Abstract
On-site severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) serological assays allow for timely in-field decisions to be made regarding patient status, also enabling population-wide screening to assist in controlling the coronavirus disease 2019 (COVID-19) pandemic. Here we propose a rapid microfluidic serological assay with two unique functions of nanointerstice filling and digitized flow control, which enable the fast/robust filling of the sample fluid as well as precise regulation of duration and volume of immune reaction. Developed microfluidic assay showed enhanced limit of detection, and 91.67% sensitivity and 100% specificity (n = 152) for clinical samples of SARS CoV-2 patients. The assay enables daily monitoring of IgM/IgG titers and patterns, which could be crucial parameters for convalescence from COVID-19 and provide important insight into how the immune system responds to SARS CoV-2. The developed on-site microfluidic assay presented the mean time for IgM and IgG seroconversions, indicating that these titers plateaued days after seroconversion. The mean duration from day 0 to PCR negativity was 19.4 days (median 20 d, IQR 16–21 d), with higher IgM/IgG titres being observed when PCR positive turns into negative. Simple monitoring of these titres promotes rapid on-site detection and comprehensive understanding of the immune response of COVID-19 patients.
Keywords: SARS-CoV-2, COVID-19, POCT, Microfluidics, Immunoassay
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the aetiological agent of coronavirus disease 2019 (COVID-19), has spread rapidly worldwide. Different from that in other viral infections, identification and hospitalization of patients with COVID-19 may result from government-led active testing and surveillance, with potential disease severity ranging from asymptomatic to critical (Lee and Lee, 2020). Therefore, rapid and quantitative on-site serological assay investigating the dynamic nature of antibody titres for SARS-CoV-2 is urgently needed. This study proposes a microfluidic assay with rapid on-site properties similar to lateral flow assay (LFAs) and quantitative aspects similar to enzyme-linked immunosorbent assays (ELISAs), for use in monitoring immunological responses of patients to SARS-CoV-2 under various clinical environments. Developed microfluidic assay was finally commercialized (Absoludy COVID-19 IgM/IgG Combo; Absology, Korea). Although molecular diagnostics, such as reverse transcription polymerase chain reaction (RT-PCR), largely enable reliable COVID-19 detection with high sensitivity and specificity (Fang et al., 2020; Lan et al., 2020), they impose financial burdens when applied for point-of-care monitoring of populations on a large scale. Furthermore, the sensitivity of these techniques reportedly vary according to infection duration, the site and quality of specimen collection, and the viral load (Weissleder et al., 2020) For instance, RT-PCR false-negative rates are reportedly approximately 30% for patients with COVID-19 (Ai et al., 2020; Weissleder et al., 2020).
As an alternative, serological assays measuring antibody responses in the serum (blood) of patients with COVID-19 include (ELISAs), LFAs, and Western blot analysis (GeurtsvanKessel et al., 2020; Krammer and Simon, 2020a). Serological assays first serve as triage/screening tools, mainly detecting IgM/IgG antibodies in blood serum (Krüttgen et al., 2020), and complementing the clinical diagnosis of suspected patients with negative RT-PCR results and asymptomatic patients with COVID-19 (Long et al., 2020a, 2020b; Wu et al., 2020). The World Health Organization (WHO) reported that 80% of COVID-19 infections are mild or asymptomatic (Long et al., 2020b; Oran and Topol, 2020), highly indicating the need for regular monitoring of individual immune responses in patients with COVID-19. In addition to the triage/screening application, serological assays are also expected to be beneficial in determining the level of community immunity, in enabling sero-surveillance at the population level to monitor COVID-19 prevalence, and for use in the field testing of therapeutic agents/vaccines (Krammer and Simon, 2020a; Shen et al., 2020). Serological assays using whole-blood collection processes, compared to oropharyngeal (OP)/nasopharyngeal (NP) approaches, significantly reduce the potential of infection, also providing better accessibility to asymptomatic or suspected-infected individuals with negative RT-PCR results (Amanat et al., 2020; Li et al., 2005; Long et al., 2020a).
