Abstract
The myeloproliferative neoplasms frequently progress to blast phase disease, an aggressive form of acute myeloid leukemia. To identify genes that suppress disease progression, we performed a focused CRISPR/Cas9 screen and discovered that depletion of LKB1/Stk11 led to enhanced in vitro self-renewal of murine MPN cells. Deletion of Stk11 in a mouse MPN model caused rapid lethality with enhanced fibrosis, osteosclerosis and an accumulation immature cells in the bone marrow, as well as enhanced engraftment of primary human MPN cells in vivo. LKB1 loss was associated with increased mitochondrial ROS and stabilization of HIF1a, and downregulation of LKB1 and increased levels of HIF1a were observed in human blast phase MPN specimens. Of note, we observed strong concordance of pathways that were enriched in murine MPN cells with LKB1 loss with those enriched in blast phase MPN patient specimens, supporting the conclusion that STK11 is a tumor suppressor in the MPNs.
Keywords: JAK2, HIF1a, acute myeloid leukemia, leukemia transformation
Introduction
Philadelphia-negative myeloproliferative neoplasms (MPNs) are a group of blood stem cell disorders affecting the myeloid lineage that include polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF). Each MPN phenotype is characterized by increased proliferation of differentiated myeloid lineages including red blood cells, megakaryocytes and granulocytes and is caused by driver mutations in JAK2, MPL and CALR that constitutively activate the JAK/STAT pathway. MPN patients transform to acute leukemia with rates of 10–20%, 2–5%, and 1–4% in PMF, PV and ET, respectively (1–4). Post-MPN AML, also known as blast phase MPN (MPN-BP), is an aggressive malignancy with a median survival of 3–5 months (5–7). Although several studies involving sequencing of MPN and MPN-BP samples identified mutations or variants in several genes related to myeloid malignancies such as ASXL1, EZH2, SRSF2, LNK, TET2, and TP53 (3,8) and recurring deletions of JARID2 (9), the mechanisms of transformation remain largely unclear.
MPN-BP is characterized by the presence of 20% or more peripheral blood or bone marrow blasts suggesting the emergence of a malignant myeloid clone with impaired differentiation and increased self-renewing capacity consistent with the transformation to AML. To better understand the genetic alterations that cause a switch from the chronic phase to MPN-BP, we performed a focused CRISPR/Cas9 screen to identify tumor suppressor genes whose deletion enhanced self-renewal of hematopoietic stem and progenitor cells (HSPCs) harboring MPN driver mutations in a serial replating assay. We identified Liver Inducible Kinase (LKB1)/Serine threonine kinase 11 (STK11) as a driver of leukemic transformation in MPN and reveal that its downregulation is accompanied by an increase in mitochondrial reactive oxygen species (ROS) and stabilization of hypoxia inducible factor 1 alpha (HIF-1a). These results are surprising because, although LKB1 is widely known as a tumor suppressor in solid cancer, its loss leads to depletion of hematopoietic stem and progenitor cells and bone marrow failure rather than expansion (10–12). Our results suggest that enhanced JAK/STAT signaling is required for survival and subsequent transformation of hematopoietic cells lacking LKB1.
Results
A focused CRISPR/Cas9 screen reveals Stk11 loss as a driver of HSPC self-renewal
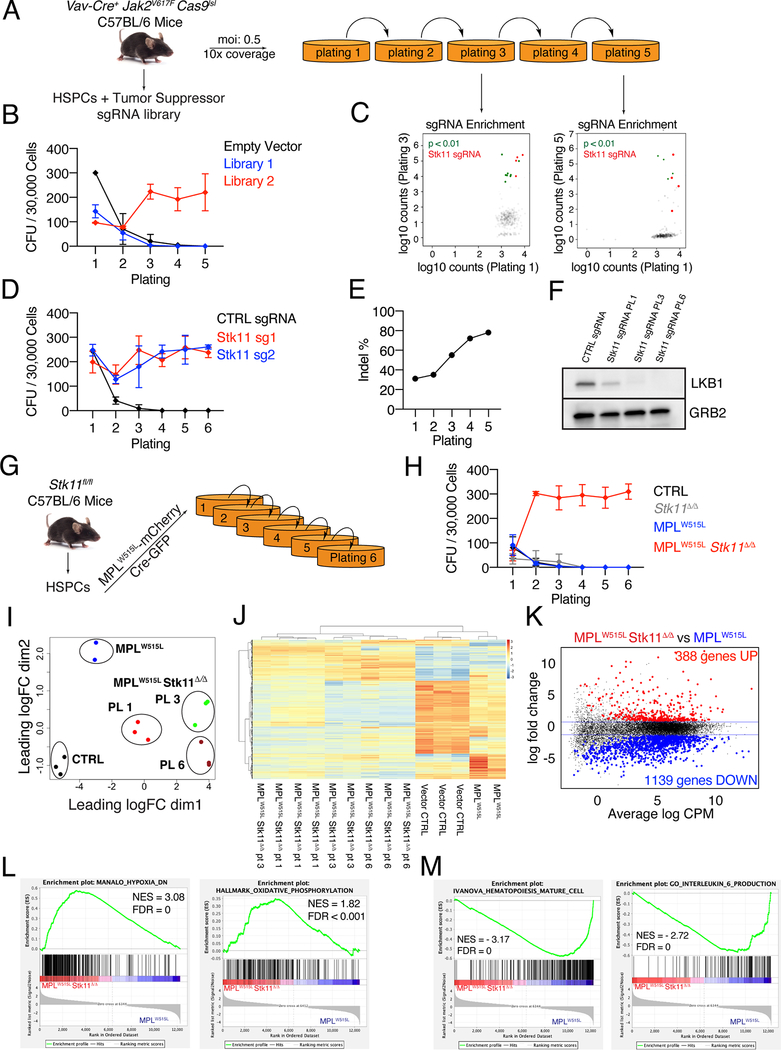
To investigate tumor suppressor genes that drive leukemic transformation in cells with activated JAK/STAT signaling, we performed a positive CRISPR/Cas9 screen by transducing two curated lentiviral single-guide RNA (sgRNA) libraries, each containing four sgRNAs targeting ∼100 unique annotated tumor suppressors and several MPN associated genes as well as 50 non-targeting control sgRNAs (Table S1), in Jak2V617F/Vav-Cre c-kit+ HSPCs with a Lox-STOP-Lox allele of Cas9 (Cas9lsl) and assessed their serial replating capacity in methylcellulose media (Fig 1A). Trp53 was intentionally omitted from both libraries due to previous reports already identifying it as a transformation factor (8) for JAK2 mutant MPNs. Transduction of either library 1 or 2 in Vav-Cre/Cas9lsl lin- HSPCs did not result in enhanced replating (Fig S1A), while CRISPR editing of Trp53 in Jak2V617F/Vav-Cre/Cas9lsl c-kit+ HSPCs as a positive control resulted in enhanced serial replating as expected (Fig S1B, C). By contrast, transduction of library 2 led to robust serial replating of JAK2V617F cells over 5 generations while transduction of library 1 or a control empty vector failed to induce this phenotype (Fig 1B). Next generation sequencing (NGS) of DNA obtained from cells at plating 1 as a baseline versus plating 3 showed enrichment of all four guides present in the library that target Serine/Threonine Kinase 11 (Stk11), which encodes LKB1 (Fig 1C), while two sgRNAs targeting Stk11 were significantly enriched at plating 5 (Table S2). Although there were 9 other genes that had significant guide enrichment in platings 3 and/or 5 none of these genes showed enrichment of more than one guide (Table S2). Of note, out of 50 non-targeting sgRNAs in library 2, only one was significantly enriched at plating 3, but was not enriched at plating 5. To confirm the Stk11 result of the screen, we electroporated ribonucleotide complexes (RNPs) containing Cas9 and two independent sgRNAs targeting Stk11 in Jak2V617F/Vav-Cre c-kit+ HSPCs and performed a serial replating assay. Cells electroporated with either Stk11 sgRNA induced a strong serial replating over 6 generations while cells electroporated with the control sgRNA failed to replate (Fig 1D). To evaluate the efficiency of sgRNA targeting, we sequenced a region of Stk11 flanking the Cas9 cut site and performed Inference of CRISPR Edits (ICE) analysis (13) on the resulting chromatograms from Sanger sequencing. The percentage of productive indels increased from plating 1 to plating 5 indicating progressive enrichment of Stk11 knockout cells during replating (Fig 1E). Finally, we performed western blot analysis for LKB1 levels in cells at different stages and observed strong down-regulation of LKB1 expression (Fig 1F). Together, these results show that loss of the tumor suppressor Stk11 is a driver of in-vitro self-renewal in Jak2V617F MPN cells.
