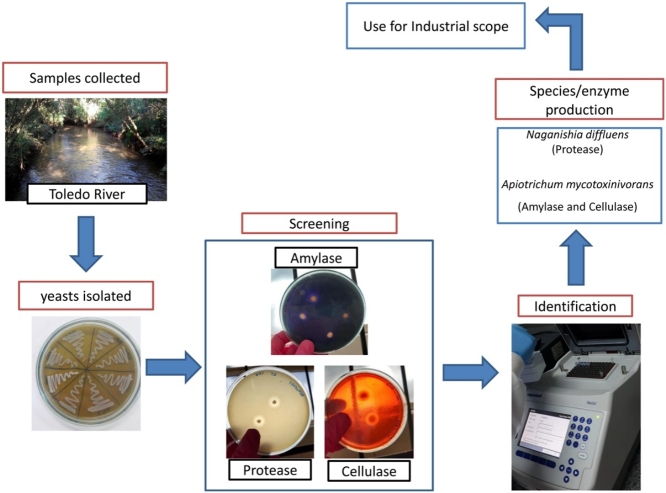
Graphical abstract
Keywords: Biotechnology, Enzyme industry application, Amylase, Cellulase, Protease
Highlights
-
•
Yeast isolated from a lotic ecosystem in Brazil show high enzymatic production.
-
•
One third of the individuals tested have demostrated the ability to secrete more than one enzyme.
-
•
Naganishia diffluens and Apiotrichum mycotoxinivorans show potential applicability as biocatalysts.
-
•
Individuals from Apiotrichum mycotoxinivorans were the best producers of amylase and cellulase.
Abstract
Yeasts have wide applicability in the industrial field, as in the production of enzymes used in biocatalysts. Biocatalysts are more efficient when compared to chemical catalysts, with emphasis on hydrolytic enzymes, such as amylase, cellulase and protease. Here we focused on prospecting yeasts, with a high capacity to synthesize hydrolytic enzymes, from a continental lotic ecosystem environment in Brazil. 75 yeasts were grown in Yeast Extract-Peptone-Dextrose (YPD) medium supplemented with antibacterial and their capacity for enzymatic production was tested in specific media. Accordingly, 64 yeasts showed enzyme production capacity. From those, six showed good enzyme indexes, 3 for amylase, 2 for cellulase and 1 for protease. All showed at least one hydrolytic enzyme activity for the tested enzymes (amylase, cellulase and protease), which suggested that the yeasts are metabolically active. By sequencing the 26S gene, we identified Naganishia diffluens and Apiotrichum mycotoxinivorans as the species with highest enzyme production activities. Those species showed potential for application as biological catalysts in the biotechnological scope, collaborating in a sustainable way for the development of industrial products.
1. Introduction
In industrial microbiology, microorganisms are commonly used in the large scale production of compounds with high added value [16,23]. Not only these microorganisms are cultivated to obtain cells, but also cellular components and their final products, are being used directly (using the microorganism itself) in the production of food, animal ration, cosmetics, biofuel, beverages, drugs, paper, cellulose and in the textile industry, etc. [33,32].
Among those microorganisms, fungi naturally produce a large number of enzymes that decompose organic matter for their growth [20,23]. Enzymes are proteins that catalyze a chemical or biological reaction. They are classified according to the catalysis reactions in: oxidoreductases (oxide-reduction), transferases (transfer of groups of molecules), hydrolases (hydrolysis), lyases (breaking bonds), isomerases (intramolecular change) and ligases (forming a covalent bond between two molecules with high energy consumption) [10]. Among these, we can highlight that hydrolases are widely used in the industrial scope [5].
In fact, the global enzymes market is divided into three main segments: technical enzymes (detergents, paper and cellulose, leather and pharmaceuticals), enzymes used in the food industry and enzymes used for animal ration production [46]. It is estimated that industrial applications using enzymes will generate more than 6 billion dollars in 2021 [5]. That being said, there is an urgent need for discovering and testing microbial species, including those from natural environments, which may have a high capacity to produce and secrete enzymes.
Yeasts have great biotechnological potential in extracellular enzymatic activities, which makes it important to study species mainly from environmental sources [9]. Indeed, enzymes-derived from yeast tend to be eco-friendly and economically relevant than animal or plant-derived enzymes. Yeasts are easy to obtain, have simple nutritional requirements, short fermentation cycles, greater thermal stability and diversity in catalytic activities. Furthermore, they show seasonal independence and can be obtained in large concentrations through genetic manipulation [48], being considered as an ideal biocatalyst.
