digestion, the protein precipitate was dissolved in lysis buffer containing 7 M urea, 2 M thiourea, 40 mM Tris‐base, 25 mM dithiothreitol (DTT) and the protein concentration was detected using the Bradford method. For ELISA assay, the protein precipitate was dissolved in lysis buffer (100 mM Tris, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X‐100, 0.5% sodium deoxycholate) and the protein concentration was detected using Pierce BCA assay (Thermo Fisher Scientific). For mice urinary protein extraction, the protein was precipitated with acetone from the samples and dissolved in lysis buffer.
2.5. Protein digestion and DIA MS
Quantitative proteome analysis of the differential urine proteins was performed using an Orbitrap Fusion Lumos (Thermo Fisher Scientific) mass spectrometer. For human urine analysis, 4 pairs of samples from 4 lung cancer patients and 4 healthy people as controls were analyzed. For mouse analysis, 3 mouse samples from the EGFR L858R mutant, EGFR DEL, and EGFR WT (as control) were mixed and protein digestion, MS analysis, and quantitative comparison were performed.
Protein digestion was performed as previously described. 20 Briefly, urinary proteins were reduced in buffer containing 8 M urea and 50 mM DTT at 37°C for 1 h and alkylated with 100 mM iodoacetamide (IAA) at room temperature for 30 min. Samples were then transferred to 10 kDa centrifugal devices (Lot FC1233, PALL) for in‐solution protein digestion. Protein was mixed with trypsin at a mass ratio of 30:1 and digested at 37°C for 16 h. Finally, peptides were collected, freeze dried and desalted.
Tryptic peptides were reconstituted using deionized water containing 0.1% formic acid. Ion libraries were constructed by collecting the same amounts of tryptic peptides from all samples. The iRT‐standard (Biognosys) was added into the pooled sample at a ratio of 1/10 by volume for retention time calibration. Samples were then analyzed in data‐dependent acquisition (DDA) mode using an EASY‐nLC 1200 (Thermo Fisher Scientific) HPLC system and Orbitrap Fusion Lumos (Thermo Fisher Scientific) mass spectrometer. For LC separation, peptides were separated on an Acclaim PepMap 100 C18 column (100 μm × 2 cm, 5 μm, Thermo Fisher Scientific, P/N:164564) and an Acclaim PepMap 100 C18 column (50 μm × 15 cm, 2 μm, Thermo Fisher Scientific, Waltham, MA, USA, P/N:164943). Four runs were performed to construct the precursor ion library in DDA mode. For DIA MS analysis, individual tryptic peptide samples were mixed with the iRT‐standard (1/10 by volume) and analyzed on the same LC‐MS/MS system. Global settings for DDA and DIA were according to the reference. 21
2.6. Database searches
DDA raw data files were searched using the SEQUEST‐HT engine of Proteome Discoverer (PD) v.2.1 (Thermo Fisher Scientific) against a combined database of neXtProt (23 January 2017 release, 20 159 entries; the reverse sequence was used to generate the decoy database) and the iRT‐standard peptides sequence. Common contaminants trypsin and keratins were included in the database. Confident protein identifications should meet the following criteria: (a) protein level FDR ≤ 1%; (b) unique peptides ≥ 2; (c) peptide length ≥ 9 aa. To build the ion spectral library, the pdResult files generated by the PD software were imported into Spectronaut™ software v.10 (Biognosys). The parameters of the BGS Factory Settings were according to the reference. 21 For individual DIA MS files analysis, the DIA raw files were converted into HTRMS files and analyzed with the review section of Spectronaut by choosing the matched database fasta file and spectral library. Protein identification and quantitation were exported from Spectronaut software for further analysis.
For comparison of the differential proteins between patients with cancer and healthy people, expression differential fold change >2 and T Test P‐value < .05 were used to identify the differentially expressed proteins (DEPs), and quantitative proteins were delineated using a volcano plot.
2.7. GTEx and TCGA gene expression, correlation, and outcome analysis
The comparison of the MDH2 level between normal tissue, para‐carcinoma tissue, and primary tumor tissues was performed with the “Compare tumor vs normal within or across tissue types” function of the UCSC Xena tool (http://xena.ucsc.edu/compare‐tissue/). The databases of lung cancer samples from the UCSC RNA‐seq Compendium, plus The Cancer Genome Atlas (TCGA) and the GTEx, (normal tissue of individuals without cancer) were re‐aligned to the hg38 genome using the same RNA‐seq pipeline. The “RSEM norm_count” dataset, which was normalized by the upper quartile method, was chosen for the gene expression comparison. For survival analysis, data were collected from the Human Protein Atlas (https://www.proteinatlas.org/). Patients were ordered by expression level and split into the top 50% and bottom 50%. Using this information, a 2‐condition (high expression vs low expression) Kaplan‐Meier survivability plot was generated.
2.8. Enzyme‐linked immunosorbent assay
The MDH2 concentrations of urine samples were measured using ELISA kits (EK1911, Signalway Antibody). Urinary protein was extracted and quantified as described above. To each well, 100 µg urinary protein was added. The optical density of each well was simultaneously determined using a microplate reader set to 450 nm.
