Abstract
Functional circuits consist of neurons with diverse axonal projections and gene expression. Understanding the molecular signature of projections requires high-throughput interrogation of both gene expression and projections to multiple targets in the same cells at cellular resolution, which is difficult to achieve using current technology. Here, we introduce BARseq2, a technique that simultaneously maps projections and detects multiplexed gene expression by in situ sequencing. We determined the expression of cadherins and cell-type markers in 29,933 cells, and the projections of 3,164 cells in both the mouse motor cortex and auditory cortex. Associating gene expression and projections in 1,349 neurons revealed shared cadherin signatures of homologous projections across the two cortical areas. These cadherins were enriched across multiple branches of the transcriptomic taxonomy. By correlating multi-gene expression and projections to many targets in single neurons with high throughput, BARseq2 provides a potential path to uncovering the molecular logic underlying neuronal circuits.
Introduction
Neural circuits are comprised of neurons diverse in many properties, such as morphology 1,2, gene expression 3,4, and projections 5,6. Although recent technological advances have made it possible to characterize the diversity in individual neuronal properties, associating multiple properties in single neurons with high throughput remains difficult to achieve. Investigating the relationship between multiple neuronal properties is essential for understanding the complex organization of neural circuits.
Of particular interest is the relationship between endogenous gene expression and long-range projections in the cortex. Cortical neurons have diverse patterns of long-range projections 5,6 and diverse patterns of gene expression 3,4. The full diversity of neuronal projection patterns can often only be appreciated by assessing multiple projection targets simultaneously (Fig. 1A) 2,6. For example, Han, et al. 5 showed that neurons in mouse visual area V1 that project to area PM tend not to project to area AL and vice versa, a projection “motif” that involves the relative probability that a single neuron projects to two targets and hence could not have been discovered by assessing projection targets one at a time. Gene expression patterns are also complex, and although the diversity in gene expression can be described by clustering neurons into transcriptomic types, these transcriptomic types have limited power in explaining the diversity of cortical projections beyond the major classes of projection neurons 3,6(but also see 7,8). Moreover, because the determination of a transcriptomic type relies on the expression of only a subset of genes, the inability of transcriptomic type to predict projection patterns raises the possibility that the expression of other genes—potentially in gene co-expression motifs— might be better correlated with projection patterns. Although transcriptomic methods can be combined with retrograde labeling 3,9, retrograde labeling is limited to one or at most a few brain areas at a time. Resolving the relationship between gene expression and projection patterns in the adult cortex thus requires high-throughput techniques that allow simultaneous multiplexed gene detection with projection mapping to multiple target areas at single-neuron resolution, which remains difficult to achieve.
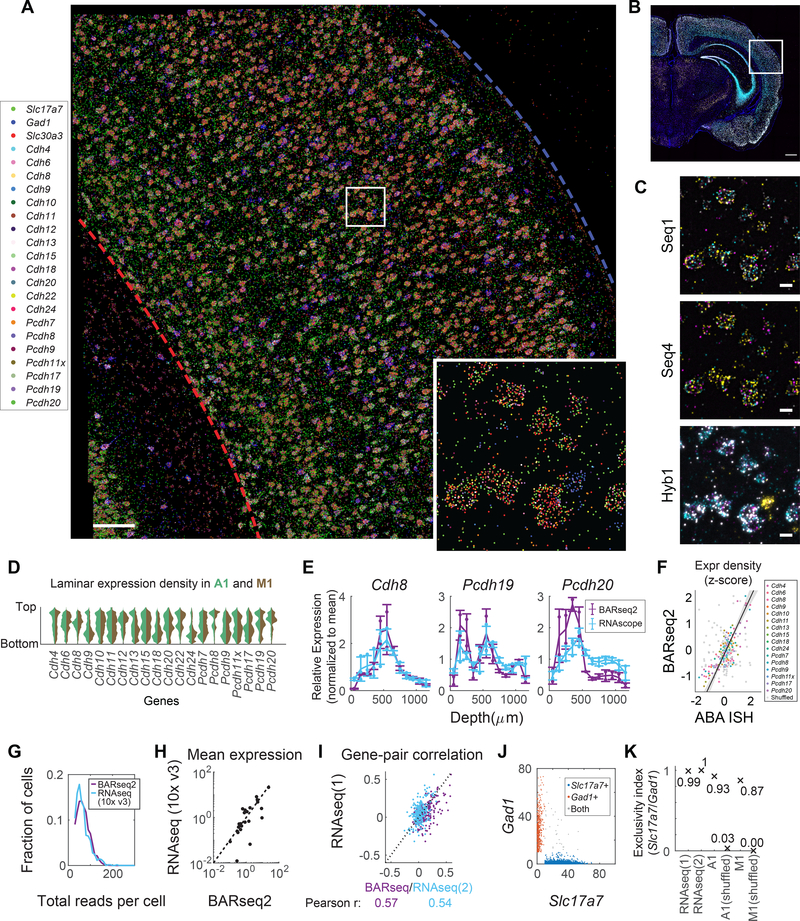
Figure 1. In situ sequencing of endogenous mRNAs using BARseq2.
(A) Cartoon of an example model in which the relationship between projections and gene expression can only be correctly inferred by multiplexed interrogation of both projections and gene expression. In this model (Top), neurons that express both genes project to both targets A and B, whereas neurons that express only one of the two genes project randomly to either A or B, but not both. (Bottom left) Methods that combine multiplexed single neuron gene expression with data about only a single projection target will conclude that all three gene expression patterns project to target A, and thus fail to detect the underlying “true” relationship between gene expression and projections. (Bottom right) Similarly, methods that combine multiplexed single neuron projections with data about only a single gene will also fail to detect any relationship between gene expression and projections. (B)(C) BARseq2 correlates projections and gene expression at cellular resolution. In BARseq2, neurons are barcoded with random RNA sequences to allow projection mapping, and genes are also sequenced in the same barcoded neurons. RNA barcodes and genes are amplified and read out using different strategies (C). (D) Theoretical imaging cycles using combinatorial coding (BARseq2), 4-channel sequential coding, or 4-channel sparse coding as used by Eng, et al. 50. Imaging cycles assumed 3 additional cycles for BARseq2, 1 additional round for sparse coding, and no extra cycle for sequential coding for error correction. (E) Mean and individual data points of the relative sensitivity of BARseq2 in detecting the indicated genes using different numbers of padlock probes per gene. The sensitivity is normalized to that using one probe per gene. n = 2 slices for each gene. (F) Representative images of BARseq2 (bottom) detection of the indicated genes using the maximum number of probes shown in (E) compared to RNAscope (top). Scale bars = 10 μm.
To achieve high-throughput mapping of projections to many brain areas, we recently introduced BARseq (Barcoded Anatomy Resolved by sequencing), a projection mapping technique based on in situ sequencing of RNA barcodes 6. In BARseq, each neuron is labeled with a unique virally-encoded RNA barcode that is replicated in the somas and transported to the axon terminals. The barcodes at the axon terminals located at various target areas are sequenced and matched to somatic barcodes, which are sequenced in situ, in order to determine the projection patterns of each labeled neuron. Because BARseq preserves the location of somata with high spatial resolution, in principle it provides a platform to combine projection mapping with other neuronal properties also interrogated in situ, including gene expression. We have previously shown 6 that BARseq can be combined with fluorescent in situ hybridization (FISH) and Cre-labeling to uncover projections across neuronal subtypes defined by gene expression. However, these approaches can only interrogate one or a few genes at a time, which would be insufficient for unraveling the complex relationship between the expression of many genes to diverse cortical projections (Fig. 1A).
Here we aim to develop a technique to simultaneously map projections to multiple brain areas and detect the expression of dozens of genes in hundreds to thousands of neurons from a cortical area with high throughput, high spatial resolution, and cellular resolution. To achieve this goal, we combine the high throughput and multiplexed projection mapping capability of BARseq with state-of-the-art spatial transcriptomic techniques with high imaging throughput and multiplexing capacity 10,11. This second-generation BARseq (BARseq2) greatly improves the ability to correlate the expression of many genes to projections to many targets in the same neurons. As a proof-of-principle, we first demonstrate multiplexed gene detection using BARseq2 by mapping the spatial pattern of up to 65 cadherins and cell-type markers in 29,933 cells. We then correlate the expression of 20 cadherins to projections to up to 35 target areas in 1,349 neurons in mouse motor and auditory cortex. Our study reveals novel sets of cadherins that correlate with homologous projections in both cortical areas. BARseq2 thus bridges transcriptomic signatures obtained through spatial transcriptional profiling with sequencing-based projection mapping to illuminate the molecular logic of long-range projections.
Results
To investigate how cadherin expression relates to diverse projections, we developed BARseq2 to combine high-throughput projection mapping with multiplexed detection of gene expression using in situ sequencing (Fig. 1B, C). BARseq2 is based on BARseq (Fig. 1C, left), which achieves high-throughput projection mapping by in situ sequencing of RNA barcodes 6. Projection patterns observed using BARseq are consistent with those obtained using conventional neuroanatomical techniques in multiple circuits2,5, but it can achieve throughput that is at least two to three orders of magnitude higher than the state-of-the-art single-cell tracing techniques 2. Possible technical concerns, including distinguishing fibers of passage from axonal termini, sensitivity, double labeling of neurons, and degenerate barcodes, have previously been addressed2,6,12,13 and will not be discussed in detail again here. Combining barcoded single-cell projection mapping with in situ detection of endogenous mRNAs exploits the unique advantage of BARseq in throughput to efficiently interrogate both neuronal gene expression and long-range projections simultaneously.
To detect gene expression using BARseq2, we used a non-gap-filled padlock probe-based approach to amplify target endogenous mRNAs 10,11(Fig. 1C, right). The elimination of gap-filling, necessary for reading out extremely diverse sequences of barcodes, increases the sensitivity for endogenous gene detection. In this approach, the identity of the target is read out by sequencing a gene-identification index (GII) using Illumina sequencing chemistry in situ. Because the GII is a nucleotide barcode sequence that uniquely encodes the identity of a given gene, the multiplexing capacity increases exponentially as 4N, where N is the number of sequencing cycles. This combinatorial coding by sequencing readout thereby allows simultaneous detection of a large number of genes using only a few cycles of imaging (Fig. 1D). Although sequencing readout offers many advantages, BARseq2 is also compatible with hybridization-based readout when necessary. The combination of non-gap-filling in situ sequencing of endogenous genes and the gap-filling approach for sequencing barcodes allows many genes to be detected simultaneously with projections using BARseq2.
In the following, we first demonstrate that, by optimizing targeted in situ sequencing, BARseq2 could achieve sufficient sensitivity for detection of endogenous mRNAs. We next combined in situ sequencing of endogenous mRNAs with in situ sequencing of RNA barcodes to associate the expression of cadherins with projection patterns at cellular resolution. We then validated BARseq2 by demonstrating that it could be used to recapitulate projection patterns specific to transcriptomic neuronal subtypes and to identify cadherins that were differentially expressed across major projection classes. Finally, we identified a set of cadherins shared between the mouse auditory cortex and motor cortex that correlate with homologous projections of intratelencephalic (IT) neurons in both cortical areas.
BARseq2 robustly detects endogenous mRNAs
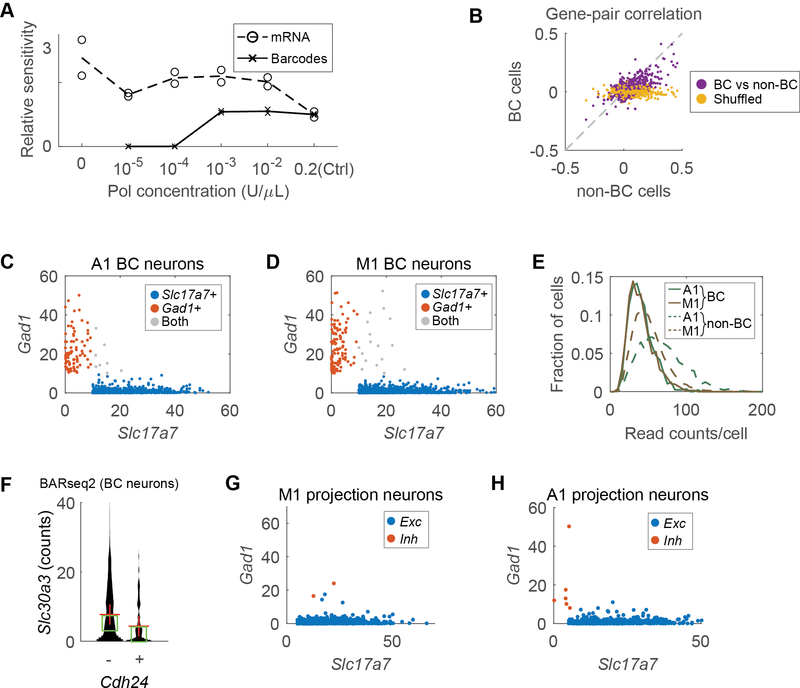
To adequately detect genes using BARseq2, we sought to improve the detection sensitivity. In most in situ hybridization methods, high sensitivity is achieved by using many probes for each target mRNA 14,15. We reasoned that increasing the number of padlock probes per gene might similarly improve the sensitivity of BARseq2. Indeed, we observed that tiling the whole gene with additional probes resulted in as much as a 46-fold increase in sensitivity compared to using a single probe (Fig. 1E; see Methods). Combined with other technical optimizations (Extended Data Fig. 1A, B), we increased the sensitivity of BARseq2 to 60 % of RNAscope, a sensitive and commercially available FISH method (Fig. 1F; Extended Data Fig. 1C, D; see Methods). We further optimized in situ sequencing to robustly read out GIIs of single rolonies over many sequencing cycles (Extended Data Fig. 1E–J; see Methods). These optimizations allowed BARseq2 to achieve sufficiently sensitive, fast, and robust detection of mRNAs.
BARseq2 allows multiplexed detection of mRNAs in situ
To assess multiplexed detection of cadherins in situ using BARseq2, we examined the expression of 20 cadherins, along with either three (in auditory cortex) or 45 (in motor cortex) cell-type markers (Fig. 2A–C). We chose to focus on the cadherins because of their known roles in cortical development, including projection specification 16,17, and their differential expression among cardinal cell types defined by multiple properties 18. These cadherins included most classical cadherins and non-clustered protocadherins expressed in auditory cortex and motor cortex. We successfully resolved and decoded 419,724 rolonies from two slices of mouse auditory cortex (1.7 mm2 × 10 μm per slice) and 1,445,648 rolonies from four slices of primary motor cortex (2.8 mm2 × 10 μm per slice). We recovered 20 rolonies in auditory cortex and 115 rolonies in motor cortex that matched two GIIs that were not used in the experiment, corresponding to an estimated error rate of 0.1 % and 0.2 %, respectively, for rolony decoding.
Figure 2. Multiplexed detection of mRNAs using BARseq2.
