Abstract
In asthma, most of the identified biomarkers pertain to the Th2 phenotype and no known biomarkers have been verified for severe asthmatics. Therefore, identifying biomarkers using the integrative phenotype-genotype approach in severe asthma is needed. The study aims to identify novel biomarkers as genes or pathways representing the core drivers in asthma development, progression to the severe form, resistance to therapy, and tissue remodeling regardless of the sample cells or tissues examined. Comprehensive reanalysis of publicly available transcriptomic data that later was validated in vitro, and locally recruited patients were used to decipher the molecular basis of asthma. Our in-silicoanalysis revealed a total of 10 genes (GPRC5A, SFN, ABCA1, KRT8, TOP2A, SERPINE1, ANLN, MKI67, NEK2, and RRM2) related to cell cycle and proliferation to be deranged in the severe asthmatic bronchial epithelium and fibroblasts compared to their healthy counterparts. In vitro, RT qPCR results showed that (SERPINE1 and RRM2) were upregulated in severe asthmatic bronchial epithelium and fibroblasts, (SFN, ABCA1, TOP2A, SERPINE1, MKI67, and NEK2) were upregulated in asthmatic bronchial epithelium while (GPRC5A and KRT8) were upregulated only in asthmatic bronchial fibroblasts. Furthermore, MKI76, RRM2, and TOP2A were upregulated in Th2 high epithelium while GPRC5A, SFN, ABCA1 were upregulated in the blood of asthmatic patients. SFN, ABCA1 were higher, while MKI67 was lower in severe asthmatic with wheeze compared to nonasthmatics with wheezes. SERPINE1 and GPRC5A were downregulated in the blood of eosinophilic asthmatics, while RRM2 was upregulated in an acute attack of asthma. Validation of the gene expression in PBMC of locally recruited asthma patients showed that SERPINE1, GPRC5A, SFN, ABCA1, MKI67, and RRM2 were downregulated in severe uncontrolled asthma. We have identified a set of biologically crucial genes to the homeostasis of the lung and in asthma development and progression. This study can help us further understand the complex interplay between the transcriptomic data and the external factors which may deviate our understanding of asthma heterogeneity.
Subject terms: Immunology, Molecular biology, Systems biology
Introduction
Identifying novel markers that can help in the patient classification, clinical outcome prediction, and therapy response was and will remain the ultimate goal for many research projects 1, 2. These markers should meet precise specifications and criteria such as reliability, ease of collection and measurement, and non-invasive. Using reliable biomarkers to predict asthma patients’ response to steroid therapy early in the disease course is particularly important and essential to have an optimal response and avoid undesirable side effects 3. Using such biomarkers is not routinely requested in severe asthma 4 as the biomarkers most extensively studied at present in asthma are Th2 related biomarkers 5. Peripheral blood mononuclear cells (PBMCs) (which include lymphocytes, monocytes, and dendritic cells) constitute an essential source of mediators of an allergic response in asthma and serve as an excellent alternative to the costly and challenging approach of obtaining airways samples in severe asthma 6. The integrative phenotype–genotype approach is a novel, simple, and powerful tool to identify clinically relevant potential biomarkers. We recently used this approach to identify clinically essential biomarkers in complex and heterogeneous diseases like diabetes 7.
Among the omics, bronchial epithelium transcriptomics-driven phenotyping of asthmatic patients showed the potential to discover gene expression profiles characteristic in asthma 8. Bronchial epithelium transcriptomics can identify different molecular mechanisms underlying divergent asthmatic phenotypes and have the power to identify novel clinically efficient biomarkers 9, 10. It is especially crucial in comparing different groups of adult-onset severe asthma where conventional single gene expression may not capture underlying pathways11. Transcriptomics, in comparison to genomics, are more dynamic in terms of changes in response to the environment, treatment, age and is cell-specific, which is very important to understand active heterogeneous diseases like asthma and help in its “prediction” and “diagnosis” 12.
There was no link between the identified transcriptomic data and clinical findings in some well-designed transcriptomic analyses in asthma samples, indicating a missing biological relevance if those studies are taken separately13. At present, this lost biological relevance can be identified if more comprehensive designs and data analysis are tried14. This relatively expensive approach can be more informative if a large number of samples are combined to extract meaningful information by using a large number of datasets available in public databases 15. However, such an approach in a broader range of patients has not yet been performed 16.
Current asthma research aims to identify novel biomarkers as genes or pathways representing the core drivers in asthma development, progression to the severe form, resistance to therapy, and tissue remodeling. We hypothesized that such genes and pathways should be altered or deranged in lung resident cells and infiltrating immune cells of asthmatics when compared to healthy control to be selected as a novel biomarker. Our research team conducted a comprehensive extensive data analysis from the publicly available transcriptomic datasets to achieve this aim using stringent in-house bioinformatics workflow to identify confounding biological and nonbiological factors that might interfere with the proper identification of such biomarkers. Extensive in silico, in vitro, and in vivo validation of the diagnostic value of the identified genes was followed to explore their translational potentials.
Our research team identified ten genes that were deranged in the severe asthmatic bronchial epithelium and fibroblasts related to cell cycle, cell proliferation, and survival-related pathways17. Therefore, further exploration of these genes was done in silico on larger datasets, in vitro on primary cell lines from healthy controls and asthmatic patients, and in clinical samples from locally recruited patients.
Materials and methods
In silico exploration of lung and blood transcriptomics dataset
Publicly available transcriptomics datasets of the bronchial epithelium (GSE67472, GSE64913, and GSE76227), bronchial fibroblasts (GSE27335), whole blood, and PBMCs (GSE137394, GSE123750, GSE115823, GSE69683, GSE31773, GSE16032, and GSE73482) from asthmatic patients versus healthy subjects in different settings (n = 1780 sample) were explored to extract the normalized gene expression of the identified genes, as shown in Supplement Table S1 and Fig. S1.
Genome-wide association studies (GWASs)
We explored the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) (v1.0) of published genome-wide association studies 18. The file (gwas_catalog_v1.0-associations_e96_r2019-07-12) was downloaded, and we searched for SNPs associated with the strongest Asthma-related SNP-risk allele.
Methylomes
DNA methylation dataset (GSE85568) of primary airway epithelial cells from 74 asthmatic and 41 nonasthmatic adults was used to investigate the differential methylation of genes of interest between the two groups. Another dataset was explored to analyze the DNA methylation profiles in peripheral blood monocytes (GSE59339) from eosinophilic asthma (n = 21), paucigranulocytic asthma (n = 22), neutrophilic asthma (n = 9), and healthy controls (n = 10) for the same purpose. Normalized methylation scores were extracted from the datasets; accordingly, results are shown in the Supplement (S2).
Prediction of the specificity of the identified genes to lung cells
The lung gene expression analysis (LGEA) integrative web portal (https://research.cchmc.org/pbge/lunggens/mainportal.html) was used to check the specificity of these shortlisted genes to the different components of the lung 19.
In vivo validation
Patient population
From January 2017 to May 2019, individuals were recruited consecutively from the Asthma Clinic in Rashid Hospital, Pulmonary Medicine Department. Thirty-two asthmatic patients were included, 19 were nonsevere asthmatic patients (mild to moderate), and 13 were severe asthmatic subjects (based on the criteria for asthma as per American Thoracic Society). These patients were compared to 12 nonasthmatic volunteer subjects who had no recent infection of the respiratory tract and no histories of allergy or asthma. Participants completed the Asthma Control Test self-assessment. The Ethics Committee of Dubai Health Authority and the University of Sharjah approved the study with REC (Research Ethics Committee) approval number DSREC-11/2017_04, and each subject gave written informed consent after a thorough explanation by the treating physician and the researchers. This study was conducted following the Declaration of Helsinki. The demographic characteristics of the asthmatic patients and control subjects are shown in Supplementary Table S2. Blood was collected, as previously described 20.
Blood collection protocol and plasma isolation
Twelve milliliters of whole blood were collected from each patient in EDTA-containing blood collection tubes (3 ml each) and transferred immediately within 2 h to Sharjah Institute for Medical Research (SIMR), Sharjah for further processing to isolate PBMCs as previously described 20. Twelve milliliters of Histopaque-1077 (Sigma, #10771, Germany) were added to a 50 ml centrifuge tube and brought to room temperature, then 12 ml of whole blood were carefully layered on top of the Histopaque and centrifuged at 400 × g for precisely 30 min at room temperature. After centrifugation, the plasma layer and the buffy layer interface were carefully collected with separate Pasteur pipettes and transferred to clean 15 ml conical centrifuge tubes separately, to be frozen at − 80 °C till future use.
In vitro validation
Primary cell lines
Primary cells from healthy and asthmatic patients were isolated from bronchial biopsies in Meakins-Christie Laboratories, The Centre for Respiratory Research at McGill University, and the Research Institute of McGill University Health Centre as previously described 21. We compared healthy bronchial epithelium (NHLE, n = 3) to asthmatic cells (DHLE, n = 3), and healthy lung fibroblasts (NHLF, n = 3) were compared to asthmatic lung fibroblasts (DHLF, n = 3). Epithelial cells were revived and maintained using epithelial growth medium PneumaCul-Ex Medium (Stem Cell Technology, Canada), supplemented with 100 units/ml penicillin/streptomycin (Gibco, USA). Primary fibroblasts were maintained in complete Dulbecco’s Modified Eagle’s medium (DMEM) (Sigma-Aldrich, Germany) with 10% fetal bovine serum (FBS) (Sigma-Aldrich, Germany) supplemented with 100 units/ml penicillin/streptomycin (Gibco, USA).
mRNA gene expression using qRT-PCR
RNA was extracted using RNAeasy mini kit (Qiagen, Germany) as per the manufacturer’s instructions. RNA purity (OD260/OD280) and quantity were measured using Nanodrop 2000 (ThermoFisher Scientific, USA). The purified RNA was reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcription (Applied Biosystems, USA) as per the manufacturer’s instructions. 5X Hot FIREPol EvaGreen qPCR Supermix (Solis BioDyne, Estonia) was used to quantify mRNA expression of the selected genes using QuantStudio3 (Applied Biosystems, USA). Details of the used primers are in the Supplementary Table S3.
Statistical analysis
GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla California USA) was used for statistical analysis. First, the D’Agostino–Pearson normality test was used to determine whether to perform parametric or nonparametric tests. A one-way ANOVA test was performed to determine whether there were any statistically significant differences between the mean values of the controls and different asthma groups for the gene expression. Prism was used to compute a multiplicity-adjusted P-value for each comparison when ANOVA was used. The same software was used to examine the correlations between the different parameters using linear regression. The student t-test was used to determine the difference between two groups under a given experiment or treatment. A p-value < 0.05 was considered to be statistically significant.
Results
ANLN, ABCA1, MKI67, and NEK2 were upregulated in asthmatic epithelial cells but were downregulated in severe asthmatic fibroblasts when compared to healthy controls
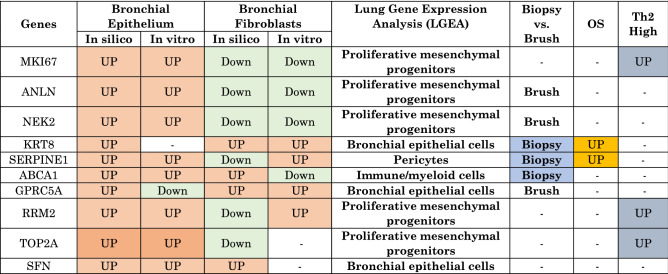
Our research team identified ten genes that were deranged in the severe asthmatic bronchial epithelium and fibroblasts related to cell cycle, cell proliferation, and survival-related pathways17 shown in Fig. 1. Four genes (GPRC5A, SFN, ABCA1, and KRT8) were upregulated in both asthmatic epithelium and fibroblasts, while the rest six genes (TOP2A, SERPINE1, ANLN, MKI67, NEK2, and RRM2) were downregulated in asthmatic fibroblasts despite being upregulated in the asthmatic epithelium. The list of genes and their details are shown in Table 1.
Figure 1.
Schematic representation of the filtering of Genes Identified using our bioinformatic pipeline showing the resultant gene list that shares a common pathway mainly related to cell cycle, cell proliferation, and survival.
Table 1.