As a matter of fact, the use of biocatalysts became a viable option for several reasons, as low cost, enantioselectivity and regioselectivity capacities [26,19], enhanced biodegradability and applicability in milder conditions, capacity of being recovered for turnover use, etc [13,21,42].
With the great increase in the applicability of enzymes of microbial origin, we focused on prospecting yeasts with the ability to synthesize amylase, cellulase and protease. Here, we collected water samples from two points of a continental lotic ecosystem in Brazil and tested their enzyme production capacities. We observed that the big majority of yeasts produced at least one enzyme, but only few showed good enzymatic activities. Specifically, Naganishia diffluens showed the best protease activity and Apiotrichum mycotoxinivorans showed the best amylase and cellulase production. Our results suggest the importance of more in-depth studies in N. diffluens and A. mycotoxinivorans and their possible application in biotechnology, due to their high enzyme indexes. Manuscripts dealing with prospecting tests that includes proteolytic activities were already published [8,9,11,12,40]. However, we have found only few articles in the literature regarding the species characterized here. Therefore, our work collaborates with advances in biotechnology following the principle of green chemistry, seeking sustainable alternatives for the production of catalysts.
2. Materials and methods
2.1. Collection and isolation of microorganisms
The yeasts were obtained from water samples collected in the Toledo River, located in Toledo, Parana (PR), Brazil. 200 mL sterilized bottles were used for collection, in duplicates, at each point. Point 1 is located at the geographical coordinates 24°45′10.86″S 53°45′3.52″W in the urban perimeter of the city of Toledo, close to the bridge of the southern contour of the city (area characterized with little agricultural activity and with preserved forest). Point 2 is located 300 m from the river mouth in an urban perimeter and close to a quarry, with geographical coordinates 24°45′14.35″S 53°46′33.91″W (Fig. 1).
Fig. 1.
Geographical location of the water collection points in the Toledo River - PR. (Source: Google Earth).
Aliquots of 250 μL of the water samples were shown by surface spreading method in autoclaved Petri dishes filled with YPD medium (1% of yeast extract, 2% of glucose, 2% of peptone and 2% agar) supplemented with 0.05 % chloramphenicol and 0.05 % tetracycline hydrochloride. The plates were incubated at 28 °C for 48 h.
2.2. Physico-chemical water analysis
PH and temperature were measured at the time of collection using the protocol of Apha [1], with slight modifications. Biochemical Oxygen Demand (BOD) and Chemical Oxygen Demand (COD) were evaluated later on laboratory. For BDO, the 5210-B method (5 days at 20 °C) were monitored and the 5220-D method (reflux closed) were used for COD.
2.3. Screening to identify amylolytic activity
To identify amylase activity, nutrient agar (NA) was used (0.5 % NaCl, 0.5 % peptone, 0.2 % yeast extract, 0.3 % meat extract, 1% glucose and 2% agar) supplemented with 0.5 % soluble and autoclaved starch for 20 min at 121 °C [45]. After point inoculation, the samples were incubated for 96 h at 28 °C followed by aqueous iodine solution staining for 5 min. After discharding the solution, amylase-producing yeasts were identified by the presence of a clear halo around them, indicating starch degradation.
2.4. Screening to identify cellulolytic activity
To identify the cellulase production activity, the methodology of Ruegger and Tauk-Tornisielo [36] with modifications were followed, using a base medium (0.02 % MgSO4, 0.01 % NaCl, 0.04 % yeast extract, 0.04 % KH2PO4 and 0.01 % K2HPO4) supplemented with 1% carboxymethylcellulose and autoclaved for 20 min at 121 °C. After point inoculation, the samples were incubated for 96 h at 28 °C. For staining, 0.01 % Congo red dye was used for 30 min before discharging the excess. After another 30 min, cellulase-secreting yeasts were identified by the presence of a pale red color around the colonies [36].
2.5. Screening to identify proteolytic activity
To identify protease-producing yeasts, NA medium (0.5 % NaCl, 0.5 % peptone, 0.2 % yeast extract, 0.3 % meat extract, 1% glucose and 2% agar) were supplemented with 2% skimmed-milk powder. NA was autoclaved for 20 min at 121 °C, while the powdered milk was autoclaved separated for 5 min at 121 °C, to prevent casein denaturation. After point inoculation, the samples were incubated for 96 h at 28 °C. For identification, clear areas were observed around the positive yeasts [6].
2.6. Enzyme index calculation
The enzyme index (EI) of the yeasts was estimated using the equation between the diameter of the hydrolysis halo and the diameter of the colony (Eq. (1)), according to the methodology of Hankin and Anagnostakis [24].
| (1) |
2.7. Molecular identification of selected yeasts
For genetic indentification of the selected yeasts, we performed the following molecular analysis (Fig. 2).
