Abstract
KRAS mutations are among the most common drivers of human carcinogenesis, and are associated with poor prognosis and an aggressive disease course. With the advent of KRASG12C inhibitors, the RAS protein is now targetable, with such inhibitors showing marked clinical responses across multiple tumor types. However, these responses are short-lived due to the development of resistance. Preclinical studies now suggest MAPK reactivation, stimulation of CDK4/6-dependent cell cycle transition, and immune defects as possible mechanisms of resistance. Devising strategies to overcome such resistance mechanisms, which are a barrier to long-term clinical response, remain an active area of research.
Introduction
The KRAS (Kirsten rat sarcoma) gene is the prototypical oncogene, and is among the most frequently mutated in human cancer. Indeed, three of the five leading causes of cancer deaths in the United States, non-small cell lung cancer (NSCLC), colorectal cancer (CRC), and pancreatic cancer (PDAC), are also among the most frequently associated with KRAS mutations, at rates of approximately 30%, 42%, and 80%, respectively (1–3). Additionally, KRAS mutations, when present, are associated with poorer prognosis from these cancers than non-KRAS oncogenic drivers (4–6).
Nearly all oncogenic mutations in KRAS are activating missense mutations that center on three codons: 12, 13, and 61. Of these, missense mutations in the glycine residue at codon 12 are far and away the most common (7). Interestingly, tissue of origin predicts the likely KRAS missense mutation, with KRASG12D representing approximately 25–40% of all KRAS mutations in CRC and PDAC, and, in contrast, KRASG12C representing approximately 40% of all KRAS mutations in lung adenocarcinomas (LUAC) (1,3,7).
As the primary intracellular secondary messenger of epidermal growth factor receptor (EGFR), KRAS constantly cycles between the inactive GDP-bound and the active GTP-bound states (7,8). When activated by a ligand-bound receptor tyrosine kinase (RTK) like EGFR, KRAS triggers multiple proliferative signaling cascades, including the mitogen-activated protein kinase/extracellular-regulated kinase (MAPK/ERK) and phosphoinositide-3-kinase (PI3K) pathways, to induce cell growth, division, and differentiation (9–11). In cases of oncogenic activating KRAS mutations, GTP hydrolysis is impaired and the KRAS protein is preferentially held in the active GTP-bound state (7,12), which, in turn, drives proliferative MAPK/PI3K signaling and ultimately carcinogenesis, as classically modeled by Vogelstein and Fearon (7,12). We note that although KRAS was historically considered to be a necessary stage in the adenoma-carcinoma sequence in CRC, this concept has been revolutionized with the identification of microsatellite instability (13) to now encompass tissue-, mutation-, and pathway-specific mechanisms of carcinogenesis.
Unfortunately, targeting mutated KRAS have been unsuccessful despite 40 years of sustained research and development, with generally limited response rates and short durations of response (14,15). Accordingly, KRAS had long been considered “undruggable” due to high affinity for GTP and lack of large binding pockets for allosteric inhibitors to occupy (14,15). However, the groundbreaking discovery by Shokat et al. of small molecules that covalently bind to the acquired cysteine residue within the switch II region in KRASG12C laid the first viable foundational steps to therapeutic KRAS blockade (16). Although these molecules selectively target KRASG12C alone, their potential impact is significant across many common cancer types, including in approximately 12% of all LUAC and 3% of all CRC. Furthermore, these agents provide an option to patients for whom there had been a lack of targeted treatments.
Recently published and ongoing early-phase clinical trials of the KRASG12C inhibitors sotorasib (AMG 510) and adagrasib (MRTX849) demonstrate clear clinical benefit, with tumor response rates approximating 30–40% with little toxicity (17,18). However, the duration of response for most patients is short, with the most recent data from the CodeBreaK100 trial showing median progression-free survival of only 6.3 months (17). Now, in this new era of targeting KRASG12C, the next research obstacle will be to understand and overcome mechanisms of resistance. Thus, we will review the current preclinical understanding of resistance to KRASG12C-targeted therapies, and present possible treatment approaches to combat such resistance.