2.9. Quantitative real‐time PCR
Total RNA was extracted from the tissues and cells using TRIzol reagent (Lot 250404, Ambion) and was quantified and qualified using NanoDrop and Agilent 2100 bioanalyzers (Thermo Fisher Scientific). cDNA was synthesized using oligo(dT) primers and PrimeScript II (Lot AJ50605A). The MDH2 level was determined using qRT‐PCR and TB Green™ Premix Ex Taq™ II (Lot RR820) on the CFX96 Real‐time PCR Detection System (Bio‐Rad Laboratories). The 2–ΔΔCT method was used to calculate the relative expression levels of RNA. The following primers (BGI, Beijing Genomic Institute) were used: 5′‐CCCACGGGTTCATAGTTCAG‐3′ and 5′‐CATCAGGGTTCGGTCAGAAG‐3′.
2.10. Western blot
Tissues were lysed in RIPA lysis buffer and centrifuged at 14 000 g for 15 min. Pierce BCA protein assay was used to determine protein concentration. Protein was resolved on 10% SDS‐PAGE gels and transferred onto PVDF membranes. After blocking for 1 h, membranes were incubated with primary antibody (1:10 000, Lot ab181857, Abcam) at 4°C overnight, followed by incubation with secondary antibody (1:10 000 dilution, Lot Z0219, Ray Antibody) for 1 h at room temperature. The immune reactive bands were visualized using ECL Prime Western Blotting Detection Reagent and exposed using a Chemiluminescence Western Blot Scanner (C DIGIT, LI‐COR).
2.11. Immunohistochemical staining
In total, 197 formalin‐fixed, paraffin‐embedded (FFPE) tissue slides, including lung cancer tissue and normal lung tissue, were used for immunohistochemical staining. FFPE tissue slides were dehydrated and rehydrated, then antigen retrieval was carried out using high‐pressure heating. The slides were incubated with primary antibodies (1:200, Lot ab181857, Abcam) overnight at 4°C. After incubation with secondary antibody (Origene) for 1 h at room temperature, the sections were exposed to 3,3′‐diaminobenzidine (DAB) substrate (Origene) and counterstained with hematoxylin (Solarbio).
Slides were scored by 2 impartial technicians for overall staining intensity and the percentage of cells stained. The proportion scores ranged from 0 to 4 (0, none; 1, 1%‐25%; 2, 26%‐50%; 3, 51%‐75%; and 4, >75%), and the intensity scores ranged from 0 to 3 (0, none; 1, weak; 2, intermediate; and 3, strong). These scores were added to obtain a final score ranging from 0 to 7. Cases scoring 0‐4 were considered to have low expression; cases scoring 5‐8 were considered to have high expression.
2.12. Cell proliferation assay
The A‐549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines with knockdown MDH2 were used to perform a cell proliferation assay. Briefly, in total, 1000 cells were seeded into a 96‐well plate, and doxycycline (1 μg/mL) was added to induce MDH2 knockdown. Cell activity was monitored with CCK8 reagent every day following the manufacturer's instruction (Lot PJ762, DOJINDO Laboratories).
2.13. Colony‐forming assay
We used the A‐549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines with doxycycline‐induced MDH2 knockdown to perform the colony‐forming assay. Briefly, in total, 200 cells were seeded into a 6‐well plate and cultured for 7 d before being fixed and dyed with 0.5% crystal violet staining solution. Cells were transferred to flash medium every 2 d and doxycycline (1 μg/mL) was added to induce MDH2 knockdown. Images of cell clones were acquired and colonies were counted using ImageJ software.
2.14. Mice
Animal experiments were approved by the Institutional Committee for Animal Care and Use of Jinan University (approval number: 20180709‐01). The experiments were conducted in accordance with approved institutional guidelines and regulations. All the EGFRL858R‐driven and EGFR19DEL‐driven lung cancer model mice were on the C57BL6 background, c. 6‐8 wk old, with no restrictions on sex, and bred in a pathogen‐free environment. The lung tumor burdens of the mice were recorded using CT scanning (PINGSENG Healthcare) after c. 2 mo of Dox diet feeding. Urine samples were collected for further mass spectrometry analysis. EGFRWT mice were used as the control.
2.15. Statistical analysis
IBM SPSS software (SPSS Inc) and GraphPad Prism (GraphPad Software Inc) were used for statistical analysis. The Mann‐Whitney U test was applied to explore the differences between different groups. ROC analysis was performed to assess diagnostic effectiveness. Cut‐off values were determined by calculating the Youden index. Correlation analysis was performed using Fisher exact test. Interobserver concordance was evaluated by Cohen's kappa for two‐rater comparisons. All tests were two‐sided, and a P‐value <.05 was considered to indicate a significant difference.