Figure 1: CRISPR/Cas9 screening reveals loss of LKB1 is a driver of serial re-plating of Jak2V617F hematopoietic cells.
A) Schematic of the experimental workflow. B) Number of hematopoietic colonies formed at each generation of plating for Jak2V617/Cas9/Vav-Cre cells infected with library 1 or 2. The average of 2 biological replicates plus/minus SD are shown. C) sgRNA enrichment of plating 1 versus 3 or 5 as determined by DNA sequencing. sgRNAs targeting Stk11 are highlighted in red. The P value represents significance by Fisher’s exact test. D) Number of hematopoietic colonies formed over 6 generations for Jak2V617F/Vav-Cre cells targeted with two independent sgRNA targeting Stk11. The average plus/minus SEM are shown, n=3. E) ICE analysis of the extent of Stk11 targeting in cells from (B). F) Western blot for LKB1 levels at the various stages of re-plating of Jak2V617F/Vav-Cre cells electroporated with RNPs against Stk11. GRB2 is displayed as the loading control. G) Schematic of the experimental workflow for the MPLW515L studies. H) Number of hematopoietic colonies formed for up to six generations for murine Stk11-floxed HSPCs expressing various combinations of MPLW515L and Cre. The average plus/minus SD are shown, n=6 I) MDS plot of RNA-seq data comparing MPLW515L/Stk11-null to MPLW515L/Stk11+/+ cells after one, three and six rounds of plating. J) Unsupervised clustering analysis comparing RNA-seq data from cells collected after the first round of plating for control and MPLW515L/Stk11+/+ and at first, third and sixth plating for MPLW515L/Stk11-null cells. K) MA plot of the comparison MPLW515L/Stk11-null versus MPLW515L/Stk11+/+ at plating one showing differentially expressed genes. L,M) Pathways that are correlated with the MPLW515L/Stk11-null phenotype (L) or correlated with the MPLW515L phenotype (M) by GSEA analysis.
LKB1 loss in MPN cells induces transcriptional changes related to hypoxia, oxidative phosphorylation and the stem cell program
Hematopoietic specific depletion of Stk11 in mice leads to bone marrow failure due to an initial burst of proliferation of Stk11-deficient HSCs followed by their depletion (10–12). To confirm the phenotype induced by Stk11 loss with a different MPN driver mutation, we obtained Stk11fl/fl mice (12). We co-transduced Stk11fl/fl c-kit+ HSPCs with a retroviral vector encoding the MPN driver mutation MPLW515L and an mCherry reporter (Migr1-MPLW515L-mcherry) along with a retroviral vector encoding CRE recombinase and a GFP reporter (Migr1-CRE-GFP) to induce Stk11 deletion, sorted mCherry-GFP double positive cells and cultured them in methylcellulose media (Fig. 1G). As with the Jak2V617F genotype, hematopoietic progenitor cells expressing MPLW515L alone failed to serially replate (Fig 1H). By contrast, homozygous deletion of Stk11 in combination with MPLW515L (MPLW515L/Stk11Δ/Δ) led to a robust replating phenotype similar to the Jak2V617F /Stk11 targeted cells while those with empty vector control (CTRL), Stk11 deletion alone, or MPLW515L overexpression alone failed to replate (Fig. 1H). Of note, MPLW515L Stk11Δ/Δ cells also grew in cytokine free methylcellulose media and in liquid culture in a cytokine independent fashion with an immature morphology when transferred from cytokine free methylcellulose to liquid culture (Figure S2A-C).
To investigate the transcriptional changes that characterize the replating phenotype, we performed RNA-seq on CTRL and MPLW515L cells at plating 1 along with MPLW515L/Stk11Δ/Δ cells at platings 1, 3 and 6. Multidimensional Scaling (MDS) plot showed a distinct clustering of MPLW515L/Stk11Δ/Δ from CTRL and MPLW515L cells (Fig. 1I). Differential expression analysis across all the conditions resulted in 5463 genes differentially expressed genes, and unsupervised hierarchical clustering of these genes highlighted a transcriptional signature separated from CTRL and MPLW515L cells (Fig 1J). To identify genes specifically associated with deletion of Stk11 in the context of JAK/STAT activation, we performed differential expression analysis on MPLW515L Stk11Δ/Δ cells versus MPLW515L cells at plating 1. Stk11 deletion had a profound effect on the transcriptome of MPLW515L cells and resulted in 1527 genes being differentially expressed (Fig. 1K, Table S3). Gene Set Enrichment Analysis (GSEA) revealed enrichment of pathways related to hypoxia, embryonic stem cells, and regulation of oxidative phosphorylation (OXPHOS) in the MPLW515L/Stk11Δ/Δ phenotype, while pathways related to myeloid differentiation, maturation of hematopoietic cells and interleukin-6 production were enriched in the MPLW515L MPN cells (Fig 1L,M, Fig S3A,B). Full GSEA pathway analysis is available in Table S4. Together, these data demonstrate that loss of Stk11 results in the same self-renewal phenotype in both JAK2 and MPL mutant settings and induces profound transcriptional changes that separate immature MPLW515L Stk11Δ/Δ cells from the differentiated state of MPLW515L MPN cells.
Enhanced self-renewal of Stk11-deficient cells with activated JAK/STAT signaling is associated with HIF1a stabilization
We then investigated the mechanism by which loss of LKB1 contributes to enhanced self-renewal. First, we collected cells from the first-generation methylcellulose cultures of murine HSPCs of four different genotypes: wild-type (WT), MPLW515L, Stk11Δ/Δ, and MPLW515L/Stk11Δ/Δ. Whole cell lysates were then assessed for levels of LKB1 and its well-studied downstream substrate AMPK (Fig 2A). Previous studies have shown that loss of LKB1 did not induce HSC failure through AMPK (10–12), therefore we were not surprised to see that levels of total and p-AMPK at threonine 172 were not substantially depleted by loss of Stk11. Of note, overexpression of MPLW515L also did not result in changes in AMPK levels, nor did the combination of Stk11 loss and MPLW515L expression.
