Abstract
Drought is a major factor which reduces cane growth and productivity. In the present study, we sequenced drought susceptible (V1) and drought tolerant (V2) sugarcane varieties using high-throughput miRNA deep sequencing method to study the regulation of gene expression by miRNAs during drought stress in sugarcane. A total of 1224 conserved miRNAs which belong to 89 miRNA families were identified and 38% of the differentially regulated miRNAs were common for both varieties. Additionally 435 novel miRNAs were also identified from four small RNA libraries. We identified 145 miRNAs that were differentially expressed in susceptible variety (V1–31) and 143 miRNAs differentially expressed in the tolerant variety (V2–31). Target prediction revealed that the genes mainly encoded transcription factors, proteins, phosphatase and kinases involved in signal transduction pathways, integral component of membrane and inorganic ion transport metabolism, enzymes involved in carbohydrate transport and metabolism and drought-stress-related proteins involved in defense mechanisms. Pathway analysis of targets revealed that “General function prediction only” was the most significant pathway observed in both tolerant and susceptible genotypes followed by “signal transduction mechanisms”. Functional annotation of the transcripts revealed genes like calcium-dependent protein kinase, respiratory burst oxidase, caffeic acid 3-O-methyltransferase, peroxidase, calmodulin, glutathione S-transferase and transcription factors like MYB, WRKY that are involved in drought tolerant pathways. qRT-PCR was used to verify the expression levels of miRNAs and their potential targets obtained from RNA sequencing results.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02857-x.
Keywords: Sugarcane, Drought stress, miRNAseq, Target regulation, Differential expression
Introduction
Sugarcane is an important crop in the tropical and subtropical regions and it is responsible for almost 86% of sugar production in the world (OECD-FAO 2019). Among the various abiotic stress affecting sugarcane drought is considered to be the most detrimental stress, affecting crop production worldwide (Wang et al. 2003; Rampino et al. 2006).In recent years it has been the major limitation for sugarcane production and sugarcane growth is highly sensitive to water deficit (Companhia Nacional de Abastecimento) (Lakshmanan and Robinson 2014; CONAB 2017). Drought stress decreases stomatal conductivity reducing water loss in the leaf and reduction in photosynthesis takes place leading to low sugarcane yields (Namwongsa et al. 2019). Yield losses due to drought stress goes up to 60% (Gentile et al. 2015), depending on the severity and duration of water stress (Machado et al. 2009). Development of sugarcane varieties with increased drought resistance has been regarded as an efficient strategy to stabilize and improve production. Studies have demonstrated that the morphological, physiological and biochemical responses of plants to drought stress are controlled by various molecular mechanisms that are regulated by gene expression (Farooq et al. 2009; Osakabe et al. 2014). Plant responses to stress are modulated by relative expression of genes as well as their regulation at different levels, post transcriptional or post translational.
Among the eukaryotes RNA silencing, which is facilitated by small non-coding RNAs of 20–35 nucleotides is an important form of gene regulation apart from transcriptional factors. In recent years, regulatory small RNAs (sRNAs) have received wide attention for their roles in the post-transcriptional and translational regulation of gene expression and form an universal component of endogenous plant transcriptome (Axtell 2013). SmallRNAs can be classified into four major categories based on their origin, processing mode and effector protein association. They are microRNA (miRNA), small interfering RNA (siRNA), transfer RNA-derived small RNAs (tsRNAs) and PIWI-interacting RNA (piRNA, animals only) (Borges and Martienssen 2015; Zhu et al. 2018; Czech et al. 2018). miRNAs are small non-coding RNAs of 18–25 nucleotides that occur in most eukaryotic genomes. Mature microRNAs generated from primary miRNAs with the aid of dicer-like enzymes (DCL1, RNA III enzymes) could inactivate the expression of target RNAs possessing complementary sequences in plants by binding and guiding their effector proteins (Rogers and Chen 2013).They play significant roles by negatively regulating target genes in many biological processes such as growth and hormone signal transduction of the organism (Bartel 2004; Jin et al. 2008; Sunkar 2010; Sunkar et al. 2012).
There are several miRNAs that have been identified in a wide array of species which play important role in response to biotic and abiotic stresses (Khraiwesh et al. 2012; Pei et al. 2013; Sailaja et al. 2014; Shriram et al. 2016).Very few studies are reported in identifying mature miRNA sequences and their expression during drought stress in sugarcane plants (Ferreira et al. 2012; Thiebaut et al. 2012; Gentile et al. 2013, 2015). In the present study, high-throughput miRNA deep sequencing method was used to study the regulation of gene expression by miRNAs and validate their expression during drought stress in sugarcane. We sequenced sRNA from leaf tissues of sugarcane varieties Co 8021 (drought susceptible) and Co 06022 (drought tolerant) which were subjected to drought treatment to identify differentially expressed miRNAs and their interaction with predicted target mRNAs. The results suggested the differential regulation of miRNAs that are associated with drought and the varied roles of their target mRNAs that will help to establish the molecular basis of drought stress tolerance in sugarcane plants.
Materials and methods
Selection of plant samples and RNA isolation
Two sugarcane varieties Co 8021 (Variety1-V1) and Co 06022 (Variety2-V2) were selected based on differential drought tolerance potential (Devi et al. 2018) from the fields of ICAR-Sugarcane Breeding Institute, Coimbatore and were planted in pots under normal irrigation for 2 months. After 60 days of planting, gradual drought stress was imposed by withholding water for 10 days, while the control plants were normally irrigated throughout the experiment (Devi et al. 2019). Tissue was collected (Leaf + 1, the highest unfolded leaf with a visible dewlap) in triplicates after 10 days drought treatment along with the respective controls. The samples were flash-frozen and total RNA was extracted using TRI reagent (Sigma-Aldrich, USA) and purified using Spectrum™ Plant Total RNA kit (Sigma-Aldrich, USA). The quantification of the total RNA samples was done using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The quality and quantity of RNA was also analysed using Agilent RNA bioanalyzer chip. The samples were named as V1–11 (Co 8021-control), V1–31 (Co 8021-treated) and V2–11 (Co 06022-control) and V2–31 (Co 06022-treated).
Analysis of small RNAs
Small RNA libraries were prepared from total RNA of four selected samples using the Illumina Truseq small RNA library prep (Illumina, USA). The libraries were sequenced using IlluminaNextSeq500.751platforms (Illumina, Inc. USA) (according to manufacturer’s instructions). The libraries were sequenced by the Genotypic Technology Pvt. Ltd., Bangalore. The raw reads from the libraries were first cleaned by removing 5′ and 3′ adaptors, low quality reads and reads with length < 16 or length > 35 bp were filtered using the srna-workbenchV3.0_ALPHA1 (University of East Anglia (UEA)). Only trimmed reads 16–35 nt in length was considered for further analysis.
Known miRNA and novel miRNA prediction
The clean reads were used for BLASTn search against Sanger RNA database (Rfam) (http://www.sanger.ac.uk/software/Rfam). All reads were checked for ncRNA (rRNA, tRNA, snRNA and snoRNA) contamination. The unaligned reads to ncRNAs were clustered based on the 95% coverage and 90% similarity to generate the read count. To obtain known miRNAs, clustered reads were used for homology search of the miRNAs against all matured Viridiplantae miRNA sequences retrieved from miRbase-21 database using ncbi-blast-2.2.30. To predict novel miRNAs in the samples, sRNAseq reads not aligning to miRBase were given as input to MIREAP v0.2 software (http://sourceforge.net/projects/mireap/). MIREAP identified novel microRNA based on alignment, secondary structure, free energy and location on the precursor arm. Stem-loop hairpins were retained only when 1. The mature miRNAs-associated reads, mapped in the arm region of the precursors and 2. The free energy of the secondary structure calculated by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was lower than − 18 kcal/mol. Other parameters that were set included a minimal miRNA length of 18, maximal miRNA length of 26; minimal reference miRNA length of 20; maximal reference miRNA length of 24; uniqueness of miRNA was 20; maximal free energy for the miRNA precursor was − 25 kcal/mol; maximal space between miRNA and miRNA* was 300, minimal base pairs of miRNA and miRNA* was 14, maximal bulge of miRNA and miRNA* was 4, maximal asymmetry of miRNA/miRNA* duplex was 4 and flank sequence length of miRNA precursor was 20. The free energy of the secondary structure of the miRNAs calculated by RNAfold is lower than − 5 kcal/mol.
Differential gene expression (DGE) analysis
Read count table for all the samples were generated. Read counts of each miRNAs were normalized to calculate the normalized miRNA abundance values using following formula RPM = RmiR/Rall*10^6 where: RmiR—number of reads mapped to a particular miRNA reference in the sample, Rall—total number of reads mapped in the sample. DGE analysis was carried out using DESeq5 tool and list of miRNAs expressed only in control, both in control and stress and only in stress were generated. miRNAs that were significantly expressed under drought stress were screened with p value ˂ 0.05 and fold change ˃ 1 or ˂ − 1.
Target prediction for miRNA
Differentially expressed miRNA's with copy number ≥ 10 were considered for target prediction. The miRNA target sequences were predicted using miRanda-3.3 (http://www.microrna.org/microrna/getGeneForm.do) using the “strict” option. This option ensures strict alignment in the seed region in position between 2 and 8 nucleotides. This option prevents the detection of target sites which contain gaps or non-canonical base pairing in this region. Other criteria were conservation, free energy, and site accessibility. miRNA hits having minimum free energy ≤ − 25 were assumed to be the targets for reported miRNA.
KOG annotation and KEGG pathway analysis of miRNA target
All the miRNA targets were searched against KOG protein sequences. KOG classification analysis was conducted to reveal the biological functions of the miRNA targets. Pathway analysis was performed using KEGG pathway database (https://www.Genome.jp/kegg).