Fig. 2.
Flowchart of the steps of the molecular analyzes carried out for the genetic identification of the yeasts selected as good producers of amylase, cellulase and protease.
DNA were extracted in house, following the procedure described by Brandão et al., [8], with modifications. Samples were grouped based on enzyme production capacities (itens 2.2–2.6). Subsequently, yeasts were characterized by DNA fingerprints, obtained by the PCR (Polymerase Chain Reaction) methodology using the microsatellite primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′).
Identification of the yeasts were confirmed by amplification with the primers NL1 (5′GCATATCAATAAGCGGAGGAAAAG’3) and NL4 (5′GGTCCGTGTTTCAAGACGG’3), that ring in the D1-D2 domains in the 26S region, which is a conserved gene used to identify species [8]. PCR purification products were used for sequencing (used in the purification process: 47.0 μL of amplicon, 11.75 μL of EDTA (125 mM), 141 μL of absolute ethanol) using a MegaBACE ™ 1000 automatic sequencer (Amersham Biosciences, USA). The DNA sequences were compared with those available in the GenBank database using a basic local alignment search tool (BLAST available at: http://www.ncbi.nlm.nih.gov) [28]. The FASTA sequences were added in Supplemental Material 1.
2.8. Phylogenetic analysis
Phylogenetic analyses were conducted using MEGAX software [27]. Sequences deposited as Supplemental Material 1 were aligned using the ClustalW algorithm. The phylogenetic tree was constructed with the Neighbour-joining algorithm [38] using the Kimura w-parameter method [25] as the DNA substitution model.
3. Results and discussion
3.1. Isolation of the yeasts
First of all, 75 yeasts were obtained from the two collection points. Point 1 showed a higher incidence of isolated yeasts compared to point 2 (Table 1).
Table 1.
Number of isolated yeasts per collection point in the Toledo River located in Toledo, Paraná, Brazil.
| Collection points | Quantity of isolated yeasts |
|---|---|
| Point 1 | 48 |
| Point 2 | 27 |
| Total | 75 |
No significant differences were observed in the the quantity of yeasts / mL between points 1 and 2 (48 yeasts/mL for point 1; 54 yeasts/mL for point 2). Indeed, the two points are located in the urban perimeter and have a distance of approximately 3.5 km between them. According to Morais et al., [29], the distribution and specificity of yeasts depend on the characteristics present in the substrate, enabling their feeding and reproduction, whereas different substrates may show different types of colonies. Thus, the distance between the points proved to be irrelevant because they are relatively close, not drastically changing the characteristics of the substrates, nor the amount of yeasts / ml isolated.
3.2. Physicochemical characteristics of water
The data of the physical-chemical characterization of the water samples are shown in Table 2.
Table 2.
Results of physical-chemical analyzes of water samples collected at two points on the Toledo River, Paraná, Brazil.
| Physiochemical aspects | Point 1 (24°45′10.86″S 53°45′3.52″O) | Point 2 (24°45′14.35″S 53°46′33.91″O) |
|---|---|---|
| pH | 6,96 | 6,93 |
| Temperature (°C) | 22,3 | 22,9 |
| BOD (mg.L−1) | 4,26 | 3,32 |
| COD (mg.L−1) | 34,80 | 38,50 |
The pH remained constant along the two collection points, with values between 6.93 and 6.96. The water temperature also did not show significant variation between the points. According to Hankin and Anagnostakis [24], lotic continental environments that have a pH between 4.0 and 7.0 are the most suitable for finding enzyme-producing microorganisms. Also, authors suggested that pH 6.0 enhances the growth of microorganisms that produce most of the enzymes, corroborating our results. Regarding temperature, aquatic microorganisms can adapt in a range of 4 °C–30 °C, which vary seasonally. However, they have an ideal temperature that favors their growth and survival (in the range of 30 °C). Given this context, we observed that the temperature during collection was in accordance with the ideal described in literature, with no significant variation between them.
Following CONAMA resolution 357/05 [14], BOD values at 20 °C of up to 3 mg.L−1 O2 are considered class I freshwater; 3.1−5 mg.L−1 O2 applies to class II freshwater; and up to 10 mg.L−1 O2 freshwater class III. As shown in Table 2, both collection points of this study can be classified as BOD class II. Also, according to the CONAMA Resolution 357/05 [14], COD values >50 mg.L-1 of O2 apply to polluted and effluent waters (industrial and domestic) and COD < 50 mg.L-1 of O2 to unpolluted waters. Our results for the 2 points of the Toledo River did not exceed 50 mg.L-1 and did not show major variation, characterizing unpolluted waters.