ELISAs are a popular quantitative serological assay for determining the serological kinetics of antibody responses to SARS-CoV-2. Strong correlation between ELISA results and virus neutralization was reported (Amanat et al., 2020; Okba et al., 2020). However, ELISAs are not rapid, and their use requires specialized laboratories with well-trained personnel. In contrast, LFAs can be easily implemented for quick point-of-care situations but mainly provide only qualitative binary results (positive/negative), while several research groups have developed semi-quantitative LFA assays through the image-based analysis algorithm and additional equipment (GeurtsvanKessel et al., 2020, Lee et al., 2019, Li et al., 2019). Therefore, rapid and quantitative on-site serological assay investigating the dynamic nature of antibody titres for SARS-CoV-2 is urgently needed. This study proposes a microfluidic assay with rapid on-site properties similar to LFAs and quantitative aspects similar to ELISAs, for use in monitoring immunological responses of patients to SARS-CoV-2 under various clinical environments. Developed microfluidic assay was finally commercialized (Absoludy COVID-19 IgM/IgG Combo; Absology, Korea).
2. Material and methods
2.1. Design and fabrication of microfluidic chip
The microfluidic chip used for the COVID-19 serological assay (Fig. 1 A) has two unique functions of nanointerstice (NI) filling and digitized flow control. As the first key technique, the nanointerstices (NIs) were formed at the both sides of the microfluidic channel during bonding procedure of bottom and top substrates (Fig. 1B). Meniscus in the small gap of the NI (~500 nm) was formed with extremely small diameter and large pressure difference at its air-liquid interface. It enables fast and robust filling of the sample liquid without any hydrophilic surface treatment, creating robust sample filling into the main channel even after long-term storage (Chung et al., 2009). The NI-driven flow is then regulated by flow digitization control (Fig. 1C&D). The NI-driven flow stopped at a desired position and duration by controlled vent opening/closing, providing sufficient reaction time only with the fixed sample volume and significantly enhancing the limit of detection (LOD) (Kim et al., 2020; Yoon et al., 2017), Top and bottom substrates of the microfluidic chip were fabricated by injection moulding of Polymethyl methacrylate (PMMA) (Incyto, Korea) and bonded by acetone injection using an in-house plate press machine with a pressure of 0.5 MPa (Absology, Korea). We formed dots spotted with detection antibodies or fluorescent beads in the bottom substrate before the bonding process. The microfluidic chip finally consists of an inlet, conjugate region, main channel (3 mm wide, 40 μm high, and 52 mm long), waste region and outlet. Height of the waste region was designed inclined from 100 to 400 μm toward venting outlet. The nanointerstices at both sides of the main channel were formed during bonding process, which provides fast and robust filling of the fluids into the main channel. The bonded microfluidic devices were stored in a dry chamber for one week and packaged.
Fig. 1.
Schematics on the on-site quantitative point-of-care microfluidic assay for SARS-CoV-2.
A) Schematic of the microfluidic chip and fluorescence reader. The reader has a display and vent controller for flow digitization, as well as optical unit for detecting fluorescent signals. B) Schematic of microfluidic chip fabrication with NI and NI-driven filling mechanism. C) NI-driven flow digitized by vent control. In 1 and 3, the vent was closed to stop the flow for reaction. In 2 and 4, the vent was open to allow the sample flow to the reaction zone (2) and fully removed for washing (4). D) Process chart for flow digitization, consisting of a camera, controller, and micromotor. E) Schematic of the microfluidic chip to detect anti-Cov19 IgM or anti-Cov19 IgG. The chip consisted of five parts, i) sample inlet (yellow box), ii) conjugated zone (green box) with dots of anti-IgM or anti-IgG conjugated fluoresce beads (FB), iii) test zone (red box) with dots of SARs-CoV2 antigen-immobilized IgG/IgM, iv) control zone (blue box) with dots immobilized anti-FBs as a control, and v) vents. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Conjugation of fluorescent beads
Fluorescent beads were provided by the BioNano Health Guard Research Center (Daejeon, Korea). The surface of the fluorescent beads (diameter: 450 nm) was activated with 3 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC; cat #77149, Pierce) and 3 mM N-hydroxysuccinimide (NHS; cat #56485, Sigma) in 50 mM 2-(4-morpholino)ethanesulfonic acid (MES; cat #2933, Sigma) buffer for 1 h. Activated fluorescent beads were centrifuged at 15,000 rpm for 15 min. After removing the supernatant, the fluorescent beads were mixed with 125 μg/mL mouse anti-human IgG and mouse anti-human IgM (Thermo Fisher scientific, USA) for 2 h. Next, 1/10 volume of 20% weight/volume skim milk (cat #232100, Gibco) was added, along with 1/10 volume of the second blocking solution (cat #ABF2BS, Absology, Korea), and incubated for 30 min at room temperature. The fluorescent beads were washed three times with storage buffer (cat #IBFSB, Absology), centrifuged, and the supernatant then removed. Pellets were resuspended in MES buffer, and the concentration of dAb-conjugated fluorescent beads was determined using a UV-1800 spectrometer (Shimadzu). Fluorescent beads at a concentration of 0.2% weight/volume in 1.5 μL conjugate buffer (cat #ABCB; Absology) were loaded onto the dAb deposition zone of the bottom substrate (Fig. 1E green box) using a nanoliter dispenser (Musashi, Japan) and then dried.