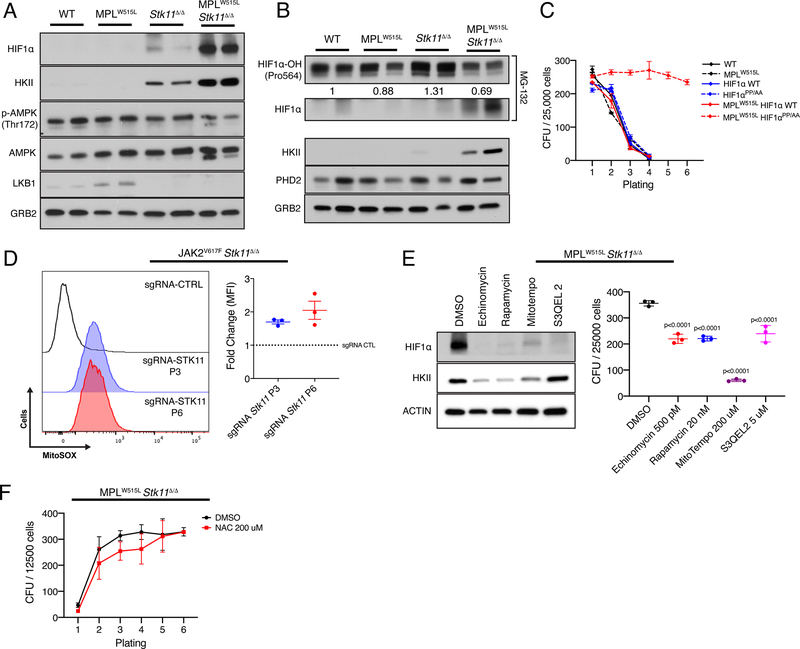
Figure 2: Stabilization of HIF1a promotes colony formation in cells with MPLW515L that are deficient for Stk11.
A) Western blot analysis of protein levels in hematopoietic cells after the first plating. GRB2 is shown as the loading control. n=2 B) Western blot analysis of HIF1a hydroxylation and PHD2 after the first plating. Densitometry is showed relative to WT. GRB2 is displayed as the loading control. n=2 C) Colony forming units over 6 generations in wild type cells transduced with MPLW515L, HIF1a WT, HIF1a PP/AA mutant and the various combinations. The average plus/minus SEM are shown, n=2 D) Comparison of mitochondrial ROS in cells at platings 1, 3 and 6. Representative flow and the average plus/minus SEM are shown. E) (Left) Western blot analysis of protein levels in MPLW515L/Stk11-null cells after treatment with various small molecules known to target HIF1a stabilization. Actin is shown as a loading control. (Right) Effect of small molecules on colony formation capacity of MPLW515L/Stk11-null cells. The average plus/minus SD are shown. P values by Dunnet’s multiple comparisons test against DMSO control. F) Effect of N-acetylcysteine (NAC) on the colony re-plating phenotype. No significant differences were observed. The average plus/minus SEM are shown, n=3
Given the gene expression data which revealed that there was enrichment of the hypoxia geneset by GSEA and substantial upregulation of hexokinase II (HKII), we assessed the expression of these factors in cells of the various genotypes. Consistent with the RNA-seq data, we saw that HKII was markedly upregulated by deletion of Stk11 and expression of MPLW515L (Fig 2A). In accordance with GSEA, we also observed that HIF1a, the key transcriptional regulator of HKII, was also substantially elevated in the LKB1 deficient cells that expressed MPLW515L. Moreover, stabilization of HIF1a persisted even in cells expanded in liquid culture (Fig S4). These results strongly suggest that activation of JAK/STAT signaling in combination with Stk11 deletion leads to stabilization of HIF1a under normoxic conditions and increases its downstream target gene expression. To investigate how HIF1a is stabilized in MPLW515L/Stk11Δ/Δ cells, we assayed for levels of prolyl hydroxylase 2 (PHD2) and hydroxylated HIF1a. In the mutant cells, hydroxylation of HIF1a was reduced compared to controls while PHD2 was largely unchanged, suggesting an impairment of PHD activity in these cells (Fig 2B).
Next, to determine whether stabilization of HIF1a is sufficient to promote enhanced self-renewal, we transduced wild-type murine bone marrow cells with combinations of MPLW515L, wild-type HIF1a and an allele of HIF1a (HIF1aPP/AA) that cannot be hydroxylated and targeted for VHL-mediated ubiquitination (14). Consistent with a key role for HIF1a in the replating phenotype of cells with activated JAK/STAT signaling, we found that HIF1a stabilization was sufficient to confer serial replating of MPLW515L cells with wild-type STK11 expression (Fig 2C). Together, these data confirm that HIF1a stabilization is sufficient for enhanced self-renewal of cells expressing MPLW515L. Prior studies have indicated that mitochondrial function through OXPHOS and production of mitochondrial reactive oxygen species (mitoROS) has been linked to the stabilization of HIF1a (15–17). Consistent with our prediction that HIF1a is stabilized through increased mitoROS, we also observed that the levels of mitoROS were elevated in Jak2V617F/Stk11Δ/Δ cells (Fig 2D).
Finally, we assayed whether the replating of MPLW515L/Stk11Δ/Δ cells was sensitive to drugs known to target HIF1a through various mechanisms: disruption of HIF1a DNA binding (echinomycin), inhibition of mTOR (rapamycin), and reduction of mitochondrial ROS (mitotempo and S3QEL 2). We observed that colony formation was inhibited by all four compounds, with the most significant decrease caused by mitotempo (Fig 2E). Furthermore, targeting mitochondrial superoxide with mitotempo resulted in both strong inhibition of colony formation and HIF1a destabilization while suppression of complex III-derived superoxide without alteration of OXPHOS with S3QEL-2 (18) was still sufficient to inhibit colony formation and HIF1a protein stabilization (Fig 2E). By contrast, sequestration of cellular ROS by N-acetylcysteine failed to impede replating (Fig 2F). Taken together our data are consistent with the model that loss of LKB1 leads to increased mitochondrial ROS signaling which increases stabilization of HIF1a and enhances self-renewal.
Loss of Stk11 transforms MPLW515L induced MPN to spent phase with immature blasts in vivo
Serial replating is often used as an in vitro surrogate for leukemic transformation, but there are examples which show that these are not always inexorably linked. To study whether the potent re-plating phenotype seen with deletion of Stk11 is associated with in vivo transformation, we modeled heterozygous and homozygous loss in animal models of MPNs. First, we bred Jak2V617F inducible mice with Vav-Cre and the Stk11-floxed strains. As expected, Jak2V617F/Vav-Cre animals developed an MPN characterized by polycythemia and succumbed to disease by 220 days, with 50% lethality seen at day 150 days (Fig S5A,B). Loss of Stk11 did not appreciably alter the peripheral blood counts, but significantly impaired survival, with all mice succumbing to disease by 120 days with 50% lethality seen at day 75. Of note, heterozygous loss of Stk11 had no phenotype, consistent with prior reports (10–12). Furthermore, activation of JAK/STAT signaling in vivo led to an increase in the Lin-Sca+c-Kit+ HSC fraction and a modest decrease in Lin-Sca+ progenitors, with no change in the proportion of myeloid progenitors, simultaneous deletion of one allele of Stk11 did not alter these phenotypes (Fig S5C-E).
