Abstract
In nutrient-poor conditions, autophagy buffers metabolic stress and counteracts the effects of chemotherapy and radiation on cancer cells, which depend on autophagy for survival. However, clinical trials targeting autophagy have failed to produce successful anti-cancer treatments using currently available inhibitors. Recent studies have shown that PIKfyve kinase inhibitors disrupt lysosome function in autophagy and can selectively kill certain cancer cells. Analysis of biochemical changes caused by PIKfyve inhibition revealed that resistant cells contain significantly higher levels of cellular p38MAPK protein and phosphorylation. Expression of the lysosomal LAMP2 protein carrying phosphomimetic mutations of the p38MAPK phosphorylation sites prevented all effects caused by PIKfyve inhibition-induced lysosome dysfunction. Thus, the activation of p38MAPK in response to PIKfyve inhibition revealed a novel compensatory role in maintaining lysosome function in autophagy. The functional cooperation between the cellular PIKfyve and p38MAPK pathways in regulating lysosome homeostasis was especially important in cancer cells. Combined inhibition of PIKfyve and p38MAPK activities synergistically blocked autophagy-mediated protein degradation, prevented cathepsin maturation, and markedly reduced the viability of multiple cancer cell types without affecting the viability of normal cells. Furthermore, combined PIKfyve and p38MAPK inhibitors synergistically reduced tumor growth in mice bearing xenografts of human colorectal adenocarcinoma, suggesting a novel way to target cancer cells by prolonged inhibition of autophagy using lower drug concentrations.
Keywords: colorectal cancer, combination chemotherapy, autophagy, PIKfyve, p38MAPK, LC3, p62/SQSTM1, LAMP2, Cathepsin D
INTRODUCTION
Macroautophagy, or simply autophagy, is a complex cellular system that degrades damaged or unnecessary cellular organelles and proteins (1). Targeted components are initially engulfed into autophagosomes, which then fuse with lysosomes to become autolysosomes (2), where the sequestered material is degraded to basic components that are re-used for macromolecular synthesis and energy (3). This recycling system is particularly important during starvation to buffer against metabolic stress. Cancer cells growing in low-nutrient conditions, commonly found in the tumor microenvironment, become autophagy-dependent for survival (4,5). Furthermore, autophagy is frequently activated in tumor cells during chemotherapy or radiation (6). Inhibition of autophagy in such cancer cells led to cell death and tumor regression in a mouse model of lymphoma (7,8). However, no successful clinical application in cancer treatment has been identified so far using chloroquine derivatives, the only clinically-approved autophagy inhibitors (9).
Recent studies have demonstrated that autophagy can be inhibited using PIKfyve kinase inhibitors (10). PIKfyve phosphorylates the 5-site of the lipid signaling molecule phosphatidylinositol, involved in the control of membrane trafficking (11). Newly-discovered chemical inhibitors demonstrate that PIKfyve activity is essential for lysosome fission and heterotypic fusion with autophagosomes to form autolysosomes (12,13). Consequently, PIKfyve inhibition leads to accumulation of enlarged lysosomes and autophagy disruption. PIKfyve inhibitors have been shown to be cytotoxic against certain cancer cells in vitro and in vivo (12,14,15).
To improve the therapeutic potential of PIKfyve inhibitors, we sought to investigate the mechanisms of cellular sensitivity to these inhibitors. The results revealed that the p38 mitogen-activated protein kinase (p38MAPK) stress response pathway activity modulates the autophagy blockage caused by PIKfyve inhibition. p38MAPKs are evolutionarily conserved serine/threonine protein kinases that are activated by cellular stress to induce compensatory biochemical changes (16). They may suppress (17) or promote (18) tumorigenesis and play an important role in the balance of growth and death signals in response to therapy-induced cellular stress (19).
The studies described here demonstrate that higher basal p38MAPK activity in normal cells protects them from PIKfyve inhibition-induced autophagy blockage. Moreover, p38MAPK was activated in response to PIKfyve inhibition, suggesting a compensatory function since the induced lysosome dysfunction effects could be alleviated by expression of the lysosomal LAMP2 protein carrying phosphomimetic mutations of the p38MAPK phosphorylation sites. Accordingly, inhibition of p38MAPK stress response dramatically and synergistically exacerbated PIKfyve inhibition-induced autophagy blockage and reduced viability of autophagy-dependent cancer cells. The combined inhibition of PIKfyve and p38MAPK synergistically killed cancer cells with diverse tissues of origin and genetic backgrounds in vitro, and in vivo in a xenograft model with colorectal adenocarcinoma cells.
MATERIALS & METHODS
Cell Culture
Cell lines A375 (ATCC Cat# CRL-1619, RRID: CVCL_0132), A549 (ATCC Cat# CCL-185, RRID:CVCL_0023), BJ (ATCC Cat# CRL-2522, RRID:CVCL_3653), CCD-841CoN (ATCC Cat# CRL-1790, RRID:CVCL_2871), Daudi (ATCC Cat# CCL-213, RRID:CVCL_0008), G361 (ATCC Cat# CRL-1424, RRID:CVCL_1220), HCT116 (ATCC Cat# CCL-247, RRID:CVCL_0291), HeLa (ATCC Cat# CCL-2, RRID:CVCL_0030), Hs27 (ATCC Cat# CRL-1634, RRID:CVCL_0335), IMR90 (ATCC Cat# CCL-186, RRID:CVCL_0347), MRC5 (ATCC Cat# CCL-171, RRID:CVCL_0440), OVCAR3 (ATCC Cat# HTB-161, RRID:CVCL_0465), SHP77 (ATCC Cat# CRL-2195, RRID:CVCL_1693), SW480 (ATCC Cat# CCL-228, RRID:CVCL_0546), U2OS (ATCC Cat# HTB-96, RRID:CVCL_0042), VMM39 (ATCC Cat# CRL-3230, RRID:CVCL_A739), WI38 (ATCC Cat# CCL-75, RRID:CVCL_0579), HCT116(TP53-) (provided by Dr. Bert Vogelstein)] were cultured in high glucose DMEM medium supplemented with L-glutamine, sodium pyruvate and 10% fetal calf serum at 37°C with 5% CO2. FHC (ATCC Cat# CRL-1831, RRID:CVCL_3688) and MCF10A (ATCC Cat# CRL-10317, RRID:CVCL_0598) cells were cultured in Mammary Epithelial Growth Medium (Lonza Cat# CC-3150). SW480, A549, U2OS, HCT116 and Hs27 were most recently authenticated in 2019 by short tandem repeat profiling (performed by ATCC, RRID:SCR_001672). All cell lines were passed for fewer than 10 passages following recovery from the frozen vials, regularly authenticated by Western blots analysis, growth, and morphology observation, and stained with DAPI (Sigma Cat# D8417) to rule out Mycoplasma contamination.
Expression of recombinant proteins
Full-length human PIKfyve and LAMP2 open reading frames were cloned by RT-PCR, and mutations were generated using PCR. Cells were transduced with viral particles of retroviral vector pOP (20) constructed to direct the expression of wild-type or mutant proteins fused with Flag and HA-tag peptides at the N-terminus. Stable expressing clones were selected using 2 μg/ml puromycin (Sigma Cat# P8833). For transient expression of EGFP-p38MAPK fusion protein, cells were transfected with a mix of expression plasmid (Addgene plasmid #86832; RRID:Addgene_86832) and empty pEGFP-C1 plasmid to maintain the same DNA amount using FuGENE6 (Promega) per manufacturer’s instructions.
Chemical Inhibitors
Vacuolin-1 (#20425), YM201636 (#13576), Apilimod (#19094), SB202190 (#10010399), BIRB-796 (#10460), Skepinone-L (#16974), LY2228820 (#23259), Tak-715 (#26170) from Cayman Chemical, and WX8 and NDF from SPECS were dissolved in DMSO (Sigma Cat# D2650) at 20 mM stock concentration and diluted as necessary to be added to cell culture medium at 1:1,000 dilution. An equal volume of DMSO (vehicle) was added to control cells.
Immunoblotting
Cells were harvested by trypsinization, combined with the unattached cells, lysed in SDS sample buffer, and subjected to electrophoresis and transfer to Nitrocellulose membrane using NuPAGE gel system (ThermoScientific). The membranes were probed with specific primary antibodies and peroxidase-linked secondary antibodies (Amersham Biosciences), the signals visualized using West-DURA chemiluminescence kit (ThermoScientific) and quantified by densitometry of low exposure images. Protein molecular weight was determined using Mark12 protein standards (ThermoScientific). Primary antibodies were: Caspase-3 (Cell Signaling Technology Cat# 9661, RRID:AB_2341188), LC3 (Cell Signaling Technology Cat# 12741, RRID: AB_2617131, 1:1,000), p62 (Cell Signaling Technology Cat# 5114, RRID:AB_10624872, 1:1,000), MKK3/6-p (Cell Signaling Technology Cat# 12280, RRID:AB_2797868, 1:1,000), HSP27-p (Cell Signaling Technology Cat# 12741, RRID: AB_2617131, 1:1,000), p38MAPK-p (Cell Signaling Technology Cat# 9211, RRID:AB_331641, 1:1,000), p38MAPK (Cell Signaling Technology Cat# 9212, RRID:AB_330713, 1:1,000), Cathepsin B (Cell Signaling Technology Cat# 31718, RRID:AB_2687580, 1:1,000), H2AX-p (Cell Signaling Technology Cat# 2577, RRID:AB_2118010, 1:1,000), TFEB (Cell Signaling Technology Cat# 4240, RRID:AB_11220225, 1:1,000), Ki-67 (Cell Signaling Technology Cat# 9449, RRID:AB_2715512, 1:1,000), p53 (Abcam Cat# ab1101, RRID:AB_297667), p53-p (Cell Signaling Technology Cat# 9284, RRID:AB_331464), PARP (Cell Signaling Technology Cat# 9542, RRID:AB_2160739), PIKfyve (Santa Cruz Biotechnology Cat# sc-100408, RRID:AB_2164550, 1:500), LAMP2 (Abcam Cat# ab25631, RRID:AB_470709, 1:500), Cathepsin D (Abcam Cat# ab75852, RRID:AB_1523267, 1:6,000). Ponceau S (Sigma Cat# P7170) staining was used to quantify histone proteins to correct for sample loading differences.
siRNA Transfection
Cells were seeded in 6-well plates, transfected with siRNA one day later using RNAiMAX (ThermoScientific) per manufacturer’s instructions. All cells were collected 3 days after transfection. p38MAPK (GAAGCUCUCCAGACCAUUUtt and GUCCAUCAUUCAUGCGAAAtt) and control (CGUACGCGGAAUACUUCGAtt) siRNA sequences were reported (21,22).
