Abstract
Acute myeloid leukemia (AML) patients frequently relapse after chemotherapy, yet the mechanism by which AML reemerges is not fully understood. Herein, we show that primary AML cells enter a senescence-like phenotype following chemotherapy in vitro and in vivo. This is accompanied by induction of senescence/inflammatory and embryonic diapause transcriptional programs, with downregulation of MYC and leukemia stem cell genes. Single-cell RNA-seq suggested depletion of leukemia stem cells in vitro and in vivo, and enrichment for subpopulations with distinct senescence-like cells. This senescence effect was transient and conferred superior colony forming and engraftment potential. Entry into this senescence-like phenotype was dependent on ATR, and persistence of AML cells was severely impaired by ATR inhibitors. Altogether, we propose that AML relapse is facilitated by a senescence-like resilience phenotype that occurs regardless of their stem cell status. Upon recovery, these post-senescence AML cells give rise to relapsed AMLs with increased stem cell potential.
INTRODUCTION
AML is the most lethal of all blood cancers, killing 3 out of 4 patients within 5-years (https://seer.cancer.gov/statfacts/). Even though 70–80% of AML patients (<60 years) achieve clinical complete remission, the majority of patients will relapse (1) through a largely cryptic mechanism. One prevailing idea is that relapse of AML emerges from an immature drug resistant subpopulation, the so-called leukemia stem cells (LSCs) (2). The LSC concept derived from the observation that only a subpopulation of AML cells with an immature immunophenotype (CD34+, CD38−) can reconstitute leukemia in engraftment experiments using NOD/SCID mice (3). Yet, recent analysis of AML cells surviving chemotherapy in mice and patients at nadir (remaining fraction of chemotherapy-persistent cancer cells after treatment) did not reveal an enrichment of LSCs (4,5), suggesting additional nuances are involved in relapse. Thus, there is an urgent need to better understand the emergence of relapse in AML.
Genotoxic stress by chemotherapy induces, besides apoptosis, senescence (6,7), a cellular stress response characterized by aberrant metabolic activity in absence of proliferation (8). Senescent cells display phenotypic alterations including enlarged cell morphology with increased cytoplasmic granularity and are most commonly characterized by the induction of senescence-associated beta-galactosidase (SA-β-gal) (9). Senescent cells undergo epigenetic changes (10) and secrete extracellular matrix (ECM) modulators as well as pro-inflammatory cytokines, generally termed as the senescence-messaging secretome (11), senescence-associated inflammatory response (SIR) (12) and senescence-associated secretory phenotype (SASP) (13). SASP factors include ECM proteins (e.g. fibronectin, collagen), proteases (e.g. cathepsins, matrix metalloproteinases), interleukins (e.g. IL-1, IL-6), chemokines (e.g. IL-8/CXCL8, MCP-1) and growth factors (e.g. FGF, PDGF) (11,14,15). The SASP factors mediate wound healing (16,17), developmental senescence (18,19), chronic inflammation (20) and cellular plasticity (21,22). Reconciling these contradictory features of senescent cells from an evolutionary standpoint has been challenging.
The initial studies on cellular senescence were performed on non-malignant cells describing a natural barrier to unlimited proliferation, termed as “replicative senescence” or the so-called Hayflick limit (23). Yet, cells that have not reached their replicative lifespan can also undergo cell arrest in response to sublethal stress, often referred to as (stress-induced) premature senescence (24). A landmark study in the late 90’s demonstrated that expression of oncogenes in primary cells induced premature senescence, defined as oncogene-induced senescence (OIS) that led to the concept that senescence might function as a tumor-suppressive mechanism (25). However, more recent studies have shown that expression of oncogenes in even fully transformed cancer cell lines activates OIS, and that both malignant and non-malignant cells can escape OIS after a prolonged period in the senescence arrested state (26,27). Thus, the premature senescence phenotype is not as static and irreversible as initially thought, particularly in malignant cells (27–30).
Overall, cellular senescence is a complex phenomenon, initiated by a wide range of stimuli including DNA damage, oncogene activation, telomere erosion, protein misfolding, oxidative damage, extracellular signaling by mitogens or cytokines, and high-fat diet (8). One interpretation that potentially reflects the common effect of these various senescence inducers could be that senescence acts as a conserved stress-response mechanism. Given the frequency with which cells encounter environmental stress, it is not unreasonable to expect cells to possess evolutionary conserved mechanisms to tolerate harmful conditions. For example, the embryonic diapause is an evolutionary strategy in many mammals that describes a reversible growth arrest (up to many months) of blastocysts (31,32), presumably enabling survival during stressful environmental conditions. We hypothesized that AML relapse is dependent on these cells undergoing a senescence-like resilient state, potentially mimicking a diapause state, to survive genotoxic stress caused by chemotherapy and eventually repopulate the disease.
RESULTS
Chemotherapy induces cellular senescence in patient-derived AMLs.
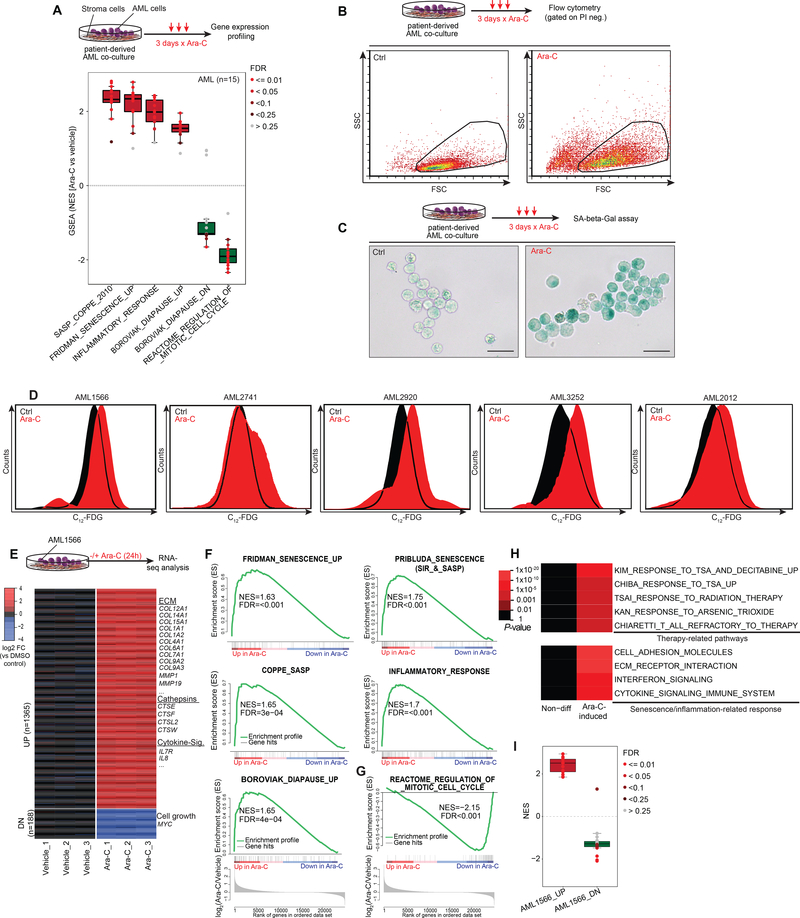
To determine if chemotherapy induces a senescence-like state, we analyzed published gene expression microarray profiles of 15 primary patient-derived AML specimens exposed to Ara-C (a standard component of AML therapy) for three days in a co-culture system (33) (Fig. 1A). Gene set enrichment analysis (GSEA) (34) showed significant enrichment of senescence and SASP/inflammatory signatures in >90% of primary patient-derived cases (Fig. 1A). We also observed significant enrichment for diapause-associated genes (Table S1) (35) after chemotherapy (Fig. 1A). Reciprocally we observed significant negative enrichment for genes that are downregulated during diapause, as well as genes associated with cell division, consistent with the growth arrest linked to senescence.
Figure 1. Chemotherapy-induced senescence in human patient-derived AML.
A, Box-and-whisker plot showing NES scores derived from GSEA using senescence and relevant gene signatures in 15 primary AMLs (33) after ex vivo Ara-C treatment (3 days x 100 nM) compared to their corresponding vehicle-treated conditions. Data is presented with median (bisecting line in the box) and whiskers representing the 1.5 x of the interquartile range (IQR; 25th – 75th percentile, length of the box). Red box indicates that the majority of samples are positively enriched, while green reflects negative enrichment. Each dot represents a primary case, color of dots reflect FDRs calculated by GSEA. NES, Normalized enrichment score; FDR, False discovery rate. B, Upper panel illustrates Ara-C regimen of patient-derived AML cells in an ex vivo co-culture model (36) before subjecting the cells to flow cytometry. FSC vs SSC plots show increased cell size and granularity of patient-derived AML cells after Ara-C exposure for three days. C, Representative images of SA-β-Gal activity in patient-derived AML1566 exposed to Ara-C for three days and stained with X-gal. Scale bar corresponds to 25 μm. D, Flow cytometry histogram depicting SA-β-Gal activity using the fluorogenic β-galactosidase substrate C12-FDG in five patient-derived AML cases, gated on viable cells (PI- exclusion), after 3 days of using a case-dependent moderate-to-high dose of Ara-C exposure (1566: 1000 nM, 2741: 50 nM, 2920: 250 nM, 3252 & 2012: 100 nM). E, RNA-seq analysis of differentially expressed genes (at least 2-FC rel. to mean of vehicle-treated cells and q<0.01) induced by Ara-C treatment (1000 nM) in patient-derived AML1566 cells within 24 hours (n=3 per group). Genes of interest are exemplarily highlighted. F, G, GSEA showing positive enrichment for senescence, diapause and SASP/inflammatory-associated gene signatures unlike a signature of mitotic cell cycle (G) showing negative enrichment after Ara-C exposure. H, Heat map of overrepresented pathways induced by Ara-C treatment in patient-derived AML1566 cells. I, Box-and-whisker plot showing NES scores derived from GSEA using up and down DEG signatures from panel E in the 15 primary patient-derived AMLs (33) from panel A, relative to their corresponding vehicle-treated condition.
To validate whether chemotherapy induces a senescence-like phenotype in myeloid leukemia cells, we examined induction of X-gal-based β-galactosidase staining as a canonical biomarker for this state (9). Using an ex vivo co-culture system that allows reliable expansion of primary AML cells collected from patients (36), we observed key features of senescence including increased cell size, granularity and SA-β-gal activity after three-days exposure to Ara-C or anthracyclines, the two chemotherapy drugs used for standard treatment of this disease, among cell lines and patient specimens (Fig. 1B, C and Fig. S1A) (6). The X-gal based assay has two main limitations: the requirement for chemical fixation, precluding viable cells for further analysis, and limited ability to quantify populations of cells through microscopic assessment. In order to quickly and quantitatively assess SA-β-Gal activity within viable cells, we used the fluorogenic substrate 5-dodecanoylaminofluorescein di-β-D-galactopyranoside (C12-FDG) (37) (Fig. S1B–C). Using this assay, we confirmed induction of this senescence marker as well as increased granularity in additional patient-derived AML specimens (Fig. 1D, Fig. S1D). Because of the inherent fluorescence property of anthracyclines (38) that would interfere with the C12-FDG signal in our subsequent analyses, we focused largely on chemotherapy treatment with Ara-C. Chemotherapy-induced senescence (CIS) occurred in myeloid leukemia cells regardless of p16 or p53 mutation or expression (Fig. S2A, B), consistent with previous reports (12,39,40). Senescence was not simply a marker of differentiation, since a LSD1 inhibitor (36,41,42) induced strong expression of CD11b without an increase of beta-Gal activity (Fig. S2C, D).