(A) A representative image of rolonies in auditory cortex (out of two slices sequenced). Scale bar = 100 μm. The inset shows a magnified view of the boxed area. (B) Low magnification image of the hybridization cycle showing the location of the area imaged in A. Scale bar = 100 μm (C) Representative images of the indicated sequencing cycle and hybridization cycle of the boxed area in A. Scale bars = 10 μm. (D) Violin plots showing the laminar distribution of cadherin expression in neuronal somata. Expression in auditory cortex and motor cortex is shown in different colors as indicated. (E) Laminar distribution of gene expression as detected by BARseq2 or FISH. Lines indicate means, error bars indicate standard deviations, and dots show individual data points. n = 2 slices for BARseq2 and n = 3 slices for FISH. (F) Relative gene expression observed using BARseq2 and in Allen gene expression atlas. Each dot represents the expression of a gene in a 100 μm bin in laminar depth. Gray dots indicate correlation between data randomized across laminar positions. A linear fit and 95 % confidence intervals are shown by the diagonal line and the shaded area. n = 2 slices for BARseq2 and n = 1 slice for ABA ISH. (G) Distribution of total read counts per cell in BARseq2 and single-cell RNAseq in auditory cortex. Only genes used in the panel detected by BARseq2 were included. (H) Mean expression for each gene detected using BARseq2 or single-cell RNAseq. Each dot represents a gene. The dotted line indicates equal expression between BARseq2 and single-cell RNAseq. (I) The correlation between pairs of genes observed in BARseq2 and single-cell RNAseq (purple dots), or in two single-cell RNAseq datasets (blue dots). (J) Expression of Slc17a7 and Gad1 in single neurons. Color codes indicate whether the neuron dominantly expressed Slc17a7 (blue) or Gad1 (red), or expressed both strongly (gray). (K) Exclusivity indices (see Methods) of Slc17a7 and Gad1 in neurons in two single-cell RNAseq datasets, BARseq2 in auditory or motor cortex, and shuffled BARseq2 data.
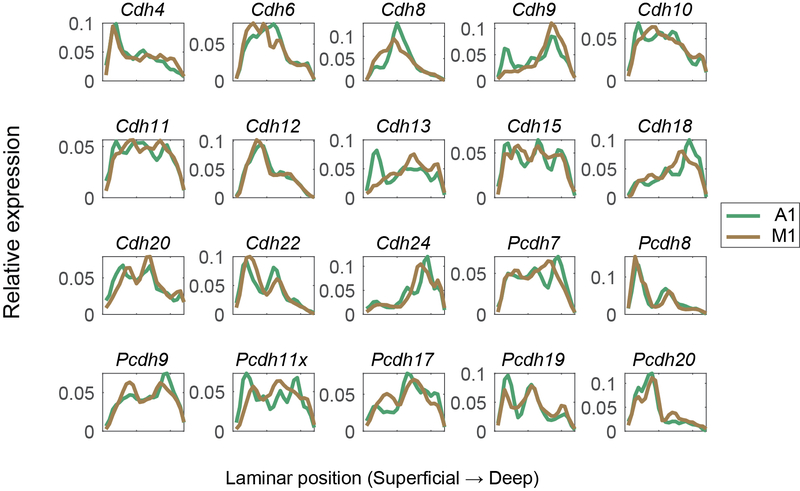
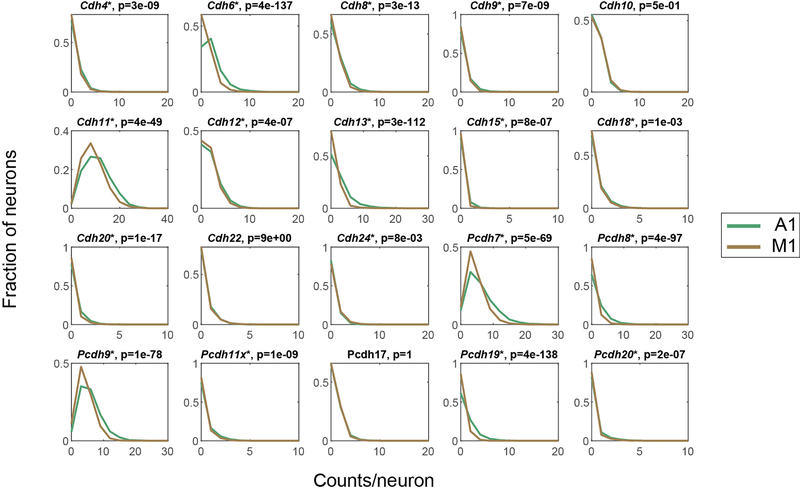
Consistent with previous reports 19,20, many cadherins were enriched in specific layers and sublayers in the cortex (Fig. 2D). Interestingly, although most cadherins had similar laminar expression in both auditory cortex and motor cortex, some cadherins were differentially expressed across the two areas. For example, Cdh9 and Cdh13 were enriched in L2/3 in auditory cortex, but not in motor cortex (Fig. 2D; Extended Data Fig. 2). The laminar positions of peak cadherin expression were consistent with those obtained by other methods, including RNAscope (Fig. 2E) and the Allen ISH atlas 21(Fig. 2F; Extended Data Fig. 3; see Methods). Thus, BARseq2 accurately resolved the laminar expression patterns of cadherins.
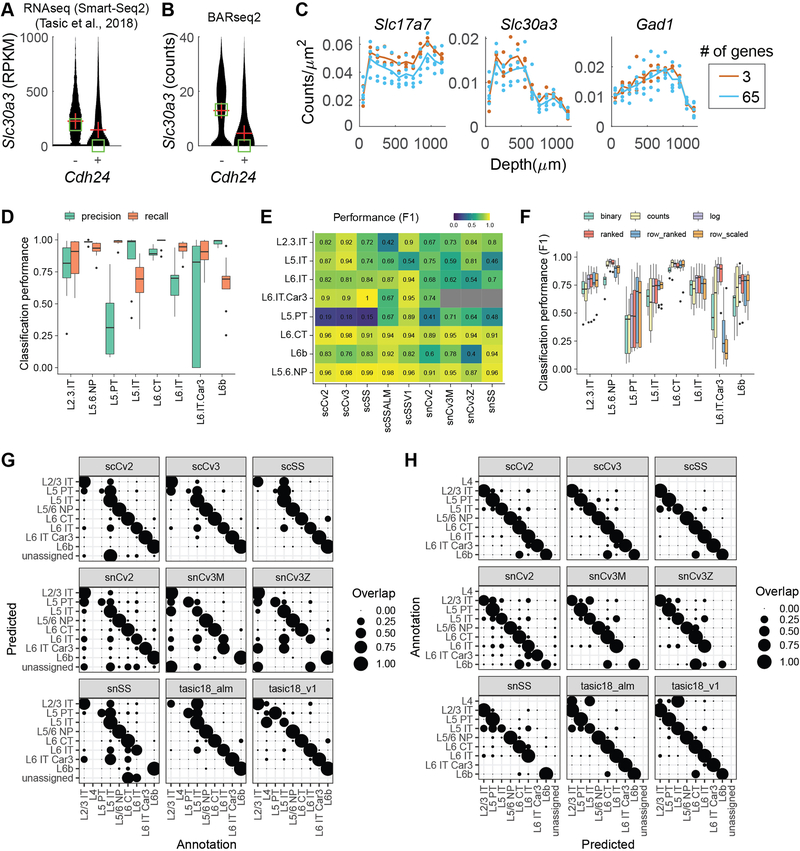
We then characterized gene expression obtained by BARseq2 at single-cell resolution (see Methods). We assigned 228,371 rolonies to 3,377 excitatory or inhibitory neurons [67.6 ± 28.8 (mean ± standard deviation) rolonies per neuron] in auditory cortex, and 752,687 rolonies to 11,492 excitatory or inhibitory neurons [65.5 ± 26.0 (mean ± standard deviation) rolonies per neuron] in motor cortex. Most cadherins showed slight differences in single-cell expression levels in these two cortical areas (Extended Data Fig. 4). In auditory cortex, the total read counts per cell was higher in BARseq2 than in single-cell RNAseq using 10x Genomics v3 (Fig. 2G; median read counts 64 for BARseq2, n = 3,337 cells compared to 57 for single-cell RNAseq, n = 640 cells, p = 5.3×10−5 using rank sum test). Thus, even using a limited number of probes, BARseq2 achieved sensitivity at least equal to single-cell RNAseq using 10x v3. For experiments requiring better quantification of low-expressing genes, the sensitivity could potentially be further improved by using more probes.
Further analyses showed that detection of mRNA by BARseq2 was specific. The mean expression of genes determined by BARseq2 was highly correlated with that determined by single-cell RNAseq using 10x v3 (Fig. 2H; Pearson correlation r = 0.88). A few outliers had significantly more counts in BARseq2 than in single-cell RNAseq, which likely reflected sampling differences across cell types, area-specific gene expression, and differences in RNA accessibility in situ. For example, Cdh6 expression observed by BARseq2 was 26 times of that observed by single-cell RNAseq. This difference could be attributed to under-sampling of Cdh6 expressing PT (pyramidal tract) neurons in our single-cell RNAseq data 6 and potentially variable sampling of neighboring cortical areas in which Cdh6 is differentially expressed 22. Furthermore, correlations between pairs of genes in single neurons determined by BARseq2 were consistent with single-cell RNAseq using 10x v3 to a similar extent as two independent 10x v3 experiments (Fig. 2I–K, Extended Data Fig. 5A, B; see Methods). These results indicate that the single-cell gene expression patterns observed by BARseq2 were comparable to those of single-cell RNAseq.
We wondered if BARseq2 could detect more genes in parallel, and thus be potentially useful in associating projections with larger gene panels. Because BARseq2 imaging time scales logarithmically with the number of genes detected (Fig. 1D), the multiplexing capacity of BARseq2 is not limited by imaging time. Furthermore, targeting up to 65 genes did not significantly affect the detection sensitivity of each gene (Extended Data Fig. 5C; see Methods). The detection of this 65-gene panel in motor cortex (Fig. 3A) allowed us to classify neurons to one of nine transcriptomic neuronal types defined by single-cell RNAseq 23 (Fig. 3B; see Methods and Extended Data Fig. 5D–H). Consistent with previous studies 3,9, these transcriptomic neuronal types displayed distinct laminar distributions (Fig. 3B, C; see Methods) and cadherin expression (Fig. 3D). Most transcriptomic types were found in the expected layers with the notable exception of L5 PT and L6 IT Car3, which were seen in additional layers (e.g. L2/3). These inaccuracies in cell typing likely resulted from suboptimal choice of marker genes (see Methods for a detailed discussion), and could potentially be improved in the future by optimizing the gene panels. These optimizations, however, are out of the scope of this study. These results demonstrate that BARseq2 can be applied to probe gene panels consisting of high dozens of genes, with minimal decrease in sensitivity and minimal increase in imaging time.
Figure 3. Cadherin expression across transcriptomic neuronal types in motor cortex.
(A) A representative image of rolonies in motor cortex (out of four slices sequenced). mRNA identities are color-coded as indicated. The top and the bottom of the cortex are indicated by the blue and red dashed lines, respectively. Scale bar = 100 μm. (B) Transcriptomic cell types called based on gene expression shown in (A). (C) Laminar distribution of transcriptomic neuronal types based on marker gene expression observed by BARseq2. Layer identities are shown on the right. (D) Differential expression of cadherins across transcriptomic neuronal types identified by BARseq2. Over-expression is indicated in yellow and under-expression is indicated in blue. Only differential expression that was statistically significant was shown. Statistical significance was determined using two-tailed rank sum test with Bonferroni correction for each gene between the indicated transcriptomic type and the expression of that gene across all other neuronal types.
BARseq2 correlates gene expression to projections
Previous studies of the relationship between projection patterns and gene expression have largely focused on revealing the projection patterns of transcriptomic neuronal types. Although this approach has identified some projection patterns biased in certain transcriptomic types 6,8, the diversity of projections in IT neurons remains largely unexplained by transcriptomic types 3,6. To further understand the relationship between gene expression and projections, we demonstrate an alternative approach that screens a targeted panel of genes for correlates of diverse projections. This approach relies on the ability of BARseq2 to interrogate both the expression of many genes and projections to many targets simultaneously, and thus would have been difficult to achieve using existing transcriptomic approaches that could only interrogate one or a small number of projections (e.g. Retro-seq 3,9) or barcoding-based projection mapping approaches that could only interrogate a small number of genes (e.g. BARseq 6).
As a proof-of-principle, we examined long-range axonal projections and the expression of 20 cadherins, along with three marker genes, in motor cortex and auditory cortex in three mice. We optimized BARseq2 to detect both endogenous mRNAs and barcodes in the same barcoded neurons without compromising sensitivity (Extended Data Fig. 6A; see Methods). In each barcoded cell, we segmented barcoded cell bodies (Fig. 4A, middle) using the barcode sequencing images (Fig. 4A, left). We then assigned rolonies amplified from endogenous genes that overlap with these pixels to the barcoded cells (Fig. 4A, right). This allowed us to map both projection patterns (Fig. 4B, left) and gene expression (Fig. 4B, right) in the same neurons. We matched barcodes in these target sites to 3,164 well-segmented barcoded neurons (1,283 from auditory cortex and 1,881 from motor cortex) from 15 slices of auditory cortex and 16 slices of motor cortex, each with 10 μm thickness. Of the barcoded neurons, 624 and 791 neurons had projections above the noise floor in auditory cortex and motor cortex, respectively. Most neurons [53 % (329/624) in auditory and 89 % (703/791) in motor cortex] projected to multiple brain areas. We then focused on 598 neurons in auditory cortex and 751 neurons in motor cortex, which also had sufficient endogenous mRNAs detected in each cell, for further analysis (Fig. 4C). These observations were largely consistent with previous BARseq experiments in auditory and motor cortex performed without assessing gene expression 2,6, confirming that the modifications for BARseq2 did not compromise projection mapping.
Figure 4. Correlating gene expression to projections using BARseq2.
(A) False-colored barcode sequencing images (left), soma segmentations (middle), and gene rolonies (right) of three representative neurons from the motor cortex. The segmentation and gene rolony images correspond to the white squared area in the barcode images. In the gene rolony images, the areas corresponding to the soma segmentations of the target neurons are in black. All scale bars = 20 μm. (B) Projections (left) and gene expression (right) of the target neurons shown in (A). The dots indicating gene expression are colored using the same color code as that in the gene rolony plots in (A). The neurons shown in the first two rows are excitatory projection neurons, whereas the neuron shown in the bottom row is an inhibitory neuron without projections. See Supp. Table S2 for the brain areas corresponding to each abbreviated target area. (C) Projections (left) and gene expression (right) of neurons in auditory cortex (top) and motor cortex (bottom). Each row represents a barcoded projection neuron. Both projections and gene expression are shown in log scale. Major projection neuron classes determined by projection patterns are indicated on the right. (D) (E) The number of excitatory neurons (blue) or inhibitory neurons (red) in all barcoded neurons (D) or barcoded projection neurons (E). Neurons in auditory cortex are shown in the top row and those in motor cortex are shown in the bottom row.
BARseq2 recapitulates known projection biases
Although BARseq2 can read out gene expression and projections in the same neurons, one might be concerned that barcoding neurons using Sindbis virus could disrupt gene expression 24. To determine the relationship between genes and projections, one would require that the gene-gene relationship in Sindbis-infected single neurons reflects that in non-infected neurons, and that any change in absolute gene expression level would have little effect. Reassuringly, previous reports have shown that the relationship among genes in single neurons is indeed largely preserved despite a reduction in the absolute expression of genes in Sindbis-infected cells 6,25. Furthermore, correlations between transcriptomic types and projections revealed in Sindbis-infected neurons were corroborated by other methods that did not require Sindbis infection 6,26. In agreement with these previous reports, we observed that the correlations between pairs of genes in the barcoded neurons were consistent with those in non-barcoded neurons despite an overall reduction in gene expression (Extended Data Fig. 6B–F; see Methods). Therefore, the relationship between gene expression and projections resolved by BARseq2 likely reflects that in non-barcoded neurons.