3. RESULTS
3.1. Screening of aberrantly expressed urinary proteins in patients with lung cancer
We used DIA MS to analyze the differentially expressed urinary proteins between patients with lung cancer and healthy participants as well as between EGFRL858R‐driven and EGFR19DEL‐driven lung cancer model mice and control mice; 3 female patients and 1 male patient all with stage I LUAD were included. The characteristics of the 4 NSCLC patients are shown in Table S1. Candidate urinary proteins, which were abundant both in patients with lung cancer (Figure 1B,C; Tables S1 and S2) and in lung cancer model mice, were screened out (Figure 1D; Table S3). MDH2 expression was aberrantly high both in patients with lung cancer and in lung cancer model mice.
3.2. Evaluation of the urinary MDH2 level as a diagnostic biomarker of early‐stage NSCLC
We compared by ELISA assay the MDH2 levels in urine samples from 239 healthy participants and 318 patients with stage I NSCLC. Because the MDH2 concentration was not normally distributed in each group, the urinary MDH2 concentration is shown as the median of each group in the following context. The urinary MDH2 concentration was significantly higher in patients compared with in healthy participants (patients vs controls: 18.78 vs 4.31 ng/100 µg, P <.0001; Figure 2A). The AUC for the urinary MDH2 concentration to distinguish patients with stage I NSCLC from healthy participants was 0.7679 (95% CI, 0.7291‐0.8066), with 70.13% sensitivity and 66.11% specificity at the cut‐off value of 9.523 ng/100 µg (Figure 2B). The FPR, FNR, and PPV were 33.89%, 29.87% and 0.7311.
FIGURE 2.
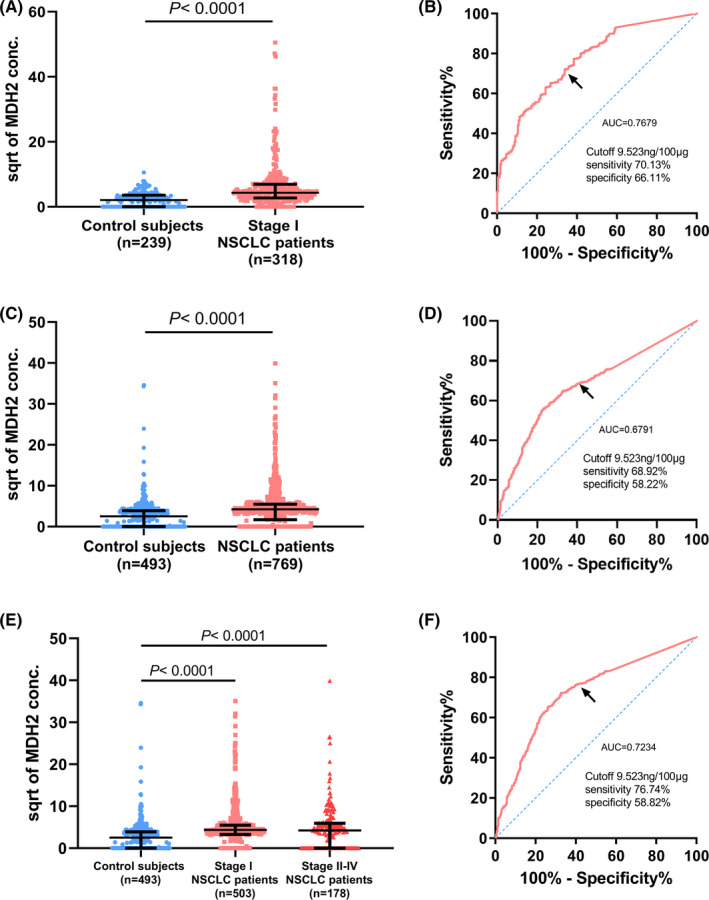
Malate dehydrogenase 2 as a diagnostic biomarker in distinguishing stage I patients with NSCLC from control subjects. A, Comparison by ELISA of the MDH2 concentration in urine between control subjects (n = 239) and patients with stage I NSCLC (n = 318) of the retrospective cohort P <.0001 by Mann‐Whitney U tests. B, ROC analysis of the detectable efficiency of MDH2 in the retrospective cohort in control subjects vs patients with stage I NSCLC (AUC = 0.7679, 95% CI:0.7291‐0.8066, P <.0001). C, Comparison of the MDH2 concentration in urine between control subjects (n = 493) and patients with NSCLC (n = 769) of the prospective cohort by ELISA P <.0001 by Mann‐Whitney U tests. D, ROC analysis of the detectable efficiency of MDH2 in the prospective cohort in control subjects vs patients with NSCLC (AUC = 0.6791, 95% CI: 0.6495‐0.7088, P <.0001). E, Comparison of the MDH2 concentration in urine between control subjects (n = 493) and patients with stage I NSCLC (n = 503) of the prospective cohort using ELISA P <.0001 by Mann‐Whitney U tests. F, ROC analysis of the detectable efficiency of MDH2 in the prospective cohort in control subjects vs stage I NSCLC patients (AUC = 0.7234, 95% CI: 0.6916‐0.7551, P <.0001). Scatter diagrams are represented as median with interquartile range and the vertical axis shows the square root (sqrt) of urinary MDH2 concentration. Black arrows show the cut‐off point
Urine samples were collected prospectively from 493 healthy participants and 769 patients with NSCLC to validate the performance of the urinary MDH2 level as a diagnostic biomarker. Similarly, patients showed a higher urinary MDH2 concentration compared with control subjects (patients vs controls: 17.78 vs 6.295 ng/100 µg, P <.0001; Figure 2C). The AUC was 0.6791 (95% CI, 0.6495‐0.7088), with 68.92% sensitivity and 58.22% specificity at the same cut‐off value, and the FPR, FNR, and PPV were 41.78%, 31.08% and 0.7317 (Figure 2D). When comparing urinary MDH2 concentrations of patients at different stages, patients with stage I NSCLC had higher MDH2 concentrations compared with control subjects, but there was no significant difference in MDH2 concentrations between stage I and stages II‐IV (patients with stage I vs controls: 19.04 vs 6.295 ng/100 µg, P <.0001; Figure 2E). The AUC for discrimination patients with stage I NSCLC from healthy participants was 0.7234 (95% CI, 0.6916‐0.7551), with 76.74% sensitivity and 58.82% specificity at the same cut‐off value, and the FPR, FNR, and PPV were 41.18%, 23.26%, and 0.7787, respectively (Figure 2F).