To explore the gene signatures induced by chemotherapy exposure at an earlier timepoint that might further indicate the role of senescence as a primary effect on these cells, we performed RNA sequencing on patient-derived AML cells exposed to Ara-C and harvested 24 hours later. We identified 1553 differentially expressed genes (DEG; at least 2-FC and adjusted p value < 0.01) with approximately 88% of genes being upregulated (Fig. 1E, Table S2). Among the upregulated genes were many that encoded typical SASP factors including several members of the cathepsin family and cytokines like IL-8. GSEA demonstrated significant enrichment for gene signatures associated with senescence, inflammation, and diapause (Fig. 1F) (12,13,35,43,44). Reciprocally to the senescence phenotype, cell cycle genes were significantly downregulated (Fig. 1G). Among the downregulated genes was MYC (Table S2), depletion of which was shown to induce a dormant state mimicking diapause (45). Additional pathway analysis of genes upregulated by Ara-C exposure showed enrichment for gene signatures associated with ECM modulation and transcriptional response to genotoxic stress (Fig. 1H). We observed significant enrichment of our early upregulated signature in 14/15 of the patient specimens profiled at day 3 post-chemotherapy exposure (33), suggesting a conserved response mechanism in AML (Fig. 1I).
Senescence-like AML cells can repopulate AMLs after a latent state.
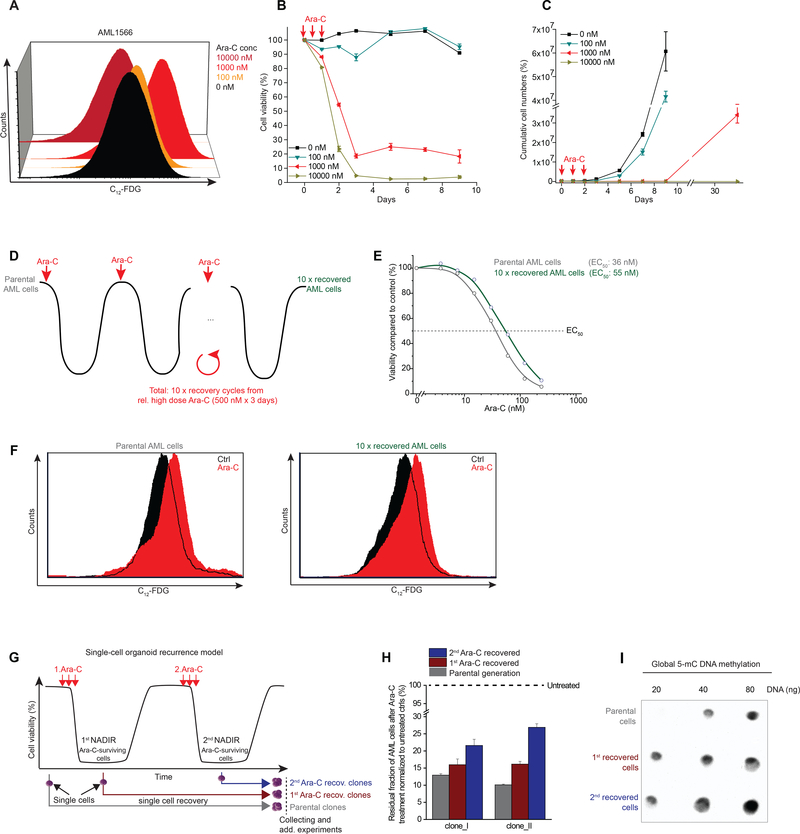
Relapse of AML frequently occurs following a variable nadir phase. To mimic this, we first seeded primary human AML cells in our co-culture organoid system and exposed them to increasing Ara-C concentrations in the range of clinically-relevant concentrations (ranging between the induction vs close to consolidation doses of this drug used in the clinic) (46). We determined the association between the degree of genotoxic stress and senescence induction by SA-β-Gal activity assessed by flow cytometry after 3 days of Ara-C treatment, and observed little induction of senescence at 100 nM of drug, but much stronger induction upon exposure to 1000 nM (Fig. 2A). In contrast, senescent cells were markedly diminished after exposure to 10,000 nM Ara-C (Fig. 2A).
Figure 2. Chemotherapy-induced senescence as a cellular resilience mechanism.
A, Dose-dependent induction of chemotherapy-induced senescence (CIS) after three days of treatment at the indicated doses measured by C12-FDG. B, C, Ex vivo AML recurrence model with patient-derived AML1566 cells measured by flow cytometry for viability (B; by PI exclusion) and cell numbers (C; total viable cells) at indicated Ara-C doses. High dose Ara-C-treated conditions (≥ 1,000 nM) were measured after one month of recovery (C). D, Scheme demonstrating that patient-derived AML cells (AML2928) treated with a case-dependent high dose of Ara-C (3 days x 500 nM) and retreated after recovery from nadir. E, Dose-response curves for Ara-C (3 days treatment) in parental vs 10 x recovered AML cells including their EC50 values. F, Flow histogram showing C12-FDG signal in parental and 10 x recovered AML cells following Ara-C treatment for three days. G, Patient-derived AML cells (AML2741) were treated with a case-dependent high dose of chemotherapy (3d x 100 nM Ara-C). Single cells were picked at indicated time points before and after chemotherapy. Single cells from untreated leukemia were defined as “parental clones”, while cells derived from Ara-C-treated leukemia at nadir were defined as 1st or 2nd Ara-C recovered clones depending on the treatment cycle. To exclude time-dependent epigenetic variations, all recovered monoclonal populations were processed together. H, Bar diagram shows the viability of parental and Ara-C recovered AML monoclonal populations following re-exposure to the same Ara-C regimen (3d x 100 nM Ara-C) that was used to enrich these cells (G). I, Dot plot for 5-mC methylation in parental and Ara-C recovered clones.
To model the nadir phase ex vivo and how leukemia regenerates after exposure to chemotherapy, we determined cell viability by flow cytometry (dye exclusion) over the course of 10 days after treating the cells with increasing doses of Ara-C for 3 days (Fig. 2B). There was very little effect on viability with the 100 nM dose, but close to 100% cell death with the 10,000 nM dose. In contrast the intermediate dose yielded a significant fraction of cells (~20%) that persisted in a viable state (Fig. 2B). To determine if these cells were end-stage or could recover and regenerate the leukemic population we performed serial viable cell counts over an extended time period by flow cytometry up to 30 days post Ara-C treatment (Fig. 2C). Cells treated with 100 nM Ara-C manifested only a slight delay in their expansion, whereas the few cells remaining after the highest dose of Ara-C remained in a quiescent phase throughout the experiment. In contrast, those AMLs treated with 1000 nM Ara-C remained in a latent state until day 9 and after which they were able to re-expand (Fig. 2C). These results suggest that induction of senescence is dose-dependent and reversible, allowing cells to potentially recover and restore the disease.
Chemotherapy-induced senescence as a cellular resilience mechanism.
To explore whether the senescence-like phenotype is a reversible resilience response as opposed to a process of selecting pre-existing chemotherapy-resistant C12-FDGhigh subpopulations, we first sorted C12-FDGhigh vs C12-FDGlow AML cells in three different patient-derived cases (Fig. S3A). Exposure to increasing doses of Ara-C or Daunorubicin did not reveal evidence of differential sensitivity in these C12-FDGhigh cells (Fig. S3B). To address this question in a more rigorous fashion, we next performed sequential cycles of Ara-C treatment and recovery in primary patient-derived AML cells, using a dose (500 nM) that kills >90% of cells from this patient (Fig. 2D). These cells required approximately 1–2 months to recover after each Ara-C treatment and these cycles were repeated a total of ten times (Fig. 2D). Notably, after the tenth cycle the AML cells manifested only a small increase in their EC50 (Fig. 2E). Moreover, Ara-C induced a similar degree of senescence in AML cells after the 10th recovery as compared to the initial parental cells (Fig. 2F).
In addition, we created an ex vivo AML clonal recurrence model in which single primary human cells were placed in culture before, or after serial Ara-C treatment from the residual fraction of chemotherapy-persistent cells at nadir (Fig. 2G). Single cells from each time point formed a monoclonal population thereby excluding genetic heterogeneity within the recovered population. This procedure revealed a modest and gradual increase in the fraction of surviving cells at nadir after each Ara-C recovery cycle (Fig. 2H), indicating that the single cell-derived clones are slightly more resilient to chemotherapy, consistent with progressive stress adaptation (47,48) rather than chemotherapy resistance, as there was no major shift in the EC50 of previously exposed cells to Ara-C (Fig. 2H). Of note, consistent with previous data linking DNA methylation with population fitness in AML patients (49), we observed increased abundance of methylcytosine in recovered cell populations (Fig. 2I). Collectively, these findings support that the senescence-like phenotype is a reversible resilience state instead of a manifestation of an inherently chemotherapy-resistant subpopulation that would have selected Ara-C-resistant clones (Fig. 2F).
Induction of senescence-like phenotype is dose-dependent based on chemotherapy sensitivity.
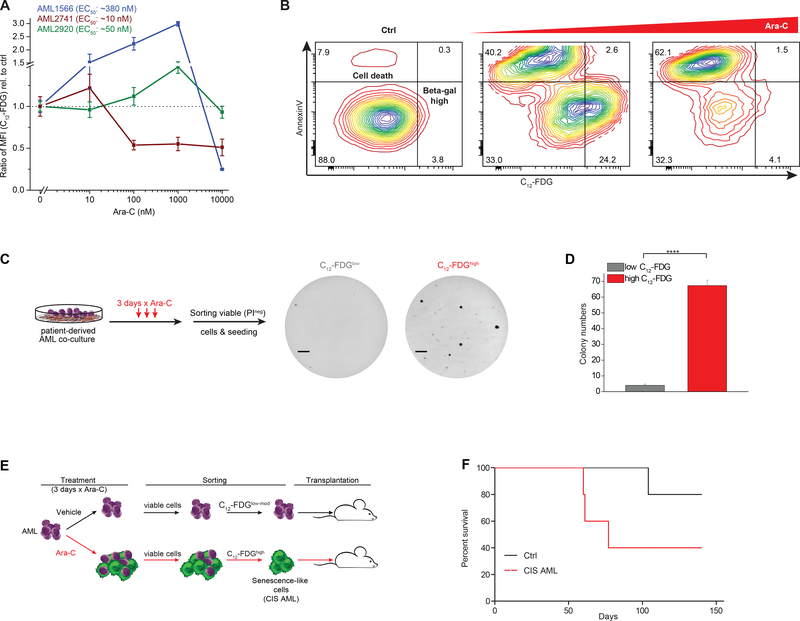
Patient-derived AML cells manifest significant variation in sensitivity (EC50) to Ara-C (Fig. S4A). To explore the link between chemotherapy sensitivity and senescence, we next examined the senescence response at four different doses of Ara-C in three patient specimens with variable degree of sensitivity. Notably, the patient that was most insensitive to Ara-C manifested the greatest induction of senescence that peaked at 1000 nM dose of drug (Fig. 3A), but did not display senescence at the highest dose (where these cells were virtually all killed). The most sensitive case (AML 2741) peaked at 10 nM, after which there was loss of senescent populations. The intermediate case manifested a weaker degree of senescence induction at lower doses than the resistant patient (Fig. 3B). Hence in general we observed that higher concentrations of Ara-C induced increased SA-β-Gal activity in AML cells until reaching a critical toxic dose that killed these cells (Fig. 3A, B).
Figure 3. Chemotherapy-induced senescence is dose-dependent and capable of repopulating AML.
A, Three different patient-derived AML cases (including their EC50 values for Ara-C) were exposed from relatively low (10 nM) to highly toxic (10,000 nM) doses of Ara-C. SA-β-Gal activity was assessed by the ratio of median fluorescence intensity (MFI) of C12-FDG relative to untreated cells set as 1. B, Exemplary of AML cells measured by flow cytometry for SA-β-Gal activity with C12-FDG and Annexin V co-staining following moderate or high dose Ara-C exposure. Shown is the separation of apoptotic cells with reduced C12-FDG levels and the C12-FDGhigh viable fraction following Ara-C. C, D, Ara-C-treated patient-derived AML cells (AML1566) were sorted for low and high SA-β-gal signal and plated in methylcellulose. Colonies were counted after 3 weeks (F, line = 4 mm). Bar graph indicates mean (n=3) of colony numbers (G). (****P<0.0001, Student’s t-test, means of triplicate measurements ± s.d.). E, Schematic showing the treatment of patient-derived AML cells (AML1566) with Ara-C (1000 nM) for 3 days with subsequent sorting of non-senescent cells (untreated condition) and chemotherapy-induced senescence-like cells (CIS AML cells). F, Overall survival of mice after sorting leukemia cells that were untreated or exposed to Ara-C. Each group consisted of 5 NSG mice.