2.3. Capture antigen on a bottom substrate
SARS-CoV-2 nucleocapsid protein (NP), provided by the BioNano Health Guard Research Center, was used as the capture antigen. Briefly, SARS-CoV-2 NP (1 mg/mL) was biotinylated using EZ-link™ Sulfo–NHS–LC-Biotin (cat #21335, Thermo Scientific) for 1 h, according to the manufacturer's protocol. Surplus biotin was removed via three 2 h cycles of dialysis. Concentration of the biotinylated antigen (A/g) was determined using a spectrometer. Streptavidin (0.3 mg/mL; cat #SA10, Prozyme, USA) was mixed with 3 mM EDC and 3 mM NHS in 50 mM MES buffer overnight. Next, 2 μL of streptavidin solution was loaded onto the bottom substrate of the microfluidic chip (Fig. 1E red and blue box) using a nanoliter dispenser. The plates were then incubated for 1 h in a humid chamber at room temperature. Streptavidin was washed with 1 M tris(hydroxymethyl)aminomethane (Tris) buffer and then dried. Biotinylated A/g (2 μL), which was diluted with 0.2% weight/volume sucrose in 1 × sodium phosphate dibasic, was loaded onto the streptavidin-loaded spot of the microfluidic chip. In 1 h of drying, the immobilized capture A/g was washed using the second washing buffer solution. Washed bottom substrate was then dried overnight and ready for bonding.
2.4. Microfluidic assay preparation
The intensity profiles of test and control zones were measured using a reader (Absology, Korea) 10 min after sample injection. Fluorescence signals were measured at an excitation of 660 nm and an emission of 680 nm. During the reaction time, a solenoid vent valve was connected to the venting outlet of the microfluidic chip to stop the NI-driven serum sample in the main channel at the desired position covering the reaction zones (Fig. 1E). The digitized flow control pauses the sample flow filling and therefore provides sufficient and controlled immune reaction time of 3 min, which enables accurate and precise quantification irrespective of serum viscosity variation between individuals. We calculated the cut-off index, the ratio of assay signal to cut-off signal, defined as 20% excess value at the top 99% from the negative control serum samples (NC). NC samples were tested from patients before the COVID-19 pandemic emerged.
2.5. Definition of cut-off and titre plateau values of IgM/IgG
Seroconversion was defined as the transition period (time) of SARS-CoV-2 IgG or IgM signal from negative to positive. We also defined antibody levels with the relative value of measured one divided by the cut-off index (S/COI) and let S/COI > 1 positive (Fig. 2 B). Titre plateau was set where signal slope showed a first flattening according to the following equation. All the IgG/IgM titer curves with log2(S/COI) versus days after infections were depicted (Fig. 3 B and Fig. 4 D–E).
Fig. 2.
Performance of the developed microfluidic assay
A) Cut-off index(COI) reproducibility of the assay on concentration. B) COI of IgM/IgG for negative control using sera collected prior to the COVID-19 pandemic. C) IgG (S/COI) versus IgM (S/COI) plot showing the sensitivity (91.67%) and specificity (100%) of the developed microfluidic assay. D) Specificity versus sensitivity plot of other US Food and Drug Administration serological tests (LFA, CLIA, ELISA, Microsphere immunoassay). Additional details are provided in Supplementary Table 1.
Fig. 3.