Stem-loop reverse transcription and RT-qPCR validation
Reverse Transcription stem-loop primers (STL), sequence-specific forward PCR primers and universal reverse primers were designed following Chen et al. (2005) for reverse transcription and qPCR amplification of sugarcane miRNAs. miRNA-cDNA synthesis was carried out using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, USA) and reverse transcription reactions were performed as described by Varkonyi-Gasic et al. (2007). Each reaction contained 10 ng of DNA-free total RNA and 1 µl of each RT-loop primer (1 µM). The mix was incubated for 10 min at 65 °C and then placed on ice for 2 min. Subsequently, 2.5 µl of 10 × Reverse transcription buffer, 0.25 µl of dNTPs (100 mM), 0.3 µl of RNAse inhibitor (40 U/µl), and 1 µl of Multiscribe™ Reverse Transcriptase (100 U/µl) enzyme (Taqman® MicroRNA Reverse Transcription Kit, Thermo Fisher Scientific) and 13.95 µl of nuclease free water were added. This reaction was incubated in a Thermal Cycler (MJ Research Inc. model: PTC 100) for 30 min at 16 °C, followed by 42 °C for 30 min and finally at 85 °C for 5 min at for the enzyme inactivation. To validate the expression levels of miRNA, 25 s rRNA was used as internal control. qRT-PCR amplification of the miRNAs was performed using SYBR Premix Ex Taq II (Takara, Dalian, China) in Rotor gene Q real time PCR machine (Qiagen, Germany).The reaction mixture included 8 µl of SYBR green master mix (1 ×), 2 µl of cDNA, 1 µl of specific forward primer (10 mM), 1 µl of universal reverse primer (10 mM). The volume was made upto 20 µl with 8 µl of nuclease free water. The reactions were performed at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min with a final dissociation curve analysis. All reactions were run in triplicates. The relative expression values were calculated using the 2−∆∆CT method as described by Livak and Schmittgen (2001). qRT-PCR for selected miRNA and their targets was also performed to check the miRNA target regulation. Ten miRNAs and their target genes were used for expression analysis with cDNA synthesized from leaf samples of 10 days stress of both varieties.
Results
Sequencing results of small RNA libraries
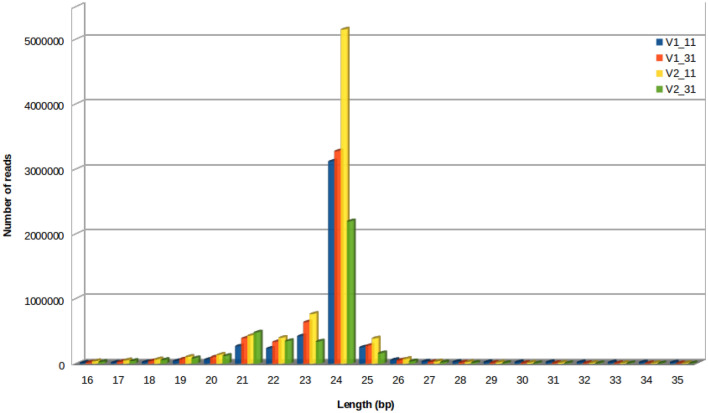
Four sRNA libraries were constructed and sequenced from control and drought stressed samples of the susceptible variety Co 8021 (V1–11, V1–31) and tolerant variety (V2–11, V2–31) to identify drought responsive miRNAs in sugarcane plants. Each library generated 11–18 million raw reads (Additional file 1: Table S1).The data has been submitted in the National Center for Biotechnology Information (NCBI) with accession number PRJNA593909 (https://www.ncbi.nlm.nih.gov/sra/PRJNA593909). Clean reads were obtained by removing adaptors, junk and low-quality reads and small RNAs within 16–35 nt were subjected to further analysis. The lengths of miRNA were mainly distributed between 21 and 25 nt, which account for an average of 88% of the small RNAs. However, there was a considerable difference for the length distribution among different small RNA libraries. Among all sRNAs, 24 nt miRNAs had the highest number of reads in all of the four libraries (Fig. 1).
Fig. 1.
Read length distribution of small RNAs in four libraries of sugarcane cultivars. The most abundant class of sRNA sequence length was 24 nt. V1_11—Co 8021 control, V1_31—Co 8021 subjected to 10 days drought, V2_11—Co 06022 control and V2_31—Co 06022 subjected to 10 days drought
Identification of known miRNAs in sugarcane and their response to drought stress
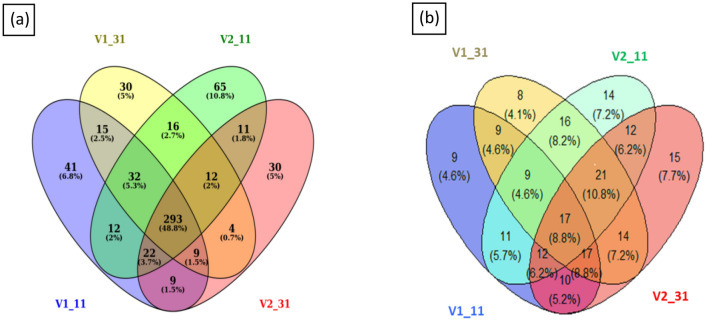
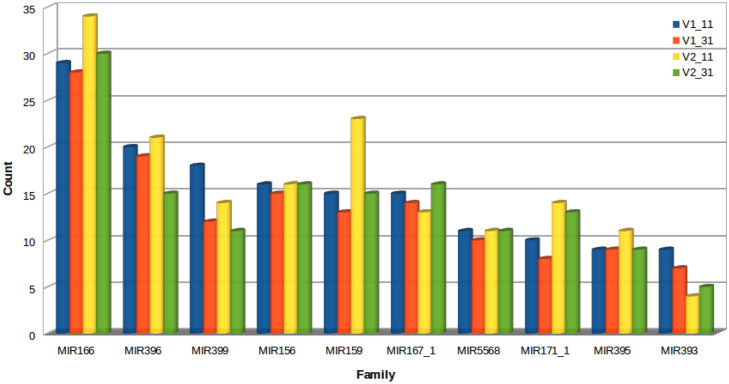
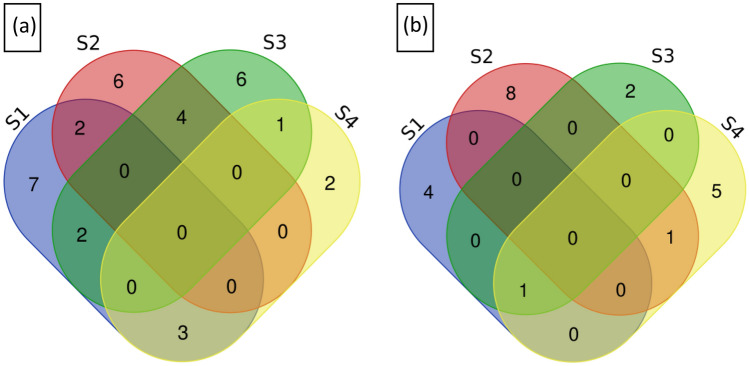
To identify the known miRNAs, the clean reads were aligned with miRNAs in miRBase. A total of 1224 known miRNAs were identified, which belong to 89 miRNA families. There were 313, 296, 331 and 284 known miRNAs identified in the V1–11, V1–31, V2–11 and V2–31 small RNA libraries, respectively (Additional file 1: Table S2). The distribution of the miRNAs revealed that 293 miRNAs were common in all libraries. The numbers of miRNAs specific to each library was 41, 30, 65 and 30 in V1–11, V1–31, V2–11 and V2–31 respectively (Fig. 2a). The numbers of miRNAs varied in different miRNA families, with the maximum number of 34 in MIR166 followed by 23 miRNAs in the family of MIR159 and 21 miRNAs in the family of MIR396 in V2–31, when compared to other libraries (Fig. 3). The expression of the miRNAs in the different libraries varied to a great extent. miRNAs that showed higher expression levels (more than 10,000 reads) in the control and drought stressed libraries of Co 8021 were bdi-miR166e-3p, osa-miR168A-5p and zma-miR396g-3p respectively and in Co 06022 were pta-miR166a, osa-miR168a-5p and sbi-miR396d respectively. Around 14 and 11 miRNAs had more than 1000 reads in Co 8021 and Co 06022 libraries respectively, which belonged to ten miRNA families. Differentially regulated miRNAs were identified across different comparison sets to obtain up and down regulated miRNAs. Comparison of drought susceptible Co 8021 (V1–11, V1–31) revealed 14 miRNAs (log2 ratio ≥ 1 up and ≤ 1 down, p ≤ 0.05) that were differentially expressed including seven upregulated and seven downregulated. Similar comparison in the tolerant cultivar (V2–11, V2–31) revealed 12 miRNAs that were differentially expressed including seven upregulated and five downregulated. Further comparison of drought stressed libraries of both varieties identified six differentially expressed including five up and one downregulated (Additional file 2: Tables S1, S2, S3, S4). Among the different comparison sets (Fig. 4a) of the differentially regulated miRNAs no miRNA was common among different comparisons. Seven were specific to V1–11 vs V1–31 comparisons of which 5 (sbi-miR5568g-3p, bna-miR156a, zma-miR393c-3p, tae-miR5384-3p and zma-miR393a-3p) were up regulated and two (hvu-miR444b, smo-miR408) were downregulated. Similarly six miRNAs were found specific to V2–11 vs V2–31 of which four (ath-miR845a, gma-miR166k, ppt-miR390c-5p, sbi-miR6235-5p) were upregulated and two (ath-miR394b-5p, bdi-miR166e-3p) were downregulated. Common miRNAs that were differentially expressed in both the varieties were sbi-miR397-3p that was downregulated in both V1–11 vs V1–31 and V2–11 vs V2–31 and csi-miR166a that was downregulated in V1–11 vs V1–31 and upregulated in V2–11 vs V2–31. Two miRNAs, vvi-miR394c and bdi-miR159b-3p.3 were uniquely expressed when the treated libraries (V1–31 and V2–31) of both varieties were compared.
Fig. 2.