Our co-culture experiments suggested that acquisition of the senescence-like phenotype was associated with persistence and retention of leukemia repopulating potential. To further explore if this was the case, we treated patient-derived AML cells in co-culture with 1000 nM Ara-C for 3 days, after which the specimens were sorted into C12-FDGhigh and C12-FDGlow viable (dye exclusion flow cytometry) fractions and plated on methylcellulose for colony forming cell (CFC) assays. Two of our cases were able to form colonies, and in both cases, the senescence-like cells had significantly higher clonogenic potential (P<0.001) compared to cells with low SA-β-gal activity after Ara-C treatment (Fig. 3C, D and Fig. S4B–D). To determine if senescence was associated with true leukemia-initiating potential, we next treated patient-derived AML cells ex vivo with Ara-C 1000 nM or vehicle for three days in co-culture, after which we sorted viable C12-FDGhigh cells from the Ara-C-treated condition, as well as viable control cells, and implanted them in NSG mice (n=5 mice each, Fig. 3E, F). Contrary to the expectation that senescence-like cells would be terminally non-replicative and unable to reconstitute disease in vivo, we found that mice transplanted with senescence-like cells readily repopulated leukemia (Fig. 3F). We observed a trend to lower survival in mice transplanted with senescence-like cells as compared to untreated AML cells (P=0.15; Fig. 3F). This observation is consistent with recent reports showing increased tumor repopulation properties of tumor cells that recover from chemotherapy-induced senescence (28,50).
Senescence-like AML cells are depleted of leukemia stem cell populations.
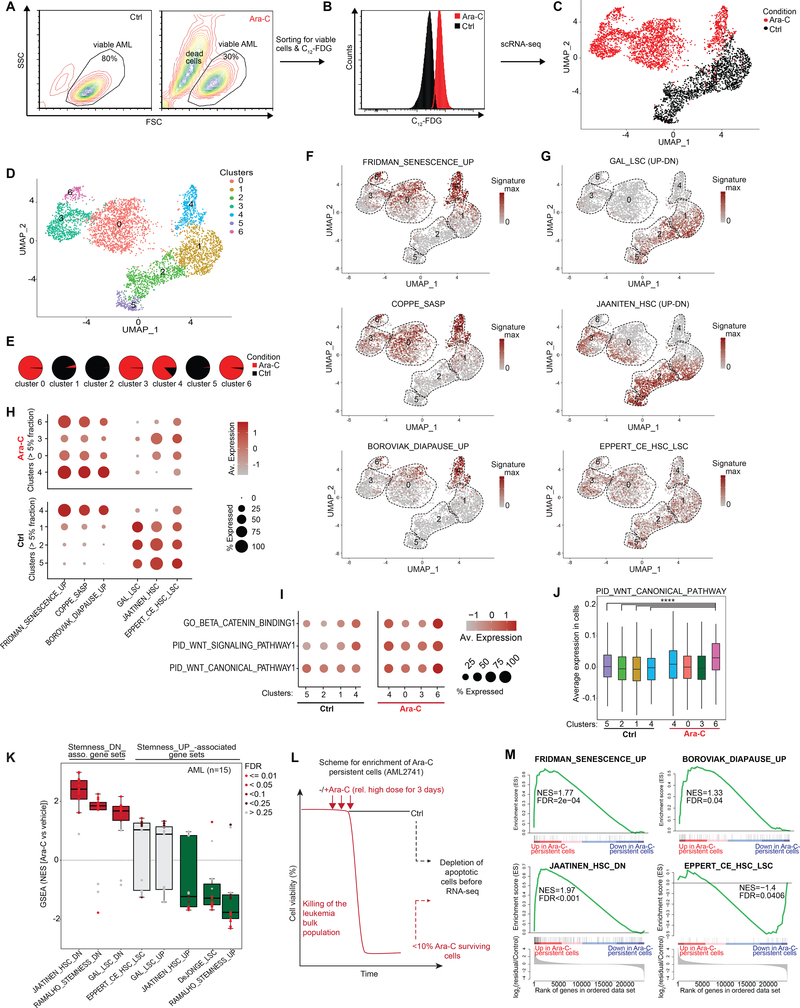
Cancer stem cells are considered inherently resistant to cytotoxic drugs (2) and are hence, expected to be enriched following chemotherapy. Given the ability of senescent cells to repopulate primary AMLs, we next examined whether leukemia stem cells are enriched in this population. For this, we used sorted viable C12-FDGhigh senescence-like primary patient-derived AML cells collected three days after exposure to 1000 nM Ara-C (that killed the majority cells) as well as untreated viable control cells (Fig. 4A–B). These two populations were subjected to single cell droplet-based RNA-seq. Plotting the variance of individual cell gene expression profiles on 4627 cells using UMAP embedding (51) demonstrated separation between the C12-FDGhigh vs untreated AML cells (Fig. 4C). As expected, projecting the bulk RNA-seq gene expression signature derived from Ara-C-treated cells in Fig. 1E showed predominant enrichment among the sorted senescent cells (Fig. S5A). Genes linked to SASP upregulated upon Ara-C treatment (e.g. cathepsins, IL-8) were confirmed to be highly expressed in the senescent population (Fig. S5B, Table S3). On the other hand, MYC and genes associated with ribosome biosynthesis (e.g. RPL38) were among the most downregulated genes (Fig. S5C). MYC target genes were depleted in AML cells that highly expressed senescence- and diapause gene signatures (Fig. S5D). Although there was some degree of heterogeneity among AML cells, we found significant inverse correlation of expression between MYC target genes and chemotherapy-induced stress genes (CISG) that we identified in this study (Fig. S5E, Table S1).
Figure 4. Chemotherapy-surviving AML cells are not enriched for LSCs.
A, Flow cytometry plot displaying primary patient-derived AML1566 cells with and without daily treatment with 1000 nM Ara-C for 3 days. B, Sorted viable C12-FDGhigh senescence-like AML cells collected following three days exposure to 1000 nM Ara-C as well as untreated viable control cells. Sorted viable untreated control cells and Ara-C-treated C12-FDGhigh AML cells were subjected to single-cell RNA-seq. C, UMAP plot of untreated (black; n=1952) and Ara-C-induced senescent AML cells (red, n=2675). D, Same as (C), colored according to the identified 7 clusters. E, Pie chart illustrating the relative distribution within each cluster by untreated control and Ara-C-induced senescent AML cells across the clusters. F, G, UMAP plot displaying the signature scores for senescence and diapause-associated gene signatures (F) as well as LSC/HSC-associated gene signatures (G). H, Dot plot showing the average expression levels (color scale) and the proportion of expressing cells (circle size) per cluster for the indicated gene signatures. Clusters were selected by at least containing >5% of Ara-C-induced senescent cells or untreated control cells, respectively. I, J, Dot plot (I) and box plot (J) showing average expression levels for WNT/β-catenin-associated gene signatures in the AML1566 clusters. P values calculated by Wilcoxon signed-rank test using cluster 6 (Ara-C-treated cells) with any of the cluster in the untreated cells (**** P<0.0001). K, Box-and-whisker plot showing NES scores derived from GSEA using hematopoietic stem cell (LSC/HSC)-relevant gene signatures in 15 primary AMLs (33) after Ara-C treatment (3 days x 100 nM) compared to their corresponding vehicle-treated condition. Data is presented with median (bisecting line in the box) and whiskers representing the 1.5 x of the interquartile range (IQR; 25th – 75th percentile, length of the box). Red box indicates that the majority of samples are positively enriched, while green reflects negative enrichment. Each dot represents a primary case, color of dots reflect FDRs calculated by GSEA. NES, Normalized enrichment score; FDR, False discovery rate. L, Schematic showing the selection of the residual Ara-C-persistent AML cells and depletion of apoptotic cells before RNA-seq analysis. A regimen consisting of 3 days with a case-dependent high dose of Ara-C (100 nM) left ~10% viable cells in AML2741 after chemotherapy that was used to analyze the chemotherapy-persistent leukemia cells. M, GSEA results for senescence and diapause signatures (upper panel) as well as stem cell-related ones (lower panel) in the residual fraction of chemotherapy-persistent leukemia cells vs untreated control cells of AML2741.
To further delineate subpopulations among these cells we performed a k-nearest-neighbor-based analysis, which yielded seven clusters (Fig. 4D and Table S4), that were largely mutually exclusive between control and senescent cells, with the exception of senescence cluster 4, which consisted of ~17% untreated control cells (Fig. 4E and Fig. S4F). We next projected gene sets linked to senescence, diapause or LSC/HSCs on the UMAP plots (Fig. 4F–G). As expected, genes associated with senescence and SASP were highly expressed within Ara-C-treated cells that compose clusters: 0, 3, 4, and 6 (Fig. 4F, H). Diapause genes were most highly expressed in cells composing clusters 4 and 6. Among untreated cells, only the small fraction contained in cluster 4 manifested strong association with senescence signatures, consistent with the presence of small numbers of senescent cells under basal conditions (Fig. 4E, H). Analysis of stem cell signatures showed the highest expression among control cell clusters 1, 2 and 5 (Fig. 4G, H). In contrast there were reduced numbers of cells and lower expression of these signatures within senescent cell populations, although a sizable fraction of cells in clusters 3 and 0 still retained moderate expression of these gene sets (Fig. 4G, H). scRNA-seq analysis of the senescence-like AML cells showed no enrichment of LSC markers that could explain the association with increased AML engraftment (Fig. 3F, Fig. S5G). WNT/beta-catenin pathways were reported to become de novo activated during senescence-associated reprogramming in cancer cells (28) and were likewise induced among cluster 6 post-treatment subpopulations compared to control cells (P<0.0001) (Fig. 4I, J). Altogether, we did not observe an enrichment for expression of LSC-associated genes nor an increase of the abundance of LSCs among chemotherapy-induced senescent cells.
To further confirm that we do not select for LSCs, we performed GSEA for sets of genes that are preferentially expressed or repressed in leukemia stem cells in the set of 15 AML patients treated with Ara-C from Klco et al. 2013 (33) (Fig. 1A). This analysis revealed significant enrichment of genes that are downregulated in stem cells in Ara-C-treated cells, with reciprocal depletion or lack of enrichment of genes that are induced in leukemia stem cells (Fig. 4K). GSEA also suggested induction of WNT/beta-catenin programs after Ara-C exposure (Fig. S5H) although not always significant, perhaps due to the cellular heterogeneity seen at the single-cell level (Fig. 4I, J). In addition, we treated an Ara-C-sensitive primary AML specimen (AML2741, EC50: ~10 nM) with Ara-C of 100 nM (a toxic dose for this patient specimen) for three days, and then performed RNA-seq on the small number of purified viable Ara-C-persistent AML cells after treatment, as well as purified viable cells exposed to vehicle only (Fig. 4L). GSEA analysis showed significant enrichment for senescence, diapause and genes induced by Ara-C at 24 hours (Fig. 4M and Fig. S5I). Genes downregulated in HSC/LSCs were positively enriched in these Ara-C-persisting cells, whereas genes upregulated in HSC/LSCs were significantly downregulated (Fig. 4M).
We next sought to examine which pre-existing AML subpopulations might preferentially undergo senescence and treated six independent patient-derived cases (Fig. S6A, B) that were measured for CD34, CD38, and C12-FDG by flow cytometry. Apart from the expected differences in the cellular composition between AML samples, we found that immature (CD34+CD38-) as well as mature AML cells (CD34-CD38+) can undergo senescence (Fig. S6A, B).