Quantification of cell proliferation and survival
Cells were seeded routinely in 6-well plates at a density of 7×104 cells/well. One day after plating, the medium was replaced with fresh medium containing the indicated inhibitor at the indicated concentration(s), and the cells were then cultured for 3 days unless otherwise indicated. Cells were harvested routinely by trypsinization, combined with the unattached cells, and counted using a hemocytometer to determine the total number of cells in each well. Cells (1×105) were analyzed using a FACSCalibur flow cytometer (BD FACSCalibur Flow Cytometry System, RRID:SCR_000401), as described (23), and the data analyzed using FCS Express software (FCS Express, RRID:SCR_016431). The number of viable cells was determined by subtracting the number of cells with <2N DNA content considered as dead. Cell divisions per day were calculated as a binary logarithm of the fold increase in cell number.
Statistical analysis.
Average values, error bars, P-value cutoffs, number of replicates, and statistical tests are identified in the corresponding figure legends. Replicates represent separate treatments of cultures from the original stock unless otherwise specified. The statistical significance of results of two samples was assessed using Student’s paired two-tailed t test.
Chou-Talalay Combination Index
Results from drug dosage matrices were analyzed using CompuSyn software (http://www.combosyn.com) to generate Chou-Talalay Combination Indexes (24).
Mouse xenograft experiments
Outbred athymic nude mice (#007850) were purchased from Jackson Laboratory. All procedures were performed under general anesthesia with isoflurane. Mice were injected subcutaneously on their rear flanks with 1 million SW480 cells in 100 μL 1:1 Matrigel/PBS (Corning). Tumors were allowed to establish for ten days and then the mice were randomly separated into four groups with similar average tumor sizes. Groups received (1) DMSO, (2) 20 mg/kg WX8, (3) 12.5 mg/kg SB202190, and (4) 20 mg/kg WX8 + 12.5 mg/kg SB202190, adjusted for individual animal weight. Injections were prepared by mixing 6 ul DMSO or drugs dissolved in DMSO with 150 μL sunflower seed oil and injected into the intraperitoneal cavity. Tumor volume measured using caliper and weights calculated as (Length x Width x Width)/2 were recorded daily. All procedures were conducted under protocols approved by the NICHD Animal Care and Use Committee.
Histopathology and Immunohistochemistry
Tumor specimens were fixed in 10% buffered formalin (Sigma Cat# HT501128). For histopathological analysis, tumor tissues were embedded in paraffin, sectioned and stained with Hematoxylin and Eosin, or with anti-Ki-67 antibody and Hematoxylin (performed by Histoserv, Inc., Gaithersburg, MD). Ki-67 proliferation index was assessed by counting at least 1000 cells and was reported as percentage positive cells.
RESULTS
Cancer Cells are More Sensitive to PIKfyve Inhibition Than Normal Cells
PIKfyve inhibition affects human cell survival to varying degrees (14). To identify a model system for studying PIKfyve inhibition, human cell lines were tested for their sensitivity to WX8, characterized previously as a novel specific inhibitor of PIKfyve (12). Cell survival in response to WX8 concentrations ranging from 0.005 to 10 μM was measured after 3 days of treatment (Figs. 1A, S1A). The number of cells in each well was counted, and an aliquot was analyzed by fluorescence-activated cell sorting (FACS) to identify dead cells. Cell death can be initiated through different mechanisms that eventually lead to cellular and nuclear lysis and DNA degradation (25). Thus, cells with less than 2N DNA content were considered dead and removed from the total number to obtain the number of viable cells. The tested 20 lines could be separated into two main groups. The 12 cancer lines demonstrated relatively high sensitivity to WX8, with median IC50 (concentration that cause 50% reduction in cell survival) of 0.2 μM. The 8 normal fibroblast cell lines were relatively resistant, with median IC50 value of 2.7 μM.
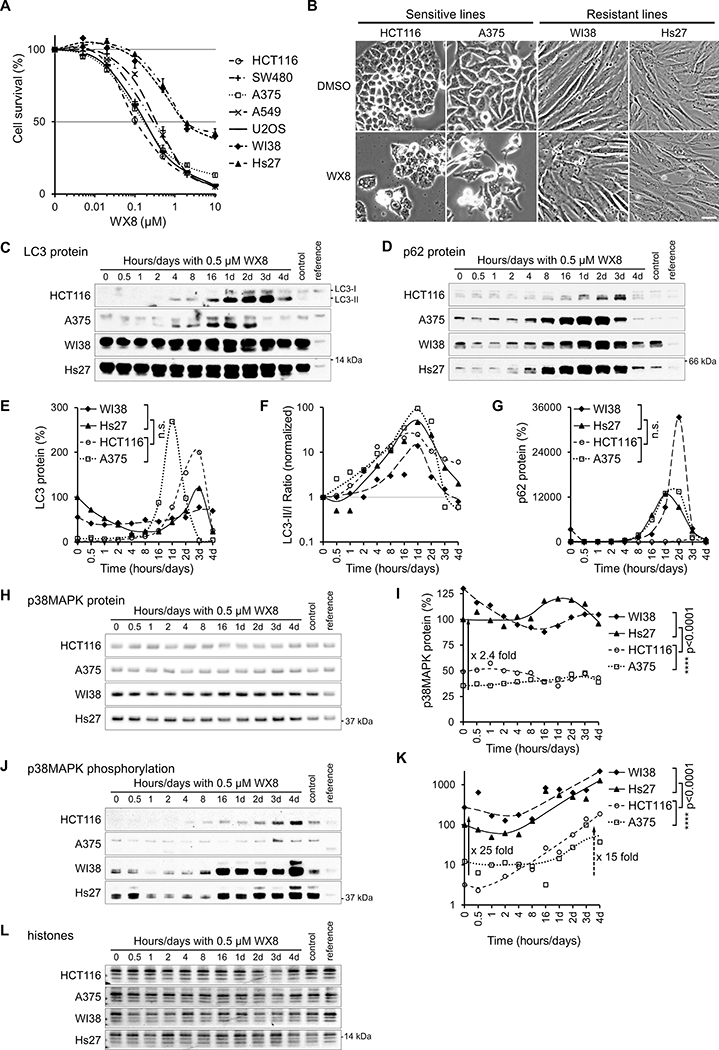
Figure 1. Biochemical differences between PIKfyve inhibitor sensitive and resistant cells.
(A) Cells were cultured for 3 days in the presence of the indicated concentration of WX8, collected, counted and analyzed by FACS. The number of viable cells for each treatment was presented as a percentage of the vehicle-treated viable cells “Cell Survival (%)”. The vehicle was set to 0.001 μM instead of 0 μM. Values are mean ±SEM (n = 3). Only results with 7 cell lines are plotted, the full set of 20 lines can be found in supplemental figure S1.
(B) Sensitive (HCT116 and A375) and resistant (WI38 and Hs27) cell lines were cultured with 0.5 μM WX8, collected at indicated times, and imaged on day 3. Scale bar: 20 μm. Total cell lysates were assayed for (C) LC3 (H), p62 (D), p38MAPK (H) and p38MAPK phosphorylation (J) by immunoblot. Samples of vehicle-treated cells (control) and U2OS cells (reference) were included to compare protein levels between cell lines. (E,F,G,I,K) The proteins were quantified, normalized by sample histone content (L), and results plotted as a percentage of the protein present at time zero. ns nonsignificant p >0.05; **** p<0.0001 (Student t-test).
Low p38MAPK Phosphorylation Levels Correlate with High Sensitivity to PIKfyve and p38MAPK Inhibition
We hypothesized that differences in cellular biochemistry determine sensitivity to PIKfyve inhibition and compared the dynamic of protein changes induced by a selective concentration of WX8 to identify potential differences. To eliminate unique genetic background effects, two sensitive (HCT116 colon carcinoma and A375 melanoma) and two resistant (WI38 lung and Hs27 skin fibroblasts) cell lines, which differ approximately 16-fold in sensitivity to WX8 (Fig. 1A,B), were analyzed. We first searched for differences in autophagy. The conversion of microtubule-associated protein 1 light chain, LC3-I, to the faster migrating form, LC3-II, has been used to monitor the abundance of cellular autophagosomes. p62 (SQSTM1) is an ubiquitin binding protein that is normally degraded in the autolysosome but accumulates when autophagy is inhibited (26). Total LC3 and the LC3-II/I ratio increased in all four cell lines after treatment with WX8, consistent with formation and/or accumulation of active autophagosomes (Fig. 1C–G). Concomitant p62 accumulation was consistent with the amassing of autophagosomes, but pointed to a blockage of autophagy-mediated protein degradation rather than activation.