List of genes related to cell cycle displayed significant differential expression between asthmatic and healthy epithelium in both genders, bronchial brush versus the biopsy sample, and between asthmatic and healthy fibroblasts, from publicly available datasets (GSE64913, GSE76227, and GSE27335).
| Genes | Number of enriched pathways | Gender | Biopsy vs brush | In asthmatic epithelium | In asthmatic fibroblasts |
|---|---|---|---|---|---|
| ABCA1 | 117 | Both | Both | Upregulated | Upregulated |
| GPRC5A | 108 | Both | Both | Upregulated | Upregulated |
| KRT8 | 106 | Both | Both | Upregulated | Upregulated |
| SFN | 102 | Both | Both | Upregulated | Upregulated |
| TOP2A | 175 | Both | Both | Upregulated | Downregulated |
| MKI67 | 171 | Both | Both | Upregulated | Downregulated |
| RRM2 | 147 | Both | Both | Upregulated | Downregulated |
| ANLN | 109 | Both | Both | Upregulated | Downregulated |
| NEK2 | 108 | Both | Both | Upregulated | Downregulated |
| SERPINE1 | 103 | Both | Both | Upregulated | Downregulated |
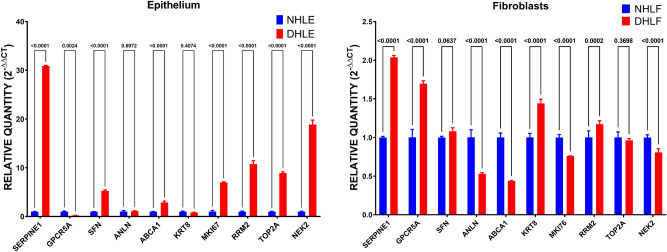
The expression of the ten genes was examined on representative structural cells from healthy controls versus asthmatic patients to validate the preliminary bioinformatics findings. mRNA expression of (SERPINE1, GPRC5A, SFN, ANLN, ABCA1, KRT8, MKI67, RRM2, TOP2A, and NEK2) was examined in healthy bronchial epithelium (NHLE, n = 3) versus asthmatic cells (DHLE, n = 3), and healthy lung fibroblasts (NHLF, n = 3) compared to asthmatic lung fibroblasts (DHLF, n = 3) as shown in Fig. 2.
Figure 2.
mRNA expression of cell cycle-related genes in healthy bronchial epithelium (NHLE, n = 3) versus asthmatic cells (DHLE, n = 3), and healthy lung fibroblasts (NHLF, n = 3) compared to asthmatic lung fibroblasts (DHLF, n = 3).
SERPINE1, SFN, ABCA1, MKI67, RRM2, TOP2A, and NEK2 were significantly upregulated in asthmatic epithelium compared to healthy ones; these results match the bioinformatics data while GPRC5A was decreased considerably in asthmatic epithelium compared to a healthy one (p < 0.05). On the other hand, asthmatic fibroblasts showed higher expression of (SERPINE1, GPRC5A, KRT8, and RRM2) and lower (ANLN, ABCA1, MKI67, and NEK2) compared to healthy.
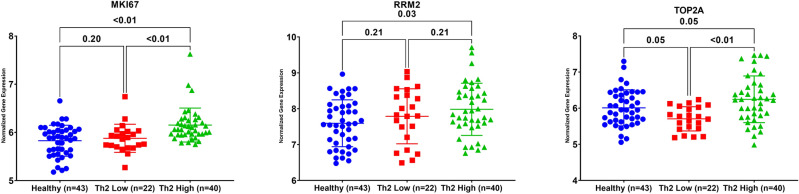
MKI67, RRM2, and TOP2A showed higher expression in Th2-high asthmatics
Asthmatics patients can be categorized according to the levels of type 2 inflammation into ‘T helper 2 (Th2)-high’ and ‘Th2-low’ 22. The type 2 immune response-driven endotype consists of T2-high asthma endotype encompasses several related subtypes in both children and adults23. At the same time, T2-low endotype included neutrophilic or paucigranulocytic airway inflammation with greater resistance to steroids24. The differential expression for the identified genes between Th2-high and Th2-low asthma phenotypes was investigated using the GSE67472 dataset, which includes the airway epithelial brushings transcriptomics data of 62 subjects with mild-to-moderate asthma not on inhaled steroids and 43 healthy controls. Asthma subjects were stratified into two subgroups, Th2-high, and Th2-low asthma, based on their expression of a three-gene signature of Type 2 inflammation: POSTN, SERPINB2, and CLCA1. Interestingly, our analysis showed that MKI67, RRM2, and TOP2A expression in Th2-high asthmatics were higher than controls, as shown in Fig. 3. On the other hand, MKI67 and TOP2A were higher in Th2-high compared to Th2-low asthmatics. Interestingly, TOP2A showed lower expression in Th2-low asthmatics when compared to healthy controls too.
Figure 3.
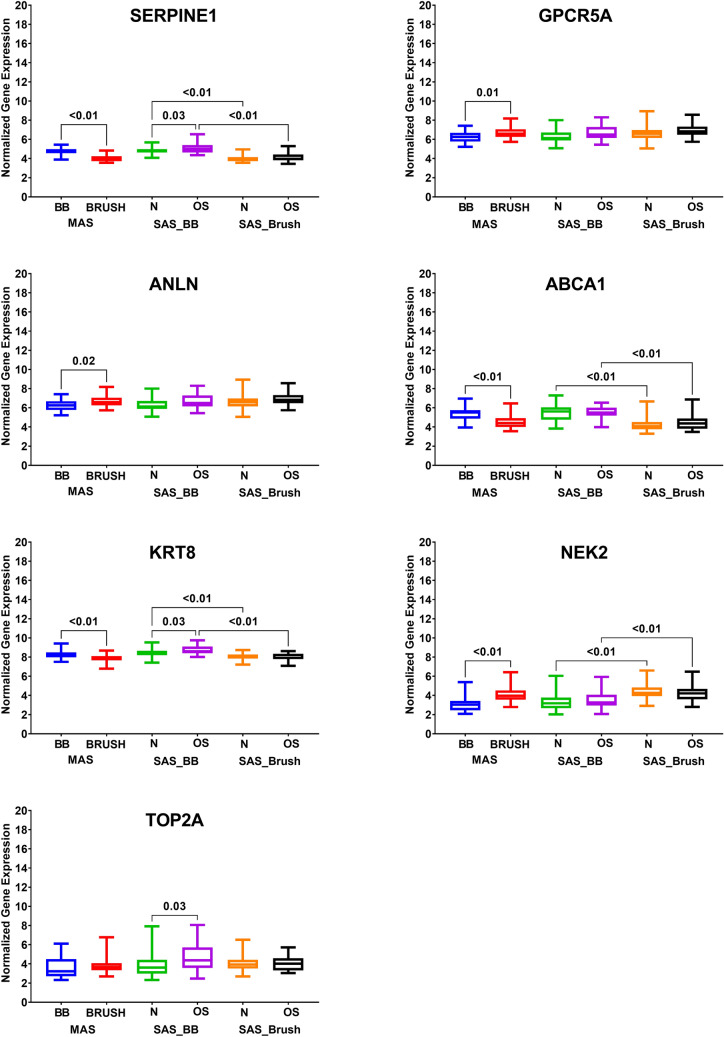
mRNA expression of the ten genes in bronchial epithelium using, GSE76227 transcriptomic dataset that contains the expression data of 190 bronchial biopsies (BB) and epithelial brushing (BRUSH) from Unbiased BIOmarkers in Prediction of REspiratory Disease outcomes (U-BIOPRED) Project. The normalized gene expression of each of the identified genes was extracted and compared between different subgroups. The datasets were subdivided into nonsevere asthmatic (MAS), severe asthmatics (SAS) (oral steroid naïve (N) vs. oral steroid users (OS).
Oral steroids upregulated the SERPINE1 and KRT8 mRNA expression in the bronchial biopsy from severe asthmatics but not in the bronchial brush tissue
The mRNA expression of the identified genes can indicate the difference in the cells of the given sample examined in terms of pure epithelial cells or mixed mesenchymal and/or infiltrating immune cells; this necessitates the examination of such genes in biopsies of lung rather than just bronchial brushes. To examine that transcriptomic dataset (GSE76227) was used. This dataset contains the expression data of 190 bronchial biopsies and epithelial brushing from Unbiased BIOmarkers in Prediction of REspiratory Disease outcomes (U-BIOPRED) Project. The normalized gene expression of each of the identified genes was extracted and compared between different subgroups. The datasets were subdivided into nonsevere asthmatic (bronchial brush vs. biopsy), severe asthmatics (oral steroid naïve (bronchial brush vs. biopsy) vs. oral steroid users (bronchial brush vs. biopsy)).
As shown in Fig. 4, SERPINE1, ABCA1, and KRT8 showed higher expression in bronchial biopsies than the pure bronchial epithelium, in moderate and severe asthma, with and without oral steroid use. Oral steroids upregulated the SERPINE1, and KRT8 mRNA expression in the bronchial biopsy from severe asthmatics but not in the bronchial brush tissue indicating the source of their upregulation might be from stromal cells rather than epithelial cells in response to steroids. Another possible source of this increase can be redistributing T cells away from the epithelium25. Steroids did not affect the mRNA expression of ABCA1 in both types of samples in severe asthmatics.
Figure 4.
mRNA expression of (MKI67, RRM2, and TOP2A) genes in bronchial epithelium using GSE67472 dataset to compare healthy controls (n = 43) to Th2-high asthmatics (n = 40) and Th2 low asthmatics (n = 22).
GPCR5A and ANLN showed higher expression in nonsevere bronchial epithelium than bronchial biopsy, while NEK2 showed higher expression in nonsevere and severe bronchial epithelium compared to bronchial biopsy. Interestingly, TOP2A showed upregulation in bronchial biopsies of asthmatics who were on oral steroids only.
Summary of in silico and in vitro analysis of the ten genes in bronchial epithelium and fibroblasts of asthmatic patients
The summary of the selected genes mRNA expression in the lung showed that in vitro RT qPCR results confirmed the in-silico findings for most of the genes identified. The Lung Gene Expression Analysis (LGEA) showed that KRT8, GPRC5A, and SFN have the highest specificity to bronchial epithelial cells exclusively. While RRM2, TOP2A, ANLN, MKi67, and NEK2 showed the highest correlation to proliferative mesenchymal progenitors. NEK2 was expressed by many cell types, while SERPINE1 was specific to pericytes, and ABCA1 was specific to immune/myeloid cells. The summary of the ten genes in silico and in vitro results in the lung are listed in Table 2.
Table 2.
Summary of in silico and in vitro analysis of the ten genes in bronchial epithelium and fibroblasts of asthmatic patients.
If their expression is specific to bronchial brush or biopsy or not, their response to oral steroids (OS) and their expression level in Th2 high versus Th2 low asthma.
MKI67, ANLN, and NEK2 were upregulated in asthmatic bronchial epithelium while they were downregulated in asthmatic bronchial fibroblasts, although according to LGEA, those genes are specific proliferative mesenchymal progenitors who might indicate the loss of this characteristic in severe asthmatic fibroblasts. Among the three, MKI67 was shown to be upregulated in Th2 High asthmatic epithelium along with RRM2 and TOP2A, which might be an EMT marker in asthmatic epithelium too.
Protein’s products of the identified genes are specific to NK and T regulatory cells
After elucidating the role of the ten genes in the lung, the next question was whether these genes are altered in the circulating immune cells to check their feasibility as a biomarker in asthmatic patients. Human Protein Atlas database was explored https://www.proteinatlas.org/, to validate if the identified genes can be detected as mRNA in the blood or not and what cells express them as detailed in the Table 3. Proteins of two genes were documented to be secreted from cells either to blood (SERPINE1) or other tissues (SFN) and can be detected in the blood. The rest genes’ products were intracellular (MKI67, KRT8, TOP2A, ANLN, and NEK2), intracellular, and membranous (GPRC5A, RRM2, and ABCA1). Some of these proteins were specific to the blood cell lineage of dendritic cells “plasmacytoid DC,” granulocytes, NK-cells (KRT8), granulocytes “eosinophil and basophil,” and for T-cells “naive CD4 T-cell, T-reg” and NK-cells (MKI67). On the other hand, some gene products showed lineage enrichment for basophil (SFN), eosinophil and neutrophil (ABCA1), NK-cell (SERPINE1), and T-reg (RRM2, TOP2A, and ANLN). NEK2 showed Low lineage specificity and low cell type specificity as its mRNA was detected in many blood cells.
Table 3.
Gene’s products and blood or cell lineage using the Human Protein Atlas database were explored https://www.proteinatlas.org/.
| Genes | Secretome annotation | RNA blood cell lineage specificity | RNA blood cell lineage distribution | RNA blood cell type specificity | RNA blood cell type distribution |
|---|---|---|---|---|---|
| SFN | Secreted in other tissues | Lineage enriched (granulocytes) | Detected in single | Cell type enriched (basophil) | Detected in single |
| SERPINE1 | Secreted to blood | Lineage enriched (NK-cells) | Detected in single | Cell type enriched (NK-cell) | Detected in single |
| ANLN | Intracellular | Low lineage specificity | Detected in single | Cell type enhanced (T-reg) | Detected in single |
| KRT8 | Intracellular | Group enriched (dendritic cells, granulocytes, NK-cells) | Detected in many | Cell type enhanced (plasmacytoid DC) | Detected in some |
| MKI67 | Intracellular | Group enriched (T-cells, NK-cells) | Detected in many | Cell type enhanced (naive CD4 T-cell, T-reg) | Detected in some |
| GPRC5A | Intracellular, Membrane | Lineage enriched (granulocytes) | Detected in single | Cell type enhanced (basophil) | Detected in some |
| TOP2A | Intracellular | Low lineage specificity | Detected in many | Cell type enhanced (T-reg) | Detected in some |
| ABCA1 | Intracellular, Membrane | Lineage enriched (granulocytes) | Detected in many | Group enriched (eosinophil, neutrophil) | Detected in many |
| RRM2 | Intracellular, Membrane | Lineage enriched (T-cells) | Detected in many | Cell type enhanced (T-reg) | Detected in many |
| NEK2 | Intracellular | Low lineage specificity | Detected in many | Low cell type specificity | Detected in many |
The generated shortlist genes were differentially expressed in bronchial epithelium and bronchial mesenchymal cells like fibroblasts and were lineage and cell-specific to individual immune cells, so we tried to explore their dynamic expression in the blood of asthmatics under different settings and the PBMCs of locally recruited patients.