Venn diagram showing number of overlapping a known and b novel miRNAs in the four libraries of sugarcane cultivars. V1_11—Co 8021 control, V1_31—Co 8021 subjected to 10 days drought, V2_11—Co 06022 control V2_31—Co 06022 subjected to 10 days drought. The numbers within the intersection are the number of miRNAs common between the libraries compared and the numbers outside the intersection are the miRNAs specific to control and water stressed libraries of the two cultivars
Fig. 3.
Top 10 abundant miRNA families in the four libraries of sugarcane cultivars. V1_11—Co 8021 control, V1_31—Co 8021 subjected to 10 days drought, V2_11—Co 06022 control, V2_31—Co 06022 subjected to 10 days drought
Fig. 4.
Venn diagram showing number of overlapping differentially expressed mature miRNAs (p value ˂ 0.05 and fold change ˃ 1 or ˂ − 1) between comparisons a known miRNAs. b Novel miRNAs. The comparisons are S1—Co 8021 control vs treated (V1_11 vs V1_31), S2—Co 06022 control vs treated (V2_11 vs V2_31), S3—Co 8021 control vs Co 06022 control (V1_11 vs V2_11), S4—Co 8021 treated vs Co 06022 treated (V1–31 vs V2–31). The numbers within the intersection are the number of differentially expressed miRNAs common between the different comparisons and the numbers outside the intersection are the differentially expressed miRNAs specific to each comparison
Identification of novel miRNAs and their expression under drought stress
The putative novel miRNAs detected in the present study using MIREAP software ((http://sourceforge.net/projects/mireap/), Li et al. 2012) were evaluated for their secondary structure. The length of the novel miRNAs precursor identified ranged from 62 to 224 nt that folded to form hairpin structures with ≤ 4 symmetrical mismatches in the duplex. The free energy of the secondary structure ranged from − 20 to − 134.9 kcal/mol as calculated by RNAfold from ViennaRNA Package 2.0 (Lorenz et al. 2011). Also the mature miRNA reads mapped onto the arm region of the stemloop hairpins. The free energy of the mature miRNAs were less than − 5 kcal/mol as calculated by RNAfold. Example of secondary structure of four precursor miRNAs along with their minimum free energy values are given in Additional file 3: Fig. S1.
The number of novel miRNAs detected varied among the four libraries (Additional file 4: Tables S1, S2, S3, S4). In the drought susceptible variety Co 8021, 94 and 111 novel miRNAs were discovered in V1–11 and V1–31 libraries and in the drought tolerant variety Co 06022, 112 and 118 novel miRNAs were discovered in V2–11and V2–31 libraries. Seventeen miRNAs were common among the different libraries (Fig. 2b). Most novel miRNAs had a relatively low expression and only mireap-m0040-5p had reads over 1000 in the library V1–11. A total of 11, 9, 14 and 10 novel miRNAs were detected with reads count over 50 in the V1–11, V1–31, V2–11and V2–31 libraries respectively. Differential expressed novel miRNAs (log2 ratio ≥ 1 up and ≤ 1 down, p ≤ 0.05) were identified in this study (Additional file 4: Tables S5, S6, S7, S8). In the drought-susceptible variety Co 8021, five novel miRNAs were found to be differentially expressed between V1–11 and V1–31, of them, four were upregulated (mireap-m0095-5p, mireap-m0049-5p, mireap-m0099-3p, mireap-m0024-3p) and one was downregulated (mireap-m0045-5p). In the drought-tolerant variety Co 06022, nine novel miRNAs were found to be differentially expressed between V2–11 and V2–31, of them, three (mireap-m0017-3p, mireap-m0087-5p, mireap-m0082-3p) were upregulated while six (mireap-m0021-3p, mireap-m0016-5p, mireap-m0086-5p, mireap-m0049-5p, mireap-m0104-3p, mireap-m0061-3p) were downregulated. Comparisons of the stressed libraries of the two varieties V1–31 vs V2–31, revealed five downregulated (mireap-m0043-3p, mireap-m0103-3p, mireap-m0020-5p, mireap-m0062-3p, mireap-m0073-3p) and two upregulated novel miRNAs (mireap-m0011-3p, mireap-m0067-5p). There were no common miRNAs that was differentially expressed in the two varieties (Fig. 4b).
Functional analysis of target genes of the drought induced conserved miRNAs in sugarcane
A total of 10,312 target genes were found to be regulated by the 145 known miRNAs expressed in the susceptible variety and 11,357 target genes were regulated by 143 known miRNAs expressed in the tolerant variety (Additional file 5: Tables S1, S2). The predicted target genes mainly encoded transcription factors, proteins, phosphatase and kinases involved in signal transduction pathways, integral component of membrane and inorganic ion transport metabolism, enzymes involved in carbohydrate transport and metabolism, drought-stress related proteins involved in defense mechanisms, and cell wall/membrane biogenesis. For instance, the target of sbi-miR397-3p, tae-miR5384-3p, hvu-miR444b and sbi-miR5568g-3p were genes encoding protein phosphatase type 2-C, calmodulin-dependent protein kinase phosphatase, calcium-dependent protein kinase, mitogen-activated protein kinase, receptor protein kinase CRINKLY4, Histidine kinase 3, and serine/threonine-protein kinase which are involved in signal transduction pathways. Some targets encode drought responsive transcription factors such as MYB transcription factor, AP2-EREBP, Dof-type zinc finger protein, DRE-binding protein, GNAT, GATA-4/5/6, ALFIN-like transcription factor, GRF transcription factor, MADS-box transcription factor, heat shock factor protein 7 predicted by miRNAs tae-miR5384-3p, hvu-miR444b, smo-miR408 and sbi-miR396d. Some of the miRNAs like csi-miR166a, sbi-miR397-3p, osa-miR393b-3p, zma-miR393c-3p and cme-miR156j have targets that are involved in integral component of membrane modification. The targets of tae-miR5384-3p encode several genes like phenylalanine ammonia-lyase, amine oxidase and laccase gene which are involved in secondary metabolites biosynthesis, transport, catabolism and genes encoding calcium-transporting ATPase, catalase, zinc transporter 4, potassium channel which function in inorganic ion transport and metabolism.
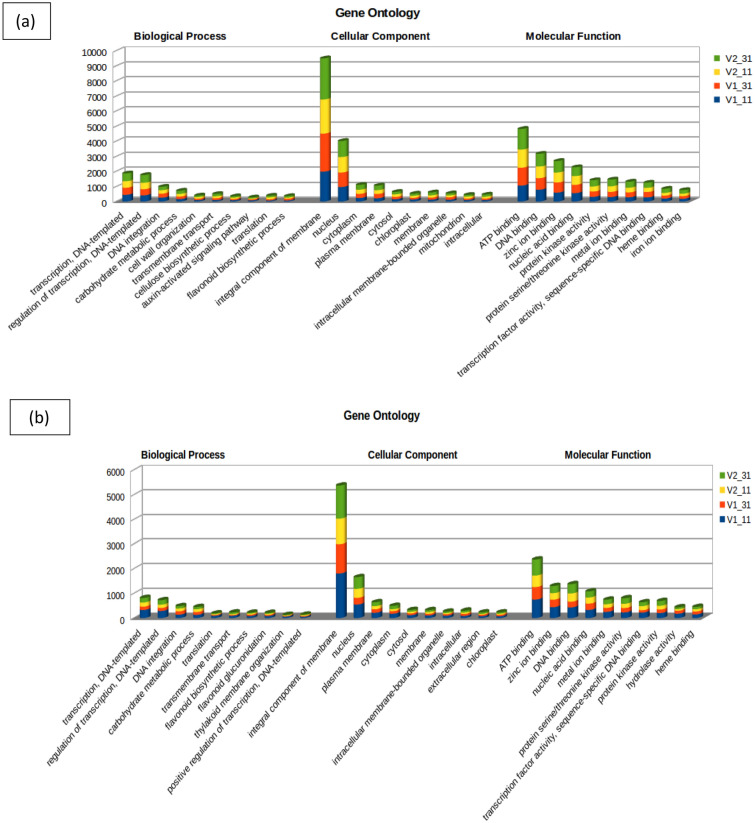
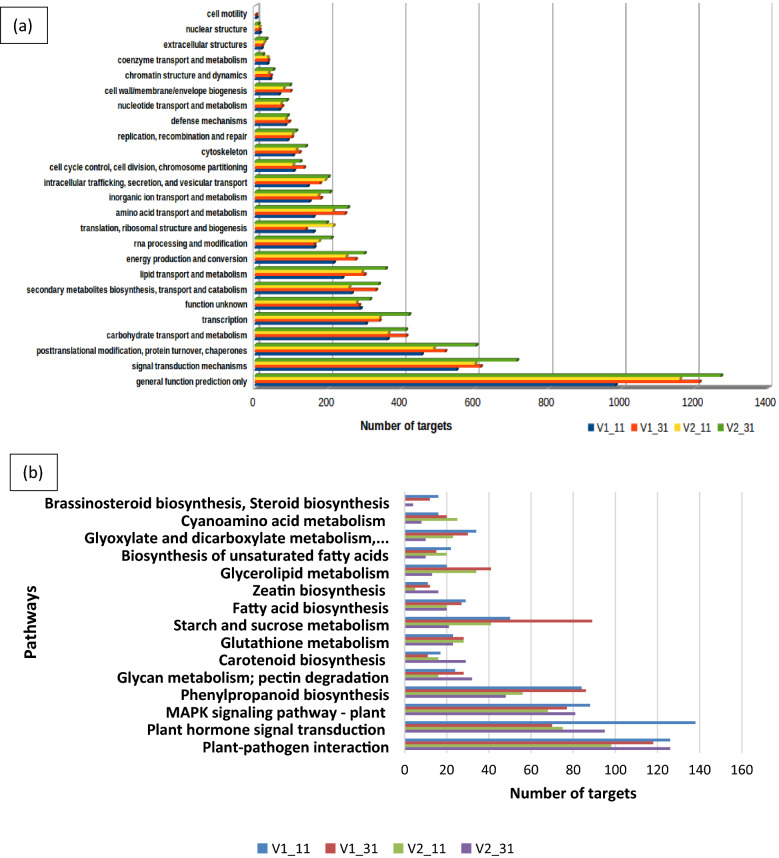
The target genes of the conserved miRNAs were further subjected to GO and KEGG pathway analysis. Target genes of known miRNAs of V1–31 were classified into 1092, 264 and 676 GO terms and targets of V2–31 were classified into 1105, 264 and 688 GO terms for biological process, cellular component and molecular function ontology, respectively (Additional file 6: Tables S1, S2, S3). GO analysis showed that the target genes of many differentially-expressed known miRNAs enriched in the GO term cellular component, were related to integral component of membrane and nucleus. The GO term biological process was enriched with targets for transcription, DNA integration and carbohydrate metabolic processes and the GO term molecular function was enriched with targets for ATP binding, DNA binding, zinc ion binding, protein kinase activity and transcription factor activity (Fig. 5a). To identify the pathways in which the target genes were involved, they were mapped on KEGG pathway database. The top enriched pathways of both genotypes were studied to identify genes involved in major metabolic activities (Table 1). “General function prediction only” was the most significant category observed in both tolerant and susceptible genotypes followed by signal transduction mechanisms and posttranslational modification, protein turnover and chaperones (Fig. 6a). Pathways such as secondary metabolite biosynthesis, transport and catabolism, inorganic ion transport and metabolism, defence mechanisms and cell wall/membrane/envelope biogenesis were other pathways that are involved in sugarcane stress tolerance.