3.3. Screening of cellulase, amylase and protease
Of the 75 isolated yeasts, 83 % of them demonstrated the ability to produce and secrete amylase, cellulase and protease. Furthermore, 62 % were collected in Point 1 and 38 % were collected in Point 2 (Supplemental Material 2). Many yeasts produced more than one enzyme (Supplemental Material 2). Fig. 3A represents the amount of yeasts capable of producing and synthesizing the enzymes mentioned above, taking into consideration that 35 % yeasts produced two or three enzymes (Fig. 3B).
Fig. 3.
Yeast quantity isolated from the Toledo River producing amylase, cellulase and/or protease. (A) Yeast quantity separated by produced enzyme. (B) Venn diagram of number of individuals producing one or more enzymes.
The enzymatic activity was estimated by the relationship between the diameter of the hydrolysis halo and the diameter of the colony. The diameter of the hydrolysis halo is considered a faster and simpler parameter to qualitatively assist in the selection of strains with the highest rate of degradation. According to Hankin and Anagnostakis [24], yeasts with EI ≥ 2.0 are considered good enzyme producers for industrial application. Here we consider some individuals with close values once exogenous factors as temperature and incubation time may cause a slight variation in the halo measurement. In sum, this first stage is a qualitative way of selecting individuals to later confirm their enzymatic potential through quantitative tests, so we included individuals with sub-optimal EI.
Table 3 shows the average growth diameters of microorganisms in the plates, as well as the average of their degradation halos and the values found for the enzyme activity indexes of amylase, cellulase and protease.
Table 3.
Index of enzymatic activity of yeasts that showed better production of amylase, cellulase and protease.
| Enzyme | Yeast code | Colony diameter (mm) | Halo diameter (mm) | Enzymatic index |
|---|---|---|---|---|
| Amylase | JC072 | 11 | 6 | 1,85 |
| JC074 | 10 | 5 | 2,00 | |
| JC079 | 11 | 6 | 1,85 | |
| Cellulase | JC032 | 12 | 6 | 2,00 |
| JC033 | 11 | 6 | 1,85 | |
| Protease | JC008 | 14 | 4 | 3,50 |
Table 3 shows the best-individuals for enzyme production capacity. Among these, 3 were isolated from point 1 (JC072, JC074, JC079) and produced amylase, and 3 were isolated from point 2 and produced cellulase (JC032 and JC033) and protease (JC008).
3.4. Molecular identification of isolated yeasts
Through the characterization of the bands by fingerprint, we classified the individuals in 6 groups, and 5 groups presented identical DNA band patterns. A representative of each group was selected for genetic sequencing of the 26S gene (JC008, JC032, JC074, JC079). Table 4 shows the obtained species.
Table 4.
Identification of selected yeasts with better enzyme indexes.
| Yeast code | Species | Produced enzyme (Enzyme index) | Collection Point |
|---|---|---|---|
| JC008 | Naganishia diffluens | Protease (3.50) | Point 1 |
| JC032 | Apiotrichum mycotoxinivorans | Cellulase (2.00) | Point 1 |
| JC033 | Apiotrichum mycotoxinivorans | Cellulase (1.85) | Point 1 |
| JC072 | Apiotrichum mycotoxinivorans | Amylase (1.85) | Point 2 |
| JC074 | Apiotrichum mycotoxinivorans | Amylase (2.00) | Point 2 |
| JC079 | Apiotrichum mycotoxinivorans | Amylase (1.85) | Point 2 |
First, JC008, in which a protease enzyme index of 3.50 was obtained, was identified as Naganishia diffluens. This species belonged to the genus Cryptococcus and is currently classified in the genus Naganishia [17], being an organism commonly found in colder environments such as Antarctica [41,43,4,7] and polluted water or air [17]. Indeed, finding this species in the evaluated lotic environment suggests high adaptative capacities of this species. The enzyme index of the aforementioned species proved to be expressive, which corroborates previous studies of aquatic yeasts with capacity in the production and proteolytic synthesis [31,8]. In this context, the synthesis of the protease enzyme, mainly coming from less-studied species as N. Diffluens becomes necessary, as this enzyme is one of the three most important in the industrial sector [22,30]. Its biotechnological applicability occurs in the feed, textile, pharmaceutical industries, in leather treatment [44], detergent manufacturing [39], in food industries [42] and bioremediation of industrial effluents [42,49].