Multiple major cellular pathways, including autophagy regulators and effectors, cellular growth and differentiation, cell cycle control, DNA damage, stress response and apoptosis, were surveyed to identify trends explaining the differential sensitivity to PIKfyve inhibition by WX8. The p38MAPK cellular stress response pathway emerged as the only significant and consistent difference between sensitive and resistant cell lines. The tested resistant cells contained on average 2.4-fold more total cellular p38MAPK protein, which was phosphorylated at 25-fold higher levels at the activating T180/Y182 site (27) (Fig. 1H–L). p38MAPK phosphorylation increased on average 15-fold, suggesting that the p38MAPK pathway was activated in response to PIKfyve inhibition. Consequently, we determined the survival of all cell lines in response to concentrations ranging from 0.1 to 40 μM of the specific p38MAPK inhibitor SB202190 (28) (Fig. S1B).
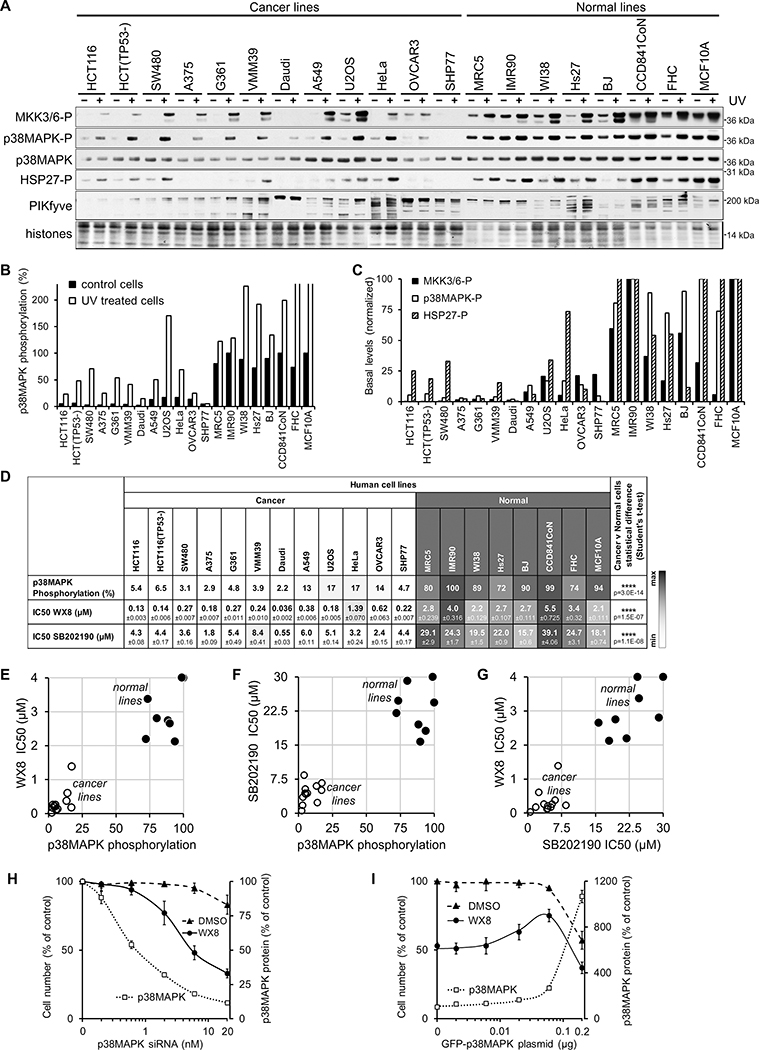
To determine if the p38MAPK pathway was defective or less active in sensitive cancer cell lines, the set of 20 lines tested initially was analyzed further under normal conditions and under stress. The p38MAPK pathway is a cascade of kinases that relays extracellular stress signals to downstream effector proteins (27). In response to UV-induced DNA damage, as indicated by histone H2AX phosphorylation, the p38MAPK cascade kinases were activated in all cell lines. Specifically, p38MAPK as well as MKK3/6, the upstream protein kinases that activate p38MAPK by T180/Y182 phosphorylation, and HSP27, a downstream target of p38MAPK, were all hyperphosphorylated (Fig. 2A–C). While the p38MAPK stress response was functional in all lines, the WX8-resistant lines had consistently higher steady-state levels of p38MAPK protein and phosphorylation (Fig. 2D) suggesting a protective function of p38MAPK activity.
Figure 2. p38MAPK phosphorylation inversely correlates with sensitivity to PIKfyve inhibition.
(A) Immunoblot analysis of MKK3/6, p38MAPK, HSP27 and PIKfyve proteins amount and/or phosphorylation in exponentially growing and UV-irradiated cells (30 minutes after 80 Joules) from 20 human lines. (B) Quantification of the basal and UV-induced p38MAPK phosphorylation level presented as a percentage of the level in IMR90 cells after normalization by histone content. (C) Quantification of the basal levels of MKK3/6, p38MAPK, and HSP27 phosphorylation presented as a percentage of the level in IMR90 cells after normalization by histone content.
(D) Calculated IC50 concentrations (50% reduction in survival) of WX8 and SB202190 (Fig. S1), and p38MAPK basal phosphorylation levels for 20 cell lines. (E) Plot of the p38MAPK basal phosphorylation levels versus WX8 IC50 values. (F) Plot of the p38MAPK basal phosphorylation levels versus SB202190 IC50 values. (G) Plot of WX8 versus SB202190 IC50 values.
(H) p38MAPK was depleted in MCF10A cells with the indicated concentration of p38MAPK siRNA or control siRNA and incubated with 0.5 μM WX8 or DMSO. Cells were imaged and collected after 3 days, and the counts plotted as a percentage of the control siRNA-treated cells. The amount of p38MAPK protein determined by immunoblot analysis (Fig. S3), quantified and the results presented as a percentage of the vehicle-treated cells. Values are mean ±SEM (n = 3).
(I) GFP-p38MAPK fusion protein and/or GFP was expressed transiently in U2OS cells by transfection with 2 μg plasmid. One day later, the media was replaced with fresh containing 0.5 μM WX8 or DMSO. Cells were collected after 3 days and the counts plotted as a percentage of vehicle-treated GFP transfected cells. The amount of p38MAPK protein was determined by immunoblot analysis (Fig. S3), quantified and the results presented as a percentage of the vehicle-treated GFP transfected cells. Values are mean ±SEM (n = 3).
Sensitivity to WX8 did not correlate with p62, PIKfyve or TFEB expression (Figs. 2A, S2A). While TFEB was a previously reported driver of sensitivity to the PIKfyve inhibitor Apilimod, only Daudi B-cell lymphoma expressed ultra-high levels of the protein, indicating it could not account for the sensitivity of other cancer lines (14). In addition, no correlation was observed between PIKfyve inhibitor sensitivity and cellular protein and phosphorylation levels of the other two major MAPK stress response signaling pathways, JNK and p44/p42 (Erk1/2), supporting specific involvement of p38MAPK (Fig. S2B).
In summary, cells with higher p38MAPK phosphorylation demonstrated greater resistance to SB202190 (Fig. 2D), perhaps because higher inhibitor concentration is necessary to inhibit a more active enzyme. Furthermore, a consistent correlation between sensitivities to PIKfyve and p38MAPK inhibitors was observed for each line, (Fig. 2E–G) suggesting that the two cellular pathways share common functions or mechanisms.
Cellular p38MAPK Protein Levels Directly Modulate the Effects of PIKfyve Inhibition
Since higher p38MAPK amount and phosphorylation is correlated with resistance to WX8, we hypothesized that direct modulation of cellular protein levels would affect sensitivity. First, the sensitivity of normal epithelial breast MCF10A cells was determined after depletion of the cellular p38MAPK protein using siRNAs. Reduction of p38MAPK protein led to dramatic increase in sensitivity to WX8 that was inversely proportional to the amount of cellular p38MAPK content (Figs. 2H, S3A–D). While 0.5 μM WX8 reduced MCF10A cell survival by 20% in control siRNA-transfected cells, survival reduction reached 65% for the highest levels of p38MAPK depletion. Moreover, while WX8-induced vacuolization dissipated after 1 day in control siRNA-transfected cells, vacuoles persisted in cells treated with p38MAPK siRNA for up to 3 days in a dose-dependent manner.
Conversely, overexpression of p38MAPK in sensitive osteosarcoma U2OS cells decreased their sensitivity to PIKfyve inhibition. Although a narrow window of efficacy existed due to toxicity with dramatic overexpression, GFP-p38MAPK expression significantly reduced sensitivity and vacuolization of U2OS cells (Figs. 2I, S3E–G). These results confirmed that p38MAPK plays a critical role in modulating the cytotoxic effects of PIKfyve inhibition.