All identified genes were expressed in peripheral blood lymphocytes
GSE31773 dataset was extracted as it showed the comparison of mRNA expression in circulating T-cells from patients with severe asthma compared to healthy controls. All identified genes were expressed in CD4+ and CD8+ PBMCs, indicating their feasibility to serve as potential biomarkers in blood. Next, we explored the mRNA expression level of GPRC5A, SFN, ABCA1, KRT8, TOP2A, SERPINE1, ANLN, MKI67, NEK2, and RRM2 in the blood of asthmatic patients compared to healthy controls.
NEK2 and GPRC5A mRNA expression were lower in severe asthmatic patients compared to healthy controls
The mRNA expression of GPRC5A, SFN, ANLN, and NEK2 was higher in CD4+ T lymphocytes than CD8+ T lymphocytes (p < 0.05). Only KRT8 mRNA expression was higher in CD8+ T lymphocytes in contrast to CD4+ T lymphocytes (p < 0.05). Interestingly, only GPRC5A and NEK2 mRNA expression in CD4+ T lymphocytes were lower in severe asthmatic patients compared to healthy controls (p < 0.05), as shown in Fig. 5.
Figure 5.
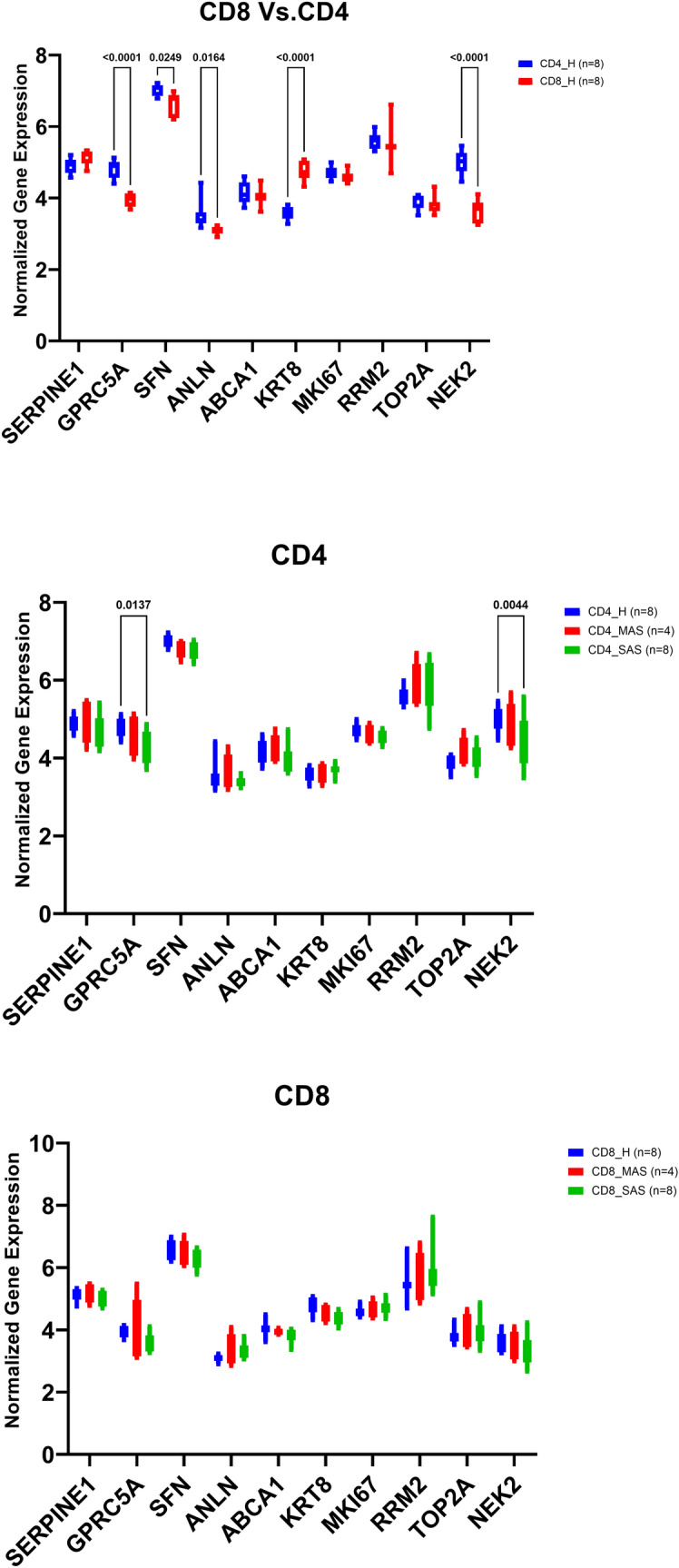
Normalized gene expression of genes identified in circulating T-cells (CD4 versus CD8) of asthmatic patients “MAS = mild to moderate and SAS = severe” compared to healthy (H) controls extracted from the expression profile of publicly available datasets (GSE31773).
GPRC5A, SFN, ABCA1, and NEK2 showed specific and significant differential expression in asthmatic patients’ blood compared to healthy control
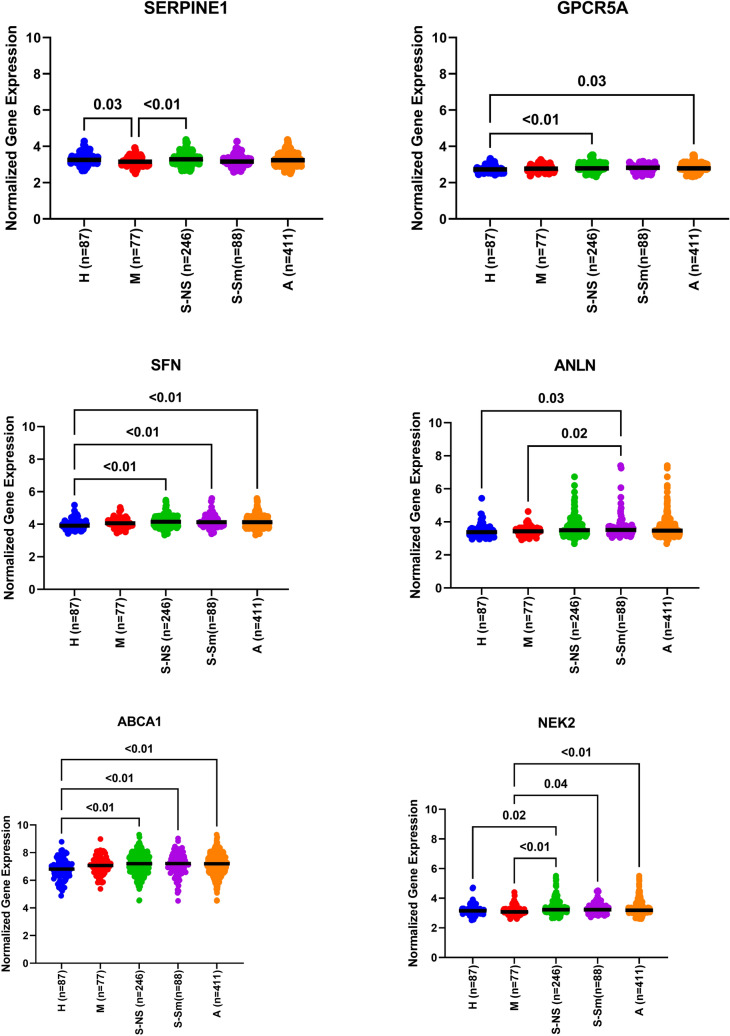
The expression profile of the publicly available dataset (GSE69683) of blood transcriptomic from a large number of participants was extracted. A total number of 498 participants divided into 87 healthy controls, 77 moderate asthma, 246 nonsmoker’s severe asthma, and 88 smoker’s severe asthma collected in the U-BIOPRED study, as shown in Fig. 6. Our reanalysis showed that GPRC5A, SFN, ABCA1, and NEK2 mRNA expression was upregulated in asthmatic patients compared to healthy controls (P < 0.05), specifically the nonsmoker severe asthmatics. SERPINE1 mRNA expression was lower in nonsevere asthmatics compared to severe and healthy controls.
Figure 6.
Normalized gene expression of genes identified in the blood of asthmatic patients compared to healthy controls extracted from the expression profile of publicly available datasets (GSE69683) collected in the U-BIOPRED study, where (H) represents the 87 healthy controls, (M) represents the 77-moderate asthma, (S) represents the 246 nonsmoker’s severe asthma, and 88 smoker’s severe asthma, (A) represents the sum of all asthmatic patients in all severities.
SFN and ABCA1 were higher while MKI67 was lower in severe asthmatic with wheeze compared to nonasthmatics with wheezes
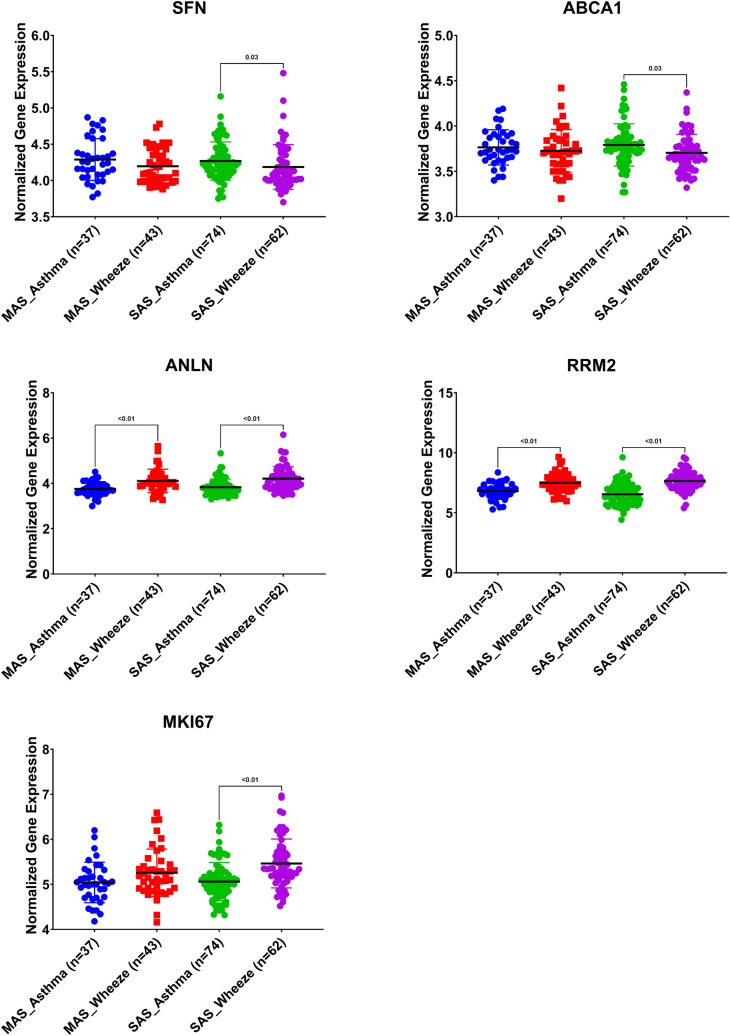
The correlation of the shortlisted genes to wheeze in asthmatic versus nonasthmatic children was investigated using (GSE123750) dataset. GSE123750 is a cross-sectional study of school-age children with severe asthma or mild, moderate asthma, and pre-school age children with severe wheeze or mild, moderate wheeze. Blood was collected at baseline. mRNA expression of the identified genes in the whole blood of children was compared between the two groups. As shown in Fig. 7, SFN, ABCA1 mRNA expression was significantly higher, while MKI67 mRNA expression was lower in severe asthmatic compared to severe wheezing children. On the other hand, ANLN and RRM2 mRNA expression were significantly lower in asthmatics than nonasthmatic wheezers (both moderate and severe forms).
Figure 7.
Normalized gene expression of genes identified in whole blood of children with nonsevere and severe wheezes were compared to nonsevere and severe asthmatics extracted from the expression profile of publicly available datasets (GSE123750), where nonsevere asthmatic (MAS_Asthma) and severe asthmatic (SAS_Asthma) patients were compared to patients presented with nonsevere wheezes (MAS_Wheeze) and severe wheezes (SAS_Wheeze).
