Summary
The Parkin Co-Regulated Gene (PACRG) binds at the inner junction between doublet microtubules of the axoneme, a structure found in flagella and cilia. PACRG binds to the adaptor protein meiosis expressed gene 1 (MEIG1), but how they bind to microtubules is unknown. Here, we report the crystal structure of human PACRG in complex with MEIG1. PACRG adopts a helical repeat fold with a loop that interacts with MEIG1. Using the structure of the axonemal doublet microtubule from the protozoan Chlamydomonas reinhardtii and single-molecule fluorescence microscopy, we propose that PACRG binds to microtubules while simultaneously recruiting free tubulin to catalyze formation of the inner junction. We show that the homologous PACRG-like protein also mediates dual tubulin interactions but does not bind MEIG1. Our findings establish a framework to assess the function of the PACRG family of proteins and MEIG1 in regulating axoneme assembly.
eTOC Blurb
Khan et al. solved the crystal structure of human PACRG:MEIG1, a protein complex essential for flagella maturation in sperm. PACRG and its homolog PACRGL can recruit tubulin to microtubules via multiple binding sites, suggesting a mechanism for formation of the axonemal doublet microtubule found in flagella and cilia.
Graphical Abstract
Introduction
Loss-of-function mutations in Parkin cause early-onset recessive Parkinson’s disease (PD) in humans (Kitada et al., 1998). Parkin is an E3 ubiquitin ligase that ubiquitinates proteins selectively on damaged mitochondria following activation by PINK1, a mitochondrial ubiquitin kinase (Bayne and Trempe, 2019). Parkin and PINK1 are conserved across nearly all metazoans and appear to play a key role in mitochondrial quality control. While Parkin or PINK1 knockout (KO) mice do not exhibit significant muscle or neuronal degeneration on their own (Gautier et al., 2008; Goldberg et al., 2003), mitochondria-linked stress and infection-induced inflammation can induce neurodegeneration in those strains (Matheoud et al., 2019; Pickrell et al., 2015; Sliter et al., 2018). Parkin KO mice are more susceptible to intracellular bacterial pathogens (Manzanillo et al., 2013), and missense mutations in Parkin are associated with an increased susceptibility to a type-1 reaction in leprosy, a disease caused by a chronic Mycobacterium leprae infection (Fava et al., 2019). Furthermore, a previous study showed that susceptibility to leprosy was strongly associated with genetic variants in the promoter region of Parkin (Mira et al., 2004). This promoter is shared with the Parkin Co-Regulated Gene (PACRG), which is linked head-to-head with Parkin on the opposite DNA strand, with only 204 bp separating the two transcripts (West et al., 2003). While the function of Parkin has been intensively studied over the last 20 years, far less is known about PACRG.
PACRG encodes a 29 kDa protein that is conserved in all metazoans, as well as flagellated protozoans (Lorenzetti et al., 2004). Immunostaining shows that Lewy bodies, which are intracellular aggregates of α-synuclein found in the brain of PD patients, contain PACRG (Imai et al., 2003; Taylor et al., 2007). However, the normal physiological role of PACRG is in the formation of the axoneme, a microtubule doublet structure found in motile and non-motile cilia and flagella. Early proteomics studies showed that cilia axonemes isolated from human epithelial bronchial cells contain PACRG (Ostrowski et al., 2002). Loss of PACRG causes male sterility in the quakingviable (qkv) mouse, a recessive deletion of the Parkin, PACRG and QK genes with a severe neurological phenotype (Lorenzetti et al., 2004; Stephenson et al., 2018). Mouse PACRG is highly abundant in testis and is required for spermiogenesis (Lorenzetti et al., 2004). Loss of PACRG results in hydrocephalus in both PACRG KO and qkv mice (Stephenson et al., 2018; Wilson et al., 2009); PACRG levels are indeed elevated in ependymal cells lining the cerebral ventricles, suggesting that PACRG is required for cilia motility. Knockdown of the two PACRG paralogues in trypanosome, a protozoan parasite, results in paralysis of the flagellum and loss of outer doublet microtubules (Dawe et al., 2005). Consistent with its role in stabilizing the axoneme outer-doublet, recombinant human PACRG co-sediments with microtubules and can bundle microtubules in vitro, thus showing that it binds directly to tubulin (Ikeda, 2008). The PACRG protein localizes to the axoneme in trypanosome as well as in Chlamydomonas reinhardtii, a flagellated green alga (Ikeda et al., 2007). Electron microscopy of C. reinhardtii axoneme doublet microtubules showed that PACRG localizes at the inner junction of the two tubules, in alternance with FAP20 (Dymek et al., 2019; Khalifa et al., 2020; Ma et al., 2019). In addition to its role in the formation of motile cilia and flagella, PACRG also localizes to a subset of non-motile cilia in sensory neurons from the nematode Caenorhabditis elegans, where it regulates signaling processes linked to gustatory plasticity (Loucks et al., 2016). However, PACRG is not required for ciliogenesis per se, nor for intraflagellar transport. Thus, PACRG is critical for the formation of functional motile and non-motile cilia.
In addition to its interaction with tubulin, PACRG also binds to the meiosis expressed gene 1 product (MEIG1), a 11 kDa protein that is required for spermiogenesis (Salzberg et al., 2010; Zhang et al., 2009b). MEIG1 KO mice display male sterility, but don’t have other phenotypic traits found in PACRG KO mice, suggesting a more specific role in sperm maturation. PACRG and MEIG1 colocalize to the manchette of elongating spermatids, and both are required for the manchette localization of the cargo protein SPAG16, a sperm central apparatus protein (Li et al., 2015). Furthermore, the manchette localization of MEIG1 requires PACRG, whereas the localization of PACRG is unaffected by the loss of MEIG1. PACRG is thus upstream of MEIG1, with the latter playing a more specific role in sperm-specific cargo recruitment. The NMR structure of MEIG1 revealed a small domain with four exposed aromatic side-chains that are required for binding and stabilization of PACRG in bacterial cells (Li et al., 2016). Thus, PACRG and MEIG1 form a stable heterodimer, but the structure of PACRG and how it binds MEIG1 are unknown.
Here, we report the crystal structure of the C-terminal domain of human PACRG bound to MEIG1. Using the high-resolution cryo-electron microscopy structure of the axoneme doublet microtubule, we gain insight into how PACRG:MEIG1 interacts with tubulin protofilaments. We validate those interactions using single molecule fluorescence microscopy and crosslinking assays and identify rare human variants that interfere with the functions of PACRG. We also characterize the interactions mediated by the PACRG-like protein, a homolog of PACRG. We explore the evolution and expression patterns of axonemal and Parkinson-associated genes and discuss the implications for their functions.
Results
Crystal structure of the human PACRG:MEIG1 complex at 2.1 Å resolution
Since previous reports indicated that mouse PACRG was stabilized by MEIG1 in E. coli (Li et al., 2016), we co-expressed the two human orthologues with a cleavable hexahistidine (His6) tag on MEIG1 for immobilized metal affinity chromatography (IMAC). We confirmed that human PACRG is indeed stabilized by co-expression with MEIG1 (Figure S1). PACRG co-purifies with MEIG1 and the two proteins co-elute on size-exclusion chromatography (SEC) at ~40 kDa, consistent with the predicted molecular weight of the heterodimer (Figure S2). However, the complex did not produce crystals. To facilitate crystallization, we removed the N-terminal 69 amino acids of PACRG, which are predicted to be disordered and are poorly conserved in other PACRG orthologues (Figure S3). However, PACRGΔ1–69 did not co-purify with His6-MEIG1 following co-expression. We had previously noted that glutathione-S-transferase (GST)-PACRGΔ1–69 forms inclusion bodies. The fusion protein was resolubilized in urea and then diluted and dialyzed to refold the GST domain (Li et al., 2004). The partially refolded fusion protein was then incubated with purified His6-MEIG1, and the complex was affinity purified. The affinity tags were cleaved and the products resolved by SEC, which confirmed that the two proteins form a heterodimer complex of ~33 kDa (Figure 1A).
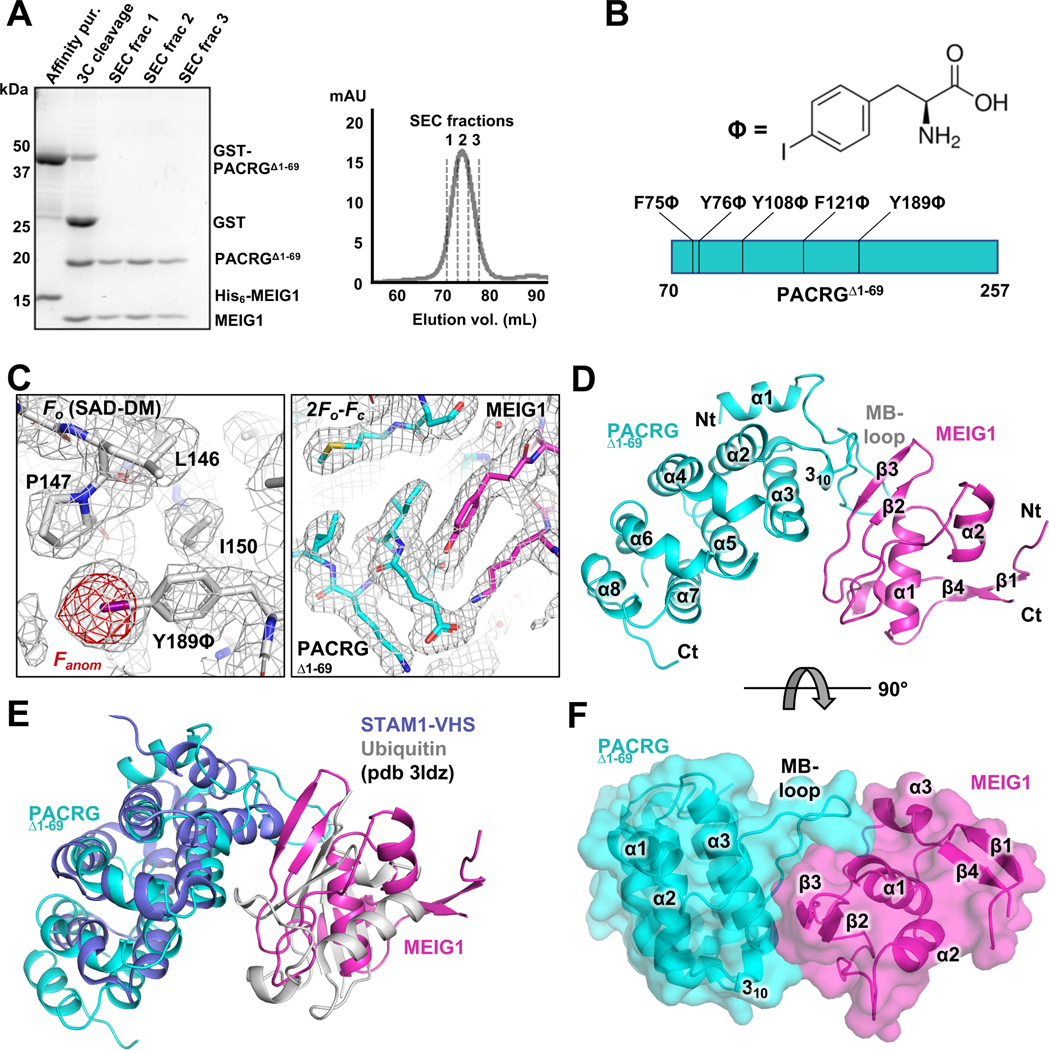
Figure 1. Crystal structure of the PACRG:MEIG1 complex.
(A) Purification of PACRGΔ1–69: MEIG1 by affinity and size-exclusion chromatography (SEC). Left, SDS-PAGE stained with Coomassie Blue. Lanes of the SEC fractions contain both MEIG1 and PACRGΔ1–69. (B) Chemical structure of p-iodo-L-phenylalanine and sites of incorporation in PACRG. (C) Left, electron density maps after SAD phasing and density modification (grey mesh), with the final refined model of the Y189ϕ derivative. The anomalous difference density map (red mesh) shows the position of iodine. Right, refined 2Fo-Fc electron density maps for the native complex of PACRGΔ1–69 (cyan) and MEIG1 (magenta). (D) Cartoon representation of the native complex of PACRGΔ1–69 and MEIG1. (E) Superposition of the STAM1-VHS domain bound to ubiquitin (violet and grey) on the PACRGΔ1–69:MEIG1 complex. (F) Surface representation of PACRGΔ1–69:MEIG1, rotated at 90° from the view in panel D.