Fig. 5.
Gene ontology classification of predicted targets of a known and b novel miRNAs. The results are summarized in three main categories: biological process, cellular component and molecular function. V1_11—Co 8021 control, V1_31—Co 8021 subjected to 10 days drought, V2_11—Co 06022 control V2_31—Co 06022 subjected to 10 days drought
Table 1.
Predicted target genes of miRNAs involved in top enriched pathways of drought tolerance in sugarcane
| S. No. | Pathway | miRNAs | Target genes | Gene ID |
|---|---|---|---|---|
| 1 | Plant-pathogen interaction |
Smo-miR408 Mireap-m0077-3p Mireap-m0073-3p Mireap-m0049-5p Mireap-m0016-5p Mireap-m0016-5p Mireap-m0073-3p |
Serine/threonine protein kinase transcription factor MYB CERK1; chitin elicitor receptor kinase 1 RPM1; disease resistance protein RPM1 CPK; calcium-dependent protein kinase RBOH; respiratory burst oxidase Carboxypeptidase |
31_c37860_g1_i2 41_c56809_g2_i6 31_c35993_g1_i3 51_c41883_g1_i4 51_c45744_g2_i2 41_c55871_g2_i1 41_c57068_g1_i4 |
| 2 | Plant hormone signal transduction |
Tae-miR5384-3p Smo-miR408 Tae-miR5384-3p Tae-miR5384-3p Tae-miR5384-3p Tae-miR5384-3p Tae-miR5384-3p Tae-miR5384-3p Tae-miR5384-3p Sbi-miR396d Sbi-miR397-3p Tae-miR5384-3p Mireap-m0020-5p Mireap-m0043-3p Mireap-m0020-5p Mireap-m0067-5p Mireap-m0049-5p Mireap-m0020-5p Mireap-m0049-5p Mireap-m0049-5p Mireap-m0049-5p |
Mitogen-activated protein kinase Phosphoribulokinase Pyruvate dehydrogenase 2C-type protein phosphatase protein (uncharacterized protein) ACC synthase 2 Rhomboid-like protein Putative CRINKLY4-like receptor protein kinase family protein (uncharacterized protein) Protein kinase domain containing protein Calmodulin (uncharacterized protein) Histidine-containing phosphotransfer protein 4 ATP binding protein (uncharacterized protein) 1-Aminocyclopropane-1-carboxylate synthase Jasmonate ZIM domain-containing protein GH3; auxin responsive GH3 gene family TGA; transcription factor TGA putative bZIP transcription factor superfamily protein ARR-B; two-component response regulator ARR-B family JAZ; jasmonate ZIM domain-containing protein Putative snRK/SAPK family protein kinase SNRK2; serine/threonine-protein kinase SRK2 Serine/threonine-protein kinase SAPK6 |
41_c61679_g1_i3 51_c35422_g1_i2 21_c47811_g1_i4 41_c61886_g1_i2 51_c31538_g1_i1 21_c46681_g2_i1 51_c40570_g1_i2 21_c44921_g3_i1 11_c45593_g1_i2 21_c33465_g1_i2 51_c34821_g1_i2 11_c50950_g2_i1 51_c31421_g1_i1 11_c35859_g1_i5 11_c40163_g1_i1 31_c22010_g1_i2 41_c57376_g1_i1 21_c39304_g1_i2 51_c47267_g3_i5 21_c46764_g1_i2 51_c47267_g3_i4 |
| 3 | MAPK signaling pathway—plant |
Tae-miR5384-3p Mireap-m0075-5p Mireap-m0011-3p Mireap-m0020-5p Mireap-m0067-5p Mireap-m0043-3p Mireap-m0020-5p Mireap-m0016-5p |
Mitogen-activated protein kinase WRKY transcription factor 33 ERECTA-like 2 Chitinase 1 ACC synthase 1 PREDICTED: abscisic acid receptor PYL4-like RBOH; respiratory burst oxidase peroxidase activity |
41_c89678_g1_i1 41_c34210_g1_i1 21_c51388_g2_i1 41_c49938_g1_i3 31_c40137_g1_i3 31_c25419_g2_i1 41_c47344_g2_i1 41_c55871_g2_i1 |
| 4 | Phenylpropanoid biosynthesis |
Tae-miR5384-3p Tae-miR5384-3p Mireap-m0077-3p Mireap-m0043-3p Mireap-m0049-5p |
Peroxidase Glycosyltransferase Peroxidase Peroxidase Laccase |
41_c55109_g2_i1 11_c45559_g1_i2 21_c52014_g1_i5 11_c33778_g2_i6 11_c35815_g1_i1 |
| 5 | General function prediction only |
Sbi-miR397-3p Sbi-miR397-3p Osa-miR393b-3p Osa-miR393b-3p Sbi-miR5564c-5p Tae-miR5384-3p Tae-miR5384-3p |
Putative polyprotein Caffeic acid 3-O-methyltransferase (EC 2.1.1.6) (caffeic acid-O-methyltransferase) Putative RING zinc finger domain superfamily protein (uncharacterized protein) S-acyltransferase (EC 2.3.1.225) (palmitoyltransferase) Putative gag-pol polyprotein NAD dependent epimerase/dehydratase family protein (uncharacterized protein) Carboxypeptidase |
41_c50099_g1_i1 31_c31301_g1_i1 51_c28098_g1_i1 41_c62565_g1_i2 21_c53598_g1_i2 41_c59268_g5_i4 41_c57369_g4_i1 |
| 6 | Posttranslational modification, protein turnover, chaperones |
Sbi-miR397-3p Sbi-miR397-3p Tae-miR5384-3p Tae-miR5384-3p Mireap-m0020-5p Mireap-m0043-3p |
Heat shock protein MYB-like transcription factor DIVARICATA Carboxypeptidase (EC 3.4.16.-) Thioredoxin M-type HSP20; HSP20 family protein HSP90B; heat shock protein 90 kDa beta |
51_c29439_g1_i4 21_c49879_g1_i2 41_c62042_g1_i4 51_c25511_g1_i1 21_c36141_g1_i1 41_c54140_g1_i3 |
| 7 | Carbohydrate transport and metabolism |
Tae-miR5384-3p Sbi-miR397-3p Sbi-miR397-3p Tae-miR5384-3p Mireap-m0073-3p |
Trehalose-6-phosphate synthase component TPS1 and related subunits Glycosyltransferase (EC 2.4.1.-) Fructose-6-phosphate 2-kinase/fructose-2,6-biphosphatase Alpha-galactosidase (EC 3.2.1.22) (melibiase) GAPDH; glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] |
21_c4416_g1_i1 11_c18089_g1_i1 11_c53370_g1_i1 31_c37634_g2_i2 21_c47885_g2_i1 |
| 8 | Secondary metabolite biosynthesis transport and catabolism |
Sbi-miR397-3p Tae-miR5384-3p Tae-miR5384-3p Mireap-m0052-3p |
Cytochrome P450 Acc oxidase S-glutathione dehydrogenase/class III alcohol dehydrogenase Cytochrome P450 |
31_c38488_g1_i2 11_c49592_g2_i2 11_c48315_g1_i1 51_c36473_g1_i1 |
| 9 | Glutathione metabolism |
Tae-miR5384-3p Csi-miR166a Tae-miR5384-3p Mireap-m0086-5p |
Multicopper oxidases Glutathione S transferase Glutathione S transferase Glutathione S-transferase |
21_c49637_g2_i3 51_c29924_g1_i2 41_c51347_g1_i1 51_c34512_g1_i1 |
Pathway: pathway in which protein is involved; miRNAs: differentially expressed miRNAs with log2 ratio ≥ 1 up and ≤ 1 down, p ≤ 0.05; target genes: description of genes obtained from GenBank by BLAST search; gene ID: target mRNA ID (sugarcane transcript list available in NCBI BioProject ID—PRJNA590595) to which miRNA binds
Fig. 6.
KEGG enrichment analysis of predicted target genes of a known and b novel miRNAs. The x-axis shows the number of unigenes involved in each pathway and the y axis denotes the different metabolic pathways. V1_11—Co 8021 control, V1_31—Co 8021 subjected to 10 days drought, V2_11—Co 06022 control V2_31—Co 06022 subjected to 10 days drought
Functional analysis of target genes of the drought induced novel miRNAs in sugarcane
A total of 5308 targets were identified by 50 novel miRNAs expressed upon drought stress in susceptible variety (V1–31) and 5979 targets were identified for 49 novel miRNAs in the tolerant variety (V2–31) (Additional file 7: Tables S1, S2). The novel target genes mainly encoded drought-stress related genes involved in plant defense mechanisms. For instance, the novel miRNAs mireap-m0020-5p, mireap-m0104-3p, mireap-m0086-5p, mireap-m0043-3p, mireap-m0049-5p, mireap-m0017-3p targeted genes encoding, ALDH (aldehyde dehydrogenase), calmodulin binding protein, carboxypeptidase, cytochrome P450, disease resistance protein RPM1, peroxidase, sucrose-phosphate synthase, heat shock 70 kDa protein, HSP20 family protein and xyloglucan endotransglucosylase/hydrolase, respectively. Some targets of novel miRNAs, were drought responsive transcription factors like MYB, TGA transcription factor, putative bZIP transcription factor super family protein encoded by mireap-m0043-3p, mireap-m0020-5p and mireap-m0067-5p respectively.