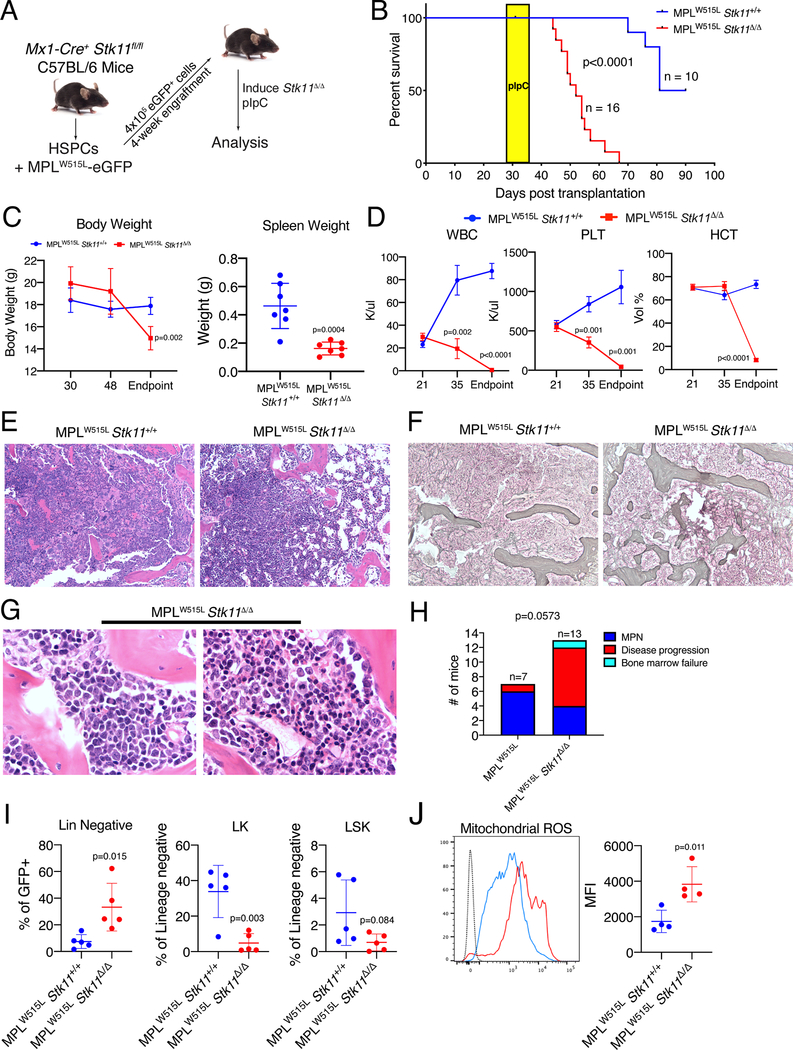
Next, to assess the effect of complete LKB1 loss after MPN development, we collected HSPCs from Mx1-Cre/Stk11fl/fl or Stk11fl/fl control animals without Cre, transduced with MPLW515L-eGFP and transplanted the cells to irradiated congenic animals (Fig 3A). After three weeks, when the animals displayed features of an MPN, we confirmed that the presence of two floxed Stk11 alleles did not alter the engraftment efficiency (Fig S6A). We then treated the mice with three doses of pIpC to induce Stk11 deletion and observed that loss of Stk11 led to rapid lethality, reduced body and spleen weights and pancytopenia (Fig 3B-D). Histological analysis of the MPLW515L/Stk11Δ/Δ mice at the time of sacrifice revealed that loss of Stk11 was associated with much more intense fibrosis and osteosclerosis of the bone marrow compared to those mice which retained Stk11 (Fig 3E,F). Furthermore, in contrast to the MPLW515L group, in which 6 of 7 mice developed MPN, the majority of the MPLW515L/Stk11 mice (8 of 13) developed a malignancy characterized by a variably hypoceullular bone marrow containing multiple foci of blasts comprising >20% of the mononuclear cells (Fig 3G,H). This disease was also accompanied by minimal to no residual normal bone marrow hematopoiesis and splenic infiltration (Fig S6B). By contrast to the heterozygous deletion, complete absence of Stk11, confirmed by Western blot (Fig S6C), altered the hematopoietic stem and progenitor cell populations, with a prominent decrease of LK and LSK compartments (Fig 3I). This change was accompanied by an overall decrease in GFP+ hematopoietic cells in the bone marrow of MPLW515L/Stk11Δ/Δ mice (Fig S6D). Finally, we saw increased mitochondrial ROS in the whole bone marrow of MPLW515L/Stk11Δ/Δ mice (Fig 3J). Although these data are somewhat reminiscent of the Stk11 knockout bone marrow aplasia phenotype, in the context of mutant JAK2 or MPL, loss of Stk11 recapitulates MPN disease progression in patients.
Figure 3: Deletion of Stk11 drives progression of MPN in vivo.
A) Schematic of experimental workflow. B) Survival curve of animals transplanted with Stk11fl/fl/MX1-Cre/MPLW515L or Stk11fl/fl/MPLW515L bone marrow cells. Yellow box indicates timing of pIpC treatment. P value (log-rank test) represents the difference in survival between the two groups of mice. C) Body and spleen weights of the two groups of mice. The average plus/minus SD is shown, n=7. D) White blood cell, platelet counts and hematocrit of mice in the two groups. P values were derived by Student’s t-test with Benjamini and Yekutieli correction for multiple hypothesis testing. The average plus/minus SEM is shown, n=5. E,F) Representative H&E and reticulin stained sections of the bone marrow recipient animals. Original magnification 100X (C,D). G) Higher power magnification of the bone marrow of two MPLW515L/Stk11Δ/ Δ mice highlighting the accumulation of immature blasts. Original magnification 500X. H) Comparison of the malignant phenotypes of MPLW515L versus MPLW515L/Stk11 Δ/ Δ mice. MPN, animals with a hypercellular bone marrow with mature cells; Disease progression, animals with >20% blasts in in an otherwise hypocellular bone marrow; Bone marrow failure, animals with a hypocellular marrow and no blasts. Differences in the proportions of the phenotypes in the two groups were evaluated by Fisher’s exact test. I) Flow cytometry data for hematopoietic progenitor cells. The average plus/minus SD is shown. P values by student’s t-test J) Measurement of mitochondrial ROS in whole bone marrow cells from the mice as determined by intracellular flow cytometry. Representative plot (left) and individual data (right) are shown. The average plus/minus SEM is shown. P value determined by Student’s t-test.
Deletion of STK11 enhances engraftment of human MPN primary cells
We next assayed the consequences of STK11 depletion of the growth of primary human MPN cells in vivo. We collected peripheral blood CD34+ from patients diagnosed with an MPN, introduced sgRNA targeting STK11 by nucleofection of RNP particles with Cas9 and then transplanted these cells to NSGS animals (Fig S7A). After 12 weeks, we analyzed engraftment and evaluated the degree of editing of the human cells. Remarkably, we observed a consistent increase in engraftment of cells that showed successful STK11 gene editing (Fig S7B,C). This was reflected by an increase of human cells in peripheral blood for 2 out of 3 patient samples and an increase of human CD45+ cells in the bone marrow and an increase of total bone marrow cells (Fig S7D). These data indicate that loss of STK11 increases engraftment of human MPN cells in a xenotransplantation model.
Loss of STK11 is associated with leukemia progression in human MPNs
We performed a number of studies to determine whether the murine phenotypes observed with Stk11 loss in cells with activated JAK/STAT signaling are relevant to patients. First, we assayed the mRNA expression level of STK11 in matched bone marrow specimens from patients at chronic and blast phase MPN. We observed downregulation of STK11 in all cases, with several patients showing more than a 50% decrease in expression at the blast phase (Fig S8). Second, to determine whether the level of LKB1 was altered with progression we performed immunohistochemistry on matched bone marrow samples form patients in chronic and blast phase. We observed a consistent and significant decrease in LKB1 staining in the bone marrow (Fig 4A). Together these results confirm that downregulation of LKB1 is a feature of MPN progression. We further stained the same chronic and blast phase sections for HIF1a and observed increased staining in the blast phase of the disease consistent with LKB1 loss leading to the stabilization of HIF1a (Fig 4B).
Figure 4: LKB1 loss is a feature of human MPN.