To further test whether BARseq2 can capture the relationship between gene expression and projections, we asked if we could identify differences in projection patterns across transcriptomic neuronal types that could also be validated by previous studies and/or other experimental techniques. We performed these validation analyses at three different levels of granularity. First, BARseq2 confirmed that most barcoded neurons with long-range projections were excitatory, not inhibitory: Whereas about 8–9% of all barcoded neurons were inhibitory (100 of 1,047 in auditory cortex and 140 of 1,689 in motor cortex; Fig. 4D), only 7 of 240 (3 %) inhibitory neurons (5 in auditory cortex and 2 in motor cortex) had detectable projections (Fig. 4E; see Methods and Extended Data Fig. 6G, H). Second, BARseq2 identified many cadherins (8 for auditory cortex and 12 for motor cortex) that were differentially expressed across intratelencephalic (IT) neurons, pyramidal tract (PT) neurons, and corticothalamic (CT) neurons 27 (Fig. 5A–D); the differential expression of these genes was consistent with the expression observed by single-cell RNAseq 3(Extended Data Fig. 7A; see Methods). Finally, BARseq2 confirmed known biases in projection patterns across transcriptionally defined IT subtypes in auditory cortex (Extended Data Fig. 7B, C; see Methods). Thus, BARseq2 recapitulated known projection differences across transcriptomic subtypes of IT neurons.
Figure 5. Differential cadherin expression across major classes and cortical areas.
(A) Vertical histograms of the expression (raw counts per cell) of cadherins that were differentially expressed across major classes in either auditory or motor cortex. Y-axes indicate gene expression level (counts per cell) and x-axes indicate number of neurons at that expression level. The numbers of neurons are normalized across plots so that the bins with the maximum number of neurons have equal bar lengths. Gene expression in auditory cortex (green) are shown on the left in each plot, and gene expression in motor cortex (brown) are shown on the right in each plot. Lines beneath each plot indicate pairs of major classes with different expression of the gene (FDR < 0.05). (B)(C) Volcano plots of cadherins that were differentially expressed across pairs of major classes in auditory cortex (B) or motor cortex (C). Y-axes indicate significance and x-axes indicate effect size. The horizontal dashed lines indicate significance level for FDR < 0.05, and the vertical dashed lines indicate equal expression. (D) Volcano plots of cadherins that were differentially expressed across auditory and motor cortex in the indicated major classes. Y-axes indicate significance and x-axes indicate effect size. The horizontal dashed lines indicate significance level for FDR < 0.05, and the vertical dashed lines indicate equal expression. For all panels, p values are calculated using two-tailed rank sum tests.
BARseq2 identifies cadherin correlates of IT projections
Having established that BARseq2 identified gene correlates of projections that were consistent with previous studies, we then asked whether cadherin expression correlates with projection patterns within the IT class of neurons. Although cadherins and other cell adhesion molecules are involved in projection specification and axonal growth during development 16,28, many take on other functions unrelated to projection specification in later development stages 29,30. In addition, other mechanisms such as axonal pruning could further shape the projection patterns of neurons independent of initial genetic programs. Therefore, any correlation between cadherins and projections is likely a remnant, or “echo,” of the developmental program that initially specified projections, and may thus be weak and further obscured by gene expression associated with later developmental stages. To overcome the challenges of identifying potentially weak relationships between gene expression and projections, we used BARseq2 to identify correlations between projections and cadherins using a module-based strategy inspired by similar approaches in transcriptomics 31. Projection modules and gene modules average over the noise in the measurement of individual projections and genes, respectively, and are thus easier to detect when there is considerable biological and/or technical noise in the measurements. This approach requires knowing the projections to many brain areas from individual neurons, a unique advantage of barcoding-based projection mapping techniques (i.e. BARseq and BARseq2) compared to retrograde labeling-based approaches 3,9. In the following section, we identify modest associations between cadherin expression and projections in IT neurons, including several associated pairs of cadherins/projections that were shared across cortical areas.
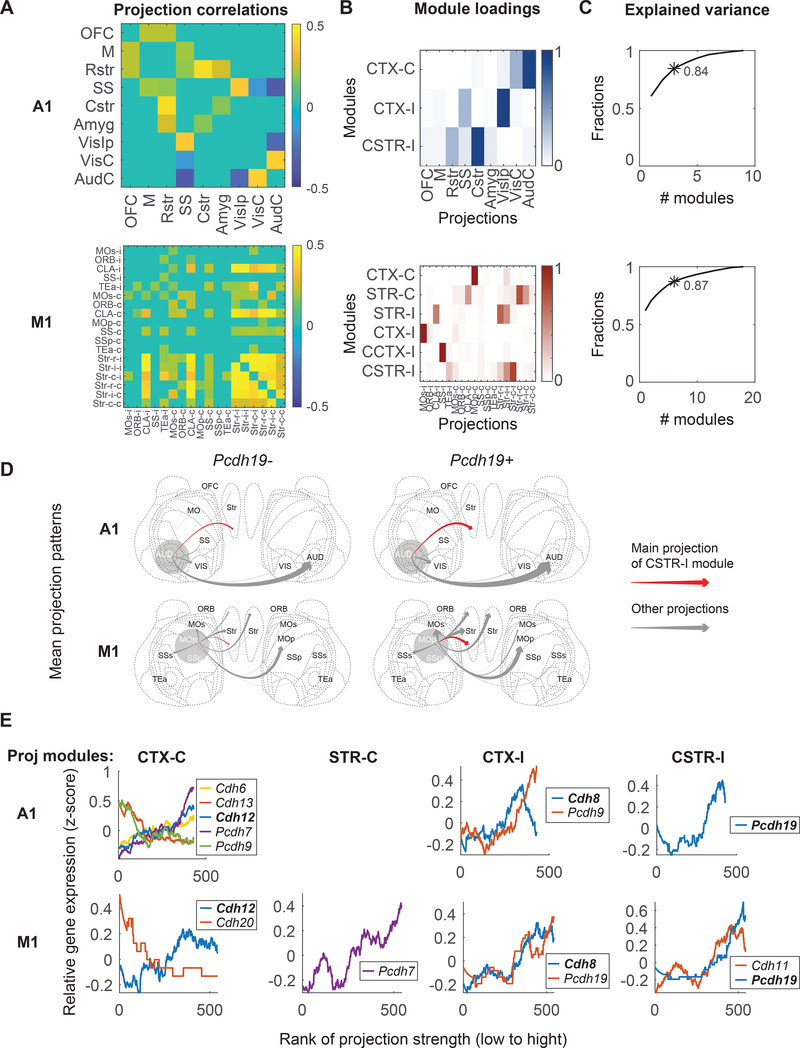
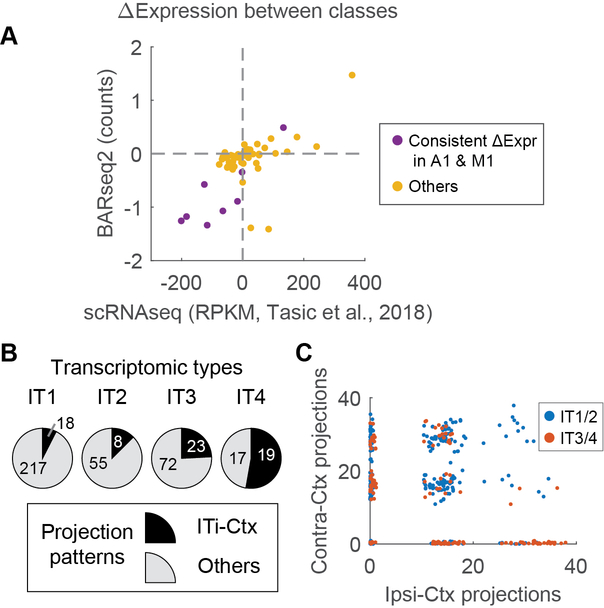
The projections of an IT neuron to its targets are not random. Rather, in both auditory cortex and motor cortex, these projections are organized and show statistical regularities that can be uncovered within the large datasets obtained by BARseq 2,6(Fig. 6A). For example, neurons in the auditory cortex that projected to the somatosensory cortex (SS) were also more likely to project to the ipsilateral visual cortex (VisIp), but not the contralateral auditory cortex (AudC). To exploit these correlations, we used non-negative matrix factorization (NMF)32 to represent the projection pattern of each neuron as the sum of several “projection modules.” (NMF is an algorithm related to PCA, but imposes the added constraint that projections be non-negative). Each of these modules (six modules for the motor cortex and three for the auditory cortex; Fig. 6B) consisted of subsets of projections that were likely to co-occur. We named these modules by the main projections (cortex, CTX, or striatum, STR) followed by the side of the projection (ipsilateral, -I, or contralateral, -C). For some modules, we further indicated that the projections were to the caudal part of the structure by prefixing with “C” (e.g. CSTR-I or CCTX-I). A small number of projection modules could explain most of the variance in projections (three modules and six modules explained 84 % and 87 % of the variance in projections to nine areas in auditory cortex and 18 areas in motor cortex that IT neurons project to, respectively; Fig. 6C).
Figure 6. Cadherins correlate with diverse projections of IT neurons.
(A) Pearson correlation of projections to different brain areas in IT neurons of auditory cortex (top) or motor cortex (bottom). Only significant correlations are shown. (B) Projection modules of IT neurons in auditory cortex (top) or motor cortex (bottom). Each row represents a projection module. Columns indicate projections to different brain areas. (C) The fractions of variance explained by different numbers of projection modules in auditory cortex (top) and motor cortex (bottom). The numbers of projection modules that correspond to those in (B) are labeled with an asterisk with the fraction of variance explained indicated. (D) Mean projection patterns of neurons in A1 (top) and M1 (bottom) with or without Pcdh19 expression. The thickness of arrows indicates projection strength (barcode counts). Red arrows indicate projections that correspond to the strongest projection in the CSTR-I projection modules. (E) The expression of cadherins (y-axes) that were rank correlated with the indicated projection modules in auditory cortex (top row) and motor cortex (bottom row). Neurons (x-axes) are sorted by the strengths of the indicated projection modules. Only genes that were significantly correlated with projection modules are shown (FDR < 0.1 using two-tailed rank sum tests). Genes that were correlated with the same projection modules in both areas are shown in bold.
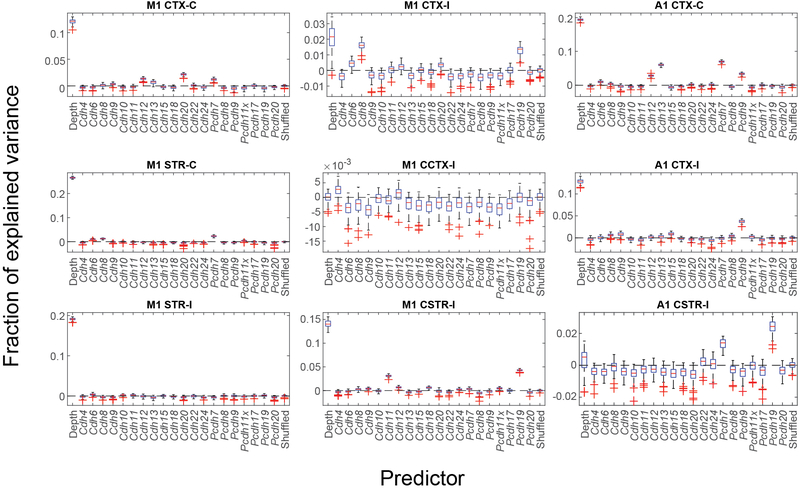
Because both the projection patterns of neurons 2,27 and their transcriptomic types 3,9 are well correlated with laminae, we first asked how well cadherins explained the diversity of projections in IT neurons compared to the laminar positions of neurons (see Methods). Although most cadherins had no predictive power on the projection modules, some individual cadherins could explain a significant fraction of the variance in projections compared to that explained by the laminar positions of neurons (Extended Data Fig. 8). For example, Cdh13 and Pcdh7 explained 6.0 ± 0.3% and 7.0 ± 0.3% (mean ± std) of the variations in CTX-C in auditory cortex, compared to 19.4 ± 0.3% (mean ± std) explained by the laminar positions of neurons. Strikingly, Pcdh19 and Pcdh7 were predictive of CSTR-I in auditory cortex whereas the laminar positions were not. These results indicate that some but not all cadherins were modestly predictive of projections, and that the predictive power of these cadherins could be comparable in magnitude to the laminar positions of neurons, one of the strongest known predictors of projection patterns.
To further understand how cadherin expression relates to projections, we examined how it co-varied with projection modules (Supp. Fig. S1). Interestingly, the expression of several cadherins co-varied with similar projection modules in both cortical areas. For example, auditory cortex neurons expressing Pcdh19 were stronger in the CSTR-I projection module than those not expressing Pcdh19 [Fig. 6D, top; p = 5 × 10−4 comparing the CSTR-I module in neurons with (n = 83) or without (n = 346) Pcdh19 expression using rank sum test]; the same association between Pcdh19 and the CSTR-I projection module was also seen in motor cortex (Fig. 6D, bottom; p = 4 × 10−6 using rank sum test, n = 31 for Pcdh19+ neurons and n = 512 for Pcdh19- neurons). Similarly, Cdh8 was correlated with the CTX-I module and Cdh12 was correlated with the CTX-C module (Fig. 6E, FDR < 0.1) in both auditory and motor cortex. These correlations were independently validated by retrograde tracing using cholera toxin subunit B (CTB) and FISH (Extended Data Fig. 9A–E; see Methods). Pcdh19, together with Cdh8 and Cdh11, respectively correlated with both CTX-I and CSTR-I modules in motor cortex (Fig. 6E; Extended Data Fig. 8), consistent with a potential combinatorial nature of cadherin correlates of projections. Although the correlations between individual cadherins and projections were relatively modest, our observations that the same cadherins correlated with similar projection modules in both areas suggest that a common molecular logic might underscore the organization of projections across cortical areas beyond class-level divisions.
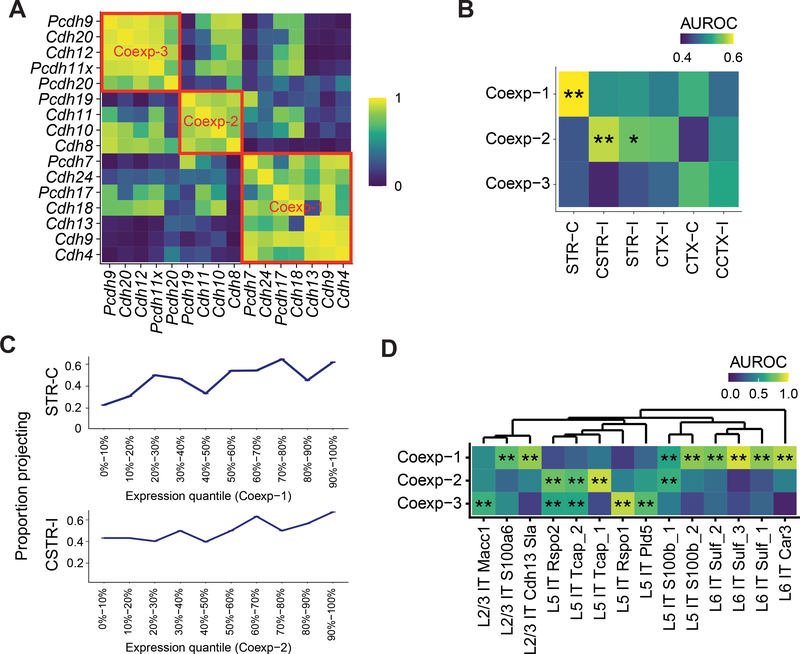
Analyses based on the expression of single genes suffer from biological and technical noise of gene expression in single neurons. We reasoned that the correlations among genes might allow us to identify additional relationships between gene expression and projections that were missed by analyzing each gene separately. This ability to leverage the relationship among genes represents an advantage of BARseq2 over the original BARseq because of the improved capacity of BARseq2 for multiplexed gene detection. To exploit the correlations among genes, we grouped 16 cadherins into three meta-analytic co-expression modules based on seven single-cell RNAseq datasets of IT neurons in motor cortex (Fig. 7A; Extended Data Fig. 10A, B) 23. To obtain the modules, we followed the rank-based network aggregation procedure defined by Ballouz, et al. 33 and Crow, et al. 34 to combine the seven dataset-specific gene-gene co-expression networks into an aggregated network, and then grouped together genes showing consistent excess correlation using the dynamic tree cutting algorithm 31. Two co-expressed modules were associated with projections: Module 1 was associated with contralateral striatal projections (STR-C projection module), and Module 2 was associated with ipsilateral caudal striatal projections (CSTR-I; Fig. 7B, C; Extended Data Fig. 10C, D). These associations between the co-expression modules and projections were consistent with, but stronger than, associations between individual genes contained in each module and the same projections (Extended Data Fig. 10E). Interestingly, these co-expression modules were enriched in multiple transcriptomic subtypes of IT neurons, but these transcriptomic subtypes were found in multiple branches of the transcriptomic taxonomy (Fig. 7D; Extended Data Fig. 10F). For example, Module 1 is associated with transcriptomic subtypes of IT neurons in L2/3, L5, and L6. This result is consistent with previous observations 3,6 that first-tier transcriptomic subtypes of IT neurons (i.e. subtypes of the highest level in the transcriptomic taxonomy within the IT class) appeared to share projection patterns, and further raises the possibility that transcriptomic taxonomy does not necessarily capture differences in projections. Taken together, our finding that projections correlate with cadherin co-expression modules independent of transcriptomic subtypes demonstrates that BARseq2 can reveal intricate relationships between gene expression and projection patterns.