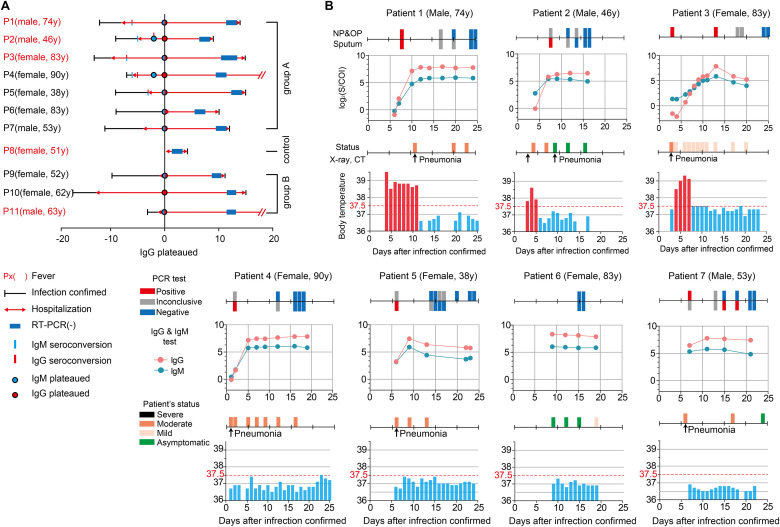
Daily monitoring of nucleic acid amplification tests (RT-PCR), patient status, X-ray/CT imaging, and body temperature compared with the serological assay (IgM/IgG antibody) results. A) Summary of the selected 11 patients in the cohort. Red letters indicate patients who have a fever. Infection confirmed date, hospitalization duration (red line), date of negative PCR results based on tests for 2 consecutive days relative to days after initial confirmation of the infection by PCR (blue box). All patients were temporally aligned to the day of IgG levels plateauing (blue circle). IgM seroconversion, IgG seroconversion, and IgM plateau were are indicated by the light blue bar, red bar, and red circle, respectively. B) Daily timeline graphs. Patient age, sex, and COVID-19 testing results are provided, as well as, PCR test results (Positive: red box, Inconclusive: gray box, Negative: blue box), and the serological assay including the sampling time-points. Patient status severity is indicated (severe: black box, moderate: orange box, mild: ivory, asymptomatic: green) with body temperature (red bar ≥ 37.5 °C, light blue bar < 37.5 °C) observed during hospitalization. Pneumonia was confirmed during hospitalization by chest CT or X-ray and are indicated with black arrow. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Serological assay assessment.
A) Box graph of confirmed infection dates for antibody patterns and PCR negative results for Group A patients. Bar graph of the average S/COI level of IgM (turquoise) and IgG (red) expression and average body temperature (green) in B) the fever group and C) the non-fever group in Group A during hospitalization. All patients are temporally aligned to the day of the IgG plateau. Daily monitoring time-line of D) patients negative for COVID-19 and E) a three selected patient from cohort (Group B). . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.6. Clinical samples of COVID-19 patients
Fresh blood samples were obtained from patients at Chungbuk National University Hospital (Cheongju, Korea) and Seoul Clinical Laboratory (SCL, Korea), in accordance with guidelines of the institutional review board (CBNUH 2020-03-025-001 at CNUH and MDCTC-20-027 at SCL). Informed consent was obtained from each patient prior to the study. Collected blood samples were centrifuged at 3000 rpm for 15 min to separate the serum. 10 μL of separated serum was mixed with 200 μL of buffer, and then applied to the inlets of the microfluidic channels.
2.7. RT-PCR analysis
RT-PCR SARS-CoV-2 tests were performed using Allplex 2019-nCoV assays (Seegene Inc, Korea), according to the manufacturer's protocol. Viral RNA was isolated from pharyngeal swabs using KingFisher Flex (ThermoFisher, USA) and reverse transcribed into complementary DNA (cDNA). Viral cDNA was then amplified, and Ct values determined, using a CFX96™ Dx System (Bio-Rad, USA). Ct values less than 40 were classified as COVID-19 positive; those with a Ct value > 40 were classified as COVID-19 negative. According to PCR results, the group in which all three viral genes (E/RdRP/N) were detected was classified as positive (P); the group in which only one or two genes were detected was classified as inconclusive (I). If none of the three genes were detected, the sample was classified as negative (N).