The target genes of the novel miRNAs were further classified into GO terms (Fig. 5b). Novel miRNAs of V1–31 identified 790, 179 and 475 GO terms and of V2–31 identified 853, 201 and 520 GO terms for the biological process, cellular component and molecular function ontology, respectively (Additional file 8: Tables S1, S2, S3). In biological process, more number of targets were involved in transcription, regulation of transcription, DNA integration and carbohydrate metabolic process. However, cellular component had more targets that were related to stress response such as integral component of membrane and nucleus. Furthermore, molecular function had many novel miRNAs related to ATP binding, DNA binding, zinc ion binding, protein kinase activity and transcription factor activity. The targets were enriched in stress response pathways such as plant pathogen interaction, plant hormone signal transduction, MAPK signaling pathway, phenylpropanoid biosynthesis etc. (Fig. 6b). Other pathways such as starch and sucrose metabolism, glycan metabolism; pectin degradation, glutathione metabolism, carotenoid biosynthesis, fatty acid biosynthesis, glycerolipid metabolism, zeatin biosynthesis, biosynthesis of unsaturated fatty acids, glyoxylate and dicarboxylate metabolism, cyanoamino acid metabolism and brassinosteroid biosynthesis were also identified.
Network analysis of drought responsive miRNA and their targets
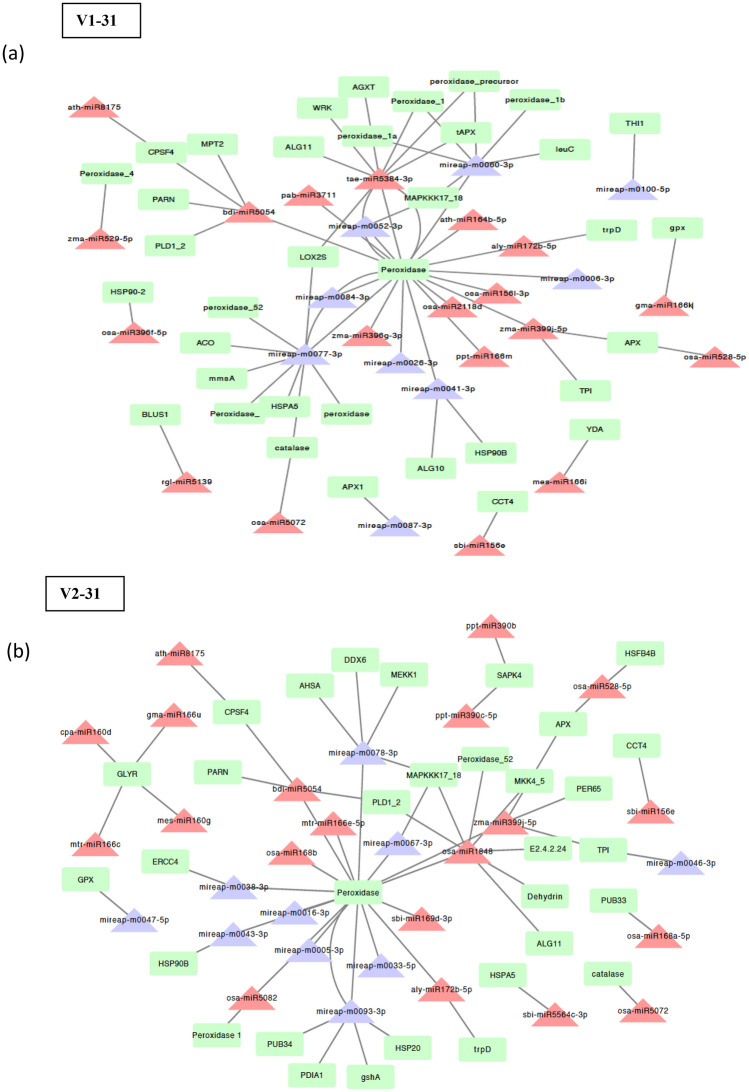
The network analysis of drought responsive miRNA and their targets upon stress in sugarcane was investigated. Drought tolerant variety (V2–31) had 144 conserved miRNAs belonging to 32 families encoding 28 genes and 89 novel miRNAs belonging to 14 families encoding 19 genes for stress tolerance. Drought susceptible variety (V1–31) had 128 conserved miRNAs belonging to 34 families encoding 27 genes and 89 novel miRNAs belonging to 17 families encoding 20 genes involved in stress tolerance. The genes were mostly encoding stress responsive transcription factors, plant hormone signal transduction and phenylpropanoid biosynthesis. Combined network analysis of most abundant miRNAs revealed that 9 novel miRNAs and 19 known miRNAs interacted with 36 genes in V1–31, and in V2–31, 10 novel miRNAs and 20 known miRNAs interacted with 32 genes (Fig. 7a, b). It was found that different miRNAs targeted different number of stress-responsive genes. For instance, the conserved miRNA osa-miR396f-5p targeted only one gene (heat shock protein 90), while bdi-miR5054 targeted five genes (peroxidase, phospholipase D1/2, poly(A)-specific ribonuclease, mitochondrial phosphate carrier protein 3 and cleavage and polyadenylation specificity factor subunit 4). Similarly, the novel mireap-m0100-5p targeted only one gene (thiazole biosynthetic enzyme), mireap-m0087-3p targeted two genes (l-ascorbate peroxidase and WRKY transcription factor 26) but mireap-m0077-3p targeted seven genes (aconitatehydratase, catalase, peroxidase, WRKY, heat shock 70 kDa protein 5, lipoxygenase and malonate-semialdehyde dehydrogenase). Among the different targets, peroxidase gene was targeted by most of the known and novel miRNAs.
Fig. 7.
Network analysis of abundant conserved (in red triangle) and novel miRNAs (in blue triangle) and predicted target genes (in green box) of a drought susceptible Co 8021 (V1–31) and b drought tolerant Co 06022 (V2–31)
Expression analysis of miRNAs and their targets by qRT-PCR
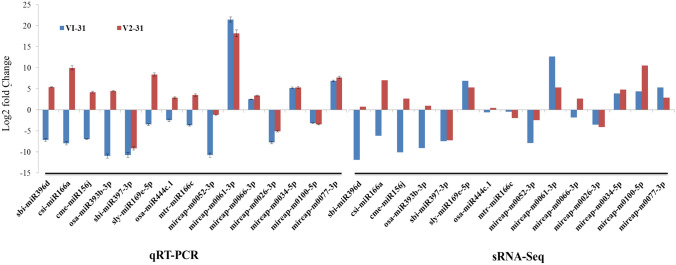
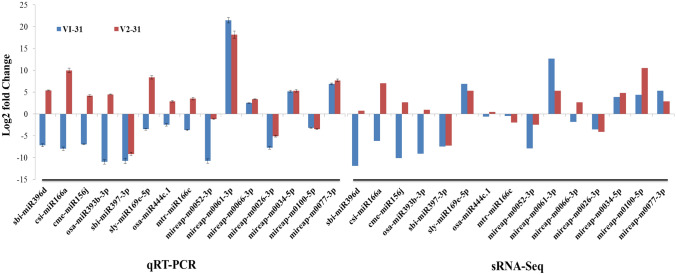
qRT-PCR was used to verify the expression levels of miRNAs obtained from sRNA sequencing results. Eight known miRNAs (sbi-miR396d, csi-miR166a, cme-miR156j, osa-miR393b-3p, sbi-miR397-3p, sly-miR169e-5p, osa-miR444c.1 and mtr-miR166c) and seven novel miRNAs (mireap-m0052-3p, mireap-m0061-3p, mireap-m0066-3p, mireap-m0026-3p, mireap-m0034-5p, mireap-m0100-5p and mireap-m0077-3p) were selected for the validation (Additional file 9: Table S1). Among the 15 miRNAs, 11 miRNAs were found to be consistent with the expression pattern obtained through small RNA deep sequencing data (Fig. 8). miRNAs can regulate post-transcriptional gene expression by targeting mRNAs for degradation. To explore the potential extent of miRNA-directed regulation of mRNA levels, qRT-PCR expression analysis of the targets of differentially expressed miRNAs was done (Additional file 9: Table S2). The results of the qRT-PCR analysis revealed that all miRNAs negatively regulate their target genes except hvu-miR444 which positively regulated defensin gene in both V1 and V2. Similarly miRNAs, gma-miR166k and cpa-miR167d revealed positive regulation of peroxidase and WRKY transcription factor in V1 and V2 respectively (Fig. 9). The target gene expression of putative cellulose synthase, disease resistant protein RGA2, AP2-EREBP transcription factor, xyloglucan endotransglucosylase hydrolase, heat shock protein, zinc finger protein and abscisic acid showed consistent up and down regulation by the concomitant expression of miRNA in both V1 and V2 showing that these are highly regulated by the miRNA.
Fig. 8.
Expression profiling of 15 randomly selected miRNAs by qRT-PCR and comparison with sRNA-Seq data of 10 days drought stress in drought susceptible Co 8021 (V1–31) and drought tolerant Co 06022 (V2–31). For qRT-PCR each bar represents the average of three replicates and error bars indicate SE
Fig. 9.