The MEIG1:PACRGΔ1–69 complex formed crystals that diffracted to 2.1 Å resolution in a tetragonal space group (Table 1). However, the structure could not be solved by molecular replacement using the mouse MEIG1 solution structure or homology models of PACRG. Thus, we produced p-iodo-L-phenylalanine derivatives of PACRGΔ1–69 using site-specific incorporation at the amber TAG codon, for iodine phasing (Bryson et al., 2017; Xie et al., 2004). We selected five aromatic residues in human PACRGΔ1–69 for site-specific incorporation, with either low sequence conservation or potential surface exposure to alleviate loss of interaction with MEIG1 or disruption of the fold (Figure 1B and Figure S3). All five mutants expressed and formed complexes with MEIG1, and mass spectrometry confirmed incorporation of a single p-iodo-L-phenylalanine (ϕ) residue (Figure S4). Three mutants produced diffracting crystals (F75ϕ, Y108ϕ, and Y189ϕ), and the Y189ϕ mutant produced the best diffraction for phasing by singlewavelength anomalous dispersion (Figure 1C and Table 1). The resulting mutant structure was then used as a search model for molecular replacement of the native data set. The native and mutant structures are very similar (0.15 Å backbone rmsd), confirming that the mutation does not disrupt the fold of PACRG. Finally, to confirm that the structure obtained was not an artefact from the refolding procedure, we obtained crystals by limited proteolysis with subtilisin of co-expressed full-length PACRG:MEIG1. These crystals diffracted to 2.6 Å (Table 1) and the structure is nearly identical to MEIG1:PACRGΔ1–69 (0.15 Å backbone rmsd). No additional residues were observed upstream of Thr70, which is consistent with the N-terminus being disordered.
Table 1.
X-ray crystallography data collection and refinement statistics
| Native MEIG1:PACRGΔ1–69 | iodo-Y189Φ MEIG1:PACRGΔ1–69 | Proteolysed MEIG1:PACRGFL | |
|---|---|---|---|
| Wavelength (Å) | 0.9793 | 1.4760 | 0.9795 |
| Resolution range (Å) | 47.6 – 2.1 (2.175 – 2.1) | 47.64 – 2.09 (2.16–2.09) | 47.3 – 2.6 (2.72 – 2.6) |
| Space group | P41212 | P41212 | P41212 |
| Unit cell dimensions (Å) | 67.30 67.30 158.26 | 67.37 67.37 159.72 | 66.85 66.85 158.92 |
| Unit cell angles (deg) | 90 90 90 | 90 90 90 | 90 90 90 |
| Total reflections | 138796 (14193) | 253522 (9944) | 148977 (17761) |
| Unique reflections | 21980 (2139) | 22406 (1769) | 11731 (1395) |
| Multiplicity | 6.3 (6.6) | 11.3 (5.6) | 12.6 (12.7) |
| Completeness (%) | 99.79 (99.95) | 99.8 (98.4) | 100 (100) |
| Mean I/sigma(I) | 20.25 (1.93) | 13.7 (1.1) | 9.1 (1.1) |
| Wilson B-factor | 47.87 | 43.83 | 54.77 |
| R-merge | 0.050 (1.15) | 0.093 (1.11) | 0.274 (2.87) |
| CC1/2 | 0.999 (0.644) | 0.998 (0.733) | 0.996 (0.557) |
| Reflections used for R-free | 1113 (5.1%) | 2049 (9.1%) | 634 (5.4%) |
| R-work | 0.190 (0.280) | 0.229 (0.359) | 0.214 (0.300) |
| R-free | 0.227 (0.323) | 0.249 (0.340) | 0.284 (0.363) |
| # non-hydrogen atoms | 2166 | 2156 | 2112 |
| macromolecules | 2083 | 2076 | 2066 |
| ligands | 6 | 6 | 25 |
| water | 77 | 89 | 21 |
| Protein residues | 254 | 254 | 253 |
| RMS (bonds, Å) | 0.008 | 0.009 | 0.007 |
| RMS (angles, deg) | 1.08 | 0.953 | 0.883 |
| Ramachandran favored (%) | 96.0 | 98.4 | 97.2 |
| Ramachandran allowed (%) | 4.0 | 1.6 | 2.8 |
| Ramachandran outliers | none | none | none |
| Clashscore | 1.96 | 3.67 | 6.57 |
| Average B-factor | 60.1 | 52.0 | 58.9 |
| macromolecules | 60.1 | 52.1 | 58.9 |
| ligands | 93.9 | 51.6 | 59.6 |
| solvent | 58.4 | 48.9 | 58.8 |
Statistics for the highest-resolution shell are shown in parentheses.
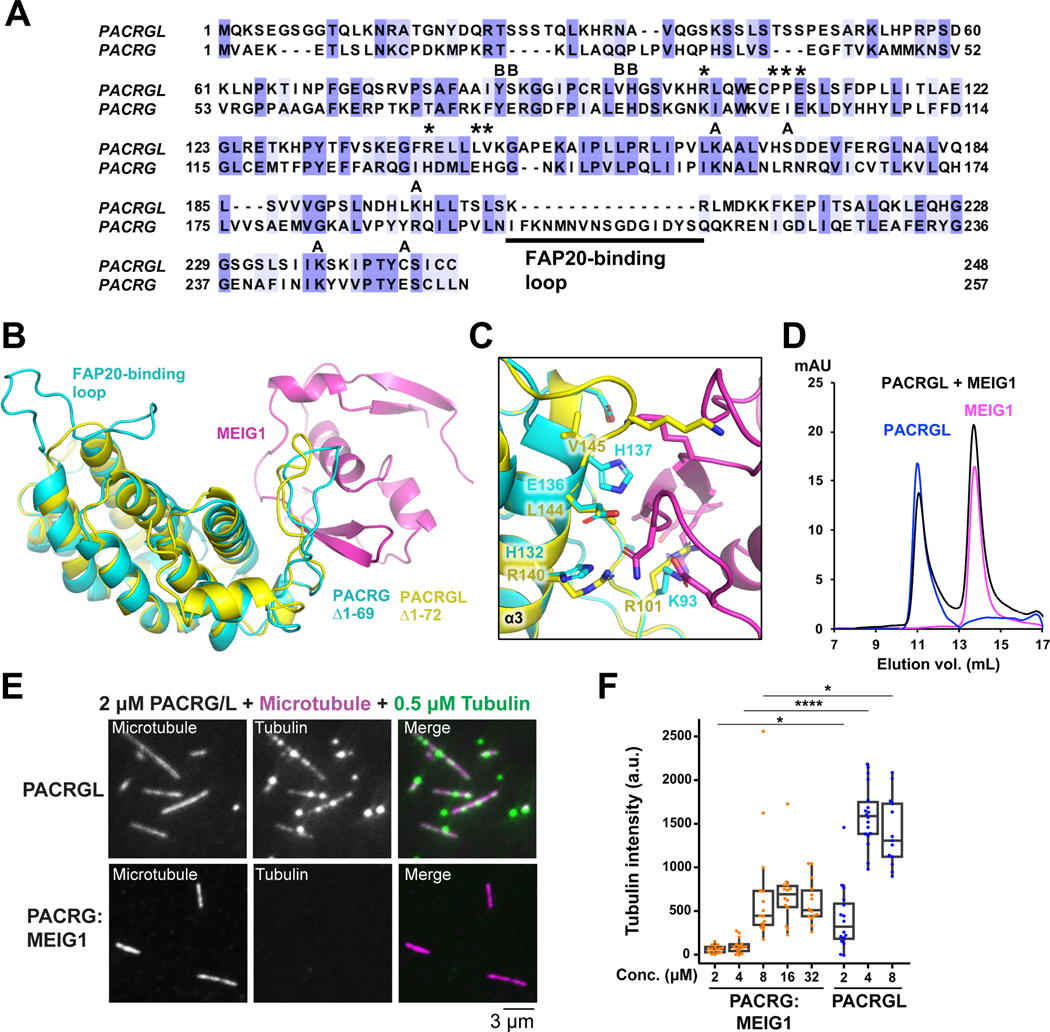
The structure of PACRGΔ1–69 in the complex consists of a core of 7 α-helices that fold into parallel repeats, preceded by an N-terminal helix and a 310 helix (Figure 1D). Continuous electron density is observed in PACRG from a.a. 70 to 255, except for a.a. 203–219, a disordered loop located between α6 and α7 at the C-terminus. Intriguingly, a longer isoform of PACRG resulting from an alternative splicing event results in a longer insertion in that same loop (Figure S5). Fold analysis with the PDBeFold server reveals similarity between PACRG and HEAT repeat proteins such as VHS domains and the RNA polymerase II-binding domain of Pcf11 (Meinhart and Cramer, 2004). The overall structure of the complex is notably similar to the structure of the STAM1 VHS domain bound to ubiquitin, where MEIG1 occupies a position similar to ubiquitin (Figure 1E). This is consistent with this type of fold being involved primarily in protein-protein interactions. However, co-expression with ubiquitin did not allow co-purification of PACRG, suggesting that PACRG does not bind ubiquitin.
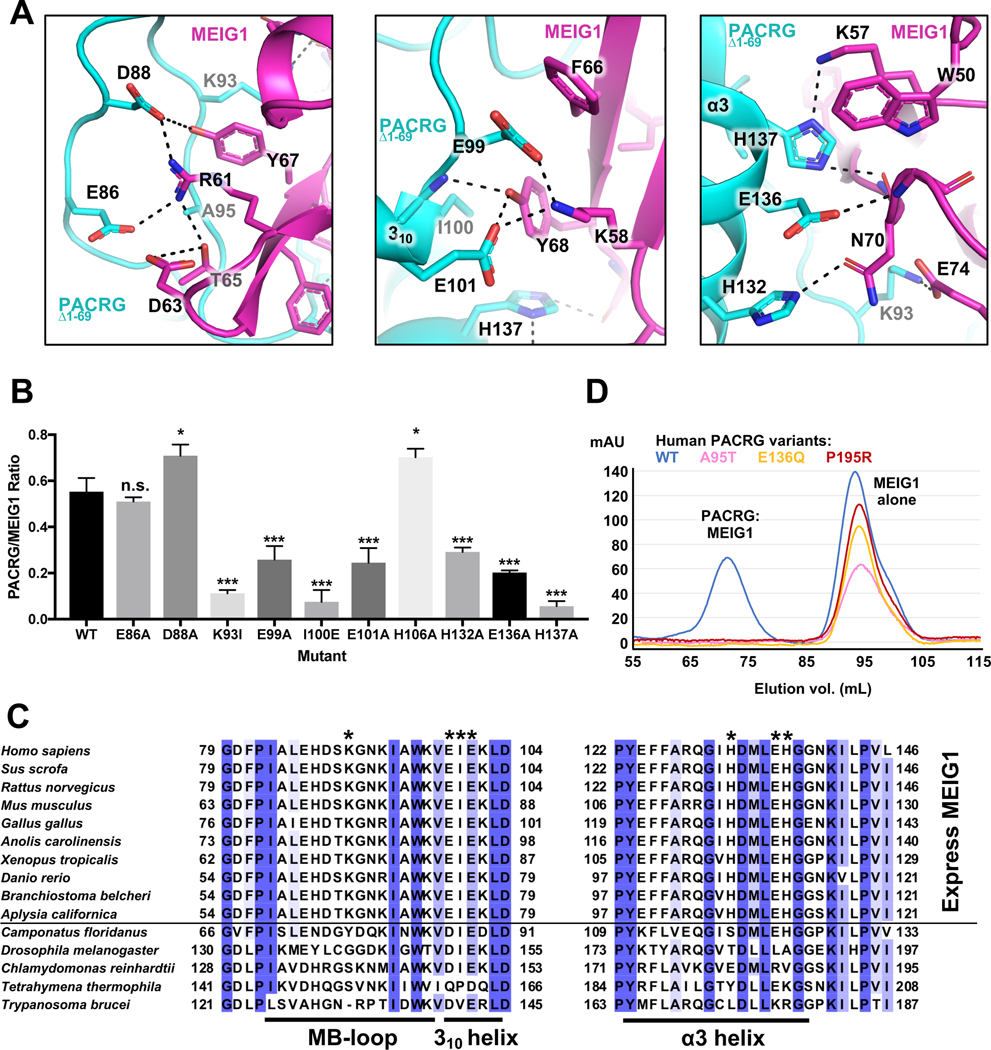
The structure of human MEIG1 is similar to the NMR structure of mouse MEIG1 (1.34 Å backbone rmsd) and shows electron density for a.a. 5 to 88. MEIG1 interacts with a long loop in PACRG (which we name the MB-loop) located between helices α1 and α2, as well as residues in the α3 helix (Figure 1F). The buried surface area between the two proteins (694 Å2) is relatively small but involves 12 hydrogen bonds and 6 salt bridges (Figure 2A). Contacts notably involve the MEIG1 residues Trp50, Lys57, Phe66 and Tyr68, which are essential for binding PACRG (Li et al., 2016). To determine which amino acids in PACRG are essential for binding to MEIG1, we co-expressed full-length PACRG mutants with His6-MEIG1. PACRG mutants that don’t bind should not co-purify with His6-MEIG1 following IMAC. The results show that Lys93, Glu99, Ile100, Glu101, His132, Glu136, and His137 are required for the interaction (Figure 2B and Figure S6). These residues are conserved in all animals that express MEIG1, which comprise all vertebrates as well as some invertebrates such as Aplysia, but not insects nor single-celled organisms (Figure 2C). Overall, these results confirm that PACRG forms a stable complex with MEIG1, as observed in the crystal structure.
Figure 2. The PACRG:MEIG1 interface reveals a conserved network of polar interactions.
