Abstract
Epithelial-mesenchymal transition (EMT) is reported to involve in the crosstalk between tumor cells and tumor-associated macrophages (TAMs). Exosomes are considered as important mediators of orchestrating intercellular communication. However, the underlying mechanisms by which EMT-colorectal cancer (CRC) cells promote the M2 polarization of TAMs remain less understood. In this study, we found that EMT-CRC cells promoted the M2-like polarization of macrophages by directly transferring exosomes to macrophages, leading to a significant increase of the microRNA-106b-5p (miR-106b) level in macrophages. Mechanically, an increased level of miR-106b activated the phosphatidylinositol 3-kinase (PI3K)γ/AKT/mammalian target of rapamycin (mTOR) signaling cascade by directly suppressing programmed cell death 4 (PDCD4) in a post-transcription level, contributing to the M2 polarization of macrophages. Activated M2 macrophages, in a positive-feedback manner, promote EMT-mediated migration, invasion, and metastasis of CRC cells. Clinically, miR-106b was significantly elevated in CRC tissues and negatively correlated with the levels of PDCD4 in CRC specimens, and high expression of exosomal miR-106b in plasma was significantly associated with the malignant progression of CRC. Taken together, our results indicate that exosomal miR-106b derived from EMT-CRC cells has an important role in intercellular communication for inducing M2 macrophage polarization, illuminating a novel mechanism underlying CRC progression and offering potential targets for prevention of CRC metastasis.
Keywords: exosome, macrophages, EMT, miR-106b, PDCD4, colorectal cancer
Graphical abstract
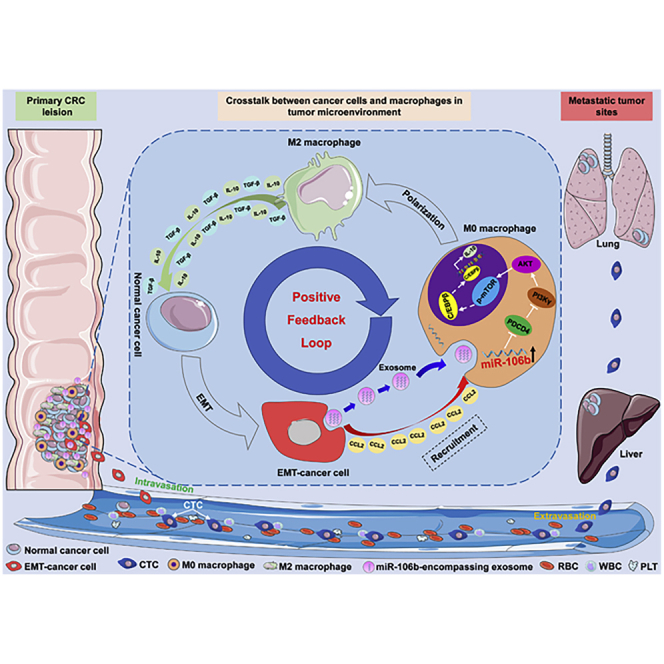
Herein, Bin Xiong and colleagues uncover the important role of tumor-derived exosomes in crosstalk between CRC cells and TAMs, by which, EMT-CRC cells deliver miR-106b-5p to induce macrophage M2-subtype polarization, thereby promoting CRC metastasis. These findings illuminate a novel mechanism for cancer progression and provide potential targets for combating CRC.
Introduction
Colorectal cancer (CRC) is the third frequently diagnosed malignancy and the second leading cause of cancer death worldwide in 2018.1 Metastasis, a multi-step complex process involving multi-factors, remains the main cause for CRC-associated mortality.2 Nowadays, increasing evidence indicates that the interplay among various cells in the tumor microenvironment (TME) represents the necessary prerequisite for cancer metastasis.3,4 Tumor-associated macrophages (TAMs), as the prominent immune population in TME, have been showed to be frequently related to the metastasis of human cancers.5 They can respond to various signals delivered from tumor cells to polarize the M1 or M2 phenotype, thereby exerting different functions in tumor progression.6 Numerous studies have demonstrated that M2-like TAMs exhibit various pro-tumoral activities, including invasion promoting, immunosuppression, and angiogenesis, to mediate the process of caner metastasis.4,6 Therefore, the exploration of the potential mechanisms of TAM M2 polarization holds important value for in-depth understanding the metastatic process of CRC.
Epithelial-mesenchymal transition (EMT) in cancer, as known to increase cell motility and invasive potential, is a crucial metastasis-associated event.7 Currently, the intercellular communication between M2 TAMs and tumor cells has been proposed to play an important role in inducing EMT of tumor cells and promoting EMT-mediated metastasis.5,8 Recently, our group also revealed the EMT-mediated interaction between CRC cells and M2-subtype TAMs: on one hand, M2-like TAMs could induce EMT of tumor cells by secreting interleukin (IL)-6 to promote CRC metastasis;9 on the other hand, EMT-programmed CRC cells could activate the expression of different cytokines, such as CCL2 and IL-4, in cancer cells to enhance the macrophages’ recruitment and promote their M2-like polarization.9,10 However, the cross-linking among CRC cells, EMT, and macrophages is a complex process involved in multiple factors, in addition to soluble cytokines, the underlying mechanisms still needed for further exploration.
Exosomes, characterized by small, disk-shaped extracellular vesicles with a diameter of 30−150 nm, are nowadays considered as other important mediators of orchestrating intercellular communication.11,12 They originated from multivesicular bodies and are secreted by many cell types and reported to play important functions in multiple physiological and pathological processes.13 A growing body of studies have indicated that tumor-derived exosomes could modulate the interaction between tumor cells and stroma cells in the TME through transmitting functional molecules, including messenger RNAs (mRNAs), microRNAs (miRNAs), proteins, and lipids.12,14 Recently, emerging and accumulating studies pointed out that cancer-derived exosomes could deliver miRNAs to macrophages to induce their M2 polarization, consequently promoting EMT-medicated cancer metastasis.15, 16, 17, 18, 19 Given the important role of tumor-derived exosomal miRNAs in the cross-link among cancer cells, EMT, and macrophages, our group previously performed miRNA sequencing to analyze the expression profiling of miRNAs in EMT-CRC cell-secreted exosomes (EMT-Exos) and identified multiple differentially expressed miRNAs in EMT-Exos compared to that in normal CRC cell-secreted exosomes (Nor-Exos) (Sequence Read Archive [SRA]: SRP227899). Combined with our previous findings,9,10 we hypothesized that EMT-CRC cells might modulate macrophages to the M2 subtype through delivering exosomal miRNAs to then promote the malignant progression of CRC. However, the potential mechanisms by which EMT-CRC-derived exosomal miRNAs facilitate M2 macrophage polarization are yet to be elucidated.
In the present study, we found that EMT-Exos could be transferred into macrophages and induced their M2-like polarization in vitro and in vivo. Then, microRNA-106b-5p (miR-106b) was identified to be the critical exosomal miRNA to induce M2 macrophage polarization via comparing the miRNA differences between EMT-Exos and Nor-Exos by miRNA sequencing. Mechanistically, exosomal miR-106b induced M2 macrophage polarization by directly suppressing programmed cell death 4 (PDCD4) to activate the phosphatidylinositol 3-kinase (PI3K)γ/Akt/mammalian target of rapamycin (mTOR) signaling pathway. Moreover, activated M2 macrophages maintain the EMT of CRC cells in a positive-feedback manner, thereby enhancing the ability of migration, invasion, and metastasis in vitro and in vivo. Further clinical data demonstrated that plasma exosomal miR-106b could serve as a promising biomarker in CRC patients. Collectively, our findings indicate that exosomal miR-106b derived from EMT-CRC cells has an important role in intercellular communication for inducing M2 macrophage polarization, illuminating a novel mechanism underlying CRC progression and offering potential targets for prevention and treatment of cancer metastasis.
Results
EMT-CRC cell-derived exosomes induce M2 macrophage polarization in vitro
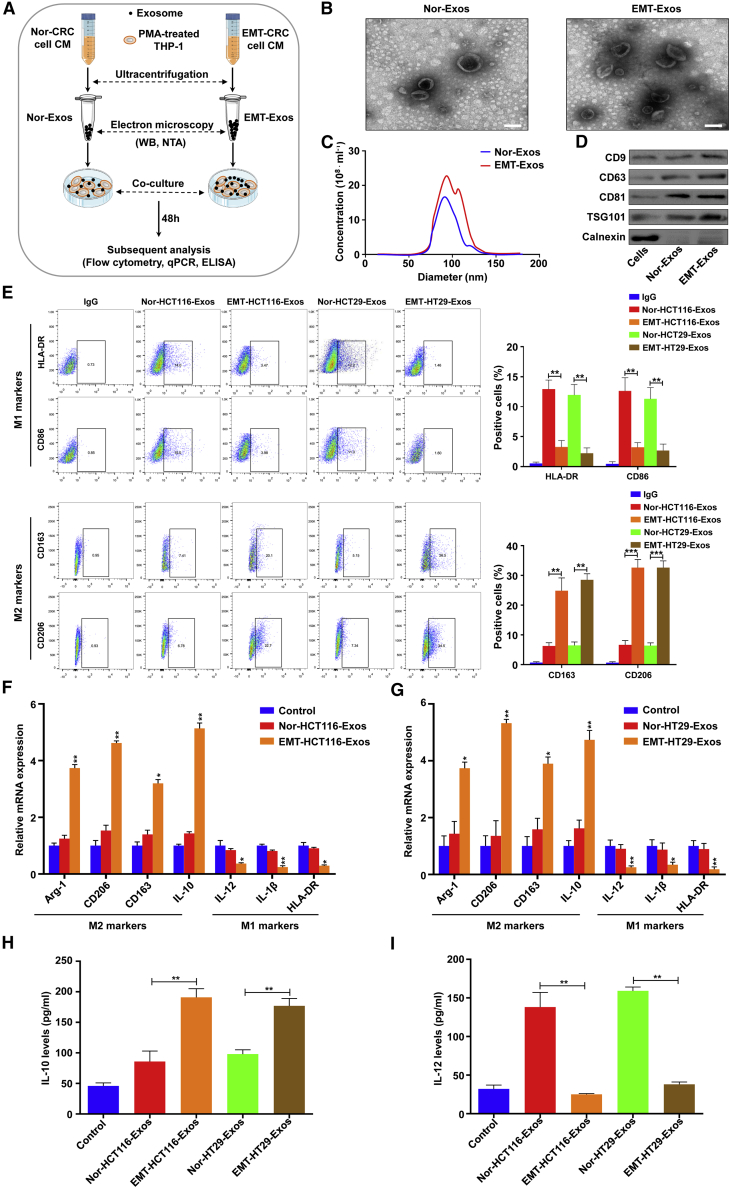
The bilateral crosstalk between EMT-programmed tumor cells and M2 TAMs has been demonstrated to actively participate in the tumor progression.9,10,19 Previously, our clinical analyses have shown that the infiltration degree of M2-like TAMs was closely associated with the EMT program of tumor cells in CRC tissues.9,20 Thus, it is important to further explore whether EMT-CRC cells can induce M2 macrophage polarization. In light of the fact that exosomes are important mediators for cell-cell communication, it rationally asks the question of whether exosomes participate in the above process. To make it clear, two CRC cell lines (HCT116 and HT29) with epithelial characteristic were chosen, and identical concentrations of CRC cells were seeded and treated without and with IL-6 (100 ng/mL), according to our previous report.9 After 48 h, cells were harvested for morphology, western blot, and qRT-PCR analyses. The results found that compared with cells without IL-6 treatment, both HCT116 and HT29 cells exhibited a spindle-shaped morphology and increased formation of pseudopodia (Figures S1A and S1B). Further western blot and qRT-PCR results showed that the expression of the epithelial marker (E-cadherin) was reduced, whereas the mesenchymal marker (N-cadherin and vimentin) was upregulated in HCT116 (Figures S1C and S1D) and HT29 (Figures S1E and S1F) cells treated with IL-6. These data indicate that IL-6 treatment can induce CRC cells to exhibit an EMT phenotype. Then, Nor-Exos and EMT-Exos were isolated from the conditional media (CM) by ultracentrifugation (Figure 1A). Transmission electron microscopy (TEM) showed that the exosomes were typical rounded particles ranging from 30 to 150 nm in diameter (Figure 1B), and NanoSight analysis exhibited a similar size distribution of exosomes (Figure 1C). Further western blot analysis demonstrated the presence of exosomal proteins CD9, CD63, CD81, and TSG101 and the absence of the endoplasmic reticulum marker calnexin in exosomes (Figure 1D). Intriguingly, all of the above results demonstrated that much more exosomes were secreted from EMT-CRC cells than normal ones (Figures 1B−1D). These results indicate that the EMT program can enhance the exosome secretion from CRC cells.