House dust mite downregulates SERPINE1 and ABCA1 mRNA expression in CD lymphocytes
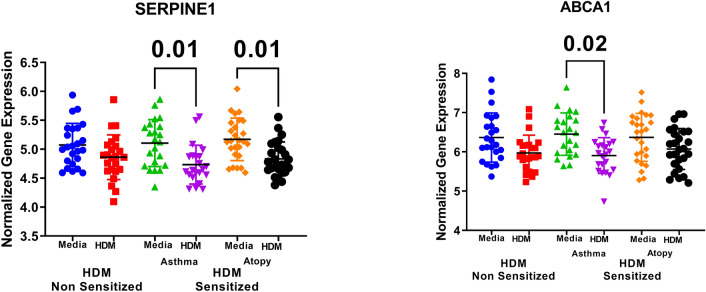
HDM is a significant allergen source and a significant cause of allergic rhinitis and allergic asthma 26. We explored whether the identified genes were stimulated by HDM or not using GSE73482 transcriptomic dataset. SERPINE1 mRNA expression was significantly downregulated in asthmatic or atopic patients CD4+ T Lymphocytes when challenged with HDM, while ABCA1 was downregulated in asthmatic samples only, as shown in Fig. 8. Our analysis showed that HDM did not alter the gene expression of the other genes identified, indicating that they are related to pathways where HDM had no role.
Figure 8.
Normalized gene expression of genes identified in CD4 lymphocytes stimulated with HDM compared to media stimulation only in healthy, atopic, and asthmatic patients extracted from the expression profile of publicly available datasets (GSE73482).
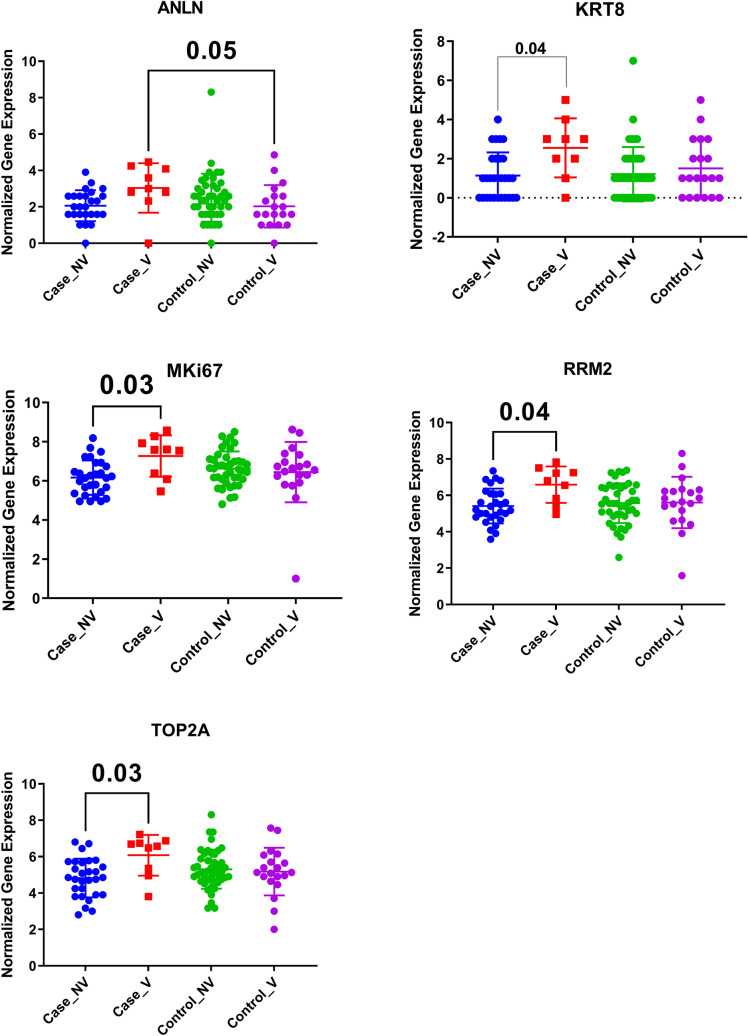
Viral respiratory tract infections upregulated KRT8, MKI67, RRM2, TOP2A mRNA expression in asthmatic patients
To examine the effect of viral infection on the gene expression of the identified genes, we extracted the normalized gene expression of the GSE115823 transcriptomic dataset where 208 children with exacerbation prone asthma were enrolled with baseline samples collected and were prospectively monitored for the onset of cold symptoms (Events) for six months. KRT8, MKI67, RRM2, and TOP2Agene expression were upregulated significantly in asthmatic who developed cold symptoms, as shown in Fig. 9. ANLN was upregulated in asthmatic who got cold compared to healthy who got colds, indicating its specificity to asthma exacerbation to viral infections.
Figure 9.
Normalized gene expression of genes identified in blood taken from asthmatic children at baseline (Case_NV) and after they develop viral infections (Case_V) compared to healthy controls (Control_NV and Control_V) extracted from the expression profile of publicly available datasets (GSE115823).
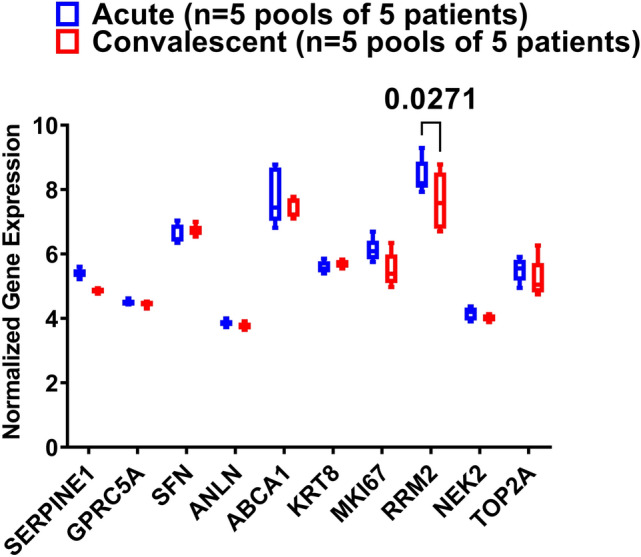
PBMCs of asthmatic patients during the exacerbation phase express higher RRM2 compared to the convalescent phase
GSE16032 dataset was chosen to identify the differential expression of the selected genes in PBMCs taken from asthmatic patients during the acute/exacerbation phase and the convalescent phase, as shown in Fig. 10. Interestingly, only RRM2 was upregulated in the acute/exacerbation phase of asthma and returned to the basal level when the attack subsides.
Figure 10.
Normalized gene expression of genes identified in PBMCs taken from asthmatic patients during the acute/exacerbation phase and the convalescent phase extracted from the expression profile of publicly available datasets (GSE16032).
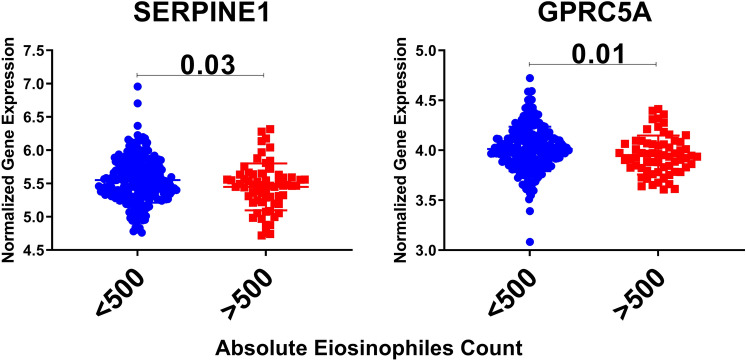
SERPINE1 and GPRC5A mRNA expression in whole blood of eosinophilic asthma was significantly lower compared to non-eosinophilic
To explore the correlation of the shortlisted genes to the eosinophilic phenotype of asthma, (GSE137394) dataset was reanalyzed. mRNA expression of the identified genes in whole blood of eosinophilic asthma (> 500 cells) compared to non-eosinophilic (eosinophils count less than 500) were explored, as shown in Fig. 11. Only SERPINE1 and GPRC5A were significantly lower in eosinophilic asthmatics (p = 0.03, and 0.01, respectively). This indicates that SERPINE1 and GPRC5A mRNA levels can identify the eosinophilic subtype of a severe asthma phenotype.
Figure 11.
Normalized gene expression of genes Identified in whole blood of eosinophilic asthma (> 500 cells) compared to non-eosinophilic (eosinophils count less than 500) extracted from the expression profile of publicly available datasets (GSE137394).
The ten genes have never been linked to SNPs in asthma
Genome-wide association studies (GWASs) of asthma have identified many risk alleles and loci that can be a potential source for diagnostic biomarkers. NHGRI-EBI GWAS Catalog was explored to identify 1174 SNPs with the strongest Asthma-related SNP-risk allele. Interestingly, none of these SNPs were reported in any of the ten genes identified.
Methylomes
Another explanation of differential expression of mRNA of the shortlisted genes in asthmatic after excluding genetic alterations in terms of SNPs was epigenetic modifications. DNA methylation dataset (GSE85568) in primary airway epithelial cells from 74 asthmatic and 41 nonasthmatic adults was used to investigate the differential methylation of genes of interest between asthma and healthy controls. There was no significant difference between the methylation status of identified genes in lung epithelium, indicating that epigenetic changes are not responsible for the differential expression seen in severe asthma.
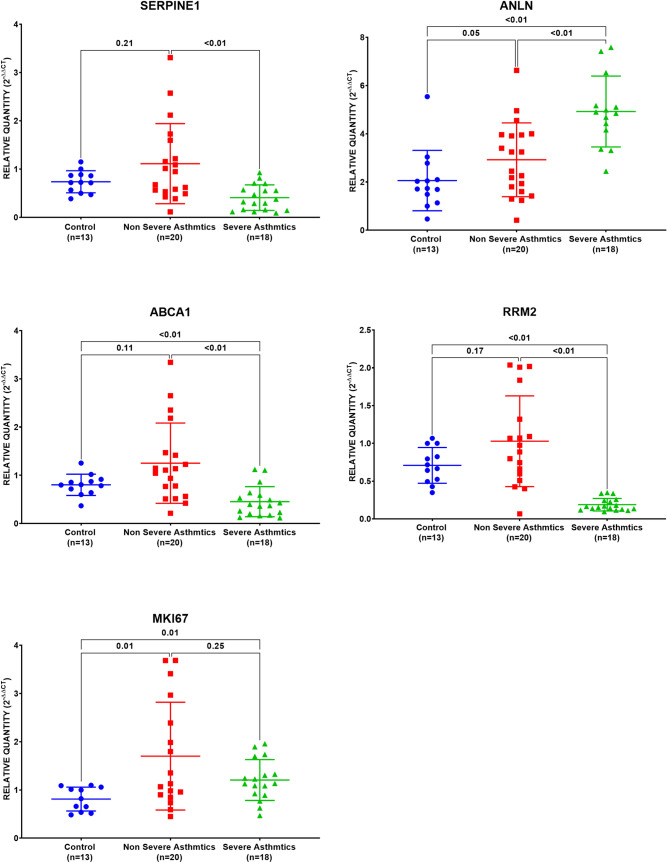
SERPINE1, ABCA1, and RRM2 were downregulated in severe asthmatics
mRNA expression of the ten genes was assessed in a locally recruited cohort of asthmatic patients compared to healthy controls; the genes with a significant difference are plotted in Fig. 12. ANLN was the only gene that showed increased expression levels with the severity of the disease. ANLN mRNA expression was higher in asthmatics (nonsevere and severe) and was higher in severe compared to nonsevere asthmatics PBMCs (p < 0.05). The rising level of ANLN with the disease might be used to monitor the disease progression. MKI67 and SERPINE1 expression was higher in nonsevere asthma but was downregulated in severe asthmatics compared to healthy controls (p < 0.05). ABCA1 and RRM2 expression were lower in severe asthmatics than nonsevere asthmatics and healthy controls, making them potential biomarkers to differentiate the asthma severity.
Figure 12.
mRNA gene expression using RT qPCR of the ten genes in PBMC of the locally recruited cohort, healthy controls (n = 13), Nonsevere asthmatics (n = 20), and severe asthmatic (n = 18).
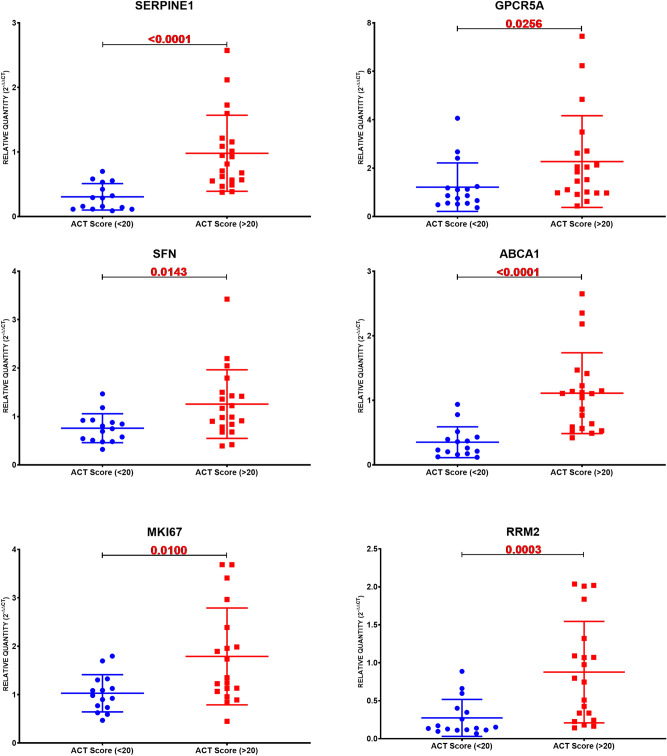
SERPINE1, GPCR5A, SFN, ABCA1, MKI67, and RRM2 are upregulated in well-controlled asthma
Asthma Control Test (ACT) is a self-administered patient tool for identifying those with poorly controlled asthma. The scores range from 5 (poor control of asthma) to 25 (complete control of asthma), with higher scores reflecting greater asthma control. An ACT score > 20 indicates well-controlled asthma. When we divided our patients accordingly, SERPINE1, GPCR5A, SFN, ABCA1, MKI67, and RRM2 are upregulated in well-controlled asthma, as shown in Fig. 13. This shows that dividing the patients according to ACT, a more objective evaluation of the condition and management compliance, is more accurate to have a homogenous expression of biomarkers in blood samples.