Based on sequencing of the 26S gene and DNA fingerprint, all other samples were identified as Apiotrichum mycotoxinivorans and showed cellulase and amylase production capacities. This microorganism was classified in the genus Trichosporon and was first described in 2004 [3]. Although the 5 yeasts were identified as the same species and showed similar phenotypes and fingerprint band profiles, they showed different behavior for amylase and cellulase production. In fact, samples JC032 and JC033 were considered good producers for cellulase and also produced amylase, but with a low enzyme index (> 0.1 for both), while JC072, JC074 and JC079 did not show production for cellulase, only for amylase. These results corroborates [18], who performed the tests with A. mycotoxinivorans for the cellulase enzyme and obtained positive results for the species. Corroborating the aforementioned work, [47], isolated six strains of A. mycotoxinivorans from a natural environment and obtained individuals with good enzymatic capacity for cellulose. Thus, yeasts JC032 and JC033 identified as Apiotrichum mycotoxinivorans showed cellulolytic production capacities and represent a major importance in the biotechnology industry, due to its thermal stability and potential to develop sustainable products [35]
Regarding amylolytic activity, Pretscher et al. [34] tested two A. mycotoxinivorans individuals and observed high EI (> 10) for amylase production. No detectable activity was observed in the cellulase enzyme tests. This can be explained due to the expression of the phenotype that varies from each individual, depending on nutrients availability in the environment. Indeed, we observed 3 individuals identified as A. mycotoxinivorans as showing best amylolytic activity. Strains of A. mycotoxinivorans have already been isolated by other authors in the Iguaçu National Park, a region also located in western Paraná, aiming at the production of xylanolitic enzymes for the synthesis of ethanol [29]. Others described the use of this species as bioemulsifiers, accelerating the degradation process in the treatment of oily wastewater [15,37]. Therefore, this species has biotechnological interest in the use of the biodegradation of drugs such as tetracycline [2], in addition to its use in biocontrol, degrading mycotoxins, using it as a probiotic food additive [34], etc.
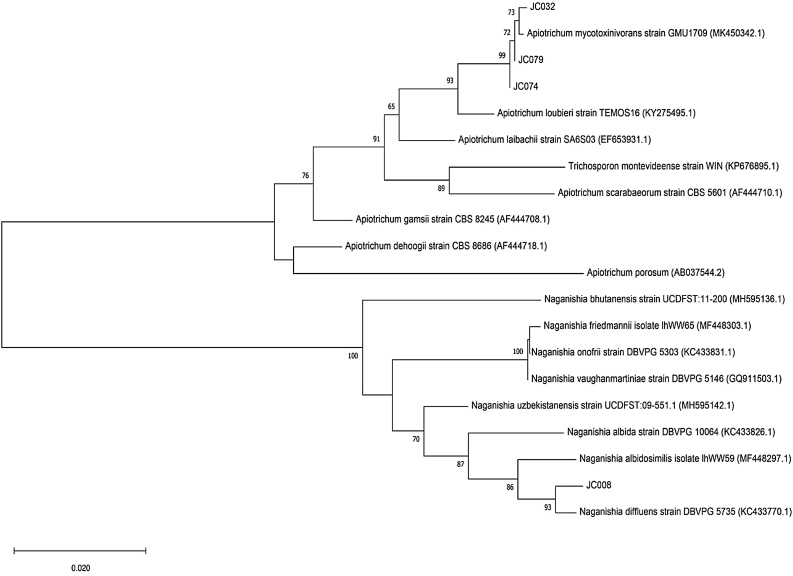
Nevertheless, we performed a phylogenetic analysis comparing JC008, JC032, JC074 and JC079 with available data. Indeed, JC032, JC079 and JC074 were clustered together with Apiotrichum mycotoxinivorans strain GMU1709, while JC008 was clustered in the other extremity of the tree with Naganishia diffluens strain DBVPG 5735 (Fig. 4). These results confirm the identification of the individuals presented in this work.
Fig. 4.
Phylogenetic analysis using the 26S ribosomal RNA sequence from our individuals vs related species.
Boostrap values were made by 1000 replicate runs (shown as %, greater than 70 % are listed). The optimal tree is shown and GenBank accession numbers are listed after species name
4. Conclusion
The method used in this work sought alternatives that included the use of natural resources to isolate good producers of amylase, cellulase and protease. As result, the best producers were identified as Apiotrichum mycotoxinivorans and Naganishia diffluens. It is interesting to speculate the potential biotechnological interest of these findings in the environmental and industrial areas. For example, biodegradability and turn over capacity reduces operational costs and might be highlighted as advantages over chemical catalysts, combining sustainability with enhancer development in the industrial scope.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
The authors are thankful for the research grants and financial support of the National Council for Scientific and Technological Development (CNPq) and the Coordination for the Improvement of Higher Education Personnel (CAPES) and the Post-Graduation program in Environmental Sciences (PPGCA) in Universidade Estadual do Oeste do Paraná (Unioeste) and Universidade Federal de Minas Gerais (UFMG).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2021.e00630.