Figure 7. Gene co-expression modules correlate with diverse projections of IT neurons.
(A) Correlation among cadherins as identified using single-cell RNAseq in IT neurons in motor cortex 23. Three co-expression modules are marked by red squares. Cadherins that did not belong to any module were not shown. (B) Association between cadherin co-expression modules and projection modules (AUROC). Significant associations are marked by asterisks (*FDR < 0.1, **FDR < 0.05). (C) Fractions of neurons with the indicated projection modules as a function of co-expression module expression. Neurons are binned by gene module quantiles as indicated. (D) Association of the three co-expression modules in transcriptomic IT neurons in the scSS dataset (AUROC, significance shown as in B).
Discussion
BARseq2 combines high-throughput mapping of projections to many brain areas with multiplexed detection of gene expression at single-cell resolution. Because BARseq2 is high-throughput, we are able to correlate gene expression and projection patterns of thousands of individual neurons in a single experiment, and thereby achieve statistical power that would be challenging to obtain using other single-cell techniques. By applying BARseq2 to two distant cortical areas—primary motor and auditory cortex—in the adult mouse, we identified cadherin correlates of diverse projections. Our results suggest that BARseq2 provides a path to discovering the general organization of gene expression and projections that are shared across the cortex.
High-throughput and multiplexed gene detection by BARseq2
To correlate panels of genes to projections, we designed BARseq2 to detect gene expression with high throughput, multiplex to dozens of genes, have sufficient sensitivity, and be compatible with barcoding-based projection mapping. To satisfy these needs, we based BARseq2 on padlock probe-based approaches 10,11. With additional optimizations for sensitivity, sequencing readout, and compatibility with barcode sequencing, we successfully used BARseq2 to identify gene correlates of projections.
One of the critical requirements for BARseq2 is high throughput when reading out many genes. Through strong amplification of mRNAs, combinatorial coding, and robust readout using Illumina sequencing chemistry 6,35, BARseq2 achieves fast imaging at low optical resolution compared to many other imaging-based spatial transcriptomic methods 14,36. Further optimizations, including computational approaches for resolving spatially mixed rolonies 37, have the potential to increase imaging throughput even further. Although the gene multiplexing capacity of BARseq2 may ultimately be limited by other physical constraints, such as crowding of rolonies and reduced detection sensitivity, these factors are unlikely to be limiting when multiplexing to dozens to hundreds of genes 11.
Another critical optimization was increasing the low sensitivity that early versions of the padlock probe-based technique suffered from, unless special and expensive primers were used 10. Inspired by other spatial transcriptomic methods, we and others 11 have found that tiling target genes with multiple probes could greatly improve the sensitivity. This design allowed variable sensitivity for different experimental purposes. Although in the present work we identified cadherin correlates of projections using only a modest number of probes per gene to achieve sensitivity similar to single-cell RNAseq using 10x Genomics v3, the sensitivity of BARseq2 can be considerably higher when more probes are used (Fig. 1E). This high and tunable sensitivity, combined with the fact that the gene multiplexing capacity of BARseq2 is not limited by imaging time, opens potential application of BARseq2 to a wide range of questions that require high-throughput interrogation of gene expression in situ.
BARseq2 reveals gene correlates of projections
BARseq2 exploits the high-throughput axonal projection mapping that BARseq offers to identify gene correlates of diverse projections. BARseq has sensitivity comparable to single neuron tracing 5. Although the spatial resolution of BARseq for projections is lower than conventional single neuron tracing, it offers throughput that is several orders of magnitude higher than the state-of-the-art single-cell tracing techniques 1,2. This high throughput allows BARseq to reveal higher-order statistical structure in projection patterns that would have been difficult to observe using existing techniques, such as single-cell tracing 5,6. The increased statistical power of BARseq, obtained at the cost of some spatial resolution, is reminiscent of different clustering power across single-cell RNAseq techniques of varying throughput and read depth 23,38. The high throughput of BARseq thereby provides a powerful asset for investigating the organization of projection patterns and their relationship to gene expression.
BARseq2 enables simultaneous measurement of multiplexed gene expression and axonal projections to many brain areas, at single neuron resolution and at a scale that would be difficult to achieve with other approaches. For example, Cre-dependent labeling allows interrogation of the gene expression and projection patterns of a genetically defined subpopulation of neurons 6. However, this approach lacks cellular resolution, is limited by the availability of Cre lines, and requires that a neuronal population of interest be specifically distinguished by the expression of one or two genes. The combination of single-cell transcriptomic techniques with retrograde labeling does provide cellular resolution, but can only interrogate projections to one or at most a small number of brain areas at a time 3,9. The inability to interrogate projections to many brain areas from the same neuron would miss higher-order statistical structures in projections, which are non-random 5 and provide additional information regarding other properties of the neurons, such as laminar position and gene expression 2,6. The projections of individual neurons to multiple brain areas can be obtained using multiplexed single-cell tracing 1, but the throughput of these methods remains relatively low. Moreover, many advanced single-cell tracing techniques require special sample processing that hinders multiplexed interrogation of gene expression in the same sample. The throughput of single-cell projection mapping was addressed by the original BARseq 6, but the small number of genes (up to three) that could be co-interrogated with projections limited its use in identifying the general relationship between gene expression and projections. BARseq2 thus addresses limitations of existing techniques and provides a powerful approach for probing the relationships between gene expression and projection patterns.
Cadherins correlate with diverse projections of IT neurons.
As a proof-of-principle, we used BARseq2 to identify several cadherins that correlate with homologous IT projections in both auditory and motor cortex, two spatially and transcriptomically distant areas with distinct cortical and subcortical projection targets. In addition, cadherin co-expression modules that correlated with projections were associated with multiple branches of the transcriptomic taxonomy. This type of correlation between neuronal connectivity and variations in gene expression independent of transcriptomic types is not unique to the cortex and has previously been observed in other brain areas, such as the hippocampus 39. Therefore, our findings are consistent with the hypothesis that a shared cell adhesion molecule code might underlie the diversity of cortical projections independent of transcriptomic types 18,39.
Even though the power of some cadherins to predict projections was comparable in magnitude to that of laminar position, a strong predictor of projection patterns, these cadherins could only explain a small fraction of the overall variance in projections. This noisy association between cadherin expression and projection patterns contrasts with the known roles of cadherins in specifying neuronal connectivity in the cortex and other circuits 20,40, but the relatively small magnitude of these associations is not surprising for a few reasons. First, gene expression programs and signaling cues needed for specifying projections are usually transient in development 41, so it is likely that these cadherins only represent the remnants of a common developmental program that establish projections 42, or may be needed for ongoing functions or maintenance of projections. Second, non-cadherin cell adhesion molecules (e.g. IgCAMs 43,44) and other cell surface molecules (e.g. Plexins, Semaphorins 45, Teneurins 46) are also involved in specifying projections, so cadherins likely only represent a fraction of the molecular programs that specify projections. Finally, cortical projections undergo extensive activity-dependent modifications after the initial specification, so the overall diversity in cortical projections is likely much higher than that produced by the initial molecular program. These possibilities can be better resolved by applying BARseq2 to reveal gene expression in both the projection neurons and the areas they project to during development, in combination with perturbation experiments. BARseq2 thus provides a path to discovering the myriad genetic programs that specify and/or correlate with long-range projections in both developing and mature animals.
BARseq2 builds a unified description of neuronal diversity
Neuronal barcoding was originally proposed as a method for untangling circuit connectivity at synaptic resolution 47,48. Solving neuronal connectivity with barcode sequencing not only has the potential to achieve high throughput and single-cell resolution by exploiting advances in sequencing technology, but also provides a path to integrate measurements of multiple neuronal properties in single neurons—toward the “Rosetta brain” 49. BARseq2 is a step toward this goal. Although BARseq2 currently only resolves projections at relatively low spatial resolution (brain areas, i.e. hundreds of microns), this limitation can be addressed in the future by using in situ sequencing to read out axonal barcodes (Yuan et al., unpublished data), which would resolve axonal projections at subcellular spatial resolution. Further combining in situ sequencing of axonal barcodes with synaptic labeling, expansion microscopy, and/or trans-synaptic viral labeling could yield information regarding the synaptic connectivity of neurons. Because BARseq2 integrates neuronal properties using spatial information, it is potentially compatible with other in situ assays, such as immunohistochemistry, two-photon calcium imaging, and dendritic morphological reconstruction. By spatially correlating various neuronal properties in single neurons, BARseq2 represents a feasible path towards achieving a comprehensive description of neuronal circuits.
Methods
Animal processing and tissue preparation
All animal procedures were carried out in accordance with the Institutional Animal Care and Use Committee protocol 19–16-10–07-03–00-4 at Cold Spring Harbor Laboratory. The animals were housed at maximum of 5 in a cage on a 12 hrs on/12 hrs off light cycle. The temperature in the facility was kept at 22 ˚C with a range not exceeding 20.5 ˚C to 26 ˚C. Humidity was maintained at around 45–55% not exceeding a range of 30–70%. A list of animals used is provided in Supp. Table S1.
For samples used for only endogenous mRNA detection, 8–10 week old male C57BL/6 mice were anesthetized and decapitated. We immediately embedded the brain in OCT in a 22 mm2 cryomold and snap-froze the tissue in an isopentane bath submerged in liquid nitrogen. Sections were cut into 10 μm-thick slices on Superfrost Plus Gold Slides (Electron Microscopy Sciences). Unlike in the original BARseq, the sections were directly melted onto slides without the use of a tape transfer system. This change in mounting methods allowed increased efficiency in gene detection. The slides were stored at −80 °C until use.
For BARseq2 samples, 8–10 week old male C57BL/6 mice were injected as indicated in Supp. Table S1. After 24 hrs, we anesthetized and decapitated the animal, punched out the injection site, and snap-froze the rest of the brain on a razor blade on dry ice for conventional MAPseq 6. The injection site was embedded, cryo-sectioned, and stored as described above.
To prepare samples for BARseq2 experiments, we immersed slides from −80 °C instantly into freshly made 4 % PFA (10mL vials of 20 % PFA; Electron Microscopy Sciences) in PBS for 30 mins at room temperature. We washed the samples in PBS for 5 mins before installing HybriWell-FL chambers (22 mm × 22 mm × 0.25 mm; Grace Bio-labs) for subsequent reactions on the samples. We then dehydrated the samples in 70 %, 85 %, and 100 % EtOH for 5 mins each, and then washed in 100 % EtOH for at least 1 hr at 4 °C. Finally, we rehydrated the samples in PBST (0.5 % Tween-20 in PBS).
For retrograde labeling experiments, we prepared 1.0 mg/mL of Cholera Toxin subunit B (CTB) in PBS from 100 μg for injections (see Supp. Table S1 for a list of animals and coordinates used). We perfused the animals with fresh 4 % PFA 96 hrs after injection, post-fixed for 24 hrs in 4 % PFA, and cryo-protected in 10 % sucrose in PBS for 12 hrs, 20 % sucrose in PBS for 12 hrs, and 30 % sucrose in PBS for 12 hrs. The brain was then frozen in OCT and cryo-sectioned to 20 μm slices using a tape transfer system.
BARseq2 detection of endogenous genes
We prepared a master mix of reverse transcription primers at 0.5 μM each for all target mRNAs. For volumes exceeding the amount required for reverse transcription, we speed-vacuumed to concentrate the primer mix into a smaller volume. We then prepared the reaction [0.5 μM per gene RT primer (IDT), 1 U/μL RiboLock RNase inhibitor (Thermo Fisher Scientific), 0.2 μg/μL BSA, 500 μM dNTPs (Thermo Fisher Scientific), 20 U/μL RevertAid H-Minus M-MuLV reverse transcriptase (Thermo Fisher Scientific) in 1× RT buffer]. We incubated the samples in reverse transcription at 37 °C overnight. After reverse transcription, we crosslinked the cDNAs in 50 mM BS(PEG)9 (Thermo Fisher Scientific) for 1 hr and neutralized excess crosslinker with 1 M Tris-HCl, pH 8.0 for 30 mins, and then washed the sample with PBST twice to eliminate excess Tris. We then prepared a master padlock mix with 200 nM per padlock probe for each target mRNA and speed-vacuumed the mixture for a higher concentration at a smaller volume, if necessary. We ligated the gene padlock probes on the cDNA [200 nM per gene padlock (IDT), 1 U/μL RiboLock RNase Inhibitor, 20 % formamide (Thermo Fisher Scientific), 50 mM KCl, 0.4 U/μL RNase H (Qiagen), and 0.5 U/μL Ampligase (Epicentre) in 1× Ampligase buffer] for 30 mins at 37 ˚C and 45 mins at 45 ˚C. Finally, we performed rolling circle amplification (RCA) [125 μM amino-allyl dUTP (Thermo Fisher Scientific), 0.2 μg/μL BSA, 250 μM dNTPs, 5 % glycerol, and 1 U/μL ϕ29 DNA polymerase (Thermo Fisher Scientific) in 1× ϕ29 DNA polymerase buffer] overnight at room temperature. After RCA, we again crosslinked the rolonies in 50 mM BS(PEG)9 for 1 hr, neutralized with 1 M Tris-HCl, pH 8.0 for 30 mins, and washed with PBST. We washed the sample in hybridization buffer (10 % formamide in 2× SSC) and then either added probe detection hybridization solution (0.25 μM fluorescent probe in hybridization buffer) or genes sequencing primer hybridization solution (1 μM of sequencing primer in hybridization buffer) for 10 mins at room temperature. We then washed the sample with hybridization buffer three times at two mins each, rinsed the sample in PBST twice, and proceeded to imaging or continue with Illumina sequencing.