Expression profiling of miRNAs and predicted target genes of drought susceptible Co 8021 (V1) and drought tolerant Co 06022 (V2) sugarcane cultivars subjected to 10 days drought stress. The target genes are cellulose synthase, RGA2 disease resistance protein RGA2, AP2-EREBP AP2-EREBP transcription factor, peroxidase, XET xyloglucan endotransglucosylase hydrolase, HSP 70 heat shock protein 70-kDa, ZFP zinc finger domain superfamily protein, defensin, WRKY WRKY transcription factor and ABA abscisic acid. In qRT-PCR each bar represents the average of three replicates and error bars indicate SE
Discussion
During drought stress, plant activates several defense mechanisms to withstand environmental challenge, which includes physiological, biochemical and molecular changes (Valliyodan and Nguyen 2006). microRNAs play an important role in the regulation of plant responses to numerous environmental stimuli (Bartel 2004). Several studies proved that “omic” technologies have greatly enhanced our knowledge to interpret the mechanisms of stress tolerance in plants (Zhuang et al. 2014). High throughput deep sequencing approaches have been used to discover small RNAs and is a most effective method for miRNA detection in plants (Fahlgren et al. 2007). In this study, we performed high throughput deep sequencing for drought tolerant and susceptible sugarcane varieties to study the regulation of gene expression by miRNAs during drought stress.
In the present study, the length of small RNAs was mainly distributed between 21 and 25 nt, which account for an average of 88% of the small RNAs. Among all nucleotides, 24 nt small RNAs had the highest number of reads in all of the four libraries. Ferreira et al. (2012) reported that the majority of the reads were 21–24 nt in length, with 21 nt being the most redundant species, followed by 24 nt in the drought stressed and irrigated libraries of sugarcane cultivars. Findings of Yang et al. (2017) indicate that 21 nt and 24 nt small RNAs were the most abundant under low temperature stress in sugarcane genotypes. Su et al. (2017) reported sRNA sequence length in four libraries within the range of 20–24 nt, under Sporisorium scitamineum treatment in sugarcane genotypes.
A total of 313, 296, 331 and 284 known miRNAs were identified in the V1–11, V1–31, V2–11 and V2–31 small RNA libraries, respectively. Comparison of V1–11 and V1–31 revealed 14 differentially expressed miRNAs (log2 ratio ≥ 1 up and ≤ 1 down, p ≤ 0.05) including seven upregulated and seven downregulated. Furthermore, comparison of drought stressed libraries of both varieties (V1–31 and V2–31) revealed six known differentially expressed miRNAs including five upregulated and one downregulated. Ferreira et al. (2012) detected a total of 21 miRNA candidates belonging to 18 miRNAs families in sugarcane drought stressed libraries obtained at different time points of stress. Most of the miRNAs that were differentially expressed varied between the two varieties showing varietal differences in drought responses. Differences in expression pattern of miRNAs depending on the cultivar, growth conditions, duration of stress has been previously observed in sugarcane (Gentile et al. 2015). Among the differentially expressed known miRNAs, only two miRNAs, sbi-miR397-3p and csi-miR166a were common in both varieties, where sbi-miR397 was downregulated in both varieties and csi-miR166a was upregulated in the tolerant cultivar and downregulated in the susceptible cultivar. The role of miR397-3p in drought stress has been commonly observed in several crops (Ferreira et al. 2012; Chen et al. 2020; Hamza et al. 2016). Differential expression of miR397-3p was observed in all stages of drought stress in switch grass and sorghum (Chen et al. 2020; Hamza et al. 2016) whereas in sugarcane mir397 was upregulated at 2 days stress in both tolerant and susceptible cultivars and when drought progressed to 4 days it was downregulated in both types. It is also notable that in our study sbi-miR397-3p targeted around 407 genes including several drought responsive genes like molecular chaperone HSP90 family, cytochrome P450, Myb superfamily, Ca2+/calmodulin-dependent protein kinase, Zn-finger protein among others. Similarly differential regulation of csi-miR166a was observed in tea plants subjected to drought stress (Guo et al. 2017). miR166 has been identified as a drought responsive miRNA that is conserved among several plant species (Chen et al. 2017; Aravind et al. 2017; Akdogan et al. 2016; Zhu et al. 2011) and plays an important role in post transcriptional changes related to root architecture during drought stress. Comparison of drought stressed libraries of both varieties revealed two miRNAs, vvi-miR394c and bdi-miR159b-3p.3 that were not shared in other comparisons. Drought stress studies in desert plant Ammopiptanthaus mongolicus showed that miR159 family had the highest members of miRNA that were expressed during drought stress (Gao et al. 2016). Oxidative stress responses in crops like rice, Brachypodium distachyon have also reported upregulation of bdi-miR159a/b during stress (Zhou et al. 2010; Bertolini et al. 2013) indicating involvement of bdi-miR159a/b in abiotic stress responses. Similarly miR394 family is conserved among plants and is known to regulate plant growth and biotic and abiotic stress responses. Predicted targets of vvi-miR394c include dehydration responsive proteins, MYB super family proteins, CBS domain containing protein, glycosyltransferase, which play important roles in drought and salt responsive pathways (Yang et al. 2017; Dong et al. 2020; Singh et al. 2012; Pagliarani et al. 2017). High levels of down regulation of sbi-miR396d, cme-miR156j, osa-miR393b-3p (log2 fold change − 11, − 10, − 9, respectively) in Co 8021 and osa-miR171h, osa-miR444b.2, (log2 fold change − 9.5, − 9.5, respectively) in Co 06022 was observed. MicroRNA sbi-miR396d targets growth regulating factors coordinating cell division and differentiation that affects leaf growth and orientation, conferred drought tolerance in Arabidopsis upon induction (Jones-Rhoades and Bartel 2004). Monocot abundant miRNA156g-h targeting squamosa-binding protein which positively regulates stem and internode growth was identified to be downregulated in sensitive sorghum cultivars during drought stress (Katiyar et al. 2015). Similarly upregulation of osa-miR393b-3p and down regulation of osa-miR171 has shown to improve drought tolerance in rice (Zhang et al. 2017; Cheah et al. 2015). MicroRNAs gma-miR166k, ppt-miR390c-5p were highly upregulated in Co 06022 (log2 fold change 10, 9 respectively). Enhanced expression of gma-miR166k which targets genes for root growth and elongation and ppt-miR390c targeting stress responsive leucine-rich repeat receptor like kinase imparts tolerance to several biotic and abiotic stresses (Guirao et al. 2018; Huang et al. 2018; Li et al. 2017; Ding et al. 2016).
In present study, 94 and 111 novel miRNAs were detected in the library of the drought-susceptible variety V1–11 and V1–31 respectively and in the drought-tolerant variety, 112 and 118 novel miRNAs were detected in V2–11 and V2–31 libraries respectively. Most novel miRNAs had a relatively low expression and only mireap-m0040-5p had reads over 1000 in the library V1–11. In the drought-susceptible variety Co 8021, five novel miRNAs were found to be differentially expressed (log2 ratio ≥ 1 up and ≤ 1 down, p ≤ 0.05) and in drought-tolerant variety Co 06022, nine novel miRNAs were found to be differentially expressed. All novel miRNAs expressed were cultivar specific. More number of novel miRNAs were upregulated in Co 8021 whereas in Co 06022 many were downregulated. Novel miRNAs mireap-m0016 and mireap-m0020 downregulated in Co 06022 was also observed to be differentially expressed in flag leaf and anthesis spikelet and mature root of drought tolerant Nagina 22 rice (Mutum et al. 2016). Upregulation of mireap-m0017 and mireap-m0082 was observed in Co 06022 and similar observations were made in wild rice Dongxiang during drought stress (Fantao et al. 2018). Overall novel miRNAs implicated in drought stress in this study have also been found to play important roles in crops like tomato, barley, rice etc. (Liu et al. 2017; Smoczynska et al. 2020; Mutum et al. 2016).
The predicted target genes of known and novel miRNAs mainly encode transcription factors, proteins, phosphatase and kinases involved in signal transduction pathways, integral component of membrane and inorganic ion transport metabolism, enzymes involved in carbohydrate transport and metabolism, drought-stress related proteins involved in defense mechanisms and cell wall/membrane biogenesis. Hua et al. (2019) reported that a number of target genes regulated by differentially expressed miRNAs for drought stress in wheat, included signal transducer, response to stress and antioxidant pathways. Furthermore, Li et al. (2017) reported that most of the drought-responsive miRNAs were involved in development or disease resistance, substance synthesis and transportation indicating these miRNAs play important roles during drought stress in alfalfa leaves.
According to previous reports, GO and KEGG analysis indicated that miRNAs were intricate in stress-responsive biological pathways. In our study, bdi-miR159, cpa-miR167d and sbi-miR397-3p targeting auxin responsive factors were regulated in both V1–31 and V2–31 varieties under drought stress. Studies in Ipomea indicated that miR160 and miR167 were involved in auxin response by targeting of auxin response factors (ARF) (Glazinska et al. 2013). The up-regulation of miR167 was identified in wheat genotypes, suggesting that miR167 and the trans-acting short-interfering RNA-auxin response factor had an important role in the auxin signaling pathway and developmental response to cold stress (Tang et al. 2012). Among the miRNA targets, transcription factors such as the ARF, TIR, MYB and NF-Y were identified to be involved in the response of sugarcane to abiotic stresses and these factors control a series of downstream genes by binding to cis-elements in promoter regions (Singh et al. 2002). In our study, tae-miR5384-3p, hvu-miR444b, smo-miR408 and sbi-miR396d targeted drought responsive transcription factors like heat shock factor protein 7, ScMYB32 protein, GNAT, NAC, GRAS, WRKY, DRE-binding protein 3, Dof-type zinc finger protein, AP2-EREBP and BZIP respectively. Among them HSP7, DREB3, AP2-EREBP and NAC were upregulated in both varieties and ScMYB32, GNAT, GRAS, WRKY and BZIP were downregulated in both sugarcane genotypes in response to drought stress. According to Silva et al. (2019) some miRNAs, such as miR159, miR160, miR319, and miR396 were differentially regulated in sugarcane seedlings upon aluminum stress and their targets were transcription factors associated with seed germination, embryo development, cold and drought responses.