PIKfyve and p38MAPK Inhibitors Selectively and Synergistically Reduce Cancer Cell Viability
We next sought to determine whether pharmacological inhibition of p38MAPK would further exacerbate the effects of PIKfyve inhibition and increase the therapeutic potential of PIKfyve inhibitors against cancers. Colorectal cancers (CRCs) demonstrate a high incidence of metastasis, most of which are only treatable by chemotherapy (29). Moreover, p38MAPK has been previously identified as a mediator of resistance to chemotherapy in CRC (30). Thus, CRC cells provided a fitting model to study how p38MAPK interacts with PIKfyve and autophagy inhibition. SW480 colon adenocarcinoma cells, which demonstrated an average sensitivity to WX8 for cancer cells (Fig. 2D), were cultured for 3 days with multiple concentrations of WX8 in combination with multiple concentrations of SB202190 to create a dose-response matrix. WX8 and SB202190 in combination dramatically reduced SW480 cell survival to a greater extent than either inhibitor alone over a wide range of concentrations (Figs. 3A, S5A,B).
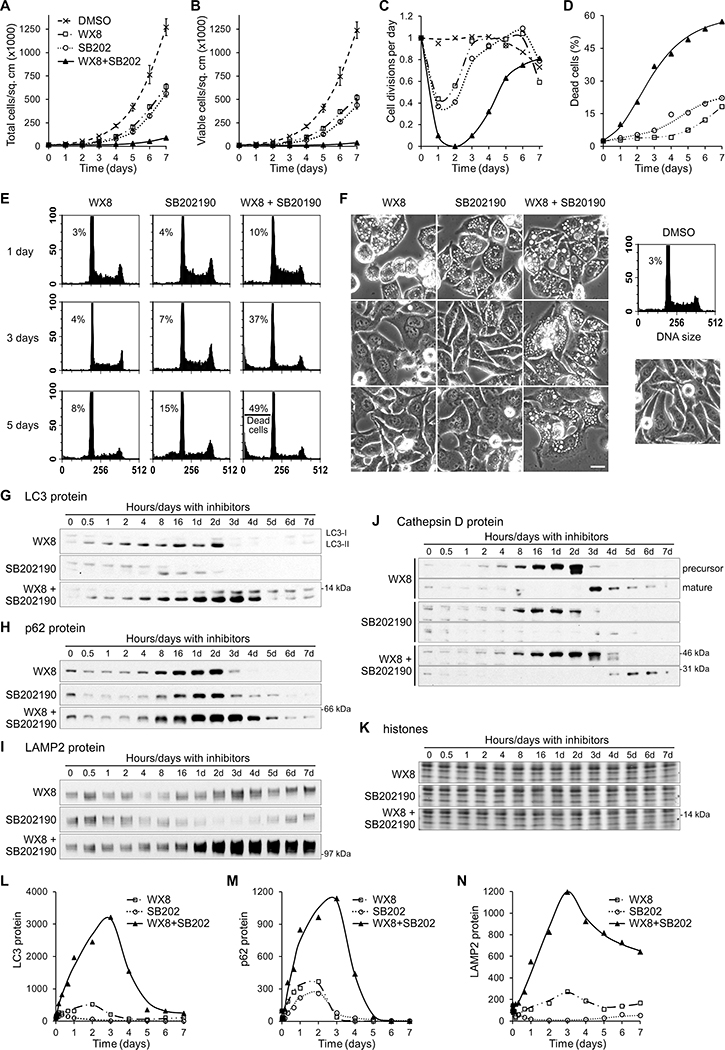
Figure 3. p38MAPK and PIKfyve inhibitors synergistically and selectively reduce cancer cell viability.
(A) WX8 and SB202190 dosage matrix was performed with SW480 cells cultured for 3 days with the indicated drug concentrations. All cells were collected, counted and analyzed by FACS. The number of viable cells for each treatment was presented as a percentage from the control viable cells “Cell Survival (%)”. (B) “Synergy above additive” was calculated by subtracting the individual toxicities of WX8 and SB202190 from the toxicity of their combinations. Positive numbers indicate a synergistic effect above the expected additive effect. (C) The Chou-Talalay Combinational Index (CI) was calculated for each WX8 and SB202190 combination in panel A. The smaller the CI, the larger the synergistic effect. (D) “Dead cells (%)” indicate the percentage of dead cells in the population for each treatment. (E) Examples of FACS generated DNA size histograms with the percentage of dead cells and phase contrast images of cells treated with the indicated drugs. Scale bar: 20 μm. (F) “Cell Survival (%)” and (G) “Dead cells (%)” results from a WX8 and SB202190 dosage matrix generated with Hs27 cells under the same conditions used in panels A. (H to K) Results from WX8 and SB202190 dosage matrices performed with the indicated cell lines. Values are mean (n = 3). Total cell count and SEM for each treatment is presented in Fig. S5.
To determine whether the combined inhibition was synergistic, the reductions in cell survival caused by individual inhibitors and their combination were compared. First, synergy was identified when a combined effect was above the expected additive effect ([combined effect] - [WX8 effect] - [SB202190 effect] >0). Multiple combinations resulted in reductions in survival over 30% above the expected, indicating impressive synergy between WX8 and SB202190 (Fig. 3B). Still, the combined effect of drugs is not simply the arithmetic sum of individual effects, especially for sums exceeding 100%. Therefore, the data were also analyzed by the Chou-Talalay method, utilizing a median effect equation based on the mass-action law (24), to calculate a Combinational Index (CI) for each combination. CI values of <1 prove synergistic effect, with higher synergy producing lower values. Practically all combinations of WX8 and SB202190 resulted in very low CI values <1 indicating strong synergistic effects (Fig. 3C). In both methods, the dosage matrix of WX8 and SB202190 revealed a broad range of concentrations that synergistically reduced cell survival.
To differentiate the effects on proliferation and survival (31), the changes in cell viability (Fig. 3A) and dead cell fractions (Fig. 3D) were compared. Mild concentrations of both inhibitors alone and in combination resulted only in cellular arrest, not death. However, higher concentrations of the inhibitors in combination resulted in substantial cell death reaching 94%, indicating synergistic cytotoxicity, as evidenced from FACS profiles and images (Figs. 3E, S4A). Remarkably, combined PIKfyve and p38MAPK inhibitors did not notably affect the viability (Figs. 3F,G, S5C,D) of Hs27 normal human fibroblasts, demonstrating potential anti-cancer selectivity.
To determine if this paradigm was limited to SW480 and Hs27, dose response matrices were performed using cell lines derived from different tissues with varied genetic backgrounds and cancer driver mutations. When combined, WX8 and SB202190 induced synergistic reduction of survival (using both calculation methods) in HCT116 colon carcinoma, A549 lung carcinoma and U2OS osteosarcoma (Figs. 3H–J, S5E–G). Again, viability of normal colon epithelial CCD-841CoN cells (Figs. 3K, S5H) was not significantly affected under the same treatments, confirming the selective anti-cancer potential of combined PIKfyve and p38MAPK inhibition.
Combined PIKfyve and p38MAPK Inhibition Significantly Extends the Proliferation Arrest, Cell Death, and Vacuolization of Cancer Cells
To investigate the dynamic of cancer cell arrest and death, proliferation curves were created for SW480 cells incubated with DMSO vehicle, WX8, SB202190, or both inhibitors together. Results were reported as total cells/cm2, as viable cells/cm2, and as cell divisions per day (Fig. 4A–C). As expected, vehicle-treated cells consistently divided every day, but proliferation slowed after day 5, likely due to media exhaustion. WX8 or SB202190 alone reduced proliferation for up to two days, but normal division rates recovered by day 3. The combination treatment practically halted proliferation until day 4, demonstrating long-lasting anti-proliferative effects. The percentage of dead cells as determined by FACS at each day was also plotted (Fig. 4D,E). The individual inhibitors caused increased cell death only after day 4, when media exhaustion can be expected. In contrast, the combined inhibitor treatment caused rapid accumulation of dead cells, approaching 40% of the population by day 3 and 60% at day 7. The reduction of cancer cell viability was not mediated by p53 stress response or apoptosis (Fig. S6A). Incubation for up to 5 days with WX8 and SB202190, alone or in combination, did not induce noticeable phosphorylation and accumulation of p53 protein, or cleaved Caspase 3 and PARP proteins which are characteristic of apoptosis.
Figure 4. p38MAPK and PIKfyve inhibitors synergistically block autophagy-mediated protein degradation and maturation of lysosomal enzymes.
SW480 cells were cultured with either 0.125 μM WX8 or 5 μM SB202190 or both together for the times indicated, imaged (F), collected, counted and analyzed by FACS (E). Results are reported as (A) total cells per sq. cm, (B) viable cells per sq. cm, (C) calculated cell divisions per day, and (D) percentage of dead cells in the sample. Values are mean ±SEM (n = 3).
The amounts of LC3 (G,L), p62 (H,M), LAMP2 (I,N), precursor and mature Cathepsin D (J) proteins were determined by immunoblot, normalized by histone content (K), and plotted as a percentage of the protein present at time zero.
Images of treated cells demonstrated that at day 1, all treatments caused cell vacuolization, consistent with lysosome dysfunction (Fig. 4F). Vacuolization dissipated in cells treated with individual inhibitors by day 3, but persisted through day 5 with the combination. Interestingly, vacuole disappearance coincided with the beginning of proliferation recovery (Fig. 4C), suggesting that the induced vacuolization is related to the reduction of cancer cell viability.