Figure 1.
EMT-CRC cell-derived exosomes induce M2 macrophage polarization in vitro
(A) Schematic description of the experimental design. (B) Transmission electron microscopy images of exosomes isolated from the medium of HCT116 cells under normal (Nor-Exos) and EMT status (EMT-Exos). Scale bars, 100 nm. (C) Nanosight particle tracking analysis of Nor-Exos and EMT-Exos. (D) Western blot analysis of the expression of exosome markers (CD9, CD63, CD81, TSG101) in cells, Nor-Exos, and EMT-Exos. (E) Flow cytometry for analyzing the expression of HLA-DR, CD86, CD163, and CD206 in macrophages incubated with Nor/EMT-HT29-Exos or Nor/EMT-HCT116-Exos for 48 h. (F and G) qRT-PCR analyses of the macrophage-associated markers in macrophages cocultured with Nor/EMT-HT29-Exos or Nor/EMT-HCT116-Exos for 48 h. (H and I) ELISA for analyzing the secretion of IL-10 and IL-12 in Nor/EMT-HT29-Exos or Nor/EMT-HCT116-Exos incubated macrophages for 48 h. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01.
To further evaluate the different abilities of educating macrophages between Nor-Exos and EMT-Exos in vitro, macrophages were treated with Nor-Exos or EMT-Exos in vitro (Figure 1A). Flow cytometry was performed and showed that the increased surface M2 markers (CD163 and CD206) and reduced M1 markers (human leukocyte antigen [HLA]-DR and CD86) in EMT-Exos-treated macrophages compared with the Nor-Exos ones (Figure 1E; Figure S2A). qRT-PCR showed that compared with Nor-Exos, EMT-Exos significantly increased the expression of M2 markers (Arg-1, CD206, CD163, and IL-10), and markedly decreased the expression of M1 markers (IL-12, IL-1β, and HLA-DR) in macrophages (Figures 1F and 1G), which were consistent with the flow cytometry results. Furthermore, we measured the secretion levels of IL-10 and IL-12 in macrophage culture supernatants by ELISA, and the results demonstrated that increased IL-10 and reduced IL-12 secretion were noted in macrophages treated with EMT-Exos (Figures 1H and 1I). Moreover, to further confirm that EMT-Exos could induce M2 macrophage polarization, exosomes were depleted from EMT-CRC cell (HCT116 and HT29) CM by microfilter, and the Exos-depletion CM were used to coculture with macrophages for 48 h (Figure S2B). Flow cytometry and qRT-PCR results demonstrated that EMT-Exos depletion significantly decreased the surface M2 markers (CD163 and CD206) and the expression of M2 markers (Arg-1, CD206, CD163, and IL-10) (Figures S2C−S2F), suggesting that EMT-Exos contribute to the induction of M2 macrophage polarization. Taken together, the above results suggest that EMT-Exos contribute to the M2 polarization of macrophages.
miR-106b is highly expressed in EMT-Exos and can be transferred to macrophages via exosomes
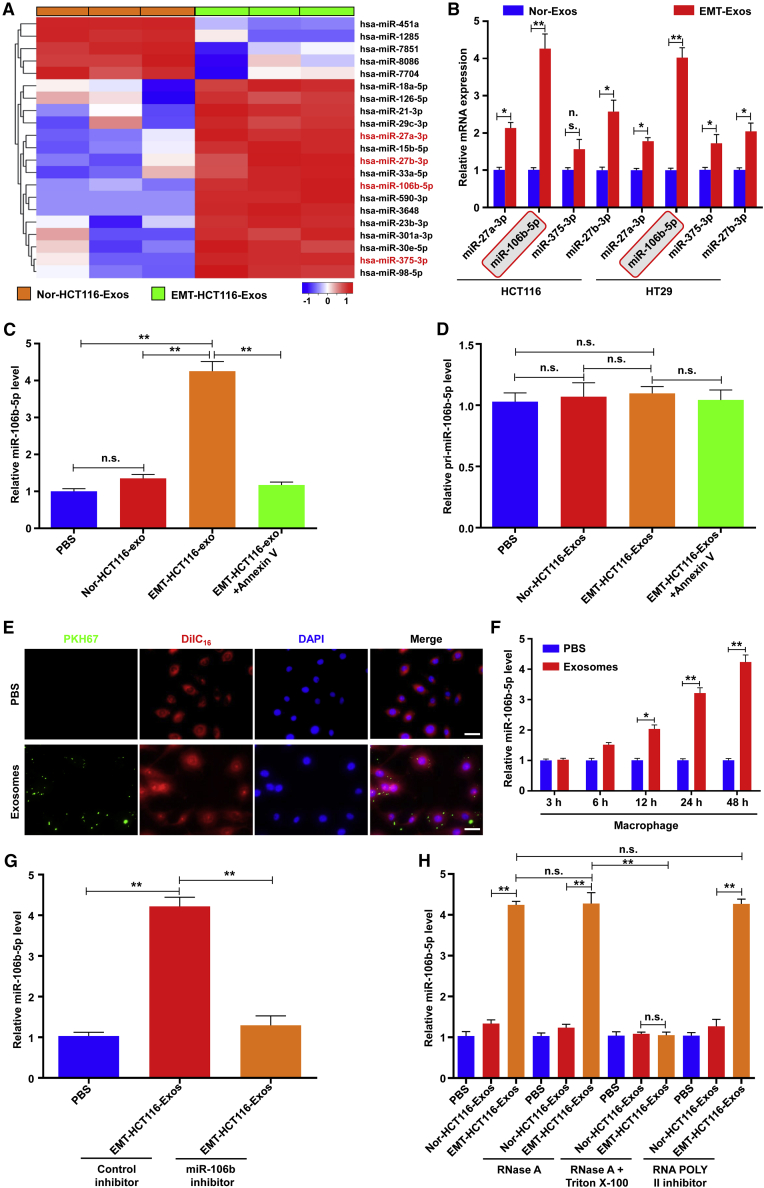
We next explored how EMT-Exos induce M2 macrophage polarization. It has been widely accepted that miRNAs encapsulated in exosomes are the most abundant RNA content and play an important role in intercellular communication.21 Recently, emerging evidence has demonstrated that exosomal miRNAs are involved in the polarization of macrophages.15, 16, 17, 18, 19,22 Therefore, we hypothesized that EMT CRC cell-derived exosomal miRNAs mediate M2 macrophage polarization. To identify the specific miRNAs involved, we performed a miRNA sequencing to identify the differentially expressed miRNAs in Nor-Exos and EMT-Exos. The results were shown as heatmaps in Figure 2A; among them, four miRNAs (miR-27a-3p, miR-106b, miR-375-3p, and miR-25b-3p) were significantly upregulated in EMT-Exos compared with Nor-Exos. To further confirm the sequencing results, RT-PCR analysis was performed to validate the expression of the above four miRNAs in five independent sets of Nor-Exos and EMT-Exos, and the results found that miR-106b was emerged as the most prominently upregulated miRNA in EMT-Exos compared with Nor-Exos from both HCT116 and HT29 cells (Figure 2B). Thus, we thought that miR-106b was a key factor in inducing M2 macrophage polarization by EMT-Exos. Then, we investigated whether miR-106b could be delivered into macrophages via exosomes. Macrophages incubated with EMT-Exos (both HCT116 and HT29) expressed higher levels of mature miR-106b than that with Nor-Exos (Figure 2C; Figure S3A). However, primary (pri)-miR-106b was not significantly different in the Nor-Exos or EMT-Exos from both HCT116 and HT29 (Figure 2D; Figure S3B). Moreover, with the treatment of Annexin V, an inhibitor of exosome internalization,23 EMT-Exos failed to induce increased pri/mature miR-106b in macrophages (Figures 2C and 2D; Figures S3A and S3B). When macrophages were incubated with PKH67-labeled EMT-Exos derived from HCT116 and HT29 cells, PKH67 lipid dye spots were observed in the recipient macrophages, suggesting that labeled exosomes released by different EMT-CRC cells could be delivered to macrophages (Figure 2E; Figure S3C). Further qRT-PCR results demonstrated that EMT-Exos were gradually internalized by macrophages over time (Figure 2F; Figure S3D). Besides, when macrophages were incubated with EMT-Exos derived from HCT116 or HT29 cells that were transfected with the miR-106b inhibitor, EMT-Exos did not significantly increase the level of miR-106b in macrophages (Figure 2G; Figure S3E). Furthermore, a more significant reduction of miR-106b was observed in the exosomes treated with both RNase A and Triton X-100/proteinase K compared with the control and RNase A-alone groups, and the RNA polymerase II inhibitor did not affect the increase of miR-106b in recipient macrophages, suggesting that miR-106b indeed existed in EMT-Exos and increased cellular miR-106b in macrophages that arose from EMT-Exos-mediated miRNA transferring but not endogenous miR-106b induction (Figure 2H; Figure S3F). Overall, these findings suggest that miR-106b is highly expressed in EMT-Exos and can be transferred to macrophages via exosomes.