3. Results
3.1. Microfluidic assay for on-site serological quantitative screening
The developed on-site quantitative point-of-care (POC) microfluidic assay for SARS-CoV-2 requires 50 μL aliquot of serum (10 μL) of a patient mixed with buffer (200 μL) loaded into the inlet of each microfluidic channel (Fig. 1A). Each microfluidic channel has two NIs at both sidewalls as presented ( Fig. 1B) (Kim et al., 2015; Yoon et al., 2017). The microfludic chip was designed to have two microchannels; one for IgM and the other for IgG (See Fig. 1E). Test and control zones (Fig. 1E red and blue box) in the two channels were immobilized with stippled dots of SARS-CoV-2 nucleocapsid protein (NP) for test and anti-fluorescent beads as control on the bottom substrate. Conjugation parts were dotted with anti-IgM- (diameter:450 nm) (upper channel in Fig. 1E) and anti-IgG-conjugated fluorescent beads (diameter:450 nm). To check the reproducibility of the microfluidic assay, we repeated assaying three times for three titers: low (COI < 1), medium (COI 1–2), and high (COI > 15). We observed small standard deviations, representing good reproducibility on the mean values: 0.25(std 0.14), 1.8(std 0.1) and 18.01(std 0.97) for IgM and 0.26(std 0.14), 2.0(std 0.12), and 16(std 0.96) for IgG (Fig. 2A).
Fluorescence intensity cut-off values for IgM and IgG in the developed microfluidic serological platform were determined using negative control serum samples (n = 244) collected prior to Oct 30, 2019, which preceded emergence of the COVID-19 pandemic ( Fig. 2B). A total of 152 samples, including 60 from patients with COVID-19 and 92 from healthy volunteers, were used to verify the sensitivity and specificity of the microfluidic platform. All serum samples were confirmed using RT-PCR and ELISA IgG/IgM analyses. Evaluation of the microfluidic platform revealed a sensitivity and specificity of 91.67% and 100%, respectively ( Fig. 2C). Accuracy (overall agreement) was 96.7%. The results are shown in Fig. 2D and Supplementary Table 1, together with sensitivity and specificity data from previously reported ELISA, LFA, chemiluminescence immunoassay (CLIA), and microsphere immunoassay (MIA) from the US Food and Drug Administration (FDA) (FDA, EUA authorized serology test performance, 2020). The microfluidic platform performed comparably to other serological assays, with results provided within 5 min.
3.2. Microfluidic assay for on-site serological quantitative monitoring of patients' immune response
Eleven patients (6 female and 5 male), aged 38–90 years, at Chungbuk National University Hospital were randomly selected and monitored during their hospitalization following clinical confirmation of COVID-19 using RT-PCR. Day of onset (day 0) was defined as the first day of a positive RT-PCR result. IgM/IgG titres were monitored during hospitalization until the time of discharge, which was based on confirmation with a final negative RT-PCR result. The IgM/IgG evaluation revealed the kinetics of seroconversion and the plateauing of titre levels, which were then compared with clinical severity, categorized as either severe, moderate, mild, or asymptomatic, as defined by WHO (World Health Organization, 2020), chest X-ray/CT radiologic findings, body temperature, and RT-PCR results from the analysis of upper/lower respiratory specimens collected using NP/OP and sputum ( Fig. 3A). Three patients (P1, P4, and P11) had pre-existing medical conditions (Supplementary Table 2). These included hypertension, diabetes mellitus, parathyroid disease (five months previous), renal failure, and myocardial infarction (stent insertion 15 years previous) in patient P1 (male, 74 yr), postherpetic neuralgia in patient P2 (female, 90 yr), and diabetes mellitus in patient P11 (male, 63 yr). Six patients (P1, P2, P3, P8, P9, and P11) had fevers during their hospitalizations. Patient P8 (female, 51 yr) was hospitalized with fevers but tested RT-PCR negative and was considered a control. Fever was defined as a body temperature ≥37.5 °C, as defined by the Korean government's response system guidelines (Guidelines on the Management and Operation of Temporary Living/Testing Facilities For Inbound Travelers, 2020); the day of seroconversion was defined when the log absorbance (S)/COI value exceeded zero. The day on which the titre plateaued was indicated when the change of log2 (S/COI) was <0.2. Cut-off values of IgM and IgG were set as zero on the y-axis.