Combined PIKfyve and p38MAPK Inhibition Synergistically Disrupts Autophagy-mediated Protein Degradation and Lysosomal Function in Cancer Cells
We next sought to determine how vacuole accumulation is related to PIKfyve and p38MAPK inhibition cytotoxicity. To address this question, time courses of SW480 cells were analyzed for changes in autophagy-associated proteins. The lysosomal-associated membrane protein 2 (LAMP2), a transmembrane protein specific for lysosomes (32), and the autophagosomal LC3-II and p62 proteins are degraded through autophagy, but accumulate when protein degradation is blocked (26). With WX8 or SB202190 alone, total LC3 protein (corresponding to LC3-II since LC3-I was in negligible amounts) accumulated 5- and 2-fold above initial levels, respectively, but increased 32-fold when the inhibitors were combined (Fig. 4G–N). Similarly, p62 and LAMP2 levels increased about 3-fold with individual inhibitors, but reached almost 12-fold with the combination. The duration of protein accumulation was also significantly prolonged with combined inhibitors. LC3 and p62 protein levels peaked on day 2 with WX8 alone, and returned to baseline by day 3. When SB202190 was added to WX8, LC3 and p62 protein levels remained high through day 5, and LAMP2 levels through day 7. In summary, WX8 and SB202190 together induced synergistic accumulation of autophagy degraded proteins, indicating a severe blockage of autophagy.
The effect of combined inhibition on the lysosomal proteolytic cathepsin enzymes was also investigated. Cathepsins are synthesized as inactive, longer precursor proteins that are subsequently cleaved to become active during lysosome maturation. WX8 alone impaired cathepsin maturation leading to abnormal accumulation of Cathepsin D precursor (52 kDa) (33) that was only converted to active form (34 kDa) after day 2 (Fig. 4J). WX8 and SB202190 together prolonged the accumulation of Cathepsin D precursor until day 4, when the precursor was finally processed. Active Cathepsin D present at time zero was depleted by 8 hours and did not reappear until day 3 with WX8 alone and day 4 when with WX8 and SB202190, suggesting that prior to that time, lysosomes did not contain active Cathepsin D. Analogous results were obtained for Cathepsin B (34) maturation. The recovery of cathepsin maturation, and therefore their proteolytic activity, coincided with the precipitous degradation of p62, LC3, and LAMP2 proteins, suggesting restoration of autophagy protein processing.
While WX8 alone inhibited autophagy in both sensitive and resistant cells (Fig. 1), an outstanding question remained of whether combined PIKfyve and p38MAPK inhibition influenced autophagy in normal cells. Time courses performed with A549 cancer and Hs27 normal cells (Fig. S7A–I) revealed that SB202190 synergistically enhanced WX8-induced accumulation of p62, LC3 and LAMP2 proteins, consistent with increased autophagy inhibition in all cells, regardless of their sensitivity to WX8. However, only the viability of autophagy-dependent cancer cells was affected by this synergistic inhibition.
Combined PIKfyve and p38MAPK Inhibition Causes Complete and Persistent Blockage of Autophagy
The ability of PIKfyve inhibition to block autophagy-mediated protein degradation decreased over time (Fig. 4) – to determine why this occurred, SW480 cells were incubated with WX8 and SB202190, alone or in combination, for 3 days with or without resupplying the inhibitors every day. The extent of LC3-II protein accumulation was surveyed by immunoblot (Fig. 5A,B). Resupplying the inhibitors daily caused greater accumulation of LC3-II, proposing that the cells do not adapt or compensate, but the chemical compounds likely degrade with time. Strikingly, while resupplying individual inhibitors led to decreased cell survival after 3 days, a single dosage of the combined treatment was as effective as resupplying it daily (Fig. 5C). Near-maximum efficacy was achieved by the combined inhibitors in this time frame, demonstrating long-lasting and efficient blocking of autophagy through PIKfyve and p38MAPK kinase inhibition.
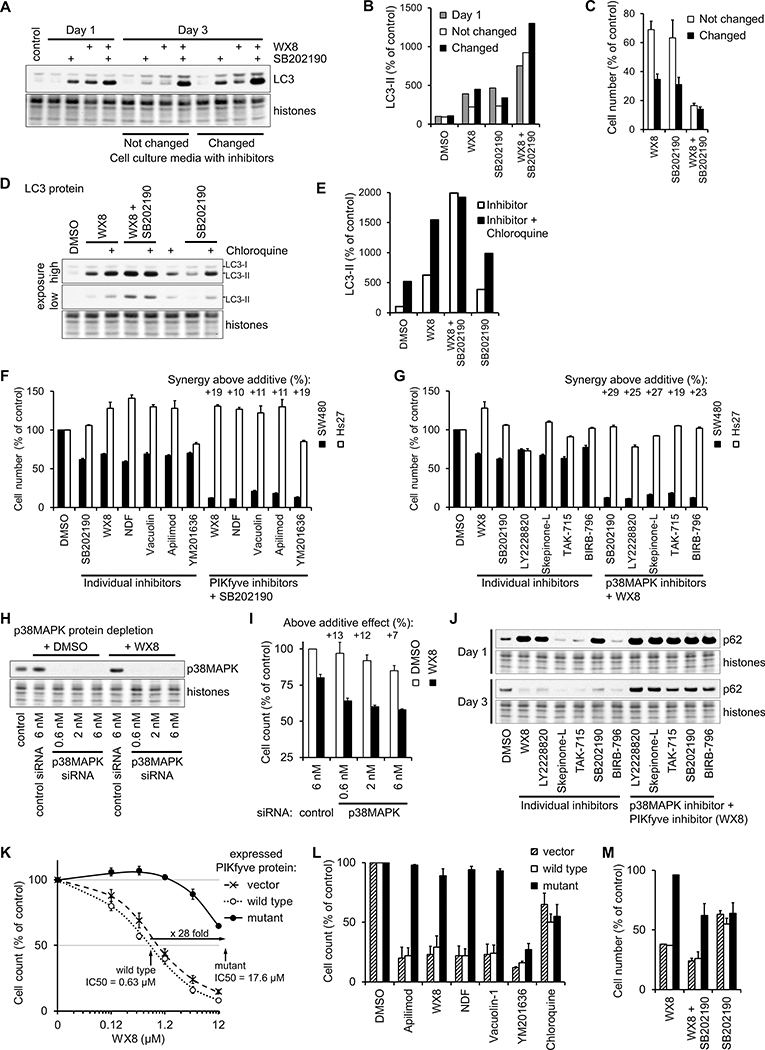
Figure 5. Specific inhibition of PIKfyve and p38MAPK activities are responsible for the effects on cellular viability and autophagy.
(A) SW480 cells cultured for 3 days with 0.125 μM WX8 and/or 5 μM SB202190 with or without changing the medium every day to resupply the inhibitors. The amount of LC3-II form was determined by immunoblot, quantified, and presented as a percentage of the amount in vehicle-treated cells after normalization by histone content (B). The number of cells at day 3 was reported as a percentage of the cells cultured with vehicle (C). Values are mean ±SEM (n = 3).
(D) SW480 cells cultured for 12 hours with 0.125 μM WX8 and/or 5 μM SB202190, 30 μM Chloroquine was added as indicated and the cells collected 8 hours later. The amount of LC3-II form was determined by immunoblot, quantified and presented as a percentage of the amount in vehicle-treated cells after normalization by histone content (E).
SW480 and Hs27 cells were cultured for 3 days with the indicated inhibitors and the number of cells cultured with inhibitor reported as a percentage of the cells cultured with vehicle. (F) PIKfyve inhibitors (0.125 μM WX8, 0.3 μM NDF, 0.25 μM Vacuolin 1, 30 nM Apilimod, 0.6 μM YM201636) were applied either alone (individual inhibitors) or together with 5 μM SB202190. (G) p38MAPK inhibitors (5 μM SB202190, 5 μM LY2228820, 5 μM Skepinone-L, 3.5 μM TAK-715, 7.5 μM BIRB-796) were applied either alone or together with 0.125 μM WX8. “Synergy above additive” was calculated as in Fig. 3. Values are mean ±SEM (n = 3).
(H,I) p38MAPK in A549 cells was depleted with the indicated concentration of p38MAPK siRNA or control siRNA and incubated with 0.375 μM WX8. Cells were collected after 3 days, and the counts plotted as a percentage of the control siRNA-treated cells. Values are mean ±SEM (n = 3).
(J) The amount of p62 protein was determined by immunoblot of cells cultured for 1 or 3 days with the indicated inhibitors.
A375 cells, stably expressing wild-type or N1939K mutant PIKfyve protein, or vector transduced, were cultured for 3 days in the presence of the indicated concentration of WX8 (K), a set of autophagy inhibitors (3.75 μM WX8, 6 μM NDF, 5 μM Vacuolin-1, 0.3 μM Apilimod, 12.5 μM YM201636, 30 μM chloroquine) (L) or WX8 and SB202190 (M). The cells were collected and the counts presented as a percentage of the vehicle-treated cells. The vehicle was set to 0.01 μM instead of 0 μM. Values are mean ±SEM (n = 3).
The lysosomal detergent chloroquine is often used to block autophagic protein degradation and determine the effect of compounds on autophagy flux (26). Previous studies have determined that PIKfyve inhibition blocks autophagic flux (10). To determine how efficiently WX8 and SB202190, alone or together, block autophagic flux, SW480 cells were incubated with the inhibitors alone or in combination for 12 hours, and then chloroquine was added for 8 hours. When chloroquine was added to either inhibitor alone, LC3-II further accumulated (Fig. 5D,E), indicating that autophagic processing was not completely blocked. Addition of chloroquine did not increase LC3-II accumulation in cells treated with WX8 and SB202190 together, demonstrating that the inhibitor combination blocked autophagic flux completely. In summary, combining WX8 and SB202190 caused complete and long-lasting inhibition of autophagy with lower concentrations, demonstrating remarkable synergistic effect on autophagy.