(A) Close-up views and interactions in the PACRGΔ1–69:MEIG1 complex. Dashed lines indicate hydrogen bonds. (B) Interaction assay for the binding of PACRG mutants to His6-MEIG1. The ratio of PACRG to His6-MEIG1 was calculated by densitometry on SDS-PAGE (n=4). One-way ANOVA and Dunnett’s tests were performed for comparison with the wild-type (WT) interaction (* for p ≤ 0.05, *** for p ≤ 0.001). (C) Sequence alignment of PACRG orthologues across different organisms. Species above the horizontal line also express MEIG1. The asterisks indicate residues in human PACRG that are required for binding MEIG1. Figure generated with Jalview. Blue shading indicates degree of conservation. (D) Size-exclusion chromatography of WT and rare human variants of PACRG, following co-expression with His6-MEIG1 and IMAC.
Model of PACRG:MEIG1 bound to the inner junction of the axonemal doublet microtubule
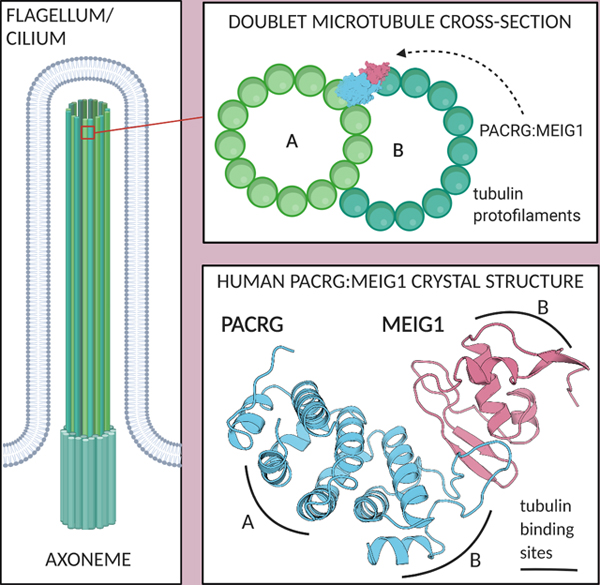
Electron microscopy studies showed that PACRG localizes outside the A-tubule of the axonemal doublet and positions itself at the inner junction of the two tubules (Dymek et al., 2019; Ikeda, 2008). However, it is unclear how human PACRG interacts with tubulin. Furthermore, while there is no evidence that MEIG1 is located in the mature axoneme, it would be important to determine how MEIG1 may be involved in coordinating interactions with tubulin protofilaments leading to formation of the axoneme inner junction. The structure of the axonemal doublet microtubule from C. reinhardtii was recently solved by cryo-electron microscopy by two different groups (Khalifa et al., 2020; Ma et al., 2019). High-resolution electron density was observed at the inner junction, which allowed model-building of PACRG, FAP20 and a number of other axonemal proteins. The structure revealed that C. reinhardtii PACRG (CrPACRG) and FAP20 alternate at the inner junction of the A and B-tubules (Figure 3A). The CrPACRG C-terminal helical domain, corresponding to human (Hs)PACRGΔ1–69, interacts simultaneously with the doublet microtubule A1 and B10 protofilaments and makes contacts with both α- and β-tubulin (Figure 3B,C). Electron density was observed for the N-terminus of CrPACRG, which folds into an α-helix that binds β-tubulin on the B-tubule. This helix is then followed by a long segment that adopts a “triple-helix”-like conformation that interacts with β-tubulin subunits in protofilaments A1 and A13.
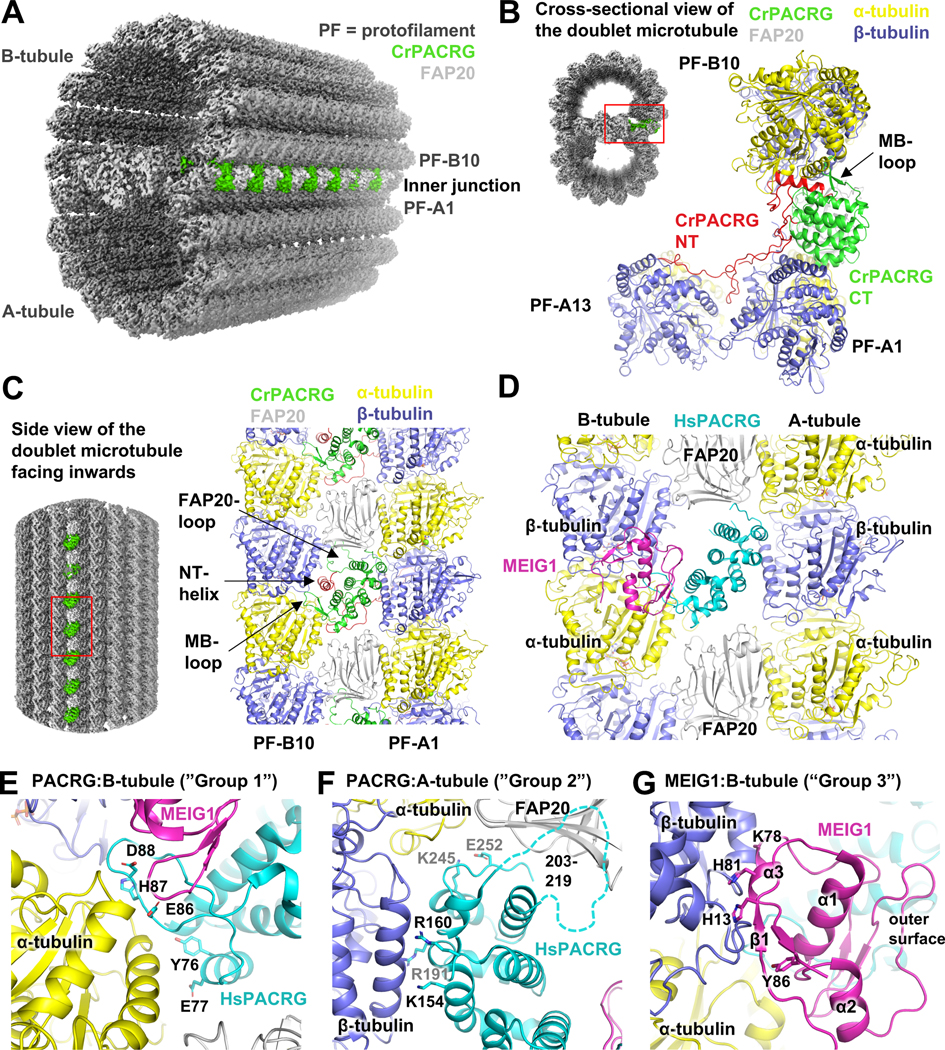
Figure 3. Structural model of the doublet microtubule reveals potential roles of PACRG:MEIG1 in axoneme assembly.
(A) CryoEM structure of the Chlamydomonas reinhardtii axonemal doublet microtubule (Khalifa et al., 2020). PACRG is colored at the inner junction between the A- and B-tubules. Tubulin protofilaments (PF) are in gray. (B) Cross-sectional view of the axonemal doublet showing the interaction of PACRG with the A1 and B10 PFs. The N-terminus (NT) of PACRG is red. (C) Side-view of the axonemal doublet viewed from the outside facing inwards, as viewed from the central pair. (D) Side-view of the homology model (facing inwards) of the inner junction of the human axonemal doublet, showing the hypothetical position of MEIG1. (E-G) Close-up views of the interactions mediated by HsPACRG:MEIG1 in the homology model of the inner junction. The dashed line in panel F indicates the position of the FAP20-binding loop in HsPACRG.
In order to understand how the human PACRG:MEIG1 complex would interact with tubulin protofilaments such as those found in the doublet microtubule, we have built a homology model by superposing our crystal structure on the CrPACRG coordinates in the cryo-EM structure (Khalifa et al., 2020), as well as the structure of porcine tubulin dimers (Nogales et al., 1998). HsPACRG70–257 and CrPACRG119–307 are very similar (0.71 Å backbone rmsd) and thus the two proteins likely mediate very similar types of interactions with tubulin (Figure 3D). The MB-loop is located on the B-tubule side and interacts primarily with α-tubulin. Notably, we find that residues Glu86 and Asp88, which interact with MEIG1 but whose mutations do not affect MEIG1 binding (see Figure 2), are in very close proximity with a basic patch in α-tubulin comprising Lys163 and Lys164 (Figure 3E). This would position MEIG1 very close to β-tubulin, with potential interactions between the α3 helix of MEIG1 and the H3 helix of β-tubulin (Figure 3G). This would place MEIG1 on the outer surface of the inner junction, or the outer surface of a manchette microtubule. HsPACRG would also interact with the A-tubule, notably via three invariant basic residues (Lys154, Arg160, Arg191) that would bind to an acidic patch in β-tubulin (Figure 3F). Two highly conserved residues may also interact with α-tubulin from the A-tubule (Lys245, Glu252). Finally, the CrPACRG structure shows that the loop connecting α6 and α7 (a.a. 203–219), which is disordered in the HsPACRG crystal structure, interacts with FAP20.
PACRG:MEIG1 recruit tubulin to microtubules through multiple binding sites
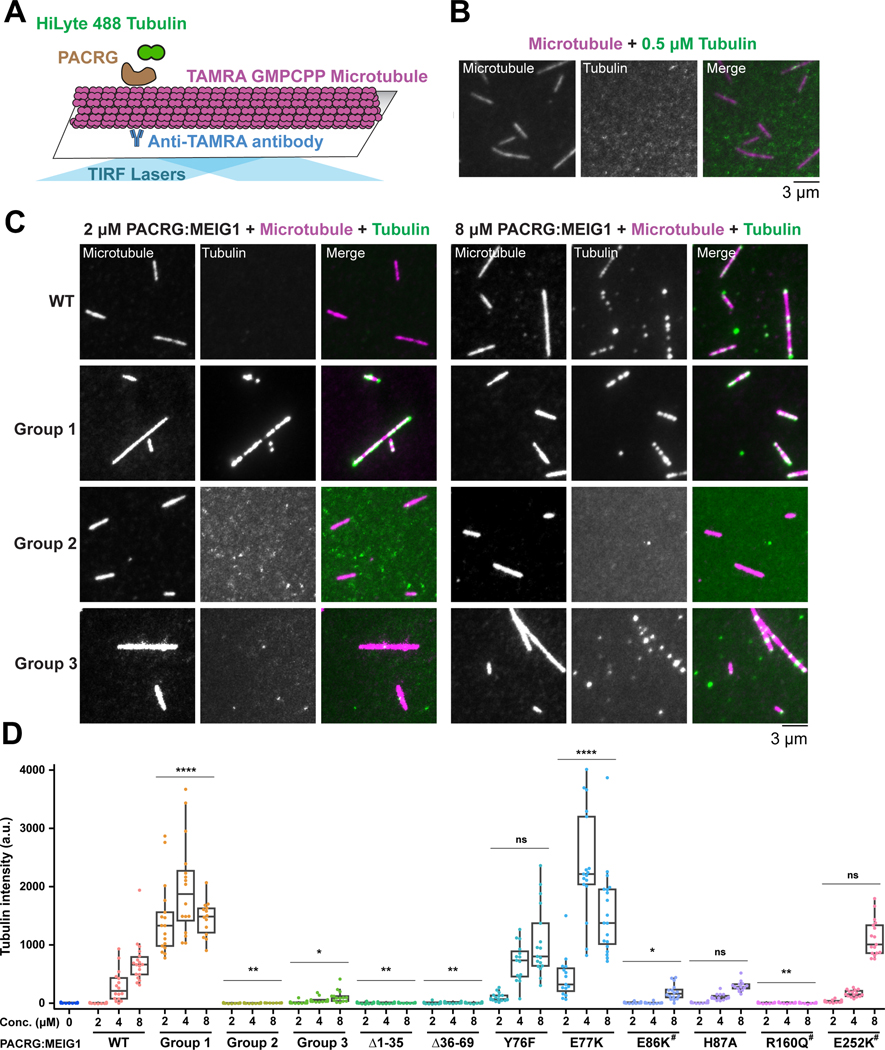
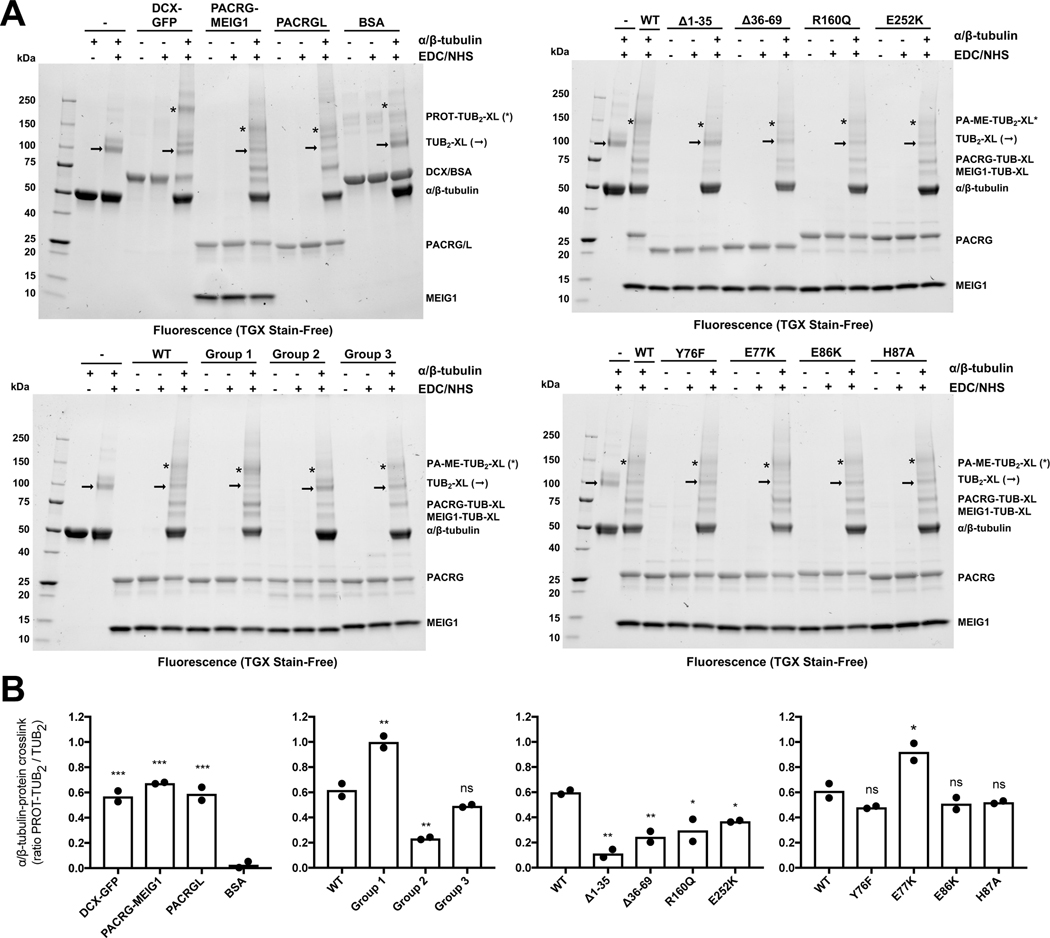
To characterize the proposed tubulin-binding activity of PACRG:MEIG1, we used a single molecule fluorescence microscopy assay for microtubule-associated proteins (Bechstedt and Brouhard, 2012; Gell et al., 2010). Briefly, single fluorescent-labeled microtubules are immobilized on a glass cover slide, and a solution of free fluorescent α/β-tubulin is added along with the candidate protein (Figure 4A). At 0.5 μM α/β-tubulin and in the absence of tubulin-binding protein, no α/β-tubulin is recruited to the microtubules (Figure 4B). However, upon addition of 2–8 μM wild-type (WT) human PACRGFL:MEIG1, we observed the formation of α/β-tubulin puncta that colocalize with microtubules (Figure 4C). Quantification shows a dose-dependent increase in colocalization intensity, suggesting that PACRG:MEIG1 binds to microtubules and recruits α/β-tubulin (Figure 4D). To test which predicted tubulin-binding site(s) was required for recruitment, we first introduced groups of site-specific mutations in PACRG that would perturb interactions with the B-tubule (Y76F-E77K-E86K-H87A; group 1) or the A-tubule (K154E-R160Q-R191E; group 2). These mutants co-purified with MEIG1, confirming that these residues are not involved in binding MEIG1 (Figure S7A). The group 2 mutations abolished recruitment, as expected, suggesting this interface is critical (Figure 4C,D). However, the group 1 mutations increased the signal intensity compared to WT. To determine which mutation(s) in group 1 is/are responsible for this increase, we tested individual mutations in the same assay, and found that the E77K mutation increases recruitment activity, whereas other mutations either decrease (E86K) or do not affect (Y76F, H87A) recruitment. In our homology model, Glu77 is adjacent to Glu55 in α-tubulin, and thus the mutation E77K may induce formation of a favorable interaction (Figure S8). On the other hand, the E86K mutations would perturb polar interactions. Together these results show that the PACRG C-terminal domain has two tubulin-binding sites that participate to the recruitment of free α/β-tubulin to microtubules.
Figure 4. PACRG:MEIG1 recruits tubulin to microtubules through multiple tubulin binding sites.
(A) Schematic of the total internal reflection fluorescence (TIRF) experiment used to characterize the dual tubulin-binding activity of PACRG:MEIG1. Microtubule seeds prepared using TAMRA-labeled polymerized α/β-tubulin (bound to GMPPCP) are immobilized and incubated with free HiLyte 488-labeled α/β-tubulin in the presence of PACRG:MEIG1 variants. (B) Control TIRF microscopy images showing that free α/β-tubulin is not recruited to microtubules. (C) TIRF microscopy images showing that WT PACRG:MEIG1 recruits α/β-tubulin to microtubules. (D) Quantification of HiLyte 488-labeled α/β-tubulin that colocalize with the microtubules in the presence of PACRG:MEIG1 variants (WT and mutants). Mutants with # indicate rare human variants. Two-way ANOVA and Dunnett’s tests were performed for comparison with the wild-type (WT) concentration series. * indicates p ≤ 0.05, ** for p ≤ 0.01, **** for p ≤ 0.0001, ns, not significant (n=14–19).
