Abstract
Lung squamous cell carcinoma (LSCC) is the second most prevalent type of lung cancer. Despite extensive genomic characterization, no targeted therapies are approved for the treatment of LSCC. Distal amplification of the 3q chromosome is the most frequent genomic alteration in LSCC, and there is an urgent need to identify efficacious druggable targets within this amplicon. We identify the protein kinase TNIK as a therapeutic target in LSCC. TNIK is amplified in approximately 50% of LSCC cases. TNIK genetic depletion or pharmacological inhibition reduces the growth of LSCC cells in vitro and in vivo. In addition, TNIK inhibition showed antitumor activity and increased apoptosis in established LSCC patient-derived xenografts. Mechanistically, we identified the tumor suppressor Merlin/NF2 as a novel TNIK substrate and showed that TNIK and Merlin are required for the activation of focal adhesion kinase. In conclusion, our data identify targeting TNIK as a potential therapeutic strategy in LSCC.
Keywords: 3q amplicon, targeted therapies, lung squamous cell carcinoma, Merlin, FAK
Introduction
Lung squamous cell carcinoma (LSCC) accounts for one-third of all lung cancer cases. Despite extensive genomic sequencing, the identification of oncogenic drivers in LSCC has remained challenging, and actionable alterations are unknown in the majority of LSCC patients (1,2). As a result, no targeted therapies have been approved to treat LSCC, and treatment still relies on chemotherapy or radiotherapy. Genomic characterization of LSCC tumors shows that distal chromosome 3q amplification (3q26–29) is the most prevalent genomic alteration in LSCC (3). This region contains very well-characterized oncogenes, including the transcription factors SOX2 and TP63, and the kinase genes PIK3CA and PRKCI (4). While the former are not druggable to date, results in head and neck squamous cell carcinoma (HNSCC) suggest that PIK3CA amplification does not predict sensitivity to PI3K inhibitors, and PI3K inhibitors have not shown significant clinical benefit as monotherapies in HNSCC (5,6). Evaluation of PKCι is still in pre- or early-phase clinical testing (7).
The protein kinase TNIK (Traf2- and Nck- interacting kinase) has been identified as a potential genetic dependency in tumors with distal amplification of the 3q chromosome (8); however, an in-depth evaluation of its functional role in LSCC has never been performed. Here we tested the potential of TNIK as a therapeutic target in LSCC. Our results suggest that targeting amplified TNIK could serve as a new intervention strategy for a subset of LSCC patients.
Results
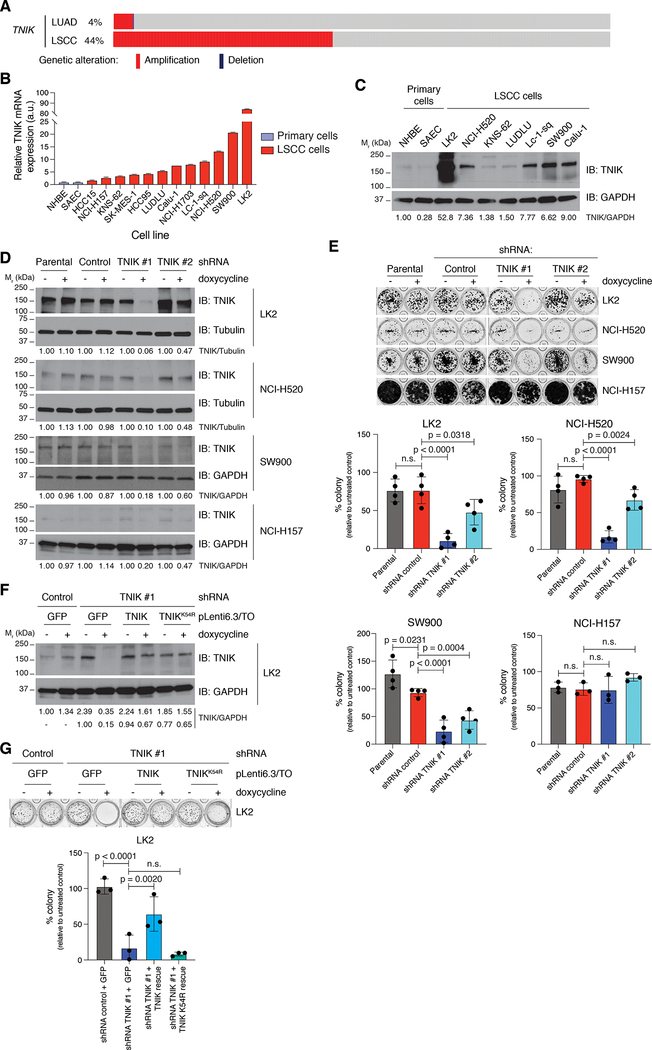
To investigate the consequences of targeting TNIK in LSCC, we queried The Cancer Genome Atlas (TCGA) LSCC data sets to determine the frequency of TNIK amplification in LSCC samples. We observed that TNIK is amplified (>3 gene copies) in 35–44% of cases, with an additional 46–54% of cases displaying copy-number gains (Fig. 1A, Supplementary Fig. S1A). In contrast, TNIK was amplified in 3–4% of lung adenocarcinoma cases (Fig. 1A, Supplementary Fig. S1A). Consistent with TCGA data, we observed increased expression of TNIK mRNA in a panel of LSCC cell lines compared to non-transformed primary lung cells (Fig. 1B). Although TNIK expression was high in LSCC cell lines with 3q amplification, some cell lines lacking TNIK amplification (SW900 and Calu-1) also showed enhanced TNIK mRNA levels. Analogous expression patterns were observed when TNIK levels were assayed by immunoblot (Fig. 1C).
Figure 1. Identification of the protein kinase TNIK as a genetic dependency in LSCC.
A. TNIK amplification in LSCC in comparison to lung adenocarcinoma cases from the TCGA data set. Graph from cBioPortal (34). B. Analysis of TNIK mRNA expression by RT-qPCR in a panel of LSCC cell lines and two primary lung cells (NHBE and SAEC). Data represent mean ± SD. NHBE were used as control (TNIK expression = 1.00). n = 2. C. Analysis of TNIK expression by immunoblot. The blot is representative of three independent replicates. D. Doxycycline (dox) induction (1 μg/ml, 72 h) of TNIK shRNA reduces TNIK expression in LK2, NCI-H520, SW900, and NCI-H157 cells. The blot is representative of three independent experiments (LK2, NCI-H520, and SW900 cells) and two independent experiments (NCI-H157). Tubulin or GAPDH were used as loading controls. E. Colony formation assay (14 days) following dox-inducible (1 μg/ml, replaced every 48 h) TNIK knockdown. The image is representative of at least three independent experiments. Data are represented as mean value ± SD, one-way ANOVA, Tukey’s multiple comparisons post-test. F. Validation of a dox-inducible (1 μg/ml, 72 h) TNIK shRNA rescue system in LK2 cells. pLenti6.3-TO-GFP or shRNA-resistant TNIK or TNIK K54R (kinase dead) containing lentiviruses were transduced in corresponding LK2 cells, and rescue of TNIK expression was evaluated by immunoblot. The blot is representative of three independent experiments. GAPDH was used as loading control. TNIK levels relative to GAPDH were normalized to untreated pLKO-shRNA negative / GFP LK2 cells (top) or untreated pLKO-shRNA TNIK#1 / GFP LK2 cells (bottom). G. Rescue of clonogenic growth (14 days; doxycycline 1 μg/ml, replaced every 48 h) in LK2 cells was evaluated in 3D Matrigel cultures. The image is representative of three independent experiments. Data are represented as mean value ± SD, one-way ANOVA, Tukey’s multiple comparisons post-test. n.s., not significant. Immunoblot band intensity was quantified with ImageJ.