Collectively the data argue against selective enrichment for LSCs immediately following Ara-C treatment, and suggest that survival post-chemotherapy may be linked instead to stochastic induction of senescence-like programs in cells independent of whether they correspond to LSC or bulk AML populations.
Chemotherapy-induced senescence gene signatures suggest potential therapeutic vulnerabilities.
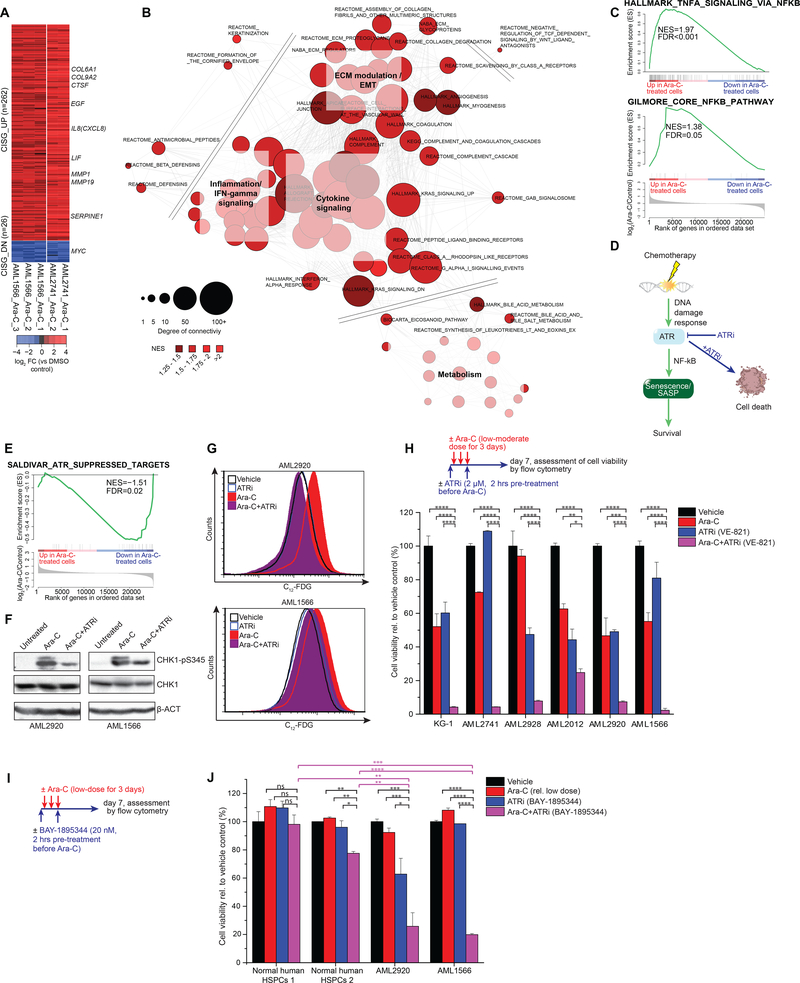
To gain deeper insight into biological functions affected by the AML chemotherapy senescence response, we merged the RNA-seq profiles obtained in the two different primary patient-derived AML specimens shown in Figs 1 and 3, and identified the subset of genes most robustly differentially expressed after Ara-C exposure. This procedure yielded a gene expression signature consisting of 262 upregulated and 26 downregulated genes (2-FC, q<0.05, Fig. 5A, Table S5). A number of the upregulated genes have been previously noted in SASP phenotypes such as IL8, ECM modulators (MMPs, collagens, and cathepsin genes), which were also noted above. We also noted significant induction of diapause regulating genes such as LIF and EGF as well as robust downregulation of MYC (32,45) (Fig. 5A).
Figure 5. Chemotherapy-induced senescence program enables survival of AML.
A, Shown are differentially expressed genes in chemotherapy-persistent AML2741 that were also differentially expressed in AML1566 cells after Ara-C treatment compared to their respective untreated controls. Fold change (FC) scores were calculated for each AML case separately. Genes consistently regulated after chemotherapy were defined as chemotherapy-induced stress genes (CISG). Selected up- and downregulated genes are exemplarily shown for genes of interest. B, Network analysis of leading edge of significantly enriched pathways. The length of each edge reflects the Jaccard index between respective leading edge pairs and the size of each node reflects its degree of connectivity among other nodes. C, GSEA demonstrating enrichment for NF-κB signaling programs after Ara-C-treated patient-derived AMLs. D, Schematic showing the ATR kinase controlling DNA damage response upstream of chemotherapy-induced senescence. E, GSEA demonstrating ATR suppressing target genes (55) are downregulated in Ara-C-treated patient-derived AMLs. F, Immunoblot for phospho-CHK1 (Ser345), total CHK1, and beta actin (β-ACT) in patient-derived AML cells after 24 hours of treatment. Cells were pre-treated with an ATR inhibitor (ATRi; 2 μM VE-821) for 2 hours before Ara-C exposure (1000 nM). G, Patient-derived AML cells were pretreated with an ATR inhibitor (ATRi; 2 μM VE-821) for 2 hours on day 0 and 2 prior exposure of Ara-C for three consecutive days starting at day 0 (AML2920: 125 nM, AML1566: 500 nM). Flow cytometry histogram showing ATR inhibition reduced the formation of the SA-β-Gal activity in response to Ara-C. H, Upper panel illustrates the treatment regimen: AML cells were pre-treated with 2 μM ATRi for 2 hours on day 0 and 2 prior exposure of a case-specific low-to-moderate Ara-C dose, treated from day 0 till day 3. Cell viability was measured by flow cytometry using PI exclusion one week after treatment start. (Mean of three replicate wells ± s.d.). I, Scheme illustrating the treatment regimen: Cells were pre-treated with 20 nM BAY-1895344 for 2 hours on day 0 and 2 prior exposure of a case-specific low Ara-C dose, treated from day 0 till day 3. Cells were measured by flow cytometry using PI exclusion one week after treatment start. J. Cell viability in normal human HSPCs and patient-derived AML cells normalized to their corresponding vehicle control set as 100%. (****P<0.0001, ***P<0.001, **P<0.01, *P<0.05; Student’s t-test, mean ± s.d.).
To further understand the biological functions induced in these cells upon chemotherapy-induced senescence, we performed GSEA and a leading edge network analysis using canonical pathways from the Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/search.jsp) (Fig. 5B, Table S6). Analysis of these gene networks strongly point to induction of inflammatory genes that are known to be induced by NF-κB activation upon induction of SASP (52). Indeed, we observed significant enrichment for NF-κB target genes among genes that are upregulated in primary AML cells exposed to Ara-C (Fig. 5C). Recent reports place ATR and ATM as upstream mediators of senescence associated NF-κB activation (53,54) If the senescence-like phenotype observed in AML after chemotherapy is induced by ATR and is required for the resilience of these cells, then ATR targeted therapy would be expected to prevent AML cells from achieving this protective state (Fig. 5D). Supporting this notion, we observed significant downregulation of genes suppressed by ATR, when examined by GSEA in Ara-C-treated AML cells (FDR=0.02, Fig. 5E) (55).
To determine whether ATR activity was induced by Ara-C in AML cells, we measured the abundance of phospho-CHK1 as a biochemical read-out for ATR function. These experiments confirmed induction of CHK1 phosphorylation vs. total CHK1 in AML specimens after exposure to Ara-C. This effect was severely impaired when cells were exposed to the specific ATR inhibitor VE821 (56) just prior to treatment with Ara-C for 24 hours (Fig. 5F). Since genetic deletion of ATR protein is lethal to dividing cells (57,58), we therefore examined the effect of catalytic inhibition with VE821, a selective ATR inhibitor chemically related to VX-970 (M6620), which is currently in clinical trials (59,60). AML cells were pretreated for 2 hours with VE821 (2 μM) or vehicle and then exposed to Ara-C or vehicle for 3 days, followed by C12-FDG flow cytometry. Strikingly, the VE821 severely impaired C12-FDG induction, suggesting that ATR activation plays a critical role in inducing the senescent/SASP phenotype in AML context (Fig. 5G). If the senescent phenotype has a protective role then it would be expected that VE821 would prevent survival of AML cells after exposure to Ara-C. For this we exposed a panel of five primary AML specimens as well as the KG-1 AML cell line to Ara-C, VE821, or the combination of both for 3 days to assess cell viability one week after treatment (Fig. 5H). In half of the cases Ara-C and VE821 killed roughly 50% of the AML cells, whereas the combination killed 95% of cells (Fig. 5H). However, in the other three cases killing was supra-additive since these cells were relatively resistant to either Ara-C or VE821, yet still manifested >95% killing in the presence of the combination. These results were confirmed using a second, more potent ATR inhibitor BAY-1895344, which is also in clinical trials (61) (Fig. 5I–J). In contrast these drugs did not induce a marked killing effect in normal donor hematopoietic stem/progenitor cells (HSPCs) (Fig. 5J). Fordham et al. recently demonstrated that in vivo treatment with ATR inhibitor and chemotherapy also yields dramatically enhanced activity against patient-derived AML in mice (62). Altogether, these results suggest that ATR targeted therapy strongly potentiates the efficacy of chemotherapy in AML by preventing these cells from being rescued through induction of the senescence resilience phenotype.
Senescence-like AML cells are enriched at post induction chemotherapy nadir in AML patients.
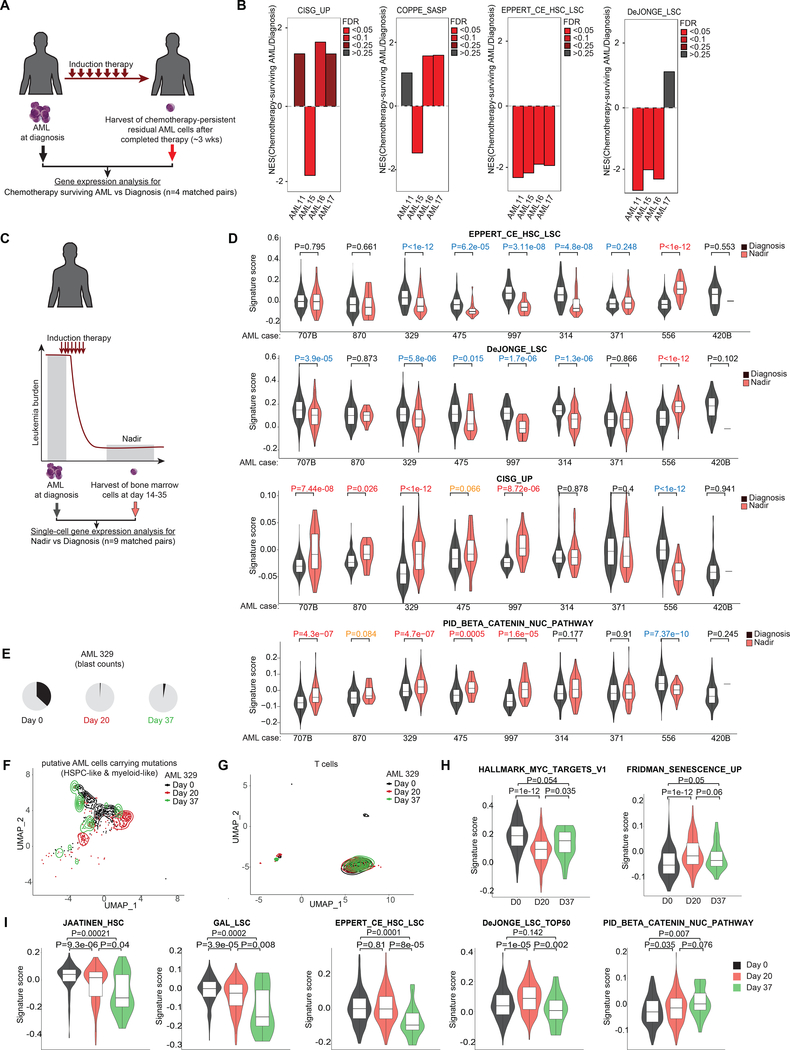
If it is true that AMLs enter a senescence-like state that enables them to persist after exposure to cytotoxic drugs, then we would expect that such cells would be enriched in the bone marrow of AML patients after induction chemotherapy. To determine if this was the case, we investigated gene expression profiles of residual AML cells that persist in the bone marrow of four human subjects approximately three weeks following induction therapy, as compared to their matched diagnostic specimens (Fig. 6A) (4). This analysis revealed enrichment of our CISG_UP senescence and SASP signatures in the residual AML cells in 3 of these patients (Fig. 6B). In contrast there was depletion of leukemia stem cell signatures in the patients.