Proof of therapeutic targeting of KRASG12C
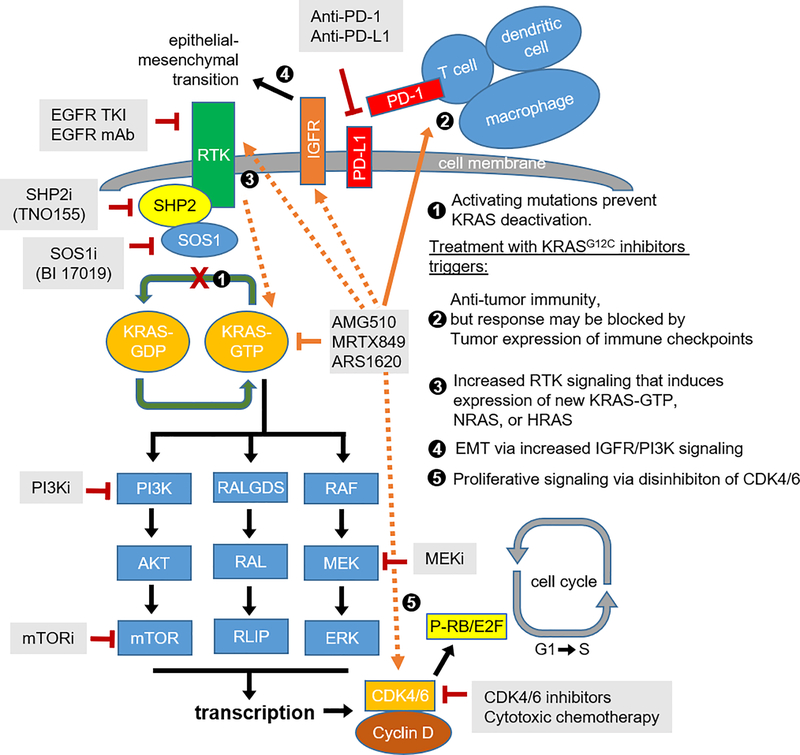
KRAS is a small, 21-kDa monometric guanosine 5’-triphosphatase. It consists of six beta strands and five alpha helices with a G-domain and a C-terminal membrane-targeting region (9,10). Wild-type KRAS constantly cycles between active GTP-bound and inactive GDP-bound states depending upon stimuli from upstream RTKs, most importantly EGFR. Upon activation, KRAS interacts with a complex set of downstream effectors in intricate and, in many cases, redundant pathways (7) (Figure 1). Notable interactions include those with proliferation-associated pathways such as RAF-MEK-ERK and PI3K-AKT-mTOR, which reinforce cyclin/CDK-dependent Rb phosphorylation and drive cellular differentiation, growth, and oppose apoptosis (7,19). The clinical relevance of KRAS signaling to human cancer is highlighted not only by frequent mutations in KRAS, but also by frequent (targetable) alterations in nearly every other downstream protein across multiple cancer types. Unfortunately, the redundancy in KRAS signaling and its centrality to cancer development foreshadows secondary resistance to KRAS blockade, as was seen with EGFR-mutated LUAC treated with osimertinib (20), and with BRAF-mutated melanoma and CRC treated with a combination of targeted BRAF inhibitors (21–23).
Figure 1.
KRAS signaling, mechanisms of resistance to KRASG12C drugs, and targeted therapies.
The oncogenicity of various KRAS mutations, including KRASG12C, arises from chronic KRAS activation due to reduced GTPase activity and prolonged residence in the GTP-bound active state. However, the specific targetability of KRASG12C relies upon the placement of the acquired cysteine within the P2 pocket of the switch II region. The resulting protein conformation with this specific missense mutation is accessible to small molecules that covalently bind the cysteine residue and hold KRASG12C in the inactive GDP-bound state, irreversibly switching off downstream signaling and inducing apoptotic cell death (16,24,25). Multiple small molecules have been developed against KRASG12C, including ARS-1620, sotorasib (AMG 510), and adagrasib (MRTX849). ARS-1620, the first KRASG12C inhibitor, has little clinical activity, but remains an important translational research tool to study mechanisms of resistance (8).
In contrast, sotorasib, developed by Amgen, and adagrasib, developed by Mirati, were the first and second KRASG12C inhibitors to reach the clinic, with recently completed or ongoing Phase I clinical trials (17,18,26). Both agents were demonstrated in vitro to covalently bind the acquired cysteine within the switch II region and inhibit downstream MAPK signaling, as evidenced by diminished phosphorylation of ERK (p-ERK), S6 (p-S6), and, in the case of sotorasib, MEK (p-MEK). Additionally, both drugs diminished the viability of KRASG12C human cancer cell lines, including and most critically, both lung and pancreatic cancer cell lines. Notably, a non-KRASG12C mutation was insensitive to treatment, as evidenced by lack of effect on p-ERK or on cell viability, highlighting the specificity of these inhibitors to KRASG12C. When tested in vivo in murine models, both agents inhibited downstream MAPK effectors and shrank tumors (18,26).
Finally, the recently completed Phase I CodeBreaK 100 trial investigated the initial safety and efficacy profile of sotorasib in patients with locally advanced or metastatic KRASG12C NSCLC (n = 59), CRC (n = 42), and other solid tumors. This trial, which enrolled patients who progressed after at least one line of systemic therapy but excluded those with active brain metastases, showed that sotorasib is well tolerated, with no dose-limiting toxicities or grade 4 therapy-related adverse events, and also efficacious, with overall response rate (ORR), disease control rate (DCR), and median progression free survival (mPFS) of 32.2%, 88.1%, and 6.3 months, respectively, in NSCLC; and 7.1%, 73.8%, and 4.0 months, respectively, in CRC (17). Remarkably, this result was achieved even in heavily pre-treated patients, with nearly all NSCLC patients having previously progressed on both platinum-doublet and anti-PD1/anti-PDL1 immunotherapy and CRC patients having previously failed at least two lines of systemic therapy. A Phase I/II clinical trial (NCT03785249) of adagrasib is ongoing; however, preliminary results presented at the EORTC-NCI-AACR Annual Symposium in 2020 similarly demonstrated little drug toxicity and ORR and DCR of 45% and 96%, respectively, in NSCLC; and 17% and 96%, respectively, in CRC.