Seven patients (P1–P7, Group A) presented typical patterns of virus-specific IgM/IgG antibody titres, with their levels increasing and then plateauing ( Fig. 3B). The body temperature of three Group A patients with fever (P1–P3) returned to normal prior to seroconversion. Graphs of IgM/IgG levels in six patients (P1–P5 and P7) in Group A diagnosed with typical COVID-19-associated pneumonia, based on chest X-ray/CT imaging, exhibited close associations with RT-PCR results, also presenting symptoms consistent with those previously reported (Long et al., 2020a). Graphs of IgM/IgG levels of an asymptomatic patient in Group A (P6) revealed high IgM/IgG titres, indicating that virus clearance occurred after antibody levels plateaued (World Health Organization, 2020). As previously described, seroconversion is the time period after the development of a specific antibody when that antibody becomes detectable in the blood (Cooper et al., 1985; Long et al., 2020a). Even with recent studies on COVID-19 and seroconversion, the duration and nature of immune responses against SARS-CoV-2 infection remain unclear (Bentivegna et al., 2020; Long et al., 2020a; Yongchen et al., 2020).
The mean time of IgM seroconversion in Group A patients was 4.2 days (median 4 d, interquartile range (IQR) 2–6.5 d), relative to PCR-confirmation of infection (day 0). The mean time of IgG seroconversion in Group A patients was 5.6 days (median 6 d, IQR 2–7 d) after day 0. These results were similar to the seroconversion patterns reported in other studies investigating 47 hospitalized patients who exhibited the median time to IgM and IgG seroconversion of 7 (IQR 5.4–9.8 d) and 8.2 days (IQR 6.3–11.3 d), respectively (Conklin et al., 2020). However, there were differences in the range of time between our current findings and those previously reported, with a variation of 2.2–3 days. These differences may have been caused by differences in the reference dates (day 0). The referred group was counted from the onset of symptoms, while our group was counted from the time of infection being confirmed by RT-PCR; the onset of symptoms is typically earlier than the RT-PCR confirmation date (Yong et al., 2020).
As shown in Fig. 4A, IgM and IgG titres plateaued on days 9.4 (median 9 d, IQR 6–12 d) and 10 (median 9 d, IQR 7–12 d), respectively. When plateaued values of both IgM and IgG were kept higher than the cut-off value we found that patients were confirmed to be RT-PCR negative on average 19.4 days (median 20 d, IQR 16–21 d) after the initial confirmation of infection. This was consistent with a previous study reporting that serum IgM and IgG plateaued 7 and 8.2 days, respectively, after infection, based on the average time between infection and RT-PCR confirmation (Conklin et al., 2020). Two of the patients in our current study, P6 and P7, did not have IgM/IgG levels measured during the first week of infection, only having four time points in which IgM and IgG were measured due to delayed hospitalization. However, data from these patients supported the inference for the final confirmation of a negative RT-PCR result. We also observed that IgM/IgG antibody responses of three patients with fever (P1–P3) and four patients without fever (P4–P7) in Group A were similar ( Fig. 4B and C). In the fever group, the body temperature of the patients returned to less than 37.5 °C when IgG levels plateaued. IgG titres in the non-fever group decreased, which was consistent with a previous report stating that 93.3% of asymptomatic individuals had a reduction in IgG levels (A. Liu et al., 2020).
4. Discussions
In discussion, we hope to describe some unique patient cases to show the need for the monitoring of IgG/IgM titres in field. Control patient P8 was hospitalized with a fever, although confirmation testing of all samples from this patient was negative. Please note all IgM/IgG titres were kept below the cut-off value (Fig. 4D). Patient P9 (female, 52 yr) in Group B had only three IgM/IgG data points owing to limited hospitalization. However only from the limited samples, both IgM and IgG titres were found plateaued above their respective cut-off values. The patient could be carefully inferred to be SARS-CoV-2-infected but currently in recovering phase (Fig. 4E). Similar infer could be applied to the patient 6 (Fig. 4B), who's infection was confirmed by precede hospital. Please note that the patient was completely asymptomatic in this hospital without any confirmation of virus infection. Only IgG/IgM titres measured by the on-site serological assay presented her infection in recovering phase. Diversity of the clinical situations therefore requires the rapid quantitative on-site serological assay. Approximately 12% of patients with COVID-19 were known to have IgG titres that plateaued within seven days of symptom onset (Long et al., 2020a). The serological assay could confirm response to SARS-CoV-2 infection in individuals not hospitalized during the ideal window for monitoring.