Figure 2.
MicroRNA (miR)-106b is highly expressed in EMT-Exos and can be transferred to macrophages via exosomes
(A) Heatmap of differential microRNA (miRNA) expression between Nor-Exos and EMT-Exos. (B) The expression levels of the top 4 enriched miRNAs in EMT-Exos were further verified by using qRT-PCR to detect their levels in Nor-Exos and EMT-Exos of HCT116 and HT29 cells. (C and D) qRT-PCR analysis of mature and pri-miR-106b-5p expression in macrophages incubated with Nor-HCT116-Exos, EMT-HCT116-Exos, and EMT-HCT116-Exos + Annexin V groups. (E) Representative immunofluorescence images show the internalization of PKH67-labeled EMT-HCT116-Exos (green) by DilC16-labeled macrophages (red). Scale bars, 20 μm. (F) qRT-PCR analysis of miR-106b-5p expression in macrophages incubated with EMT-HCT116-Exos for 3 h, 6 h, 12 h, 24 h, and 48 h. (G) qRT-PCR analysis of miR-106b-5p expression in macrophages incubated with EMT-HCT116/mock-Exos and EMT-HCT116/miR-106b inhibitor-Exos for 48 h. (H) qRT-PCR analyses of miR-106b-5p expression in macrophages incubated with Nor/EMT-HCT116-Exos that treated by RNase A, RNase A + Triton X-100, or RNA POLY II inhibitor. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. n.s., not significant; ∗p < 0.05, ∗∗p < 0.01.
EMT-Exos-derived miR-106b mediates M2 macrophage polarization
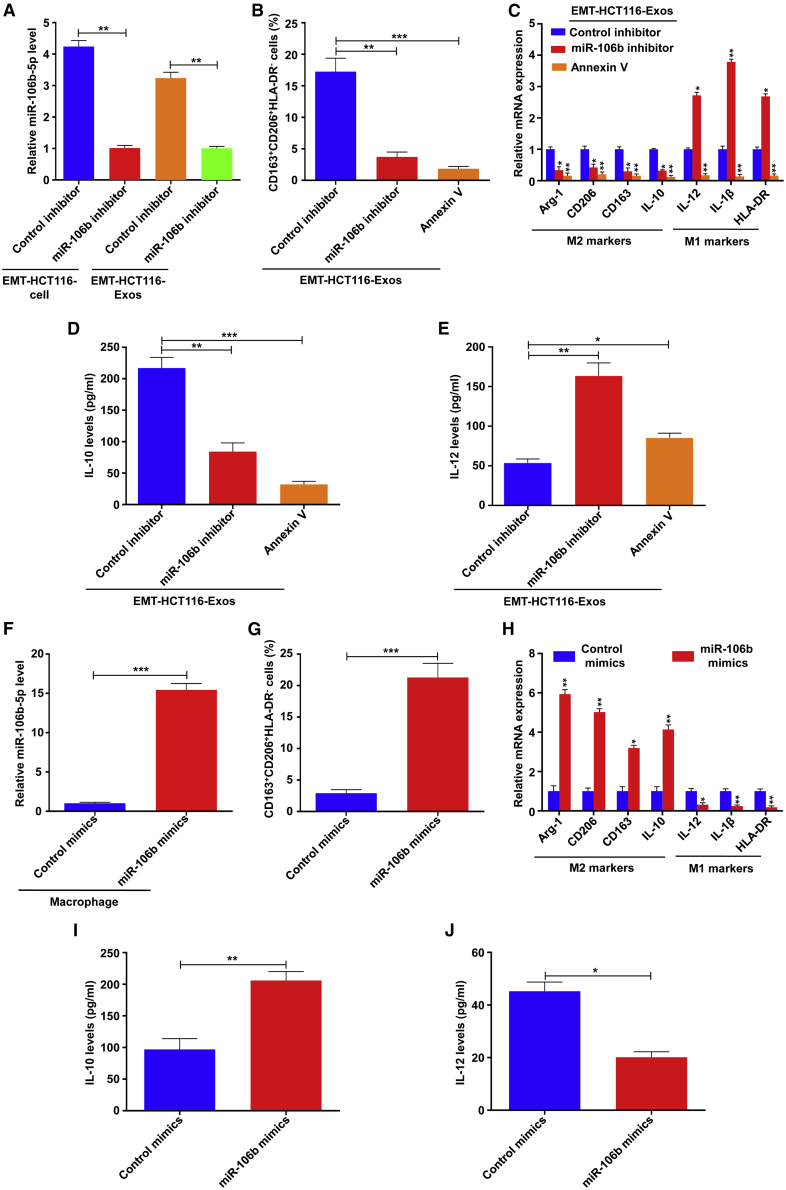
To explore the role of exosomal miR-106b in M2 polarization of macrophages, we first detected whether exosomes derived from EMT-CRC cells (HCT116 and HT29) that transfected with the miR-106b inhibitor could offset the M2 polarization of macrophages induced by EMT-Exos. As expected, qRT-PCR showed that the expression of miR-106b in EMT-HCT116 cells and exosomes derived from EMT-HCT116 cells that transfected with the miR-106b inhibitor was markedly downregulated (Figure 3A), and similar results were observed in HT29 cells (Figure S4A). Then, macrophages were treated with the exosomes derived from EMT-CRC cells that transfected with the miR-106b inhibitor. Flow cytometry showed the population of the CD163highCD206highHLA-DRlow phenotype of macrophages was strikingly decreased in this condition (Figure 3B; Figure S4B). qRT-PCR analyses were also in accordance with the above results, which found that the EMT-miR-106b-5p inhibitor-Exos significantly decreased the expression of M2 markers (Arg-1, CD206, CD163, and IL-10) and markedly increased the expression of M1 markers (IL-12, IL-1β, and HLA-DR) in macrophages (Figure 3C; Figure S4C). Furthermore, ELISA demonstrated that decreased IL-10 and increased IL-12 secretion levels were observed in macrophages treated with the EMT-miR-106b inhibitor-Exos (Figures 3D and 3E; Figures S4D and S4E). Similarly, pretreatment of EMT-Exos with Annexin V alleviated their function in promoting M2 polarization of macrophages (Figures 3B−3E; Figures S4B and S4E).
Figure 3.
EMT-Exos-derived miR-106b mediates M2 macrophage polarization
(A) qRT-PCR analysis of miR-106b-5p expression in cells and exosomes of EMT-HCT116 transfected by control inhibitor or miR-106b inhibitor. (B) Flow cytometry for analyzing the population of CD163highCD206highHLA-DRlow macrophages incubated with EMT-HCT116/mock-Exos, EMT-HCT116/miR-106b inhibitor-Exos, and EMT-HCT116-Exos + Annexin V groups. (C) qRT-PCR analyses of the macrophage-associated markers in macrophages cocultured with EMT-HCT116/mock-Exos, EMT-HCT116/miR-106b inhibitor-Exos, and EMT-HCT116-Exos + Annexin V groups. (D and E) ELISA for analyzing the secretion of IL-10 and IL-12 in macrophages incubated with EMT-HCT116/mock-Exos, EMT-HCT116/miR-106b inhibitor-Exos, and EMT-HCT116-Exos + Annexin V groups. (F) qRT-PCR analysis of miR-106b-5p expression in macrophages transfected by control mimics or miR-106b mimics. (G) Flow cytometry for analyzing the population of CD163highCD206highHLA-DRlow macrophages transfected by control mimics or miR-106b mimics. (H) qRT-PCR analyses of the macrophage-associated markers in macrophages transfected by control mimics or miR-106b mimics. (I and J) ELISA for analyzing the secretion of IL-10 and IL-12 in macrophages transfected by control mimics or miR-106b mimics. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Next, we further investigated the effect of miR-106b on M2 macrophage polarization by transfecting macrophages with miR-106b mimics. As shown in Figure 3F, miR-106b mimics transfection markedly upregulated the expression of miR-106b in macrophages. Flow cytometry analysis demonstrated that macrophages transfected with miR-106b mimics exhibited a higher proportion of the CD163highCD206highHLA-DRlow phenotype compared to those transfected with control mimics (Figure 3G). In accordance with the alteration in phenotype, qRT-PCR results showed that the miR-106b mimics significantly increased the expression of M2 markers (Arg-1, CD206, CD163, and IL-10) and markedly reduced the expression of M1 markers (IL-12, IL-1β, and HLA-DR) in macrophages (Figure 3H). Concordantly, miR-106b mimics also enhanced the secretion of IL-10, whereas inhibited the level of IL-12 in macrophages (Figure 3J). Altogether, these results demonstrate that miR-106b-containing exosomes derived from EMT-CRC cells can reprogram macrophages to the M2 subtype.