To assess if LSCC cells require TNIK to maintain cancer cell viability, we depleted TNIK with doxycycline (dox)-inducible TNIK-targeting shRNAs, which led to a significant reduction (60–80%) in the clonogenic growth of LSCC cells with high TNIK expression (LK2, NCI-H520, and SW900 cells; Fig. 1D, 1E). No significant effect on clonogenic growth was observed in low-expressing TNIK LSCC cells when TNIK was depleted with dox-inducible shRNAs (NCI-H157 and HCC15 cells; Fig. 1D, 1E, Supplementary Fig. S1B). Next, we assessed the importance of TNIK in maintaining cancer cell survival in Matrigel 3D cultures, which more accurately mimic the conditions of the tumor environment. TNIK depletion abrogated LK2 cell growth in Matrigel cultures, which was rescued by re-expression of a catalytically competent TNIK shRNA-resistant mutant (Fig. 1F, 1G). However, a TNIK kinase-dead (K54R) mutant was unable to rescue the effects of TNIK knockdown on cell growth (Fig. 1F, 1G).
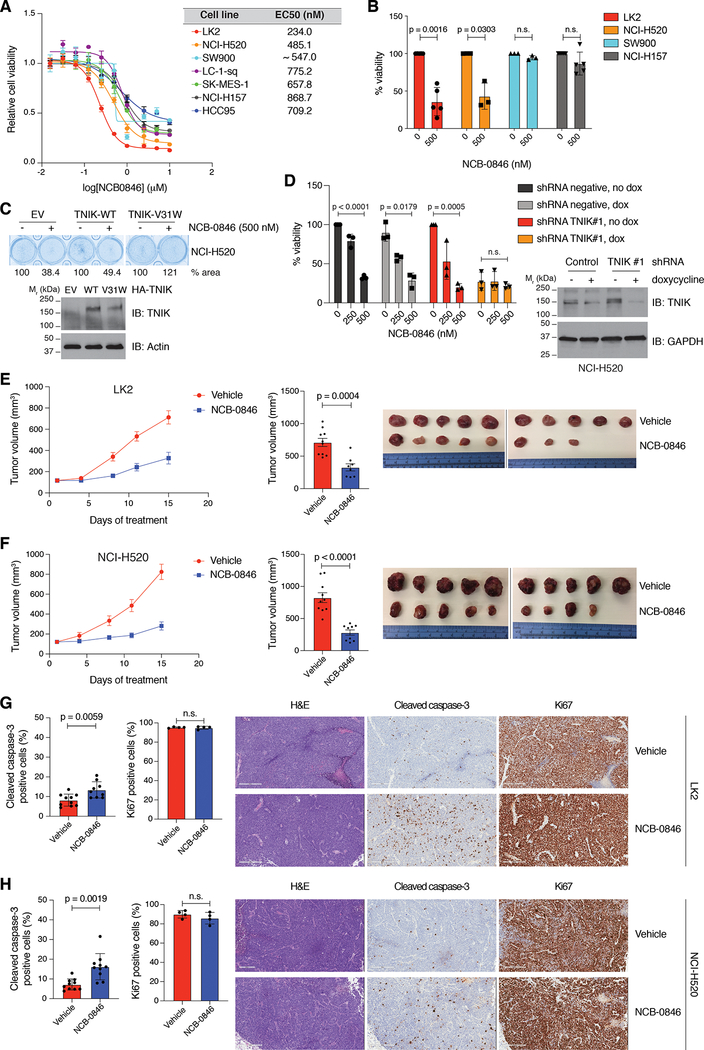
Since TNIK catalytic activity is a requirement for maintaining the growth of the LK2 LSCC cell line, we assessed the effect of a TNIK inhibitor, NCB-0846, on LSCC cell growth. This compound is a well-characterized TNIK small-molecule catalytic inhibitor (9) that traps TNIK in an inactive conformation and impedes TNIK autophosphorylation in an in vitro kinase assay (Supplementary Fig. S2A). The TNIK inhibitor reduced cell viability in all LSCC cell lines tested, with EC50 values ranging from 230 nM in LK2 cells to 870 nM in NCI-H157 cells, and TNIK-high-expressing cells displayed greater sensitivity to NCB-0846 in most cases (Fig. 2A, 2B). Interestingly the SW900 cells were not sensitive to the TNIK inhibitor, indicating TNIK will have some tumorigenic functions independent of the catalytic activity of the kinase. As kinase inhibitors are promiscuous, we identified a TNIK drug-resistant mutant that we could use as a tool to validate the NCB-0846-induced toxicity is specifically due to inhibiting the catalytic activity of TNIK. We tested numerous mutants that would be predicted to be resistant to an ATP competitive inhibitor (10,11, Supplementary Table S1) and identified that mutation of the TNIK pocket protector residue (−1 GXGXXG site, V31 in TNIK) to tryptophan (V31W) retained catalytic activity in the presence of NCB-0846 (as evaluated by phosphorylation of the TNIK substrate Merlin, Supplementary Fig. S2B). We then assessed this mutant in cells treated with NCB-0846 and observed that expression of TNIK V31W could rescue the inhibitor-induced decreases in viability (Fig. 2C). To further validate NCB-0846 effects were due to specifically inhibiting TNIK, we treated cells in which TNIK had been knocked down with increasing concentrations of NCB-0846. As expected, almost complete depletion of TNIK in NCI-H520 cells reduced short-term cell viability in this cell line, but cell viability was not diminished further after treatment with NCB-0846 in cells in which TNIK was depleted, indicating that NCB-0846 is not targeting other kinases to suppress the viability of LSCC cells (Fig. 2D). These results support that the observed effects of NCB-0846 on cell viability are due to specifically targeting TNIK.
Figure 2. TNIK inhibition significantly reduces viability of LSCC cells.