Figure 6: Chemotherapy-persistent cells are enriched for CIS cells instead of LSCs in patients.
A, Scheme showing AML samples from patients whose bone marrow disease persisted post-treatment despite successful blast reduction in peripheral circulation (4) (n=4). B, GSEA of chemotherapy-persistent AML shown in (A) using indicated gene sets. C, Scheme illustrating scRNA-seq (63) of bone marrow cells collected from AML patients at diagnosis and nadir following induction therapy. D, Violin plot showing signatures scores from (C) for selected gene sets in single cells composed of myeloid, myeloid-like, hematopoietic stem/ progenitor cells (HSPCs) and HSPCs-like cells characterized by Galen et al. 2019 (63). P values calculated by Wilcoxon signed-rank test between cells at nadir vs diagnosis. P values in red reflect significant upregulation in nadir samples vs diagnosis (orange depict tendency for upregulation), whereas values in blue represent significant downregulation. E, Pie charts showing the increase of blast counts of AML patient 329 at day 37 following induction chemotherapy (63). F, UMAP plot showing the distribution at diagnosis, day 20, and day 37 of putative AML cells composed of HSPC- and myeloid- like cells carrying AML-related mutant genes identified by van Galen et al. (63). G, UMAP plot showing the distribution at diagnosis, day 20, and day 37 of T cells. H, Violin plots showing the signature scores of MYC targets and senescence gene sets at single cell level in AML cells at diagnosis, day 20, and day 37 in #pt329 following induction therapy. I, Plots showing signature scores for selected LSC/HSC and β-catenin-associated gene sets in AML cells at indicated time points in #pt329.
We next determined the distribution of the senescence signature across the population of residual AML cells persisting 14 to 35 days post induction chemotherapy using single-cell RNA-seq data collected from nine AML patients, as compared to their matched diagnostic specimens (Fig. 6C) (63). Analyzing the gene expression profiles of myeloid, myeloid-like, HSPCs, and HSPCs-like cells collected from the bone marrow of AML patients, we found generally depletion or no enrichment of LSCs in 7 out of 9 AML patients at nadir compared to diagnosis (Fig. 6D and Fig. S7). On the other hand, we found around half of the patients (n=4 with P<0.05 and n=1 with P=0.066) exhibited significant enrichment for the CISG_UP signature at nadir (Fig. 6D). Similarly, we also observed that many cases displayed increased abundance of cells expressing SASP, senescence, diapause, inflammation, and beta-catenin signatures (Fig. 6D and Fig. S7). Of note, we found upregulation of several differentiation markers in these cases at nadir. This is coherent with the observation that differentiation markers can be induced by DNA damaging agents (64). Still, we did not observe the same set of differentiation markers higher expressed across different AML cases, which we expected for this heterogenous disease (Fig. S8).
Single-cell genotyping in one of these patients yielded sufficient cells to examine gene expression in genetically defined AML cells at diagnosis, day 20 after beginning induction therapy (<1% blasts) and a subsequent time point with increased blast counts at day 37 (3% blasts) (Fig. 6E). This allowed us to exclude potential normal cells from our analysis (63). AML cells at day 37 revealed closer clustering to diagnosis samples as compared to nadir cells (Fig. 6F), whereas T cells did not manifest detectable transcriptional difference among the three timepoints (Fig. 6G, controlling for variability between specimens). We observed strong MYC target gene signature expression in AML cells at diagnosis and significantly lower levels at nadir (P<1e-12), which were then significantly reversed at day 37 (P=0.035) (Fig. 6H). Conversely, we found significantly higher expression of senescence-related genes at nadir compared to diagnosis (P<1e-12) and partial reversal by day 37 (P=0.06) (Fig. 6H). Most LSC/HSC signatures still showed significant reduction at day 37 compared to diagnosis, except for one of these signatures (Fig. 6I), whereas beta-catenin/WNT pathway genes were enriched at nadir (Day 20) compared to diagnosis (P=0.035, respectively), and remained elevated as this patient was relapsing (P=0.007) (Fig 6I).
Overall, the data indicate preferential enrichment of senescence-like resilient AML cells in patient specimens collected at nadir after induction therapy.
Although often depleted at nadir, LSC signatures are enriched at relapse.
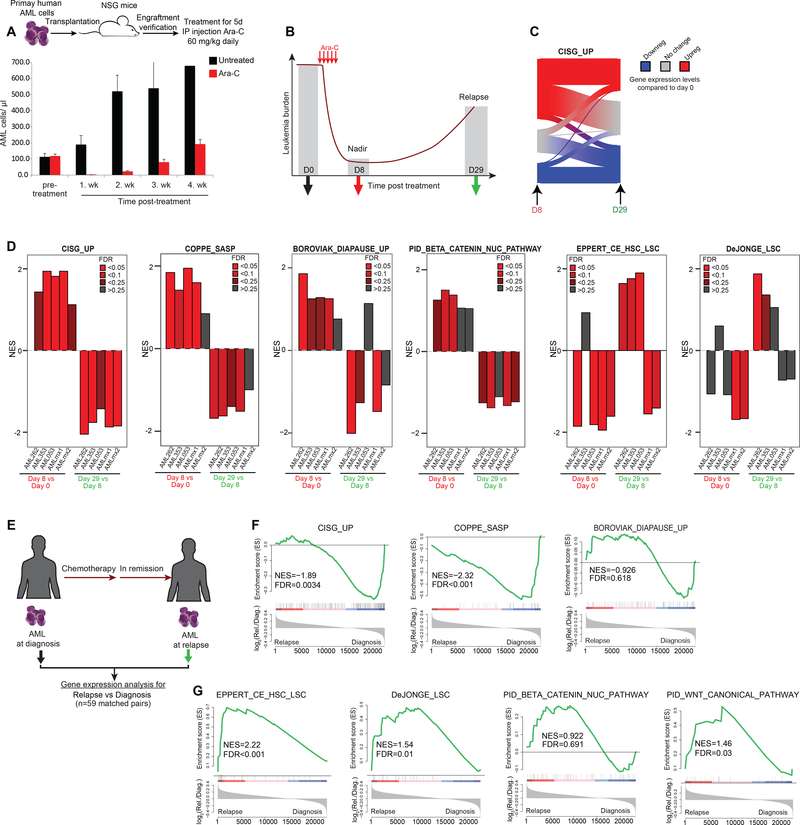
Our data suggest that senescence signatures are enriched at nadir and are associated with persistence of AML cells that can repopulate the disease when they recover. To explore senescence and LSC/HSC signatures upon recovery of primary AMLs in vivo after nadir while excluding normal cells from our analysis, we established an AML relapse model using primary human AML specimens engrafted in NSG mice (Fig. 7A). After validation of engraftment of human blasts, mice were treated with a clinically relevant dose of Ara-C (60 mg/kg/day, equivalent to induction therapy in humans (5) for five consecutive days, which resulted in severe cytoreduction and nadir by day 8, followed by leukemic relapse occurring after 4 weeks (Fig. 7A). We performed gene expression profiling on AML cells collected from NSG mice at three timepoints: just prior to treatment, at nadir (8 days post-treatment), and after leukemia relapse (day 29 post-treatment) (Fig. 7B). We found that the genes composing the CISG_UP-induced signature were upregulated at nadir, but were partially reverted to their pre-treatment levels at day 29 (Fig. 7C). We then engrafted four additional independent human primary AMLs into NSG mice, treated them with the same Ara-C regimen, and performed gene expression analysis on AML cells purified cells pre-treatment, at nadir and upon relapse (Fig. 7D). Using GSEA on these gene expression profiles revealed enrichment for the CISG_UP, SASP senescence and diapause signatures as well as partial enrichment for beta-catenin at nadir (day 8) vs pre-treatment (day 0) (Fig. 7D). In contrast there was no enrichment for LSC signatures at day 8, which were instead often significantly depleted. However, when comparing gene expression profiles at day 29 (relapse) vs day 8 nadir, we observed the opposite effect: depletion of CISG_UP, SASP, diapause and beta-catenin signatures, and enrichment for LSC profiles in at least three of the five cases. These data suggest senescent programs resolve as AML cells recover from chemotherapy-induced senescence-like state, and as they emerge from this state they feature enrichment for more stem-like transcriptional programming. These findings are consistent with a recent report showing that senescence is accompanied by epigenetic plasticity and reactivation of stem cell functionalities (28). To further examine this point, we performed RNA-seq analysis in 59 matched AML specimens collected at diagnosis vs relapse (Fig. 7E). GSEA analysis demonstrated that AML cells exhibit a depletion for senescence gene signatures and an enrichment for LSC-signatures at relapse (Fig. 7F, G). Relapsed AML compared to diagnosis samples demonstrated partial enrichment of WNT/beta-catenin programs (Fig. 7G). Overall, these data suggest that survival after exposure to chemotherapy is most likely stochastic, involves a diapause/SASP-like resilience response, and that subsequent enrichment for stem-like features at relapse could reflect the consequence, rather than the cause of leukemic survival after exposure to chemotherapy.
Figure 7. Relapse following chemotherapy-induced senescence is linked to the LSC program.
A, Representative plot showing the patient-derived xenograft (PDX) AML relapse model. Upper panel illustrates the process of engrafting human primary AML in NSG mice with subsequent chemotherapy administration. Clearance of human leukemia blast in the blood is achieved one week after Ara-C treatment but shows dominant recurrence of the disease at week 4. B, Scheme demonstrating the time points of sample extraction for gene expression analysis in our PDX AML relapse model. C, Sankey diagram depicting gene expression changes of CISG_UP- associated genes at nadir vs recovery. Results from primary AML cells at nadir demonstrated upregulation of many CISG_UP genes. Average of gene expression changes (normalized to untreated day 0) from three biological replicates at given time points. D, Plots showing NES results from GSEA using the indicated gene sets in five different engrafted primary AML samples. GSEA was performed by comparing gene expression profiles from AML cells harvested at day 8 vs day 0 and day 29 vs day 8. E, Scheme of matched AML samples collected from patients at diagnosis and relapse (n=59). F, GSEA showing the enrichment for senescence-, diapause- and SASP/inflammatory-associated gene signatures in relapsed AML vs diagnosis. G, GSEA results for LSC and WNT/β-catenin signatures in relapsed AML vs diagnosis.
DISCUSSION
AML relapse occurs in a majority of patients after chemotherapy and portends incurable disease. While selection of clones containing drug resistance mutations (e.g. TP53) following chemotherapy are relatively rarely observed in relapsed AML, a number of studies suggest that relapse is explained by the presence of inherently more resistant leukemia stem cells as the origin of relapse (65,66). Yet, most of these studies analyzed samples at diagnosis and relapse but not at nadir. Although LSC gene expression signatures are predictive of relapse (67,68), it does not necessarily mean that these cells are enriched during exposure to chemotherapy. Indeed, while we also observed an enrichment of LSC gene signatures at relapse, we did not find LSC gene signatures in the residual fraction of chemotherapy-persistent leukemia cells at nadir. This is consistent with other recent studies reporting no enrichment of LSCs at nadir (4,5). Instead, we found that AML cells surviving initial chemotherapy manifest a senescence-like resilient phenotype, with potential to repopulate leukemia. Previous studies analyzing the transient state of chemotherapy-persistent AML at nadir indicate the expression of genes related to SASP/inflammation and senescence-like cells (4,5). Upon recovery these cells manifested positive enrichment of LSC-gene signatures.