When the sensitivity was fixed at 90% to screen the more susceptible population, the MDH2 concentration was 1.866 ng/100 μg with a specificity of 41.42% in the retrospective cohort. In this case, the sensitivity and specificity of all prospective cohorts and patients with stage I NSCLC of the prospective cohort was 70.09% and 54.56%, 83.00% and 43.22%, respectively (Table S4).
Urinary MDH2 concentrations may not distinguish BPNs from malignant nodules. Urinary MDH2 concentrations were slightly higher in patients with stage I NSCLC compared with in patients with BPNs, but the difference was not significant (stage I NSCLC vs benign: 18.87 vs 10.55 ng/100 µg, P =.1801; Figure S1A, Table S5). The AUC for the urinary MDH2 concentration to distinguish patients with stage I NSCLC from patients with BPN was 0.6082 (95% CI, 0.4666‐0.7497, P =.1804), with 65.65% sensitivity and 61.54% specificity at the cut‐off value of 13.27 ng/100 µg (Figure S1B). FPR was 38.46% (5/13) and FNR was 35.35% (282/821).
3.3. Correlation between the urinary MDH2 concentration and clinicopathological characteristics in early‐stage NSCLC
The correlation between the urinary MDH2 concentration and clinicopathological characteristics of NSCLC patients in the 2 studied cohorts was explored (Table 1). The urinary MDH2 concentration did not significantly correlate with age (retrospective cohort: ≤65 y, 17.66 ng/100 µg, >65 y, 22.15 ng/100 µg, P =.420; prospective cohort: ≤65 y, 17.99 ng/100 µg, >65 y, 16.99 ng/100 µg, P =.740), EGFR mutation (retrospective cohort: wild‐type, 16.87 ng/100 µg, mutation, 23.18 ng/100 µg, P =.096; prospective cohort: wild‐type, 17.99 ng/100 µg, mutation, 20.87 ng/100 µg, P =.436) and TNM stage (stage I, 19.04 ng/100 µg, stages II‐IV, 17.94 ng/100 µg, P =.112), but correlated with gender and smoking history. Urinary MDH2 concentrations were higher in females (retrospective cohort: female, 27.72 ng/100 µg, male, 12.43 ng/100 µg, P <.001; prospective cohort: female, 24.54 ng/100 µg, male, 9.96 ng/100 µg, P <.001) and patients without a smoking history (retrospective cohort: without smoking history, 23.18 ng/100 µg, with smoking history, 13.91 ng/100 µg, P <.001; prospective cohort: without smoking history, 22.12 ng/100 µg, with smoking history, 13.32 ng/100 µg, P <.001). In prospective cohorts, the urinary MDH2 concentration revealed a correlation with histology (LUAD, 19.71 ng/100 µg, LUSC, 4.94 ng/100 µg, others, 8.47 ng/100 µg, P <.001).
TABLE 1.
Characteristics of enrolled subjects
| Variables | Retrospective cohort | Prospective cohort | ||||
|---|---|---|---|---|---|---|
| n |
Urine MDH2 Conc. (ng/100 ug) (Median) |
P‐value | n |
Urine MDH2 Conc. (ng/100 ug) (Median) |
P‐value | |
| Control subjects | ||||||
| Gender | ||||||
| Male | 99 | 0.00 | <.001 | 202 | 7.80 | .155 |
| Female | 140 | 8.90 | 291 | 4.52 | ||
| Age | ||||||
| Median, 37 y (range, 23‐65) | 239 | 4.31 | ||||
| Median, 38 y (range, 22‐79) | 474 | 6.29 | ||||
| NSCLC patients | ||||||
| Gender | ||||||
| Male | 163 | 12.43 | <.001 | 394 | 9.96 | <.001 |
| Female | 155 | 27.72 | 375 | 24.54 | ||
| Age | ||||||
| ≤65 | 243 | 17.66 | .420 | 583 | 17.99 | .740 |
| >65 | 75 | 22.15 | 186 | 16.99 | ||
| Smoking history | ||||||
| Yes | 149 | 13.91 | <.001 | 313 | 13.32 | <.001 |
| No | 169 | 23.18 | 380 | 22.12 | ||
| EGFR | ||||||
| Wild‐type | 72 | 16.87 | .096 | 107 | 17.99 | .436 |
| Mutation | 47 | 23.18 | 113 | 20.87 | ||
| Untested | 199 | 20.12 | 549 | 17.15 | ||
| Pathological type | ||||||
| Adenocarcinoma | 268 | 19.07 | .552 | 613 | 19.71 | .112 |
| Squamous carcinoma | 41 | 17.14 | 111 | 4.94 | ||
| Other malignant types | 9 | 12.07 | 45 | 8.47 | ||
| TNM stage | ||||||
| I | ‐ | ‐ | 503 | 19.04 | ||
| II‐IV | ‐ | ‐ | 178 | 17.94 | ||
The bold values are an emphasis of the P values which are less than 0.05.