Figure 13.
mRNA gene expression using RT qPCR of the ten genes in PBMC of a locally recruited cohort, the asthmatic patients were divided according to the ACT score into two groups: a well-controlled group (ACT > 20) and those with poor control (ACT < 20).
Discussion
There is currently an imminent need to discover new biomarkers to classify different asthmatic phenotypes, predict their clinical behavior and therapeutic response, and identify new therapeutic targets 8, 9. Those markers need to be less invasive and more technically convenient than the currently available 10. Despite that necessity, only a few markers are implemented in clinical practice 11. The use of transcriptomic-driven biomarker identifications is a promising approach. However, many studies using microarrays showed extreme method-to-method variations in the results of the significant differentially expressed genes (DEG) by using different approaches that seek to shorten the list of identified genes 27. These limitations affect the proper identification of truly DEG from the equally expressed genes between disease and control 28.
Based on that, some of these successful biomarkers showed significant limitations in different populations and settings due to the lack of a unified system of patients’ classification and lack of complete understanding of confounding comorbidities, co-existing conditions, sampling time, and type besides the treatment history of asthmatic patients enrolled in the biomarker assessment 12. For example, atopic status, smoking, and the use of inhaled corticosteroids (ICS) are known confounders for the interpretation of asthma biomarkers 13. Another layer of obstacles that block the appropriate utilization of transcriptomic datasets is countless elusive experimental factors called confounding factors that can be biological and nonbiological 29. Identification of confounding factors is essential for accurate transcriptomic analysis30, but this task is not straightforward as many of these confounding factors are hidden and are difficult to detect before causing systematic bias31.
Cell cycle-related pathways are significantly enriched in the DEG identified
Our research team identified ten genes that were deranged in the severe asthmatic bronchial epithelium and fibroblasts related to cell cycle, cell proliferation, and survival-related pathways17. In subjects with severe asthma, there is an increased cellular proliferation in the airway contributing to a thickened epithelium leading to progressive decline in lung function32. After a dusty hay challenge in the bronchial epithelium, activation of cell cycle genes shows underlying impaired regeneration of the epithelial barrier in asthmatics 33. In severe eosinophilic asthma, fibroblasts can secrete exosomes to control epithelial cell proliferation, further increasing airway remodeling in asthma34. Interestingly, such asthmatics myofibroblasts showed reduced proliferative responses but increased cytokine production confirming their inflammatory profile35.
Our results showed that cell cycle-related pathways are significantly enriched in the top DEG identified. This finding goes with the previous report where increased cellular proliferation was documented in severe asthmatic airways compared to other phenotypes and healthy control, leading to a progressive decline in lung function and airway remodeling in those patients32. Cell cycle-related genes were differentially expressed in different tissues from children with asthma, indicating the importance of such a pathway in disease progress36. Upon injury, the bronchial epithelium can secrete inflammatory and fibrogenic cytokines when it shows asynchronous mitosis37. The effect of the underlying subepithelial fibroblasts can increase the proliferation of epithelial cells through TGF‐β signaling38. Most of these genes that we identified were reported to be related in a way or another to asthma and lung inflammation.
In lung
Our results showed that MKI67, ANLN, and NEK2 were upregulated in asthmatic bronchial epithelium while they were downregulated in asthmatic bronchial fibroblasts, although according to LGEA, those genes are specific proliferative mesenchymal progenitors who might indicate the loss of proliferative characteristic in severe asthmatic fibroblasts. Of note, their expression was higher in the bronchial brush than biopsy, which might reveal the possible contribution of the epithelium to airway remodeling through epithelial-mesenchymal transition (EMT) expression proliferative mesenchymal progenitors genes39. Such EMT is essential in asthma airway remodeling40 and inflammation-driven fibrosis in asthma through the TGF-β1 axis41. Such tissues with increased and prolonged EMT in response to chronic damage will increase fibroblast proliferation in healthy tissues42. In asthma, it was previously reported that bronchial epithelium differentiation would shift towards the basal epithelial, and those are the most susceptible to TGF-β1-induced EMT, which can give rise to 30–50% of lung fibroblasts in lung fibrosis and allergic asthma43.
Increased epithelial expression of MKI67 is linked to increased cycling basal bronchial cells that express higher (MKI67 and TOP2A) to compensate for damaged epithelium44. MKI67 suppression in fibroblasts might indicate the loss of proliferative mesenchymal phenotype or population in asthma with specific enrichment in Th2-high phenotype only. Loss of such mesenchymal stem cells can induce lung inflammation and airway remodeling45.
The anillin actin-binding protein (ANLN) gene encodes a protein that serves a vital role in cell-cycle progression and controls mitotic progression46. ANLN is a crucial mediator of cytokinesis as it binds to actin filaments and cytoskeletal components, serving as a scaffolding protein to coordinate cytokinesis 47. When ANLN is nonfunctional, it could disrupt the cellular organization of the bronchial epithelium and predispose it to acute respiratory distress48.
Never in Mitosis (NIMA) Related Kinase 2 (NEK2) plays a crucial role in regulating mitotic processes and an essential regulator of the centrosomal cycle besides other diverse cell cycle events 49–51. The increased NEK2 kinase levels in bronchial epithelium can increase the inflammatory phenotype of cells as NEK2 was shown to activate the canonical NFκB signaling pathway52. On the other hand, the decreased expression of NEK2 in asthmatic fibroblasts can be linked to their resistance to TNFa. NEK2 expression was shown to be suppressed when the TNFa blocker is used in inflammatory diseases53. It is recently proved that NEK2 regulates the accumulation of Wnt scaffolding protein Dishevelled (DVL) toward mitosis to remove linker proteins from the centrosome51. Its downregulation might derange Wnt signaling and fibroblasts proliferation, as we documented recently 17.
Among the three(MKI67, ANLN, and NEK2), MKI67 was shown to be upregulated in Th2 High asthmatic epithelium along with RRM2 and TOP2A, which might be an EMT marker in asthmatic epithelium too.
Our in silico and in vitro results showed that RRM2 was upregulated in asthmatic bronchial epithelium and fibroblasts, specifically in Th2-high asthma. It was previously shown that IL4 could upregulate its expression in resident macrophages 54. RRM2 showed differential expression in children with asthma 55 and was related to in utero OVA exposure in developing lungs of atopic rats 56. Besides M1, RRM2 is the M2 subunit of the ribonucleotide reductase enzyme that regulates the production of 2′-deoxyribonucleoside 5′-triphosphates (dNTP) required for DNA synthesis57, and when the RRM2 gene is knocked out, the cell will go to senescence 58. RRM2 is only expressed during the late G1/early S phase and degraded in the late S phase to control DNA synthesis and cell proliferation 59. RRM2 regulates the expression of the antiapoptotic protein Bcl-2 in apoptosis signaling60. Primary epithelial cell maintenance for 3–5 passages can alter the gene expression of NEK2 and RRM2 61. RRM2 is a stress response factor and, if silenced, can inhibit target gene expression in the Wnt/β‑catenin signaling pathway 62. It is upregulated in macrophages after smoking63. In the context of lung inflammation and repair, when epithelial cells are exposed to LPS, they translocate RRM2 to the nucleus to bind Urokinase-type plasminogen activator (uPA) mRNA 3′UTR and stabilize it to regulate fibrinolysis consequently64.
Chromosome untangling during sister chromatids segregation is catalyzed by DNA Topoisomerase II Alpha, the protein encoded by TOP2A 65. TOP2A catalyzes topological changes in DNA 66 and peaks in the G2/M phase to decatenate DNA during replication 67. The expression level of human TOP2A is controlled by its promoter region68, and SNP rs11650680 in this promoter was significantly inversely associated with asthma69, 70. TOP2 was one of the genes that were changed in offsprings of pregnant mice exposed to TiO2, increasing their susceptibility to asthma71.
KRT8, SERPINE1, and ABCA1 were upregulated in asthmatic bronchial epithelium with variable expression in asthmatic bronchial fibroblasts. Of note, their expression was higher in bronchial biopsies than in the brush, indicating a mixed cell origin like mesenchymal Pericytes (SERPINE1) and immune cells (ABCA1). Of note, KRT8 and SERPINE1 were upregulated by oral steroids.
KRT8 It is a known marker of basal luminal precursors cells in the lung 72, and such KRT5+, KRT8+ parabasal cell population are the precursors of the luminal cells and act to maintain epithelial homeostasis 73. KRT8 previously was shown to be significantly increased in different tissues and secretions of asthmatic patients 74. Epithelial KRT8 was shown to be markedly upregulated during viral and nonviral asthma exacerbations in children 75.
Plasminogen activator inhibitor‐1 (PAI‐1, also known as SERPINE1) is a serine protease inhibitor that regulates urokinase‐type plasminogen activator (uPA) and tissue‐type plasminogen activator (tPA) activity76. Polymorphisms spanning SERPINE1 have shown an association with asthma susceptibility77. The SERPINE1 expression is upregulated in response to wounding and, if impaired, can lead to delayed wound healing78. SERPINE1 inhibition decreases TGFβ1-induced profibrotic network79. SERPINE1 expression was found to be upregulated in hypoxic fibroblasts 80, in epithelial cells undergoing EMT, and myofibroblasts at the invasive front81. It is a part of the senescence-associated secretory phenotype (SASP) to regulate fibrinolysis and cellular senescence 82. SERPINE1 links epithelial cells and inflammatory activity by controlling key inflammatory modulators83. LPS induces SERPINE1 secretion, which in turn modulates the immune response to LPS 84. Systemic inflammatory TNFα and IL1β can upregulate PAI‐1 synthesis, especially in sepsis‐induced systemic inflammation by gram‐negative bacteremia 76. When in contact with virally infected asthmatic bronchial epithelial cells, macrophages can secrete more PAI-1 77.
Our results showed that SERPINE1 expression was more specific to Th2-high asthmatic bronchial epithelium and was upregulated in patients using oral steroids. After the allergen challenge, higher sputum PAI-1 concentration in sputum was documented 85. Mast cells in asthmatic airways are a significant source of increased SERPINE1 levels 86; these high levels were substantial in tissue repair and remodeling, ultimately affecting asthma severity response to steroids 87.
ABCA1 showed higher expression in bronchial biopsies than the pure bronchial epithelium that might indicate the role of infiltrating immune cells or other mesenchymal cells as a source of ABCA1 upregulation. The ABCA1 gene is a member of the ATP-binding cassette family that transports cholesterol and phospholipids across cell membranes. In the lung, ABCA1 regulates reverse cholesterol transport to prevent lipid accumulation and inflammatory cytokines induction 88. ABCA1 in alveolar macrophages plays an important role in attenuating the severity of neutrophilic airway inflammation in asthma89.
Our results showed that GPRC5A was downregulated in asthmatic bronchial epithelium and upregulated in asthmatic fibroblasts compared to their healthy counterparts. This gene encodes G protein-coupled receptors involved in signaling pathways in development, cellular growth, and epithelial cell differentiation. GPRC5A can block EGFR expression and activation90. GPRC5A is expressed in lung type I and type II epithelial cells constitutively91. Its deficiency can turn the lung to be more susceptible to endotoxin-induced pulmonary injury92 through NFκB and STAT3 signaling pathway81, indicating its vital role in lung homeostasis92. Cells deficient with GPRC5A produce higher chemokines and cytokines that recruit macrophages through their conditioned medium in an NFκB dependent manner93. Repression of GPRC5A is associated with activated IL6 induced STAT3 signaling in tracheal epithelial cells94.
Our results showed that Stratifin (SFN) was upregulated in the asthmatic epithelium and was specific to bronchial epithelial cells. SFN, also called 14–3–3 is a p53-inducible gene due to DNA damage to stop cell cycle progression95. 14–3–3σ is expressed primarily in epithelial cells as a cell cycle regulator and tumor suppressor96. On the other hand, suppression of SFN significantly reduced cell proliferation 95. Stratifin plays a significant role in desmosome dynamics in squamous epithelial cells97, and epithelial cells with 14–3–3σ loss can undergo epithelial to mesenchymal transition and loss of the epithelial phenotype98. SFN can be released from epithelial cells to stimulates matrix metalloproteinase (MMP)-1 expression, and this can affect tissue remodeling99. Neighboring epithelial cells releasable 14–3–3σ bind the APN cell surface receptor fibroblasts to act as a potent MMP-1-stimulatory factor 100, through c-Fos and c-Jun activity101. Such cross-talk should be highly controlled as the ECM continues to accumulate until fibroblasts receive epidermal cells signal to slow down 102; any disturbance can lead to excessive fibrosis proving the anti-fibrogenic effect of a controlled release form of stratifin 103. SFN was shown to be overexpressed in tissue biopsies of severe equine asthma 104.