A. Dose-response curves of a panel of LSCC cell lines to NCB-0846. The cells were treated with increasing concentrations of NCB-0846 (72 h). The lines represent the fitted curves, where each dot indicates the mean value ± SD of at least three independent experiments each conducted in triplicate. B. Cell viability (MTS assay) of selected LSCC cell lines treated with NCB-0846 (500 nM) for 72 h. At least three independent experiments with triplicates were conducted for each cell line. Data are represented as mean value ± SD, two-tailed t-test with Welch’s correction. C. Cell viability (crystal violet assay) of NCI-H520 cells treated with NCB-0846 (500 nM, 48 h) is rescued by expression of a TNIK-inhibitor resistant mutant (TNIK V31W). n = 3 independent experiments with triplicates. D. Reduced effect of NCB-0846 treatment (72 h) on NCI-H520 cell viability after TNIK knockdown (72 h doxycycline 2 μg/ml induction followed by 72 h co-treatment with doxycycline and NCB-0846). Data are represented as mean value ± SD, two-way ANOVA, Tukey’s multiple comparison post-test. n = 3 independent experiments with triplicates. E. (Left) Tumor growth curve of LK2-cell-derived xenografts treated with NCB-0846 (100 mg/kg, b.i.d., five days on/two days off). Mean tumor volumes ± SEM are shown. (Center) Average tumor volume at the end of treatment, mean ± SEM. n = 10 mice in the vehicle-treated group; 8 mice in the NCB-0846-treated group; two-tailed t-test. (Right) Tumor images from the study at the end of treatment. F. (Left) Tumor growth curve of NCI-H520-cell-derived xenografts treated with NCB-0846 (100 mg/kg, b.i.d., five days on/two days off). Mean tumor volumes ± SEM are shown. (Center) Average tumor volume at the end of treatment, mean ± SEM. n = 10 mice in the vehicle-treated group; 9 mice in the NCB-0846-treated group; two-tailed t-test. (Right) Tumor images from the study at the end of treatment. G. (Left) Immunohistochemistry (IHC) analysis of apoptosis (cleaved caspase-3; n = 10 in vehicle group, n = 8 in NCB-0846-treated group) and proliferation (Ki67; n = 4/group) in LK2 cell xenografts. Data are represented as mean ± SD; two-tailed Mann–Whitney test. (Right) Representative H&E and cleaved caspase-3 and Ki67 IHC images from each treatment group. H. (Left) IHC analysis of apoptosis (cleaved caspase-3, n = 10 in vehicle-treated group; n = 9 in NCB-0846-treated group) and proliferation (Ki67; n = 4/group) in NCI-H520 cell xenografts. Data are represented as mean ± SD; two-tailed Mann–Whitney test. (Right) Representative H&E and cleaved caspase-3 and Ki67 IHC images from each treatment group. n.s., not significant. Scale bar = 300 μm.
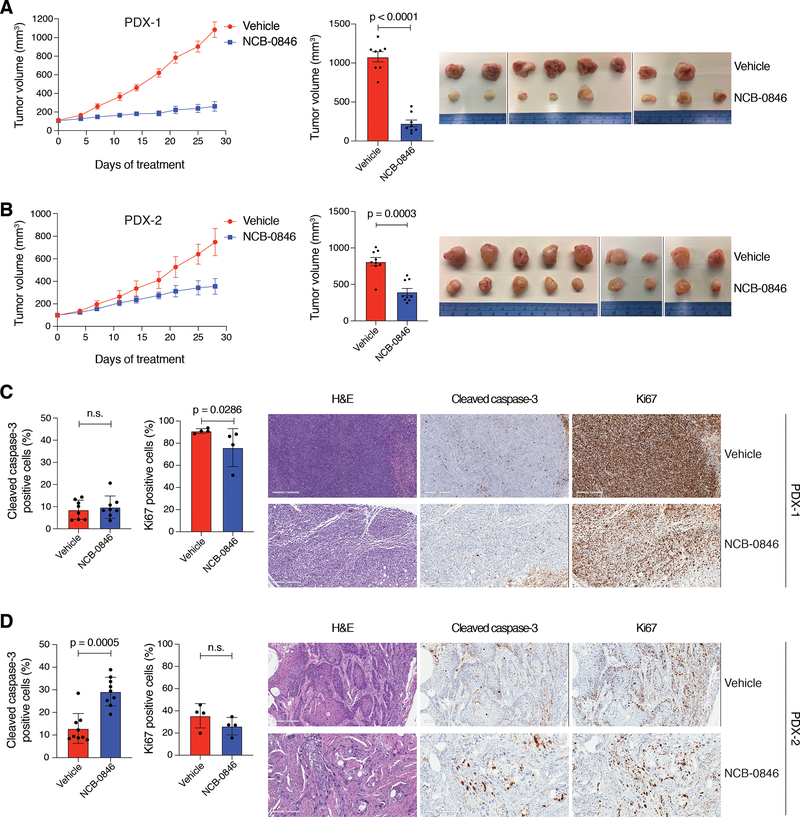
To determine if TNIK was essential for maintaining LSCC growth in vivo, we generated mouse xenograft models derived from the LK2 and NCI-H520 cell lines. Treatment of the mice with 100 mg/kg of NCB-0846 significantly reduced tumor growth in vivo without effects on the overall weight of the mice (Fig. 2E, 2F, Supplementary Fig. S2C), consistent with our results indicating that TNIK is essential for maintaining LSCC clonogenic growth in culture. Tumor analysis by immunohistochemistry showed that NCB-0846 treatment significantly increased cell apoptosis but did not affect cell proliferation (Fig. 2G, 2H and Supplementary Fig. S2D). We then established LSCC mouse xenograft models derived from two treatment-naïve LSCC patients (PDX), as these models more accurately recapitulate patients’ pharmacological responses to cancer drugs (12). A similar response to NCB-0846 was observed in these PDX models. In PDX model 1 (PDX-1), TNIK inhibition significantly halted tumor growth from the very onset of the treatment, while in the second model (PDX-2), TNIK inhibition effectively stabilized tumor growth (Fig. 3A, 3B). There was a minor overall decrease in the relative weight of the mice treated with NCB-0846 in the PDX-1 model (5.84 ± 3.92 %, Supplementary Fig. S3A). Analysis of tumor samples by immunohistochemistry demonstrated that NCB-0846 treatment increased cell apoptosis in PDX-2 but did not affect cell proliferation, however we observed that NCB-0846 suppressed proliferation in PDX-1 (Fig. 3C, 3D, Supplementary Fig. S3B). These results indicate that TNIK represents a pharmacological target in LSCC and that TNIK inhibitors could represent new treatment options for LSCC patients.
Figure 3. Preclinical evaluation of TNIK inhibitor NCB-0846 in LSCC PDX.
A. (Left) Tumor growth curve of PDX-1 treated with NCB-0846 (100 mg/kg, b.i.d., five days on/two days off). Mean tumor volumes ± SEM are shown. (Center) Average tumor volume at the end of treatment, mean ± SEM. n = 8 mice/group; two-tailed t-test. (Right) Tumor images from the study at the end of treatment. B. (Left) Tumor growth curve of PDX-2 treated with NCB-0846 (100 mg/kg, b.i.d., five days on/two days off). Mean tumor volumes ± SEM are shown. (Center) Average tumor volume at the end of treatment, mean ± SEM. n = 9 mice/group; two-tailed Mann-Whitney test. (Right) Tumor images from the study at the end of treatment. C. (Left) Immunohistochemistry (IHC) analysis of apoptosis (cleaved caspase-3; n = 8/group) and proliferation (Ki67; n = 4/group) in PDX-1 cell xenografts. Data are represented as mean ± SD; two-tailed Mann–Whitney test. (Right) Representative H&E and cleaved caspase-3 and Ki67 IHC images from each treatment group. D. IHC analysis of apoptosis (cleaved caspase-3; n = 9/group) and proliferation (Ki67; n = 4/group) in PDX-2 cell xenografts. Data are represented as mean ± SD; two-tailed Mann–Whitney test. (Right) Representative H&E and cleaved caspase-3 and Ki67 IHC images from each treatment group. n.s., not significant. Scale bar = 300 μm.