EMT-Exos-derived miR-106b induces M2 macrophage polarization by directly downregulating PDCD4
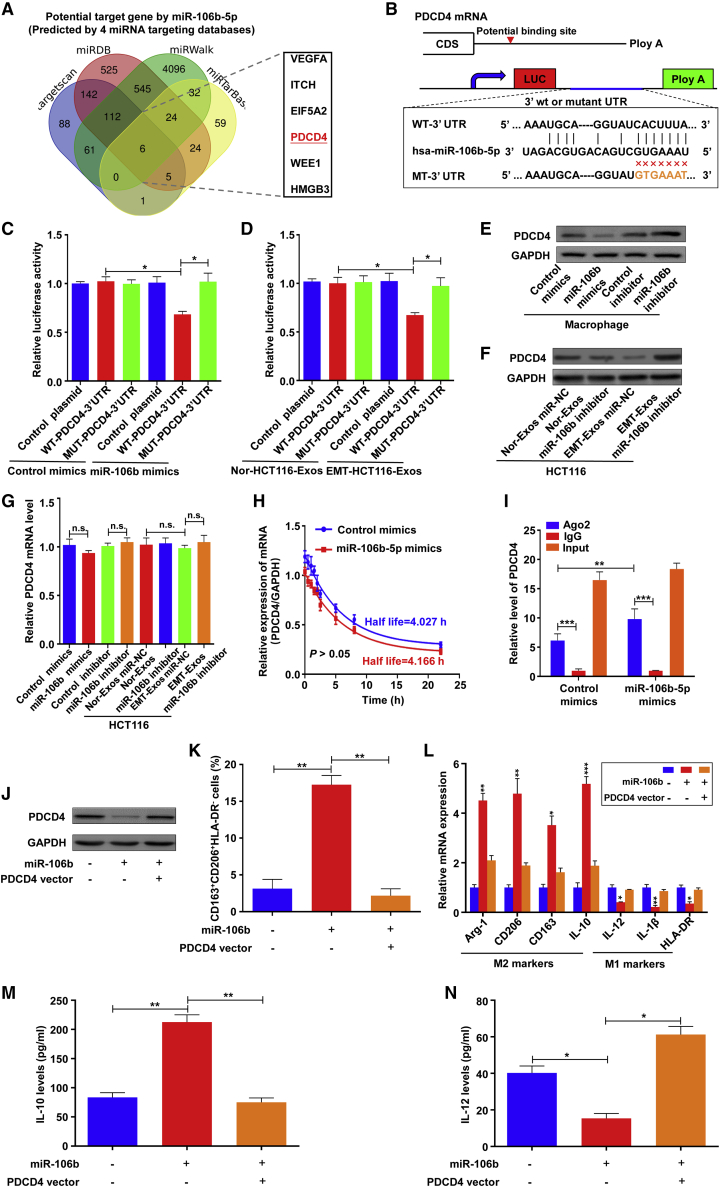
To explore how exosomal miR-106b could regulate M2 macrophage polarization, four mRNA target-predicting algorithms, including TargetScan, miRWalk, miRDB, and miRTarBase, were used to jointly identify the putative downstream targets of miR-106b (Figure 4A). Among the predicted targets, PDCD4, which has been demonstrated to be an important molecule in regulating M1 macrophage polarization,24,25 was overlapped among all databases and was chosen for further validation (Figure 4A). After confirming the potential binding site of miR-106b in the 3′ UTR of PDCD4 (Figure 4B), we performed a luciferase reporter assay in which wild-type (WT)-PDCD4-3′ UTR or mutant (MUT)-PDCD4-3′ UTR was cotransfected with miR-106b mimics or control mimics in macrophages. The results showed that cotransfection of WT-PDCD4-3′ UTR and miR-106b mimics significantly attenuated the luciferase activity, whereas cointroduction of MUT-PDCD4-3′ UTR and miR-106b mimics had no significant effect (Figure 4C). In addition, EMT-Exos-derived miR-106b also effectively decreased luciferase activity in macrophages expressing WT-PDCD4-3′ UTR reporters (Figure 4D; Figure S5A). The above results suggest that exosomal miR-106b can directly target PDCD4 by binding to its 3′ UTR sites. Moreover, the PDCD4 mRNA and protein were also assessed in macrophages transfected with miR-106b mimics or inhibitors. Western blot analysis revealed that the expression of PDCD4 was strongly suppressed by overexpressed miR-106b, whereas the inhibitors of miR-106b relatively enhanced PDCD4 expression (Figure 4E). Similarly, EMT-Exos-derived exosomal miR-106b could also functionally downregulate the expression of the PDCD4 protein in macrophages (Figure 4F; Figure S5B). miRNAs are well known to suppress the expression of downstream target genes at the post-transcriptional level, and as expected, PDCD4 mRNA remained unchanged with the effects of miR-106b mimics or inhibitors or EMT-Exos (Figure 4G; Figure S5C). Furthermore, we explored the specific role of miR-106b-5p for PDCD4 expression. Macrophages were added with actinomycin D after 24 h after transfection with miR-106b-5p mimics or control mimics. Then, the PDCD4 mRNA levels were detected at designated time points. The PDCD4 mRNA levels were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the decay model was fit in a one-phase exponential decay model.26 The results showed that the half-life of PDCD4 mRNA in the miR-106b-5p mimics or control mimics group was 4.027 h and 4.166 h, respectively (Figure 4H; not significant), which revealed that miR-106b-5p did not affect PDCD4 mRNA stability. Moreover, AGO2-RNA immunoprecipitation (RIP) assay demonstrated that PDCD4 mRNA was strongly enriched in the AGO2 immunoprecipitates compared with negative control (NC) immunoglobulin G (IgG) immunoprecipitates, and the transfection of miR-106b-5p mimics significantly increased PDCD4 mRNA enrichment compared with the control mimics group (Figure 4I). Taken together, these data reveal that EMT-Exos-derived miR-106b regulates PDCD4 expression in macrophages at a post-transcriptional level by directly targeting the 3′ UTR of PDCD4 mRNA.
Figure 4.
EMT-Exos-derived miR-106b directly targets PDCD4 in macrophages to induce M2 macrophage polarization
(A) Target gene prediction of miR-106b-5p with four bioinformatics tools. (B) Schematic diagram of the wild-type and mutate-type binding site between miR-106b-5p and the PDCD4 3′ UTR. (C) Relative luciferase activity of reporter plasmids carrying wild-type or mutant PDCD4 3′ UTR in macrophages cotransfected with miR-106b mimics or control mimics. (D) Relative luciferase activity of reporter plasmids carrying wild-type or mutant PDCD4 3′ UTR in macrophages treated with Nor-HCT116-Exos or EMT-HCT116-Exos. (E) Western blot analysis of PDCD4 expression in macrophages transfected with miR-106b mimics/inhibitor. (F) Western blot analysis of PDCD4 expression in macrophages treated with exosomes from normal and EMT-HCT116 cells that transfected control inhibitor or miR-106b inhibitor. (G) qRT-PCR analysis of PDCD4 mRNA expression in macrophages treated with exosomes from normal and EMT-HCT116 cells that transfected control inhibitor or miR-106b inhibitor. (H) PDCD4 mRNA decay curves of macrophages carrying miR-106b-5p mimics or control mimics. The decay model was fit in a one-phase exponential decay model. (I) AGO2-RNA immunoprecipitation (RIP) assay showing that miR-106b-5p interacted with PDCD4 mRNA in macrophages. (J) PDCD4 expression in macrophages transfected with miR-106b mimics or/and PDCD4-overexpression plasmids. (K) Flow cytometry for analyzing the population of CD163highCD206highHLA-DRlow macrophages transfected with miR-106b mimics or/and PDCD4-overexpression plasmids. (L) qRT-PCR analyses of the macrophage-associated markers in macrophages transfected by miR-106b mimics or/and PDCD4-overexpression plasmids. (M and N) ELISA for analyzing the secretion of IL-10 and IL-12 in macrophages transfected with miR-106b mimics or/and PDCD4-overexpression plasmids. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To further clarify the function of miR-106b-induced M2 macrophage polarization through PDCD4 repression, the PDCD4 vector and control vector were transfected into miR-106b-overexpressing macrophages. As expected, western blot showed that PDCD4 was significantly downregulated by miR-106b transfection but was rescued after transfection with the PDCD4 vector (Figure 4J). Functionally, flow cytometry, qRT-PCR, and ELISA analyses demonstrated that the promoting effects of miR-106b on M2 macrophage polarization were abolished by ectopic PDCD4 expression (Figures 4K−4N). Taken together, these results suggest that EMT-Exos-derived miR-106b promotes M2 macrophage polarization by directly downregulating PDCD4 in macrophages.
EMT-Exos-derived miR-106b induces M2 macrophage polarization via the PDCD4/PI3Kγ/AKT/mTOR axis
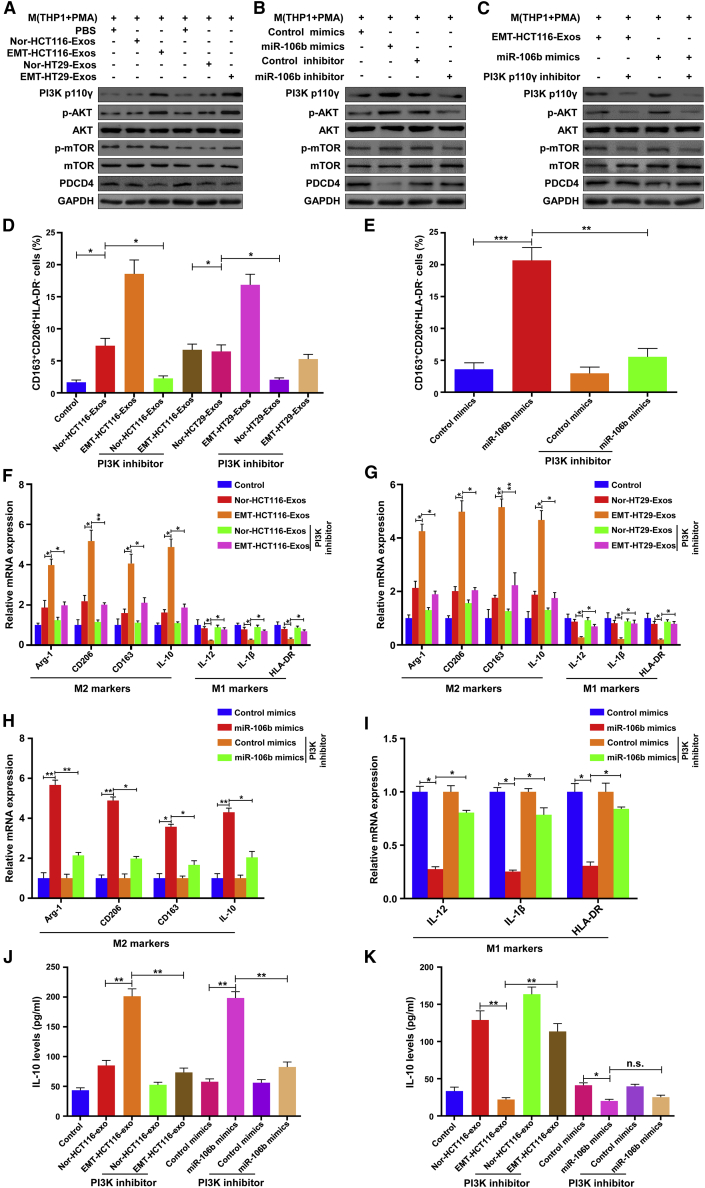
Numerous studies have indicated that the activation of the PI3Kγ/AKT signaling axis in macrophages is involved in the M2 polarization of macrophages.16,27,28 Moreover, it has been reported that PDCD4 participated in the regulation of the PI3K/AKT/mTOR pathway in multiple cellular activities.29,30 With the combination of previous findings with our above results, we speculated that the PI3Kγ/Akt/mTOR signaling pathway might contribute to M2 macrophage polarization mediated by the EMT-Exos-derived miR-106b/PDCD4 axis. To verify the above assumption, western blot was performed to detect the expression of PI3Kγ, AKT, and mTOR in macrophages treated with Nor- and EMT-Exos derived from CRC cells (HCT116 and HT29). The results showed that the stimulation of EMT-Exos decreased PDCD4 expression and increased the expressions of the phosphorylation level of PI3Kγ (p-PI3Kγ), AKT (p-AKT), and mTOR (p-mTOR) in macrophages (Figure 5A). To investigate that EMT-Exos activate the PI3Kγ/AKT/mTOR pathway in macrophages through delivering miR-106b, miR-106b mimics or inhibitors were transfected into macrophages to examine the activation of the PDCD4/PI3Kγ pathway. As shown in Figure 5B, overexpression of miR-106b in macrophages significantly downregulated the expression of PDCD4 and upregulated the expression of p-PI3Kγ, p-AKT, and p-mTOR, whereas inhibition of miR-106b exhibited the opposite effects. To further confirm whether EMT-Exos derived miR-106b via the PDCD4/PI3Kγ pathway, macrophages were treated with TG100-115, a PI3K p110γ inhibitor, prior to coculture with EMT-Exos, or transfected with miR-106b mimics (Figure 5C). The following results found that the expression of p-AKT and p-mTOR was downregulated in PI3Kγ inhibition groups. Furthermore, flow cytometry, qRT-PCR, and ELISA analyses were performed and determined that EMT-Exos-derived miR-106b induced M2 macrophage polarization via regulating the PDCD4/PI3Kγ pathway (Figures 5D−5I). Collectively, these results indicate that EMT-Exos-derived miR-106b activates the PI3Kγ/AKT/mTOR signaling pathway to promote M2 macrophage polarization through inhibiting PDCD4 expression in macrophages.