A) IHC for LKB1 in bone marrow sections from chronic and blast phase MPN. Left, representative IHC; right, Area of staining quantification for 5 paired samples. Original magnification, 400X. The average plus/minus SD are shown. *, p value <0.05 by Student’s t-test. B) IHC for HIF1a in bone marrow sections from chronic and blast phase MPN. Left, representative IHC; right, Area of staining quantification for 5 paired samples. Original magnification, 400X. The average plus/minus SD are shown. *, p value <0.05 by Student’s t-test. Images from A and B are from the same patient. C) Principal component analysis of RNA-sequencing data from 5 paired samples of chronic phase myelofibrosis (MF) and blast phase disease (AML). D) Unsupervised clustering of differentially expressed genes between chronic phase (MPN) and blast phase (AML). E) GSEA analysis of downregulated genes in mouse RNA-seq against human RNA-sequencing dataset. F) GSEA analysis revealing enrichment of a published hypoxia related geneset (19). G) Comparison of normalized enrichment scores (NES) of all pathways between the mouse and human RNA-seq datasets. Significance at FDR < 0.1. H) Number of hematopoietic colonies generated by CD34+ progenitors from healthy donors treated with echinomycin, mitotempo, PP242, PT2977 and ruxolitinib at various concentrations versus DMSO control. Dotted line represent mean of the DMSO control. **p=0.0044 for mitotempo at 500 μM, ****p<0.0001 for ruxolitinib at 300 nM. All comparisons by Dunnet’s multiple comparisons test versus DMSO control I) Number of hematopoietic colonies generated by peripheral blood mononuclear cells from MPN-BP patient samples treated with echinomycin, mitotempo, PP242, PT2977 and ruxolitinib at various concentrations versus DMSO control. Dotted line represent mean of the DMSO control. *p=0.0119 and ***p=0.0001 for echinomycin at 500 pM and 1 nM, respectively. **p=0.0020 and ****p<0.0001 for mitotempo at 200 μM and 500 μM, respectively. *p=0.0312 for PP242 at 100 nM. *p=0.0161 for PT2977 at 300 nM. *p=0.0136 for ruxolitinib at 300 nM. All comparisons by Dunnet’s multiple comparisons test against DMSO control. The average plus/minus SEM are shown.
Next, to better understand the molecular signatures associated with progression to the blast phase, we performed RNA-seq on matched peripheral blood mononuclear cells from 11 patients who progressed from chronic to blast phase MPN over a period of time ranging from 12 months to 13 years (Table S5). We observed variable separation between the groups by principal component analysis (PCA) and unsupervised clustering (Fig S9A,B), but also some significant variability in both groups. The variation may be due to differences in the genetic composition at the blast stage among the patients, differential timing of disease evolution, or other factors. Both PCA and unsupervised clustering indicated a subset of chronic samples and a subset of blast samples that were clearly well-separated from each other and a smaller number of samples from each group that exhibited intermediate, mixed expression patterns with features of both of the two well-separated subsets. We chose to remove those sample pairs for which one or both of the samples fell in the middle of the distribution with intermediate expression patterns where blast and chronic phase appeared to overlap and focused on 5 well separated pairs (Fig 4C) in order to avoid potential confounders which might not be related to the transition between these two disease states. Unsupervised clustering of these 5 pairs revealed major differences in the transcriptome between the chronic and blast phase of MPN (Fig 4D). To compare the mouse transcriptome of cells overexpressing MPLW515L alone or with Stk11 loss and the human transcriptome of chronic and blast phase MPN, we performed GSEA using differentially expressed genes in the mouse MPLW515L versus MPLW515L/Stk11Δ/Δ comparison and in the human chronic phase versus blast phase MPN as genesets. GSEA showed a strong concordance between human and mouse downregulated genes in the two datasets (Fig 4E). Moreover, GSEA analysis of a published geneset of hypoxia induced genes in CD34+ cells (19) revealed enrichment of the hypoxia signature in human blast-phase MPN (Fig. 4F). We also compared normalized enrichment scores from the GSEA analysis in both comparisons. Strikingly, we observed extensive concordance between the pathways enriched in the blast phase of the disease and the ones in MPLW515L cells lacking Stk11, including pathways related to hypoxia, oxidative phosphorylation and translation (Fig 4G, Table S6, Fig S10A,B). Finally, we returned to the 11 pairs and confirmed that there was extensive concordance between the pathways identified by GSEA in this complete set with the subset of 5 pairs, including the Manalo hypoxia geneset (Fig S9C,D). Together these results confirm that loss of LKB1 is a common feature and potential driver, of blast phase MPN progression in humans.
Finally, we treated primary MPN-BP samples and healthy CD34+ controls with drugs targeting the HIF, mTOR and JAK/STAT pathways. Echinomycin and mitotempo were tested based on our observation that they led to destabilization of HIF1a in cells with MPLW515L that lack LKB1 (Fig 2E). We also tested PT2977, a small molecule that inhibits HIF2a (20), based on our observation that HIF2a is also stabilized by expression of MPLW515L in cells lacking Stk11 (Fig. S11), as well as the mTOR pathway inhibitor PP242 and the JAK2 inhibitor ruxolitinib. With the exception of the highest doses of mitotempo and ruxolitinib, the drugs had no significant effect on colony formation by healthy CD34+ cells (Fig 4H). By contrast, we observed consistent, dose-dependent inhibition of colony formation of the five primary MPN-BP specimens with all compounds, with the most striking effects caused by echinomycin and mitotempo (Fig 4I). Of note, the cells were largely resistant to ruxolitinib with little discrimination between the MPN-BP and healthy CD34+ cells, consistent with the poor activity of this drug in MPN-BP patients. Together, these results suggest that drugs which target the HIF pathway should be considered in this disease.
Discussion
Loss of Stk11 is a key event in the progression of several solid malignancies as well as the dominant genetic mutation that leads to Peutz-Jeghers syndrome, characterized by increased risk of cancer development (21,22). Prior studies have shown that loss of LKB1 in hematopoietic cells leads to an initial burst of proliferation of HSPCs followed by bone marrow failure as opposed to tumor development (10–12). Our findings that LKB1 loss imparts enhanced self-renewal of cells with activated JAK/STAT signaling strongly suggest that continued stimulation of this pathway allows for growth of HSPCs that lack LKB1. Of note, we did not detect mutations of STK11 in a cohort of chronic or blast phase MPN patients, thus downregulation of this tumor suppressor is likely the driving event.
Deletion of Stk11 in MPN cells induced expression of genes related to hypoxia and mitochondrial function. At the molecular level, stabilization of HIF1a under normal oxygen tension was a striking feature of Stk11 deleted MPN cells: this effect is shared with a mouse model of Peutz-Jeghers syndrome and mouse embryonic fibroblasts (23,24). HIF1a is a transcriptional factor widely known for its role in solid malignancies where it is consistently stabilized to promote tumor cell survival under hypoxia, tumor angiogenesis and cell proliferation. In hematopoietic cells with enhanced JAK/STAT signaling, HIF1a promotes self-renewal of MPN cells likely through induction of genes related to stemness and proliferation. With respect to the link between LKB1 loss and HIF1 stabilization, increased mitochondrial ROS produced by the electron transport chain during OXPHOS has been linked to the stabilization of HIF1a in hypoxia (15,16). We surmise that the observed increase in mitochondrial ROS impairs the normal prolyl hydroxylase function needed for VHL-mediated HIF1a ubiquitination and degradation by the proteasome.