BARseq2 simultaneous detection of endogenous genes and barcodes
We prepared a master mix of reverse transcription primers at 0.5 μM each for all target mRNAs. For volumes exceeding the amount required for reverse transcription, we speed-vacuumed to concentrate the primer mix into a smaller volume. We then prepared the reaction [0.5 μM per gene RT primer (IDT), 1 μM barcode LNA RT primer (Qiagen), 1U/μL RiboLock RNase inhibitor (Thermo Fisher Scientific), 0.2 μg/μL BSA, 500 μM dNTPs (Thermo Fisher Scientific), 20 U/μL RevertAid H-Minus M-MuLV reverse transcriptase (Thermo Fisher Scientific) in 1× RT buffer], adding the barcode LNA primer last into the reaction mix to reduce cross-hybridization due to the LNA strong binding affinity. We incubated the samples in reverse transcription at 37 °C overnight. After reverse transcription, we crosslinked the cDNAs in 50 mM BS(PEG)9 (Thermo Fisher Scientific) for 1 hr and neutralized excess crosslinker with 1 M Tris-HCl, pH 8.0 for 30 mins, and then washed the sample with PBST twice to eliminate excess Tris. We then prepared a master padlock mix with 200 nM per padlock probe for each target mRNA and speed-vacuumed the mixture for a higher concentration at a smaller volume, if necessary. We ligated the gene padlock probes on the cDNA [200 nM per gene padlock (IDT), 1 U/μL RiboLock RNase Inhibitor, 20 % formamide (Thermo Fisher Scientific), 50 mM KCl, 0.4 U/μL RNase H (Qiagen), and 0.5 U/μL Ampligase (Epicentre) in 1× Ampligase buffer] for 30 mins at 37 ˚C and 45 mins at 45 ˚C. After ligating padlock probes for our target genes, we ligated the padlock probe for the barcode cDNA [100 nM barcode padlock (IDT), 50 μM dNTPs, 5 % glycerol, 1 U/μL RiboLock RNase Inhibitor, 20 % formamide (Thermo Fisher Scientific), 50 mM KCl, 0.4 U/μL RNase H (Qiagen), 0.001 U/μl Phusion DNA polymerase (NEB), and 0.5 U/μL Ampligase (Epicentre) in 1× Ampligase buffer] without any wash in between, and incubated the reaction for 5 mins at 37 ˚C and 40 mins at 45 ˚C. We then washed the sample twice with PBST and once with hybridization buffer (10 % formamide in 2× SSC), before hybridizing 1 μM of RCA primer in hybridization buffer for 15 mins at room temperature. We washed the sample with hybridization buffer three times at two mins each. Finally, we performed rolling circle amplification (RCA) [125 μM aadUTP (Thermo Fisher Scientific), 0.2 μg/μL BSA, 250 μM dNTPs, 5 % glycerol, and 1 U/μL ϕ29 DNA polymerase (Thermo Fisher Scientific) in 1× ϕ29 DNA polymerase buffer] overnight at room temperature. After RCA, we again crosslinked the rolonies in 50 mM BS(PEG)9 for 1 hr, neutralized with 1 M Tris-HCl, pH 8.0 for 30 mins, and washed with PBST. We washed the sample in hybridization buffer (10 % formamide in 2× SSC) and then added genes sequencing primer hybridization solution (1 μM of sequencing primer in hybridization buffer) for 10 mins at room temperature. We then washed the sample with hybridization buffer three times at two mins each, rinsed the sample in PBST twice, and proceeded to Illumina sequencing.
In situ sequencing of endogenous genes
To sequence the endogenous genes using Illumina sequencing chemistry, we used the HiSeq Rapid SBS Kit v2 reagents to reduce cost from the original sequencing protocol 6. For the first cycle, we incubated samples in Universal Sequencing Buffer (USB) at 60 °C for 3 mins, then washed in PBST, and then incubated in iodoacetamide (9.3 mg in 2 mL PBST) at 60 °C for 3 mins. We washed the sample in PBST again, rinsed with USB twice more, and then incubated in Incorporation Mix (IRM) at 60 °C for 3 mins. We repeated the IRM step again to ensure as close to 100 % complete reaction as possible. We then washed the sample in PBST once and then continued to wash in PBST four more times at 60 °C for 3 mins each time. To reduce bleaching during imaging, we imaged the sample in Universal Scan Mix (USM).
For subsequent cycles, we first washed samples in USB, then incubated in Cleavage Reagent Mastermix (CRM) at 60 ˚C for 3 mins. We repeated the CRM step to ensure complete reaction and washed out residual CRM twice with Cleavage Wash Mix (CWM). We then washed the sample with USB, and then with PBST, before incubating in iodoacetamide at 60 ˚C for 3 mins. We repeated this step again to ensure we block as many of the free thiol-groups as possible to reduce background. We then continued with IRM and PBST washes as described for the first cycle and imaged after each cycle. We performed four sequencing cycles and seven sequencing cycles in total for our cadherins panel of 23 genes and our motor cell type markers and cadherins panel of 65 genes, respectively.
To visualize high expressors, we cleaved the fluorophores in the last sequencing cycle and washed the sample with CWM and PBST. We then washed our sample in hybridization buffer and added probe detection solution (0.5 μM each probe in hybridization buffer) for four different fluorescent probes detecting Slc17a7, Gad1, Slc30a3, and all previously sequenced genes, respectively, for 10 mins at room temperature. We washed the sample in the same hybridization buffer three times for two mins each, washed in PBST, before adding DAPI stain (ACDBio) for 2 mins at room temperature. We rinsed in PBST again and finally in USM for imaging.
In situ sequencing of barcodes
After sequencing and hybridizing for endogenous genes as described above, we stripped the sample of all hybridized oligos and sequenced bases by incubating twice in strip buffer (40 % formamide in 2× SSC with 0.01 % Triton-X) at 60 ˚C for 10 mins. We washed with PBST, then washed with hybridization buffer, and then incubated samples in barcode sequencing primer hybridization solution (1 μM sequencing primer in hybridization buffer) for 10 mins at room temperature. We washed with hybridization solution three times for two mins each, before rinsing twice in PBST. We sequenced barcodes with the same sequencing procedure as described for endogenous genes but for 15 cycles in total. Around cycle 4 or 5, we eliminate the iodoacetamide blocker incubation for the rest of sequencing because iodoacetamide blockage is irreversible, so further incubation in this blocker becomes unnecessary after several cycles.
Target area barcode sequencing
Barcode sequencing in target brain areas was performed by the Cold Spring Harbor Laboratory MAPseq core following procedures used in a previous study 6. The target areas were dissected to match two other studies in A1 6 and in M1 2, resulting in 11 and 35 projection targets for neurons in auditory cortex and motor cortex, respectively; these projection targets corresponded to most of the major projection targets based on bulk tracing 51. Detailed description of each dissected area and correspondence to the Allen reference atlas are shown in Supp. Table S2. Example annotated images from the dissected brain slices are provided at Mendeley Data (see Data Availability).
Fluorescent in situ hybridization (FISH)
FISH experiments were performed using RNAscope Fluorescent Multiplex Kit v1 according to the manufacturer’s protocols with minor modifications to sample preprocessing. For FISH experiments in comparison to BARseq2 endogenous mRNA detection (Fig. 1F; Fig. 2E), the samples were fresh-frozen in isopentane bath as described above. From −80 °C storage, the samples were immediately submerged in freshly-made 4 % PFA (Electron Microscopy Sciences) for 15 mins at 4 °C, then dehydrated in 75 %, 85 %, and 100 % ethanol twice for 5 mins each. After air-drying, we assembled HybriWell-FL chambers (22 mm × 22 mm × 0.25 mm; Grace Bio-Labs) and digested the samples in Protease IV for 30 mins at room temperature. We washed the samples in PBST, and then proceeded with probe hybridization and subsequent amplification and visualization steps following the manufacturer’s protocol, and mounted the samples with coverslips finally for imaging.
For FISH experiments in retrogradely labeled samples, we first imaged the samples before performing FISH. The samples were then dehydrated in 50 %, 75 % and 100 % ethanol twice for 5 mins each. After air-drying the samples, we either assembled HybriWell-FL chambers (22 mm × 22 mm × 0.25 mm; Grace Bio-Labs) or drew a barrier around the samples using a ImmEdge hydrophobic barrier pen. The samples were then digested in Protease III for 30 mins at 40 °C, and washed in nuclease-free H2O twice. We then proceeded to probe hybridization and subsequent amplification and visualization steps following the manufacturer’s protocol, and mounted the samples with coverslips finally for imaging.
For Fig. 1F, the FISH probes used were Mm-Slc17a7-C1, Mm-Slc30a3-C2, and Mm-Cdh13-C3 visualized with Amp4 A It A. For Fig. 2E, the FISH probes used were Mm-Pcdh19-C1, Mm-Cdh8-C2, and Mm-Pcdh20-C3 visualized with Amp4 A It A. For retrograde labeling experiments in Extended Data Fig. 9A–E, the FISH probes used for the cadherins were Mm-Cdh12-C1 (custom-ordered no. 842531), Mm-Cdh8-C1, or Mm-Pcdh19-C1, in addition to Mm-Slc30a3-C2 and Mm-Slc17a7-C3, visualized with Amp4 A It C.
Imaging
All sequencing experiments were performed on an Olympus IX81 microscope with Crest X-light 2 spinning disk confocal, a Photometrics BSI prime camera, and an 89North LDI 7-channel laser bank. Retrograde labeling experiments were imaged either on the same microscope or on an LSM 710 laser scanning confocal microscope. Filters and lasers used for imaging are listed in Supp. Table S3. Images were acquired using micro-manager v1.4.2352 on the spinning disk confocal and Zeiss Zen 2012 SP5 FP2 (Version 14.0.0.0) on the laser scanning confocal.
For all BARseq2 experiments, we imaged endogenous genes using an Olympus UPLFLN 40× 0.75 NA air objective and tiled 5 × 5 or 7 × 5 with 15 % overlap between tiles for all sequencing cycles and the hybridization cycles. For each sequencing cycle, the four sequencing channels (G, T, A, and C) and the DIC channel was captured. For hybridization cycles, GFP, RFP, TexasRed, Cy5, and DIC channels were captured. At the last cycle (usually the hybridization cycle for high expressors), we also imaged the DAPI channel.
For barcode sequencing, we imaged the first three cycles using the same imaging settings described above at 40×. The third sequencing cycle was additionally reimaged at 10× using an Olympus UPLANAPO 10× 0.45 NA air objective without tiling. All subsequent barcode sequencing cycles were imaged at 10×.
On the spinning disk confocal, all 40× BARseq2 and FISH images were acquired as z-stacks with 1 μm step size and 0.16 μm xy pixel size, and all 10× images were acquired as z-stacks with 5 μm step size.
On the LSM 710, CTB labeled samples were first imaged using a Plan-Apochromat 10× 0.45 NA objective without a coverslip as a z-stack with 7 μm z-step size and 0.7 μm xy pixel size. After FISH, the same samples were imaged using a Plan-Apochromat 20× 0.8 NA objective as a z-stack with 2 μm step size and 0.35 μm xy pixel size.
Probe design
A detailed description of probe sets used for each experiment and their sequences is provided in Supp. Table S4.
To design reverse transcription primers and padlock probes, we tried to design as many probe sets as possible on each transcript while avoiding the end (~20 nt) of the mRNA transcripts and ensuring at least a 3 nt gap between two adjacent probe sets. Specific reverse transcription primers were designed to be 25 to 26 nt with amino modifier C6 at the 5’ end and HPLC purified. In addition, we avoided sequences that contained G/C quadruplexes and/or had a low melting temperature (below 55 ˚C). Padlock probes were designed to have two arms of 21 to 23 nt with minimum Tm of 58 ˚C, GC contents between 40 % and 60 %, and high complexity. The two arms were connected by a backbone consisting of a 32 nt sequencing primer or detection probe target site, a 7 nt gene-specific index, and a 3 nt 3’ linker. For padlock probes designed for hybridization readout, different backbone sequences were used for different genes. We further filtered out padlock probe sequences with potential non-specific binding. To find potential non-specific binding targets, we blasted the ligated padlock arm sequences against the mouse genome and identified all targets with (1) 3 nt of perfect match on either side of the ligation junction, (2) no gap and/or insertion within 7 nt of the ligation junction, and (3) melting temperatures of at least 37 ˚C for non-specific binding of each arm.
We maximized the number of padlock probe sets for Slc17a7 (23 probes), Slc30a3 (19 probes), Gad1 (24 probes), and Cdh13 (30 probes). These probe sets were used to evaluate the relationship between detection sensitivity and probe numbers. For the cadherin panels and the cell-type marker panels, we selected a subset of probes for each gene so that we have at most 12 probe sets per gene. Some shorter genes had fewer than 12 probes. These panels resulted in sensitivity that was sufficient for the present experiments, albeit somewhat below the maximum achievable with more probes. All but three genes (Slc17a7, Slc30a3, and Gad1) were visualized using combinatorial GII codes (4-nt in auditory cortex and 7-nt in motor cortex; see Supp. Table S4); only a small subset of all possible GIIs were used, ensuring a Hamming distance of at least two bases between all pairs of GIIs in auditory cortex (out of 4-nt) and three bases in motor cortex (out of 7-nt) for error correction. The three remaining genes with high expression (Slc17a7, Gad1, and Slc30a3) were detected by hybridization.
Optimization of endogenous mRNA detection
We optimized padlock probes, tissue pretreatment, and reverse transcription to maximize detection sensitivity. We found that using multiple padlocks per mRNA transcript, each padlock targeting a different site on the mRNA coding sequence, increased detection efficiency significantly (Fig. 1E). The increase in sensitivity varied across genes, but this is likely caused by differences in sensitivity of the single probe to which we normalized the sensitivities. For tissue pretreatment, we found that thin fresh frozen tissue cryosections fixed with 4% PFA for 30 mins to 1 hour (Extended Data Fig. 1A) yielded higher mRNA sensitivity than shorter fixation or other pretreatments, such as PFA-perfused tissue slices with or without post-fixation. For reverse transcription, we found that reverse transcription primers specific to the targets at a concentration of 0.5 – 5 μM each yielded higher sensitivity than using random primers at concentrations up to 50 μM (Extended Data Fig. 1B). Altogether, these optimizations were crucial for increased mRNA detection sensitivity comparable to hybridization-based techniques.
To quantify the sensitivity of BARseq2 compared to conventional FISH methods, we detected two genes, Slc30a3 and Cdh13, using both BARseq2 and RNAscope (Fig. 1F). We also probed for a third gene, Slc17a7, but at the resolution we imaged at, we were unable to fully resolve the signals from both BARseq2 and RNAscope. We therefore only used Slc30a3 and Cdh13, not Slc17a7, to evaluate the sensitivity of BARseq2. Linear regression between BARseq2 and RNAscope counts of Slc30a3 and Cdh13 genes in these two genes resulted in a slope of 1.65 (Extended Data Fig. 1C, D; dashed line R2 = 0.73), indicating that BARseq2 achieved about 1 / 1.65 ≈ 60% sensitivity compared to RNAscope.
To multiplex gene detection with high imaging throughput, we optimized in situ sequencing to robustly read out GIIs of single rolonies over many sequencing cycles. We had previously adapted Illumina sequencing chemistry to sequence neuronal somata filled abundantly with RNA barcode rolonies, i.e. DNA nanoballs generated by rolling circle amplification 6,35. However, directly applying this method to sequence single rolonies generated from individual mRNAs proved difficult due to heating cycles and harsh stripping treatments that led to loss and/or jittering of rolonies (Extended Data Fig. 1E). To allow robust sequencing of single rolonies, we optimized cryo-sectioning and amino-allyl dUTP concentration 53 to crosslink rolonies more extensively, achieving less spatial jitter of single rolonies between imaging cycles (Extended Data Fig. 1E–H) and stronger signals (Extended Data Fig. 1I, J) retained over cycles. This robust in situ sequencing of combinatorial GII codes allowed BARseq2 to achieve fast imaging critical for high throughput correlation of gene expression with projections.