Specific Inhibition of PIKfyve and p38MAPK is Responsible for Synergistic Effects on Cell Viability and Autophagy
To determine if the phenomena described in this study so far were unique to WX8 and SB202190, multiple compounds characterized as specific PIKfyve and p38MAPK inhibitors were tested. The PIKfyve inhibitors WX8, NDF, Apilimod, Vacuolin-1 and YM201636 (12,35,36), were tested in combination with the p38MAPK inhibitors SB202190, BIRB-796, Skepinone-L, LY2228820 and Tak-715 (28,37–39). SW480 and Hs27 cells were cultured with inhibitor concentrations that individually reduced SW480 cell viability by about 30%. Since total and viable cells analysis of SW480 and Hs27 cells sensitivity produced practically the same results (Figs. 3A,F, S5A,C), only total cells counts were analyzed. When SW480 cells were cultured with SB202190 plus any PIKfyve inhibitor or with WX8 plus any p38MAPK inhibitor, viability was reduced by 10% to 29% above the expected additive effect (Fig. 5F,G). Furthermore, siRNA targeted against p38MAPK in SW480 cells also acted synergistically with WX8 to inhibit proliferation (Fig. 5H,I). Notably, none of the combinations significantly affected the viability of normal Hs27 cells (Fig. 5F,G). Thus, the synergistic and selective effects of WX8 and SB202190 on cancer cell viability were not dependent on a particular chemical compound.
Some p38MAPK inhibitors affect autophagy independently of p38MAPK inhibition (40). To ascertain whether direct autophagy inhibition is important, SW480 cells were treated with five different p38MAPK inhibitors alone and in combination with WX8 (Fig. 5J). Alone, only SB202190 and LY2228820 caused accumulation of p62, but when combined with WX8, all five p38MAPK inhibitors caused similar synergistic accumulation of p62 that persisted for at least 3 days.
To confirm the specificity of the WX8-family of PIKfyve inhibitors, we utilized the PIKFYVE missense mutation N1939K reported to confer resistance to the PIKfyve inhibitor Apilimod in lymphoma cells (14) and stably expressed PIKfyve proteins in A375 melanoma cells (Fig. S8A). Cells expressing PIKfyveN1939K were resistant to cytotoxicity and vacuolization induced by WX8, NDF, Apilimod and Vacuolin-1 (Figs. 5K,L, S8B,C), confirming that these effects are mediated exclusively through inhibition of PIKfyve. Interestingly, the mutant did not prevent effects caused by the structurally different PIKfyve inhibitor YM201636 or the general autophagy inhibitor chloroquine, confirming that the mutation only affects activities directly mediated by PIKfyve. Moreover, the PIKfyve mutant could not prevent any of the effects caused by p38MAPK inhibitors. When treated with the combination of WX8 and SB202190, the mutant-expressing cells appeared to be affected only to the extent of the p38MAPK inhibitor alone (Fig. 5M). These results provide strong evidence that specific inhibition of PIKfyve and p38MAPK is responsible for the synergistic effects on cell viability and autophagy described in this study.
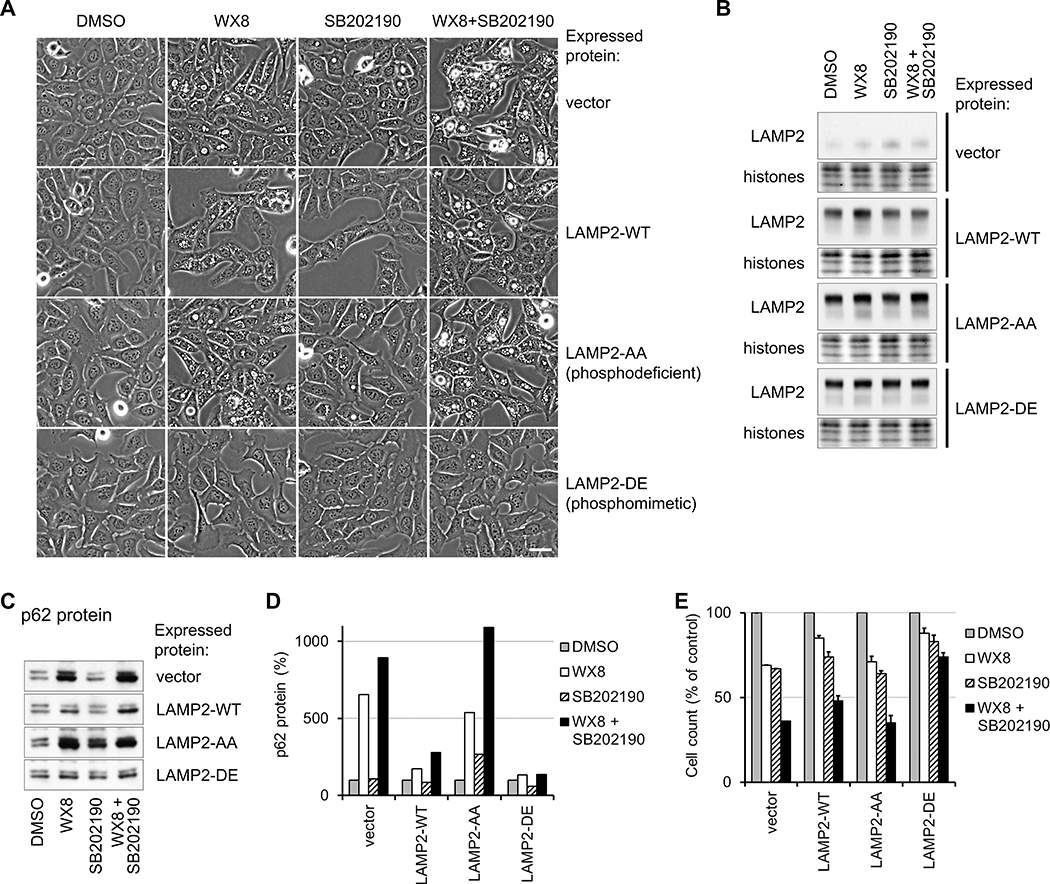
Expression of Phosphomimetic LAMP2 Mutant in p38MAPK-targeted Sites Prevents Lysosome Dysfunction Caused by PIKfyve Inhibition
p38MAPK can directly phosphorylate amino acids T211 and T213 of the LAMP2 protein (41) to activate chaperone-mediated autophagy at the lysosomes (41). To determine if LAMP2 phosphorylation by p38MAPK can affect lysosome fusion with autophagosomes, we generated U2OS cell lines which stably express epitope-tagged wild-type LAMP2 (LAMP2-WT) or mutants that were either phosphodeficient (LAMP2-AA) or phosphomimetic (LAMP2-DE) at the p38MAPK-phosphorylated sites. Inhibition of p38MAPK should prevent T211 and T213 phosphorylation of endogenous and recombinant wild-type LAMP2. Overexpression of the phosphomimetic mutant should rescue functions that require p38MAPK phosphorylated LAMP2, but the phosphodeficient mutant should not.
Expression of the phosphomimetic mutant (LAMP2-DE) prevented vacuolization, p62 protein accumulation, and reduction in cell viability (Fig. 6A–E) caused by WX8 and SB202190 individually and in combination. Expression of the phosphodeficient mutant (LAMP2-AA) did not prevent the effects, indicating that LAMP2 phosphorylation was necessary for rescuing lysosomal dysfunction. Overexpression of wild-type LAMP2 partially ameliorated effects of treatment, suggesting that simply increased LAMP2 activity can compensate for the effects of PIKfyve and p38MAPK inhibition. These data demonstrated that phosphorylation of LAMP2 by p38MAPK promoted lysosome function in autophagy. Consequently, inhibition of p38MAPK exacerbated the lysosome dysfunction caused by PIKfyve inhibition.
Figure 6. Phosphorylation of LAMP2 by p38MAPK modulates lysosome dysfunction caused by PIKfyve inhibition.
U2OS cells, stably expressing epitope-tagged wild-type LAMP2 protein, phosphodeficient LAMP2-AA or phosphomimetic LAMP2-DE mutants, or vector transduced, were cultured in the presence of 0.06 μM WX8, 5 μM SB202190 alone or together. Cells treated for one day were imaged (A) and total cell lysates were assayed for LAMP2 (B) and p62 (C) by immunoblot. p62 amounts were quantified and the results presented as a percentage of the vehicle-treated cells (D) after normalization by histone content. Cells treated for 3 days were collected and the counts presented as a percentage of the vehicle-treated cells (E). Values are mean ±SEM (n = 3). Scale bar: 20 μm.