Collectively, these data suggest that targeting KRASG12C is efficacious and well tolerated, and has prompted the development of multiple new KRASG12C agents, as summarized in Table 1. Nonetheless, these clinical responses, while significant and exciting relative to historical attempts at KRAS targeting, are highly variable between different tumor types, with markedly different overall response rate between NSCLC and CRC. Additionally, no patient in any study achieved complete response, and the observed clinical responses are not durable, lasting only 4–6 months for most patients. Thus, resistance to treatment is evident, and necessitates further investigation to guide future treatment approaches.
Table 1.
Activity of KRASG12C inhibitors in early-phase clinical trials.
| KRASG12C inhibitor | Sotorasib (AMG 510) | Adagrasib (MRTX849) | JNJ-74699157 (ARS-3248) | LY3499446 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sponsor | Amgen | Mirati Therapeutics | Janssen | Eli Lilly | ||||||||||
| Clinical Trial | Phase I/II (CodeBreaK 100) | Phase I/II (NCT03785249) *Last update: 2019 & 2020 AACR-NCI-EORTC Conference |
Phase I (NCT04006301) | Phase I/II (NCT04165031) | ||||||||||
| Patient population | KRASG12C-mutated advanced cancer | KRASG12C-mutated advanced cancer | KRASG12C-mutated advanced cancer | KRASG12C-mutated advanced cancer | ||||||||||
| Dose (mg) | All doses | 960 mg daily | All doses | 600 mg b.i.d | Terminated | Terminated | ||||||||
| NSCLC | CRC | AC | NSCLC | CRC | AC | NSCLC | CRC | AC | NSCLC | CRC | AC | NR | NR | |
| Study population (N) | 59 | 42 | 28 | 34 | 25 | 26 | 6 | 4 | 2 | 51 | 18 | 6 | NR | NR |
| ORR (%) | 32.2 | 7.1 | 14.3 | 35.3 | 12 | NR | 50 | 25 | 0 | 45 | 17 | 66 | NR | NR |
| DCR (%) | 88.1 | 73.8 | 75 | 91.2 | 80 | NR | 100 | 75 | 100 | 96 | 94 | 100 | NR | NR |
| mDOR (mo.) | 10.9 | 5.4 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| mPFS (mo.) | 6.3 | 4 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
Mechanisms of resistance to KRASG12C therapy and attempts at co-targeting
Bypass signaling resistance mechanisms
Pre-clinical studies have hinted at multiple possible mechanisms of resistance including innate, acquired, and adaptive tumor responses that diminish the therapeutic efficacy of KRASG12C inhibitors. One frequently identified mechanism is induction of bypass MAPK signaling to overcome KRAS blockade. Indeed, multiple studies have now demonstrated that KRASG12C inhibition can be overcome via feedback activation of either upstream or downstream mediators of the RTK-KRAS-MAPK cascade, as was observed with selective targeting of BRAF and EGFR (8,18,26,27).
Among the first suggestions of this bypass signaling was made by Hallin et al. in initial studies of adagrasib (MRTX849) (18), which demonstrated that cell line-derived mouse xenografts can be highly sensitive (i.e., PaCa-2, H1373), partially sensitive (i.e., H358, H2122), or refractory to KRASG12C inhibition. Gene set enrichment analysis on all tumor models regardless of response demonstrated that the most differentially expressed genes encompassed those mediating KRAS signaling, including MYC and mTOR, confirming that the drug specifically and efficiently inhibits KRASG12C and its downstream effectors. However, RNA sequencing of MAPK feedback pathways revealed that KRASG12C inhibition also elicits significant suppression of DUSP, SPRY, and PHLDA family genes, which are known negative regulators of MAPK signaling (28). Indeed, this finding was further corroborated by immunohistochemistry for p-ERK and p-S6, which significantly diminished (by >90%) in both highly sensitive (PaCa-2 and H1373) and partially sensitive (H358 and H2122) tumor models soon after exposure to adagrasib, only to subsequently recover in the latter, but not in the former, even after five days of continuous treatment. Together, these experiments demonstrate that ERK-dependent signaling is reactivated to bypass KRASG12C treatment. A survey of in vitro and in vivo models using a CRISPR/cas9 knockout screen with short guide RNAs targeting approximately 400 genes revealed that in the partially sensitive H2122 xenograft model, guide RNAs targeting SHP2 (a phosphatase that mediates signaling between activated RTK and KRAS), MYC, and mTOR pathway genes, all mediators of the RTK-KRAS-MAPK/PI3K cascade, were among the most depleted after two weeks of exposure to MRTX849, while guide RNAs targeting KEAP1, a tumor suppressor, were notably enriched.