In addition, we mutated MEIG1 residues that we predict would interact with β-tubulin in the B-tubule of our hypothetical model (H13A-K78A-H81A-K82A-Y86A; group 3). These mutations impaired recruitment of α/β-tubulin to microtubules (Figure 4C,D), confirming that MEIG1 also has tubulin-binding activity. Finally, we tested two deletions in the N-terminus of PACRG (Δ1–35 and Δ36–69), which form extensive interactions with both the A- and B-tubules in the Chlamydomonas axoneme (Figure 3B). Both deletions abolish recruitment, implying that the N-terminus of PACRG is essential for the recruitment of α/β-tubulin to microtubules (Figure 4D).
To determine whether PACRG:MEIG1 mutants bind directly to tubulin, we also established a chemical crosslinking assay with taxol-polymerized α/β-tubulin. For this purpose, we used the zero-length crosslinker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) combined with N-hydroxysuccinimide (NHS), which efficiently crosslink microtubules (Rafiei and Schriemer, 2019). While the disuccinimidyl suberate (DSS) crosslinker efficiently crosslinks PACRG and MEIG1, it poorly crosslinks α/β-tubulin and does not allow observation of tubulin:PACRG interactions (Figure S7B). EDC/NHS leads to the formation of a 100 kDa α/β-tubulin crosslink (Figure 5A). Addition of GFP-fused doublecortin (DCX-GFP), a high-affinity microtubule-associated protein (Bechstedt and Brouhard, 2012), causes formation of a ternary complex at 170 kDa and a decrease in the intensity of the 100 kDa band, which can be quantified (Figure 5B). The DCX ternary complex is specific because the bovine serum albumin (BSA), a control protein, does not form a ternary complex. PACRG:MEIG1 crosslinks with tubulin dimers at 150 kDa, as well as monomeric tubulin between 60–75 kDa (Figure 5A). The 150/100 kDa ratio is higher in the group 1 and E77K mutants, and lower in the group 2 and deletion mutants (Figure 5B), in agreement with the single-molecule microscopy assay. The other single mutants from group 1 (Y76F, E86K, H87A) do not change the ratio, suggesting that the A-tubule interface likely dominates formation of the ternary complex. The group 3 mutant also shows no significant change in the 150/100 kDa ratio, but noticeably reveals the loss of the monomeric tubulin-MEIG1 crosslink band at 65 kDa (Figure 5A, right). Overall, these results are consistent with PACRG possessing two tubulin interaction sites in its C-terminal domain, as well as via its N-terminus.
Figure 5. Crosslinking studies show that PACRG:MEIG1 and PACRGL bind to tubulin.
(A) SDS-PAGE analysis of crosslinking reactions between taxol-polymerized tubulin (5 μM) and different candidate microtubule-associated proteins (12.5 μM, except DCX at 1.5 μM). Gels were imaged by UV-induced fluorescence. Interactions between tubulin and candidate proteins are detected by the loss of tubulin α/β dimers (→) and appearance of tubulin-protein trimers or tetramers (*). (B) Densitometry analysis of crosslinked bands highlighted in A, shown as a ratio of the two bands, following background subtraction (tubulin alone). One-way ANOVA and Dunnett’s tests were performed for comparison with either BSA (gel 1) or wild-type (WT) PACRG:MEIG1 (gels 2–4). * indicates p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, ns, not significant (n=2).
Rare human PACRG genetic variants impair MEIG1 binding and tubulin interactions
In order to explore the impact of the PACRG interactions in human health, we searched the GnomAD database for missense variants in the protein at residues involved in binding MEIG1 or tubulin (Lek et al., 2016). By excluding variants with only one allele, we identified 7 rare variants with allele frequencies ranging between 1 and 10 per 100,000 individuals (Table 2). We introduced these mutations in the full-length PACRG:MEIG1 complex for expression and purification. Three of those missense mutations failed to form a complex with MEIG1 on gel filtration (Figure 2D). Two of those missense mutations, A95T and E136Q, are in proximity to MEIG1 in the crystal structure and likely disrupt this interaction (Figure 2A). The P195R mutation is located at the interface of β-tubulin in the B-tubule, but the mutation likely disrupts the α6 helix and thus unfolds PACRG. Thus, the A95T, E136Q and P195R rare variants disrupt the PACRG:MEIG1 complex. The remaining four variants are potentially involved in making interactions that stabilize the inner junction of the axonemal doublet. Notably, the variants Y76C and E86K may affect the interaction of PACRG with α-tubulin in the B-tubule (Figure 3E) and the variants R160Q and E252K would affect interaction with the A-tubule (Figure 3F). Three of the variants above co-purified with His6-MEIG1 and were characterized using the single-molecule microscopy assay. As reported above, the E86K variant showed a decrease in α/β-tubulin recruitment, whereas the E252K variant did not (Figure 4D). However, the R160Q variant abolished recruitment, highlighting the critical importance of the A-tubule interaction for α/β-tubulin recruitment. Finally, crosslinking studies with those variants are generally consistent with previous results, with R160Q showing the strongest reduction in formation of the 150 kDa complex (Figure 5). In our homology model, Arg160 interacts with a helix in β-tubulin that carries two acidic residues (Asp427 and Asp437, Figure S8), and those potential salt bridges would be disrupted by the mutation. We have thus identified five rare human PACRG variants that interfere with MEIG1 or tubulin binding.
Table 2.
Human PACRG variants that affect MEIG1 or tubulin interactions
| Human PACRG variants | Allele count | Total alleles | Allele frequency | Role of residue |
|---|---|---|---|---|
| Y76C | 8 | 282714 | 2.8E-05 | Binding to α-tubulin (B-tubule) |
| E86K | 2 | 251328 | 8.0E-06 | Binding to α-tubulin (B-tubule) and MEIG1 |
| A95T | 26 | 251066 | 1.0E-04 | Binding to MEIG1 |
| E136Q | 2 | 282826 | 7.1E-06 | Binding to MEIG1 |
| R160Q | 10 | 282322 | 3.5E-05 | Binding to β-tubulin (A-tubule) |
| P195R | 8 | 282736 | 2.8E-05 | Binding to β-tubulin (A-tubule) |
| E252K | 10 | 282336 | 3.5E-05 | Binding to α-tubulin (A-tubule) |
Data obtained from the GnomAD database
The PACRG-like protein recruits tubulin to microtubules and does not bind MEIG1.
Sequence homology search identifies the PACRG-like protein (PACRGL), a poorly characterized protein with 23% sequence identity with PACRG (Figure 6A). Using the crystal structure of PACRG, we have created a homology model of PACRGL (Figure 6B). The model shows that PACRGL has a very short loop between α6 and α7, strongly suggesting that it cannot interact with FAP20. Furthermore, five residues in PACRG that are essential for MEIG1 binding (Lys93, Ile100, His132, Glu136 and His137) are not conserved in PACRGL, suggesting that it does not bind MEIG1 (Figure 6C). To test this, we expressed human His6-PACRGL and found that it could be purified in the absence of MEIG1. Furthermore, size-exclusion chromatography of PACRGL and MEIG1 shows no co-elution, confirming that they do not interact (Figure 6D). We find that the tubulin-interacting PACRG residues Tyr76, His87, Lys154, Arg191, and Lys245 are conserved in PACRGL (Tyr84, His95, Lys164, Lys198, and Lys237, respectively), and both PACRG and PACRGL have an extended N-terminus (Figure 6A), suggesting PACRGL may also have multiple tubulin binding sites. To test this, we used the single-molecule microscopy assay and found that PACRGL was able to recruit tubulin to microtubules more efficiently than PACRG:MEIG1 at all concentrations tested (Figure 6E,F). Furthermore, PACRGL forms crosslinks with tubulin dimers (Figure 5). These results confirm that PACRGL binds microtubules and tubulin in a manner similar to PACRG:MEIG1.
Figure 6. The PACRG-like protein recruits tubulin to microtubules and does not bind MEIG1.