Figure 5.
EMT-Exos-derived miR-106b induces M2 macrophage polarization via the PDCD4/PI3Kγ/AKT/mTOR axis
(A) Western blot analysis of the expression of PDCD4, PI3Kγ, and phosphorylation of AKT and mTOR in macrophages coincubated with Nor/EMT-Exos derived from HCT116/HT29 cells. (B) Western blot analysis of the expression of PDCD4, PI3Kγ, and phosphorylation of AKT and mTOR in macrophages transfected with miR-106b mimics or miR-106b inhibitors. (C) Western blot analysis of the expression of PDCD4, PI3Kγ, and phosphorylation of AKT and mTOR in macrophages treated with EMT-HCT116-Exos or transfected with miR-106b mimics and post-treated with the PI3K p110γ inhibitor. (D and E) Flow cytometry analyses show the PI3Kγ inhibitor decreases the population of CD163highCD206highHLA-DRlow macrophages induced by EMT-Exos or miR-106b mimics. (F−I) qRT-PCR analyses show that the PI3Kγ inhibitor decreases the expression of M2 markers, whereas increase the expression of M1 markers in macrophages induced by EMT-Exos or miR-106b mimics. (J and K) ELISA shows that the PI3Kγ inhibitor decreases the secretion of IL-10, whereas increase IL-12 levels in macrophages induced by EMT-Exos or miR-106b mimics. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Activated M2 macrophages promote EMT, migration, and invasion of CRC cells in vitro
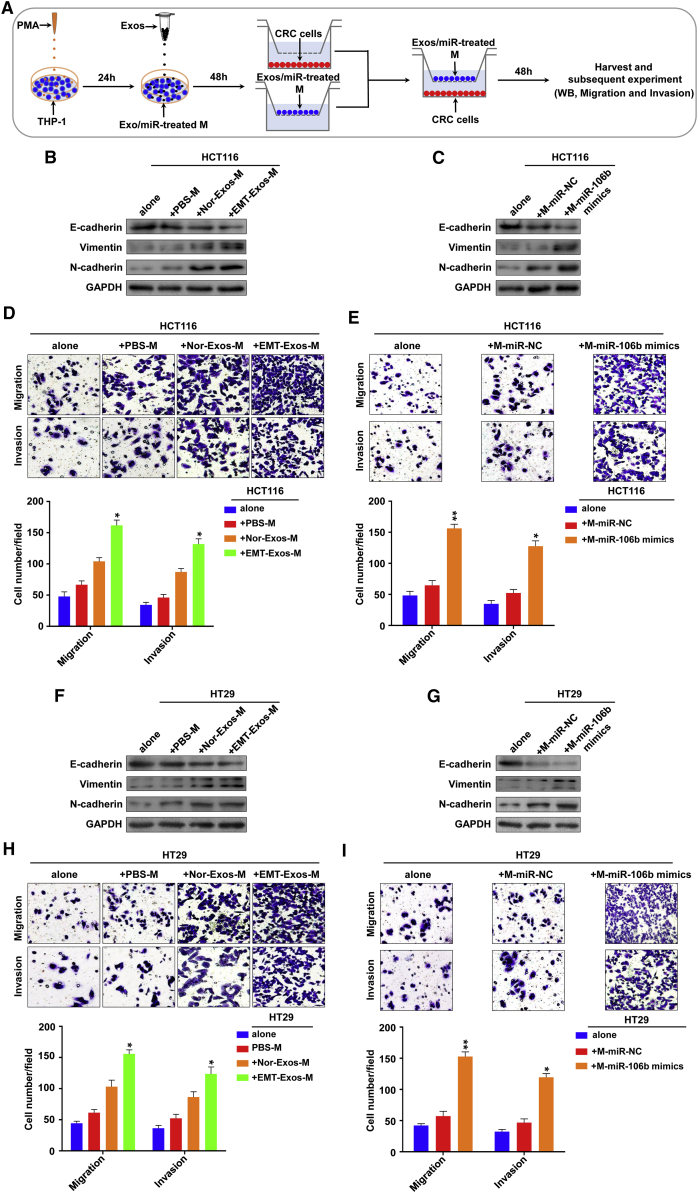
Previously, our and other groups have demonstrated that M2 TAMs promote the EMT of tumor cells by secreting immunosuppressive factors, such as transforming growth factor (TGF)-β and IL-6 to facilitate tumor progression.10,20 To determine whether M2 macrophages activated by exosomal miR-106b contribute to the promotion of EMT, migration, and invasion of CRC cells, we first conducted a set of experiments in vitro. Macrophages were divided into two groups: one group was treated with EMT-HCT116-Exos or EMT-HT29-Exos; the other group was transfected with miR-106b mimics or control mimics. These macrophages were indirectly cocultured with HCT116 or HT29 cells by using an in vitro noncontact Transwell coculture system, as we previously used9,10,20 (Figure 6A). The results demonstrated that after coculturing with the macrophages that were treated by EMT-HCT116-Exos or transfected with miR-106b mimics for 48 h, the expression levels of E-cadherin were significantly decreased, whereas the expression levels of N-cadherin and vimentin markedly increased in HCT116 cells at both mRNA (Figures S6A and S6B) and protein (Figures 6B and 6C) levels. Further Transwell migration and invasion assays showed that macrophages treated by EMT-HCT116-Exos or transfected with miR-106b mimics significantly increased the migration and invasion of HCT116 cells (Figures 6D and 6E). Consistently, similar results were found in HT29 cells (Figures 6F−6I; Figures S6C and S6D).Together, these data suggest that activated M2 macrophages by EMT-Exos-derived miR-106b promote CRC cells’ EMT in a positive-feedback manner, thereby enhancing the migration and invasion abilities of CRC cells in vitro.
Figure 6.
Activated M2 macrophages promote EMT, migration, and invasion of CRC cells in vitro
(A) Schematic representation of the in vitro model for cell coculture. (B and C) Western blot analysis of the expression of the epithelial marker (E-cadherin) and the mesenchymal marker (N-cadherin and vimentin) in HCT116 cells that cocultured with the macrophages treated by EMT-HCT116-Exos or transfected with miR-106b mimics for 48 h. (D and E) Transwell assays for evaluating the migration and invasion capacity of HCT116 cells cocultured with the macrophages treated by EMT-HCT116-Exos or transfected with miR-106b mimics for 48 h. (F and G) Western blot analysis of the expression of the epithelial marker (E-cadherin) and the mesenchymal marker (N-cadherin and vimentin) in HT29 cells that cocultured with the macrophages treated by EMT-HT29-Exos or transfected with miR-106b mimics for 48 h. (H and I) Transwell assays for evaluating the migration and invasion capacity of HT29 cells cocultured with the macrophages treated by EMT-HT29-Exos or transfected with miR-106b mimics for 48 h. Experiments were performed in triplicates. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. ∗p < 0.05, ∗∗p < 0.01.
EMT-Exos-derived miR-106b induces M2 macrophage polarization to facilitate CRC metastasis in vivo
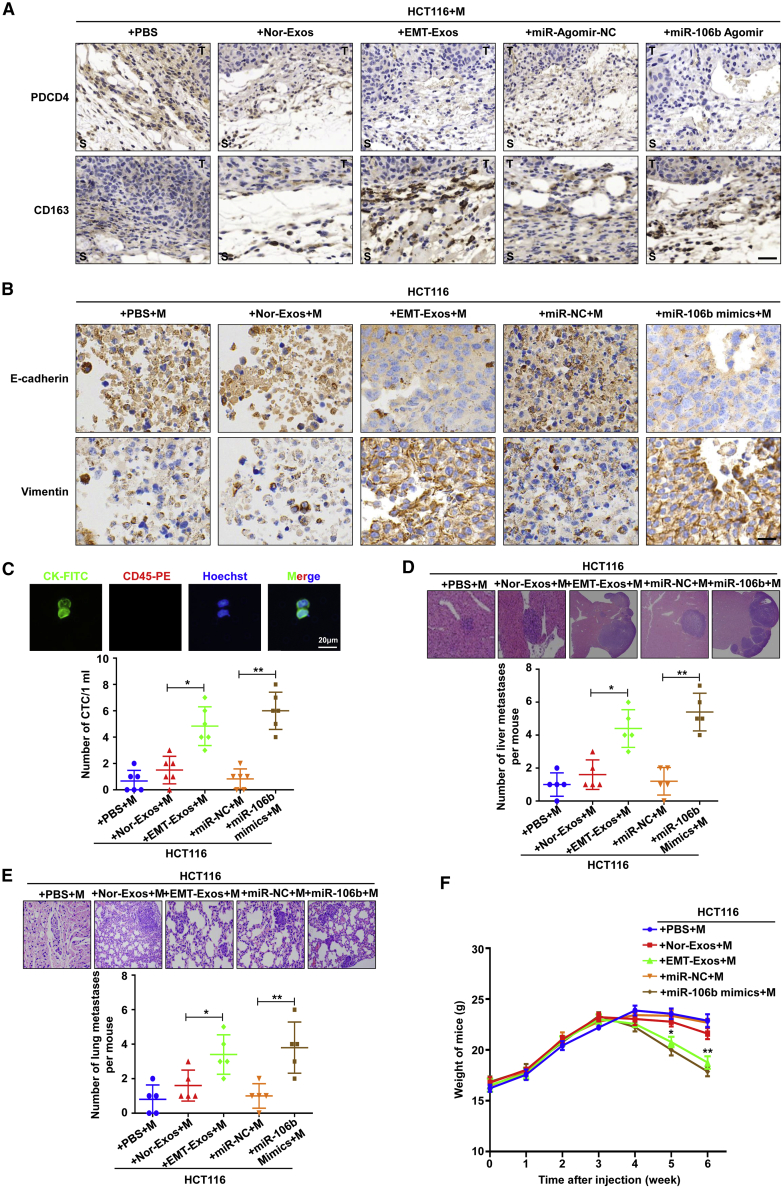
Then, we explored the role of EMT-Exos-derived miR-106b in M2 macrophage polarization by in vivo experiments. HCT116 cells were subcutaneously injected into the right flank region of mice; meanwhile, THP1-induced macrophages were injected into the mice via tail-vein interval for 3 days. Then, implanted tumors were treated with Nor-Exos, EMT-Exos, miR-106b agomir, or miR-agomir-NC, respectively. The results analyzed by immunohistochemistry (IHC) and qRT-PCR demonstrated that EMT-Exos-derived miR-106b could induce M2 polarization of TAMs by post-transcriptionally suppressing PDCD4 expression (Figure 7A; Figure S7A). These findings indicate that EMT-Exos-derived miR-106b induces M2 macrophage polarization.