Further clarification of resistance mechanism was provided by Xue et al. (8), who hypothesized that as novel KRASG12C inhibitors solely inhibit the inactive GDP-bound conformation of KRASG12C, only those cells with KRASG12C in the inactive conformation would be strongly inhibited in any population of cells with non-uniform rates of inactive to active KRASG12C cycling. As such, those cells with KRASG12C preferentially held in the active conformation would be insensitive to treatment and could mediate reactivation of MAPK signaling (8). In human lung adenocarcinoma cell lines previously shown to be either partially sensitive or refractory to adagrasib, namely H358, H2122, and SW1573, ARS-1620 was found to induce a quiescent (G0) state in most, but not all cells, as defined by abundant expression of p27 and as analyzed by single-cell RNA sequencing. However, those cells with low-level expression of p27 do not become quiescent and express active GTP-bound KRAS more abundantly, and are not eliminated by re-challenge with ARS-1620. Differential expression analysis and genome-wide knockout screening subsequently revealed two candidate genes that can mediate escape from KRASG12C inhibition: heparin-binding epidermal growth factor (HBEGF) and aurora kinase (AURKA). Specifically, HBEGF is downregulated soon after exposure to ARS-1620, but is then rapidly upregulated after 48 hours within a subpopulation of quiescent cells, suggesting a role in adaptive resistance. Corroborating this, small interfering RNAs knocking down HBEGF augment the anti-proliferative effect of ARS-1620. Conversely, stimulation with epidermal growth factor (EGF) induces KRAS activation in quiescent, ARS-1620-treated cells, strongly suggesting that EGFR signaling mediates adaptive resistance to KRASG12C drugs. On the other hand, AURKA accumulates in adapting cells as opposed to quiescent ARS-1620-treated cells, suggesting a relationship with overcoming quiescence. Alternatively, induction of AURKA in ARS-1620-treated H358 cells elicits accumulation of KRAS-GTP and p-ERK, and lowers the potency of ARS-1620 as assessed by cell viability.
In an elegant experiment, quiescent/p27-expressing H358 cells were then engineered to inducibly express siRNA-resistant KRASG12C. To mimic the initial quiescence phase following exposure to ARS-1620, cells were treated with a KRASG12C-targeted siRNA, but were then induced to express siRNA-resistant KRASG12C to mimic the adaptive phase. As seen with cells exposed to ARS-1620, these cells became initially quiescent following exposure to siRNA targeting KRASG12C, but a sub-population induced to express siRNA-resistant KRASG12C subsequently escaped this state. Accordingly, the adaptive response to KRASG12C inhibition was hypothesized to arise from newly synthesized KRASG12C that undergo immediate nucleotide change to an active GTP-bound conformation before being trapped by KRASG12C inhibitors, with EGF being the likely driver of new KRAS transcription and AURKA maintaining KRAS in the active GTP-bound conformation.
Moreover, Ryan et al. recently observed that reactivation of MAPK signaling after treatment with ARS-1620 coincided with increased expression of wild-type GTP-bound RAS (i.e. HRAS, NRAS) and phosphorylated-RTK (i.e. EGFR, FGFR, HER2, c-MET) in KRASG12C-driven lung, pancreatic, and colon cancer cells, suggesting secondary resistance via upregulated RTK signaling to wild type RAS isoforms (29). Notably, the RTK specifically activated by ARS-1620 was not the same across cell lines, which suggests that different RTKs may drive MAPK reactivation, and these differences may be histology- or even tumor-specific. Indeed, tissue of origin predicts responsiveness of KRASG12C inhibition, as seen in the divergent clinical responses to AMG 510 between LUAC and all other advanced cancer types, likely via differences in adaptive resistance. In fact, Amodio et al. demonstrated that although both NSCLC and CRC cells treated with AMG 510 exhibit equivalent reduction in cell viability, the latter show rapid upregulation of p-MEK and p-ERK, suggestive of early development of adaptive resistance (30). Further, CRC cells show increased basal phosphorylation (activation) of EGFR and respond to EGF stimulation by activating RAS-MAPK signaling even in the presence of an activating KRASG12C mutation, behaviors not seen in NSCLC cells. These findings strongly suggest that EGFR specifically mediates the adaptive resistance response in CRC, as previously observed in BRAF-mutant CRC (31), and could explain the poor ORR to single-agent KRASG12C inhibition.
Finally, Adachi et al. observed that among cell lines previously sensitive to AMG 510 (i.e. H358), induction of epithelial-to-mesenchymal transition (EMT), either by treatment with TGF-β or conditional expression of Twist or Snail, was associated with intrinsic and acquired resistance to KRASG12C inhibition (32). Resistance via EMT occurred in conjunction with increased PI3K/AKT signaling due to upregulated IGFR signaling, and led to increased MAPK signaling via FGFR.