(A) Sequence alignment of the human PACRG-like protein (PACRGL) with human PACRG. Blue shading indicates degree of conservation. Asterisks indicate residues important for binding MEIG1. Black circles highlight residues that contact tubulin in the axonemal doublet tubule structure. (B) Homology model of PACRGLΔ1–72 (yellow) superposed on the crystal structure of PACRGΔ1–69:MEIG1 (cyan and magenta). (C) Close-up view of the PACRG:MEIG1 interface, showing the side-chain of residues critical for the interaction. The side-chain of corresponding PACRGL residues are also shown. (D) Size-exclusion chromatography of PACRGL, MEIG1, and an equimolar mixture of 50 μM PACRGL and MEIG1. (E) TIRF microscopy images showing that PACRGL recruits free HiLyte 488-labeled α/β-tubulin to TAMRA-labeled microtubules more efficiently than PACRG:MEIG1 at 2 μM. (F) Quantification of HiLyte 488-labeled α/β-tubulin that colocalize with the microtubules in the presence of PACRG:MEIG1 or PACRGL. Brown-Forsythe and Welch ANOVA and Dunnett’s tests were performed for multiple comparisons. * indicates p ≤ 0.05, **** for p ≤ 0.0001, (n=14–18).
Conservation of the PACRG-Parkin gene structure and transcription regulation
Gene pairs such as Parkin and PACRG, which share bi-directional promoters, may also share a common function and have partially overlapping expression patterns (West et al., 2003). Furthermore, their relative orientation is not random in mammals, with the head-to-head orientation being preferred for gene duos separated by less than 600 bp (Franck et al., 2008). To establish whether Parkin and PACRG function in a common biological pathway, we have analyzed the conservation of the PACRG and Parkin gene structure. In all amniotes (terrestrial animals), PACRG and Parkin are found in a head-to-head orientation, with most species having less than 600 bp between the two genes, except in a few mammals and birds (Table S1). The relationship breaks in amphibians, which do not have a Parkin gene. Fishes and invertebrates have Parkin and PACRG, but they are typically located on different chromosomes. This is consistent with head-to-head gene duos starting to accumulate in tetrapods (Franck et al., 2008). Finally, it is worth noting that single-celled organisms do not have Parkin but have PACRG if they are ciliated or flagellated.
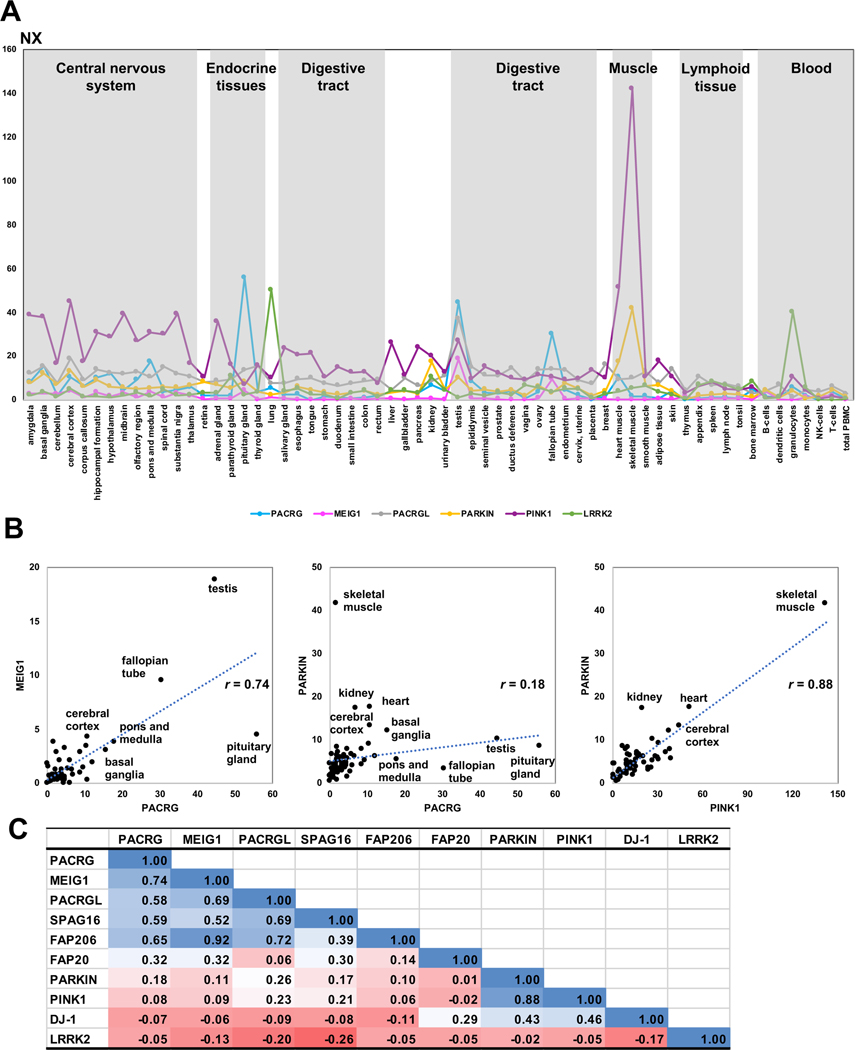
Given that the head-to-head structure of the PACRG-Parkin genes is conserved in amniotes, we sought to determine whether their expression was co-regulated as a result of acting in related pathways. We therefore analyzed their mRNA expression profiles across different tissues using the Protein Atlas consensus dataset (Uhlen et al., 2015). We extended our analysis to MEIG1, PACRGL, and other axonemal proteins (SPAG16, FAP20, and FAP206), as well as PINK1 and DJ-1, two proteins implicated in PD and which regulate mitochondrial homeostasis (Trempe and Fon, 2013). Furthermore, we also included LRRK2, a kinase associated with both leprosy and PD (Fava et al., 2019). We found a strong correlation (r = 0.88) between the expression of Parkin and PINK1, with both expressing at very high levels in mitochondria-rich tissues such as muscles and the central nervous system (Figure 7). Likewise, we found a significant correlation between PACRG and MEIG1 (r = 0.74), with high expression in reproductive tissues such as testis and Fallopian tubes as well as some brain areas. PACRG expression is notably very high in the pituitary gland, which contains multiciliated cells (Correr and Motta, 1981; Nakakura et al., 2017). PACRG and MEIG1 levels both correlate well with other axonemal proteins, specifically FAP206 and SPAG16. However, there was little correlation between PACRG and Parkin (r = 0.18), with some overlap for expression in the basal ganglia and cerebral cortex. LRRK2 expression did not correlate with Parkin nor with PACRG, although the two tissues with the highest levels of LRRK2, namely lung and granulocytes, show significant levels of PACRG. On the other hand, the expression of PACRGL correlates with PACRG (r = 0.58), MEIG1 (r = 0.69), and other axonemal proteins except FAP20, suggesting that PACRGL may also be an axonemal protein.
Figure 7. Transcriptomics analysis of PACRG and other genes across human tissues.
Data retrieved from the Protein Atlas consensus dataset for mRNA expression in different human tissues and blood cells (A). Consensus Normalized eXpression (NX) levels for each gene, sorted by tissue type. (B) Correlation graphs of NX levels for three selected pairs of genes (PACRG/MEIG1, PACRG/PARKIN, PARKIN/PINK1). Tissues showing high levels of expression are labeled. Correlation coefficients (r) are indicated. (C) Table of pairwise correlation coefficients between tissue mRNA expression levels, color-coded from low (red) to high (blue) values.
Discussion
Using the crystal structure of PACRG:MEIG1, we built a homology model of the human inner junction of an axonemal doublet microtubule (Figure 3), which we validated using a microscopy tubulin recruitment assay (Figure 4). While we don’t know how PACRG:MEIG1 binds to the microtubule seeds, we speculate that it binds primarily through the N-terminus and the basic residues forming the group 2 mutations, since these sites are predicted to bind the A-tubule, which is structurally similar to a standard microtubule lattice. This step is critical because mutations at those sites completely abrogate recruitment of tubulin. Once bound to a microtubule lattice, PACRG:MEIG1 can then catalyze formation of the B-tubule by recruiting free tubulin through their other tubulin-binding site (group 1). Here, the E77K mutation is intriguing because it increases the absolute intensity of tubulin recruitment at saturating protein concentrations compared to WT. This mutation likely increases the affinity of the “group 1” site for tubulin, perhaps by making it bind to the A-tubule as well, thus initiating the recruitment of multiple layers of tubulin. Indeed, the group 1 and E77K mutants increase not only the number of tubulin puncta, but also the intensity of each punctum. Glu77 is highly conserved (Figure S3), implying that the binding affinity for this site has been selected to be in a weak range. PACRGL also appears to have a higher tubulin-binding affinity than PACRG, although more experiments will be required to dissect out the contribution from its different binding sites.
PACRG residues that are essential for complex formation are conserved in all organisms that also express MEIG1 (Figure 2), which comprises all vertebrates as well as invertebrates that express motile sperm cells such as mollusks (Salzberg et al., 2010). However, other metazoans as well as ciliated protozoans do not have MEIG1 but have PACRG, hinting to the latter playing a more fundamental role in axonemal doublet formation, whereas MEIG1 evolved later to have a function specifically related to transport of cargo critical for axoneme assembly. While there is no evidence that MEIG1 localizes to the axoneme in mature sperm cells, MEIG1 colocalizes with PACRG and microtubules in the manchette and acts as an adaptor protein for SPAG16 (Li et al., 2015), which is itself interacting with central pair proteins (Zhang et al., 2006). Furthermore, MEIG1 is required for flagella formation. Thus MEIG1 is at the very least involved in the assembly of the axoneme. Our structural model shows how MEIG1 may coordinate recruitment of a second tubulin protofilament and SPAG16 on a microtubule, which could be a critical step for axoneme assembly. Using our microscopy and crosslinking assays, we indeed find that MEIG1 contributes to the recruitment of tubulin to microtubules by PACRG (Figures 4 and 5, group 3). However, it is unclear whether MEIG1 dissociates from PACRG following recruitment of tubulin. Future studies will aim to understand how PACRG and MEIG1 are recruited and retained on microtubules using TIRF microscopy. Thus, while our microscopy tubulin recruitment assay is a simplified version of a far more complex process involving several other axonemal proteins, its sensitivity and modularity will make it a useful tool to characterize the steps involved in axoneme assembly.
The PACRG:MEIG1 structure enables us to predict how naturally-occurring variants in PACRG might affect its function. While the phenotype of humans carrying these variants is unknown, we would predict that they display phenotypes similar to those found in ciliopathies, at least in the homozygous state (Ibanez-Tallon et al., 2003). Loss-of-function mutations in PACRG would likely impair male and female fertility and might also lead to neurodevelopmental and respiratory defects associated with loss of motile cilia/flagella. Furthermore, it has not yet been established whether mammalian PACRG localizes at the primary cilia, a non-motile axoneme-based structure that coordinates a plethora of cellular signalling pathways (Anvarian et al., 2019). Thus, if PACRG is essential to the function of primary cilia, loss-of-function PACRG mutations would have long-ranging effects on Sonic hedgehog signalling, G-protein signalling and so on, which could have a profound impact on human health. Case in point, primary cilia are essential for the development midbrain dopamine neurons, which are affected in Parkinson’s disease (Gazea et al., 2016). Future studies should aim to determine the potential pathogenicity of PACRG variants in human.
The significance of the head-to-head arrangement of the Parkin and PACRG genes remains unclear. In mouse, PACRG and Parkin are both expressed in testis and brain, and the single knockout of PACRG leads to an increase in Parkin levels, suggesting some cross-talk (Stephenson et al., 2018). A potential route of interaction between the two genes might be in sperm maturation. Mitochondria play an important role in sperm development as they form an array with microtubules that promotes the elongation of the tail of the sperm, which eventually becomes the flagella (Noguchi et al., 2011). The mature spermatid tail is filled with mitochondria, which are in contact with the flagellar axoneme to provide the energy required for motility. Parkin is a well-established mitochondrial quality control protein, and Parkin-null Drosophila have mitochondrial dysfunction that results in a defect of spermatid individualization at the late stage of sperm maturation (Greene et al., 2003). Other cells with motile cilia, such as ependymal cells, also have high energy requirements, where coupling between mitochondria and axoneme would be essential. Thus, we speculate that the two genes are co-regulated only in a subset of tissues, under the control of a tissue-specific transcription factor that can transcribe both genes.
Another intriguing link between Parkin and PACRG is their involvement in immunity and inflammation (Mira et al., 2004). Both proteins promote tumor necrosis factor (TNF)–induced NF-κB signalling, suggesting a role in innate immunity (Meschede et al., 2020; Müller-Rischart et al., 2013). Intriguingly, SNPs and missense variants in LRRK2, a gene associated with autosomal dominant PD, have also been linked to susceptibility, inflammatory responses and type-1 reactions in leprosy (Fava et al., 2016; Fava et al., 2019; Zhang et al., 2009a). LRRK2 is a large protein of the ROC-COR family of protein kinases, which regulates intracellular trafficking via phosphorylation of Rab GTPases, which are key mediators of pathways implicated in PD (Bonet-Ponce and Cookson, 2019). Intriguingly, LRRK2 phosphorylates two Rab GTPases, Rab8A and Rab10, which regulate ciliogenesis (Dhekne et al., 2018; Steger et al., 2017). Pathogenic mutations in LRRK2, which increase its kinase activity, impair primary cilia formation as a result of Rab hyper-phosphorylation. In adult mice, LRRK2 is highly expressed in the lung and lymph node (Biskup et al., 2007), which is consistent with the expression pattern observed in human, where it is found in the lung and granulocytes (Figure 7). While there is no overall correlation between the expression of LRRK2 and any other genes we investigated, PACRG is expressed in lung and granulocytes, which point to a role in immunity.