Contributor Information
Jéssyca Ketterine Carvalho, Email: jessycakcarvalho@gmail.com.
Andressa Alves Silva Panatta, Email: andressabio91@gmail.com.
Maruhen Amir Datsch Silveira, Email: maruhen-amir.datsch-silveira@crchudequebec.ulaval.ca.
Christophe Tav, Email: christophe.tav@crchudequebec.ulaval.ca.
Susana Johann, Email: susjohann@yahoo.com.br.
Maria Luiza Fernandes Rodrigues, Email: mlmfernandes@hotmail.com.
Cleide Viviane Buzanello Martins, Email: cvbmartins@gmail.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Apha A. American Public Health Association/American Water Works Association/Water Environment Federation; Washington, DC, USA: 1995. WPCF, Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- 2.Al-Dhabi N.A., Esmail G.A., Arasu M.V. Effective degradation of tetracycline by manganese peroxidase producing Bacillus velezensis strain Al-Dhabi 140 from Saudi Arabia using fibrous-bed reactor. Chemosphere. 2021;268:128726. doi: 10.1016/j.chemosphere.2020.128726. [DOI] [PubMed] [Google Scholar]
- 3.Almeida J.N.D., Jr., Francisco E.C., Barberino M.G.M., Da Silva Filho L.V.R.F., Brandão O.M., Colombo A.L., Padovan A.C.B. Emergence of Trichosporon mycotoxinivorans (Apiotrichum mycotoxinivorans) invasive infections in Latin America. Memórias do Instituto Oswaldo Cruz. 2017;112:719–722. doi: 10.1590/0074-02760170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenz B.E., Blanchette R.A. Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region and McMurdo dry valleys. Soil Biol. Biochem. 2011;43:308–315. doi: 10.1016/j.soilbio.2010.10.016. [DOI] [Google Scholar]
- 5.Amadi O.C., Egong E.J., Nwagu T.N., Okpala G., Onwosi C.O., Chukwu G.C., Moneke A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon. 2020;6:e04566. doi: 10.1016/j.heliyon.2020.e04566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Gigirey B., de Sousa J.M.V.B., Villa T.G., Barros-Velazquez J. Characterization of biogenic amine-producing Stenotrophomonas maltophilia strains isolated from white muscle of fresh and frozen albacore tuna. Int. J. Food Microbiol. 2000;57:19–31. doi: 10.1016/S0168-1605(00)00240-3. [DOI] [Google Scholar]
- 7.Białkowska A., Turkiewicz M. Cold-adapted Yeasts. Springer; Berlin, Heidelberg: 2014. Miscellaneous cold-active yeast enzymes of industrial importance; pp. 377–395. 0.1007/978-3-642-39681-6_17. [Google Scholar]
- 8.Brandão L.R., Libkind D., Vaz A.B., Espírito Santo L.C., Moliné M., de García V., Rosa C.A. Yeasts from an oligotrophic lake in Patagonia (Argentina): diversity, distribution and synthesis of photoprotective compounds and extracellular enzymes. FEMS Microbiol. Ecol. 2011;76:1–13. doi: 10.1111/j.1574-6941.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 9.Buzzini P., Martini A. Extracellular enzymatic activity profiles in yeast and yeast‐like strains isolated from tropical environments. J. Appl. Microbiol. 2002;93:1020–1025. doi: 10.1046/j.1365-2672.2002.01783.x. [DOI] [PubMed] [Google Scholar]
- 10.Cai Y.D., Chou K.C. Predicting enzyme subclass by functional domain composition and pseudo amino acid composition. J. Proteome Res. 2005;4:967–971. doi: 10.1021/pr0500399. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco M., Rozas J.M., Barahona S., Alcaíno J., Cifuentes V., Baeza M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012;12:1–9. doi: 10.1186/1471-2180-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavello A.I., Martinez A., Garmendia G., Vero S., Cavalitto S. Yeasts from Tierra Del Fuego Province (Argentina): biodiversity, characterization and bioprospection of hydrolytic enzymes. Geomicrobiol. J. 2019;36:847–857. [Google Scholar]
- 13.Czaja R. Cluster-Screening – eine effiziente Methode zur Identifizierung neuer Enzyme. BioSpektrum. 2015;21:344–346. doi: 10.1007/s12268-015-0580-0. [DOI] [Google Scholar]
- 14.CONAMA 357/2005 do Ministério do Meio Ambiente. Ministério do Meio Ambiente. Available in: http://www2.mma.gov.br/port/conama/legiabre.cfm?codlegi=459. Access in: 08 set. 2020.