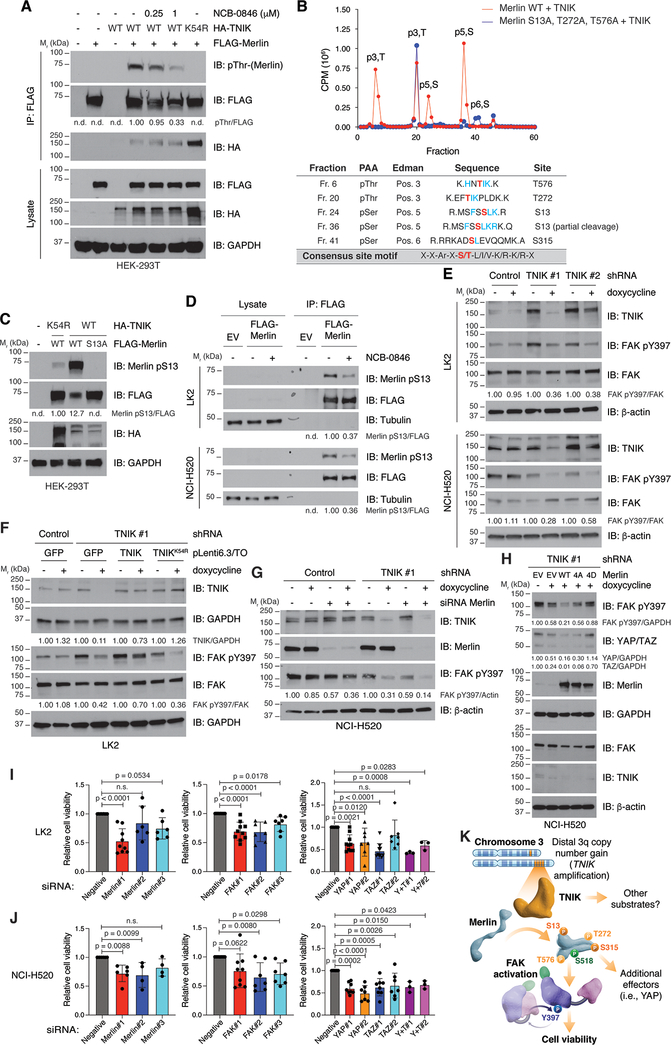
We next investigated the mechanisms through which TNIK could promote LSCC cell viability. In colon cancer cells, TNIK has been reported to contribute to cell viability through regulation of the Wnt/β-catenin pathway (13,14); however, analysis of β-catenin expression and activity did not differ in LSCC cells depleted of TNIK (Supplementary Fig. S4A). To identify alternative mechanisms by which TNIK could modulate LSCC viability, we scanned established oncogene and tumor suppressor gene products for occurrences of the consensus TNIK phosphorylation site motif (15). We found that the tumor suppressor Merlin (moesin-ezrin-radixin-like protein; NF2) harbored a potential TNIK phosphorylation site at T576, a position analogous to known regulatory sites on the related ERM proteins (ezrin, radixin and moesin). Moreover, Merlin had several additional sites (S13, T23, and T272) conforming to the TNIK consensus sequence. Of note, these phosphorylation sites are evolutionally conserved in metazoans, suggesting that their posttranslational modification has functional consequences (Supplementary Table S2). Also, structural modeling of Merlin indicates that these sites are accessible to the solvent, and thus, exposed to be phosphorylated. These data suggested that Merlin may be a novel substrate of TNIK. We also performed a global reverse phase protein array analysis in cells depleted of TNIK and identified FAK as a downstream effector (Supplementary Fig. S4B). FAK has been reported to be regulated by Merlin (16), providing us with additional rationale to evaluate Merlin as a downstream substrate of TNIK. Indeed, co-expression of Merlin and TNIK resulted in increased Merlin phosphorylation as assessed with a pan-phospho-threonine antibody; this phosphorylation was sensitive to the addition of the TNIK inhibitor NCB-0846 (Fig. 4A). In addition, we observed that TNIK and Merlin co-immunoprecipitated (Fig. 4A). A TNIK kinase-dead mutant (K54R) also interacted with Merlin but did not induce its phosphorylation (Fig. 4A). We verified that TNIK directly phosphorylated Merlin in vitro in a radiolabel kinase assay, and this phosphorylation was also sensitive to treatment with NCB-0846 (Supplementary Fig. S4C). Using in vitro kinase assays followed by phosphopeptide mapping, and mass spectrometry analysis on Merlin isolated from cells co-expressing TNIK and Merlin, we determined that TNIK could phosphorylate Merlin at S13, T272, S315, and T576 (Fig. 4B, Supplementary Fig. S4D, and Supplementary Table S3). The mass spectrometry analysis of Merlin isolated from cells co-expressing TNIK and Merlin also revealed phosphorylation of Merlin at S518, a site that is phosphorylated by PKA and the PAK kinases (17), S566, and T581, which were unlikely to be phosphorylated by TNIK since they were not detected by phosphopeptide mapping, nor were these sites of phosphorylation increased upon TNIK overexpression (Supplementary Fig. S4D and Supplementary Table S3). Co-expression of TNIK and Merlin in HEK 293T cells resulted in increased phosphorylation of Merlin at S13, which did not occur when the TNIK kinase-dead mutant was expressed, indicating that TNIK was directly phosphorylating S13 (Fig. 4C). This phosphorylation was lost when expressing a Merlin mutant where the serine is exchanged for an alanine (S13A; Fig. 4C), confirming its specificity. In addition, pThr-phosphorylated Merlin was largely diminished when the non-phosphorylatable T272A mutant was expressed (Supplementary Fig. S4E); however, expression of the T576A or T581A mutants did not affect global Merlin threonine phosphorylation, indicating that Merlin pT272 is the main site recognized by the pan-phospho-threonine antibody (Supplementary Fig. S4F). Phosphorylation of Merlin at S315 has been previously reported (18) but could not be verified by immunoblot due to lack of site-specific antibodies. In agreement with the above observations, molecular dynamics simulations were performed and overall indicate that TNIK-mediated phosphorylation of Merlin at the sites identified (S13, T272, S315, and T576) will impact Merlin structure and activity (Supplementary Fig. S5 and Supplementary Table S4).
Figure 4. TNIK regulates Merlin and FAK in LSCC cells.