To our knowledge, there is no publication on the PACRGL protein, which appears to be expressed in a broad range of tissues in human (Figure 7). It is highly expressed in sperm and its tissue distribution correlates to some degree with PACRG, MEIG1 and other axonemal proteins, suggesting that their functions overlap. Although we cannot exclude that PACRGL has a tubulin-dependent function that is unrelated to axonemes, we speculate that PACRGL may be an inner junction protein that binds at sites distinct from PACRG. Inner junction proteins play a critical role not only in 9+2 axoneme structures found in motile cilia and flagella, but also in the regulation of microtubule doublets and singlet-to-doublet transitions that occur in non-motile primary cilia (Sun et al., 2019). However, the identity of inner junction proteins regulating primary cilia is unknown. While PACRGL has a loop that is similar in length to that of the MB-loop in PACRG, it does not bind MEIG1. Further experiments will be required to determine if PACRGL binds another adaptor protein. Finally, we observe that the tissue distributions of PACRG and MEIG1 are not perfectly correlated, with PACRG being expressed in tissues with little to no MEIG1 (e.g. heart and granulocytes). Thus, it is also possible that the MB-loop in PACRG binds other adaptor proteins that enable other proteins to bind at the inner junction of various axoneme microtubule structures. Likewise, while PACRG most likely does not bind ubiquitin, it is however intriguing that proteomics study of primary cilia performed by proximity labeling identified components of the ubiquitin pathway (Mick et al., 2015). Thus, we cannot exclude that PACRG binds a ubiquitin chain or a ubiquitin-like molecule. Future work should seek to identify adaptor proteins for PACRG and PACRGL and determine their respective roles in the regulation of motile and non-motile cilia via their tubulin-binding sites.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jean-François Trempe: jeanfrancois.trempe@mcgill.ca.
Materials availability
All bacterial plasmids used for expression of PACRG, MEIG1 and PACRGL are available upon request.
Data and code availability
The coordinates and maps for the three structures presented have been deposited in the Protein Databank https://www.rcsb.org: PDB 6NEP, 6NDU, and 6UCC.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All recombinant proteins were produced in BL21 (DE3) Escherichia coli cells. All cloning was performed in DH5-alpha E. coli cells. Cells were propagated in Luria broth (LB) at 37˚C with shaking at 180 rpm. Competent cell stocks were prepared in-house for heat shock transformation with plasmids of interests.
METHOD DETAILS
Plasmids, antibodies and reagents
The open-reading frame for the long isoform of human PACRG1–296 was obtained by gene synthesis following codon optimization for expression in E. coli (DNA Express Inc., Montreal). The gene was cloned directly in the pGEX-6p1 plasmid. To generate the full-length short isoform (“FL”, 257 a.a.), primers were designed to remove a.a 205–243 by PCR mutagenesis (Data S1). To generate PACRGΔ1–69, standard restriction enzyme cloning was performed using the protocol described by New England Biolabs (NEB). PACRGΔ1–69 was amplified from the GST-PACRGFL plasmid with PCR primers introduced BamHI (N-terminal) and XhoI (C-terminal) restriction sites for ligation in pGEX-6p1 with the T4 DNA ligase (NEB; Data S2). To generate the PACRG:MEIG1 co-expression vector the same protocol was performed as above using restriction enzyme cloning. His6-MEIG1 was obtained by gene synthesis from GeneArt (Thermofisher) and cloned in pRSET-A (Data S3). To generate pRSF-Duet-PACRG-His6MEIG1 (Data S4), pRSET-His6-MEIG1 was digested with NcoI and XhoI and the fragment was inserted into a pRSF-Duet vector, cleaved by the same enzymes. PACRGFL (pGEX-6P-1), digested with NcoI and XhoI restriction enzymes (NEB), was inserted into pRSF-Duet vector containing His6-MEIG1, which was digested with NcoI and SalI, which has compatible cohesive ends with XhoI. Single-point mutants in PACRG were generated by PCR mutagenesis using the QuickChange II Site Directed Mutagenesis kit (Agilent Technologies). The open-reading frame for human PACRGL was obtained by gene synthesis from GeneArt (Thermofisher) and cloned in pET-151D-TOPO (Data S5). The pDB070.iodo.5 plasmid containing the modified tRNA and tRNA synthetase for p-iodo-L-phenylalanine incorporation was obtained from Addgene (#99397) as a gift from David Liu (Bryson et al., 2017). p-iodo-L-phenylalanine was purchased from Sigma (#I8757). The PACRG mouse C-8 monoclonal antibody was from Santa Cruz Biotechnology (#sc-373851). The His-tag polyclonal rabbit antibody was from Cell Signaling Technology (#2366P).
Protein expression and purification
Purification of His6-MEIG1:PACRGFL, His6-MEIG1, and His6-PACRGL.
BL21 (DE3) E. coli cells transformed with the pRSF-DUET, pRSET-A or pET-151D plasmids were grown overnight in LB at 37˚C shaking at 180 rpm with the corresponding antibiotic. The next day, the starter culture was added to 1L or more LB, grown at 37˚C shaking at 180 rpm until OD600 reached 0.8, at which point 0.5 mM IPTG was added for induction. Protein expression was carried out for 3 hours at 25˚C. Cells were harvested at 5000 rpm for 30 min in a Sorvall type GS-3 centrifuge rotor. The cell pellet was resuspended in lysis buffer (50 mM HEPES pH 7.4, 120 mM NaCl, 5 mM MgCl2, 0.25 mg/mL DNase, 0.5 mg/ml of lysozyme, 2.5 mM DTT, 0.2% triton X-100, and 1 protease cocktail inhibitor: complete ULTRA Tablets from Roche diluted in 60 ml of lysis buffer). The resuspended cell pellet was then lysed using sonication with a process time of 2 minutes, 20 seconds on pulse and 40 seconds off time. The lysate was then clarified by centrifugation at 15,000 rpm in a Beckman JA-20 rotor. Co-NTA beads were obtained from stripping 1 mL of Ni-NTA resin from Qiagen in a Bio-rad gravity column with 50 mL of 0.5 M EDTA (pH 8.0), followed by 20 ml of water. 50 mL of 1 M Co(II) was then added to the NTA resin to produce Co(II) charged NTA resin. 1 mL of this resin was then washed in wash buffer (50 mM HEPES pH 7.4 and 300 mM NaCl). The cleared lysate was then added directly on the resin. The resin was then washed once in 3 column volume (cv) of wash buffer, followed by 4 washes, each of 1 cv of wash buffer. The resin was then washed twice with 1 cv of wash buffer that additionally contained 20 mM imidazole. Finally, the protein was eluted with 4 cv of 300 mM imidazole wash buffer. The purified complex was concentrated to ≤5 mL and cleaved at 4 °C overnight with 20 μg of GST-3C protease per mg of protein to cleave the His6 tag from MEIG1. The sample was then loaded on a 120 mL HiLoad 16/600 Superdex 75 size-exclusion chromatography column equilibrated in buffer A at 1 mL/min. PACRGL was cleaved with 10 μg His6-TEV per mg protein and reloaded on Co-NTA to remove the His6-tag and TEV.
Purification of GST-PACRGΔ1–69:MEIG1.
GST-PACRGΔ1–69 was expressed from the pGEX-6p1 plasmid using the same protocol described above. The inclusion bodies protein extraction protocol used was adapted from Li et al. (Li et al., 2004). Inclusion body pellets were washed, 5 mL/g of pellet, with a buffer made of 50 mM HEPES, 50 mM NaCl, 2 mM DTT, 1% Triton X-100 (pH 7.0). The pellet was resuspended 3 times using 7 mL Wheaton Dounce tissue grinder (Fisher Scientific) and centrifuged at 12,000 rpm at 4°C for 30 minutes (Sorvall SS-34 centrifuge rotor). The last wash was performed with a buffer that did not contain Triton X-100. To extract the recombinant protein from the inclusion bodies, the cell pellet was resuspended in a buffer of 8 M urea, 50 mM HEPES, 50 mM NaCl, 2 mM DTT at pH of 7.0. The resuspension was incubated at 4°C with constant rotation overnight. Once the pellet was fully dissolved the mixture was centrifuged at 15,000 rpm at 4°C for 30 minutes. The supernatant was slowly added dropwise to 450 mL of refolding buffer with 2 M urea, 50 mM HEPES, 50 mM NaCl, 2 mM DTT, 1% Tween20 (pH 7.0) to initiate protein refolding and dilution of urea. The refolding mixture was incubated at 4°C with constant slow stirring for 2 days. The mixture was then centrifuged to remove any insoluble material and then dialyzed against a 4 L solution of 20 mM HEPES, 50 mM NaCl, 2 mM DTT, 1% Tween20 (pH 7.0) for 2 days at 4°C using 3,500 molecular weight cutoff dialysis tubing (Fisherbrand). The dialyzed sample was centrifuged and kept at 4 °C or frozen at −80°C for later use. 4 mg of His6-MEIG1 was then added dropwise to the supernatant to induce complex formation with PACRG. The solution was left stirring overnight at 4 °C. The next day, the GST-PACRGΔ1–69:MEIG1 complex was purified by affinity chromatography using 1.5 mL of glutathione-sepharose resin (Cytiva) washed in Buffer A (50 mM of HEPES, 120 mM of NaCl, 1 mM of DTT, pH 7.4). This resin was added to dialyzed PACRGΔ1–69: MEIG1 complex and incubated at 4 °C with slow rotation for 1 h. The resin was washed Buffer A and the protein was eluted in Buffer A with 20 mM glutathione (pH 8.0). The purified complex was cleaved at 4 °C overnight with 20 μg of GST-3C protease per mg of protein to cleave the GST tag from PACRG and the His6 tag from MEIG1, and then further purified by size-exclusion chromatography as described above, except that the column was connected to a GSTrap (Cytiva) to remove GST and GST-3C.
Iodo-phenylalanine incorporation.
The procedure is a modified version of a previously described protocol (Bryson et al., 2017). Briefly, the pGEX6p1-PACRGΔ1–69 plasmid was co-transformed with the pDB070.iodo.5 plasmid into BL21 (DE3) cells. Ampicillin (100 μg/mL) and chloramphenicol (25 μg/mL) were added to the overnight culture. 25 mL of the overnight culture was added to 525 mL of LB with ampicillin and chloramphenicol, cells were grown in 37°C shaker until an optical density of 0.3. 500 μL of p-iodo-L-phenylalanine in DMSO (320 mg/mL) was added to the cells along with 110 μL of anhydrotetracycline (1 mg/mL). Once the cells reached an optical density of 0.5, IPTG was added to 0.5 mM and cells were incubated in the shaker for 3 hours at 37°C. The remainder of the protocol is identical to the inclusion body protocol described above.
PACRG:MEIG1 interaction assay
His6-MEIG1:PACRGFL WT and mutant proteins were expressed in 300 mL of LB and purified by affinity chromatography as described above. 20 μl of the imidazole elution fraction was then loaded on 12.5% acrylamide SDS-PAGE gels (Figure 2B). The protein was then stained by Coomassie Brilliant Blue and visualized using Biorad Imaging System.
Mass spectrometry
For tandem mass spectrometry of protein digests, 10 μg of samples was denatured in 6 M urea, 1 mM EDTA, 50 mM TEAB pH 8.5. Cysteine residues were then reduced with TCEP (2 mM pH 7.0) and alkylated with iodoacetamide (10 mM solution in H2O freshly prepared, Sigma). Protein samples were diluted to 1 M urea with 50 mM TEAB buffer and digested with 1:100 trypsin (Sigma) 2 h at 37°C. Digested peptides were purified using C18 spin columns (Thermo Scientific). Peptides were diluted in loading buffer (2% acetonitrile, 0.1% formic acid), and 0.5 μg of peptides was captured on a Waters C18 BEH 1.0/15 mm column, washed 5 min with 4% acetonitrile, followed by a 30-min 5–40% gradient of acetonitrile in 0.1% formic acid, with a flow rate of 40 μl/min. The eluate was analyzed on a Bruker Impact II Q-TOF mass spectrometer equipped with an Apollo II ion funnel ESI source. Data were acquired in positive-ion profile mode, with a capillary voltage of 4,500 V and dry nitrogen heated at 200°C. Spectra were analyzed using the DataAnalysis and Biotools softwares (Bruker) to confirm incorporation of p-iodo-L-phenylalanine.