- 15.De Souza Monteiro A., Domingues V.S., Souza M.V., Lula I., Gonçalves D.B., De Siqueira E.P., Dos Santos V.L. Bioconversion of biodiesel refinery waste in the bioemulsifier by Trichosporon mycotoxinivorans CLA2. Biotechnol. Biofuels. 2012;5:1–12. doi: 10.1186/1754-6834-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirel R., Sariözlü N.Y., İlhan S. Production of fungal amylase enzyme from Aspergillus species in solid-state culture. J. Biotechnol. 2008 pp. 31-31. [Google Scholar]
- 17.Fotedar R., Kolecka A., Boekhout T., Fell J.W., Anand A., Al Malaki A., Al Marri M. Naganishia qatarensis sp. Nov., a novel basidiomycetous yeast species from a hypersaline marine environment in Qatar. Int. J. Syst. Evol. Microbiol. 2018;68:2924–2929. doi: 10.1099/ijsem.0.002920. [DOI] [PubMed] [Google Scholar]
- 18.Giese E.C., Dussán K.J., Pierozzi M., Chandel A.K., Pagnocca F.C., Da Silva S.S. Cellulase production by Trichosporon laibachii. Orbital. 2017;9:271–278. doi: 10.17807/orbital.v9i4.1024. [DOI] [Google Scholar]
- 19.Girelli A.M., Astolfi M.L., Scuto F.R. Agro-industrial wastes as potential carriers for enzyme immobilization: a review. Chemosphere. 2020;244:125368. doi: 10.1016/j.chemosphere.2019.125368. [DOI] [PubMed] [Google Scholar]
- 20.Gopinath Subash C.B., Hilda A., Anbu P. Periasamy. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience. 2005;46:119–126. doi: 10.1007/S10267-004-0221-9. [DOI] [Google Scholar]
- 21.Grajales-Hernández D.A., Velasco-Lozano S., Armendáriz-Ruiz M.A., Rodríguez-González J.A., Camacho-Ruíz R.M., Asaff-Torres A., Mateos-Díaz J.C. Carrier-bound and carrier-free immobilization of type A feruloyl esterase from Aspergillus niger: Searching for an operationally stable heterogeneous biocatalyst for the synthesis of butyl hydroxycinnamates. J. Biotechnol. 2020;316:6–16. doi: 10.1016/j.jbiotec.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Homaei A., Lavajoo F., Sariri R. Development of marine biotechnology as a resource for novel proteases and their role in modern biotechnology. Int. J. Biol. Macromol. 2016;88:542–552. doi: 10.1016/j.ijbiomac.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Kango N., Jana U.K., Choukade R. Fungal enzymes: sources and biotechnological applications. In: Satyanarayana T., Deshmukh S.K., Deshpande M.V., editors. Advancing Frontiers in Mycology and Mycotechnology: Basic and Applied Aspects of Fungi. Springer Nature; 2019. [Google Scholar]
- 24.Hankin L., Anagnostakis S.L. The use of solid media for detection of enzyme production by Fungi. Mycologia. 1975;67:597. doi: 10.1080/00275514.1975.12019782. [DOI] [Google Scholar]
- 25.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlmann J., Kretschmann A.C., Bester K., Bollmann U.E., Dalhoff K., Cedergreen N. Enantioselective mixture toxicity of the azole fungicide imazalil with the insecticide α-cypermethrin in Chironomus riparius: investigating the importance of toxicokinetics and enzyme interactions. Chemosphere. 2019;225:166–173. doi: 10.1016/j.chemosphere.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medeiros A.O., Missagia B.S., Brandão L.R., Callisto M., Barbosa F.A., Rosa C.A. Water quality and diversity of yeasts from tropical lakes and rivers from the Rio. Braz. J. Microbiol. 2012:1582–1594. doi: 10.1590/S1517-83822012000400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morais P.B., Pagnocca F.C., Rosa C.A. Biodiversity and Ecophysiology of Yeasts. Springer; Berlin, Heidelberg: 2006. Yeast communities in tropical rain forests in Brazil and other South American ecosystems; pp. 461–484. [DOI] [Google Scholar]
- 30.Morellon-Sterling R., El-Siar H., Tavano O.L., Berenguer-Murcia A., Fernández-Lafuente R. Ficin: a protease extract with relevance in biotechnology and biocatalysis. Int. J. Biol. Macromol. 2020:1–53. doi: 10.1016/j.ijbiomac.2020.06.144. [DOI] [PubMed] [Google Scholar]
- 31.Ogrydziak D.M. Yeast extracellular proteases. Crit. Rev. Biotechnol. 1993;13:1–55. doi: 10.3109/07388559309069197. [DOI] [PubMed] [Google Scholar]