Several phosphatases (sspmiR394 and ssp-miR397), kinases (ssp-miR399-seq 1 and ssp-miR528), and oxidases (sspmiR1432) have been involved in signal transduction pathways and few others like glyceraldehyde-3-phosphate dehydrogenase (ssp-miR394) was involved in carbohydrate metabolism during drought stress in sugarcane genotypes (Ferreira et al. 2012). In present study, the targets of sbi-miR397-3p, hvu-miR444b and tae-miR5384-3p, sbi-miR5568g-3p which were down and upregulated respectively in Co 8021 were genes encoding protein phosphatase type 2-C, calmodulin-dependent protein kinase phosphatase, calcium-dependent protein kinase, mitogen-activated protein kinase and receptor protein kinase CRINKLY4, respectively, that are involved in signal transduction pathways. Target genes of tae-miR5384-3p, sbi-miR397-3p and hvu-miR444b which are all downregulated encode glyceraldehyde 3-phosphate dehydrogenase, invertases and alpha-galactosidase respectively, which are involved in carbohydrate transport and metabolism.
The target genes of novel miRNAs mainly encode drought-stress related genes involved in plant defense mechanisms such as mireap-m0077-3p (upregulated), mireap-m0086-5p (downregulated), mireap-m0073-3p (downregulated), mireap-m0061-3p (upregulated), mireap-m0049-5p (upregulated), mireap-m0043-3p (downregulated), mireap-m0067-5p (upregulated), mireap-m0020-5p (downregulated) and mireap-m0104-3p (downregulated) targeted genes encoding ABA responsive element binding factor (Ferreira et al. 2012), calmodulin binding protein (Liu et al. 2017), carboxypeptidase (Selvi et al. 2020), cytochrome P450 (Li et al. 2017), disease resistance protein RPM1 (Hua et al. 2019), peroxidase (Wei et al. 2009), glutathione S-transferase 4 (Devi et al. 2019), HSP20 family protein (Zhao et al. 2018) and calcium-dependent protein kinase (Selvi et al. 2020), respectively during drought stress.
GO analysis showed that the target genes of many differentially-expressed miRNAs were related to integral component of membrane and nucleus in cellular component category Yang et al. (2014) and Liu et al. (2017) reported that several differentially-expressed genes were involved in physiological, metabolic and biochemical pathways, however, most genes were involved in plant adaptation during abiotic stress by cellular metabolism. The enrichment of biological process GO term, response to abiotic stimulus and response to water were identified in tomato plants during drought stress (Liu et al. 2017). In our study, enrichment of biological process GO terms was related to stress tolerance, such as transcription factors, DNA integration and carbohydrate metabolic processes. Significantly enriched molecular function GO terms were ATP binding, DNA binding, zinc ion binding, protein kinase activity and transcription factor activity. The top enriched category “General function prediction only” was observed in both tolerant and susceptible genotypes with 1212 and 1271 genes respectively, followed by signal transduction mechanisms with 615 and 714 genes respectively and posttranslational modification, protein turnover, chaperones with 518 and 603 genes respectively. Pathways such as secondary metabolites biosynthesis, transport and catabolism, inorganic ion transport and metabolism, defense mechanisms and cell wall/membrane/envelope biogenesis were also involved in plant stress tolerance. Golldack et al. (2014) and Xia et al. (2015) reported that many differentially-expressed genes were involved in various stress-related metabolic pathways such as plant hormone signal transduction, phosphatidylinositol signalling system and oxidative phosphorylation.
Several factors may explain the regulation of miRNA and their targets. Although not widely reported, studies have suggested that miRNAs could also act as positive regulators of transcription (Vasudevan et al. 2007). Further, not all miRNAs-targets are identified by the algorithms nor the complexity of miRNA-target gene regulation is fully studied. Expression analysis done in our study indicated that most miRNAs negatively regulate their targets. However miRNAs like miR166 showed positive regulation of peroxidases in V1 and negative regulation in V2 while cellulose synthase was negatively regulated in both varieties. Abundance of miR166 was observed in the tolerant sugarcane genotype and are known to be involved in ABA dependent stress response by targeting class III homeodomain-leucine zipper genes which play a major role in growth and development (Yan et al. 2016). Similarly xyloglucan endotransglycosylase a prominent cell wall modifying enzyme that causes cell expansion, loosening/reinforcing cell walls, particularly in response to environmental stress was negatively regulated by miR6235 in both varieties. Thus miRNA:mRNA regulation is a complex phenomenon owing to the fact that each miRNA could regulate the expression of several target genes and the expression of each target gene could be potentially regulated by several miRNAs.
Conclusions
Our study was the first study that provided an insight into microRNA gene regulation of Indian tropical sugarcane cultivars that are tolerant and susceptible to drought stress at formative stage. The study provided valuable information on differentially expressed known and novel microRNAs that contribute to varietal variations in mitigating sugarcane drought. It was interesting to note that most miRNAs significantly expressed during drought were specific to the tolerant and susceptible cultivars indicating the various mechanisms of drought tolerance adopted by the varieties. The study also led to identifying target genes that are regulated by the microRNAs and their possible functions in overcoming drought. Major pathways that attributed to stress tolerance like plant hormone signal transduction, posttranslational modification protein turn over and chaperones, carbohydrate metabolism, MAPK signalling pathway, plant plant-pathogen interaction, phenyl propanoid biosynthesis were identified.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by Indian Council of Agricultural Research, New Delhi under the research project PI-14/1.2.35. The authors gratefully acknowledge the financial support and facilities provided by the Director, ICAR-Sugarcane Breeding Institute, Coimbatore and also thank Genotypic Technology Pvt. Ltd., Bangalore for small RNA sequencing.
Author contributions
AS: conceptualization, project administration, data analysis and interpretation of the data, supervision, writing, review and editing. KD: laboratory experiments, validation, writing and review. RM: methodology for RNA Seq, analysis and interpretation of the data. PTP: data analysis, methodology for validation, review and editing. VPR: methodology for validation and expression analysis. KL: data analysis.
Data availability
All the data supporting the results of this article are given in the paper and in additional files. The sequencing reads have been submitted as sequence read archive (SRA) in NCBI with the BioProject ID—PRJNA593909.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in this publication.
Contributor Information
Athiappan Selvi, Email: A.Selvi@icar.gov.in, Email: selviathiappan@yahoo.co.in.
Kaliannan Devi, Email: 20biot03@selvamtech.edu.in.
Ramaswamy Manimekalai, Email: R.Manimekalai@icar.gov.in.
Perumal Thirugnanasambandam Prathima, Email: Prathima.Pt@icar.gov.in.
Rabisha Valiyaparambth, Email: VP.Rabisha@icar.gov.in.
Kasirajan Lakshmi, Email: K.Lakshmi@icar.gov.in.
References
- Akdogan G, Tufekci ED, UranbeyS UT. MiRNA-based drought regulation in wheat. Funct Integr Genomics. 2016;16:221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- Aravind J, Rinku S, Pooja B, Shikha M, Kaliyugam S, Mallikarjuna MG, Kumar A, Rao AR, Nepolean T. Identification, characterization, and functional validation of drought-responsive MicroRNAs in subtropical maize inbreds. Front Plant Sci. 2017;8:941. doi: 10.3389/fpls.2017.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bertolini E, Verelst W, Horner DS, Gianfranceschi L, Piccolo V, Inze D, Pe ME, Mica E. Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant. 2013;6(2):423–443. doi: 10.1093/mp/sss160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah BH, Nadarajah K, Divate MD, Wickneswari R. Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage. BMC Genomics. 2015;16:692. doi: 10.1186/s12864-015-1851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33(20):1–9. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Li M, Zhang Z, Tie W, Chen X, Jin L, Zhai N, Zheng Q, Zhang J, Wang R, Xu G, Zhang H, Liu P, Zhou H. Integrated mRNA and microRNA analysis identifies genes and small miRNA molecules associated with transcriptional and post-transcriptional-level responses to both drought stress and re-watering treatment in tobacco. BMC Genomics. 2017;18:62. doi: 10.1186/s12864-016-3372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Chen J, Sun M, Yan H, Feng G, Wu B, Zhang X, Wang X, Huang L. Comparative transcriptome study of switch grass (Panicum virgatum L.) homologous autopolyploid and its parental amphidiploids responding to consistent drought. Biotechnol Biofuels. 2020;13:170. doi: 10.1186/s13068-020-01810-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Companhia Nacional de Abastecimento [CONAB] (2017) Acompanhamento da safrabrasileira de cana-de-açúcar, v.4 – SAFRA 2017/2018 n.1 – PrimeiroLevantamento
- Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi K, Gomathi R, Arun Kumar R, Manimekalai R, Selvi A. Field tolerance and recovery potential of sugarcane varieties subjected to drought. Indian J Plant Physiol. 2018;23(1):271–282. doi: 10.1007/s40502-018-0367-7. [DOI] [Google Scholar]
- Devi K, Prathima PT, Gomathi R, Manimekalai R, Lakshmi K, Selvi A. Gene expression profiling in sugarcane genotypes during drought stress and rehydration. Sugar Tech. 2019;21(5):717–733. doi: 10.1007/s12355-018-0687-y. [DOI] [Google Scholar]
- Ding Y, Ye Y, Jiang Z, Wang Y, Zhu C. MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci. 2016;7:235. doi: 10.3389/fpls.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong OX, Yu S, Jain R, Zhang N, Duong PQ, Butler C, Li Y, Lipzen A, Martin JA, Barry KW, Schmutz J, Tian L, Ronald PC. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. 2020;11:1178. doi: 10.1038/s41467-020-14981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MiRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantao Z, Yuan L, Meng Z, Yi Z, Hongping C, Biaolin H, Jiankun X. Identification and characterization of drought stress responsive novel microRNAs in Dongxiang wild rice. Rice Sci. 2018;25(4):175–184. doi: 10.1016/j.rsci.2018.06.001. [DOI] [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA. Plant drought stress: effects, mechanisms and management. Agron Sustain Dev. 2009;29:185–212. doi: 10.1051/agro:2008021. [DOI] [Google Scholar]