Figure 7.
EMT-Exos-derived miR-106b induces M2 macrophage polarization to facilitate CRC metastasis in vivo
(A) Immunohistochemistry (IHC) analyses of the expression of PDCD4 and CD163 in implanted tumor tissues that received indicated treatment (scale bar, 50 μm). (B) IHC analyses of the expression of E-cadherin and vimentin in implanted tumor tissues that received indicated treatment (scale bar, 20 μm). (C) Representative image and quantification of CTCs captured from blood of mice that received indicated treatment (scale bar, 20 μm). (D and E) Representative images and quantification of liver and lung metastasis in nude mice that received indicated treatment. (F) Weight of nude mice was monitored weekly after receiving indicated treatment. Error bars, SEM. Statistical analysis was conducted using one-way ANOVA. ∗p < 0.05, ∗∗p < 0.01.
Furthermore, we explored the role of EMT-Exos-derived, miR-106b-activated M2 macrophages in EMT of CRC cells by using an in vivo xenograft model. HCT116 cells that cocultured with the conditioned macrophages (stimulated by EMT-Exos or transfected by miR-106b mimics) were subcutaneously injected to establish the xenograft model. The results showed that the implanted tumors in the EMT-Exos + M group were significantly larger and heavier than those in the PBS + M or Nor-Exos + M group (Figure S7B). Similarly, larger and heavier tumors were also observed in the miR-106b agomir + M group compared with the miR-agomir-NC + M group (Figures S7C and S7D). More importantly, further analyses showed that activated M2 TAMs could promote the EMT program of CRC cells (Figure 7B). In addition, the enhanced EMT of tumor cells was reported to promote circulating tumor cell (CTC) generation, which is considered to be the precursor of tumor metastasis.31,32 Thus, we further detected CTC count from blood of every mouse by using our previously developed device.9,10,20,33 The representative CTC images were presented in Figure 7D, and quantitative analysis found that the count of CTC was significantly increased in the EMT-Exo + M or miR-106b mimics + M group compared with the other three groups (Figure 7C). These findings were similar to the in vitro results, substantiating that EMT-Exos-activated M2 macrophages promote the EMT of CRC cells.
To further evaluate the function of M2 macrophages activated by EMT-Exos-derived miR-106b in promoting the metastasis of CRC in vivo, HCT116 cells that cocultured with the conditioned macrophages (stimulated by EMT-Exos or transfected by miR-106b mimics) were injected into the mice via tail vein. After 6 weeks, we observed that more metastatic nodules in both liver and lung were generated in the EMT-Exos + M group than the PBS + M or Nor-Exos + M group (Figures 7D and 7E). Likewise, there were more visible liver and lung and metastatic nodules in the miR-106b mimics + M group compared with the control group (Figures 7D and 7E). Moreover, the weight of mice in the EMT-Exos + M or miR-106b mimics + M group was significantly decreased compared with that in the other three groups (Figure 7F). Overall, these results indicate that EMT-Exos-derived miR-106b contributes to induce M2 macrophage polarization to promote the EMT of tumor cells, thereby facilitating CRC metastasis in vivo.
miR-106b upregulation in plasma exosomes and tumor tissues indicates malignant transformation of CRC
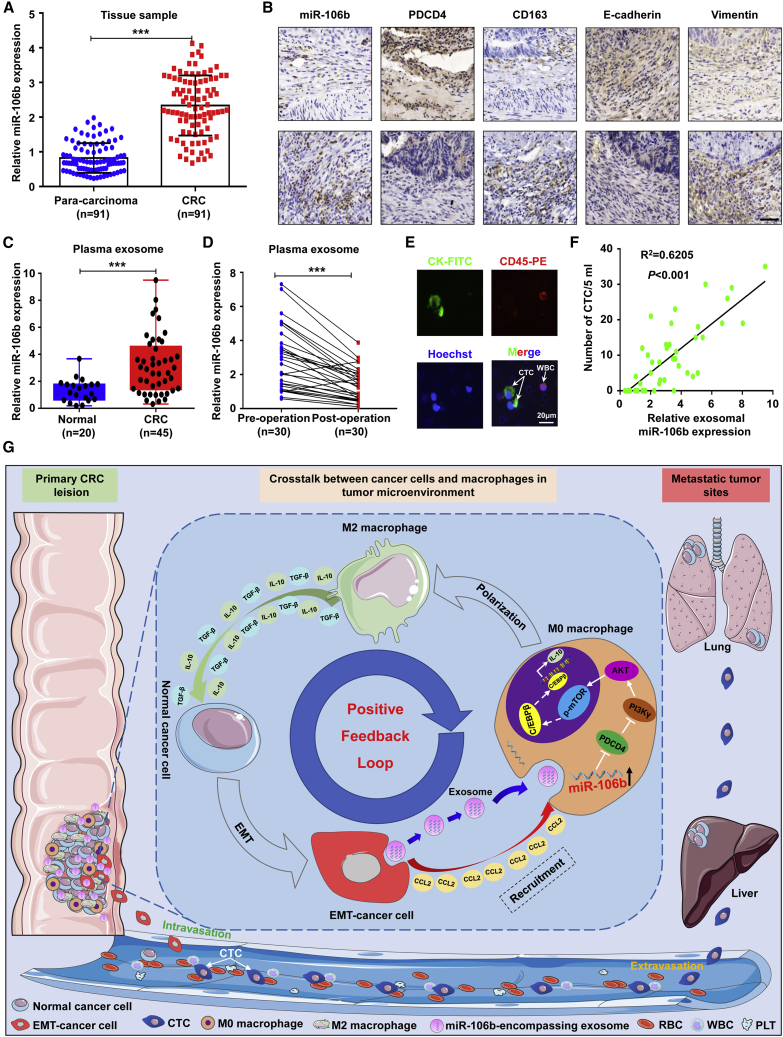
To extend current knowledge to clinical application, we first investigated whether miR-106b accounts for M2 macrophage polarization and malignant progression in clinical CRC samples. As shown in Figure 8A, the expression of the miR-106b level was upregulated in CRC tissues compared with adjacent normal tissues (n = 91; p < 0.05). Further, in situ hybridization (ISH) and IHC results confirmed that miR-106b was closely related to M2 macrophage polarization and the EMT of tumor cells in the TME (Figure 8B; Figures S8A and S8B). Then, we analyzed the correlations between miR-106b expression in tissues and clinicopathological features of CRC patients. The results showed that positive associations were identified between upregulated miR-106b levels in CRC tissues and poor tumor grade, deep tumor invasion, lymph node metastasis, advanced tumor stage, and positive preoperative CTC status (p < 0.05, respectively; Table S1). Moreover, Kaplan-Meier analysis showed that high miR-106b levels in CRC tissues were correlated with reduced recurrence-free survival (RFS) and overall survival (OS) of CRC patients (p < 0.05, respectively; Figures S8C and S8D), and Cox proportional hazards regression analysis further demonstrated that high miR-106b expression was an independent prognostic factor for affecting the RFS and OS of CRC patients (p < 0.05, respectively; Table S2).
Figure 8.
miR-106b upregulation in plasma exosomes and tumor tissues indicates malignant transformation of CRC
(A) qRT-PCR analysis of the expression of miR-106b in tumor tissues and matched para-carcinoma tissues from CRC patients (n = 91). (B) In situ hybridization of miR-106b-5p in combination with IHC staining of PDCD4, CD163, E-cadherin, and vimentin on serial sections of human hepatocellular carcinoma (HCC) tissues. Scale bar, 100 μm. (C) qRT-PCR analysis of the expression of miR-106b in plasma exosomes from CRC patients (n = 45) and healthy donors (n = 20). (D) qRT-PCR analysis of exosomal miR-106b level in matched plasma from CRC patients pre- and postoperation (n = 30). (E) Representative image of CTC from blood sample of one CRC patient. Scale bar, 20 μm. (F) Correlation analysis between miR-106 expression level in clinical plasma samples and CTC count in peripheral blood of CRC patients. (G) Proposed schematic diagram of EMT-tumor exosomal miR-106b-5p-mediating macrophage M2 polarization to promote tumor progression of CRC. Error bars, SEM. Statistical analysis was conducted using Student’s t test or one-way ANOVA or Pearson correlation analysis. ∗∗p < 0.01, ∗∗∗p < 0.001.
Furthermore, we examined the clinical significance of exosomal miR-106b that was expressed in clinical plasma samples. qRT-PCR results showed that plasma exosomal miR-106b levels were significantly elevated in CRC patients compared with healthy controls (p < 0.05; Figure 8C), similar to the results in tumor tissues. After tumor resection, plasma exosomal miR-106b levels were decreased in most CRC patients (Figure 8D), indicating that plasma exosomal miR-106b was mainly secreted by tumor tissue. In addition, we found that increased exosomal miR-106b expression in plasma samples was positively correlated with the number of CTC in the peripheral blood of CRC patients (p < 0.05, Figures 8E and 8F). Further analysis revealed that plasma exosomal miR-106b upregulation was correlated with poor tumor grade, lymphovascular invasion, deep tumor invasion, lymph node metastasis, advanced tumor stage, and positive preoperative CTC status in CRC patients (p < 0.05, respectively; Table 1). Taken together, these clinical data suggest that miR-106b is upregulated in both plasma exosomes and CRC tissues, and increased miR-106b level is closely related to tumor malignant transformation of CRC, indicating that exosomal miR-106b could serve as a valuable biomarker for CRC patients.