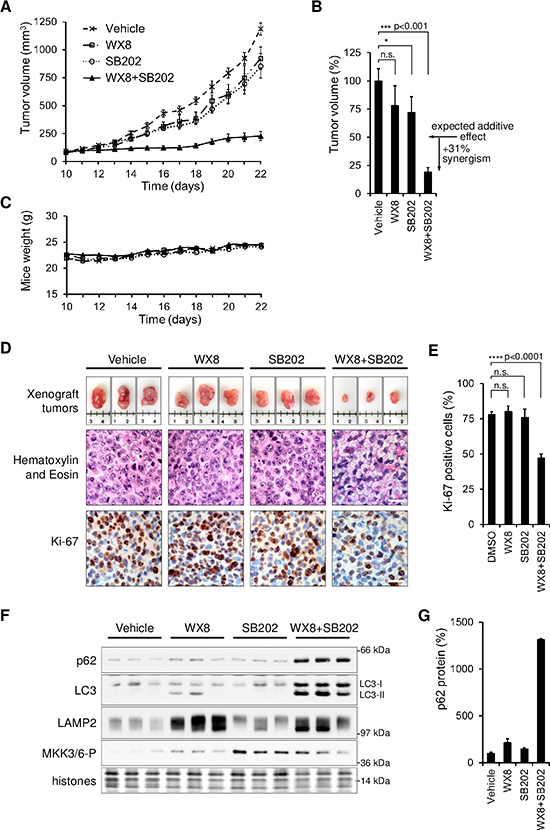
Combined PIKfyve and p38MAPK Inhibition Synergistically Reduces Tumor Growth in Mouse Xenografts
To test the therapeutic potential of combined PIKfyve and p38MAPK inhibition in vivo, mice bearing xenografts of SW480 colon adenocarcinoma cells were treated with WX8 and SB202190 alone or in combination. SW480 cells were injected in the flanks of nude mice and tumors were allowed to establish for 10 days. Mice were then treated daily via intraperitoneal injections. Dosages of 20 mg/kg WX8 and 12.5 mg/kg SB202190 caused mild reduction of tumor growth, by 22% and 28% respectively (Fig. 7A,B). However, WX8 and SB202190 in combination reduced tumor growth by 81%, or 31% greater than the expected additive effect. No adverse effects were apparent from either mouse behavior or body weight, which were tracked daily (Fig. 7C). These results confirmed that the synergistic and selective effects of WX8 and SB202190 on cancer cell viability in vitro were reproducible in vivo.
Figure 7. Combined PIKfyve and p38MAPK inhibition synergistically inhibits xenograft tumor growth.
(A) Beginning on day 10, mice bearing SW480 cells tumors were injected daily with either vehicle, 20mg/kg WX8, 12.5mg/kg SB202190 or 20mg/kg WX8+12.5mg/kg SB202190. Values are mean volume ±SEM (n = 6). (B) Relative mean tumor volumes on day 22. (C) Mean weight of mice in each group during the experiment.
(D) Representative tumors from each group with a centimeter ruler. Hematoxylin and Eosin, and Ki-67 staining of tumor sections from each group. Scale bar: 20 μm. (E) Ki-67 positive tumor cells as a percentage of those in tumor from vehicle injected mice. (F) Total cell lysates from tumors in panel D were analyzed by immunoblot for the indicated proteins. (G) p62 protein amount plotted as a percentage of vehicle sample after normalization by histone content. Error bars indicate SEM.
ns nonsignificant; * p<0.05; *** p<0.001; **** p<0.0001 (Student t-test).
Tumors were excised and subjected to histological and protein analyses. Hematoxylin and eosin staining of tumor tissue slices revealed extensive cytoplasmic vacuolization when both WX8 and SB202190 were administered together (Fig. 7D), consistent with observations in vitro. Histological staining for Ki-67, a marker of actively proliferating cells (42), showed that tumors from mice treated with DMSO vehicle, WX8 alone, or SB202190 alone were about 75% positive. Tumors from mice treated with both inhibitors together were only about 50% positive for Ki-67, consistent with the observed inhibition of tumor growth (Fig. 7D,E). Also, the combination-treated tumors were enriched for LC3, p62, and LAMP2 proteins (Fig. 7F,G), consistent with the blockage of autophagy observed in vitro. Taken together, these results confirmed that the key effects of WX8 and SB202190 observed in cultured cells were reproducible in tumors treated in vivo. The synergistic disruption of autophagy and the dramatic reduction of tumor viability in vivo confirmed the therapeutic potential of combined inhibition of PIKfyve and p38MAPK.
DISCUSSION
Combination therapy is a cornerstone of cancer treatment, used to prevent and overcome drug resistance, reduce adverse events by lower drug dosage, and enhance effectiveness through drug synergy (43). This study identified a functional cooperation between the cellular PIKfyve and p38MAPK pathways in maintaining lysosome function, which was particularly important for cancer cell survival.
Autophagy helps cells maintain homeostasis and react to environmental insults, including nutrient deprivation, hypoxia, pathogen invasion, and exposure to cytotoxic agents (44). The p38MAPK stress response pathway also helps cells respond to extracellular stressors (16). Thus, both pathways, autophagy and p38MAPK, mediate cytoprotective, compensatory metabolic changes allowing cells to adapt. Such pathways are especially useful for cancer cells that grow in nutrient-poor conditions and during chemotherapy and radiation treatments (8,30). Accordingly, studies have shown that autophagy and p38MAPK pathways can be activated in cancer cells, and inhibition of either pathway can kill tumor cells and promote the effectiveness of cancer therapies (7,45). However, clinical studies have not yet produced effective single-drug anti-cancer treatment with the currently available autophagy and p38MAPK inhibitors.
The emergence of PIKfyve inhibitors as a novel method of autophagy inhibition informed the need for improved mechanistic understanding of their activity. Accordingly, the initial goal of this study was to identify molecular mechanisms that determine why PIKfyve inhibition reduces the survival of cancer much more efficiently than normal cells (Fig. 1A). Of the cancer lines tested in this study, Daudi B-cell lymphoma demonstrated the highest sensitivity to the PIKfyve inhibitor WX8 due to the unique dependence of B-cell lymphomas on markedly high TFEB expression (14). The outstanding question was how PIKfyve inhibition can kill cancer cells that do not depend on high TFEB (Fig. S2A).
From the tested markers of diverse cellular processes, the p38MAPK protein amount and phosphorylation were the only noted consistent and dramatic differences between the tested 12 cancer and 8 normal cell lines. Interestingly, the cancer cell lines responded more aggressively to DNA damage, exhibiting higher levels of histone H2AX phosphorylation followed by higher relative increases in activating phosphorylation of p38MAPK pathway proteins (Fig. S2). However, in comparison with the PIKfyve inhibition-sensitive cancer cells, normal cells had significantly higher basal levels of phosphorylation of p38MAPK pathway proteins, consistent with higher capacity to protect against cellular stressors (Fig. 2C). Moreover, in response to PIKfyve inhibition, p38MAPK phosphorylation increased 15-fold, demonstrating activation of the pathway and suggesting a role in counteracting the detrimental effects of autophagy inhibition (Fig. 1K). Further, siRNA-mediated reduction of p38MAPK protein in resistant MCF10A cells dramatically increased their sensitivity to WX8, while overexpression of GFP-p38MAPK decreased the sensitivity of the sensitive U2OS cells suggesting that p38MAPK levels indeed modulate the cytotoxic effects of PIKfyve inhibition (Figs. 2, S3). In addition, titrations of the p38MAPK inhibitor SB202190 revealed that there is a consistent and strong correlation between cell sensitivity to PIKfyve and p38MAPK inhibition, suggesting a common mechanism or a functional link between the two cellular adaptive pathways.
Dosage matrices with WX8 and SB202190 demonstrated that reducing p38MAPK activity exacerbated the anti-proliferative and cytotoxic effects of WX8 in SW480 colon adenocarcinoma cells (Fig. 3A–E). Strong synergistic activity over a wide range of concentrations, determined by both simple additive calculations and Chou-Talalay Combination Indices, confirmed that lower p38MAPK pathway activity sensitizes cells to PIKfyve inhibition. The inhibitor combination predominantly affected the proliferation of cancer cells at low drug concentrations, but at higher concentrations, it was cytotoxic, killing as much as 94% of the population. Interestingly, the reduction of cancer cell viability was not mediated by activation of the p53 stress response or apoptosis (Fig. S6).
PIKfyve inhibition blocks autophagy (10), and based on the results in this report, p38MAPK inhibition dramatically exacerbates this phenotype. Autophagic protein degradation was blocked synergistically with the combined inhibition, in both time and magnitude. Additionally, maturation of newly synthesized cathepsin precursors was blocked until day 4, at which time active cathepsins emerged and LC3 and p62 proteins began to decrease. The recovery of autophagic protein degradation coincided with recovery of cell proliferation, proposing that the synergistic anti-proliferative and cytotoxic effects of combined inhibition were caused by enhanced autophagy inhibition. While high levels of TFEB were particularly important in B-cell lymphomas (13) and EGFR in liver cancers (14), this study identified the p38MAPK pathway as a key modulator of PIKfyve inhibition induced autophagy inhibition in a wide variety of cancer cell lines with diverse genetic backgrounds and tissue origins (Fig. 3).
Notably, the combined PIKfyve and p38MAPK inhibition-induced autophagy blockage did not significantly affect the viability of normal cells. Normal cells have been shown in this and other reports to be autophagy-independent, providing an explanation as to how they are able to proliferate normally with inhibited autophagic processing. It is also possible that non-malignant cells possess a feedback loop that compensates reduced autophagy which can explain the faster recovery of autophagy protein degradation in normal fibroblasts (Fig. S7).
Through multiple assays and methods, it was confirmed that the synergistic effects on cell survival and autophagy caused by combined PIKfyve and p38MAPK inhibition were due to specific inhibition of these kinases. Each tested PIKfyve/p38MAPK inhibitor was capable of synergistically and selectively reducing cancer cell survival when combined with a complementary inhibitor. siRNA-mediated p38MAPK (α-isoform) depletion, GFP-p38MAPK overexpression, as well as utilization of PIKfyve mutant-expressing cell lines further solidified that the observed synergy was due to specific inhibition of these kinases.