3.4. Expression of MDH2 in NSCLC tissues
We examined MDH2 RNA levels using 19 pairs of lung cancer tissues (LUAD: 12; LUSC: 5; other pathological types: 2; Table S6) and normal lung tissues collected from patients at Tianjin Medical University Cancer Institute and Hospital (P =.0067; Figure 3A). MDH2 expression at the RNA level was higher in lung cancer tissues. Protein level detection further demonstrated the higher expression of MDH2 in lung cancer tissues compared with in normal lung tissues (P =.0078; Figure 3B). By data analysis in the GTEx and TCGA database using UCSC Xena, we discovered that MDH2 expression at the RNA level was also higher in LUAD and LUSC compared with in normal lung tissues or para‐carcinoma tissues (P <.001; Figure S2A). Analysis of data from the Human Protein Atlas showed that the overall survival rate in the high MDH2 expression group was worse compared with that in the low MDH2 expression group (P =.015, Figure S2B).
FIGURE 3.
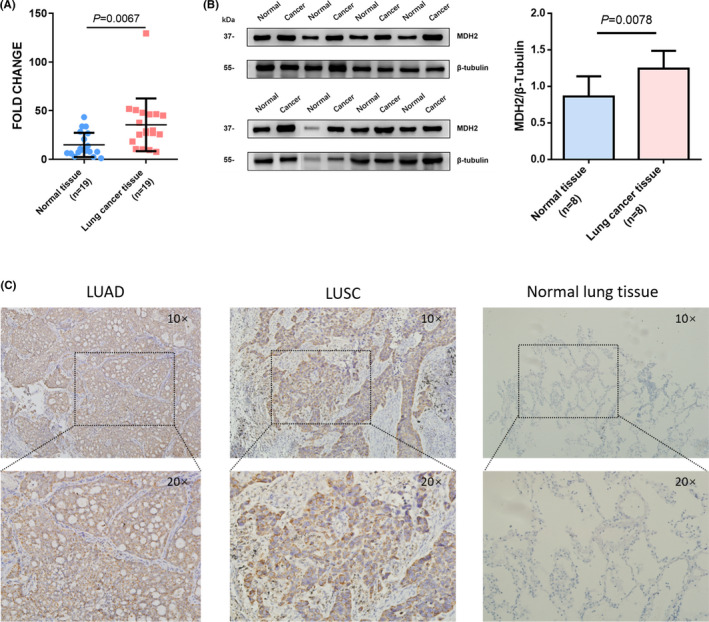
Malate dehydrogenase 2 expression is upregulated in lung cancer tissues. A, MDH2 RNA expression levels in paired normal lung tissue and cancer tissue (n = 19) were analyzed using qPCR. MDH2 expression is higher in lung cancer tissues compared with normal lung tissues. P =.0067 using Wilcoxon test. B, MDH2 protein levels in paired normal lung tissue and lung cancer tissue (n = 8) determined in triplicate using western blot analysis. The column graph indicates semi‐quantitative analysis. P =.0078 using Wilcoxon test. C, Representative images of immunohistochemical staining of MDH2 in LUAD, LUSC and normal lung tissue. Magnification ×10 (top) and ×20 (bottom). LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma
Next, we used 197 FFPE tissue samples from the prospective cohort to analyze MDH2 expression by IHC. IHC staining showed that 67.0% (132/197, Table 2) of NSCLC tissues were positive, while normal lung tissues (n = 10) were almost negative for MDH2 staining (Figure 3C). MDH2 expression was significantly associated with gender (P =.005) and smoking history (P =.002, Table 2). These results indicated upregulated MDH2 expression in lung cancer tissues.
TABLE 2.