Patient P10 (female, 62 yr) repeatedly showed symptoms of pneumonia and PCR-positive results in NP and OP specimens for more than a month without fever. In our analysis, IgM titres for this patient remained below the cut-off value, carefully allowing us to infer that patient had trouble in generating neutralizing antibodies. Another patient, patient P11 (male, 63 yr), showed severe symptom of pneumonia and high fevers. High titres of both IgM and IgG indirectly showed that the patient was in recovering phase from COVID-19, leading doctors to identify an alternative reason for the symptom, ultimately diagnosed by a bacterial infection.
Quantitative measurement of IgM/IgG titres on-site with a level of robustness and precision high enough to generate continuous titre graphs is therefore important. The developed carriable microfluidic assay proved good sensitivity and perfect specificity from 152 clinical samples, with accurate quantitative readouts in clinical field. The accuracy in quantification allows doctors to correct estimation of patients' prognosis. We also found that plateauing IgG titres strongly correlated with full recovering from COVID-19 by negative RT-PCR results for SARS-CoV-2. Accordingly, amount and change of IgG levels in the early convalescent phase may be an important indicator in serological surveys and a relevant marker for effective immunity.
5. Conclusions
The microfluidic assay provides a predictive tool for the effective surveillance of patient status and immune responses in the public. It may also be useful for cases of COVID-19 infection in which the patient has inconsistent hospitalization. As it is applicable for use in local hospitals and on-site in the field, it may be beneficial for identifying large-scale previous exposure to SARS-CoV-2 within individuals and populations. This platform may have a role in identifying highly reactive human donors for convalescent plasma therapy (Amanat et al., 2020; Duan et al., 2020). Recently, immunity to SARS-CoV-2 was reported to disappear approximately 3–6 months post-infection (A. Z. Liu et al., 2020; Long et al., 2020b). The ability to measure the dynamic nature of antibody titres in the field may allow for timely insight into vaccine efficiency by providing a relatively easy measurement of immunity within a population. A rapid and on-site quantitative COVID-19 serological test is also needed for comprehensive antibody testing after vaccination. This indicates that continuous in-field testing of antibody titres on a daily or weekly basis will allow for actual “immunity passports” (Krammer and Simon, 2020b; Voo et al., 2020). In the current study, we developed a novel microfluidic assay for monitoring antibody responses to SARS-CoV-2. It was shown to be a rapid and easy-to-use test for the on-site quantification of not only IgM/IgG titres, but also diagnostic antibody candidates for the development of therapeutic agents/vaccines and neutralizing antibodies.
Funding
H.K, Y.S, and S.C were supported by Samsung Research Funding and the Incubation Center of Samsung Electronics under Project SRFC-MA1502-51; P.K.B, S.Y.Y, J.K, and Y.B.S were supported by the BioNano Health-Guard Research Center funded by the Ministry of Science and ICT (MSIT) of South Korea as a Global Frontier Project (Grant number H-GUARD_2013M3A6B2078950).
Author contributions
J.H.L, P.K.B, H.K, and S.C wrote and edited the manuscript. J.H.L, H.K, and S.C designed the research and analysed data. Y.J.S, Y.S.S, J.K, J.-S.S, and J.L performed the experiments. H.S.P and K.S.S provided the biospecimens and performed clinical laboratory testing, including SARS-CoV-2 detection and interpretation of clinical data. H.-S.J, S.M.P, K.S.S, S.C, and Y.B.S provided the concept of the study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113406.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentivegna E., Sentimentale A., Luciani M., Speranza M.L., Guerritore L., Martelletti P. New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J. Med. Virol. n/a. 2020 doi: 10.1002/jmv.26160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Yun H., Kamm R.D. Nanointerstice-driven microflow. Small. 2009;5:609–613. doi: 10.1002/smll.200800748. [DOI] [PubMed] [Google Scholar]