We suggest that LSC programming in many AML cases occurs during the senescence-like resilient phenotype, explaining how these stem cells are enriched at relapse. This hypothesis is supported by a recent study showing that doxorubicin-induced senescence reprograms murine leukemia and lymphoma cells to cancer stemness (28). We do not exclude that in certain AMLs with initially abundant LSCs, these cells could survive chemotherapy without major harm and repopulate the disease. Given the heterogeneity between AML patients, it is also possible that other resilience mechanisms could exist that were not captured here. Yet, our data suggest that many AMLs enter a stress-adaptive and senescence-like resilient phenotype regardless of whether they are LSCs or not, and that this process plays a critical role in relapse, possibly through an acquired stemness reprogramming process mediated by the WNT/β-catenin pathway as described previously (28). Alternatively, adaptation may reflect either initial epigenetic heterogeneity or epigenetic plasticity as possible mechanisms of resilience. Fundamental to understanding this process is the inherent plasticity of cells to reprogram from one state to another. Importantly in this regard, cancer stem cell functionality is not a fixed property but rather can be induced under stress conditions (28,69). Another layer of complexity in this context is that different AMLs require different doses of chemotherapy to induce a comparable senescence phenotype in which some refractory AMLs will only manifest overt senescence phenotypes when exposed to higher drug concentrations. Furthermore, AML is a heterogenous disease and we cannot exclude that certain AML subtypes induce the senescence phenotype to a different degree than other subtypes.
Our findings are consistent with a report that investigated drug-resistance in solid tumors and revealed that the drug-persistent state shares a diapause-like dormancy signature (70). The diapause principle in theory represents a long-term dormancy state. Almost half of senescence-associated genes are also expressed in quiescent cells, potentially indicating a mechanistic overlap between senescence and dormancy (71). The concept of a transient stress-induced endurance state is a common phenomenon described in developmental biology, whereby early embryos survive adverse conditions via transition into diapause (32). MYC controls cellular growth and depletion of MYC can induce a diapause-like state (45,72). Notably, translocations leading to constitutive expression of MYC are associated with better prognosis in certain cancers (e.g. Burkitt lymphoma)(73), presumably in part because of the aggressive cell proliferation that incorporates more chemotherapy. Thus, suppression of MYC may represent a survival strategy of cells to endure stress. We have used the phrase, “senescence-like phenotype” to describe our results. As note, AML cells treated with chemotherapy induce a cell cycle arrest and growth pause similar to replicative senescence but unlike replicative senescence the growth pause is not permanent. The observed senescence transcriptional profiles are most similar to SASP, a specific phenotype of the senescent state initially described in response to genotoxic stress (13). SASP is characterized by an inflammatory response that may contribute functionally to leukemic recovery, consistent with notions on regenerative properties of AML nadir cells described by Boyd et al. (4). Among SASP genes, we observed induction of IL-8, which has been described to protect AML cells from the effects of chemotherapy (74)
Our data indicate that ATR plays a critical role in the chemotherapy-induced senescence resilient phenotype. This is consistent with previous reports showing that ATR can induce SASP through induction of NF-κB in a p53-independent manner (53,54). Indeed, we show for the first time that induction of senescence as well as tumor survival and persistence is dependent on ATR catalytic activity, through our experiments using chemotherapy treatment in primary human AML cells. Moreover, others have shown that treatment of primary AML cells with ATR inhibitor and chemotherapy is also remarkably effective in vivo (62). ATR inhibitors may thus represent a novel approach for targeting senescence resilience by preventing cells from harnessing this survival mechanism. It is intriguing to consider the potential for ATR inhibitor to potentiate the effect of drugs that directly target senescent cells such BCL2 inhibitors, which have been used to enhance the efficacy of chemotherapy and other drugs in AML patients (75,76). ATR inhibitors are currently in clinical trials both as single agent as well as in combination with various chemotherapy drugs for solid tumors. Collectively, these and our data strongly point to the rationale for initiating clinical trials of ATR inhibitor in AML patients. In summary, our findings suggest that AML relapse can be explained by entry of these cells into an ATR-dependent senescence-like resilient state of variable duration that endows them with superior fitness and enhanced ability to repopulate the disease. Although this effect appears to be independent of their initial stemness profiles, it may make these cells highly vulnerable to drugs such as ATR inhibitors, with potential to better eradicate these tumors.
MATERIAL & METHODS:
Primary AML samples.
Primary human AML specimens were collected after written informed consent and obtained from peripheral blood or bone marrow aspiration according to the declaration of Helsinki for collection and use of sample materials. The studies were approved by the institutional review board at Weill Cornell Medicine (IRB protocol # 0805009783). Information on the primary AML cases used in this study are described in Table S7 and in more detail elsewhere (36,49).
Culturing of primary patient-derived AML.
Primary AML were cultured ex vivo as previously described (36). In short, primary patient-derived AMLs were expanded on stromal feeder layers with cytokine support.
AML cell lines.
Human myeloid leukemia cell lines were purchased from ATCC or DSMZ and maintained in IMDM media containing 10–20% fetal bovine serum, 100 IU ml-1 penicillin and 100 μg ml-1 streptomycin. Cell lines were validated via short tandem repeat DNA profiling and monitored regularly for mycoplasma contamination.
Isolation of HSPCs.
Mononuclear cells (MNCs) were isolated from fresh human umbilical cord blood samples (New York Blood Center, NYBC) using Ficoll (Atlanta Biologicals, GA) density gradient centrifugation. After lysis of red blood cells, hematopoietic stem and progenitor cells (HSPCs) were selected via immunomagnetic enrichment of CD34+ MNCs using CD34 MicroBead Kit and Automacs from Miltenyi Biotec (Auburn, CA). HSPCs were propagated in our ex vivo co-culture model as previously described (36).
Drug preparation and treatment.
Stock solutions of inhibitors against ATR (VE-821, BAY-1895344) derived from Selleckchem (Houston, TX) were prepared in DMSO and stored at -80°C. Cells were exposed to 2 μmol l−1 (VE-821) or 20 nmol l−1 (BAY-1895344) in a dilution factor that did not pass 0.3% DMSO content in growth medium during whole treatment time. Cytarabine (Sigma-Aldrich, St. Louis, MO), Daunorubicin (EMD Millipore, Billerica, MA), and Adriamycin/Doxorubicin (Selleckchem) was prepared in water and kept protected from light at 4°C.
Classical SA-β-Gal activity assay using X-Gal.
SA-β-Gal staining kit from Cell Signaling was used for SA-β-Gal analyses and performed according to the manufacturer’s protocol (BD Biosciences, San Jose, CA).
Fluorogenic SA-β-Gal activity assay using C12-FDG.
Cells cultured for prolonged period were maintained in well plates that had PBS or plain media in the outer wells to reduce edge effects. Otherwise, cells cultured at outer wells exhibited stronger evaporation effects that increased intracellular stress and SA-β-Gal activity. C12-FDG procedure was performed as previously described (37) using 20 μM C12-FDG (Thermo Fisher Scientific, Waltham, MA). C12-FDG was prepared in pre-warmed media and added to cells with gently mixing of the cell suspension. Cells were protected from light and incubated for 2 hours at 37°C and 5% CO2 before flow cytometry measurement.
Flow cytometry.
Cell viability was assessed by the propidium iodide (PI)-negative cell proportion, while cell growth was measured by absolute numbers of viable cells (PIneg) including forward scatter (FSC) and sideward scatter (SSC) size discrimination. Annexin V co-stained with propidium iodide (PI) was used for apoptosis analyses and performed according to the manufacturer’s protocol (BD).
In vivo PDX relapse model.
Mononuclear cells extracted from patients were transplanted into NOD/SCID/IL2Rγnull (NSG) mice, which were obtained from The Jackson Laboratory. Mice were randomly assigned to treatment groups. The investigators were not blinded to allocation during experiments and outcome assessment. No power analysis was used to predetermine sample size. Cytosine arabinoside (Ara-C) administered using a physiologically relevant dose and schedule (60 mg/kg/day x 5 days) to AML engrafted NSG mice induced a drop in peripheral blood leukemic cells and total body leukemic burden by one week after initiation of therapy. Human AML cells were collected from 4 bones and spleen after therapy. Cells from weeks 1 and 4 were purified using human anti-CD45 antibody and analyzed for gene expression using RNA-seq or Affymetrix Human Gene 1.0 ST arrays. All studies were conducted in accordance with the guidelines of and approved by the Institutional Animal Care and Use Committee at the institution where the work was performed.
In vivo leukemia repopulation of senescence-like cells.
Untreated and treated AML cells with Ara-C for 3 consecutive days were stained with C12-FDG on the following day. Cells were washed and stained with sterile PI for cell viability discrimination. Cells were sorted with a FACSAria II system (BD) into FBS and centrifuged after collection was finished. Cell pellet was washed and resuspended in plain IMDM media. Cells were transplanted into female NSG mice via tail vein injection of 500,000 cells per condition per mouse.
Colony forming assay.
After sorting, 5,000–25,000 cells/well of primary patient-derived AML were seeded in MethoCult (H4534) medium (StemCell Technologies, Vancouver, BC) supplemented with cytokines (36). The MethoCult medium-cell mixture was plated into six-well plates with water supply in the inter-well chamber. Additionally, the six-well plate was placed inside a tub with extra water dishes to prevent evaporation. After three weeks of incubation at 37°C in 5% CO2, pictures were acquired by using ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA) or STEMvision (Stemcell Technologies) and colonies were counted using ImageJ software.
Immunoblot.
Lysates from leukemia cells were lysed using PBS containing 1% NP-40 and 1 mM DTT supplemented with 1% protease/phosphatase inhibitor cocktail (cOmplete, Roche, Indianapolis, IN; Phosphatase Inhibitor Cocktail A, Santa Cruz Biotechnology, Dallas, TX) and were sonicated for 15 seconds using a tip sonicator (Branson Sonifier 450). Protein lysates were resolved by SDS-PAGE, transferred to PVDF membrane, and probed with the indicated primary antibodies: CHK1 (clone: 2G1D5, Thermo Fisher Scientific), phospho-CHK1 Ser345 (clone: 133D3, Cell signaling, Danvers, MA), p53 (SC-126, Santa Cruz Biotechnology), p16 (ab108349, Abcam, Cambridge, MA) or beta-actin (Sigma). Membranes were then incubated with a peroxidase-conjugated correspondent secondary antibody and detected using Pierce ECL chemiluminescent substrate (Thermo Fisher Scientific).
Depletion of apoptotic cells.
When cell viability decreased below 50%, a depletion step was introduced for all relevant assays except for flow cytometry. Dead cells and cellular debris were removed via magnetic column depletion using the cell death removal kit (Miltenyi Biotec, Auburn, CA) according to the company’s instructions.
Single-cell organoid AML recurrence:
Individual clones were established before and after killing the bulk leukemia following chemotherapy exposure. Briefly, parental and nadir collected AML cells were serially diluted to a final concentration of 1 cell per two wells in a 96- or 384-well plate. Each well was checked and monitored that contained a single cell. Single cells were cultured using the ex vivo platform as previously described (36). Recovered clones were picked from the wells and sub-cultured in larger wells.
5-mC immuno dot blot.