Table 1.
Relationship between serum exosomal miR-106b level and clinicopathological features of CRC patients (n = 45)
| Parameters | No. of patients |
miR-106b expression |
χ2 value | p value | ||
|---|---|---|---|---|---|---|
| No. | % | <median | ≥median | |||
| Gender | ||||||
| Male | 27 | 60.0 | 12 | 15 | 0.534 | 0.465 |
| Female | 18 | 40.0 | 10 | 8 | ||
| Age, years | ||||||
| <60 | 17 | 37.8 | 7 | 10 | 0.650 | 0.420 |
| ≥60 | 28 | 62.2 | 15 | 13 | ||
| Tumor site | ||||||
| Colon | 24 | 53.3 | 13 | 11 | 0.573 | 0.449 |
| Rectal | 21 | 46.7 | 9 | 12 | ||
| Tumor size, cm | ||||||
| <5 | 20 | 44.4 | 8 | 12 | 1.138 | 0.286 |
| ≥5 | 25 | 55.6 | 14 | 11 | ||
| Tumor grade | ||||||
| Moderate/well | 21 | 46.7 | 14 | 7 | 4.980 | 0.026∗ |
| Poor | 24 | 53.3 | 8 | 16 | ||
| Lymphovascular invasion | ||||||
| Absence | 26 | 57.8 | 16 | 10 | 3.943 | 0.047∗ |
| Presence | 19 | 42.2 | 6 | 13 | ||
| Perineural invasion | ||||||
| Absence | 28 | 62.2 | 16 | 12 | 2.021 | 0.155 |
| Presence | 17 | 37.8 | 6 | 11 | ||
| Tumor invasion | ||||||
| T1-2 | 12 | 26.7 | 9 | 3 | 4.465 | 0.035∗ |
| T3-4 | 33 | 73.3 | 13 | 20 | ||
| Lymph node metastasis | ||||||
| N0-1 | 19 | 42.2 | 13 | 6 | 5.021 | 0.025∗ |
| N2-3 | 26 | 57.8 | 9 | 17 | ||
| TNM stagea | ||||||
| I/II | 24 | 53.3 | 16 | 8 | 6.505 | 0.011∗ |
| III | 21 | 46.7 | 6 | 15 | ||
| CEA, ng/mL | ||||||
| <5 | 32 | 71.1 | 18 | 14 | 2.402 | 0.121 |
| ≥5 | 13 | 28.9 | 4 | 9 | ||
| Preoperative CTC status | ||||||
| Negative | 12 | 26.7 | 9 | 3 | 4.465 | 0.035∗ |
| Positive | 33 | 73.3 | 13 | 20 | ||
| Overall | 45 | 10.0 | 22 | 23 | ||
∗p < 0.05. TNM, tumor-node-metastasis; CEA, carcinoembryonic antigen; CTC, circulating tumor cell.
The 8th edition of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual.
Discussion
The immunosuppressive TME, a dynamic system orchestrated by intercellular communications, is composed of diverse factors and cells.34 Macrophages, especially M2-subtype macrophages, are the most common immune components in immunosuppressive TME, which have been demonstrated to play a dramatic, significant role in tumor progression.6 EMT, as an important hallmark of solid tumors, has recently been demonstrated to be a key driving factor for macrophage polarization.7 Our previous studies have established the crosstalk between M2-TAMs and CRC cells that are mediated by chemokines and cytokines, which are responsible for maintaining tumor cells’ EMT status and tumor metastasis.9,10,20 Emerging evidence has shown that cancer cells and macrophages can interact through exosome-mediated bioactive molecule exchange.14 Therefore, it is necessary to study the relationships among tumor cells, EMT, and macrophages mediated by exosomes. In this study, we purified exosomes from the supernatant of EMT-CRC cells to investigate their role and mechanisms of M2 macrophage polarization. A series of consequent experiments showed that EMT-Exos could induce the alternative activation of macrophages toward the M2 phenotype, which facilitated the EMT of CRC cells in a feedback way and thus promoted the migration, invasion, and metastasis of CRC both ex vivo and in vivo. Our findings suggest that EMT-Exos are important components of the TME, which involve in the formation of immunosuppressive TME by acting as an important messenger to mediate the crosstalk between tumor cells and macrophages, thereby facilitating CRC progression.
Exosomes are small extracellular vesicles secreted by multiple-type cells under different conditions.13 Numerous previous studies have demonstrated that hypoxia could promote the release of exosomes from multiple tumor cells, including glioma, lung cancer, pancreatic cancer, and epithelial ovarian cancer, to regulate different cell signaling and thus exert diverse biological functions.15, 16, 17, 18 In the present study, we found that under EMT status, CRC cells secreted more exosomes than normal CRC cells, which in accordance with the previous results reported by Hsieh and colleagues,19 suggested that the EMT program could enhance the exosome secretion of tumor cells. Recently, exosome-mediated intercellular communication is gradually proposed to be an effective manner in which to regulate the biological behaviors of recipient cells via delivering proteins and genetic materials (mRNA, miRNA, long noncoding RNA [lncRNA], etc.) to recipient cells.11,12 Growing evidence has shown that tumor-derived exosomal miRNAs play an important role in the M2 polarization of macrophages.15, 16, 17,19,21,35,36 Under hypoxic condition, glioma cell-derived exosomal miR-1246, pancreatic cancer-derived exosomal miR-301a, lung cancer-derived exosomal miR-103a and epithelial ovarian cancer-derived exosomal miR-21-3p, miR-125b-5p, and miR-181d-5p could be transferred to macrophages to induce their M2-like polarization.15, 16, 17, 18 Noteworthily, a recent study showed that the EMT transcriptional factor (EMT-TF) snail-overexpressing head and neck squamous cell carcinoma cells promoted M2 macrophage polarization by delivering miR-21-abundant exosomes.19 In this study, we analyzed the profiles and patterns of miRNAs in Nor-Exos and EMT-Exos by miRNA sequencing and found that more than 20 miRNAs were upregulated in EMT-Exos compared with Nor-Exos. Among them, miR-106b was the most enriched miRNA in EMT-Exos, implying that miR-106b might play a more important role than the rest. Previously, miR-106b has been reported to act as an onco-miRNA (onco-miR) through directly regulating downstream target genes in multiple solid cancers, including gastric cancer,37 hepatocellular carcinoma,38 breast cancer,39 and renal carcinoma.40 Inconsistently, miR-106b might exert a dual role as a tumor suppressor or an oncogene in CRC development. On one hand, Zheng et al.41 reported that miR-106b overexpression could induce cell radioresistance by regulating the phosphatase and tensin homolog (PTEN)/PI3K/AKT axis and p21 expression in CRC. Zhang and colleagues42 showed miR-106b promoted the migration and invasion of CRC cells via directly targeting DLC1. Two miRNA microarray analyses also demonstrated that they were was highly expressed in CRC tissues, and high levels of miR-106b were significantly associated with poor prognosis of CRC patients.43,44 On the other hand, Zhuang et al.45 found that the expression of miR-106b in CRC was obviously downregulated, and high miR-106b levels predicted a favorable OS in CRC patients. Herein, we found that miR-106b was significantly upregulated in CRC tissues, and high miR-106b expression was an independent prognostic factor for affecting the RFS and OS of CRC patients. Moreover, for the first time, we demonstrate that miR-106b could be transferred from EMT-CRC cells to macrophages via exosomes to induce them toward an immunosuppressive M2 phenotype. In a feedback way, these activated macrophages promoted the EMT of CRC cells, thereby facilitating tumor metastasis. Interestingly, these results reveal a novel mechanism by which miR-106b indirectly plays an oncogene role in CRC progression by participating in the interplay between EMT-CRC cells and macrophages in TME.
PDCD4, a known tumor-suppressor protein, is frequently downregulated in various types of human solid cancers, including CRC, and the loss of PDCD4 contributes to tumor initiation, progression, and metastasis.46,47 Recently, PDCD4 was reported to play an important role in M1 macrophage polarization.19,24,25 Several interesting studies have shown that PDCD4 knockout mice are less susceptible to lipopolysaccharide (LPS) toxicity25 and are resistant to inflammatory disease,24 suggesting that PDCD4 possesses potent proinflammatory properties, and suppression of PDCD4 can result in M2 polarization of macrophages. This was confirmed in subsequent studies performed by Hsieh and colleagues,19 which demonstrated that tumor-derived exosomal miR-21 could promote M2-like polarization of macrophages by directly suppressing PDCD4 expression. In this study, we identified miR-106b as a key upstream regulator of PDCD4, suggesting that it is one mechanism related to M2 macrophage polarization. Our results showed that EMT-CRC cell-derived exosomal miR-106b could bind to the 3′ UTR to post-transcriptionally inhibit the expression of the PDCD4 level in macrophages, thereby promoting their M2 polarization.
PI3K p110γ, as one number of the class I PI3K lipid kinase family, involves partly promoting tumor development though stimulating neutrophil polarization and mediating immune suppression.27,28 A recent study reported that macrophage PI3Kγ signaling through AKT- and mTOR-stimulated C/EBPβ activation, which is a crucial transcription factor involved in M2 macrophage polarization, thereby drives transcriptional programming of M2-subtype macrophages that promote immune suppression during tumor growth.28 In addition, hypoxic pancreatic cancer-derived exosomal miR-301a and lung cancer-derived exosomal miR-103a mediated M2 macrophage polarization via targeting the PTEN/PI3Kγ/AKT axis to promote tumor metastasis.16,17 These findings indicate that the PI3Kγ/AKT/mTOR signaling pathway is closely associated with M2 macrophage polarization. In our study, we showed that EMT-Exos-derived exosomal miR-106b promoted the polarization of M2 macrophages via stimulating the activation of PI3Kγ/AKT/mTOR signaling, thereby promoting the migration, invasion, and EMT of CRC cells in vitro, as well as liver and lung metastasis in vivo. Previously, growing studies have demonstrated that PDCD4 loss was inversely related to constitutive activation of the PI3K/AKT signaling axis,29,30 indicating that PDCD4 leads to the inactivation of the PI3K/AKT pathway. In this study, we demonstrated that EMT-Exos-derived exosomal miR-106b could inhibit the expression of PDCD4 in macrophages and thus, induced M2 macrophage polarization by activating the PI3Kγ/AKT/mTOR pathway, thereby facilitating the malignant progression of CRC cells in vitro and in vivo. Our results first highlight the important role of the PDCD4/PI3Kγ axis in M2-subtype polarization of macrophages.