Correlations between MDH2 expression and clinicopathological characteristics
| Variables | Expression | P‐value | |
|---|---|---|---|
| Low | High | ||
| Gender | |||
| Male | 14 | 59 | .002 |
| Female | 51 | 73 | |
| Age | |||
| ≤65 | 53 | 98 | .255 |
| >65 | 12 | 34 | |
| History of smoking | |||
| Yes | 14 | 55 | .005 |
| No | 51 | 77 | |
| Pathological stage | |||
| I | 49 | 87 | .104 |
| II | 10 | 14 | |
| III | 4 | 23 | |
| IV | 2 | 8 | |
| T stage | |||
| T1 | 31 | 69 | .376 |
| T2 | 25 | 42 | |
| T3 | 7 | 9 | |
| T4 | 2 | 8 | |
| Tx | 0 | 4 | |
| N stage | |||
| N0 | 59 | 101 | .051 |
| N1 | 4 | 7 | |
| N2 | 2 | 15 | |
| N3 | 0 | 2 | |
| Nx | 0 | 7 | |
| M stage | |||
| M0 | 62 | 125 | .502 |
| M1 | 3 | 8 | |
| Pathological types | |||
| Adenocarcinoma | 62 | 115 | .072 |
| Squamous carcinoma | 0 | 10 | |
| Other NSCLC | 3 | 7 | |
The bold values are an emphasis of the P values which are less than 0.05.
3.5. The effects of MDH2 knockdown on cell proliferation in lung cancer cells
To explore the effects of MDH2 on lung cancer cell growth, we used doxycycline‐induced MDH2 knockdown A‐549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines to perform a cell proliferation assay and cell colony‐forming assay. The MDH2 expression levels of the A‐549 (RRID: CVCL_0023), HOP‐62 (RRID: CVCL_1285) and PC‐9 (RRID: CVCL_B260) cell lines were 6.3, 6.9, and 5.3 times lower after knockdown, respectively (Figure 4A,B). MDH2 knockdown reduced the cell proliferation rate and clone formation ability in all 3 cell lines (P ≤.0001; Figure 4C,D). These data suggested that inhibition of MDH2 expression suppressed cell proliferation.
FIGURE 4.
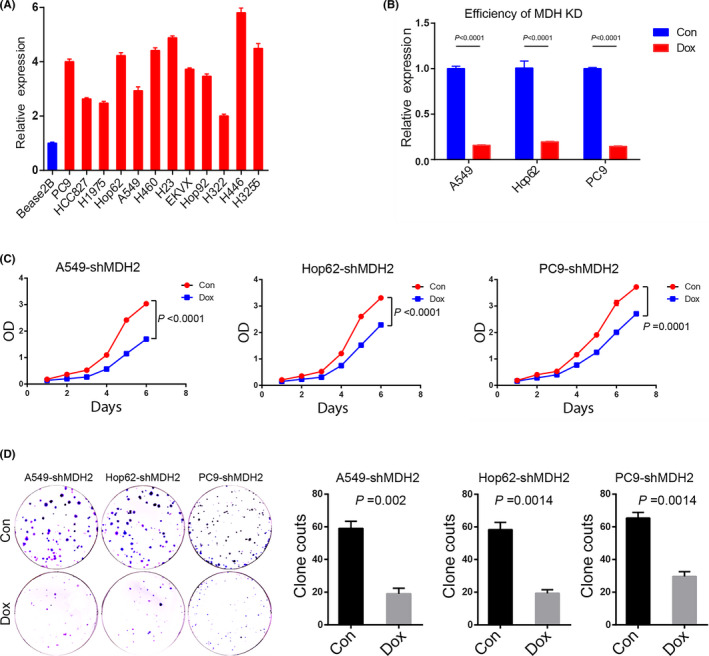
Knockdown of MDH2 inhibits lung cancer cell proliferation. A, MDH2 expression levels in different lung cancer cell lines are significantly higher compared with that in the Bease2B cell (all P <.05). B, MDH2 expression levels in the A549, PC9, and Hop 62 cell lines are 6.3‐, 6.9‐, and 5.3‐fold lower after knockdown. All P <.0001 indicate Mann‐Whitney U tests. C, Cell proliferation assay of A549 (left), Hop62 (middle) and PC9 (right) after MDH2 knockdown. Red dots show the MDH2‐knockdown groups and blue dots show the control groups. Differences were calculated on the last day. All P ≤.0001 used paired t test. D, Colony‐forming analysis of A549, Hop62, and PC9 lines with MDH2 knockdown. Representative figures are shown on the left. Comparison of the clone counts between control and MDH2 knockdown cells. Data are presented as mean ± SEM. A549: P =.002, Hop62: P =.0014, PC9: P =.0014, using one‐way ANOVA. Con, control; Dox, doxycycline; KD, knockdown; OD, optical density
4. DISCUSSION
In this study, the level of urinary protein MDH2 was found to be higher in patients with NSCLC compared with in the healthy population. The ability of urinary MDH2 concentration to discriminate patients with stage I NSCLC from healthy participants using a large‐sized sample was confirmed. AUC was 0.7234 in a prospective cohort. MDH2 expression levels were also higher in lung cancer tissues compared with in normal tissues. Knockdown of MDH2 in lung cancer cell lines inhibited cell proliferation, therefore MDH2 could be a potential noninvasive diagnostic biomarker for early‐stage NSCLC.