Genomic DNA was extracted using the Purelink Genomic DNA kit (Thermo Fisher Scientific) and quantified via NanoDrop (Thermo Fisher Scientific). Dilution of DNA samples were denatured with 0.1 mol l−1 NaOH at 95°C for 5 min. Subsequently, samples were rapidly chilled for 5 min on ice, spun down and neutralized with 0.1 volume of 6.6 mol l−1 ammonium acetate. DNA samples were spotted onto a positively charged nylon membrane (Immobilon-NY+, EMD Millipore, Bedford, MA), air dried for 15 min and UV-crosslinked for 5 min. Membranes were blocked in PBS containing 0.05% Tween-20 and 5% non-fat milk powder (PBS-TM) for 1h at room temperature (RT). After blocking, membranes were washed in PBS-T for 3×5 min at RT and incubated with the 5-mC antibody (NA81, EMD Millipore) in a 1:1000 dilution in PBS-TM for 1 h. Subsequently, membranes were washed with PBS-T for 3×5 min at RT and incubated with a secondary antibody for 1 h. After final washing, membranes were incubated in Pierce ECL chemiluminescent substrate (Thermo Fisher Scientific).
Single-cell RNA-seq library preparation.
AML cells were sorted with a FACSAria II system (BD) into FBS and washed with PBS after collection was finished. Sorted viable untreated AML cells and Ara-C-induced senescent AML cells were processed together on the same day on a 10× Chromium instrument (10× Genomics) to generate barcoded single-cell GEMs according to manufacturer’s protocol. Single-cell RNA-Seq libraries were prepared using Chromium Single Cell 3’ v3 Reagent Kit (10× Genomics) following the directions of the manufacturer. Briefly, the initial step consisted in performing an emulsion where individual cells were isolated into droplets together with gel beads coated with unique primers bearing 10× cell barcodes, unique molecular identifiers (UMI), and poly(dT) sequences. GEM-Reverse Transcription (53 °C for 45 min, 85 °C for 5 min; held at 4 °C) was performed in a C1000 Touch Thermal cycler with 96-Deep Well Reaction Module (Bio-Rad, Hercules). After RT reaction, GEMs were broken and the single-strand cDNA was cleaned up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific, Waltham, MA). The cDNA was amplified for 12 cycles (98 °C for 3 min; 98 °C for 15 s, 63°C for 20 s, 72 °C for 1). Quality of the cDNA was assessed using an Agilent Bioanalyzer 2100 (Santa Clara, CA), obtaining a product of about 1400bp. This cDNA was enzymatically fragmented, end repaired, A-tailed, subjected to a double-sided size selection with SPRIselect beads (Beckman Coulter, Indianapolis, IN) and ligated to adaptors provided in the kit. A unique sample index for each library was introduced through 12 cycles of PCR amplification using the indexes provided in the kit (98 °C for 45 s; 98 °C for 20 s, 54 °C for 30 s, and 72 °C for 20 s x 14 cycles; 72 °C for 1 min; held at 4 °C). Indexed libraries were subjected a second double-sided size selection, and libraries were then quantified using Qubit fluorometric quantification (Thermo Fisher Scientific, Waltham, MA). The quality was assessed on an Agilent Bioanalyzer 2100, obtaining an average library size of 455bp. Libraries were pooled and normalized to 2 nM and clustered on a HiSeq4000 high output mode on a pair end read flow cell and sequenced for 28 cycles on R1 (10x barcode and the UMIs), followed by 8 cycles of I7 Index (sample Index), and 98 bases on R2 (transcript), with a coverage around 130M reads per sample. Primary processing of sequencing images was done using Illumina’s Real Time Analysis software.
Single-cell RNA-seq data analysis.
Single-cell expression was analyzed using the Cell Ranger Single Cell Software Suite (v3.0.2) to perform quality control, sample de-multiplexing, barcode processing, and single-cell 3’ gene counting (77). Sequencing reads were aligned to the refdata-cellranger-hg19–3.0.0 transcriptome using the Cell Ranger suite with default parameters. Single cells from control and Ara-C-treated conditions, a total of 6278 single cells with a mean of ~3200 genes per cell were imported into Seurat 3 package (78) in R statistical software. Only genes detected in at least two cells were considered for further analysis. Apoptotic cells were filtered out by any cell containing >15% of mitochondrial UMI counts. To detect changes in gene signatures more robustly, any cell with <1500 genes was filtered out. To reduce outlier effects, cells with a total count of >40000 UMIs or >6000 genes per cell were filtered out. Overall, a total of 4627 cells were kept for downstream analyses. After merging control and Ara-C treated cells using the merge function, cell expression levels were normalized and scaled using default settings in Seurat. Principal component analysis (PCA) for dimensionality reduction was then run on the normalized gene-barcode matrix. UMAP (51) embedding was applied to visualize the cells in the two-dimensional space after selecting the first 20 principal components for the clustering analysis with a resolution parameter of 0.4. This graph-based clustering method relies on a clustering algorithm based on shared nearest neighbor (SNN) modularity optimization, which first calculates k-nearest neighbors to construct the SNN graph. Cluster-specific genes were identified by running the Seurat “FindMarkers/FindAllMarkers” function with the Wilcoxon Rank Sum test. Signature scores were computed using the Seurat function “AddModuleScore” and the corresponding gene set. A positive score suggests that the module of genes in a particular cell is higher expressed than randomly expected, given the average expression of this module across the population. To identify more robustly transcriptional programs for certain phenotypes (JAATINEN HSC; GAL LSC) that exhibited a large fraction (>1/3) of DEG both up- and downregulated, we collapsed the two sets into one score by taking the difference of the signature scores for up- and down-regulated gene signatures. Gene signature sets were compiled from the Molecular Signatures Database (MSigDB) or elsewhere (described in GSEA section) and summarized in Table S1. UMAP, dot and violin/box plots for single cell analysis were plotted using the Seurat package and R.
Single-cell RNA-seq analysis of bone marrow samples of AML patients.
Processed scRNA-seq data of bone marrow aspirates before and shortly after induction therapy (day 14 – 35, defined as nadir) including the annotation files were downloaded as tab-delimited text files from the GEO series GSE116256 (63) (Table S8). The annotation file denoted the cell types for each cell and the expression matrix contained the number of UMIs for each gene (rows) and cell (columns). The files were converted into a Seurat object using the functions “CreateSeuratObject” and “AddMetaData” in the Seurat package. After normalization and scaling, gene enrichment scores were calculated using designated gene sets with the “AddModuleScore” function. For downstream analysis, we first focused on single cells composed of the following cell types in van Galen et al. (63): HSC, GMP, Prog, Prog-like, ProMono, Mono, Mono-like, ProMono-like, HSC-like, or GMP-like. To further focus on putative AML cells in the bone marrow, we used hematopoietic cells that carried AML-associated mutant genes identified by van Galen et al. (63) and defined as: Prog-like, Mono-like, ProMono-like, HSC-like, or GMP-like cells.
Gene expression of persistent AML cells in patients.
Gene expression data were downloaded from the GEO series GSE75086 (Table S8) (4). AML samples were obtained from patients belonging to intermediate- and high-risk prognostic groups. Bone marrow aspirates of persistent disease were collected approximately 3 weeks following the completion of standard induction chemotherapy. Whenever leukemic blasts did not compose the majority of the mononuclear cells, Boyd et al. (4) sorted leukemic blasts from both pre- and post-treatment cells based on CD45 intensity and side scatter gating.
Bulk RNA-seq.
High quality RNA of treated samples was isolated using guanidinium thiocyanate-phenol-chloroform extraction (Trizol; Thermo Fisher Scientific) with subsequent purification on silica-membrane columns using Qiagen RNeasy kit. Quality of was RNA was validated using the 2100 Bioanalyzer system (Agilent, Santa Clara, CA). Samples were multiplexed and single read 50 bp sequenced on HiSeq 2500 sequencing system (Illumina, San Diego, CA). Reads were aligned to hg19 using STAR aligner (79).
Bulk RNA-seq analysis of ex vivo cultured AML cells.
Hg19 annotated RefSeq transcripts were counted with FeatureCounts (80) in the Rsubread package using the union-exon based approach. Mapped counts were normalized by the trimmed mean of M-values (TMM) method and DEGs were calculated using a generalized linear model in the edgeR (81) package in R.
Bulk RNA-seq of clinical AML samples at diagnosis and relapse.
Collecting specimens and processing them for RNA-seq analysis are described in detail elsewhere (49). Information of patients that were previously published and deposited at GEO under GSE83533 and at dbGaP under accession number phs001027.v1.p1. Additional cases of previously unpublished AMLs used in this study were added the same dbGaP number.
Gene set enrichment analysis (GSEA).
Gene set enrichment analysis was performed using GSEA (34) software (http://www.broadinstitute.org/gsea/) and R software. Senescence-, inflammation-, diapause, mitotic cell cycle, H/LSC-associated gene sets were obtained or compiled from previous publication (12,15,35,55,82) and the MSigDB (http://software.broadinstitute.org/gsea/msigdb/search.jsp). The diapause signature was defined by selecting differentially expressed genes (2-fold log change and adjusted p-value <0.05) in diapaused epiblasts vs pre-implantation E4.5 epiblasts in Boroviak et al., 2015 (35). Compiled gene sets are listed in Table S1. In general, given the lack of coherence in most expression datasets and the relatively small number of gene sets being analyzed, an FDR of 0.25 is reasonable in the setting of exploratory discovery. Published data samples used for data analysis are listed in Table S8.
Gene Category Enrichment Analysis.
Unsupervised pathway analysis was performed using information-theoretic pathway analysis approach (83). Briefly, pathways that are informative about non-overlapping gene groups were identified. Pathways annotations were used from the Biological Process annotations of the Gene Ontology database (http://www.geneontology.org). This pathway analysis estimates how informative each pathway is about the target gene groups and applies a randomization-based statistical test to assess the significance of the highest information values. We use the default significance threshold of P<0.005. We estimated the false discovery rate (FDR) by randomizing the input profiles iteratively on shuffled profiles with identical parameters and thresholds, finding that the FDR was always less than 5%. For each informative pathway, we determined the extent to which the pathway was over-represented in the target gene group, using the hypergeometric distribution.
Leading edge Network analysis.
GSEA was run on log2 fold change ranking, correcting for individual AML cases using edgeR glm with TMM normalization (81,84), using the Canonical Pathways and Hallmark signatures from MSigDB (34,85). Leading edge network analysis was performed using the Jaccard indices of leading edge of all pathways from GSEA analysis with FDR <0.05.
Microarray analysis.
Affymetrix Human Gene 1.0 ST arrays were processed with the oligo package in R using the RMA normalization method with CEL files derived from the primary AMLs collected on day 0, 8, and 29 post Ara-C treatment. IDs were annotated using the pd.hugene.1.0.st.v1 library. Differential regulation of genes was calculated for each case separately as the ratio of a sample at a given time point compared to the average of all time points.
Data availability.
Raw RNA-seq data of matched AML patient samples at diagnosis vs relapse were added to dbGaP accession number phs001027.v1.p1. Raw and processed data of patient-derived AML cells were deposited in GEO under accession number GSE146544 and accessible using the password kpkliqwopjwbhwj.
Supplementary Material
SIGNIFICANCE.
Despite entering complete remission after chemotherapy, relapse occurs in many AML patients. Thus, there is an urgent need to understand the relapse mechanism in AML and the development of targeted treatments to improve outcome. Here, we identified a senescence-like resilience phenotype through which AML cells can survive and repopulate leukemia.
Acknowledgements:
We acknowledge the ECOG-ACRIN Cancer Research Group and the Weill Cornell Medicine Epigenomics Core for technical services. We thank Yaseswini Neelamraju for assistance with data submission to dbGap. This work is supported by the National Cancer Institute of the National Institutes of Health under the following award numbers: NIH/NCI R01CA198089 (AM and MC), NIH/NCI UG1 CA233332 (AM). This work was supported by grants NIH R01 CA050947 (C.S.M.), CA196664 (C.S.M.), U01CA225730 (C.S.M); Leukemia and Lymphoma Society (LLS) Scholar Award (C.S.M.); and Ludwig Center at Harvard (C.S.M). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was further supported by LLS SCOR 7013–17 (AM) and U10CA180820 (ECOG). CD was supported by an LLS fellow award (LLS 5486) in partnership with The Jake Wetchler Foundation, and is a Forbeck scholar.