- Conklin S.E., Martin K., Manabe Y.C., Schmidt H.A., Keruly M., Klock E., Kirby C.S., Baker O.R., Fernandez R.E., Eby Y.J., Hardick J., Shaw-Saliba K., Rothman R.E., Caturegli P.P., Redd A.R., Tobian A.A., Bloch E.M., Larman H.B., Quinn T.C., Clarke W., Laeyendecker O. Evaluation of serological SARS-CoV-2 lateral flow assays for rapid point of care testing. medRxiv Prepr. Serv. Heal. Sci. 2020 doi: 10.1101/2020.07.31.20166041. 2020.07.31.20166041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D., Maclean P., Finlayson R., Michelmore H., Gold J., Donovan B., Barnes T., Brooke P., Penny R. Acute aids retrovirus infection: definition of a clinical illness associated with seroconversion. Lancet. 1985;325:537–540. doi: 10.1016/S0140-6736(85)91205-X. [DOI] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Yong, Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Yeqin, Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2020. EUA authorized serology test performance.https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance [Google Scholar]
- GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J.P.C., van Kampen J.J.A., van der Eijk A.A., van Binnendijk R.S., Haagmans B., Koopmans M. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020;11:3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidelines on the Management and Operation of Temporary Living/Testing Facilities for Inbound Travelers. 2020. [Google Scholar]
- Kim J., Han S., Yoon J., Lee E., Lim D.W., Won J., Byun J.-Y., Chung S. Nanointerstice-driven microflow patterns in physical interrupts. Microfluid. Nanofluidics. 2015;18:1433–1438. doi: 10.1007/s10404-014-1513-9. [DOI] [Google Scholar]
- Kim J., Hong K., Kim H., Seo J., Jeong J., Bae P.K., Shin Y.B., Lee J.H., Oh H.J., Chung S. Microfluidic immunoassay for point-of-care testing using simple fluid vent control. Sensor. Actuator. B Chem. 2020;316 doi: 10.1016/j.snb.2020.128094. [DOI] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science (80-. ) 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128:104394. doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. J. Am. Med. Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Hwang J., Kim K., Jeon J., Lee S., Ko J., Lee J., Kang M., Chung D.R., Choo J. Quantitative serodiagnosis of scrub typhus using surface-enhanced Raman scattering-based lateral flow assay platforms. Anal. Chem. 2019;91:12275–12282. doi: 10.1021/acs.analchem.9b02363. [DOI] [PubMed] [Google Scholar]
- Lee D., Lee J. Testing on the move: South Korea's rapid response to the COVID-19 pandemic. Transp. Res. Interdiscip. Perspect. 2020 doi: 10.1016/j.trip.2020.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen H., Wang P. Lateral flow assay ruler for quantitative and rapid point-of-care testing. Analyst. 2019;144:3314–3322. doi: 10.1039/c9an00374f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(80):1864. doi: 10.1126/science.1116480. LP – 1868. [DOI] [PubMed] [Google Scholar]
- Liu A., Wang W., Zhao X., Zhou X., Yang D., Lu M., Lv Y. Disappearance of antibodies to SARS-CoV-2 in a -COVID-19 patient after recovery. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Long W., Tu M., Chen S., Huang Y., Wang S., Zhou W., Chen D., Zhou L., Wang M., Wu M., Huang Q., Xu H., Zeng W., Guo L. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;1–15 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020 doi: 10.7326/M20-3012. [DOI] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Yang, Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Yan, Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo T.C., Clapham H., Tam C.C. Ethical implementation of immunity passports during the COVID-19 pandemic. J. Infect. Dis. 2020;222:715–718. doi: 10.1093/infdis/jiaa352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12:eabc1931–e1932. doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2020. Population-based Age-Stratified Seroepidemiological Investigation Protocol for Coronavirus 2019 (COVID-19) Infection. [Google Scholar]
- World Health Organization . 2020. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected.https://doi.org/WHO/2019-nCoV/clinical/2020.5 [Google Scholar]
- Wu X., Fu B., Chen L., Feng Y. Serological tests facilitate identification of asymptomatic SARS-CoV-2 infection in Wuhan, China. J. Med. Virol. 2020;n/a doi: 10.1002/jmv.25904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong S.E.F., Anderson D.E., Wei W.E., Pang J., Chia W.N., Tan C.W., Teoh Y.L., Rajendram P., Toh M.P.H.S., Poh C., Koh V.T.J., Lum J., Suhaimi N.A.M., Chia P.Y., Chen M.I.C., Vasoo S., Ong B., Leo Y.S., Wang L., Lee V.J.M. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., Chen Y., Gu B. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microb. Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Lee E., Kim J., Han S., Chung S. Generation of digitized microfluidic filling flow by vent control. Biosens. Bioelectron. 2017;92:465–471. doi: 10.1016/j.bios.2016.10.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.