A number of genetic alterations have been modeled in mouse models of MPN to mimic the alterations found in the human blast phase MPN including loss of TP53 and JARID2. We show that deletion of Stk11 in the MPLW515L mouse model of MPN resulted in increased lethality with intense osteosclerosis and bone marrow fibrosis, accompanied by >20% immature blasts in an otherwise hypocellular bone marrow. Furthermore, deletion of STK11 in primary myelofibrosis (PMF) patient samples and transplantation in NSGS recipients caused an increase in human myeloid cell engraftment, rarely seen in PMF xenografts, but no overt leukemia suggesting that additional mutations present in the patient samples may cooperate with STK11 loss to produce a leukemic phenotype in humans.
Of note, the two libraries contained sgRNAs that target a number of genes associated with MPN progression and myeloid leukemia, including TET2, ASXL1, EZH2, and DNMT3A. It is notable that none of these sgRNAs were enriched in JAK2 mutant cells at platings 3 or 5. Although this may be a consequence of poor gene editing, it suggests that loss of these genes individually does not confer enhanced self-renewal and argues that STK11 is a more potent tumor suppressor in the MPNs.
Pseudo-hypoxia and stabilization of HIF1a have been recently described in MDS and MPN chronic phase (25,26). With respect to the latter observation, there was only a modest increase in HIF1a levels and this was observed primarily in MPN cell lines which have been derived from patients with blast phase disease (27). Our IHC data reveal that HIF1a is present at the chronic phase, but that it is greatly increased at the blast phase, and our RNA-seq data demonstrate that HIF-induced pathways are markedly enriched in the leukemia phase.
Our findings provide insights into potential therapeutic approaches to better treat blast phase disease. Mice that lack HIF1a did not develop any detrimental phenotype under steady state conditions, suggesting that is a viable target (28). Moreover, recent developments in structure-based design approaches for drug discovery made possible the identification of a selective HIF2a antagonist, PT2399, which can dissociate HIF2a from his partner HIF1b, preventing its translocation to the nucleus (29). It has been reported that PT2399 provides on target efficacy in both in vitro and in vivo pre-clinical studies of kidney cancer (29). Here we demonstrate that PT2977, a potent and selective analog of PT2399 that is being investigated in an open-label Phase 2 study of Clear Cell Renal Carcinoma, selectively inhibited colony formation of MPN-BP cells, which demonstrate stabilization of both HIF1a and HIF2a. Thus, our work lends strong rationale for the use of HIF inhibitors in the leukemia phase of the MPNs.
Methods
Mice
All animals were of the C57BL/6 background. Jak2V617F knock-in (IMSR Cat# JAX:031658, RRID:IMSR_JAX:031658), Stk11 floxed (IMSR Cat# JAX:014143, RRID:IMSR_JAX:014143), Vav-Cre (IMSR Cat# JAX:008610, RRID:IMSR_JAX:008610), Mx1-Cre (IMSR Cat# JAX:003556, RRID:IMSR_JAX:003556) and Rosa26-Cas9 knock-in mice (IMSR Cat# JAX:026175, RRID:IMSR_JAX:026175) were purchased from Jackson Laboratories. NSGS mice (IMSR Cat# JAX:013062, RRID:IMSR_JAX:013062) were purchased from Jackson Laboratories. All mice were genotyped for the presence of the correct alleles by PCR. Animal studies were approved by the Northwestern University IACUC and the Washington University IACUC.
Patient Samples
All specimens were studied with approval by the Institutional Review Boards of Northwestern University, Memorial Sloan Kettering, Washington University, and the Mayo Clinic. Patient data are provided in Table S5.
CRISPR/Cas9 screen
We generated a mouse sgRNA library for CRISPR-KO with 4X coverage targeted against the genes listed in the Uniprot Curated List of Tumor Suppressors as well as several MPN associated genes (Table S1). Given that loss of TP53 has been previously reported to be a transforming event in MPN to AML (8), we did not include this gene in the library. To perform the screen, lineage-negative HSPCs were isolated from bone marrow of Jak2V617F/Vav-Cre/Cas9 or Vav-Cre/Cas9 mice and transduced with either of two lentiviral sgRNA libraries or empty vector control at multiplicity of infection of 0.5. Transduced cells were selected from 24 hours with 2 ug/mL blasticidin (Sigma-Aldrich, 15205) and then plated on M3434 methylcellulose media (StemCell Technologies, M3434) with 1 ug/mL blasticidin. Every 7 days of culture, colonies were enumerated and cells recovered for serial replating over 5 generations. DNA of cells from platings 1, 3 and 5 was extracted with GenElute™ Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, G1N70) and next generation sequencing was performed to evaluate enrichment of guide RNAs of plating 3 and 5 against plating 1 as a baseline. sgRNA enrichment was determined using PinAPL-Py (30). The same approach was used to target Jak2V617F/Vav-Cre/Cas9 bone marrow cells for Trp53 with the pLX-sgRNA vector (Addgene, 50662) containing an sgRNA with protospacer sequence 5’- CATAAGGTACCACCACGCTG −3’ targeting exon 6 of Trp53.
sgRNA/Cas9 RNP delivery
Gene editing with Cas9 ribonucleoprotein complexes (RNP) was achieved using the Alt-R CRISPR-Cas9 system (IDT). Briefly, equimolar amounts of crRNAs targeting mouse Stk11 or/and trcrRNA were annealed to form the gRNA. Non-targeting control crRNA #1 (IDT, #1072544) was used for control conditions. 18 pmol of recombinant Cas9 (IDT, 1081061) were combined with 22 pmol of gRNA to form RNP complexes, which were then delivered to c-kit+ HSPCs using the NEON electroporation system (Thermo Scientific) with settings 1700 V, 20 ms, 1 pulse as described previously (31). After 24 hours, cells were plated in M3434 methylcellulose media (Stemcell Technologies) and serially replated every 7 days over 6 generations.
Viral Transduction
High titer retrovirus was obtained by transfecting Platinum E cells (Cell Biolabs, RV-101, RRID:CVCL_B488) with 12 ug of retroviral construct using X-tremeGENE 9 transfection reagent (Roche, XTG9-RO) and collecting viral supernatant after 48 hours. Viral transduction was performed by spinoculation; 2–5× 10^6 cells were mixed with virus and centrifuged for 90 minutes at 2500 RPM at 32 C. Transduction efficiency was evaluated by flow cytometry after 24 hours of culture post-transduction.
Flow Cytometry
Bone marrow cells were isolated by crushing long bones with mortar and pestle in phosphate buffered saline (PBS) and filtered through a 70 μm nylon mesh to obtain a single cell suspension. The HSPC compartment was analyzed using and anti-mouse V450-lineage negative antibody cocktail (BD Biosciences Cat# 561301, RRID:AB_10611731), anti-mouse APC-CD117 (BD Biosciences Cat# 553356, RRID:AB_398536), and anti-mouse PE-Cy7-Sca1 (Thermo Fisher Scientific Cat# 25–5981-82, RRID:AB_469669). Flow cytometry was performed on a LSRII flow cytometer (BD Biosciences). To measure mitochondrial ROS, whole bone marrow cells were incubated with 1.5 μM mitoSOX (Thermo Scientific, M36008) in Earle Balanced Salt Solution (EBSS) supplemented with 0.5% BSA in a tissue culture incubator for 30 minutes, washed with EBSS and immediately analyzed. For colony forming assays with sorted cells, GFP+ mCherry+ double transduced cells were isolated 36 hours post-transduction by FACS using a FACS Aria II (BD Biosciences).