Taken together, the data indicate that upstream RTK regulators (EGFR, HER2, FGFR, and SHP2), direct mediators of KRAS activation (AURKA), and/or effectors of MAPK and PI3K pathways (MYC and mTOR) may mediate escape from KRASG12C inhibition with escape mechanisms being notably tissue-specific. Fortunately, many of these resistance mediators can be targeted with therapeutic agents already on the market or in development, enabling rapid pre-clinical testing and now clinical translation. Nevertheless, rational, tissue-specific combination therapies are necessary to provide precise and effective disease control, in light of tissue-specific resistance mechanisms.
Among the most clinically studied upstream targets for combination therapy is EGFR. Currently, small-molecule inhibitors targeting mutationally activated EGFR are standard-of-care and have had long-standing clinical success against EGFR-mutant LUAC (33,34). Similarly, anti-EGFR monoclonal antibodies (e.g. cetuximab) (35) have been highly effective against CRC in combination with fluorouracil-based chemotherapy. Additionally, following the determination that EGFR mediates resistance to BRAF inhibition in BRAF-mutant CRC, cetuximab was found to synergize with BRAF inhibitors to prolong survival relative to standard therapy, and is now standard 2nd line therapy (31,36). Consequently, combining EGFR-targeted agents (i.e. gefitinib, afatinib) with both adagrasib and ARS-1620 was found to reduce downstream MAPK signaling and tumor volume in two mouse xenograft models of KRASG12C (8,18). Similarly, addition of cetuximab to AMG 510 blocked adaptive EGFR-driven reactivation of MAPK signaling specifically in CRC cell lines and mouse xenograft models, and led to sustained suppression of p-ERK and p-MEK, decreased cell viability, and near complete tumor regression.
On the other hand, there are indications that SHP2 inhibitors, which have very limited activity as single agents against KRAS-mutated cell lines, can restore the sensitivity of KRAS-mutant NSCLC to MEK inhibition and thereby inhibit tumor growth (37). Indeed, co-administration of SHP2 inhibitors with ARS-1620 was found to diminish adaptive reactivation of GTP-bound KRASG12C in mouse xenografts, an effect further augmented in a triplet combination of KRASG12C, EGFR, and SHP2 inhibitors (8). As SHP2 mediates signaling between RTKs and RAS, co-administration of SHP2 inhibitors with ARS-1620 was also found to decrease RTK-mediated MAPK reactivation independently of the RTK (29). Finally, SHP2 inhibitors were found to increase inactive GDP-bound KRAS and, in combination with ARS-1620, to induce suppression of p-ERK and increase T-cell infiltration, eliciting tumor regression in mouse models of PDAC and NSCLC mouse models (38). Finally, a combination of SHP2, PI3K, and KRASG12C inhibitors was found to suppress both p-AKT and p-ERK and induce durable tumor regression in EMT-induced mouse xenografts, which exhibit hallmarks of FGFR- and IGFR-induced MAPK and PI3K reactivation, respectively (32). Accordingly, early-phase clinical trials are ongoing to test KRASG12C inhibitors in combination with either EGFR inhibitors, EGFR monoclonal antibodies, or SHP2 inhibitors (Figure 1; Table 2). Finally, BI1701963, developed by Boehringer Ingelheim, is a distinct therapeutic class that acts as a pan-KRAS inhibitor by preventing SOS1 from binding inactive GDP-bound KRAS, thus inhibiting exchange of GDP to GTP (39,40) and indirectly inactivating all forms of KRAS. Early-phase clinical trials of this drug are ongoing, alone or in combination with trametinib, in patients with any KRAS mutation (Figure 1; Table 2).
Table 2.
Ongoing clinical trials targeting resistance to KRASG12C inhibitors.