A. Overexpressed Merlin was immunoprecipitated from HEK 293T co-expressing TNIK or a kinase-dead (K54R) mutant. Merlin phosphorylation was assessed with a pan-phospho-threonine antibody, and interaction with TNIK was analyzed by immunoblot. Where indicated, cells were treated with the corresponding concentration of NCB-0846 for 30 minutes. The data are representative of three independent experiments. B. Purified Merlin was incubated with the kinase domain of TNIK in the presence of γ[32P]-ATP, and phosphopeptide mapping was conducted. The identified phosphorylated amino acids are indicated in red. PAA, phosphoamino acid analysis. The chromatogram is representative of three independent experiments. The TNIK phosphorylation consensus motif is shown in the bottom row. Conserved residues with the consensus motif around the identified phosphosites are indicated in blue. C. Validation of Merlin pS13 by co-expression of TNIK and Merlin wild-type (WT) or the non-phosphorylatable mutant S13A in HEK 293T cells from two independent experiments. D. Validation of Merlin pS13 in LK2 and NCI-H520 cells overexpressing Merlin and treated with NCB-0846 (500 nM, 90 minutes; n = 3 independent experiments for each cell line). E. Dox-inducible (1 μg/ml, 72 h) knockdown of TNIK reduces activation of FAK in LK2 and NCI-H520 cells. The data are representative of three independent experiments. F. Reduced activation of FAK in dox-inducible (1 μg/ml, 72 h) TNIK-knockdown cells can be rescued by expression of shRNA-resistant TNIK but not of a kinase-dead (K54R) mutant. The data are representative of three independent experiments. G. Combined knockdown of Merlin (96 h) and TNIK (with doxycycline 1 μg/ml, 72 h) results in FAK inhibition in NCI-H520 cells (n = 2 independent experiments). H. Reduced activation of FAK and YAP levels in dox-inducible (1 μg/ml, 72 h) TNIK-knockdown cells can be rescued by expression (48 h) of a Merlin phosphomimetic mutant (4D; S13D/T272D/S315D/T576D) but not by expression of Merlin wild-type (WT) or a non-phosphorylatable mutant (4A; S13A/T272A/S315A/T576A). The data are representative of three independent experiments. I, J. Effect of siRNA-mediated knockdown on LK2 (I) or NCI-H520 (J) cell viability (MTS assay). Data are represented as mean value ± SD, one-way ANOVA, Dunnett’s multiple comparison post-test (relative to siRNA negative). n ≥ 3 independent experiments with triplicates. n.d.: not detected. GAPDH, tubulin or β-actin were used as loading controls. Immunoblot band intensity was quantified with ImageJ. K. Proposed model on how amplified TNIK contributes to LSCC progression. In LSCC cells, TNIK phosphorylates Merlin at S13 and S315. In addition, TNIK might phosphorylate Merlin at secondary sites (T272 and T576). Phosphorylated Merlin is required to maintain activation of the focal adhesion kinase (FAK). Additional TNIK substrates or Merlin effectors (i.e. YAP) might also contribute to maintain LSCC cell viability.
We next determined if endogenous TNIK could phosphorylate Merlin at these sites in LSCC cells. Pulldown of endogenous Merlin from NCI-H520 cells followed by mass spectrometry analysis of phosphopeptides identified phosphorylation of Merlin S13 and S315. Phosphorylation of S13 was not identified in NCB-0846-treated samples, indicating TNIK is directly phosphorylating this site in NCI-H520 cells (Supplementary Fig. S6A, S6B). Similarly, phosphorylation at S315 was reduced after TNIK inhibition (fold-change: 1.00 vs 0.34, DMSO/NCB-0846 samples; Supplementary Fig. S6C), indicating endogenous TNIK is directly phosphorylating S315 in NCI-H520 cells. Since we lack sensitive phospho-site specific antibodies that recognize endogenous Merlin, we validated phosphorylation of Merlin at S13 by endogenous TNIK in LK2 and NCI-H520 cells by overexpressing Merlin and showed that this phosphorylation was sensitive to treatment with the TNIK inhibitor NCB-0846 (Fig. 4D). However, the pan-pThr antibody was not sensitive enough to detect phosphorylation on Merlin in LK2 or NCI-H520 cells. Combined, these results demonstrate that Merlin is a direct TNIK substrate and indicate that S13 and S315 were the main TNIK-mediated Merlin phosphosites. We were unable to identify phosphorylation of endogenous Merlin T272 in the pull-down mass spec experiments, but cannot exclude that this site may be phosphorylated by endogenous TNIK.
Merlin is a scaffold protein regulated by phosphorylation and can modulate downstream effectors such as focal adhesion kinase (FAK) (16). We found that FAK activation (as determined by FAK pY397 levels) was reduced in LK2 and NCI-520 cells upon TNIK knockdown (Fig. 4E). Importantly, re-expression of catalytically active TNIK, but not a kinase-dead mutant, rescued the effects of TNIK knockdown on FAK activation in LK2 cells (Fig. 4F). In agreement with TNIK activity being required for FAK activation, treatment of LK2 and NCI-H520 cells with NCB-0846 reduced FAK phosphorylation at Y397 (Supplementary Fig. S7A). In addition, Merlin negatively regulates the pro-oncogenic YAP/TAZ transcription factors through activation of effectors of the Hippo pathway (16). Consistently with TNIK modulating Merlin, we also observed reduced YAP levels following TNIK knockdown in LK2 and NCI-H520 cells (Supplementary Fig. S7B).
These results indicate that TNIK phosphorylates Merlin and led us to hypothesize that phosphorylated Merlin is required to maintain FAK activation. To test this model, we knocked down Merlin, TNIK, and the combination of Merlin and TNIK. Knockdown of Merlin in LK2 and NCI-520 cells reduced FAK activation, which was further diminished with the combination of Merlin and TNIK knockdown (Fig. 4G, Supplementary Fig. S7A, S7C). Furthermore, we found that expression of a Merlin phosphomimetic mutant (Merlin 4D) restored FAK activation and YAP levels in NCI-H520 cells with TNIK knockdown (Fig. 4H), but a wild-type Merlin or a non-phosphorylatable mutant (Merlin 4A), did not effect FAK activation and YAP levels in cells depleted of TNIK (Fig. 4H). We also assessed if these pathways contributed to maintain cell survival; siRNA-mediated knockdown of Merlin, FAK, and YAP significantly reduced cell viability in LK2 and NCI-H520 cells (Fig. 4I, 4J, Supplementary Fig. S7D–F). In summary, our results suggest that phosphorylated Merlin is required for the maintenance of FAK activation and YAP stabilization and provides one possible mechanism by which TNIK maintains LSCC cell survival (Fig. 4K, Supplementary Movie 1).
Discussion
We have identified amplified TNIK as a promising therapeutic target in LSCC. Using a variety of functional assays, we determined that targeting TNIK significantly reduced cell viability in LSCC cell lines with high TNIK expression as evaluated by colony formation assays and cell growth on 3D matrices. Importantly, TNIK depletion had no significant effect on LSCC cells that lack TNIK amplification and have low TNIK expression. Moreover, we have shown that pharmacological targeting of TNIK with a small-molecule inhibitor, NCB-0846, significantly reduced viability of LSCC cells in vitro and in vivo, including in established LSCC PDXs. Our data indicate that NCB-0846 effects on cell viability are specifically due to catalytic inhibition of TNIK, as cell viability could be rescued by expression of an inhibitor-resistant mutant (TNIK V31W) or viability was not affected by NCB-0846 in cells with almost complete TNIK knockdown. Although TNIK was mainly overexpressed in LSCC cell lines with 3q amplification, we found a subset of LSCC cell lines (SW900 and Calu-1) that lacked TNIK amplification but displayed high TNIK levels, suggesting that other mechanisms can drive increased TNIK abundance in these cell lines. Importantly, these cells were also sensitive to TNIK depletion.