Transplantation
Bone marrow from Stk11fl/fl Mx1-Cre+ or Stk11fl/fl mice was isolated by crushing femurs and tibias and c-kit+ cells isolated using magnetic enrichment (Miltenyi Biotec CD117 MicroBeads, mouse # 130–091-224) and cultured overnight in Stem Span media supplemented with 50 ng/mL mouse SCF, 10 ng/mL mouse IL-6 and 10 ng/mL mouse IL-3. c-kit+ cells were then transduced with the MPLW515L-GFP retrovirus by spinoculation and 0.4 × 106 GFP+ cells transplanted into lethally irradiated CD45.1 recipients. MPN development and engraftment was confirmed by CBC and flow cytometry at 3 weeks post-transplantation and pIpC injections performed at 4 weeks post-transplantation for 3 times at increasing dose every other day (2 mg/Kg, 4 mg/Kg, 8 mg/Kg).
Xenotransplantation
Human MF peripheral blood mononuclear cells were isolated with Ficoll (GE Healthcare, Munich, Germany) and mononuclear cells cryopreserved within 24 h after collection in HBSS buffer (Corning #21021CV) containing Pen/Strep (100 Units/mL; Fisher Scientific #MT30002CI), HEPES (10uM; Life Technologies # 15630080) and FBS (2%; Sigma #14009C). For xenotransplantations, CD34+ hematopoietic stem and progenitor cells isolated were isolated using magnetic enrichment (Miltenyi Biotec # 130–100-453) from JAK2V617F (ID# 953 and 179) or CALRfs (ID# 293) positive MF patients. Enriched CD34+ cells were incubated in SFEMII media (Stemcell technologies #09605) supplemented with Pen-Strep (50 Units/mL), human stem cell factor (SCF; 50 ng/mL), human thrombopoietin (TPO; 50 ng/mL), and human Flt3L (50 ng/mL). 12–24 hours post-sort, CD34+ cells were nucleofected with Cas9/ribonucleorprotein complexed with sgRNA targeting STK11 as previously described (31). 48 hours post nucleofection, these cells were transplanted into sublethally irradiated (200 rads) NOD-scid-Il2rg-null-3/GM/SF (IMSR Cat# JAX:013062, RRID:IMSR_JAX:013062) mice via X-ray guided intra-tibial injections. Engraftment of human cells in xenotransplants were assessed by flow cytometry with antibodies targeting human CD45 (BioLegend Cat# 368511, RRID:AB_2566371) and mouse CD45 (BioLegend Cat# 103139, RRID:AB_2562341) cells.
RNA-sequencing and data analysis
The mRNA-seq on stranded libraries was conducted in the Northwestern University NUSeq Core Facility. Briefly, total RNA examples were checked for quality using RINs generated from Agilent Bioanalyzer 2100. RNA quantity was determined with Qubit fluorometer. The Illumina TruSeq Stranded mRNA Library Preparation Kit was used to prepare sequencing libraries from 100 ng of high-quality RNA samples (RIN > 7). The Kit procedure was performed without modifications. This procedure includes mRNA purification and fragmentation, cDNA synthesis, 3’ end adenylation, Illumina adapter ligation, library PCR amplification and validation. Illumina NextSeq 500 sequencer was used to sequence the libraries with the production of single-end, 75 bp reads at the depth of 20–25 M reads per sample.
Resulting raw sequencing data quality was assessed using FastQC and then the sequencing data was mapped to the mouse UCSC mm10 genome using the STAR RNA-seq aligner (STAR, RRID:SCR_015899). Feature and read summarization was performed using FeatureCounts from the Subread package (Subread, RRID:SCR_009803). Differential expression analysis was performed using EdgeR (EdgeR, RRID:SCR_012802) using the QLF test. Gene Set Enrichment Analysis (GSEA, RRID:SCR_003199) was performed on normalized transcript per million (TPM) values from all samples using the GSEA software with the MSigDB gene sets. These RNA-seq data are available in GEO (accession number GSE159737).
RNA-seq analysis of paired patient data
Raw reads were trimmed for quality (threshold of 15) and adapter sequences using version 0.4.5 of TrimGalore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore), and then aligned to human assembly hg38 with STAR v2.4 using default parameters. Post-alignment quality and transcript coverage were assessed using the Picard tool CollectRNASeqMetrics (http://broadinstitute.github.io/picard/). Raw read count tables were created using HTSeq v0.9.1. Normalization and expression dynamics were evaluated with DESeq2 using the default parameters with library size factor normalization. Potential outliers were removed based on the separation of labeled sample pairs along PC1 of the principal component analysis. Gene set enrichment analysis (GSEA, RRID:SCR_003199) was run using MSigDB v6 with ‘pre-ranked’ mode and log2 fold change for pairwise comparisons. Pathway concordance between mouse and human was evaluated by linking gene set names with the normalized enrichment scores and highlighting pathways with FDR < 0.1 in either dataset. The human sample RNA-seq data are available in dbGAP.
Western blotting
Cells were lysed in RIPA lysis buffer supplemented with halt phosphatase inhibitor cocktail (Thermo Scientific, 78420) and complete protease inhibitor cocktail (Roche). Proteins were separated on a mini protean TGX 4–15% polyacrylamide gel (Bio-Rad) and transferred on a PVDF membrane (Millipore). Antibodies included the following: anti-LKB1 (Cell signaling technologies, Cat# 3050, RRID:AB_823559, Cat# 3047, RRID:AB_2198327), anti-HIF1a (Cell signaling technologies, Cat# 36169, RRID:AB_2799095), anti-HIF2a (Novus, Cat# NB100–122, RRID:AB_10002593), anti-HKII (Cell signaling technologies, Cat# 2867, RRID:AB_2232946), anti-p-AMPKa thr172 (Cell signaling technologies, Cat# 2535, RRID:AB_331250), anti-AMPKa (Cell signaling technologies, Cat# 5832, RRID:AB_10624867), anti-HIF1a-OH Pro564 (Cell signaling technologies, Cat# 3434, RRID:AB_2116958), anti-PHD2 (Novus Biologicals, Cat# NB100–137, RRID:AB_10003054), anti-GRB2 (BD Biosciences Cat# 610112, RRID:AB_397518) and anti-ACTB (Cell signaling technologies, Cat# 3700, RRID:AB_2242334).
Histology
Mouse tissues for histology were fixed in 10% neutral buffered formalin for 24 hours and then processed for H&E staining. IHC was performed using standard protocols with anti-LKB1 (Cell signaling technologies, Cat# 13031, RRID:AB_2716796) and anti-HIF1a (Abcam, Cat# ab16066, RRID:AB_302234) antibodies.
Colony forming unit assays with human MPN-BP cells
150,000 peripheral blood mononuclear cells collected from patients at the blast phase of the disease were plated in H4434 methylcellulose media (Stemcell Technologies) supplemented with echinomycin (Sigma-Aldrich), mitotempo (Sigma-Aldrich), PP242 (Selleck Chemicals), PT2977 (MedChem Express) or ruxolitinib at 4 different concentrations. Colonies were enumerated after 14 days. Peripheral blood mobilized CD34+ cells from healthy individuals were similarly cultured with drugs to compare their effects in a non-malignant setting.
Supplementary Material
Statement of significance.
Progression of the myeloproliferative neoplasms to acute myeloid leukemia occurs in a substantial number of cases, but the genetic basis has been unclear. We discovered that loss of LKB1/STK11 leads to stabilization of HIF1a and promotes disease progression. This observation provides a potential therapeutic avenue for targeting progression.