- Ferreira TH, Gentile A, Vilela RD, Costa GGL, Dias LI, Endres L, et al. MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.) PLoS ONE. 2012;7:e46703. doi: 10.1371/journal.pone.0046703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wang N, Li H, Liu J, Fu C, Xiao Z, Wei C, Lu X, Fen J, Zhou Y. Identification of drought responsive microRNAs and their targets in Ammopiptanthus mongolicus using highthroughput sequencing. Sci Rep. 2016;6:34601. doi: 10.1038/srep34601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Ferreira TH, Mattos RS, Dias LI, Hoshino AA, Carneiro MS, Souza GM, Calsa T, Jr, Nogueira RM, Endres L, Menossi M. Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta. 2013;237(3):783–798. doi: 10.1007/s00425-012-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Dias LI, Mattos RS, Ferreira TH, Menossi M. MicroRNAs and drought responses in sugarcane. Front Plant Sci. 2015;6:1–13. doi: 10.3389/fpls.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazinska P, Wojciechowski W, Wilmowicz E, Zienkiewicz A, Frankowski K, Kopcewicz J. The involvement of InMIR167 in the regulation of expression of its target gene InARF8, and their participation in the vegetative and generative development of Ipomoea nil plants. J Plant Physiol. 2013;171:225–234. doi: 10.1016/j.jplph.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Golldack D, Li C, Mohan H, Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci. 2014;5:151. doi: 10.3389/fpls.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao RS, Yi H, San Segundo B. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front Plant Sci. 2018;9:337. doi: 10.3389/fpls.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao S, Zhu C, Chang X, Yue C, Wang Z, Lin Y, Lai Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017;17:211. doi: 10.1186/s12870-017-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza NB, Sharma N, Tripathi A, Mishra NS. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr Patterns. 2016;20(2):88–98. doi: 10.1016/j.gep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Hua Y, Zhang C, Shi W, Chen H. High-throughput sequencing reveals microRNAs and their targets in response to drought stress in wheat (Triticum aestivum L.) Biotechnol Biotechnol Equip. 2019;33(1):465–471. doi: 10.1080/13102818.2019.1586586. [DOI] [Google Scholar]
- Huang SC, Lu GH, Tang CY, Ji YJ, Tan GS, Hu DQ, Cheng J, Wang GH, Qi JL, Yang YH. Identification and comparative analysis of aluminum-induced microRNAs conferring plant tolerance to aluminum stress in soybean. Biol Plant. 2018;62(1):97–108. doi: 10.1007/s10535-017-0752-5. [DOI] [Google Scholar]
- Jin W, Li N, Zhang B, Wu F, Li W, Guo A, Deng Z. Identification and verification of microRNA in wheat (Triticum aestivum) J Plant Res. 2008;121:351–355. doi: 10.1007/s10265-007-0139-3. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Katiyar A, Smita S, Muthusamy SK, Chinnusamy V, Pandey DM, Bansal KC. Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front Plant Sci. 2015;6:506. doi: 10.3389/fpls.2015.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu JH. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochem Biophys Acta. 2012;1819(2):137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan P, Robinson N. Stress physiology: abiotic stresses. In: Moore PH, Botha FC, editors. Sugarcane: physiology, biochemistry, and functional biology. Chichester: John Wiley & Sons Inc; 2014. pp. 411–434. [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, et al. MicroRNA regulation of plant innate immune receptors. Proc Nat Acad Sci USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wan L, Bi S, Wan X, Li Z, Cao J, Tong Z, Xu H, He F, Li X. Identification of drought-responsive microRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes. 2017;8:119. doi: 10.3390/genes8040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, YuH ZG, Huang Q, Lu Y, Ouyang B. Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics. 2017;18:481. doi: 10.1186/s12864-017-3869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA package 2.0. Algorithm Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RS, Ribeiro RV, Eduardo P, Marchiori R. Biometric and physiological responses to water-deficit in sugarcane at different phonological stages. Pesqui Agropecu Bras. 2009;44(12):1575–1582. doi: 10.1590/S0100-204X2009001200003. [DOI] [Google Scholar]
- Mutum RD, Kumar S, Balyan S, Kansal S, Mathur S, Raghuvanshi S. Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci Rep. 2016;6:30786. doi: 10.1038/srep30786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namwongsa J, Jongrungklang N, Songsri P. Genotypic variation in root distribution and physiological responses of sugarcane induced by drought stress. SABRAO J Breed Genet. 2019;51(4):470–493. [Google Scholar]
- OECD-FAO . OECD-FAO agricultural outlook 2019–2028, chapter 5: sugar. Paris: OECD-FAO; 2019. pp. 1–326. [Google Scholar]
- Osakabe Y, Osakabe K, Shinozaki K, Tran LSP. Response of plants to water stress. Front Plant Sci. 2014;5:86. doi: 10.3389/Fpls.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarani C, Vitali M, Ferrero M, Vitulo N, Incarbone M, Lovisolo C, Valle G, Schubert A. The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol. 2017;173:2180–2195. doi: 10.1104/pp.16.01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei HX, Ma N, Chen JW, Zheng Y, Tian J, Li J, Zhang S, Fei ZJ, Gao JP. Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS ONE. 2013;8(5):e64290. doi: 10.1371/journal.pone.0064290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino P, Pataleo S, Gerardi C, Mita G, Perrotta C. Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006;29:2143–2152. doi: 10.1111/j.1365-3040.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen XM. Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B, Voleti SR, Subrahmanyam D, Sarla N, Prasanth VV, Bhadana VP, Mangrauthia SK. Prediction and expression analysis of miRNAs associated with heat stress in Oryzasativa. Rice Sci. 2014;21(1):3–12. doi: 10.1016/S1672-6308(13)60164-X. [DOI] [Google Scholar]
- Selvi A, Devi K, Manimekalai R, Prathima PT. Comparative analysis of drought responsive transcriptomes of sugarcane genotypes with differential tolerance to drought. 3 Biotech. 2020;10(6):236. doi: 10.1007/s13205-020-02226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriram V, Kumar V, Devarumath RM, Khare TS, Wani SH. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci. 2016;7:817. doi: 10.3389/fpls.2016.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RGd, Rosa-Santos TM, Franca SdC, Kottapalli P, Kottapalli KR, Zingaretti SM. Microtranscriptome analysis of sugarcane cultivars in response to aluminum stress. PLoS ONE. 2019;14(11):e0217806. doi: 10.1371/journal.pone.0217806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Smoczynska A, Pacak AM, Nuc P, Swida-Barteczka A, Kruszka K, KarlowskiArturJarmolowski WM, Szweykowska-Kulinska Z. A functional network of novel barley microRNAs and their targets in response to drought. Genes. 2020;11:488. doi: 10.3390/genes11050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Zhang Y, Huang N, Liu F, Su W, Xu L, Ahmad W, Wu Q, Guo J, Que Y. Small RNA sequencing reveals a role for sugarcane miRNAs and their targets in response to Sporisorium scitamineum infection. BMC Genomics. 2017;18:325. doi: 10.1186/s12864-017-3716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R. MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol. 2010;21(8):805–811. doi: 10.1016/j.semcdb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17(4):196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tang ZH, Zhang LP, Xu CG, Yuan SH, Zhang FT, Zheng YL, et al. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012;159:721–738. doi: 10.1104/pp.112.196048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut F, Grativol C, Carnavale-Bottino M, Rojas CA, Tanurdzic LOS, Farinelli L, Martienssen RA, Hemerly AS, Ferreira PC. Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics. 2012;13:290. doi: 10.1186/1471-2164-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9(2):189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Rongmei Wu, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wei L, Zhang D, Xiang F, Zhang Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci. 2009;170:979–989. doi: 10.1086/605122. [DOI] [Google Scholar]
- Xia XJ, Zhou YH, Shi K, Zhou J, Foyer CH, Yu JQ. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J Exp Bot. 2015;66(10):2839–2856. doi: 10.1093/jxb/erv089. [DOI] [PubMed] [Google Scholar]
- Yan Q, Cui X, Lin S, Gan S, Xing H, Dou D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE. 2016;11:9. doi: 10.1371/journal.pone.0162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Liu J, Dong X, Cai Z, Tian W, Wang X. Short-term and continuing stresses differentially interplay with multiple hormones to regulate plant survival and growth. Mol Plant. 2014;7(5):841–855. doi: 10.1093/mp/ssu013. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang X, Su Y, Zou J, Wang Z, Xu L, Que Y. miRNA alteration is an important mechanism in sugarcane response to low temperature environment. BMC Genomics. 2017;18:833. doi: 10.1186/s12864-017-4231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Long Y, Xue MD, Xiao XG, Pei XW. Identification of microRNAs in response to drought in common wild rice (Oryza rufipogon Griff.) shoots and roots. PLoS ONE. 2017;12(1):e0170330. doi: 10.1371/journal.pone.0170330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cheng X, Liu X, Wu H, Bi H, Xu H. The wheat MYB transcription factor TaMYB31 is involved in drought stress responses in Arabidopsis. Front Plant Sci. 2018;9:1426. doi: 10.3389/fpls.2018.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61(15):4157–4168. doi: 10.1093/jxb/erq237. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, Dong A. Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 2011;66:443–455. doi: 10.1111/j.1365-313X.2011.04504.x. [DOI] [PubMed] [Google Scholar]
- Zhu L, Ow DW, Dong ZC. Transfer RNA-derived small RNAs in plants. Sci China Life Sci. 2018;61:155–161. doi: 10.1007/s11427-017-9167-5. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Zhang J, Hou XL, Wang F, Xiong AS. Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit Rev Plant Sci. 2014;33(2–3):225–237. doi: 10.1080/07352689.2014.870420. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the results of this article are given in the paper and in additional files. The sequencing reads have been submitted as sequence read archive (SRA) in NCBI with the BioProject ID—PRJNA593909.