Malate dehydrogenase 2 is 1 of the 2 main isoforms of malate dehydrogenase, which is an enzyme that catalyzes the interconversion of malate and oxaloacetate utilizing the NAD+/NADH coenzyme system. 22 It is generally present in mitochondria, participating in the tricarboxylic acid cycle and malate‐aspartate shuttle across the mitochondrial membrane. 23 , 24 In this study, MDH2 was highly expressed in lung cancer tissue and knockdown of MDH2 markedly reduced cell proliferation in lung cancer cells. MDH2 overexpression was also reported in endometrial carcinoma and prostate cancer. 25 , 26 A previous study conducted by Luo and colleagues demonstrated that knockdown of MDH2 significantly inhibited glucose consumption in melanoma cells, resulting in cell growth suppression. 27 MDH2 may repress cancer cell growth by influencing energy metabolism. Some compounds that inhibit MDH2 showed antitumor efficacy. 28 , 29 , 30 These findings suggested that MDH2, which is likely to serve as an anticancer target protein, promotes tumorigenesis.
The MDH2 level in urine has not been previously reported. Our study revealed that the urinary MDH2 level was elevated in patients with NSCLC compared with that in healthy populations in 2 large‐sample cohorts. The AUC of the urinary MDH2 concentration used to differentiate between patients with stage I NSCLC and healthy participants was 0.7679 and 0.7234, respectively. Although the identity of urinary MDH2 was insufficient (58%‐66%), the high sensitivity will be helpful for screening the susceptible population.
We further included a few patients with BPN to test the ability of urinary MDH2 to discriminate BPN from NSCLC. This biomarker may not be useful for this purpose due to its unsatisfactory AUC and high FPR, possibly due to the wide range caused by insufficient cases. Two patients diagnosed as atypical adenomatous hyperplasia had higher urinary MDH2 level. As a precursor lesion of LUAD, it is possible that atypical adenomatous hyperplasia may lead to elevation in urinary MDH2, as it is common that biomarkers from body fluid are unable to distinguish benign disease from cancers. Brown and colleagues reported that some circulating inflammation proteins were associated with lung cancer, but produced no evidence that these proteins could distinguish malignancy from benign lung nodules. 31 From this study, more patients with BPNs should further be included to determine if urinary MDH2 can distinguish benign from malignant nodules.
Urine collection is the least invasive method of specimen collection in clinical practice, and patients cooperated well during this study. Using urinary MDH2 as a biomarker, urine specimens can be rapidly processed in batches using ELISA, which is also a highly economical method. Therefore, this study provides a potential noninvasive biomarker that may be used for lung cancer screening.
This study has some limitations. First, the demographic characteristics of the healthy participants did not completely match those of the patients with NSCLC. The healthy controls were younger and the smoking history was incomplete. Second, it was unclear whether urinary MDH2 protein was specifically elevated in NSCLC or if it is also highly expressed in other types of cancer. As most patients admitted in this hospital were cancer patients, the number of benign cases was inadequate. Third, dietary habit and drug intake could influence urinary metabolites; 32 , 33 these factors could not be ruled out, however the sample size was large, which could reduce the confounding factors. Additionally, the specificity of urinary MDH2 as a single biomarker for the early detection of NSCLC was not sufficiently high, therefore its combination with other biomarkers or LDCT should be considered. Further validation is needed not only in a larger cohort but also in diverse cancers.
In conclusion, MDH2 was significantly elevated both in the urine and tissues of patients with NSCLC. The expression level of MDH2 in the urine could serve as an assistant biomarker for the early diagnosis of NSCLC. Although further validation and research still need to be conducted, this noninvasive and easy‐to‐obtain biomarker has the potential to improve the early detection of and screening for NSCLC patient outcomes after seeking medical attention.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1‐2
Tab S2
Tab S3
App S1
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China [Grant No. 2016YFC0905501] and National Natural Science Foundation of China [Grant No. 81672304].
Ma Y‐C, Tian P‐F, Chen Z‐P, et al. Urinary malate dehydrogenase 2 is a new biomarker for early detection of non‐small‐cell lung cancer. Cancer Sci. 2021;112:2349–2360. 10.1111/cas.14845
Yu‐Chen Ma, Peng‐Fei Tian, Zhi‐Peng Chen, Dong‐Sheng Yue, Liang Chen, Bin Zhang and Chang‐Li Wang these authors contributed equally to this work.
Contributor Information
Liang Chen, Email: chenliang@jnu.edu.cn.
Bin Zhang, Email: zhangbin_09@tmu.edu.cn.