- 32.Okafor N. second edition. CRC Press; 2017. Modern Industrial Microbiology and Biotechnology. [Google Scholar]
- 33.Pirota R.D.P.B., Graminha E.B.N., Gonçalves A.Z.L., Balsalobre M.A.A., Da Silva R., Gomes E. Enzymatic production by thermophilic fungi using agricultural wastes and ruminant diet as substrates. J. Biotechnol. 2007;2:S227–S228. doi: 10.1016/j.jbiotec.2007.07.413. [DOI] [Google Scholar]
- 34.Pretscher J., Fischkal T., Branscheidt S., Jäger L., Kahl S., Schlander M., Claus H. Yeasts from different habitats and their potential as biocontrol agents. Fermentation. 2018;4:31. doi: 10.3390/fermentation4020031. [DOI] [Google Scholar]
- 35.Rai A.K., Pandey A., Sahoo D. Biotechnological potential of yeasts in functional food industry. Trends Food Sci. Technol. 2019;83:129–137. doi: 10.1016/j.tifs.2018.11.016. [DOI] [Google Scholar]
- 36.Ruegger M.J., Tauk-Tornisielo S.M. Cellulase activity of fungi isolated from the soil of the Juréia-Itatins Ecological Station, São Paulo, Brazil. Braz. J. Bot. 2004:205–211. doi: 10.1590/S0100-84042004000200001. [DOI] [Google Scholar]
- 37.Rulli M.M., Alvarez A., Fuentes M.S., Colin V.L. Production of a microbial emulsifier with biotechnological potential for environmental applications. Colloids Surf. B Biointerfaces. 2019;174:459–466. doi: 10.1016/j.colsurfb.2018.11.052. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Salwan R., Sharma V. Trends in extracellular serine proteases of bacteria as detergent bioadditive: alternate and environmental friendly tool for detergent industry. Arch. Microbiol. 2019;201:863–877. doi: 10.1007/s00203-019-01662-8. [DOI] [PubMed] [Google Scholar]
- 40.Santhi V.S., Gupta A., SAranya S., Jebakumar S.R.D. A novel marine bacterium Isoptericola sp. JS-C42 with the ability to saccharifying the plant biomasses for the aid in cellulosic ethanol production. Biotechnol. Rep. 2014;1:8–14. doi: 10.1016/j.btre.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scorzetti G., Petrescu I., Yarrow D., Fell J.W. Cryptococcus adeliensis sp. nov., a xylanase producing basidiomycetous yeast from Antarctica. Antonie Van Leeuwenhoek. 2000;77:153–157. doi: 10.1023/A:1002124504936. [DOI] [PubMed] [Google Scholar]
- 42.Shakerian F., Zhao J., Li S.P. Recent development in the application of immobilized oxidative enzymes for bioremediation of hazardous micropollutants–A review. Chemosphere. 2020;239:124716. doi: 10.1016/j.chemosphere.2019.124716. [DOI] [PubMed] [Google Scholar]
- 43.Shivaji S., Prasad G.S. Yeast Biotechnology: Diversity and Applications. Springer; Dordrecht: 2009. Antarctic yeasts: biodiversity and potential applications; pp. 3–18. [Google Scholar]
- 44.Singh R., Kumar M., Mittal A., Mehta P.K. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 2016;6:1–15. doi: 10.1007/s13205-016-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonune N., Garode A. Isolation, characterization and identification of extracellular enzyme producer Bacillus licheniformis from municipal wastewater and evaluation of their biodegradability. Biotechnol. Res. Innov. 2018;2:37–44. doi: 10.1016/j.biori.2018.03.001. [DOI] [Google Scholar]
- 46.Spohner S.C., Müller H., Quitmann H., Czermak P. Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J. Biotechnol. 2015;202:118–134. doi: 10.1016/j.jbiotec.2015.01.027. [DOI] [PubMed] [Google Scholar]
- 47.Thongekkaew J., Khumsap A., Chatsa-nga P. Yeasts in mixed deciduous forest areas of Phujong Nayoy National Park and their ability to produce xylanase and carboxymethyl cellulase. Songklanakarin J. Sci. Technol. 2012;34:157–163. [Google Scholar]
- 48.Vyas S., Chhabra M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour. Technol. 2017;223:250–258. doi: 10.1016/j.biortech.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 49.Xu H., Luo Y., Wang P., Zhu J., Yang Z., Liu Z. Removal of thallium in water/wastewater: a review. Water Res. 2019;165:114981. doi: 10.1016/j.watres.2019.114981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.