In blood
One of the significant goals of asthma research is to identify simple, reliable, and accurate biomarkers that aid in the detection of the disease initiation or progression and lead to a better selection of more precise therapeutic approaches. Serum/blood markers represent the ideal candidate, as they are the least invasive and more patient-friendly approach compared to the bronchial brush or biopsy. Table 4 showed the summary of in silico and patient samples analysis of the ten genes in Blood of asthmatic patients in different settings compared to healthy controls.
Table 4.
Summary of in silico and patient samples analysis of the ten genes in Blood of asthmatic patients in different settings compared to healthy controls.
Their selective expression on lymphocytes (CD4 vs. CD8), their expression in CD4 cells in response to House Dust Mites (HDM), their expression in asthmatic whole blood (Asth.), their hypomethylation level (Meth) in different phenotypes, in asthmatic with wheezes vs. nonasthmatics wheezers, their expression in eosinophilic asthmatic vs. noneosinophilic, in acute vs. convalescent (acute) and during viral infection and in PBMPs of asthmatic patients from locally recruited cohort compared to healthy controls.
In blood, in silico analysis showed that ABCA1 was higher in asthmatic patient’s whole blood, especially in children with severe wheezes but was downregulated in CD4 lymphocytes stimulated with HDM. ABCA1 expression is specific to eosinophil and neutrophil lineage and is needed for proper Apolipoprotein A-I (apoA-I) mediated reverse cholesterol transport that blocks granulocyte colony-stimulating factor (G-CSF) from initiating neutrophilic inflammation 89. ApoA-I mimetic peptides can decrease both neutrophilic and eosinophilic airway inflammation suggesting that the apoA-I/ABCA1 pathway may have a protective effect on asthma105.
In our cohort, PBMCs analysis showed that severe asthmatics and patients with ACT < 20 showed downregulation of ABCA1. PBMCs are devoid of eosinophil and neutrophil, so it is expected to have lower ABCA1 in PBMCs than whole blood samples. On the other hand, ABCA1 downregulation might be translated into a loss of ABCA1 anti-neutrophilic effect. Another explanation for such ABCA1 downregulation is the heavy use of steroids in severe asthmatics, as most of these drugs inhibit eosinophils.
Under conditions of excess cellular cholesterol, increases cholesterol efflux through the lipid transporters ATP-binding cassette A1 (ABCA1) is initiated in proliferating T cells to maintain cholesterol homeostasis as they cannot proliferate unless cholesterol satisfies a metabolic checkpoint106 that made Abca1A has important anti-inflammatory properties107. A significant reduction of ABCA1 mRNA level in leukocytes of patients with atherosclerosis was determined compared with controls108. The decreased ABCA1 by HDM might alter T lymphocytes proliferation and function due to deranged cholesterol levels, and that augment the inflammation.
In blood, in silico analysis showed that ANLN mRNA expression was significantly higher in CD4 lymphocytes. ANLN expression is one of the genes shown to be downregulated in T lymphocytes to be Treg cells109. ANLN was shown to be upregulated in WBC from mice that developed tolerance to self-antigens (a peptide mimicking anti-DNA IgG sequences containing MHC class I and class II T cell determinant) compared to naive mice110. So ANLN upregulation in blood might indicate a decrease of this protective immune cell in response to a stimulus.
In silico analysis showed that ANLN mRNA expression was downregulated in the blood of asthmatic children compared to patients with wheezes, and it was upregulated significantly in asthmatic who develop cold symptoms compared to healthy who developed cold. ANLN mRNA expression was higher in asthmatics (nonsevere and severe) and higher in severe than nonsevere asthmatics PBMCs. Previous reports showed that defective ANLN could result in the abnormal cellular organization of the bronchiolar epithelium, which predisposes to acute respiratory distress48.
Our bioinformatics search showed GPCR5A was higher in asthma blood as a whole, while examining lymphocytes fraction only showed GPCR5A downregulation in asthma, indicating that its high level is from cells other than lymphocytes. Interestingly, although GPCR5A expression was lower in eosinophilic patients, it was hypomethylated in the monocytes of those patients, confirming that the main source of GPCR5A expression in blood is not from PBMCs. GPCR5A mRNA expression was significantly down in our cohort of patients in uncontrolled asthma (ACT < 20). It was shown that Gprc5a-ko mice had increased expression of Th2-high biomarkers111 and GPCR5A deficiency may lead to impaired pTreg-cell induction in autoimmune diseases as Gprc5a preferentially regulates FoxP3 expression 112.
Our in-silico results in asthmatic blood showed that KRT8 mRNA expression was higher in CD8 T lymphocytes in contrast to CD4 T lymphocytes and upregulated significantly in asthmatic who develop cold symptoms due to viral infection. KRT8 showed blood cell lineage of dendritic cells “plasmacytoid DC,” granulocytes, and NK-cells. KRT is considered an anti‐inflammatory gene that is induced by primed monocytes during stress113 and increased dramatically in asthmatics who have sensitization and exposure to cockroach allergen 74. Some reports showed that the self-reactive immune response to KRT might be an important factor in the inflammatory response114.
MKI67 gene expression was downregulated in the blood of asthmatic children’s severe patients with wheezes. Its specific upregulation in response to viral infection can be linked to its T-reg” and NK-cells lineage-specificity. Interleukin-15 was shown to promote the induction of MKI67 on NK cells suggesting a role for NK cells in regulating the immune-mediated pathology in severe asthma115. In our cohort, MKI67 was upregulated in the PBMCs of nonsevere but significantly downregulated in severe asthmatic patients and uncontrolled asthma (ACT < 20) compared to healthy controls.
Ki67 was upregulated in CD4+ and CD8+ T cells that had undergone in vitro proliferation after 6-day culture of human whole blood or PBMC with antigens making intracellular Ki67 expression provides a specific, quantitative and reproducible measure of antigen-specific T cell proliferation in vitro, which is a critical function of memory T cells116.
Downregulation of MKI67 in immune cells might indicate their exhaustion due to prolonged exposure to antigenic stimulation 117, 118. Immune cells with high MKI67 expressed patterns of genes associated with T-cell proliferation and contained a mixture of CD4+ T cells, CD8+ T cells, and Tregs with profound cell proliferation signature that displayed upregulation of pathways associated response to external stimulus pathways119. Based on that MKI67 mRNA expression in PBMC can give an accurate impression if the immune cells are in active proliferative phase close to antigen exposure and if the immune cells are impaired like in severe asthma by the disease or in response to immune suppression like steroids.
MKI67 and SERPINE1 expression was higher in nonsevere asthma but was downregulated in severe asthmatics compared to healthy controls. This might indicate their role in the initiation of the disease and then downregulated in severe in response to the therapy or pathophysiological changes, although with the absence of time frame for the patient's asthma progression, such conclusion can be speculative.
Our in-silico results showed that many cell types expressing NEK2 as NEK2 showed Low lineage specificity and low cell type specificity as its mRNA was detected in many blood cells, higher in CD4 T lymphocytes than CD8 T lymphocytes. In CD4 T lymphocytes, only NEK2 was lower in severe asthmatic patients than healthy controls but upregulated in the whole blood of asthmatic, indicating that their source might not be lymphocytes.
The serine/threonine kinase Nek2 is commonly implicated in promoting cell proliferation of B cells leading to an increase in the proportion of immature B cells in the bone marrow with enhanced T cell-dependent immune response120. NEK2 is the one most significantly upregulated gene in Multiple Myeloma (MM), a malignancy of terminally differentiated B-cells homing to and expanding in the bone marrow121.
Our in-silico analysis showed that RRM2 mRNA expression was upregulated in the blood of an asthmatic patient who had upper respiratory tract infections and those who have acute exacerbation of asthmatic attack but was downregulated in those with severe asthma and wheezes. Protein Atlas data showed that RRM2 is enhanced in T cells, especially Treg. In our cohort, the RRM2 mRNA expression was downregulated in severe asthmatics and those who are not well controlled (ACT < 20). Genetic ablation of RRM2 can induce G1/S phase cell-cycle arrest 30 because RRM2 may co-express with relevant genes to regulate the process of lymphocyte infiltration 122, 123.
In blood, our in-silico analysis showed that SERPINE1 expression was downregulated in eosinophilic patients or those who were challenged with HDM despite the fact that SERPINE1 local lung expression was upregulated. The SERPINE1 polymorphisms are associated with bronchial reactivity, IgE response, and susceptibility to allergic asthma 84, 124. Flagellin was shown to induce SERPINE1 with Th17- and TLR-mediated responses125. IL17 producing γδT cells can induce Serpine1 as one of the anti‐angiogenic factors 126. On the other hand, full Th17 cell development is dependent on the activity of SERPINE1 127. Th17 cells-derived expression of the PAI-1 along with IL17A was shown to block the anti-inflammatory effect of TGF-β 83. In our cohort, SERPINE1 expression in PBMCs was lower in severe asthmatic and those with ACT < 20.
Serine Proteases Inhibitors (serpins) control T lymphocytes functions like the antigen recognition and effector function plus promoting cellular viability and development of long-lived memory T cells through their potent antiapoptotic properties on hematological progenitor cells 128. During systemic inflammation, elevated circulating levels of endogenous PAI-1 serve to inhibit IFN-gamma release129. Decreased SERPINE1 after HDM in CD4 lymphocytes might indicate a deranged functional T cell.
Our in silico analysis showed that SFN has lineage enrichment for basophil and was upregulated in the whole blood of asthmatics and those with severe asthmatic wheezes. In our cohort, SFN was significantly downregulated in PBMCs with ACT less than 20. Stratifin significantly moderated inflammation by reducing infiltrated CD3(+) immune cells103. In patients with chronic rhinosinusitis with nasal polyps, Stratifin was downregulated with dysregulated CD4+ and CD8+ T cells accumulation with impaired suppression of Foxp3+ regulatory T cells leading to a dominant infiltration of lymphocytes and neutrophils and evaluated the inflammatory gene expression130. SFN mRNA expression in our cohort was downregulated and might indicate the loss of SFN anti-inflammatory effect leading to inflammatory cells accumulation and impaired regulatory T cells.
TOP2 was upregulated significantly in asthmatic who develop cold symptoms after viral infection linked to their T-reg lineage-specific. During acute lung inflammation, inflammatory macrophages in the airspaces with high proliferative capacity (high TOP2A expression showed a higher inflammatory profile 131. TOP2A gene expression was shown previously to be upregulated in human monocytes exposed acutely to Cigarette Smoke 63. During chronic inflammatory diseases, TOP2A high expressing cells represent the proliferative subpopulation in exhausted immune cells 132.
Limitation
Because most of the data-driven approach of the manuscript is based on publicly available transcriptomic data, which has significant limitations and drawbacks, a large number of studies using microarrays showed extreme method-to-method variations in the results of the significant differentially expressed genes (DEG) by using different approaches that seek to shorten the list of identified genes 27. These limitations affect the proper identification of truly DEG from the equally expressed genes between disease and control 28. Age adjustment for the gene expression across different datasets, mainly when the datasets include children while others have adults, is crucial in analyzing omics data and in vivo and in vitro analysis. As per your work, we stressed that as confounding factors that might affect the meaningful interpretations of the biomarker's significance. One of the stringent inclusion criteria that we used to include datasets is that healthy controls and patients groups should be age and sex-matched, making the DEG between different datasets consistent as much as possible. Nevertheless, across other datasets, we might need to add this as a limitation that we did not adjust for age. Also, we tried to use a large number of datasets in different settings using the same analysis method and comparing the normalized gene expression of the identified genes. Another limitation is a relatively small number of cells and patients samples because COVID-19 restricted the patient’s recruitments, although the numbers used were based on the power of calculations to identify minimal numbers needed for meaningful results.
In conclusion
In conclusion, we used combined in silico, in vitro, and in vivo approaches in an attempt to decipher the molecular basis of asthma heterogeneity by identifying a set of ten genes that are simultaneously differentially expressed in different cells and tissues. We identified factors related to the patients, disease status, and external factors that might deviate the interpretation of the transcriptomic data and proved in vitro and locally recruited patients that the identified genes are biologically crucial to the homeostasis of the lung and in asthma development and progression.
Supplementary Information
Author contributions
M.Y.H.: conceptualization, data curation, formal analysis, investigation, methodology, validation, software, visualization, writing—original draft. N.M.E.: performed the qPCR. R.R.: patients samples collections and cell lines maintenance, manuscript initial review. L.S.: patients selection and samples collection. R.O.: resources. I.Y.H.: formal analysis, investigation, software, visualization, writing-review. T.V.: technical support for qPCR and primer design. B.M.: resources, patients selection, and samples collection, supervision. S.A.H.: manuscript initial review. Q.H.: conceptualization, funding acquisition, project administration, resources, supervision, writing–review and editing. R.H.: conceptualization, Funding acquisition, methodology, resources, software, writing—review and editing.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained errors in the Figure legends of Figure 3 and Figure 4. The legends of these Figures were inadvertently switched.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/8/2021
A Correction to this paper has been published: 10.1038/s41598-021-97816-0
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91087-5.