| KRASG12C Inhibitor | Combination Options | Phase | Study | NCT Identifier |
|---|---|---|---|---|
| AMG 510 | AMG 510 + PD-1 inhibitor | Ib | AMG 510 (pINN) Sotorasib Activity in Subjects With Advanced Solid Tumors With KRAS p.G12C Mutation (CodeBreak 101) | NCT04185883 |
| AMG 510 + MEK inhibitor | ||||
| AMG 510 + SHP2 allosteric inhibitor | ||||
| AMG 510 + pan-ErbB TKI (NSCLC only) | ||||
| AMG 510 + PD-L1 inhibitor (NSCLC only) | ||||
| AMG 510 + chemotherapy (NSCLC only) | ||||
| AMG 510 + EGFR inhibitor +/- chemotherapy or MEK inhibitor (CRC only) AMG 510 + CDK inhibitor AMG 510 + mTOR inhibitor | ||||
| MRTX849 | MRTX849 + cetuximab (CRC only) | Ib | Phase 1/2 Study of MRTX849 in Patients With Cancer Having a KRAS G12C Mutation (KRYSTAL-1) | NCT03785249 |
| MRTX849 + afatinib (NSCLC only) | ||||
| MRTX849 + pembrolizumab (NSCLC only) | ||||
| MRTX849 + TNO155 (SHP2 inhibitor) | I/II | Phase 1/2 Study in Patients With Cancer Having a KRAS G12C Mutation (KRYSTAL-2) | NCT04330664 | |
| BI 1701963 | BI 1701963 + trametinib | I | A Study to Test Different Doses of BI 1701963 Alone and Combined With Trametinib in Patients With Different Types of Advanced Cancer (Solid Tumours With KRAS Mutation) | NCT04111458 |
Additionally, combining KRASG12C inhibitors with inhibitors of multiple downstream mediators has been tested pre-clinically, considering that downstream effectors of both the MAPK (RAF-MEK-ERK) and PI3K-AKT-mTOR pathways are clearly reactivated following exposure to KRASG12C drugs. For example, MEK inhibitors, although of limited utility alone, have been combined with BRAF inhibitors with great success against melanoma, and are now standard-of-care for these tumors (41). Additionally, MEK inhibitors notably enhanced the potency of chemotherapy in mouse models of lung cancer, particularly of tumors with KRASG12C (42). Finally, Canon et al. noted synergy between sotorasib and MEK inhibitors to reduce tumor volume in H358 mouse xenografts. Based on these findings, a clinical trial of sotorasib in combination with a MEK inhibitor is ongoing (26). Similarly, combining KRASG12C inhibitors with either PI3K (27,32) or mTOR (18) inhibitors overcame the adaptive increase in PI3K signaling, increased inhibition of MAPK/PI3K signaling, and reduced tumor volume in mouse xenografts. Early-phase clinical trials of KRASG12C inhibitors and mTOR inhibitors are ongoing (Figure 1; Table 2). On the other hand, there are no clinical trials combining KRASG12C inhibitors with AURKA inhibitors at this time, although AURKA has been shown to mediate resistance to KRASG12C inhibitors and remains a valid target for future combination therapy.
Proliferative signaling as resistance mechanism
In addition to adaptive reactivation of MAPK signaling, increased proliferative signaling via disinhibition of the cell cycle transition is another source of KRASG12C therapy resistance, particularly in NSCLC. Indeed, up to 20% of KRAS-mutant NSCLC have concurrent loss-of-function mutations in CDKN2A, a cell-cycle regulator and tumor suppressor, which, in turn, leads to constitutive CDK4/6-associated RB phosphorylation and cell proliferation (43–45). Further, previous reports by Puyol et al. suggest that interphase CDKs, particularly CDK4, are necessary for lung tumor development in conditional KRASG12V mouse models, such that CDK4 inactivation, either by conditional knockout or a null allele, led to reduced tumor development and induction of senescence, as defined by expression of β-galactosidase (19). Notably, CDK4 inactivation induced senescence only in the lung, as β-galactosidase was not detected in other tissues, including colon, pancreas, and stomach, suggesting a tissue-specific role. Finally, treatment with a CDK4 inhibitor led to decreased p-RB and tumor volume.
In the context of KRASG12C, the combination of adagrasib and palbociclib, a CDK4/6 inhibitor, showed significant synergy as evidenced by p27 accumulation, decreased p-RB, and marked decrease in tumor volume in CDKN2A-deficient xenograft models (18). Similarly, combining sotorasib with carboplatin, a commonly used frontline agent in NSCLC, shrank tumors in a mouse xenograft model (26). Accordingly, there exists significant translational potential for combining KRASG12C inhibitors with either cytotoxic chemotherapy (particularly in NSCLC) or with inhibitors of interphase CDKs, as in ongoing clinical trials (Figure 1; Table 2).
Immune mechanisms of resistance
Finally, a third mechanism of KRASG12C therapy resistance is impaired antitumor immunity. In light of the growing use of immune checkpoint therapy across the cancer landscape, Canon et al. explored the impact of KRASG12C inhibitor therapy on antitumor immunity (26). Interestingly, sotorasib was able to induce durable cures against CT26 KRASG12C cells injected into immunocompetent mice. In sharp contrast, sotorasib induced only short-lived tumor regression followed by recurrence in nearly all immunodeficient Balb/c mice xenografted with the same cells, suggesting that an impaired host immune system may confer resistance independent of MAPK reactivation or proliferative signaling. Further, treatment with KRASG12C inhibitors appeared to induce immune response to tumorigenic tissue, with sotorasib inducing marked infiltration of CD8 T cells, macrophages, and dendritic cells into CT26 KRASG12C tumors after five days of treatment. Gene expression analysis also revealed increased expression of genetic signatures of interferon signaling, chemokine production, and antigen processing, suggesting that KRASG12C inhibition boosts T cell priming. Moreover, combining sotorasib with anti-PD-1 therapy augmented T cell infiltration and led to complete and durable remissions. Finally, mice treated with this combination and cured of xenografted CT26 KRASG12C rejected a subsequent re-challenge with CT26 KRASG12C and parental CT26, but not 4T1 mouse breast cancer cells. Analysis of splenocytes from re-challenged mice demonstrated marked increase in IFN-y, a marker of T cell priming, in the presence of CT26 tumor cells but not in the presence of 4T1 tumor cells (26).