Simultaneous detection of endogenous mRNAs and barcodes using BARseq2
To assess multiplex gene expression and long-range projections in the same cells, we optimized for simultaneous detection and amplification of both endogenous mRNAs and barcodes. Although both endogenous mRNAs and barcodes are amplified using padlock probe-based approaches, amplifying barcodes required the addition of a DNA polymerase to copy barcode sequences into padlock probes to allow direct sequencing of diverse barcodes (up to ~1018 diversity; Fig. 1C, left). Directly combining the two processes reduced the detection sensitivity of target mRNAs due to the addition of the DNA polymerase [Extended Data Fig. 6A; 37 ± 3 % (mean ± standard error) comparing the Ctrl condition to the zero polymerase concentration]. To preserve detection sensitivity for endogenous mRNAs while allowing the sequencing of diverse barcodes, we adjusted the concentration of the DNA polymerase to 0.001 U/μl (1/200 of the amount in the original BARseq), which doubled the sensitivity for endogenous mRNAs while also maintaining the sensitivity for barcodes (Extended Data Fig. 6A). This optimization allowed BARseq2 to detect both endogenous mRNAs and RNA barcodes together in the same neurons without compromising sensitivity.
Single-cell RNAseq of auditory cortex
To dissociate neurons for single-cell RNAseq, we anesthetized animals with isofluorane and decapicated the animals. We then used a 2 mm biopsy punch to remove the auditory cortex. The tissue was then dissected in ice cold HABG medium [40 mL Hibernate A (Brainbits), 0.8 mL B27 (Thermo Fisher Scientific) and 0.1 mL Glutamax (Thermo Fisher Scientific)] into small pieces and digested in 3 mL pre-warmed papain solution [3mL Hibernate A-Ca (Brainbits), 6 mg papain (Brainbits), and 7.5 μL Glutamax] at 30 ˚C for 40 mins. The digested tissues were then triturated in 2 mL pre-warmed HABG for 10 times using a salinized pipette with 500 μm opening. The undissociated tissues were transferred to a new tube with 2 mL HABG and triturated another 10 times. The undissociated tissues were transferred again to a new tube with 2 mL HABG and triturated for 5 times. The three tubes of HABG were combined and laid on top of a density gradient of 17.3%, 12.4%, 9.9%, and 7.4% (v/v) Optiprep (Sigma) in HABG, and centrifuged at 750 g for 15 mins. After removing the top two fractions, we collected the next two and half fractions and diluted in 5 mL HABG and centrifuged at 300 g for 5 mins. The pellet was washed in 5 mL HABG, pelleted again, and resuspended in 100 μL HABG. The cell suspension was then processed for library preparation using 10x Genomics Chromium Single Cell 3’ Kits v3 according to the manufacturer’s protocol. One of the single-cell RNAseq dataset was previously published 6, and a new dataset was obtained in this study.
BARseq2 data processing
Sequencing data for projection target areas were acquired through the MAPseq core facility at Cold Spring Harbor Laboratory. We first de-multiplexed raw sequencing reads and thresheld by read counts per molecule to remove PCR errors. This produced a list of unique barcode sequences with molecule counts in each target area. We then corrected for sequencing and amplification errors, allowing up to three mismatches. The resulting error corrected barcode molecule counts were used to generate the projection matrix. A sample script for processing target area barcodes is provided at Mendeley Data (see Data Availability).
To process in situ sequencing data for genes, we first performed max projection of the image stacks along the z-axis. Each max projection image was then corrected for sequencing channel bleedthrough and lateral shift across channels. The images were then filtered with a median filter and background subtracted using a rolling ball with a radius of 10 pixels. The sequencing cycle images were then registered to the first sequencing cycle using the sum of all four sequencing channels, and the hybridization images were registered to the first sequencing using the channel that labeled all sequenced rolonies. Registrations were performed by maximizing enhanced cross correlation 54. After all images were registered, putative rolonies were then picked from the first sequencing cycle by finding all peaks that were at least brighter than all surrounding valleys by a certain threshold determined empirically. This was achieved by first performing morphological reconstruction using the original image as mask and the image minus the threshold as marker, followed by identifying all local maxima. We then deconvolve all registered images and find the signal intensities for all rolonies across all sequencing cycles and channels.
At this point the signal for each rolony is represented by an m × 1 vector, in which m equals four (sequencing channels) times the number of cycles. To identify the gene that each rolony corresponds to, we project the signal vector onto the signal vector of all genes and find the two genes with the highest projections, I1 and I2. For rolonies whose (I1 - I2) / I1 is above a threshold, we assign the genes with the highest projections to these rolonies. The remaining rolonies are filtered out. For hybridization cycles, the channel in which the rolonies are found is used directly to identify the genes.
In experiments in which genes were detected without barcodes for projection mapping, we segmented somas based on the rolony signals, background fluorescence from somas, and nuclear stain using Cellpose 55, and assigned the rolonies to the segmented cells.
In experiments in which genes were detected in conjugation with barcodes, we further registered barcode sequencing cycles to the first sequencing cycle for genes using the DIC channel. The barcode sequencing images were then filtered with a median filter and background subtracted using a rolling ball with a radius of 50 pixels. The high-resolution images for the second and third cycles were then registered to the first sequencing cycle of barcodes using the sum of all four sequencing channels. The low-resolution images of the third sequencing cycle were then registered to the high-resolution image of the same cycle.
To segment the barcoded cells from the high-resolution images, we first identify “seed” pixels by identifying local maxima in the first sequencing cycle image as described above. These seed pixels are positions of the strongest signal within putative cell bodies. Then for each seed pixel, we calculate the projection of signal vectors for all other pixels within a local area on the signal vector of the seed pixel and the rejection of signal vectors for these pixels from the signal vector of the seed pixel. We then segment the cell bodies by finding all pixels that fulfill the following criteria: (1) the projections of their signal vectors are above a threshold, (2) the ratios between the rejections and projections are below a threshold, and (3) are connected to the seed pixel. In parallel, we perform a second segmentation using only the DAPI signals and gene sequencing images with marker-based watershedding without using the barcode sequencing images, and find the segmented cells that overlap with the barcode segmented cells. We then visually inspect the sequencing images and segmentations for each cell to determine which segmentation produced better result and to eliminate badly segmented cells. We then assign gene rolonies to the filtered segmented cells to produce the expression matrix.
To find the barcode sequences of the segmented cell, we integrate signals over the whole segmented cells and call the channel with the strongest signal as the base in both the high-resolution images and the low-resolution images. We then concatenate the sequences from the high-resolution images and the low-resolution images to produce the full barcode sequences. To find the projection patterns, these in situ sequenced barcodes are then matched to the barcodes identified in the projection areas allowing one mismatch but not ambiguous matches (i.e. one in situ barcode matching to multiple barcodes found in projection sites).
Analysis of BARseq2 gene expression data
All analyses were carried out in MATLAB. Scripts for all analyses are provided at Mendeley Data (see Data Availability). For analysis of gene-only datasets, neurons were first filtered by requiring at least 10 counts of Slc17a7 or Gad1 and were positioned within the cortex. To make the data comparable to previous studies 6, the cortical depths of neurons were normalized to a total thickness of 1200 μm for auditory cortex and 1500 μm for motor cortex. To find cadherins that were differentially expressed in cell types, the expression of cadherins in each cell type was compared to the expression of cadherins in all other cell types using rank sum tests.
Laminar distribution of cadherins
Because many genes, especially cell adhesion molecules, are differentially expressed across cortical layers, we evaluated how well BARseq2 can capture spatial organization of cadherins compared to existing methods, such as FISH. To compare laminar distribution observed by BARseq2, FISH, and Allen Brain Atlas, we quantified gene expression signal densities across 100 μm bins in laminar depth. For BARseq2 and FISH, the quantification was done by counting dots. For Allen Brain Atlas, the quantifications were done by integrating signal intensities over all pixels in each bin. Because each bin had different number of pixels sampled in our data, we then divided the gene expression signals by the area observed in the images to calculate the density. We then z-scored the densities within each gene to produce the laminar profiles for each gene.
RNAscope against Cdh8, Pcdh19, and Pcdh20 revealed laminar expression profiles that were qualitatively similar to those obtained by BARseq2 (Fig. 2E). For Pcdh20, the dynamic range of gene expression (i.e. the differences between peaks and valleys in expression) was more pronounced in the BARseq2 data than that observed by RNAscope. Because low sensitivity and/or low specificity would likely result in a reduction, not an increase, in the dynamic range of expression, it is unlikely that such quantitative differences in the laminar profiles of gene expression were caused by sensitivity and/or specificity issues with BARseq2. We suspect that the reduced dynamic range in RNAscope is caused by non-specific signals inherent to amplified FISH methods. We therefore sought to compare BARseq2 to other FISH datasets to confirm its accuracy.
We then compared the distributions of genes obtained by BARseq2 to those in the Allen gene expression atlas 21(Fig. 2F; Extended Data Fig. 3). The laminar distribution of gene expression revealed by BARseq2 was highly correlated with that in the Allen gene expression atlas (Spearman correlation ρ = 0.696, p = 3.8×10−29). Specifically, the laminar distribution of Pcdh20 obtained by BARseq2 matched very well with Pcdh20 in the Allen gene expression atlas (Extended Data Fig. 3). These results indicate that BARseq2 accurately captured the laminar distribution of cadherin expression.
Gene-pair expression in single neurons
To test whether BARseq2 accurately captures gene expression, we compared the expression of two pairs of genes in single neurons. First we compared the expression of Slc17a7 and Gad1, two genes that are expressed in two distinct classes of neurons. Second we compared the expression of Slc30a3 and Cdh24, two genes that are anti-correlated at the subtype level based on single-cell RNAseq 3.
Slc17a7 and Gad1 are expressed in excitatory and inhibitory neurons, respectively. They are thus almost never expressed in the same neuron in the cortex. To quantify the mutual exclusivity of Slc17a7 and Gad1 in neurons, we defined the exclusivity index , where indicates the probability of a cell expressing at least 10 counts of Gad1 conditioned on the expression of at least 10 counts of Slc17a7, and indicates the probability of a cell expressing at least 10 counts of Gad1 in all filtered neurons.
BARseq2 recapitulated the mutual exclusivity between these two genes (Fig. 2J, K), but a small number of neurons did express both Slc17a7 and Gad1 (grey cells in Fig. 2J). This could be caused by overlapping cells (i.e. an inhibitory neuron and an excitatory neuron at the same x/y position, but in different z planes were merged together in the max projection images) or cell segmentation errors (two adjacent cells incorrectly segmented as a single cell). Because the sections we used were 10 μm thick, which was comparable to the diameter of an average neuron, the latter source of error was likely to be more common.
This type of error was similar to doublets in droplet-based single-cell RNAseq techniques. Assuming that the mutual exclusions of Slc17a7 and Gad1 were absolute, then we could estimate the “doublet” rate as the ratio between the probability of neurons expressing both genes and the product of the probabilities of neurons expressing either gene. Using this formula, we estimated the doublet rate of BARseq2 to be 7.5%, which is in a similar range as droplet-based single-cell RNAseq techniques (usually < 5%). Further improvement in cell segmentation algorithms may further reduce the doublet rate.
In addition to cells that express both Gad1 and Slc17a7 at significant levels, most cells that expressed one of the two genes dominantly also had non-zero expression of the other gene, albeit at much lower levels. This noise floor could be caused by mRNAs in dendrites that were incorrectly assigned to other neurons. Because the expression of these genes in the somata were much higher than that in the dendrites, this type of error was unlikely to significantly affect the determination of excitatory and inhibitory neurons.
Similarly, consistent with a previous single-cell RNAseq study 3, BARseq2 also confirmed the observation that Slc30a3 was more highly expressed in subtypes of excitatory neurons that did not express Cdh24 compared to projection neurons that did express Cdh24 [Extended Data 0. 5A, B; p = 5 × 10−26 using two-tailed rank sum test on single-cell RNAseq data using Smart-Seq2 (n = 10,044 neurons) 3, and p = 4 × 10−65 on BARseq2 data (n = 2,947 neurons)].
Cell typing in BARseq2 and single cell data
To select a panel of marker genes, we chose meta-analytic markers from 7 single-cell RNAseq in the motor cortex 23, accessed from the NeMo archive as indicated in the manuscript. In each dataset and for each cell type, we extracted differentially expressed (DE) genes among excitatory neurons (“Glutamatergic” class, 1-vs-all DE, fold change > 2, Mann-Whitney FDR < 0.05). We filtered out lowly expressed genes (average Counts Per Million < 100), then ranked genes according primarily by the number of datasets where they were DE, secondarily by average fold change and selected the top 5 markers.
To examine if multiplexing affects detection sensitivity, we probed for Slc17a7, Slc30a3, and Gad1 either as a separate three-gene panel or as part of the 65-gene panel (20 cadherins and 45 marker genes). The mean expression density across laminar positions for the three genes were similar between the three-gene panel and the 65-gene panel (Extended Data Fig. 5C; p = 0.22 for Slc17a7, p = 0.49 for Slc30a3, and p = 0.66 for Gad1 using two-tailed rank sum tests, respectively), suggesting that targeting more genes did not affect detection sensitivity of each gene.
To call cell types in BARseq2 and single cell data, we used the following procedure. First we normalized counts to log(1 + CPM), then we computed the average marker expression for each cell type and assigned the cell type with the highest average expression. If two marker sets were tied for highest expression, the cell was left unassigned. This method of cell typing achieved good precision and recall for most cell types when applied to single-cell RNAseq data (Extended Data Fig. 5D). We applied the procedure across 9 datasets to check whether it is robust across technologies and sequencing depth (Extended Data Fig. 5E, F). Overall, we observed extremely high performance for NP and CT subtypes in all cases, while L6b was slightly better predicted in high depth datasets. The cell typing method always predicted IT cells correctly, but not always the correct layer (L2/3, L5, L6, Car3, Extended Data Fig. 5G). This is consistent with the observation that IT types form a continuum in single cell datasets, making it difficult to fully separate subtypes by layer. Finally, the PT type proved to be the most difficult type to predict. While all PT cells were correctly annotated as PT (Extended Data Fig. 5H), numerous L2/3 IT and L5 IT cells were wrongly annotated as PT, in particular in high depth datasets (Extended Data Fig. 5F, G). We believe that this was due to an imbalance in the marker panel, with PT markers being higher expressing than markers from other types. We tested various normalization procedure to overcome this effect but found that results were insensitive to normalization overall (Extended Data Fig. 5F).
Using this panel and cell typing method, we determined the transcriptomic types of excitatory neurons in motor cortex using BARseq2 (Fig. 3B). Most transcriptomic types were found enriched in the correct layers. One exception to this was the L6 Car3+ IT type. In general, very few L6 Car3+ IT neurons were identified by BARseq2. Furthermore, even though L6 Car3+ IT neurons were predominantly in L6, some were identified in L2/3 by BARseq2 (Fig. 3C). This result was surprising, given that L6 Car3+ IT neurons, when present, were only rarely mistyped as L2/3 in our preliminary analyses (Extended Data Fig. 5G). L6 Car3+ IT neurons were only rarely detected in the datasets used to select markers, so we expect that using additional data will lead to a more robust marker selection and better cell-typing performance with BARseq2. These optimizations, however, are beyond the scope of this paper.
Gene expression in barcoded neurons
Gene expression in Sindbis-infected barcoded neurons largely reflect the gene expression in non-barcoded neurons. For example, the expression of the excitatory marker Slc17a7 and the inhibitory marker Gad1 remained mutually exclusive in barcoded neurons in both auditory cortex and motor cortex (Extended Data Fig. 6C, D). This mutual exclusivity was preserved despite an overall reduction in mRNA expression (Extended Data Fig. 6E; median reads of 38 in barcoded cells in both auditory and motor cortex, compared to 64 and 48 in non-barcoded cells in the two cortical areas, respectively). Similarly, Slc30a3 remained differentially expressed across barcoded excitatory neurons with or without Cdh24 expression as it was in non-barcoded excitatory neurons (Extended Data Fig. 6F; p = 1 × 10−6 using rank sum test, n = 810 neurons). Although our observations cannot rule out the possibility that a small subset of genes (e.g. viral response genes) may be disrupted by Sindbis infection, these results suggest that the co-expression relationships of most genes in Sindbis-infected neurons reflect those in non-infected cells.