References
- 1.Chiappori A, et al. Biomarkers and severe asthma: A critical appraisal. Clin. Mol.Allergy. 2015;13:20. doi: 10.1186/s12948-015-0027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 3.Wan XC, Woodruff PG. Biomarkers in severe asthma. Immunol. Allergy Clin. N. Am. 2016;36(3):547–557. doi: 10.1016/j.iac.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H, et al. Asthma biomarkers in the age of biologics. Allergy Asthma Clin. Immunol. 2017;13(1):48. doi: 10.1186/s13223-017-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry A, Busse WW. Biomarkers in asthmatic patients: Has their time come to direct treatment? J. Allergy Clin. Immunol. 2016;137(5):1317–1324. doi: 10.1016/j.jaci.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Shin SW, et al. Asthma-predictive genetic markers in gene expression profiling of peripheral blood mononuclear cells. Allergy Asthma Immunol. Res. 2011;3(4):265–272. doi: 10.4168/aair.2011.3.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hachim MY, et al. An integrative phenotype-genotype approach using phenotypic characteristics from the UAE National Diabetes Study Identifies HSD17B12 as a candidate gene for obesity and type 2 diabetes. Genes. 2020;11(4):461. doi: 10.3390/genes11040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persson H, et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J. Allergy Clin. Immunol. 2015;136(3):638–648. doi: 10.1016/j.jaci.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 9.Stewart CE, et al. Evaluation of differentiated human bronchial epithelial cell culture systems for asthma research. J. Allergy (Cairo) 2012;2012:943982. doi: 10.1155/2012/943982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo CS, et al. A transcriptome-driven analysis of epithelial brushings and bronchial biopsies to define asthma phenotypes in U-BIOPRED. Am. J. Respir. Crit. Care Med. 2017;195(4):443–455. doi: 10.1164/rccm.201512-2452OC. [DOI] [PubMed] [Google Scholar]
- 11.Hekking PP, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J. Allergy Clin. Immunol. 2018;141(4):1280–1290. doi: 10.1016/j.jaci.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 12.Forno, E. and J.C. Celedón. Epigenomics and transcriptomics in the prediction and diagnosis of childhood asthma: Are we there yet? Front. Pediatr. 7(115) (2019). [DOI] [PMC free article] [PubMed]
- 13.Altman MC, Busse WW. A Deep dive into asthma transcriptomics. Lessons from U-BIOPRED. Am. J. Respir. Crit. Care Med. 2017;195(10):1279–1280. doi: 10.1164/rccm.201611-2387ED. [DOI] [PubMed] [Google Scholar]
- 14.Park HW, Weiss ST. Understanding the molecular mechanisms of asthma through transcriptomics. Allergy Asthma Immunol. Res. 2020;12(3):399–411. doi: 10.4168/aair.2020.12.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim RD, Park PJ. Improving identification of differentially expressed genes in microarray studies using information from public databases. Genom. Biol. 2004;5(9):R70. doi: 10.1186/gb-2004-5-9-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modena BD, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am. J. Respir. Crit. Care Med. 2014;190(12):1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hachim, M.Y., et al. Wnt signaling is deranged in asthmatic bronchial epithelium and fibroblasts. Front. Cell Dev. Biol. 9, 641404 10.3389/fcell.2021.641404 (2021). [DOI] [PMC free article] [PubMed]
- 18.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Y, et al. Lung Gene Expression Analysis (LGEA): An integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72(5):481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagur PK, McCoy JP., Jr Collection, storage, and preparation of human blood cells. Curr. Protoc. Cytom. 2015;73(1):511–5116. doi: 10.1002/0471142956.cy0501s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SW, et al. Amphiregulin expression in human mast cells and its effect on the primary human lung fibroblasts. J. Allergy Clin. Immunol. 2005;115(2):287–294. doi: 10.1016/j.jaci.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 22.Fahy JV. Type 2 inflammation in asthma–present in most, absent in many. Nat. Rev. Immunol. 2015;15(1):57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuruvilla ME, Lee FE-H, Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 2019;56(2):219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samitas K, Zervas E, Gaga M. T2-low asthma: Current approach to diagnosis and therapy. Curr. Opin. Pulm. Med. 2017;23(1):48–55. doi: 10.1097/MCP.0000000000000342. [DOI] [PubMed] [Google Scholar]
- 25.Synek M, et al. Cellular infiltration of the airways in asthma of varying severity. Am. J. Respir. Crit. Care Med. 1996;154(1):224–230. doi: 10.1164/ajrccm.154.1.8680684. [DOI] [PubMed] [Google Scholar]
- 26.Calderon MA, et al. Respiratory allergy caused by house dust mites: What do we really know? J. Allergy Clin. Immunol. 2015;136(1):38–48. doi: 10.1016/j.jaci.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Hansel NN, Diette GB. Gene expression profiling in human asthma. Proc. Am. Thorac. Soc. 2007;4(1):32–36. doi: 10.1513/pats.200606-132JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novianti PW, et al. An application of sequential meta-analysis to gene expression studies. Cancer Inform. 2015;14(Suppl 5):1–10. doi: 10.4137/CIN.S27718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11(10):733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toubia J, Conn VM, Conn SJ. Don't go in circles: Confounding factors in gene expression profiling. EMBO J. 2018;37(11):e97945. doi: 10.15252/embj.201797945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Zhou X. Controlling for confounding effects in single cell RNA sequencing studies using both control and target genes. Sci. Rep. 2017;7(1):13587. doi: 10.1038/s41598-017-13665-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen L, et al. Epithelial cell proliferation contributes to airway remodeling in severe asthma. Am. J. Respir. Crit. Care Med. 2007;176(2):138–145. doi: 10.1164/rccm.200607-1062OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tessier L, et al. Gene set enrichment analysis of the bronchial epithelium implicates contribution of cell cycle and tissue repair processes in equine asthma. Sci. Rep. 2018;8(1):16408. doi: 10.1038/s41598-018-34636-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haj-Salem I, et al. Fibroblast-derived exosomes promote epithelial cell proliferation through TGF-beta2 signalling pathway in severe asthma. Allergy. 2018;73(1):178–186. doi: 10.1111/all.13234. [DOI] [PubMed] [Google Scholar]
- 35.Ward JE, et al. Proliferation is not increased in airway myofibroblasts isolated from asthmatics. Eur. Respir. J. 2008;32(2):362–371. doi: 10.1183/09031936.00119307. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, et al. Identification of differentially expressed genes associated with asthma in children based on the bioanalysis of the regulatory network. Mol. Med. Rep. 2018;18(2):2153–2163. doi: 10.3892/mmr.2018.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freishtat RJ, et al. Asthmatic airway epithelium is intrinsically inflammatory and mitotically dyssynchronous. Am. J. Respir. Cell Mol. Biol. 2011;44(6):863–869. doi: 10.1165/rcmb.2010-0029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semlali A, et al. Regulation of epithelial cell proliferation by bronchial fibroblasts obtained from mild asthmatic subjects. Allergy. 2010;65(11):1438–1445. doi: 10.1111/j.1398-9995.2010.02376.x. [DOI] [PubMed] [Google Scholar]
- 39.Hackett TL. Epithelial-mesenchymal transition in the pathophysiology of airway remodelling in asthma. Curr. Opin. Allergy Clin. Immunol. 2012;12(1):53–59. doi: 10.1097/ACI.0b013e32834ec6eb. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z, et al. Epithelial–mesenchymal transition in asthma airway remodeling is regulated by the IL-33/CD146 axis. Front. Immunol. 2020;11:1598–1598. doi: 10.3389/fimmu.2020.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haddad A, et al. Neutrophils from severe asthmatic patients induce epithelial to mesenchymal transition in healthy bronchial epithelial cells. Respir. Res. 2019;20(1):234. doi: 10.1186/s12931-019-1186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rout-Pitt N, et al. Epithelial mesenchymal transition (EMT): A universal process in lung diseases with implications for cystic fibrosis pathophysiology. Respir. Res. 2018;19(1):136. doi: 10.1186/s12931-018-0834-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartis D, et al. Epithelial–mesenchymal transition in lung development and disease: Does it exist and is it important? Thorax. 2014;69(8):760–765. doi: 10.1136/thoraxjnl-2013-204608. [DOI] [PubMed] [Google Scholar]
- 44.Vieira Braga, F.A., et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med.25(7), 1153–1163. 10.1038/s41591-019-0468-5 (2019). [DOI] [PubMed]
- 45.Lin HY, et al. Mesenchymal stem cells suppress lung inflammation and airway remodeling in chronic asthma rat model via PI3K/Akt signaling pathway. Int. J. Clin. Exp. Pathol. 2015;8(8):8958–8967. [PMC free article] [PubMed] [Google Scholar]
- 46.Long X, et al. Prognostic significance of ANLN in lung adenocarcinoma. Oncol. Lett. 2018;16(2):1835–1840. doi: 10.3892/ol.2018.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, et al. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology. 2018;154(5):1421–1434. doi: 10.1053/j.gastro.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holopainen S, et al. ANLN truncation causes a familial fatal acute respiratory distress syndrome in Dalmatian dogs. PLoS Genet. 2017;13(2):e1006625. doi: 10.1371/journal.pgen.1006625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha Kim Y, et al. Nek2 localizes to multiple sites in mitotic cells, suggesting its involvement in multiple cellular functions during the cell cycle. Biochem. Biophys. Res. Commun. 2002;290(2):730–736. doi: 10.1006/bbrc.2001.6212. [DOI] [PubMed] [Google Scholar]
- 50.Fang Y, Zhang X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle. 2016;15(7):895–907. doi: 10.1080/15384101.2016.1152430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cervenka I, et al. Dishevelled is a NEK2 kinase substrate controlling dynamics of centrosomal linker proteins. Proc. Natl. Acad. Sci. USA. 2016;113(33):9304–9309. doi: 10.1073/pnas.1608783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franqui-Machin R, et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Invest. 2018;128(7):2877–2893. doi: 10.1172/JCI98765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaba LC, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J. Allergy Clin. Immunol. 2009;124(5):1022–1030.e1–395. doi: 10.1016/j.jaci.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gundra UM, et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood. 2014;123(20):e110–e122. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen E, et al. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64(1):38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carpe N, et al. Maternal allergen exposure reprograms the developmental lung transcriptome in atopic and normoresponsive rat pups. Am. J. Physiol. Lung Cell Mol. Physiol. 2012;303(10):L899–911. doi: 10.1152/ajplung.00179.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duxbury MS, et al. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23(8):1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 58.Aird KM, et al. Identification of ribonucleotide reductase M2 as a potential target for pro-senescence therapy in epithelial ovarian cancer. Cell Cycle. 2014;13(2):199–207. doi: 10.4161/cc.26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K, et al. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: Implication of RRM2 in angiogenesis. Mol Cancer. 2009;8(1):11. doi: 10.1186/1476-4598-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahman MA, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: A potential target for cancer therapy. Clin. Cancer Res. 2013;19(13):3416–3428. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reeves SR, et al. Stability of gene expression by primary bronchial epithelial cells over increasing passage number. BMC Pulm. Med. 2018;18(1):91. doi: 10.1186/s12890-018-0652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, et al. Silencing RRM2 inhibits multiple myeloma by targeting the Wnt/betacatenin signaling pathway. Mol. Med. Rep. 2019;20(3):2159–2166. doi: 10.3892/mmr.2019.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright WR, et al. Inflammatory transcriptome profiling of human monocytes exposed acutely to cigarette smoke. PLoS ONE. 2012;7(2):e30120. doi: 10.1371/journal.pone.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shetty SK, et al. Regulation of urokinase expression at the posttranscription level by lung epithelial cells. Biochemistry. 2012;51(1):205–213. doi: 10.1021/bi201293x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang X, et al. PTEN stabilizes TOP2A and regulates the DNA decatenation. Sci. Rep. 2015;5:17873. doi: 10.1038/srep17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Austin CA, et al. TOP2B: The first thirty years. Int. J. Mol. Sci. 2018;19(9):2765. doi: 10.3390/ijms19092765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang X, et al. PTEN stabilizes TOP2A and regulates the DNA decatenation. Sci. Rep. 2015;5(1):17873. doi: 10.1038/srep17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T, et al. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene. 2015;34(31):4019–4031. doi: 10.1038/onc.2014.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marinho S, et al. 17q12–21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann. Allergy Asthma Immunol. 2012;108(6):402–411.e9. doi: 10.1016/j.anai.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Miyake Y, Tanaka K, Arakawa M. Association between 17q12-21 variants and asthma in Japanese women: rs11650680 polymorphism as potential genetic marker for asthma. DNA Cell Biol. 2014;33(8):531–536. doi: 10.1089/dna.2014.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fedulov AV, et al. Pulmonary exposure to particles during pregnancy causes increased neonatal asthma susceptibility. Am. J. Respir. Cell Mol. Biol. 2008;38(1):57–67. doi: 10.1165/rcmb.2007-0124OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schilders KA, et al. Regeneration of the lung: Lung stem cells and the development of lung mimicking devices. Respir. Res. 2016;17:44. doi: 10.1186/s12931-016-0358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watson JK, et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 2015;12(1):90–101. doi: 10.1016/j.celrep.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao P, et al. Keratins: Important candidate genes for asthma and immune responsiveness to cockroach. J. Allergy Clin. Immunol. 2007;119(1):S176. doi: 10.1016/j.jaci.2006.12.052. [DOI] [Google Scholar]