Competing financial interests: A.M.M. receives research funding from Janssen and Daiichi Sankyo. A.M.M has consulted for, Kdac, Epizyme and Constellation. M.C. receives research funding from Incyte Pharmaceuticals and has consulted for Janssen Pharmaceuticals. C.S.M. discloses employment of a relative with Takeda; consultant/honoraria from Fate Therapeutics, Ionis Pharmaceuticals, and research funding from Janssen/Johnson & Johnson, TEVA, EMD Serono, Abbvie, Karyopharm, Arch, Sanofi and Nurix.
REFERENCES
- 1.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. JClinOncol 2011;29(5):487–94. [DOI] [PubMed] [Google Scholar]
- 2.Jordan CT, Guzman ML. Mechanisms controlling pathogenesis and survival of leukemic stem cells. Oncogene 2004;23(43):7178–87 doi 10.1038/sj.onc.1207935. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine 1997;3:730 doi 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B, Porras DP, et al. Identification of Chemotherapy-Induced Leukemic-Regenerating Cells Reveals a Transient Vulnerability of Human AML Recurrence. Cancer Cell 2018;34(3):483–98 e5 doi 10.1016/j.ccell.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer discovery 2017;7(7):716–35 doi 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer research 1999;59(15):3761–7. [PubMed] [Google Scholar]
- 7.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer research 2002;62(6):1876–83. [PubMed] [Google Scholar]
- 8.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, et al. Cellular Senescence: Defining a Path Forward. Cell 2019;179(4):813–27 doi 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. ProcNatlAcadSciUSA 1995;92(20):9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003;113(6):703–16 doi 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 11.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nature reviews Cancer 2009;9(2):81–94 doi 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 12.Pribluda A, Elyada E, Wiener Z, Hamza H, Goldstein RE, Biton M, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 2013;24(2):242–56 doi 10.1016/j.ccr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS biology 2008;6(12):2853–68 doi 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature cell biology 2013;15(8):978–90 doi 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology 2010;5:99–118 doi 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Developmental cell 2014;31(6):722–33 doi 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nature cell biology 2010;12(7):676–85 doi 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell 2013;155(5):1104–18 doi 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 2013;155(5):1119–30 doi 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 20.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. The journals of gerontology Series A, Biological sciences and medical sciences 2014;69 Suppl 1:S4–9 doi 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 21.Mosteiro L, Pantoja C, Alcazar N, Marion RM, Chondronasiou D, Rovira M, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 2016;354(6315):aaf4445–1 aaf-10 doi 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- 22.Ritschka B, Storer M, Mas A, Heinzmann F, Ortells MC, Morton JP, et al. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes & development 2017;31(2):172–83 doi 10.1101/gad.290635.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Experimental cell research 1961;25:585–621. [DOI] [PubMed] [Google Scholar]
- 24.Toussaint O, Medrano EE, von Zglinicki T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Experimental gerontology 2000;35(8):927–45 doi 10.1016/s0531-5565(00)00180-7. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88(5):593–602. [DOI] [PubMed] [Google Scholar]
- 26.Patel PL, Suram A, Mirani N, Bischof O, Herbig U. Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence. Proceedings of the National Academy of Sciences of the United States of America 2016;113(34):E5024–33 doi 10.1073/pnas.1602379113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Carne Trecesson S, Guillemin Y, Belanger A, Bernard AC, Preisser L, Ravon E, et al. Escape from p21-mediated oncogene-induced senescence leads to cell dedifferentiation and dependence on anti-apoptotic Bcl-xL and MCL1 proteins. The Journal of biological chemistry 2011;286(15):12825–38 doi 10.1074/jbc.M110.186437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milanovic M, Fan DNY, Belenki D, Dabritz JHM, Zhao Z, Yu Y, et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018;553(7686):96–100 doi 10.1038/nature25167. [DOI] [PubMed] [Google Scholar]
- 29.Roberson RS, Kussick SJ, Vallieres E, Chen SY, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005;65(7):2795–803. [DOI] [PubMed] [Google Scholar]
- 30.Saleh T, Tyutyunyk-Massey L, Murray GF, Alotaibi MR, Kawale AS, Elsayed Z, et al. Tumor cell escape from therapy-induced senescence. Biochemical pharmacology 2019;162:202–12 doi 10.1016/j.bcp.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Lopes FL, Desmarais JA, Murphy BD. Embryonic diapause and its regulation. Reproduction 2004;128(6):669–78 doi 10.1530/rep.1.00444. [DOI] [PubMed] [Google Scholar]
- 32.Renfree MB, Fenelon JC. The enigma of embryonic diapause. Development 2017;144(18):3199–210 doi 10.1242/dev.148213. [DOI] [PubMed] [Google Scholar]
- 33.Klco JM, Spencer DH, Lamprecht TL, Sarkaria SM, Wylie T, Magrini V, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood 2013;121(9):1633–43 doi 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. ProcNatl AcadSciUSA 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, et al. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Developmental cell 2015;35(3):366–82 doi 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duy C, Teater M, Garrett-Bakelman FE, Lee TC, Meydan C, Glass JL, et al. Rational targeting of cooperating layers of the epigenome yields enhanced therapeutic efficacy against AML. Cancer Discov 2019;9(7):872–89 doi 10.1158/2159-8290.cd-19-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nature protocols 2009;4(12):1798–806 doi 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 38.Krishan A, Hamelik RM. Flow cytometric monitoring of fluorescent drug retention and efflux. Methods in molecular medicine 2005;111:149–66 doi 10.1385/1-59259-889-7:149. [DOI] [PubMed] [Google Scholar]
- 39.Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene 1999;18(34):4808–18 doi 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 40.Olsen CL, Gardie B, Yaswen P, Stampfer MR. Raf-1-induced growth arrest in human mammary epithelial cells is p16-independent and is overcome in immortal cells during conversion. Oncogene 2002;21(41):6328–39 doi 10.1038/sj.onc.1205780. [DOI] [PubMed] [Google Scholar]
- 41.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012;21(4):473–87 doi 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 42.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015;28(1):57–69 doi 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene 2008;27(46):5975–87 doi 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008;133(6):1019–31 doi 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 45.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, et al. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 2016;164(4):668–80 doi 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braess J, Wegendt C, Jahns-Streubel G, Kern W, Keye S, Unterhalt M, et al. Successful modulation of high-dose cytosine arabinoside metabolism in acute myeloid leukaemia by haematopoietic growth factors: no effect of ribonucleotide reductase inhibitors fludarabine and gemcitabine. British journal of haematology 2000;109(2):388–95 doi 10.1046/j.1365-2141.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto Y, Takano H, Fojo T. Cellular adaptation to drug exposure: evolution of the drug-resistant phenotype. Cancer research 1997;57(22):5086–92. [PubMed] [Google Scholar]
- 48.Vander Velde R, Yoon N, Marusyk V, Durmaz A, Dhawan A, Miroshnychenko D, et al. Resistance to targeted therapies as a multifactorial, gradual adaptation to inhibitor specific selective pressures. Nature communications 2020;11(1):2393 doi 10.1038/s41467-020-16212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med 2016;22(7):792–9 doi 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Fang J, Chen J. Tumor cell senescence response produces aggressive variants. Cell Death Discov 2017;3:17049 doi 10.1038/cddiscovery.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInnes L, Healy J, Saul N, Großberger L. UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software 2018;3(29):861. [Google Scholar]
- 52.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes & development 2011;25(20):2125–36 doi 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aasland D, Gotzinger L, Hauck L, Berte N, Meyer J, Effenberger M, et al. Temozolomide Induces Senescence and Repression of DNA Repair Pathways in Glioblastoma Cells via Activation of ATR-CHK1, p21, and NF-kappaB. Cancer research 2019;79(1):99–113 doi 10.1158/0008-5472.CAN-18-1733. [DOI] [PubMed] [Google Scholar]
- 54.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 2015;349(6255):aaa5612 doi 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, et al. An intrinsic S/G2 checkpoint enforced by ATR. Science 2018;361(6404):806–10 doi 10.1126/science.aap9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. Journal of medicinal chemistry 2011;54(7):2320–30 doi 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 57.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nature reviews Molecular cell biology 2017;18(10):622–36 doi 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & development 2000;14(4):397–402. [PMC free article] [PubMed] [Google Scholar]
- 59.Yap TA, O’Carrigan B, Penney MS, Lim JS, Brown JS, de Miguel Luken MJ, et al. Phase I Trial of First-in-Class ATR Inhibitor M6620 (VX-970) as Monotherapy or in Combination With Carboplatin in Patients With Advanced Solid Tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2020;38(27):3195–204 doi 10.1200/JCO.19.02404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas A, Redon CE, Sciuto L, Padiernos E, Ji J, Lee MJ, et al. Phase I Study of ATR Inhibitor M6620 in Combination With Topotecan in Patients With Advanced Solid Tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(16):1594–602 doi 10.1200/JCO.2017.76.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-Human Trial of the Oral Ataxia Telangiectasia and RAD3-Related (ATR) Inhibitor BAY 1895344 in Patients with Advanced Solid Tumors. Cancer discovery 2021;11(1):80 doi 10.1158/2159-8290.CD-20-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fordham SE, Blair HJ, Elstob CJ, Plummer R, Drew Y, Curtin NJ, et al. Inhibition of ATR acutely sensitizes acute myeloid leukemia cells to nucleoside analogs that target ribonucleotide reductase. Blood Adv 2018;2(10):1157–69 doi 10.1182/bloodadvances.2017015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019;176(6):1265–81 e24 doi 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 2014;514(7520):107–11 doi 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho TC, LaMere M, Stevens BM, Ashton JM, Myers JR, O’Dwyer KM, et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood 2016;128(13):1671–8 doi 10.1182/blood-2016-02-695312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547(7661):104–8 doi 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- 67.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van GP, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17(9):1086–93. [DOI] [PubMed] [Google Scholar]
- 68.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540(7633):433–7 doi 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 69.Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP, et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 2015;517(7533):209–13 doi 10.1038/nature14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dhimolea E, de Matos Simoes R, Kansara D, Al’Khafaji A, Bouyssou J, Weng X, et al. An Embryonic Diapause-like Adaptation with Suppressed Myc Activity Enables Tumor Treatment Persistence. Cancer Cell doi 10.1016/j.ccell.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez-Segura A, de Jong TV, Melov S, Guryev V, Campisi J, Demaria M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Current biology : CB 2017;27(17):2652–60 e4 doi 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature reviews Cancer 2010;10(4):301–9 doi 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 73.Nguyen L, Papenhausen P, Shao H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes (Basel) 2017;8(4):1–23 doi 10.3390/genes8040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vijay V, Miller R, Vue GS, Pezeshkian MB, Maywood M, Ast AM, et al. Interleukin-8 blockade prevents activated endothelial cell mediated proliferation and chemoresistance of acute myeloid leukemia. Leukemia research 2019;84:106180 doi 10.1016/j.leukres.2019.106180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2019;133(1):7–17 doi 10.1182/blood-2018-08-868752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aldoss I, Yang D, Aribi A, Ali H, Sandhu K, Al Malki MM, et al. Efficacy of the combination of venetoclax and hypomethylating agents in relapsed/refractory acute myeloid leukemia. Haematologica 2018;103(9):e404–e7 doi 10.3324/haematol.2018.188094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 2017;8:14049 doi 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology 2015;33(5):495–502 doi 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30 doi 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40 doi 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Jonge HJ, Woolthuis CM, Vos AZ, Mulder A, van den Berg E, Kluin PM, et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011;25(12):1825–33 doi 10.1038/leu.2011.172. [DOI] [PubMed] [Google Scholar]
- 83.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. MolCell 2009;36(5):900–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research 2012;40(10):4288–97 doi 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq data of matched AML patient samples at diagnosis vs relapse were added to dbGaP accession number phs001027.v1.p1. Raw and processed data of patient-derived AML cells were deposited in GEO under accession number GSE146544 and accessible using the password kpkliqwopjwbhwj.