Chang‐Li Wang, Email: wangchangli@tjmuch.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non‐small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39‐51. [DOI] [PubMed] [Google Scholar]
- 5. Aberle DR, Abtin F, Brown K. Computed tomography screening for lung cancer: has it finally arrived? Implications of the national lung screening trial. J Clin Oncol. 2013;31(8):1002‐1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seijo LM, Peled N, Ajona D, et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14(3):343‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmidt C. Urine biomarkers may someday detect even distant tumors. J Natl Cancer Inst. 2009;101(1):8‐10. [DOI] [PubMed] [Google Scholar]
- 8. Pieper R, Gatlin CL, McGrath AM, et al. Characterization of the human urinary proteome: a method for high‐resolution display of urinary proteins on two‐dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4(4):1159‐1174. [DOI] [PubMed] [Google Scholar]
- 9. Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70(1):45‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miyake M, Morizawa Y, Hori S, et al. Diagnostic and prognostic role of urinary collagens in primary human bladder cancer. Cancer Sci. 2017;108(11):2221‐2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Radon TP, Massat NJ, Jones R, et al. Identification of a three‐biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin Cancer Res. 2015;21(15):3512‐3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arasaradnam RP, Wicaksono A, O'Brien H, Kocher HM, Covington JA, Crnogorac‐Jurcevic T. Noninvasive diagnosis of pancreatic cancer through detection of volatile organic compounds in urine. Gastroenterology. 2018;154(3):485‐7.e1. [DOI] [PubMed] [Google Scholar]
- 13. Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem. 2004;279(49):51323‐51330. [DOI] [PubMed] [Google Scholar]
- 14. Matsumura K, Opiekun M, Oka H, et al. Urinary volatile compounds as biomarkers for lung cancer: a proof of principle study using odor signatures in mouse models of lung cancer. PLoS One. 2010;5(1):e8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32(15):1976‐1983. [DOI] [PubMed] [Google Scholar]
- 16. Takahashi Y, Sakaguchi K, Horio H, et al. Urinary N1, N12‐diacetylspermine is a non‐invasive marker for the diagnosis and prognosis of non‐small‐cell lung cancer. Br J Cancer. 2015;113(10):1493‐1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu L, Gao Y, Cao Y, et al. Identification of arginine and its "Downstream" molecules as potential markers of breast cancer. IUBMB Life. 2016;68(10):817‐822. [DOI] [PubMed] [Google Scholar]
- 18. Kuwata G, Hiramatsu K, Samejima K, et al. Increase of N1, N12‐diacetylspermine in tissues from colorectal cancer and its liver metastasis. J Cancer Res Clin Oncol. 2013;139(6):925‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin W, Du Z, Gao Y. Collection and preservation of urinary proteins, using a fluff pulp diaper. Sci China Life Sci. 2018;61(6):671‐674. [DOI] [PubMed] [Google Scholar]
- 20. Chen Z, Yang L, Cui Y, et al. Cytoskeleton‐centric protein transportation by exosomes transforms tumor‐favorable macrophages. Oncotarget. 2016;7(41):67387‐67402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W, Chen X, Yan Z, et al. Detergent‐insoluble proteome analysis revealed aberrantly aggregated proteins in human preeclampsia placentas. J Proteome Res. 2017;16(12):4468‐4480. [DOI] [PubMed] [Google Scholar]
- 22. Birktoft JJ, Fu Z, Carnahan GE, Rhodes G, Roderick SL, Banaszak LJ. Comparison of the molecular structures of cytoplasmic and mitochondrial malate dehydrogenase. Biochem Soc Trans. 1989;17(2):301‐304. [DOI] [PubMed] [Google Scholar]
- 23. Dasika SK, Vinnakota KC, Beard DA. Characterization of the kinetics of cardiac cytosolic malate dehydrogenase and comparative analysis of cytosolic and mitochondrial isoforms. Biophys J. 2015;108(2):420‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaNoue KF, Williamson JR. Interrelationships between malate‐aspartate shuttle and citric acid cycle in rat heart mitochondria. Metab Clin Exp. 1971;20(2):119‐140. [DOI] [PubMed] [Google Scholar]
- 25. Liu Q, Harvey CT, Geng H, et al. Malate dehydrogenase 2 confers docetaxel resistance via regulations of JNK signaling and oxidative metabolism. Prostate. 2013;73(10):1028‐1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhuang Y, Xiang J, Bao W, et al. MDH2 stimulated by estrogen‐GPR30 pathway down‐regulated PTEN expression promoting the proliferation and invasion of cells in endometrial cancer. Translat Oncol. 2017;10(2):203‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo S, Li Y, Ma R, et al. Downregulation of PCK2 remodels tricarboxylic acid cycle in tumor‐repopulating cells of melanoma. Oncogene. 2017;36(25):3609‐3617. [DOI] [PubMed] [Google Scholar]
- 28. Naik R, Ban HS, Jang K, et al. Methyl 3‐(3‐(4‐(2,4,4‐Trimethylpentan‐2‐yl)phenoxy)‐propanamido)benzoate as a novel and dual malate dehydrogenase (MDH) 1/2 inhibitor targeting cancer metabolism. J Med Chem. 2017;60(20):8631‐8646. [DOI] [PubMed] [Google Scholar]
- 29. Lee K, Ban HS, Naik R, et al. Identification of malate dehydrogenase 2 as a target protein of the HIF‐1 inhibitor LW6 using chemical probes. Angew Chem Int Ed Engl. 2013;52(39):10286‐10289. [DOI] [PubMed] [Google Scholar]
- 30. Thornburg JM, Nelson KK, Clem BF, et al. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10(5):R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown D, Zingone A, Yu Y, et al. Relationship between circulating inflammation proteins and lung cancer diagnosis in the national lung screening trial. Cancer Epidemiol Biomarkers Prev. 2019;28(1):110‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scalbert A, Brennan L, Manach C, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99(6):1286‐1308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐2
Tab S2
Tab S3
App S1