- 75.Altman MC, et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat. Immunol. 2019;20(5):637–651. doi: 10.1038/s41590-019-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iwaki T, Urano T, Umemura K. PAI-1, progress in understanding the clinical problem and its aetiology. Br. J. Haematol. 2012;157(3):291–298. doi: 10.1111/j.1365-2141.2012.09074.x. [DOI] [PubMed] [Google Scholar]
- 77.Koppelman GH, Sayers I. Evidence of a genetic contribution to lung function decline in asthma. J. Allergy Clin. Immunol. 2011;128(3):479–484. doi: 10.1016/j.jaci.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 78.Sun H, et al. Hyperglycemia-suppressed expression of Serpine1 contributes to delayed epithelial wound healing in diabetic mouse corneas. Invest. Ophthalmol. Vis. Sci. 2015;56(5):3383–3392. doi: 10.1167/iovs.15-16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rawson R, et al. TGF-beta1-induced PAI-1 contributes to a profibrotic network in patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2016;138(3):791–800.e4. doi: 10.1016/j.jaci.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muth M, et al. Hypoxia-induced down-regulation of microRNA-449a/b impairs control over targeted SERPINE1 (PAI-1) mRNA—A mechanism involved in SERPINE1 (PAI-1) overexpression. J. Transl. Med. 2010;8(1):33. doi: 10.1186/1479-5876-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu F, Korc M. Cdk4/6 inhibition induces epithelial–mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol. Cancer Ther. 2012;11(10):2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khan SS, et al. A null mutation in SERPINE1 protects against biological aging in humans. Sci. Adv. 2017;3(11):eaao1617. doi: 10.1126/sciadv.aao1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaiko, G.E. et al. PAI-1 augments mucosal damage in colitis. Sci. Transl. Med. 11(482). 10.1126/scitranslmed.aat0852 (2019). [DOI] [PMC free article] [PubMed]
- 84.Kowal K, et al. Analysis of − 675 4 g/5 G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. J. Investig. Allergol. Clin. Immunol. 2008;18(4):284–292. [PubMed] [Google Scholar]
- 85.Kowal K, et al. Concentrations of plasminogen activator inhibitor-1 (PAI-1) and urokinase plasminogen activator (uPA) in induced sputum of asthma patients after allergen challenge. Folia Histochem. Cytobiol. 2010;48(4):518–523. doi: 10.2478/v10042-010-0075-2. [DOI] [PubMed] [Google Scholar]
- 86.Ma Z, Paek D, Oh CK. Plasminogen activator inhibitor-1 and asthma: Role in the pathogenesis and molecular regulation. Clin. Exp. Allergy. 2009;39(8):1136–1144. doi: 10.1111/j.1365-2222.2009.03272.x. [DOI] [PubMed] [Google Scholar]
- 87.Dijkstra A, et al. SERPINE1 -675 4G/5G polymorphism is associated with asthma severity and inhaled corticosteroid response. Eur. Respir. J. 2011;38(5):1036–1043. doi: 10.1183/09031936.00182410. [DOI] [PubMed] [Google Scholar]
- 88.Chai AB, Ammit AJ, Gelissen IC. Examining the role of ABC lipid transporters in pulmonary lipid homeostasis and inflammation. Respir. Res. 2017;18(1):41. doi: 10.1186/s12931-017-0526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dai C, et al. ATP-binding cassette transporter 1 attenuates ovalbumin-induced neutrophilic airway inflammation. Am. J. Respir. Cell Mol. Biol. 2014;51(5):626–636. doi: 10.1165/rcmb.2013-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin E, et al. Clinical significance of reduced GPRC5A expression in surgically resected non-small cell lung cancer. Oncol. Lett. 2019;17(1):502–507. doi: 10.3892/ol.2018.9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tao Q, et al. Identification of the retinoic acid-inducible Gprc5a as a new lung tumor suppressor gene. J. Natl. Cancer Inst. 2007;99(22):1668–1682. doi: 10.1093/jnci/djm208. [DOI] [PubMed] [Google Scholar]
- 92.Liao Y, et al. Gprc5a-deficiency confers susceptibility to endotoxin-induced acute lung injury via NF-kappaB pathway. Cell Cycle. 2015;14(9):1403–1412. doi: 10.1080/15384101.2015.1006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang X, et al. GPRC5A: An emerging biomarker in human cancer. Biomed. Res. Int. 2018;2018:1823726. doi: 10.1155/2018/1823726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu S, et al. Repression of GPRC5A is associated with activated STAT3, which contributes to tumor progression of head and neck squamous cell carcinoma. Cancer Cell Int. 2017;17(1):34. doi: 10.1186/s12935-017-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shiba-Ishii A, et al. Stratifin accelerates progression of lung adenocarcinoma at an early stage. Mol. Cancer. 2015;14:142. doi: 10.1186/s12943-015-0414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hammond NL, Headon DJ, Dixon MJ. The cell cycle regulator protein 14-3-3 sigma is essential for hair follicle integrity and epidermal homeostasis. J. Invest. Dermatol. 2012;132(6):1543–1553. doi: 10.1038/jid.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts BJ, Reddy R, Wahl JK., 3rd Stratifin (14-3-3 sigma) limits plakophilin-3 exchange with the desmosomal plaque. PLoS ONE. 2013;8(10):e77012. doi: 10.1371/journal.pone.0077012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raychaudhuri K, et al. 14-3-3sigma gene loss leads to activation of the epithelial to mesenchymal transition due to the stabilization of c-Jun protein. J. Biol. Chem. 2016;291(31):16068–16081. doi: 10.1074/jbc.M116.723767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lam E, et al. Stratifin-induced matrix metalloproteinase-1 in fibroblast is mediated by c-fos and p38 mitogen-activated protein kinase activation. J. Invest. Dermatol. 2005;125(2):230–238. doi: 10.1111/j.0022-202X.2005.23765.x. [DOI] [PubMed] [Google Scholar]
- 100.Ghaffari A, et al. 14-3-3 sigma associates with cell surface aminopeptidase N in the regulation of matrix metalloproteinase-1. J. Cell Sci. 2010;123(Pt 17):2996–3005. doi: 10.1242/jcs.069484. [DOI] [PubMed] [Google Scholar]
- 101.Medina A, et al. The role of stratifin in fibroblast-keratinocyte interaction. Mol. Cell Biochem. 2007;305(1–2):255–264. doi: 10.1007/s11010-007-9538-y. [DOI] [PubMed] [Google Scholar]
- 102.Ghahary A. Stratifin as a mediator of epithelial/mesenchymal cross talking in skin. FASEB J. 2008;22(1_supplement):87.3. [Google Scholar]
- 103.Rahmani-Neishaboor E, et al. Localized controlled release of stratifin reduces implantation-induced dermal fibrosis. Acta Biomater. 2012;8(10):3660–3668. doi: 10.1016/j.actbio.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 104.Tessier L, et al. Impaired response of the bronchial epithelium to inflammation characterizes severe equine asthma. BMC Genom. 2017;18(1):708. doi: 10.1186/s12864-017-4107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yao X, et al. The A's have it: Developing apolipoprotein A-I mimetic peptides into a novel treatment for asthma. Chest. 2016;150(2):283–288. doi: 10.1016/j.chest.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armstrong AJ, et al. ATP-binding cassette transporter G1 negatively regulates thymocyte and peripheral lymphocyte proliferation. (J. Immunol. Baltimore Md.: 1950) 2010;184(1):173–183. doi: 10.4049/jimmunol.0902372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Louwe MC, et al. Abca1 deficiency protects the heart against myocardial infarction-induced injury. Atherosclerosis. 2016;251:159–163. doi: 10.1016/j.atherosclerosis.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 108.Demina EP, et al. ABCA1 gene expression in peripheral blood lymphocytes and macrophages in patients with atherosclerosis. Mol. Biol. (Mosk) 2011;45(2):289–293. doi: 10.1134/S0026893310061019. [DOI] [PubMed] [Google Scholar]
- 109.Zhang R, et al. Tissue treg secretomes and transcription factors shared with stem cells contribute to a treg niche to maintain treg-ness with 80% innate immune pathways, and functions of immunosuppression and tissue repair. Front. Immunol. 2021;11:632239. doi: 10.3389/fimmu.2020.632239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh R, et al. Distinct gene signature revealed in white blood cells, CD4+ and CD8+ T cells in (NZBx NZW) F1 lupus mice after tolerization with anti-DNA Ig peptide. Genes Immun. 2010;11:294–309. doi: 10.1038/gene.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun B, et al. Gprc5a-knockout mouse lung epithelial cells predicts ceruloplasmin, lipocalin 2 and periostin as potential biomarkers at early stages of lung tumorigenesis. Oncotarget. 2017;8(8):13532–13544. doi: 10.18632/oncotarget.14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L, et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 2015;6(1):7639. doi: 10.1038/ncomms8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gupta A, et al. Gene expression profiles in peripheral blood mononuclear cells correlate with salience network activity in chronic visceral pain: A pilot study. Neurogastroenterol. Motil. 2017;29(6):e13027. doi: 10.1111/nmo.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mihailovic PM, et al. Keratin 8 is a potential self-antigen in the coronary artery disease immunopeptidome: A translational approach. PLoS ONE. 2019;14(2):e0213025. doi: 10.1371/journal.pone.0213025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hudspeth K, et al. Natural killer cell expression of Ki67 is associated with elevated serum IL-15, disease activity and nephritis in systemic lupus erythematosus. Clin. Exp. Immunol. 2019;196(2):226–236. doi: 10.1111/cei.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Soares A, et al. Novel application of Ki67 to quantify antigen-specific in vitro lymphoproliferation. J. Immunol. Methods. 2010;362(1–2):43–50. doi: 10.1016/j.jim.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Myers LM, et al. A functional subset of CD8(+) T cells during chronic exhaustion is defined by SIRPalpha expression. Nat. Commun. 2019;10(1):794. doi: 10.1038/s41467-019-08637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Utzschneider DT, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45(2):415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 119.Fei Q, et al. High-dimensional single-cell analysis delineates radiofrequency ablation induced immune microenvironmental remodeling in pancreatic cancer. Cell Death Dis. 2020;11(7):589–589. doi: 10.1038/s41419-020-02787-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gu Z, et al. Nek2 is a novel regulator of B cell development and immunological response. Biomed. Res. Int. 2014;2014:621082. doi: 10.1155/2014/621082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu Y, et al. Targeting NEK2 induces cellular senescence in B-cell malignancies through p53-independent signaling pathways. Blood. 2019;134(Supplement_1):3102. doi: 10.1182/blood-2019-123815. [DOI] [Google Scholar]
- 122.Wang J, et al. Potential mechanism of RRM2 for promoting Cervical Cancer based on weighted gene co-expression network analysis. Int. J. Med. Sci. 2020;17(15):2362–2372. doi: 10.7150/ijms.47356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mazzu YZ, et al. Ribonucleotide reductase small subunit M2 is a master driver of aggressive prostate cancer. Mol. Oncol. 2020;14(8):1881–1897. doi: 10.1002/1878-0261.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zheng L, et al. PAI-1 gene polymorphism was associated with an increased risk of allergic diseases: Evidence from a meta-analysis of 14 case–control studies. Int. Arch. Allergy Immunol. 2019;180(4):255–263. doi: 10.1159/000502522. [DOI] [PubMed] [Google Scholar]
- 125.Van Maele L, et al. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J. Immunol. 2010;185(2):1177–1185. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patil RS, et al. IL17 producing gammadeltaT cells induce angiogenesis and are associated with poor survival in gallbladder cancer patients. Int. J. Cancer. 2016;139(4):869–881. doi: 10.1002/ijc.30134. [DOI] [PubMed] [Google Scholar]
- 127.Solt LA, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472(7344):491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ashton-Rickardt PG. An emerging role for serine protease inhibitors in T lymphocyte immunity and beyond. ISRN Immunol. 2012;2012:354365. doi: 10.5402/2012/354365. [DOI] [PubMed] [Google Scholar]
- 129.Renckens R, Pater JM, van der Poll T. Plasminogen activator inhibitor type-1-deficient mice have an enhanced IFN-gamma response to lipopolysaccharide and staphylococcal enterotoxin B. J. Immunol. 2006;177(11):8171–8176. doi: 10.4049/jimmunol.177.11.8171. [DOI] [PubMed] [Google Scholar]
- 130.Li H. Abnormal T lymphocyte patterns in chronic rhinosinusitis with nasal polyps in a chinese population. J. Allergy Clin. Immunol. 2008;121(3):793. doi: 10.1016/j.jaci.2008.01.046. [DOI] [Google Scholar]
- 131.Mould KJ, et al. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight. 2019;4(5):e126556. doi: 10.1172/jci.insight.126556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Petrelli A, et al. PD-1+CD8+ T cells are clonally expanding effectors in human chronic inflammation. J. Clin. Invest. 2018;128(10):4669–4681. doi: 10.1172/JCI96107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.