Analysis of BARseq2 gene expression and projection dataset
For analysis of BARseq2 datasets with both gene expression and projections, we first evaluated the mutual exclusivity of Slc17a7 and Gad1 expression (see below). For this purpose, the neurons were filtered with the same thresholds as in the gene-only dataset. For all other analyses, we used a more relaxed filtering to compensate for the reduced gene expression in barcoded cells, requiring neurons to have at least 5 counts of Slc17a7 or Gad1. In this filtered set, neurons were considered excitatory if the counts of Slc17a7 were larger than the counts of Gad1, and were considered inhibitory if the counts of Gad1 were larger than the counts of Slc17a7. Projection data were log normalized as in previous studies 6. We further normalized the projection strengths of each area to two previous clustered BARseq dataset 6 and used a random forest classifier to assign neurons to projection clusters.
To find cadherins that were differentially expressed across major projection classes and between auditory and motor cortex, we performed rank sum tests for pairwise comparisons among major classes or the two areas for each cadherin and calculated the FDRs.
Projection modules were identified using non-negative matrix factorization 32. To find the variance in projections explained by cadherins and/or laminar positions (Extended Data Fig. 8), we used Gaussian process regression to predict projection modules using the laminar position of neurons as a predictor and linear regression to predict projection modules using the expression of individual cadherins. The variance explained by each predictor was reported after 100 iterations of 10-fold cross validation. To find cadherins that were associated with projection modules, we calculated the Spearman correlation between the coefficients for projection modules and gene counts. To generate the plots of differential gene expression in Fig. 6E, we sorted the neurons by the coefficients for projection modules and smoothed gene expression using a window of 101 neurons.
Projections of excitatory and inhibitory neurons
BARseq2 accurately observed the fact that projection neurons in the cortex are predominantly excitatory and express the excitatory marker Slc17a7, not the inhibitory marker Gad1. To distinguish between excitatory and inhibitory neurons, we categorized a neuron as excitatory or inhibitory if (1) the neuron had higher expression of the excitatory marker Slc17a7 or the inhibitory marker Gad1, respectively; and (2) the marker was expressed at greater than five reads in the cell. This threshold resulted in 2,496 excitatory neurons (947 in auditory and 1,549 in motor cortex) and 240 inhibitory neurons (100 in auditory cortex and 140 in motor cortex) (Fig. 4D). Consistent with previous observations, most cortical projection neurons identified by BARseq2 were excitatory (Fig. 4E). However, we also identified a small fraction of inhibitory projection neurons. Some of these neurons could be caused by “doublets” as discussed above. Consistent with this hypothesis, the inhibitory projection neurons (and some excitatory projection neurons) in motor cortex expressed both Gad1 and Slc17a7 at similar levels (Extended Data Fig. 6G). However, inhibitory projection neurons in auditory cortex expressed only Gad1, not Slc17a7 (Extended Data Fig. 6H), suggesting that these were real inhibitory projection neurons. This observation was consistent with previous reports of rare inhibitory projection neurons in the cortex 6,56. We did not further analyze these inhibitory projection neurons.
We also observed many excitatory neurons without projections (Fig. 4D, E), similar to those observed in previous BARseq experiments 6. These neurons were likely non-projecting excitatory neurons and neurons that project only locally or to neighboring cortical areas 3 that we did not sample.
Differential expression of cadherins across IT, PT, and CT neurons
BARseq2 revealed differential gene expression across major classes of neurons defined by projections. We found that many cadherins (8 for auditory cortex and 12 for motor cortex) were differentially expressed across intratelencephalic (IT) neurons, pyramidal tract (PT) neurons, and corticothalamic (CT) neurons that were defined by projections as in previous studies2,6 (Fig. 5A–C). Several cadherins were consistently differentially expressed in both cortical areas. For example, Cdh6 and Cdh13 were over-expressed in PT neurons compared to the other two classes, whereas Cdh8 was under-expressed in CT neurons compared to the other two classes (FDR < 0.05 using rank sum test). In addition, we also found nine cadherins that were differentially expressed across the two cortical areas in at least one class (Fig. 5D; FDR < 0.05 using rank sum tests).
Major classes of projection neurons (IT, PT, and CT) differ in both gene expression and projection patterns. Therefore, the differential expression of cadherins observed across these three major classes defined by projection patterns should be consistent with the differential expression across the classes defined by transcriptomic methods. To test this, we compared the differences in mean expression of cadherins in the three classes in motor and auditory cortex observed by BARseq2 to those observed using single-cell RNAseq in neighboring cortical areas (V1 and ALM) 3. Generally, differentially expressed cadherins identified by BARseq were also differentially expressed in single-cell RNAseq (Extended Data Fig. 7A; the rank correlation of the differences in cadherin expression across major neuronal types was 0.61 between BARseq and single-cell RNAseq, compared to 0.39 between auditory and motor cortex in BARseq). Importantly, all cadherins that were consistently differentially expressed in both A1 and M1 were also differentially expressed across the same pairs of major classes in V1 and ALM as shown by single-cell RNAseq (purple dots in Extended Data Fig. 7A). Several cadherins, including Pcdh7 and Cdh11, were differentially expressed with the opposite signs in single-cell RNAseq and in BARseq2 (yellow dots in lower right quadrant in Extended Data Fig. 7A). However, these cadherins were not consistently expressed across motor and auditory cortex. For example, Pcdh7 was expressed at significantly higher level in PT neurons than CT neurons in motor cortex (p < 10−8; Fig. 5C), but at lower level in PT neurons than CT neurons in auditory cortex (p = 0.0011, not statistically significant at FDR < 0.05). It is thus likely that these differences between observations by BARseq2 and by single-cell RNAseq reflect area-to-area differences, not methodological differences. These results confirm the differential expression of cadherins across major classes identified by BARseq2.
Projection differences across transcriptionally defined IT subtypes
6BARseq2 confirmed known biases in projection patterns across transcriptomic IT subtypes in auditory cortex (Extended Data Fig. 7B, C). Previous studies using both barcoding-based strategy and single-cell tracing have identified distinctive projection patterns for two transcriptomic subtypes of IT neurons, IT3 (L6 IT) and IT4 (L6 Car3+ IT) 6,26. To test if we could capture the same projection specificity of transcriptomic subtypes, we mapped projection patterns to projection clusters identified in a previous study in auditory cortex, and used a combination of gene expression and laminar position to distinguish four transcriptomic subtypes of IT neurons 6. These subtypes were defined consistently with a previous study 6 to allow easy comparison. Specifically, we defined IT1 as neurons with depths less than 590 μm, IT2 as neurons with depths between 590 and 830 μm and did not express Cdh13, IT3 as neurons between 590 and 830 μm that expressed Cdh13 or neurons deeper than 830 μm that expressed Slc30a3, and IT4 as neurons deeper than 830 μm that did not express Slc30a3.
As expected, the two transcriptomic subtypes (IT3 and IT4) predominantly found in L5 and L6 were indeed more likely to project only to the ipsilateral cortex, without projections to the contralateral cortex or the striatum (p = 4 ×10−7 comparing the fraction of neurons with only ipsilateral cortical projections in IT3/IT4 to the fraction of them in IT1/IT2 using Fisher’s test; Extended Data Fig. 7B, C). Between IT3 and IT4, IT4 neurons were more likely to project ipsilaterally (58 % IT3 neurons compared to 92 % IT4 neurons, p = 1×10−4 using Fisher’s test), whereas IT3 neurons were more likely to project contralaterally (66 % IT3 neurons compared to 14 % IT4 neurons, p = 5 ×10−8 using Fisher’s test). Thus, BARseq2 recapitulated known projection differences across transcriptomic subtypes of IT neurons.
Cadherin co-expression module analysis
To extract robust modules of co-expressed cadherins, we used a previously developed approach to combine multiple datasets meta-analytically, a crucial step to attenuate technical and biological noise 33,34. Briefly, we built co-expression networks using Spearman correlation for 7 single-cell RNAseq in the motor cortex 23, accessed from the NeMo archive as indicated in the manuscript and subset to the following subclasses: “L2/3 IT”, “L4/5IT”, “L5 IT”, “L6 IT” and “L6 IT Car3”. We ranked each network, then averaged the networks to obtain our final meta-analytic network. We then applied hierarchical clustering with average linkage and extracted modules using the dynamic cutting tree algorithm 31.
To compute the association between co-expression modules and projection patterns, we framed the association as a classification task: can we predict projection patterns from module expression? First, we generated labels by binarizing each projection pattern: cells with a projection strictly greater than the median projection strength were marked as positives. Then we generated predictors by computing gene module expression as the average Log(CPM+1) across all genes in the module. We reported the association strength (classification results) as an area under the receiver-operator characteristic curve (AUROC). To compute the association between co-expression modules and cell types, we used a similar approach, using clusters defined by the BICCN 23 as labels. For visualization, cell types are organized according to the following procedure: cell types are reduced to a centroid by taking the median expression for each gene, then cell types are clustered according to hierarchical clustering with average linkage with correlation-based distance.
Validation of cadherin correlates of IT projections using in situ hybridization and retrograde labeling.
To confirm that Cdh8, Cdh12, and Pcdh19 correlated with ipsilateral, contralateral, and striatal projections, respectively, we performed CTB retrograde labeling from the projection targets and performed FISH against Slc17a7, Slc30a3, and the cadherins in both A1 and M1 (Extended Data Fig. 9A; see Supp. Table S1 for injection coordinates). We then quantified cadherin expression and CTB labeling in IT neurons that had good DAPI signals and expressed both Slc17a7, an excitatory cell marker, and Slc30a3, which labeled the majority of IT neurons (Extended Data Fig. 9B). Neurons that had weak and/or ambiguous CTB signals were excluded from the analyses. Indeed, we saw that the three cadherins were expressed at higher levels in CTB+ neurons in both areas despite significant overlap in expression between CTB+ and CTB- neurons (Extended Data Fig. 9C–E). This overlap was expected because CTB was unlikely to have labeled all neurons that projected to the areas that we sampled with BARseq2. For example, in a previous study, we found that less than half of neurons with projections detected by BARseq were also labeled by CTB injected to the same target area 6. These results thus provide further support for the finding that cadherins correlate with similar projections in both A1 and M1.
Statistics and Reproducibility
No statistical method was used to predetermine sample size but our sample sizes are similar to those reported in previous publications6,14. No data were excluded from the analyses. Because only wild-type animals were used and the findings did not rely on comparison across animals, the experiments were not randomized and the investigators were not blinded to allocation of animals during experiments and outcome assessment. All statistical tests performed were indicated in the text. Two-tailed tests and Bonferroni correction was used for all p values reported unless noted otherwise. Wherever indicated, False Discovery Rates (FDRs) were computed according to the Benjamini-Hochberg procedure 57. All statistical tests used were non-parametric except when statistical significance is estimated for Pearson correlation (Fig. 6A). When estimating statistical significance for Pearson correlation, normal distribution was assumed but this was not formally tested.
Data availability
Raw target area sequencing data (Fig. 4C) are deposited at SRA (SRR12247894, SRR12245390, and SRR12245389). Single-cell RNAseq data (Fig. 2G–I) are deposited at SRA (SRR13716225). Raw in situ sequencing images (Fig. 2–4) are deposited at Brain Image Library (https://download.brainimagelibrary.org/06/35/0635a0b3b0954c7e/). Other data and intermediate processed sequencing data are deposited at Mendeley Data (http://dx.doi.org/10.17632/jnx89bmv4s.1).
Code availability
Processing scripts are deposited at Mendeley Data (http://dx.doi.org/10.17632/jnx89bmv4s.1).
Extended Data
Extended Data Fig. 1. Optimization of BARseq2 for detecting endogenous mRNAs.
(A) Relative sensitivity (means and individual data points) of BARseq2 in detecting Slc17a7 using the indicated fixation times, normalized to that achieved with 5 mins of fixation. n = 3 for 480 mins and n = 4 for other conditions. (B) Rolony counts for Slc17a7 using either random primers or specific primers at two different concentrations. The two concentrations used were 5 μM (low) and 50 μM (high) for random primers, and 0.5 μM (low) and 5 μM (high) for specific primers. Lines indicate means and dots/crosses represent individual samples. n = 2 slices for each condition. (C) (D) BARseq2 sensitivity compared to RNAscope. (C) Spot density detected by BARseq2 or RNAscope in each 100 μm bin along the laminar axis in auditory cortex. Error bars indicate standard errors. The dashed line indicates linear fit for Slc30a3 and Cdh13. Slope = 1.65 and R2 = 0.73. n = 5 slices for both BARseq2 and RNAscope. (D) shows the means and individual samples for each gene. (E)(F) Positions of rolonies across five sequencing cycles using the original (E) or the optimized (F) sequencing protocol. Scale bars = 10 μm. (G) The distribution of minimum distance between rolonies imaged in the first cycle and in the fifth cycle using the original or the optimized protocol. (H) Median distance between rolonies imaged in the indicated cycles and the closest rolonies imaged in the first cycle using the original or the optimized protocol. Error bars indicate standard errors. For both (G) and (H), n = 148,708 rolonies for optimized condition and n = 12,114 for original condition. (I)(J) The distribution of absolute rolony intensities for the first sequencing cycle (I) and relative rolony intensities after 6 sequencing cycles and one stripping step, normalized to the intensities in the first sequencing cycle (J). Amino-allyl dUTP concentrations used are indicated. In (I), n = 63,852 rolonies for 0.08 μM and n = 4,286 rolonies for 0.5 μM; in (J), n = 128,976 rolonies for 0.08 μM and n = 113,235 rolonies for 0.5 μM.
Extended Data Fig. 2. Laminar distribution of cadherins in auditory cortex (green) and motor cortex (brown).
In both cortical areas, cortical depth is normalized so that the bottom and the top of the cortex match between M1 and A1.
Extended Data Fig. 3. Comparison between BARseq2 and Allen gene expression atlas.
Gene expression patterns in auditory cortex identified by BARseq2 are plotted next to in situ hybridization images of the same genes in Allen gene expression atlas (ABA) and the quantified laminar distribution of the gene in both datasets. Only genes that had coronal images in the Allen gene expression atlas are shown. Blue lines indicate the boundaries of the cortex in both BARseq2 and ABA images. In the laminar distribution plots, dots represent values from two BARseq2 samples (purple) and one ABA sample (blue) per gene. Lines indicate means across samples.
Extended Data Fig. 4. The distribution of read counts per cell for the indicated genes in auditory cortex (green) and motor cortex (brown).
Asterisks indicate genes with significant difference in expression between the two areas (p < 0.05 using two-tailed rank sum test after Bonferroni correction). p values after Bonferroni correction are indicated on top.
Extended Data Fig. 5. Transcriptomic typing using BARseq2.
(A)(B) Slc30a3 expression in excitatory neurons with or without Cdh24 expression in single-cell RNAseq (A) from Tasic, et al. 3 or in BARseq2 (B). A cell is considered expressing Cdh24 if the expression is higher than 10 RPKM in RNAseq or 1 count in BARseq2. Red crosses indicate means and green squares indicate medians. (C) Expression density (means and individual data points) across laminar positions for the indicated genes. n = 3 slices for the three-gene panel and n = 5 slices for the 65-gene panel. (D) Precision and recall of cell typing using the marker gene panel across nine single cell datasets. N = 9 independent datasets shown in (E). In each box, the center shows the median, the bounds of the box show the 1st and 3rd quartiles, the whiskers show the range of the data, and points further than 1.5 IQR (Inter-Quartile Range) from the box are shown as outliers. (E) Breakdown of average performance for each cell type in each dataset. The datasets are: scSSALM and scSSV1 are single cell SmartSeq datasets from ALM and V1 respectively 3. All other datasets are BICCN M1 datasets 23 and the name indicates the technology used (sc = single cell, sn = single nuclei, Cv2/3 = Chromium v2/3, SS = SmartSeq). (F) Average cell typing performance for six normalization strategies. N = 9 independent datasets shown in (E). The box plots are generated in the same way as (D). (G) Confusion matrix showing overlap between prediction and annotations, normalized by predictions. This plot emphasizes precision; it indicates the probability that a given prediction was correct. (H) Confusion matrix showing overlap between prediction and annotations, normalized by annotations. This plot emphasizes recall; it indicates the probability that a given annotation was recovered.