Thus, KRASG12C inhibition appears to induce a pro-inflammatory transcriptional signature that primes antigen-presenting cells and cytotoxic T cells, which, in turn, have anti-tumor activity. This process can be prolonged and durable when PD-1 checkpoint is inhibited. However, co-occurring genetic alterations may modulate the immune response to tumors. For example, mutations of KEAP1 and STK11 appear to induce a colder immune microenvironment with decreased T-cell infiltration, and are associated with poor clinical outcome in NSCLC treated with frontline chemo-immunotherapy (46). In contrast, co-occurring mutations in TP53 are associated with increased intra-tumoral T-cell infiltration, PD-1 expression, and prolonged clinical benefit from anti-PD-1 immunotherapy in NSCLC (47). Hence, ongoing clinical trials are pursuing combinations of KRASG12C inhibitors and immunotherapy, particularly for advanced NSCLC where immunotherapy has already shown significant efficacy (Figure 1; Table 2).
Conclusion
Innate and acquired resistance to KRASG12C inhibitors has impeded their development and remains an obstacle to their long-term success, as seen with other targeted therapies. For example, frontline osimertinib elicits nearly 80% ORR against EGFR-mutant metastatic LUAC, but patients invariably experience eventual recurrence (34). The mechanisms of recurrence are multifold and heterogeneous, encompassing both EGFR-dependent and EGFR-independent mechanisms. These mechanisms include compensatory MET amplification, activation of MAPK signaling, and even transformation to small cell or squamous cell histology (20).
Similarly, possible resistance mechanisms to singular inhibition of KRASG12C appear to be diverse, as investigated preclinically, with primary drivers consisting of reactivation of multiple MAPK effectors both upstream and downstream of RAS, disinhibition of cell-cycle transition, and defects in immunity. Importantly, these resistance mechanisms appear to be tissue-specific, with CRC developing resistance primarily via activation of upstream EGFR and NSCLC deploying all three mechanisms, depending upon the presence of co-occurring alterations in CDKN2A, STK11, and TP53. Thus, clinical trial design warrants an understanding of these tissue-specific differences in escape from KRASG12C blockade. Multiple mechanism-driven clinical trials combining KRASG12C inhibitors with a wide array of resistance mediators are now active and recruiting. It remains to be seen how well these combination therapies will be clinically tolerated. However, these collectively represent a leap forward to combat therapy resistance.
With the discovery of the switch II region in KRASG12C, targeted agents are finally being translated to the clinic, to the benefit of many patients with KRAS mutations. Unfortunately, durable response to these novel agents is yet to be achieved due to complex and diverse mechanisms of adaptive resistance. However, there is now great promise from combination therapies, which are based on an improved understanding of resistance mediators, to elicit long-term disease control or remission.
Statement of significance.
Although KRAS-targeted cancer therapy is revolutionary, tumors rapidly develop resistance. Understanding the mechanisms driving this resistance and designing combination strategies to overcome it is integral to achieve long-term disease control.
Acknowledgments
Conflict of interest: DSH receives research funding from Amgen and Mirati Therapeutics, which are developing KRASG12C inhibitors. DSH is also a consultant/advisor for Amgen. In addition, DSH receives research funding from AbbVie, Adaptimmune, Adlai Nortye, Amgen, Astra-Zeneca, Bayer, Bristol Myers Squibb, Daiichi-Sankyo, Eisai, Eli Lilly, EMD Serono, Erasca, Fate Therapeutics, Genentech, Genmab, GlaxoSmithKline, Ignyta, Infinity, Kite, Kyowa, LOXO, Merck, MedImmune, Millenium, Mirati, miRNA, Molecular Templates, Mologen, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Seattle Genetics, Takeda, Turning Point Therapeutics, Verstatem, VM Oncology; serves as consultant or advisor for Alpha Insights, Acuta, Amgen, Axiom, Adaptimmune, Baxter, Bayer, Boxer Capital, COG, Ecor1, Genentech, GLG, Group H, Guidepoint, HCW Precision, Infinity, Janssen, Merrimack, Medscape, Numab, Pfizer, Prime Oncology, Seattle Genetics, ST Cube, Takeda, Tavistock, Trieza Therapeutics, WebMD; and has ownership interests in Molecular Match, OncoResponse, and Presagia Inc. We acknowledge support from a National Institutes of Health Cancer Center Support Grant (CA016672) awarded to Dr. Peter Pisters.