While it has been demonstrated that p38MAPK phosphorylation of LAMP2 can affect chaperone-mediated autophagy at the lysosome, the effect on lysosome fusion with autophagosomes was not known. LAMP2 protein is required for fusion of autophagosomes with lysosomes (46) and our studies revealed for the first time that LAMP2 phosphorylation by p38MAPK can affect lysosome accumulation and dysfunction. Mutant studies revealed that vacuolization, p62 protein accumulation, and cell viability reduction caused by PIKfyve inhibition can be partially ameliorated (Fig. 6) by expression of wild-type LAMP2 protein, indicating that simply having more LAMP2 protein can promote lysosomal function. However, while the phosphomimetic LAMP2 mutant of the two threonine residues phosphorylated by p38MAPK prevented virtually all effects, phosphodeficient LAMP2 could not, indicating that phosphorylation of these residues is necessary for full LAMP2 function. Consequently, the observed activation of p38MAPK by PIKfyve inhibition could counteract the induced lysosome dysfunction by LAMP2 phosphorylation. Thus, inhibition of the p38MAPK compensatory function may explain the observed synergistic autophagy inhibition when PIKfyve and p38MAPK inhibitors are combined. Further, the findings corroborate the conclusion that lysosome dysfunction caused by combined PIKfyve and p38MAPK inhibition is primarily responsible for the reduction in cancer cell survival. It also demonstrates that lysosome function is the point of convergence between the PIKfyve and p38MAPK pathways underlying the synergistic effects when the pathways are inhibited simultaneously.
A mouse xenograft model using SW480 colon adenocarcinoma cells confirmed in vivo the ability of combined PIKfyve and p38MAPK inhibitor treatment to synergistically reduce tumor growth (Fig. 7). Similar to the results in vitro, the combined treatment caused dramatic vacuolization and autophagy inhibition in tumor cells leading to substantial reduction in the fraction of proliferating cells. Importantly, the treatment was well tolerated by the mice.
Combined inhibition of p38MAPK and PIKfyve demonstrates potential to increase survival of cancer patients by more effectively inhibiting autophagy using lower drug concentrations. The synergistic long-lasting effects of the combination may be especially beneficial for clinical usage since such treatment could be administered less frequently. The efficacy of this combinational treatment against multiple cancer cell lines points to both a common mechanistic vulnerability of cancers, as well as a therapeutic window for a broad spectrum of malignancies with different genetic profiles, driving mutations, and tissues of origins.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This study demonstrates that PIKfyve and p38MAPK cooperate to regulate lysosome homeostasis and their combined inhibition synergistically blocks autophagy to reduce cancer cell viability in vitro and in vivo.
ACKNOWLEDGEMENTS
We thank Dr. Elaine Jordan for providing training and expertise with the xenograft studies.
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development intramural research program (ZIA HD000506, ZIA HD000507).
Footnotes
CONFLICT OF INTEREST
The authors declare no competing interests.
REFERENCES
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011;147(4):728–41. [DOI] [PubMed] [Google Scholar]
- 2.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res 2014;24(1):58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener 2015;4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007;21(13):1621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol 2012;4(9):a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab 2017;25(5):1037–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 2007;117(2):326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006;10(1):51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chude CI, Amaravadi RK. Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors. Int J Mol Sci 2017;18(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano O, Kazetani K, Funata M, Fukuda Y, Matsui J, Iwata H. Vacuolin-1 inhibits autophagy by impairing lysosomal maturation via PIKfyve inhibition. FEBS Lett 2016;590(11):1576–85. [DOI] [PubMed] [Google Scholar]
- 11.Balla T Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 2013;93(3):1019–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma G, Guardia CM, Roy A, Vassilev A, Saric A, Griner LN, et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy 2019:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy CH, Saffi G, Gray MA, Wallace C, Dayam RM, Ou ZA, et al. Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. J Cell Sci 2018;131(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood 2017;129(13):1768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou JZ, Xi ZQ, Niu J, Li W, Wang X, Liang C, et al. Inhibition of PIKfyve using YM201636 suppresses the growth of liver cancer via the induction of autophagy. Oncol Rep 2019;41(3):1971–79. [DOI] [PubMed] [Google Scholar]
- 16.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009;9(8):537–49. [DOI] [PubMed] [Google Scholar]
- 17.Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, et al. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev 2000;14(5):574–84. [PMC free article] [PubMed] [Google Scholar]
- 18.Bhowmick NA, Zent R, Ghiassi M, McDonnell M, Moses HL. Integrin beta 1 signaling is necessary for transforming growth factor-beta activation of p38MAPK and epithelial plasticity. J Biol Chem 2001;276(50):46707–13. [DOI] [PubMed] [Google Scholar]
- 19.Sui X, Kong N, Ye L, Han W, Zhou J, Zhang Q, et al. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett 2014;344(2):174–9. [DOI] [PubMed] [Google Scholar]
- 20.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 2001;15(10):1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv ZM, Wang Q, Wan Q, Lin JG, Hu MS, Liu YX, et al. The role of the p38 MAPK signaling pathway in high glucose-induced epithelial-mesenchymal transition of cultured human renal tubular epithelial cells. PLoS One 2011;6(7):e22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vassilev A, Lee CY, Vassilev B, Zhu W, Ormanoglu P, Martin SE, et al. Identification of genes that are essential to restrict genome duplication to once per cell division. Oncotarget 2016;7(23):34956–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullen P Flow cytometric DNA analysis of human cancer cell lines. Methods Mol Med 2004;88:247–55. [DOI] [PubMed] [Google Scholar]
- 24.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70(2):440–6. [DOI] [PubMed] [Google Scholar]
- 25.Ranjan A, Iwakuma T. Non-Canonical Cell Death Induced by p53. Int J Mol Sci 2016;17(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corre I, Paris F, Huot J. The p38 pathway, a major pleiotropic cascade that transduces stress and metastatic signals in endothelial cells. Oncotarget 2017;8(33):55684–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J 2007;408(3):297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeGolvan MP, Resnick M. Pathobiology of colorectal cancer hepatic metastases with an emphasis on prognostic factors. J Surg Oncol 2010;102(8):898–908. [DOI] [PubMed] [Google Scholar]
- 30.Grossi V, Peserico A, Tezil T, Simone C. p38alpha MAPK pathway: a key factor in colorectal cancer therapy and chemoresistance. World J Gastroenterol 2014;20(29):9744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eastman A Improving anticancer drug development begins with cell culture: misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 2017;8(5):8854–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquere S, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 2005;118(Pt 14):3091–102. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun 2008;376(1):5–9. [DOI] [PubMed] [Google Scholar]
- 34.Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer 2015;15(12):712–29. [DOI] [PubMed] [Google Scholar]
- 35.Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, Shisheva A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am J Physiol Cell Physiol 2012;303(4):C436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Dong S, Hao B, Li C, Zhu K, Guo W, et al. Vacuolin-1 potently and reversibly inhibits autophagosome-lysosome fusion by activating RAB5A. Autophagy 2014;10(11):1895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koeberle SC, Romir J, Fischer S, Koeberle A, Schattel V, Albrecht W, et al. Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor. Nat Chem Biol 2011;8(2):141–3. [DOI] [PubMed] [Google Scholar]
- 38.Miwatashi S, Arikawa Y, Kotani E, Miyamoto M, Naruo K, Kimura H, et al. Novel inhibitor of p38 MAP kinase as an anti-TNF-alpha drug: discovery of N-[4-[2-ethyl-4-(3-methylphenyl)-1,3-thiazol-5-yl]-2-pyridyl]benzamide (TAK-715) as a potent and orally active anti-rheumatoid arthritis agent. J Med Chem 2005;48(19):5966–79. [DOI] [PubMed] [Google Scholar]
- 39.Patnaik A, Haluska P, Tolcher AW, Erlichman C, Papadopoulos KP, Lensing JL, et al. A First-in-Human Phase I Study of the Oral p38 MAPK Inhibitor, Ralimetinib (LY2228820 Dimesylate), in Patients with Advanced Cancer. Clin Cancer Res 2016;22(5):1095–102. [DOI] [PubMed] [Google Scholar]
- 40.Menon MB, Kotlyarov A, Gaestel M. SB202190-induced cell type-specific vacuole formation and defective autophagy do not depend on p38 MAP kinase inhibition. PLoS One 2011;6(8):e23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Zhu J, Dou J, She H, Tao K, Xu H, et al. Phosphorylation of LAMP2A by p38 MAPK couples ER stress to chaperone-mediated autophagy. Nat Commun 2017;8(1):1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inwald EC, Klinkhammer-Schalke M, Hofstadter F, Zeman F, Koller M, Gerstenhauer M, et al. Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res Treat 2013;139(2):539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2006;2(9):458–66. [DOI] [PubMed] [Google Scholar]
- 44.Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L. Chloroquine and hydroxychloroquine for cancer therapy. Mol Cell Oncol 2014;1(1):e29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suarez-Cuervo C, Merrell MA, Watson L, Harris KW, Rosenthal EL, Vaananen HK, et al. Breast cancer cells with inhibition of p38alpha have decreased MMP-9 activity and exhibit decreased bone metastasis in mice. Clin Exp Metastasis 2004;21(6):525–33. [DOI] [PubMed] [Google Scholar]
- 46.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009;43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.