Extended Data Fig. 6. Correlating gene expression to projections using BARseq2.
(A) Relative sensitivity of BARseq2 to barcodes (solid line) and endogenous mRNAs (dashed line) using the indicated concentration of Phusion DNA polymerase. Sensitivities are normalized to the original BARseq condition (Ctrl). Circles and crosses show individual data points across n = 2 slices. (B) Correlation between pairs of genes in barcoded cells (y-axis) and in non-barcoded cells (x-axis) as determined by BARseq2. Shuffled data (yellow) are also plotted for comparison. (C)(D) Slc17a7 (x-axes) and Gad1 (y-axes) expression in barcoded neurons in auditory (C) or motor cortex (D). Only neurons with more than 10 counts in either gene are shown. (E) The distributions of read counts per barcoded neuron (solid lines) or non-barcoded neuron (dashed lines) in auditory (green) and motor (brown) cortex. (F) Slc30a3 expression in barcoded excitatory neurons with or without Cdh24 expression in BARseq2. A cell is considered expressing Cdh24 if the expression is higher than 1 count. Red crosses indicate means and green squares indicate median. (G)(H) Slc17a7 (x-axes) and Gad1 (y-axes) expression in barcoded projection neurons in motor (G) or auditory cortex (H). Excitatory and inhibitory neurons are color-coded as indicated.
Extended Data Fig. 7. BARseq2 reveals projection and gene expression differences across major classes and IT subtypes.
(A) Differential gene expression across major classes (IT, PT, and CT) observed using BARseq2 and single-cell RNAseq. Each dot shows the difference in mean expression of a gene across a pair of major classes observed using BARseq2 (y-axis) or single-cell RNAseq (x-axis). Differences in expression that were statistically significant (FDR < 0.05 using two-tailed rank sum tests) in both A1 and M1 as shown by BARseq2 are labeled purple; otherwise they are labeled yellow. The single-cell RNAseq data used were collected in the visual cortex and anterior-lateral motor cortex 3. (B) The fraction of ITi-Ctx neurons in four transcriptomic types of IT neurons in auditory cortex. ITi-Ctx neurons have only ipsilateral cortical projections and no striatal projections or contralateral projections 6. The number of ITi-Ctx neurons and neurons with other projection patterns for each transcriptomic type are labeled on top of the pie charts. (C) The projection strengths for contralateral (y-axis) and ipsilateral (x-axis) cortical projections for each IT neuron in auditory cortex. IT1/IT2 neurons are labeled blue and IT3/IT4 neurons are labeled red.
Extended Data Fig. 8. Variance in projections explained by cadherins and laminar positions.
Box plots of variance in each projection modules explained by the indicated predictors after 100 iterations of 10-fold cross validation. Boxes indicate second and third quartiles and whiskers indicate minimum and maximum values excluding outliers. Outliers are shown in red.
Extended Data Fig. 9. Validation of correlation between cadherins and IT projections.
(A) Representative images of in situ hybridization in A1 (top) and M1 (bottom) slices with CTB labeling in the caudal striatum. Three marker genes and CTB labeling are shown in the indicated colors. Scale bars = 100 μm. Arrows and arrowheads indicate example CTB+ and CTB- neurons, respectively. Experiments for each combination of targeted gene and CTB labeling condition (Cdh12 with contralateral labeling, Cdh8 with ipsilateral labeling, and Pcdh19 with striatal labeling) were performed in slices from two animals. (B) Crops of the indicated individual channels of example neurons from (A). Scale bars = 10 μm. (C)(D)(E) Cumulative probability distribution of the expression of Cdh12 (C), Cdh8 (D), and Pcdh19 (E) in neurons with or without retrograde labeling of contralateral (C), ipsilateral (D), or caudal striatal (E) projections. p values from two-tailed rank sum tests after Bonferroni correction and numbers of neurons used for each experiment are indicated. N = 2 animals for each experiment.
Extended Data Fig. 10. Cadherin co-expression modules correlate with IT projections.
(A) Correlation among cadherins in IT neurons in motor cortex identified in the indicated single-cell RNAseq datasets 3,23. The datasets included are: tasic_alm and tasic_v1 are single cell SmartSeq datasets from ALM and V1 respectively 3; all other datasets are BICCN M1 datasets 23; the name indicates the technology used (sc = single cell, sn = single nuclei, Cv2/3 = Chromium v2/3, SS = SmartSeq). (B) Modularity (EGAD AUROC) of co-expression modules in BARseq2 M1 against null distribution of modularity (node permutation). BARseq2 modularity is shown by the blue lines with the corresponding p-values. P values are calculated using a one-sided non-parametric node permutation test without multiple comparison correction. (C) Association (AUROC) between cadherin co-expression modules and the indicated projections. Significant associations are marked by asterisks (* FDR < 0.1, ** FDR < 0.05). (D) Fractions of neurons with the indicated projections as a function of co-expression module expression. (E) Distribution of associations of the indicated projection modules with gene expression. Association with significant gene module is shown by a blue line; association with single genes from that module is shown by orange lines; association with all other genes is shown by a gray density. (F) Association of the three co-expression modules in transcriptomic IT neurons in the indicated datasets (AUROC, significance shown as in C).
Supplementary Material
Acknowledgement
The authors would like to acknowledge members of the MAPseq core facility, Huiqing Zhan, Yan Li, and Nicole Gemmill, for MAPseq data production, Katherine Matho and Z. Josh Huang for dissection coordinates in motor cortex, Huiqing Zhan, Li Yuan, Henry Lee Gilbert, Katherine Matho, Justus Kebschull, and Daniel Fürth for useful discussions, and Wiktor Wadolowski, Barry Burbach, Kathleen Lucere, and Eugene Fong for technical support. This work was supported by the National Institutes of Health [NIH 5RO1NS073129, 5RO1DA036913, RF1MH114132, and U01MH109113 to A.M.Z, R01MH113005 and R01LM012736 to J.G., and U19MH114821 to both A.M.Z. and J.G.], the Brain Research Foundation (BRF-SIA-2014-03 to A.M.Z.), IARPA MICrONS [D16PC0008 to A.M.Z.], Paul Allen Distinguished Investigator Award [to A.M.Z.], Simons Foundation [350789 to X.C.], Chan Zuckerberg Initiative (2017-0530 ZADOR/ALLEN INST(SVCF) SUB awarded to A.M.Z], and Robert Lourie (to A.M.Z.). This work was additionally supported by the Assistant Secretary of Defense for Health Affairs endorsed by the Department of Defense, 1120 Fort Detrick, Fort Detrick, MD 21702 through the FY18 PRMRP Discovery Award Program W81XWH1910083 awarded to X.C. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the U.S. Army. In conducting research using animals, the investigator adheres to the laws of the United States and regulations of the Department of Agriculture.
Footnotes
Competing Interests
A.M.Z. is a founder and equity owner of Cajal Neuroscience and a member of its scientific advisory board. The remaining authors declare no competing interests.
References
- 1.Winnubst J et al. Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell 179, 268–281 e213, doi: 10.1016/j.cell.2019.07.042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muñoz-Castañeda R et al. Cellular Anatomy of the Mouse Primary Motor Cortex. bioRxiv, 2020.10.02.323154, doi: 10.1101/2020.10.02.323154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tasic B et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78, doi: 10.1038/s41586-018-0654-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeisel A et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014 e1022, doi: 10.1016/j.cell.2018.06.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y et al. The logic of single-cell projections from visual cortex. Nature 556, 51–56, doi: 10.1038/nature26159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 179, 772–786.e19, doi: 10.1016/j.cell.2019.09.023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DW et al. Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728.e717, doi: 10.1016/j.cell.2019.09.020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Economo MN et al. Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84, doi: 10.1038/s41586-018-0642-9 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Zhang M et al. Molecular, spatial and projection diversity of neurons in primary motor cortex revealed by in situ single-cell transcriptomics. bioRxiv, 2020.06.04.105700, doi: 10.1101/2020.06.04.105700 (2020). [DOI] [Google Scholar]
- 10.Ke R et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 10, 857–860, doi: 10.1038/nmeth.2563 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Qian X et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat Methods 17, 101–106, doi: 10.1038/s41592-019-0631-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kebschull JM et al. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987, doi: 10.1016/j.neuron.2016.07.036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L et al. BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals. Cell 182, 177–188.e27, doi: 10.1016/j.cell.2020.05.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090, doi: 10.1126/science.aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5, 877–879, doi: 10.1038/nmeth.1253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayano Y et al. The role of T-cadherin in axonal pathway formation in neocortical circuits. Development 141, 4784–4793, doi: 10.1242/dev.108290 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Friedman LG et al. Cadherin-8 expression, synaptic localization, and molecular control of neuronal form in prefrontal corticostriatal circuits. J Comp Neurol 523, 75–92, doi: 10.1002/cne.23666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul A et al. Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539 e520, doi: 10.1016/j.cell.2017.08.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga E, Nambu S, Oka M & Iriki A Complex and dynamic expression of cadherins in the embryonic marmoset cerebral cortex. Dev Growth Differ 57, 474–483, doi: 10.1111/dgd.12228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redies C Cadherins and the formation of neural circuitry in the vertebrate CNS. Cell Tissue Res 290, 405–413 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176, doi: 10.1038/nature05453 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Terakawa YW, Inoue YU, Asami J, Hoshino M & Inoue T A sharp cadherin-6 gene expression boundary in the developing mouse cortical plate demarcates the future functional areal border. Cereb Cortex 23, 2293–2308, doi: 10.1093/cercor/bhs221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z et al. An integrated transcriptomic and epigenomic atlas of mouse primary motor cortex cell types. bioRxiv, 2020.02.29.970558, doi: 10.1101/2020.02.29.970558 (2020). [DOI] [Google Scholar]
- 24.Fros JJ & Pijlman GP Alphavirus Infection: Host Cell Shut-Off and Inhibition of Antiviral Responses. Viruses 8, doi: 10.3390/v8060166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingler E et al. Single-cell molecular connectomics of intracortically-projecting neurons. bioRxiv, 378760, doi: 10.1101/378760 (2018). [DOI] [Google Scholar]
- 26.Wang Y et al. Complete single neuron reconstruction reveals morphological diversity in molecularly defined claustral and cortical neuron types. bioRxiv, 675280, doi: 10.1101/675280 (2019). [DOI] [Google Scholar]
- 27.Harris KD & Shepherd GM The neocortical circuit: themes and variations. Nat Neurosci 18, 170–181, doi: 10.1038/nn.3917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan X, Krishnaswamy A, De la Huerta I & Sanes JR Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807, doi: 10.1016/j.cell.2014.06.047 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Friedman LG, Benson DL & Huntley GW Cadherin-based transsynaptic networks in establishing and modifying neural connectivity. Curr Top Dev Biol 112, 415–465, doi: 10.1016/bs.ctdb.2014.11.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jontes JD The Cadherin Superfamily in Neural Circuit Assembly. Cold Spring Harb Perspect Biol 10, a029306, doi: 10.1101/cshperspect.a029306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langfelder P, Zhang B & Horvath S Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720, doi: 10.1093/bioinformatics/btm563 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Lee DD & Seung HS Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791, doi: 10.1038/44565 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Ballouz S, Verleyen W & Gillis J Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics 31, 2123–2130, doi: 10.1093/bioinformatics/btv118 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Crow M, Paul A, Ballouz S, Huang ZJ & Gillis J Exploiting single-cell expression to characterize co-expression replicability. Genome Biol 17, 101, doi: 10.1186/s13059-016-0964-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Sun YC, Church GM, Lee JH & Zador AM Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res 46, e22, doi: 10.1093/nar/gkx1206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah S, Lubeck E, Zhou W & Cai L In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92, 342–357, doi: 10.1016/j.neuron.2016.10.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Loper J, Chen X, Zador T & Paninski L BARcode DEmixing through Non-negative Spatial Regression (BarDensr). bioRxiv, 2020.08.17.253666, doi: 10.1101/2020.08.17.253666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding J et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38, 737–746, doi: 10.1038/s41587-020-0465-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris KD et al. Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics. PLoS Biol 16, e2006387, doi: 10.1371/journal.pbio.2006387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan X et al. Cadherin Combinations Recruit Dendrites of Distinct Retinal Neurons to a Shared Interneuronal Scaffold. Neuron 99, 1145–1154 e1146, doi: 10.1016/j.neuron.2018.08.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H et al. Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220 e1222, doi: 10.1016/j.cell.2017.10.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Custo Greig LF, Woodworth MB, Galazo MJ, Padmanabhan H & Macklis JD Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci 14, 755–769, doi: 10.1038/nrn3586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagri A et al. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33, 233–248, doi: 10.1016/s0896-6273(02)00561-5 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Shu T, Sundaresan V, McCarthy MM & Richards LJ Slit2 guides both precrossing and postcrossing callosal axons at the midline in vivo. J Neurosci 23, 8176–8184 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida Y Semaphorin signaling in vertebrate neural circuit assembly. Front Mol Neurosci 5, 71, doi: 10.3389/fnmol.2012.00071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berns DS, DeNardo LA, Pederick DT & Luo L Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature 554, 328–333, doi: 10.1038/nature25463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zador AM et al. Sequencing the connectome. PLoS Biol 10, e1001411, doi: 10.1371/journal.pbio.1001411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peikon ID et al. Using high-throughput barcode sequencing to efficiently map connectomes. Nucleic Acids Res 45, e115, doi: 10.1093/nar/gkx292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marblestone AH, Daugharthy ER, Kalhor R, Peikon ID, Kebschull JM, Shipman SL, Mishchenko Y, Lee JH, Kording KP, Boyden ES, Zador AM, Church GM Rosetta Brains: A Strategy for Molecularly-Annotated Connectomics. arXiv, 1404.5103 [q-bio.NC] (2014). [Google Scholar]
- 50.Eng CL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239, doi: 10.1038/s41586-019-1049-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Method References
- 51.Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214, doi: 10.1038/nature13186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edelstein AD et al. Advanced methods of microscope control using μManager software. J Biol Methods 1, e10, doi: 10.14440/jbm.2014.36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JH et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363, doi: 10.1126/science.1250212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evangelidis GD & Psarakis EZ Parametric image alignment using enhanced correlation coefficient maximization. IEEE Trans Pattern Anal Mach Intell 30, 1858–1865, doi: 10.1109/TPAMI.2008.113 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Stringer C, Wang T, Michaelos M & Pachitariu M Cellpose: a generalist algorithm for cellular segmentation. bioRxiv, 2020.02.02.931238, doi: 10.1101/2020.02.02.931238 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Rock C, Zurita H, Wilson C & Apicella AJ An inhibitory corticostriatal pathway. Elife 5, e15890, doi: 10.7554/eLife.15890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300, doi: 10.1111/j.2517-6161.1995.tb02031.x (1995). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw target area sequencing data (Fig. 4C) are deposited at SRA (SRR12247894, SRR12245390, and SRR12245389). Single-cell RNAseq data (Fig. 2G–I) are deposited at SRA (SRR13716225). Raw in situ sequencing images (Fig. 2–4) are deposited at Brain Image Library (https://download.brainimagelibrary.org/06/35/0635a0b3b0954c7e/). Other data and intermediate processed sequencing data are deposited at Mendeley Data (http://dx.doi.org/10.17632/jnx89bmv4s.1).