References:
- 1.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531(7592):47–52 doi 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 3.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48(6):607–16 doi 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiesweg M, Kasper S, Worm K, Herold T, Reis H, Sara L, et al. Impact of RAS mutation subtype on clinical outcome-a cross-entity comparison of patients with advanced non-small cell lung cancer and colorectal cancer. Oncogene 2019;38(16):2953–66 doi 10.1038/s41388-018-0634-0. [DOI] [PubMed] [Google Scholar]
- 5.Hayama T, Hashiguchi Y, Okamoto K, Okada Y, Ono K, Shimada R, et al. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int J Colorectal Dis 2019;34(8):1491–6 doi 10.1007/s00384-019-03344-9. [DOI] [PubMed] [Google Scholar]
- 6.Nadal E, Chen G, Prensner JR, Shiratsuchi H, Sam C, Zhao L, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol 2014;9(10):1513–22 doi 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 7.Vasan N, Boyer JL, Herbst RS. A RAS renaissance: emerging targeted therapies for KRAS-mutated non-small cell lung cancer. Clin Cancer Res 2014;20(15):3921–30 doi 10.1158/1078-0432.CCR-13-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577(7790):421–5 doi 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 1991;349(6305):117–27 doi 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 10.Santos E, Nebreda AR. Structural and functional properties of ras proteins. FASEB J 1989;3(10):2151–63 doi 10.1096/fasebj.3.10.2666231. [DOI] [PubMed] [Google Scholar]
- 11.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009;6(2):201–5 doi 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 12.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61(5):759–67 doi 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020;158(2):291–302 doi 10.1053/j.gastro.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13(11):828–51 doi 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol 2018;15(11):709–20 doi 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 16.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov 2016;15(11):771–85 doi 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 17.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383(13):1207–17 doi 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10(1):54–71 doi 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010;18(1):63–73 doi 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 20.Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121(9):725–37 doi 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucheit AD, Davies MA. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochem Pharmacol 2014;87(3):381–9 doi 10.1016/j.bcp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 2015;5(4):358–67 doi 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaeger R, Yao Z, Hyman DM, Hechtman JF, Vakiani E, Zhao H, et al. Mechanisms of Acquired Resistance to BRAF V600E Inhibition in Colon Cancers Converge on RAF Dimerization and Are Sensitive to Its Inhibition. Cancer Res 2017;77(23):6513–23 doi 10.1158/0008-5472.CAN-17-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov 2016;6(3):316–29 doi 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 25.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172(3):578–89 e17 doi 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575(7781):217–23 doi 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 27.Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25(2):796–807 doi 10.1158/1078-0432.CCR-18-0368. [DOI] [PubMed] [Google Scholar]
- 28.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 2012;22(5):668–82 doi 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res 2020;26(7):1633–43 doi 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discov 2020;10(8):1129–39 doi 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483(7387):100–3 doi 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 32.Adachi Y, Ito K, Hayashi Y, Kimura R, Tan TZ, Yamaguchi R, et al. Epithelial-to-Mesenchymal Transition is a Cause of Both Intrinsic and Acquired Resistance to KRAS G12C Inhibitor in KRAS G12C-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2020. doi 10.1158/1078-0432.CCR-20-2077. [DOI] [PubMed] [Google Scholar]
- 33.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13(3):239–46 doi 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 34.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378(2):113–25 doi 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 35.Qin S, Li J, Wang L, Xu J, Cheng Y, Bai Y, et al. Efficacy and Tolerability of First-Line Cetuximab Plus Leucovorin, Fluorouracil, and Oxaliplatin (FOLFOX-4) Versus FOLFOX-4 in Patients With RAS Wild-Type Metastatic Colorectal Cancer: The Open-Label, Randomized, Phase III TAILOR Trial. J Clin Oncol 2018;36(30):3031–9 doi 10.1200/JCO.2018.78.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med 2019;381(17):1632–43 doi 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 37.Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med 2018;24(7):961–7 doi 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 38.Fedele C, Li S, Teng KW, Foster CJR, Peng D, Ran H, et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J Exp Med 2021;218(1) doi 10.1084/jem.20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci U S A 2019;116(7):2551–60 doi 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gort E, Johnson ML, Hwang JJ, Pant S, Dünzinger U, Riemann K, et al. A phase I, open-label, dose-escalation trial of BI 1701963 as monotherapy and in combination with trametinib in patients with KRAS mutated advanced or metastatic solid tumors. Journal of Clinical Oncology 2020;38(15_suppl):TPS3651–TPS doi 10.1200/JCO.2020.38.15_suppl.TPS3651. [DOI] [Google Scholar]
- 41.Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N Engl J Med 2019;381(7):626–36 doi 10.1056/NEJMoa1904059. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Liu S, Deng J, Akbay EA, Hai J, Ambrogio C, et al. Assessing Therapeutic Efficacy of MEK Inhibition in a KRAS(G12C)-Driven Mouse Model of Lung Cancer. Clin Cancer Res 2018;24(19):4854–64 doi 10.1158/1078-0432.CCR-17-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res 2005;576(1–2):22–38 doi 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 44.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 2016;8:30–9 doi 10.1016/j.ebiom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8(7):822–35 doi 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5(8):860–77 doi 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]