Crystallization
Crystallization was performed using the MEIG1:PACRGΔ1–69 complex concentrated to 1.7 mg/mL. and the JCSG plus (Molecular Dimensions) screen. Using a Formulatrix NT8 robotic drop dispenser sitting drops of 0.3 μL of protein and 0.3 μL of mother liquor were dispensed into 96-well plates. Plates were stored at 4°C. Crystals had grown overnight in conditions from both screens. The condition that formed crystals that diffracted to 2.1 Å resolution was from a condition containing 16% w/v PEG 8000, 20% v/v glycerol and 40 mM KH2PO4. Crystals were harvested with loops directly from this drop and plunged in liquid nitrogen for diffraction experiments. The Y189ϕ -derivative was concentrated to 2 mg/mL and mixed 1 μL with 1 μL of 18% w/v PEG 8000, 15% v/v glycerol and 60 mM KH2PO4. Crystals grew in one day at 4°C and were cryo-protected in the same condition with 20% glycerol. Limited proteolysis was performed on the purified PACRGFL:MEIG1 complex. The protein at 2 mg/mL was digested with 1:1000 subtilisin for 30 min. 1 μL of the mixture was then incubated with 1 μL 20% w/v PEG 8000, 20% v/v glycerol and 250 mM KH2PO4. Crystals grew in one day at 4°C and were frozen directly in liquid nitrogen.
X-ray data collection and structure determination
Diffraction data for native MEIG1:PACRGΔ1–69 and protease-digested MEIG1:PACRGFL were collected at the CMCF beamline 08ID-1 at the Canadian Light Source. Diffraction data for p-iodo-L-phenylalanine derivatives was collected at the Advanced Photon Source NECAT 24ID-E beamline. For the MEIG1:PACRGΔ1–69 complex, a total of 180 images were collected with an oscillation angle of 0.5° at 0.9793 Å. For the protease-digested MEIG1:PACRGFL complex, a total of 360 images were collected with an oscillation angle of 0.5° at 0.9795 Å. For the Y189ϕ - derivative of MEIG1:PACRGΔ1–69, a total of 360 images were collected with an oscillation angle of 0.5° at 1.476 Å. Reflections were integrated with the XDS (Kabsch, 2010) and scaled with Aimless (Winn et al., 2011). The Y189X derivative structure was solved by single-anomalous dispersion using Autosol from the software package PHENIX 1.12 (Liebschner et al., 2019). The average <danom/σ> was 1.08 and 0.87 at 4.5 and 3.0 Å, respectively. Following a first round of automated model building with ARP/WARP (Langer et al., 2008), the model was manually adjusted with Coot (Emsley et al., 2010) and refined in PHENIX. Native structures were solved by molecular replacement using the Y189X derivative structure as a search model in PHASER (McCoy et al., 2007), and refined with Coot and PHENIX.
Single-molecule total internal reflection fluorescence microscopy
Data was acquired with a customized Zeiss Axio Observer 7 equipped with a Laser TIRF III and 405/488/561/638 nm lasers, and Alpha Plan-Apo 100x/1.46Oil DIC M27 Objective Heater 25.5/33 S1. Images were recorded on a Prime 95B CMOS camera (Photometrics) with a pixel size of 107 nm. Tubulin was purified from bovine brains as previously described (Ashford and Hyman, 2006) with the modification of using Fractogel EMD SO3- (M) resin (Millipore-Sigma) instead of phosphocellulose. Tubulin was labeled using HiLyte Fluor 488 acid NHS ester (Anaspec) and tetramethylrhodamine (TAMRA, Invitrogen) as described (Hyman et al., 1991). An additional cycle of polymerization/depolymerization was performed before use. Protein concentrations were determined using a DS-11 FX spectrophotometer (DeNovix, Inc.). Cover glass was cleaned in acetone, sonicated in 50% methanol, sonicated in 0.5 M KOH, exposed to air plasma (Plasma Etch) for 3 min, then silanized by soaking in 0.2% dichlorodimethylsilane (DDS) in n-Heptane. 5 μL flow channels were constructed using two pieces of silanized cover glasses (22 X 22 mm and 18 X 18 mm) held together with double-sided tape and mounted into custom-machined cover slip holders. GMPCPP seeds were prepared by polymerizing a 1:4 molar ratio of TAMRA labelled:unlabelled tubulin in the presence of guanosine-5’-[(α,β)-methyleno]triphosphate (GMPCPP, Jena Bioscience) in two cycles as described (Gell et al., 2010). Channels were first incubated with anti-TAMRA antibodies (ThermoFisher Scientific) and then blocked with 5% Pluronic F-127. Flow channels were washed 3x with BRB80 buffer (80 mM NaOH-PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.9) before incubating with GMPCPP seeds. Tubulin recruitment to GMPCPP seeds was achieved by incubating flow channels with 500 nM HiLyte 488-tubulin (25% degree of labeling) and PACRG in imaging buffer: BRB80, 0.1 mg/mL BSA, 10 mM dithiothreitol, 250 nM glucose oxidase, 64 nM catalase, and 40 mM D-glucose. Image acquisition was controlled using ZEN 2.3 (Zeiss).
Crosslinking assays
Interaction between tubulin and PACRG was assessed by polymerization of tubulin in presence of PACRG WT and mutants with addition of crosslinker. Tubulin (Cytoskeleton), diluted to 5 μM with BRB80 buffer in the presence of 1 mM GTP, 4 mM MgCl2 and 10 μM taxol (from 1 mM stock in DMSO, 1% final concentration), was incubated 10 min at 36°C with 12.5 μM protein (PACRG:MEIG1 WT and mutants, PACRGL, or BSA), 4 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 10 mM N-hydroxysuccimide (NHS) in a final volume of 10 μL. A positive control was performed with 1.5 μM doublecortin (DCX) purified as described (Bechstedt and Brouhard, 2012). Reaction was stopped by addition of SDS containing sample buffer. Crosslinking experiments were also performed using 1.654 mM disuccinimidyl suberate (DSS) (from 50 mM stock in DMSO, 3.3% final concentration). Total reaction was loaded on SDS-PAGE 4–20% stain-free TGX gradient gel (BioRad) and imaged by UV-induced fluorescence (Biorad Imaging System).
Homology modelling
The structure of the human PACRG-like protein was built with the automated I-TASSER server (Yang et al., 2015), using the structure of human PACRG as a template (this work; PDB 6NDU). A homology model of the human axonemal doublet was built by superposing the human PACRG:MEIG1 complex onto the Chlamydomonas PACRG coordinates derived from cryo-electron microscopy (Khalifa et al., 2020). Mammalian alpha and beta tubulin models were generated by superposing the coordinates of pig tubulin monomers (PDB 1TUB) (Nogales et al., 1998) on the corresponding coordinates of Chlamydomonas tubulin subunits.
Transcriptomics analysis
Consensus normalized expression levels for 55 tissue types and 7 blood cell types, created by combining the data from the three transcriptomics datasets (HPA, GTEx and FANTOM5) using an internal normalization pipeline (Uhlen et al., 2015), were retrieved for individual genes from the Protein Atlas server in TSV format.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics for all crystal structures were determined using software listed in the Key Resources Table. Statistics generated from data processing and refinement are listed in Table 1. SDS-PAGE gel and TIRF microscopy images were processed and analyzed using Fiji (Schindelin et al., 2012). Gel densitometry was performed using a constant box size from which background intensity was subtracted. Tubulin recruitment was calculated by measuring the intensity of the HiLyte-488 tubulin recruited to the GMPCPP seed. This intensity was subtracted by the background HiLyte-488 intensity nearby the given GMPCPP seed. Graphics for densitometry experiments (Figures 2B, 5B) were generated using Prism 9. Graphics for microscopy quantification were generated using home-made Python 3 scripts and Adobe Illustrator (Figures 4D, 6F). Statistical analysis was performed using Prism 9, as described in figure legends, with n is the number of microtubule seeds or the number of gel lanes analyzed. Graphics and Pearson correlation coefficients between each set of transcriptomics data for each gene were produced in Excel.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-PACRG mouse monoclonal antibody (C-8) | Santa Cruz Biotechnology | Cat: sc-373851; RRID: AB_10988029 |
| Anti-His-tag rabbit polyclonal antibody | Cell Signaling Technology | Cat: 2366; RRID: AB_2115719 |
| Anti-TAMRA mouse monoclonal antibody (11H10) | ThermoFisher Scientific | Cat: MA1-042; RRID: AB_2536729 |
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) – Novagen | Sigma-Aldrich/ Merck | Cat: 69450-3 |
| E. coli DH5α | New England Biolabs | Cat: C2987I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Restriction endonucleases: BamHI, NcoI, SalI, XhoI | New England Biolabs | Cat: BamHI R0136S, NcoI, R0193S, SalI R0138S, XhoI R0146S |
| T4 DNA ligase | New England Biolabs | Cat: M0202T |
| QuickChange II Site Directed Mutagenesis kit | Agilent | Cat: 200524 |
| p-iodo-L-phenylalanine | Sigma-Aldrich | Cat: I8757 |
| Ampicillin sodium salt | Fisher Scientific | Cat: BP176025 |
| Anhydrotetracycline | Sigma-Aldrich | Cat: 37919 |
| Chloramphenicol | Biobasic | Cat: CB0118 |
| IPTG | Biobasic | Cat: IB0168 |
| Ni-NTA agarose | Qiagen | Cat: 1000632 |
| Glutathione-sepharose 4B | Cytiva | Cat: 17-0756-01 |
| Iodoacetamide | Sigma-Aldrich | Cat: I6125 |
| Subtilisin | Sigma-Aldrich | Cat: P5380 |
| Trypsin | Sigma-Aldrich | Cat: T6567 |
| GST-3C protease | Homemade | N/A |
| His6-TEV protease | Homemade | N/A |
| JCSG plus screen | Molecular Dimensions | Cat: MD-137 |
| 50% w/v PEG 8000 | Molecular Dimensions | Cat: MD2-100-13 |
| 100% glycerol | Molecular Dimensions | Cat: MD2-100-65 |
| 1 M potassium phosphate monobasic | Molecular Dimensions | Cat: MD2-100-164 |
| Fractogel EMD SO3- (M) resin | Millipore-Sigma | Cat: 1168820010 |
| HiLyte Fluor 488 acid NHS ester | Anaspec | Cat: AS-81161 |
| Tetramethylrhodamine (TAMRA) | ThermoFisher Scientific | Cat:C1171 |
| Guanosine-5’-[(α,β)-methyleno]triphosphate (GMPCPP) | Jena Bioscience | Cat: NU-405S |
| Tubulin | Cytoskeleton | Cat: T240-A |
| PIPES | Sigma-Aldrich | Cat: 80635 |
| Paclitaxel from Taxus brevifolia, PTX; Taxol | Gold Bio | Cat: P-925-100 |
| Guanosine triphosphate (GTP) | Sigma-Aldrich | Cat: G5884 |
| 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) | ThermoFisher Scientific | Cat: PG82079 |
| N-hydroxysuccimide (NHS) | ThermoFisher Scientific | Cat: PI24500 |
| Disuccinimidyl suberate (DSS) | ThermoFisher Scientific | Cat: 21655 |
| Critical Commercial Assays | ||
| 4-20% stain-free TGX gradient gels | BioRad | Cat: 4568096 |
| Pepclean C18 spin columns | ThermoFisher Scientific | Cat: PI89873 |
| Deposited Data | ||
| Native MEIG1:PACRGΔ1–69 | This work | PDB: 6NDU |
| iodo-Y189Φ MEIG1:PACRGΔ1–69 | This work | PDB: 6NEP |
| Proteolysed MEIG1:PACRGFL | This work | PDB: 6UCC |
| Porcine tubulin dimer | (Nogales et al., 1998) | PDB: 1TUB |
| Chlamydomonas reinhardtii axonemal doublet microtubule | (Khalifa et al., 2020) | PDB: 6VE7 EMD: 20858 |
| Recombinant DNA | ||
| Human His6-MEIG1, cloned in pRSET-A | This work/Thermofisher | N/A |
| Human PACRG1–257 (FL), cloned in pGEX-6P-1 | This work/DNA Express | N/A |
| Human PACRGΔ1–69 WT, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRGΔ1–69 F75X, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRGΔ1–69 Y76X, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRGΔ1–69 Y108X, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRGΔ1–69 F121X, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRGΔ1–69 Y189X, cloned in pGEX-6P-1 | This work | N/A |
| Human PACRG-WT His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG Δ1–35 His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG Δ36–69 His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-Y76F His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E77K His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E86A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E86K His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-H87A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-D88A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-K93I His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-A95T His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E99A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-I100E His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E101A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-H106A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-H132A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E136A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E136Q His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-H137A His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-R160Q His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-P195R His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-E252K His6MEIG1, cloned in pRSF-DUET | This work | N/A |
| Human PACRG-Y76F-E77K-E86KH87A His6MEIG1, cloned in pRSF-DUET (Group 1) | This work | N/A |
| Human PACRG-K154E-R160Q-R191E His6MEIG1, cloned in pRSF-DUET (Group 2) | This work | N/A |
| Human PACRG-WT His6MEIG1-H13AK78A-H81A-K82A-Y86A, cloned in pRSF-DUET (Group 3) | This work | N/A |
| Human PACRGL1–248 (FL), cloned in pET-151D-TOPO | This work/Thermofisher | N/A |
| pDB070.iodo.5 | (Bryson et al., 2017) / Addgene | Cat: 99397 |
| Software and Algorithms | ||
| XDS | (Kabsch, 2010) | xds.mpimf-heidelberg.mpg.de |
| Aimless (CCP4 package) | (Winn et al., 2011) | www.ccp4.ac.uk/html/aimless.html |
| PHENIX 1.12 including Autosol, PHASER-MR, Refine | (Liebschner et al., 2019) | www.phenix-online.org/documentation/index.html |
| ARP/wARP 8.0 | (Langer et al., 2008) | www.embl-hamburg.de/ARP |
| Coot 0.8.9.1 | (Emsley et al., 2010) | www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| Fiji | (Schindelin et al., 2012) | imagej.net/Fiji |
| DataAnalysis and Biotools | Bruker | www.bruker.com |
| PyMOL 2.4.0 | Schrödinger LLC | pymol.org |
| Prism 9 | GraphPad | www.graphpad.com/scientificsoftware/prism |
| Excel 16.40 | Microsoft | www.microsoft.com/fr-ca/microsoft365/excel |
| GnomAD server | (Lek et al., 2016) | gnomad.broadinstitute.org |
| I-TASSER server | (Yang et al., 2015) | zhanglab.ccmb.med.umich.edu/ITASSER |
| Protein Atlas server | (Uhlen et al., 2015) | www.proteinatlas.org |
| Other | ||
| GSTrap 4B | Cytiva | Cat: 45001429 |
| HiLoad 16/600 Superdex 75 pg | Cytiva | Cat: 28989333 |
| Superdex 75 Increase 10/300 GL | Cytiva | Cat: 29148721 |
Supplementary Material
Highlights.