TNIK has been previously involved in the promotion of colorectal cancer, triple-negative breast cancer, prostate cancer, and chronic myelogenous leukemia (9,19–21). Mechanistically, we observed that TNIK did not mediate its effects by regulating the Wnt/β-catenin pathway, contrary to previous reports in other cancer types (14,19). We found that TNIK’s effects were in part mediated through regulation of Merlin/NF2, although the contribution of additional substrates cannot be ruled out. Merlin is a tumor suppressor protein, closely related to members of the ERM family. The ERM proteins are an evolutionarily conserved group of proteins that act as crosslinkers between membrane receptors and the actin cytoskeleton. ERM proteins are present in two conformations, a closed or inactive conformation, which involves the intramolecular association between the C-terminal and the FERM domain, and an open or active conformation in which this intramolecular association is disrupted (22,23). Activation of ERM is a multiple-step process that requires association to the plasma membrane through binding of the FERM domain to phosphatidyil-inositol (4,5)-bisphosphate (PIP2), and subsequent phosphorylation at a conserved C-terminal threonine (T567 in ezrin), that can be mediated by multiple protein kinases (22,23). Because of the structural conservation between Merlin and ERM proteins, a similar mechanism was thought to operate to regulate Merlin’s function. Indeed, phosphorylation of Merlin at the C-terminal S518 (equivalent to ezrin T567) has been proposed to modulate Merlin activity, but contrary to ERM proteins, several studies have shown that Merlin is inactivated by phosphorylation (16,17,24). Of note, LSCC cells with distal 3q amplification (such as the LK2 and NCI-H520 cell lines) also harbor amplification of the PAK2 gene, which encodes one of the kinases responsible for the phosphorylation of Merlin at S518. However, the functional relevance of S518 phosphorylation has been recently questioned using a CRISPR knock-in approach to generate the respective phosphomimetic and non-phosphorylatable Merlin S518 mutants (25). Similar to ERM proteins, Merlin binding to phosphoinositides, such as PIP2, is critical for its activity (25).
We have established that Merlin interacts with TNIK and is a direct TNIK substrate. In this study, we have identified S13 and S315 as the primary TNIK phosphorylation sites after mass spectrometry analysis of endogenous Merlin pulldowns. However, we cannot completely exclude the possibility that TNIK could activate a downstream protein kinase that would phosphorylate Merlin at these residues. Consistent with our observations, phosphorylation of endogenous Merlin at S13 and S315 has been detected in high-throughput proteomics analysis (26,27), including studies in non-small cell lung cancer (28). Using phosphopeptide mapping from in vitro kinase assays, we identified that Merlin could also be phosphorylated by TNIK at T272 and T576. Phosphorylation of T272 was also detected in mass spectrometry analysis from cells co-expressing TNIK and Merlin. We were unable to detect phosphorylation of endogenous Merlin at T272 in cells that expressed endogenous TNIK levels; however, our data does not exclude the possibility that Merlin T272 can be phosphorylated in cells. One possible explanation for lack of detection of T272 phosphorylation is that this sites might be easily accessible to the action of protein phosphatases and their identification is difficult under more stringent conditions. In addition, these sites fall on short tryptic peptides, which complicates their detection by mass spectrometry. AKT-mediated phosphorylation of Merlin at S315 has been described in previous studies (18), although there are conflicting data on which is the responsible upstream kinase of this site (29). Mechanistically, phosphorylation at this site triggered Merlin’s polyubiquitination and proteasomal degradation (18). However, we did not observe changes in Merlin levels after TNIK knockdown in LSCC cells, suggesting that this regulatory mechanism of Merlin might not be operative in the cell lines used in this study.
Knockdown of TNIK led to reduced FAK activation (measured as FAK Y397 phosphorylation) that was rescued upon expression of shRNA-resistant TNIK, but not of a kinase-dead mutant. Consistently with a requirement for catalytic activity, TNIK inhibition with NCB-0846 also reduced FAK pY397 levels. FAK is a nucleo-cytosolic non-receptor tyrosine kinase that is involved in several signaling pathways that promote tumorigenesis (30); some reports suggest that Merlin can regulate FAK activity (31). Our results show that knockdown of Merlin also reduces FAK activation in LSCC cells and that this effect is enhanced by TNIK depletion. Moreover, reduced FAK activation in NCI-H520 cells with TNIK knockdown could be rescued after expression of a Merlin phosphomimetic mutant, suggesting that FAK activation depends on expression of a phosphorylated form of the Merlin protein (Fig. 4K). In addition, siRNA-mediated depletion of Merlin or its downstream effectors significantly diminishes cell viability in LK2 and NCI-H520 cells, indicating that this is a relevant pathway in LSCC. Combined, our studies suggest that phosphorylation of Merlin by TNIK induces a conformation of Merlin that is needed to maintain FAK activation. This proposed model is supported by multiple lines of evidence. First, depletion of Merlin suppresses FAK activation (Fig. 4G, Supplementary Fig. S7A, S7C), and second expression of a phosphomimetic mutant of Merlin (4D) can rescue loss of FAK activation in cells depleted of TNIK (Fig. 4H). Interestingly, wild-type Merlin or a non-phosphorylatable Merlin mutant (4A) were unable to rescue FAK activation in cells depleted of TNIK, highlighting that negative charges at these positions in Merlin are required to maintain FAK in an active conformation. Furthermore, catalytic activity of TNIK was essential for maintaining viability of LK2 and NCI-H520 cells and TNIK catalytic inhibition led to loss of endogenous Merlin phosphorylation on S13 and S315. Overall, this provides a detailed mechanism whereby TNIK directly phosphorylates endogenous Merlin at S13 and S315 to promote FAK activation. How exactly phosphorylated Merlin sustains FAK activation will require further study.
In summary, we propose a model in which TNIK contributes to LSCC in part by modulating the Merlin-FAK signaling axis (Fig. 4K, Supplementary Movie 1). The lack of effective treatments for LSCC has been a large therapeutic gap. The success of TNIK inhibition in our in vivo models, including established LSCC PDXs, highlights our findings’ translational potential and confirms TNIK as a potential therapeutic target for a subset of LSCC patients that harbor TNIK copy number gains.
Methods
Ethical approval and patient details
LSCC tumor fragments from treatment-naïve patients were collected by the Manchester Cancer Center Biobank between January 2015 and December 2016. All patients provided written informed consent for the use of donated samples for this research. All studies were conducted in accordance with the Human Tissue Act 2004 (UK) and regulations by the National Institutes of Health (USA). All patient information has been anonymized.
Cell lines
Normal human bronchial epithelial cells (NHBE) and small airway epithelial cells (SAEC) were purchased from Lonza in February 2016. Cells were expanded for two passages and frozen. NCI-H520 (RRID:CVCL_1566), SW900 (RRID:CVCL_1731), and HEK 293T (RRID:CVCL_0063) cells were obtained from American Type Culture Collection (ATCC). LK2 (RRID:CVCL_1377), KNS-62 (RRID:CVCL_1335), and Lc-1-sq (RRID:CVCL_3008) were obtained from the Japanese Collection of Research Bioresources (JCBR) in February 2015. LUDLU-1 (RRID:CVCL_2582) cells were obtained from the European Collection of Authenticated Cell Cultures (ECACC) General Cell Collection. Calu-1 (RRID:CVCL_0608) cells were kindly donated by Professor R. Marais (Cancer Research UK [CRUK]-Manchester Institute). HCC15 (RRID:CVCL_2057), HCC95 (RRID:CVCL_5137), and NCI-H157 (RRID:CVCL_0463) cells were kindly donated by Professor P. Massion (Vanderbilt University) in July 2017. NCI-H1703 (RRID:CVCL_1490) and SK-MES-1 (RRID:CVCL_0630) cells were kindly donated by Dr. D.K. Morrison (National Cancer Institute) in March 2017. All cell lines were verified by Short Tandem Repeat profiling upon receipt (CRUK Manchester Institute [cell lines obtained between 2015 and 2016] or Frederick National Laboratory for Cancer Research [cell lines obtained in or after 2017]). Cells were expanded (one to two passages) upon receipt, and several vials were frozen. All cell lines were maintained at 5% CO2 at 37 °C and cultured as recommended by the vendor. In brief, NHBE and SAEC were cultured in BEGM (Lonza, cat. CC-3170) or SAGM (Lonza, cat. CC-3118), respectively. LK2, NCI-H520, SW900, LUDLU-1, HCC15, HCC95, NCI-H157, NCI-H1703, and SK-MES-1 were cultured in RPMI 1640 (Quality Biological, cat. 112–024-101) + 10% fetal bovine serum (FBS, Atlanta Biologicals) + GlutaMAX (Gibco, cat. 35050061) + 1% penicillin-streptomycin (pen-strep; Gibco, cat. 15140122). KNS-62 were maintained in MEM (Gibco, cat. 11090099) + 20% FBS + GlutaMAX + 1% pen-strep. Lc-1-sq were maintained in DMEM:F12 (Gibco, cat. 10565018) + 10% FBS + GlutaMAX + 1% pen-strep. HEK 293T and Calu-1 were cultured in DMEM (Gibco, cat. 11995073) + 10% FBS + GlutaMAX + 1% pen-strep. Cells were tested for absence of Mycoplasma contamination by PCR every three months using the One-step Quickcolor Mycoplasma Detection Kit (Clark Bioscience, cat. T102).