Acknowledgements
This work was supported by the NIH grants R01CA237039 and R01HL112792 to JDC, R01HL147978 to GAC, R35CA197532 to NSC, R01CA248770 to PN, R50CA211534 to QJW, and the MPN Research Consortium P01CA108671 to RH, RL, JDC and RKR. AV was supported by K99CA230314, RKR is supported by K08CA188529-01, and MS was supported by the Northwestern University Physician Scientist Training Program as well as by a Conquer Cancer Young Investigator Award. We also acknowledge the support of Memorial Sloan Kettering Cancer Center Support Grant NIH P30 CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. GAC is a scholar of the Leukemia and Lymphoma Society. HC was supported by an Edward P. Evans Foundation Young Investigator Award, the Leukemia Research Foundation, and the When Everyone Survives Foundation. Additional support was provided by the Janus Fund (RLL), the Samuel Waxman Cancer Research Foundation (JDC), and St. Jude /ALSAC (JDC).
Conflict of interest statement:
M.B. is an employee of Abbvie. B.S. is on the advisory boards for Pharmaessentia and Constellation Pharmaceuticals. R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Imago, Mission Bio, Zentalis, Ajax, C4 Therapeutics and Isoplexis. He receives research support from and consulted for Celgene and Roche and has consulted for Incyte, Janssen, Astellas, Morphosys and Novartis. He has received honoraria from Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. R.K.R has received consulting fees from: Constellation, Incyte, Celgene/BMS, Promedior, CTI, Jazz Pharmaceuticals, Blueprint, Stemline, Galecto, Pharmaessentia, Abbvie and research funding from Incyte, Constellation, Stemline. G.A.C. has consulted and received research funding from Incyte. N.S.C. is on the scientific advisory board of Rafael Pharmaceuticals. D.E.R. receives research funding from members of the Functional Genomics Consortium (Abbvie, BMS, Jannsen, Merck, Vir), and is a director of Addgene, Inc. The other authors have nothing to disclose.
References
- 1.Cerquozzi S, Tefferi A. Blast transformation and fibrotic progression in polycythemia vera and essential thrombocythemia: a literature review of incidence and risk factors. Blood Cancer J 2015;5:e366 doi 10.1038/bcj.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cervantes F, Tassies D, Salgado C, Rovira M, Pereira A, Rozman C. Acute transformation in nonleukemic chronic myeloproliferative disorders: actuarial probability and main characteristics in a series of 218 patients. Acta Haematol 1991;85(3):124–7 doi 10.1159/000204873. [DOI] [PubMed] [Google Scholar]
- 3.Dunbar AJ, Rampal RK, Levine R. Leukemia secondary to myeloproliferative neoplasms. Blood 2020;136(1):61–70 doi 10.1182/blood.2019000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tefferi A, Rumi E, Finazzi G, Gisslinger H, Vannucchi AM, Rodeghiero F, et al. Survival and prognosis among 1545 patients with contemporary polycythemia vera: an international study. Leukemia 2013;27(9):1874–81 doi 10.1038/leu.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heaney ML, Soriano G. Acute myeloid leukemia following a myeloproliferative neoplasm: clinical characteristics, genetic features and effects of therapy. Curr Hematol Malig Rep 2013;8(2):116–22 doi 10.1007/s11899-013-0154-5. [DOI] [PubMed] [Google Scholar]
- 6.Tam CS, Nussenzveig RM, Popat U, Bueso-Ramos CE, Thomas DA, Cortes JA, et al. The natural history and treatment outcome of blast phase BCR-ABL- myeloproliferative neoplasms. Blood 2008;112(5):1628–37 doi 10.1182/blood-2008-02-138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tefferi A, Mudireddy M, Mannelli F, Begna KH, Patnaik MM, Hanson CA, et al. Blast phase myeloproliferative neoplasm: Mayo-AGIMM study of 410 patients from two separate cohorts. Leukemia 2018;32(5):1200–10 doi 10.1038/s41375-018-0019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci U S A 2014;111(50):E5401–10 doi 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celik H, Koh WK, Kramer AC, Ostrander EL, Mallaney C, Fisher DAC, et al. JARID2 Functions as a Tumor Suppressor in Myeloid Neoplasms by Repressing Self-Renewal in Hematopoietic Progenitor Cells. Cancer Cell 2018;34(5):741–56 e8 doi 10.1016/j.ccell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 2010;468(7324):701–4 doi 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 2010;468(7324):659–63 doi 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 2010;468(7324):653–8 doi 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, et al. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv 2019:251082 doi 10.1101/251082. [DOI] [PubMed] [Google Scholar]
- 14.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG, Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 2002;1(3):237–46 doi 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 15.Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab 2005;1(6):409–14 doi 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ 2008;15(4):660–6 doi 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 17.Hamanaka RB, Weinberg SE, Reczek CR, Chandel NS. The Mitochondrial Respiratory Chain Is Required for Organismal Adaptation to Hypoxia. Cell Rep 2016;15(3):451–9 doi 10.1016/j.celrep.2016.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orr AL, Vargas L, Turk CN, Baaten JE, Matzen JT, Dardov VJ, et al. Suppressors of superoxide production from mitochondrial complex III. Nat Chem Biol 2015;11(11):834–6 doi 10.1038/nchembio.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierenga AT, Vellenga E, Schuringa JJ. Convergence of hypoxia and TGFbeta pathways on cell cycle regulation in human hematopoietic stem/progenitor cells. PLoS One 2014;9(3):e93494 doi 10.1371/journal.pone.0093494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu R, Wang K, Rizzi JP, Huang H, Grina JA, Schlachter ST, et al. 3-[(1S,2S,3R)-2,3-Difluoro-1-hydroxy-7-methylsulfonylindan-4-yl]oxy-5-fluorobenzo nitrile (PT2977), a Hypoxia-Inducible Factor 2alpha (HIF-2alpha) Inhibitor for the Treatment of Clear Cell Renal Cell Carcinoma. J Med Chem 2019;62(15):6876–93 doi 10.1021/acs.jmedchem.9b00719. [DOI] [PubMed] [Google Scholar]
- 21.Shorning BY, Clarke AR. Energy sensing and cancer: LKB1 function and lessons learnt from Peutz-Jeghers syndrome. Semin Cell Dev Biol 2016;52:21–9 doi 10.1016/j.semcdb.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Han X, Li F, Wang R, Wang H, Gao Y, et al. LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response. Cancer Cell 2015;27(5):698–711 doi 10.1016/j.ccell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A 2014;111(7):2554–9 doi 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A 2009;106(27):11137–42 doi 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi Y, Zhang Y, Yokota A, Yan X, Liu J, Choi K, et al. Pathobiological Pseudohypoxia as a Putative Mechanism Underlying Myelodysplastic Syndromes. Cancer Discov 2018;8(11):1438–57 doi 10.1158/2159-8290.CD-17-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumeister J, Chatain N, Hubrich A, Maie T, Costa IG, Denecke B, et al. Hypoxia-inducible factor 1 (HIF-1) is a new therapeutic target in JAK2V617F-positive myeloproliferative neoplasms. Leukemia 2020; 34(4):1062–1074. [DOI] [PubMed] [Google Scholar]
- 27.Uozumi K, Otsuka M, Ohno N, Moriyama T, Suzuki S, Shimotakahara S, et al. Establishment and characterization of a new human megakaryoblastic cell line (SET-2) that spontaneously matures to megakaryocytes and produces platelet-like particles. Leukemia 2000;14(1):142–52 doi 10.1038/sj.leu.2401608. [DOI] [PubMed] [Google Scholar]
- 28.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010;7(3):391–402 doi 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature 2016;539(7627):107–11 doi 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spahn PN, Bath T, Weiss RJ, Kim J, Esko JD, Lewis NE, et al. PinAPL-Py: A comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci Rep 2017;7(1):15854 doi 10.1038/s41598-017-16193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep 2016;17(5):1453–61 doi 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.