Crystal structure of human PACRG:MEIG solved at 2.1 Å resolution.
PACRG:MEIG1 recruits tubulin to microtubules via multiple binding sites.
Rare human variants in PACRG interfere with binding to MEIG1 and tubulin.
PACRGL recruits tubulin to microtubules, but does not bind MEIG1.
Acknowledgments
We thank K. Winklhofer and J. Menschede for useful discussion that contributed to the development of this study. The beamline 08ID-1 at the Canadian Light Source (CLS) is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. We express our deepest appreciation to the staff at the CLS Canadian Macromolecular Crystallography Facility, in particular P. Grochulski, M. Fodje, and S. Labiuk. The Northeastern Collaborative Access Team (NECAT) beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). We thank T.M. Schmeing and C. Fortinez for sharing their time at the APS. We thank the McGill Pharmacology SPR/MS facility (M. Hancock) and the CFI for support. This work was supported by a Canada Research Chair grant from the CIHR to J.-F.T., as well as a grant and studentship from NSERC to J.-F.T. and D.P.
Footnotes
Declaration of interests
J.-F.T. is the Director of the Proteomics platform at the Research Institute of the McGill University Health Centre, as well as a member of the scientific advisory board of Mitokinin Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, and Christensen ST (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol 15, 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford AJ, and Hyman AA (2006). Preparation of Tubulin from Porcine Brain. In Cell Biology (Third Edition), Celis JE, ed. (Academic Press; ), pp. 155–160. [Google Scholar]
- Bayne AN, and Trempe JF (2019). Mechanisms of PINK1, ubiquitin and Parkin interactions in mitochondrial quality control and beyond. Cell Mol Life Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt S, and Brouhard GJ (2012). Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev Cell 23, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Rea A, Lorenz-Deperieux B, Coombes CE, Dawson VL, Dawson TM, and West AB (2007). Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci 8, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet-Ponce L, and Cookson MR (2019). The role of Rab GTPases in the pathobiology of Parkinson’ disease. Curr Opin Cell Biol 59, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson DI, Fan C, Guo LT, Miller C, Soll D, and Liu DR (2017). Continuous directed evolution of aminoacyl-tRNA synthetases. Nature chemical biology 13, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correr S, and Motta PM (1981). The rat pituitary cleft: a correlated study by scanning and transmission electron microscopy. Cell Tissue Res 215, 515–529. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Portman N, Shaw MK, and Gull K. (2005). The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J Cell Sci 118, 5421–5430. [DOI] [PubMed] [Google Scholar]
- Dhekne HS, Yanatori I, Gomez RC, Tonelli F, Diez F, Schule B, Steger M, Alessi DR, and Pfeffer SR (2018). A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife 7, e40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Lin J, Fu G, Porter M, Nicastro D, and Smith EF (2019). PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. Mol Biol Cell, mbcE19010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava VM, Manry J, Cobat A, Orlova M, Van Thuc N, Ba NN, Thai VH, Abel L, Alcais A, Schurr E, and Canadian Lrrk2 in Inflammation T.(2016). A Missense LRRK2 Variant Is a Risk Factor for Excessive Inflammatory Responses in Leprosy. PLoS neglected tropical diseases 10, e0004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava VM, Xu YZ, Lettre G, Van Thuc N, Orlova M, Thai VH, Tao S, Croteau N, Eldeeb MA, MacDougall EJ, et al. (2019). Pleiotropic effects for Parkin and LRRK2 in leprosy type-1 reactions and Parkinson’s disease. Proceedings of the National Academy of Sciences of the United States of America 116, 15616–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck E, Hulsen T, Huynen MA, de Jong WW, Lubsen NH, and Madsen O. (2008). Evolution of closely linked gene pairs in vertebrate genomes. Mol Biol Evol 25, 1909–1921. [DOI] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, and Shen J. (2008). Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proceedings of the National Academy of Sciences of the United States of America 105, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazea M, Tasouri E, Tolve M, Bosch V, Kabanova A, Gojak C, Kurtulmus B, Novikov O, Spatz J, Pereira G, Hubner W, Brodski C, Tucker KL, and Blaess S. (2016). Primary cilia are critical for Sonic hedgehog-mediated dopaminergic neurogenesis in the embryonic midbrain. Dev Biol 409, 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. (2010). Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol 95, 221–245. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, et al. (2003). Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. The Journal of biological chemistry 278, 43628–43635. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, and Pallanck LJ (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proceedings of the National Academy of Sciences of the United States of America 100, 4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, and Mitchison T. (1991). Preparation of modified tubulins. Methods Enzymol 196, 478–485. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Heintz N, and Omran H. (2003). To beat or not to beat: roles of cilia in development and disease. Human molecular genetics 12 Spec No 1, R27–35. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ikeda T, Morikawa K, and Kamiya R. (2007). Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil Cytoskeleton 64, 814–821. [DOI] [PubMed] [Google Scholar]
- Ikeda T. (2008). Parkin-co-regulated gene (PACRG) product interacts with tubulin and microtubules. FEBS Lett 582, 1413–1418. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Murakami T, Shoji M, Abe K, and Takahashi R. (2003). A product of the human gene adjacent to parkin is a component of Lewy bodies and suppresses Pael receptor-induced cell death. The Journal of biological chemistry 278, 51901–51910. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa AAZ, Ichikawa M, Dai D, Kubo S, Black CS, Peri K, McAlear TS, Veyron S, Yang SK, Vargas J, Bechstedt S, Trempe JF, and Bui KH (2020). The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, and Shimizu N. (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, and Perrakis A. (2008). Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Yu JF, Chen YJ, Ma HB, Wang ZF, Zhu YB, and Zhang XG (2004). Refolding and characterization of recombinant human GST-PD-1 fusion protein expressed in Escherichia coli. Acta biochimica et biophysica Sinica 36, 141–146. [DOI] [PubMed] [Google Scholar]
- Li W, Tang W, Teves ME, Zhang Z, Zhang L, Li H, Archer KJ, Peterson DL, Williams DC Jr., Strauss JF 3rd, and Zhang Z. (2015). A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development 142, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Walavalkar NM, Buchwald WA, Teves ME, Zhang L, Liu H, Bilinovich S, Peterson DL, Strauss Iii JF, Williams DC Jr., and Zhang Z. (2016). Dissecting the structural basis of MEIG1 interaction with PACRG. Sci Rep 6, 18278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta crystallographica. Section D, Structural biology 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti D, Bishop CE, and Justice MJ (2004). Deletion of the Parkin coregulated gene causes male sterility in the quaking(viable) mouse mutant. Proceedings of the National Academy of Sciences of the United States of America 101, 8402–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucks CM, Bialas NJ, Dekkers MP, Walker DS, Grundy LJ, Li C, Inglis PN, Kida K, Schafer WR, Blacque OE, Jansen G, and Leroux MR (2016). PACRG, a protein linked to ciliary motility, mediates cellular signaling. Mol Biol Cell 27, 2133–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, and Zhang R. (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell 179, 909–922 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, and Cox JS (2013). The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheoud D, Cannon T, Voisin A, Penttinen AM, Ramet L, Fahmy AM, Ducrot C, Laplante A, Bourque MJ, Zhu L, et al. (2019). Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1(−/−) mice. Nature 571, 565–569. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, and Cramer P. (2004). Recognition of RNA polymerase II carboxy-terminal domain by 3’-RNA-processing factors. Nature 430, 223–226. [DOI] [PubMed] [Google Scholar]
- Meschede J, Sadic M, Furthmann N, Miedema T, Sehr DA, Muller-Rischart AK, Bader V, Berlemann LA, Pilsl A, Schlierf A, et al. (2020). The parkin-coregulated gene product PACRG promotes TNF signaling by stabilizing LUBAC. Sci Signal 13, eaav1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, and Nachury MV (2015). Proteomics of Primary Cilia by Proximity Labeling. Dev Cell 35, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, et al. (2004). Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427, 636–640. [DOI] [PubMed] [Google Scholar]
- Müller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, et al. (2013). The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell 49, 908–921. [DOI] [PubMed] [Google Scholar]
- Nakakura T, Suzuki T, Horiguchi K, Fujiwara K, Tsukada T, Asano-Hoshino A, Tanaka H, Arisawa K, Nishijima Y, Nekooki-Machida Y, Kiuchi Y, and Hagiwara H. (2017). Expression and localization of forkhead box protein FOXJ1 in S100beta-positive multiciliated cells of the rat pituitary. Med Mol Morphol 50, 59–67. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, and Downing KH (1998). Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Koizumi M, and Hayashi S. (2011). Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr Biol 21, 805–814. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, and Boucher RC (2002). A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics 1, 451–465. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, and Youle RJ (2015). Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron 87, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiei A, and Schriemer DC (2019). A microtubule crosslinking protocol for integrative structural modeling activities. Anal Biochem 586, 113416. [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Eldar T, Karminsky OD, Itach SB, Pietrokovski S, and Don J. (2010). Meig1 deficiency causes a severe defect in mouse spermatogenesis. Dev Biol 338, 158–167. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, Cai H, Borsche M, Klein C, and Youle RJ (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, Karayel O, Tonelli F, Martinez TN, Lorentzen E, Pfeffer SR, Alessi DR, and Mann M. (2017). Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6, e31012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson SEM, Aumann TD, Taylor JM, Riseley JR, Li R, Mann JR, Tomas D, and Lockhart PJ (2018). Generation and characterisation of a parkin-Pacrg knockout mouse line and a Pacrg knockout mouse line. Sci Rep 8, 7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Fisher RL, Bowser SS, Pentecost BT, and Sui H. (2019). Three-dimensional architecture of epithelial primary cilia. Proceedings of the National Academy of Sciences of the United States of America 116, 9370–9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JM, Song YJ, Huang Y, Farrer MJ, Delatycki MB, Halliday GM, and Lockhart PJ (2007). Parkin Co-Regulated Gene (PACRG) is regulated by the ubiquitin-proteasomal system and is present in the pathological features of Parkinsonian diseases. Neurobiology of disease 27, 238–247. [DOI] [PubMed] [Google Scholar]
- Trempe JF, and Fon EA (2013). Structure and Function of Parkin, PINK1, and DJ-1, the Three Musketeers of Neuroprotection. Front Neurol 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- West AB, Lockhart PJ, O’Farell C, and Farrer MJ (2003). Identification of a novel gene linked to parkin via a bi-directional promoter. Journal of molecular biology 326, 11–19. [DOI] [PubMed] [Google Scholar]
- Wilson GR, Tan JT, Brody KM, Taylor JM, Delatycki MB, and Lockhart PJ (2009). Expression and localization of the Parkin co-regulated gene in mouse CNS suggests a role in ependymal cilia function. Neuroscience letters 460, 97–101. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wang L, Wu N, Brock A, Spraggon G, and Schultz PG (2004). The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol 22, 1297–1301. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, et al. (2009a). Genomewide association study of leprosy. N Engl J Med 361, 2609–2618. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Tang W, Haig-Ladewig L, Sapiro R, Wei Z, Patel AM, Bennett J, Gerton GL, Moss SB, Radice GL, and Strauss JF 3rd (2006). Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod 74, 751–759. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shen X, Gude DR, Wilkinson BM, Justice MJ, Flickinger CJ, Herr JC, Eddy EM, and Strauss JF 3rd (2009b). MEIG1 is essential for spermiogenesis in mice. Proceedings of the National Academy of Sciences of the United States of America 106, 17055–17060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and maps for the three structures presented have been deposited in the Protein Databank https://www.rcsb.org: PDB 6NEP, 6NDU, and 6UCC.