RT-qPCR
The Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems; Thermo Scientific cat. 4389986) was used for RT-qPCR. The primers used are listed in Supplementary Table S5. Samples were run on an ABI 7900HT real-time PCR instrument with the following program: reverse transcription (30 minutes, 48 °C), activation of DNA polymerase (10 minutes, 95 °C), PCR amplification (40 cycles of denaturing at 95 °C for 15 sec and annealing/extension at 60 °C for one minute). SDS software was used to compute CT values.
Cell lysis, immunoprecipitation, and immunoblots
After appropriate treatment time, whole-cell extracts were prepared by lysing the cells on ice in RIPA (Sigma-Aldrich, cat. R0278) lysis buffer supplemented with EDTA-free protease inhibitors (Roche, cat. 05 056 489 001) and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich, cat. P2850 and P5726). Lysates were cleared (15,000 × g, 15 minutes, 4 °C), after which protein concentration was determined with the Pierce™ 660nm Protein Assay Reagent (Thermo Scientific, cat. 22660). Lysates containing equivalent amounts of protein were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) in 4–20% gradient gels (Bio-Rad) and transferred into low fluorescence polyvinylidene fluoride membranes.
For immunoprecipitation, cells were lysed in 1% Triton X-100 lysis buffer (Cell Signaling Technology, #9803) supplemented with protease inhibitors after appropriate treatment. Lysates (1 mg/ml) were incubated with corresponding antibody overnight (4 °C) in a rocking platform followed by incubation with Protein G magnetic beads (Thermo Scientific, cat. 10004D). Immunoprecipitates were washed five times with lysis buffer, denatured in 2× Laemmli buffer (15 minutes, 100 °C), and analyzed by immunoblot together with equalized lysates. Band signal intensity was quantified using ImageJ. All samples loaded in a corresponding gel derive from the same experiment and were processed in parallel. Uncropped blots are available in Supplementary Fig. S8.
Antibodies against TNIK (#32712, 1:1000, RRID:AB_2799027), pFAK Y397 (D20B1) (#8556, 1:1000), FAK (#3285, 1:1000, RRID:AB_2269034), Merlin (D3S3W) (#12888, 1:2000, RRID:AB_2650551), Non-phospho (Active) β-Catenin (Ser45) (D2U8Y) (#19807, 1:1000, RRID:AB_2650576), β-catenin (#9562, 1:1000, RRID:AB_331149), p-Thr (#9381, 1:1000, RRID:AB_330301), V5-Tag (D3H8Q) (#13202, IP: 1:100, RRID:AB_2687461), HA-Tag (6E2) (#2367, 1:2000, RRID:AB_10691311), GAPDH (14C10) (#2118, 1:2000, RRID:AB_561053), β-actin (8H10D10) (#3700, 1:5000, RRID:AB_2242334), Tubulin (DM1A) (#3873, 1:5000, RRID:AB_1904178), and anti-rat IgG HRP-linked Antibody (#7077, 1:3000, RRID:AB_10694715) were purchased from Cell Signaling Technology. Antibody against TNIK (GTX13141, 1:1000, RRID:AB_383061) was purchased from GeneTex. Antibody against YAP/TAZ (63.7) was purchased from Santa Cruz Biotechnology (sc101199, 1:2000, RRID:AB_1131430). Antibody against Merlin pS13 (cat. 13146; 1:1000) was from Signalway Antibody. Anti-FLAG (L5) antibody was from Biolegend (cat. 637301, 1:5000, RRID:AB_1134266). Anti-V5 Tag (SV5-Pk1) was from BIO-Rad (cat. MCA1360, IB: 1:2000, RRID:AB_322378). HRP-linked anti-mouse (cat. NA931, 1:3000, RRID:AB_772210) and anti-rabbit (cat. NA934, 1:3000, RRID:AB_772206) antibodies were purchased from Global Life Sciences Solutions.
Phosphorylation site mapping
pcDNA3-FLAG-Merlin (Addgene #11623, kindly donated by Professor V. Ramesh [Massachusetts General Hospital]) was transfected in HEK 293T cells (1 × 106 cells in 60-mm plates) with jetPRIME according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were treated with staurosporine and NCB-0846 (1 μM each inhibitor, one hour), after which cells were lysed and Merlin was immunoprecipitated as described above. Immunoprecipitates were washed with lysis buffer and kinase buffer and subjected to in vitro kinase assays in a final volume of 30 μL (containing 5 μM ATP, 0.5 μM γ[32P]-ATP [Perkin Elmer Life Sciences, cat. BLU502], and 50 ng of TNIK-kinase domain [Carna Biosciences, cat. 07–438-20N]) for 30 minutes at 30 °C. NCB-0846 was added at a final concentration of 5 μM as control of TNIK inhibition. Kinase assays were terminated by the addition of Laemmli buffer (2× final concentration), followed by denaturation (five minutes, 100 °C). Samples were separated by SDS-PAGE in 8.5% gels and transferred onto nitrocellulose membrane. Radiolabeled FLAG-Merlin protein was eluted from the membrane and digested with trypsin. The resulting peptides were separated by reverse-phase high-pressure liquid chromatography, and phosphopeptides were subjected to phosphoamino acid analysis and Edman degradation as previously described (32).
Mouse experiments
Frederick National Laboratory for Cancer Research is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (33). All animal studies were approved by the Animal Care and Use Committee of the National Cancer Institute at Frederick. For details, see Supplementary Methods.
Statistical analysis
Data are expressed as mean ± standard deviation (SD) or ± standard error of the mean (SEM), as indicated in the corresponding figure legend. Statistical significance was determined using Student’s t-test (with Welch’s correction when variance between groups was significantly different as analyzed with F-test), Mann–Whitney U test, one-way ANOVA, or two-way ANOVA as indicated in the corresponding figure legends. Statistical analyses were performed in Prism 8 (GraphPad Software, RRID:SCR_002798). Significance was set at p = 0.05.
Supplementary Material
Significance.
Targeted therapies have not yet been approved for the treatment of LSCC, due to lack of identification of actionable cancer drivers. We define TNIK catalytic activity as essential for maintaining LSCC viability and validate the anti-tumor efficacy of TNIK inhibition in pre-clinical models of LSCC.
Acknowledgments
We thank the patients and their families for providing samples for this research, as well as the Manchester Cancer Research Centre Biobank (Manchester, UK) and Dr. R. Booton (University Hospital of South Manchester, Manchester, UK) for logistical support to provide tissue samples for the generation of LSCC PDX. We thank Dr. E.W. Trotter (CRUK-Manchester Institute, Manchester, UK) for lab logistics. We thank members of the Laboratory of Cell and Developmental Signaling for helpful discussions.
This research was mainly funded by the National Cancer Institute, project number 1ZIABC011691-01 (J.B.). Additional funding came from a Fundacion Ramon Areces postdoctoral fellowship (P.T-A.), the Lung Cancer Research Foundation (J.B. and P.T-A.), and National Institute for General Medical Sciences R01 GM102262 (B.E.T.). This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Financial support: This research was mainly funded by the National Cancer Institute, project number 1ZIABC011691-01 (J.B.). Additional funding came from a Fundacion Ramon Areces postdoctoral fellowship (P.T-A.), the Lung Cancer Research Foundation (J.B. and P.T-A.), and National Institute for General Medical Sciences R01 GM102262 (B.E.T.). This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
References
- 1.Gold KA, Wistuba II, Kim ES. New strategies in squamous cell carcinoma of the lung: identification of tumor drivers to personalize therapy. Clin Cancer Res 2012;18(11):3002–7 doi 10.1158/1078-0432.CCR-11-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandara DR, Hammerman PS, Sos ML, Lara PN Jr., Hirsch FR. Squamous cell lung cancer: from tumor genomics to cancer therapeutics. Clin Cancer Res 2015;21(10):2236–43 doi 10.1158/1078-0432.CCR-14-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25 doi 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields AP, Justilien V, Murray NR. The chromosome 3q26 OncCassette: A multigenic driver of human cancer. Adv Biol Regul 2016;60:47–63 doi 10.1016/j.jbior.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazumdar T, Byers LA, Ng PK, Mills GB, Peng S, Diao L, et al. A comprehensive evaluation of biomarkers predictive of response to PI3K inhibitors and of resistance mechanisms in head and neck squamous cell carcinoma. Mol Cancer Ther 2014;13(11):2738–50 doi 10.1158/1535-7163.MCT-13-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaacsson Velho PH, Castro G Jr., Chung CH. Targeting the PI3K Pathway in Head and Neck Squamous Cell Carcinoma. Am Soc Clin Oncol Educ Book 2015:123–8 doi 10.14694/EdBook_AM.2015.35.123. [DOI] [PubMed] [Google Scholar]
- 7.Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 2014;25(2):139–51 doi 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagerstrand D, Tong A, Schumacher SE, Ilic N, Shen RR, Cheung HW, et al. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Discov 2013;3(9):1044–57 doi 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda M, Uno Y, Ohbayashi N, Ohata H, Mimata A, Kukimoto-Niino M, et al. TNIK inhibition abrogates colorectal cancer stemness. Nat Commun 2016;7:12586 doi 10.1038/ncomms12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey FP, Andreev VI, Eyers PA. The resistance tetrad: amino acid hotspots for kinome-wide exploitation of drug-resistant protein kinase alleles. Methods Enzymol 2014;548:117–46 doi 10.1016/B978-0-12-397918-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 11.Persky NS, Hernandez D, Do Carmo M, Brenan L, Cohen O, Kitajima S, et al. Defining the landscape of ATP-competitive inhibitor resistance residues in protein kinases. Nat Struct Mol Biol 2020;27(1):92–104 doi 10.1038/s41594-019-0358-z. [DOI] [PubMed] [Google Scholar]
- 12.Izumchenko E, Paz K, Ciznadija D, Sloma I, Katz A, Vasquez-Dunddel D, et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol 2017;28(10):2595–605 doi 10.1093/annonc/mdx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S, et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J 2009;28(21):3329–40 doi 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shitashige M, Satow R, Jigami T, Aoki K, Honda K, Shibata T, et al. Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res 2010;70(12):5024–33 doi 10.1158/0008-5472.CAN-10-0306. [DOI] [PubMed] [Google Scholar]
- 15.Miller CJ, Lou HJ, Simpson C, van de Kooij B, Ha BH, Fisher OS, et al. Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output. PLoS Biol 2019;17(3):e2006540 doi 10.1371/journal.pbio.2006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett 2014;588(16):2743–52 doi 10.1016/j.febslet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 2016;35(5):537–48 doi 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, et al. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat Cell Biol 2007;9(10):1199–207 doi 10.1038/ncb1641. [DOI] [PubMed] [Google Scholar]
- 19.Schurch C, Riether C, Matter MS, Tzankov A, Ochsenbein AF. CD27 signaling on chronic myelogenous leukemia stem cells activates Wnt target genes and promotes disease progression. J Clin Invest 2012;122(2):624–38 doi 10.1172/JCI45977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Lim SK, Liang X, Lim YP. The transcriptional coactivator WBP2 primes triple-negative breast cancer cells for responses to Wnt signaling via the JNK/Jun kinase pathway. J Biol Chem 2018;293(52):20014–28 doi 10.1074/jbc.RA118.005796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RS, Zhang L, Berger A, Lawrence MG, Song J, Niranjan B, et al. Characterization of the ERG-regulated Kinome in Prostate Cancer Identifies TNIK as a Potential Therapeutic Target. Neoplasia 2019;21(4):389–400 doi 10.1016/j.neo.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol 2002;3(8):586–99 doi 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 23.Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci 2015;128(6):1253 doi 10.1242/jcs.170027. [DOI] [PubMed] [Google Scholar]
- 24.Sher I, Hanemann CO, Karplus PA, Bretscher A. The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Dev Cell 2012;22(4):703–5 doi 10.1016/j.devcel.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong AW, Meng Z, Plouffe SW, Lin Z, Zhang M, Guan KL. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev 2020;34(7–8):511–25 doi 10.1101/gad.333435.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, et al. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J Proteome Res 2013;12(6):2414–21 doi 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res 2013;12(1):260–71 doi 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 28.Schweppe DK, Rigas JR, Gerber SA. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J Proteomics 2013;91:286–96 doi 10.1016/j.jprot.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laulajainen M, Muranen T, Nyman TA, Carpen O, Gronholm M. Multistep phosphorylation by oncogenic kinases enhances the degradation of the NF2 tumor suppressor merlin. Neoplasia 2011;13(7):643–52 doi 10.1593/neo.11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer 2005;5(7):505–15 doi 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 31.Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene 2006;25(44):5960–8 doi 10.1038/sj.onc.1209587. [DOI] [PubMed] [Google Scholar]
- 32.Morrison DK, Heidecker G, Rapp UR, Copeland TD. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem 1993;268(23):17309–16. [PubMed] [Google Scholar]
- 33.In: 8th, editor. Guide for the Care and Use of Laboratory Animals, The National Academies Collection: Reports funded by National Institutes of Health. Washington (DC): National Academies Press (US). 2011. [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.