Liquid biopsy, which refers to the molecular analysis of disease genetic phenotypes based on circulating genetic material (such as circulating tumor DNA [ctDNA], miRNA, and exosome) in body fluids, is already reported to be a useful diagnostic and prognostic tool in various kinds of solid tumors, including CRC.48,49 Recent studies have shown that exosomal noncoding RNAs (ncRNAs) present in a stable form because the exosomal vesicle can protect them from being degraded by endogenous RNase.21,50Therefore, exosome-encapsulated ncRNAs are more accurate and better biomarkers for cancer management. Especially, plasma exosomal miRNAs, as a new liquid biopsy source in free form, are promising diagnostic and prognostic biomarkers for cancers.51,52 In this study, we demonstrated that the levels of exosomal miR-106b were significantly elevated in CRC patients’ plasma compared with healthy controls, and the upregulation of serum exosomal miR-106b levels was positively correlated with unfavorable clinicopathological features of CRC patients, including poor tumor grade, lymphovascular invasion, deep tumor invasion, lymph node metastasis, advanced tumor stage, and positive preoperative CTC status. In addition, a dramatic drop of exosomal miR-106b levels was observed in most CRC patients after tumor resection. Collectively, our clinical data suggest that a quantitative plasma test for the miR-106b level in circulating exosomes would be useful for selection of the patients with a high risk of metastasis for preventive treatment. However, the clinical diagnostic and prognostic efficiency of plasma exosomal miR-106b in CRC patients needs to be further confirmed in expanded CRC cohorts.
In summary, our results show that EMT tumor-derived exosomal miR-106b-5p promotes macrophages to M2-like polarization via downregulating PDCD4 to activate the PI3Kγ/AKT/mTOR signaling pathway. Activated macrophages facilitate the intravasation of tumor cells to mediate CTC generation by promoting the EMT of tumor cells in a feedback manner, thereby facilitating the liver and lung metastasis of CRC (Figure 8G). These findings illustrate new crosstalk between tumor-macrophage and CRC cells in TME through communication with exosomes during EMT, which explains why EMT-tumor cells show increased metastasis ability and uncover a novel molecular mechanism of CRC metastasis, highlighting secreted miR-106b-5p as a promising diagnostic and therapeutic target for CRC.
Materials and methods
Detailed materials and methods are provided in Supplemental materials and methods.
Patient samples
A volume of 5 mL peripheral blood samples were collected in EDTA-containing tubes (Becton Dickinson [BD], USA) from CRC patients (n = 45) and healthy volunteers without any malignancy (n = 20) for plasma exosome isolation. Primary CRC and paired paracarcinoma (distance to cancer >5 cm) tissue samples were obtained from 91 patients, and 5 mL peripheral blood samples were also collected in EDTA-containing tubes from these patients at the time of hospitalization for CTC detection. All enrolled patients were diagnosed as adenocarcinoma of colorectal by histopathology and underwent curative resection in the Zhongnan Hospital of Wuhan University (Wuhan, China). Moreover, all patients who were devoid of neoadjuvant chemotherapy or/and radiotherapy had available preoperative CTC and survival data. This study was endorsed by the Ethics Committee of Zhongnan Hospital of Wuhan University in compliance with the Helsinki Declaration. Written, informed consents were obtained from all participants prior to sample collection.
Exosome isolation, identification, and quantification
The cell lines were cultured in the normal medium until 80%–90% confluent; thereafter, medium was replaced with RPMI 1640 with 10% exosome-depleted fetal bovine serum (FBS; 160,000 g, 16 h). After 72 h, the cell CM was harvested. The exosomes from the normal (Nor-Exos) and EMT (EMT-Exos) CRC cell CM were isolated by the ultracentrifugation method and characterized by using a transmission electron microscope (JEM-100CX-II; Jeol, Japan), NanoSight analysis (NanoSight NS-300 instrument; UK), and western blot, as previously described.35 Exosomes from patients’ plasma samples were isolated by using exoRNeasy Serum/Plasma MaxiKits (QIAGEN, Germany), according to the manufacturer’s protocol. The exosomes were dissolved with PBS and stored at −80°C. The harvested exosomes were quantified by evaluating the protein concentration of exosomes, which was determined using a bicinchoninic acid (BCA) protein assay kit (Millipore, Billerica, MA, USA). The depletion of exosomes from CM was carried out by a microfiltration ExoMir PLUS Kit (Bioo Scientific, USA).
miRNA sequencing and bioinformatics analysis
The miRNA sequencing of Nor-Exos and EMT-Exos was performed by an Illumina HiSeq 2000/2500 platform (Illumina, San Diego, CA, USA). A small RNA sequencing library used for sequencing was prepared using the TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA). Expression data were normalized to fragments per kilobase per million mapped reads (FPKM), and the differentially expressed miRNAs were determined by the DESeq2 package of R software using default values. Significantly differentially expressed miRNAs were defined by log2|fold change| >1 and adjusted p value <0.01. A heatmap was generated with all significant, differentially expressed miRNAs in EMT-Exos compared with that in Nor-Exos. The four online miRNA targeting databases, including TargetScan, miRWalk, miRDB, and miRTarBase, were used to jointly predict the potential targets of our candidate miRNA-miR-106b.
Animal experiments
To examine the effects of EMT-Exos-derived miR-106b on macrophage polarization in vivo, thirty BALB/c nude mice (female, 4−6 weeks old, and 16−20 g) were purchased from Hubei Research Center of Laboratory Animals (Wuhan, China) and were randomly divided into 5 groups (n = 6 per group). Clodronate liposomes were intravenously injected into each mouse for macrophage ablation.35 Then, HCT116 cells (5 × 106 cells per mouse) were subcutaneously injected into the right flank region of each mouse. Meanwhile, THP1-induced macrophages (106 cells/50 μL per mouse) were injected into the mice via tail-vein interval for 3 days. After 10 days, PBS, Nor-Exos, EMT-Exos, miR-106b agomir, or miR-agomir-NC (RiboBio, Guangzhou, China) was directly injected into the implanted tumor. After 30 days of cell injection, the mice were sacrificed, and the transplanted tumors were removed for further experiments. Furthermore, to explore the role of EMT-Exos-derived, miR-106b-activated M2 macrophages in EMT of CRC cells, the thirty above nude mice were randomly divided into 5 groups (n = 6 per group). HCT116 cells that cocultured with the conditioned macrophages (stimulated by PBS, Nor-Exos, and EMT-Exos or transfected by miR-NC and miR-106b mimics) were subcutaneously injected to establish a xenograft model. Then, tumor volume was measured every 5 days. After 30 days, the mice were sacrificed, and the transplanted tumors were removed for visually examination and further detection. 1 mL blood per mouse was collected into EDTA-containing tubes via cardiac puncture. The tumor and blood samples were collected for subsequent experiments. To examine the effects of the conditioned macrophages on tumor metastasis in vivo, the twenty-five aforementioned mice were randomly divided into five groups (n = 5 per group). HCT116 cells that cocultured with the conditioned macrophages (stimulated by PBS, Nor-Exos, and EMT-Exos or transfected by miR-NC and miR-106b mimics) were injected into the mice via tail vein. Then, mice weight was measured every week. After 6 weeks, all mice were euthanized. Then, the liver and lung tissues of all mice were performed with H&E staining to examine the suspicious metastases sites. All animal procedures were carried out in accordance with the Guide for the Care and Use of Laboratory Animals with the approval of Wuhan University Institute Animal Care and Use Committee.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 (SPSS, Chicago, IL, USA), and data were visualized by GraphPad Prism 6.0 (GraphPad Software, USA) for Windows. The differences between two or more groups were analyzed selectively by Student’s t test or one-way ANOVA. The associations between miR-106b level in exosomes or tumor tissues and clinicopathologic parameters were determined using the χ2 test or Fisher’s exact test. Pearson correlation analysis was performed to determine the correlation between two variables. All CRC patients were divided into a high- and low-expression group according to the median of the miR-106b level, and the Kaplan-Meier method with log-rank tests was used for survival analysis. Univariate and multivariate Cox proportional hazards regression analyses were applied to identify the independent factors for affecting CRC patients’ prognoses. In general, all in vitro experiments were performed at least three times, and data were shown as the mean ± SEM. In all cases, a two-tailed value of p < 0.05 was considered statistically significant and was denoted by ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p > 0.05 or not significant (n.s.).
Acknowledgments
The authors appreciate Wuhan YZY Medical Science and Technology Co., Ltd. (Wuhan, China), for providing equipment and excellent technical support for CTC isolation and identification. The raw exosomal miRNA sequence data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database (SRA: SRP227899). All other remaining data are included in the article and Supplemental Information or available from the authors upon reasonable request. This work was supported by grants from the National Natural Science Foundation of China (81572874 and 81872376); National Natural Science Fund Youth Fund of China (81702411); Clinical Medical Research Center of Peritoneal Cancer of Wuhan (2015060911020462); Health Commission of Hubei Province Scientific Research Project (WJ2019H012); and Zhongnan Hospital of Wuhan University, Excellent Doctor Fund Project (ZNYB2020002).
Author contributions
Conception and design, C.Y., S.W., and B.X.; experimental operation, C.Y., R.D., C.W., and K.L.; provision of materials or patients’ information, C.Y., D.S., C.Z., and Q.L.; collection and assembly of data, C.Y. and K.L.; manuscript writing, C.Y., R.D., C.W., and K.L.; manuscript revision, C.Y., S.W., and B.X.; final approval of manuscript, all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.02.006.
Contributor Information
Shuyi Wang, Email: shuyiwang@whu.edu.cn.
Bin Xiong, Email: binxiong1961@whu.edu.cn.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel K., Brakenhoff R.H. Dissecting the metastatic cascade. Nat. Rev. Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vakili-Ghartavol R., Mombeiny R., Salmaninejad A., Sorkhabadi S.M.R., Faridi-Majidi R., Jaafari M.R., Mirzaei H. Tumor-associated macrophages and epithelial-mesenchymal transition in cancer: Nanotechnology comes into view. J. Cell. Physiol. 2018;233:9223–9236. doi: 10.1002/jcp.27027. [DOI] [PubMed] [Google Scholar]
- 6.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 8.Tam W.L., Weinberg R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei C., Yang C., Wang S., Shi D., Zhang C., Lin X., Liu Q., Dou R., Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X., Wang S., Sun M., Zhang C., Wei C., Yang C., Dou R., Liu Q., Xiong B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019;12:20. doi: 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han L., Lam E.W., Sun Y. Extracellular vesicles in the tumor microenvironment: old stories, but new tales. Mol. Cancer. 2019;18:59. doi: 10.1186/s12943-019-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mashouri L., Yousefi H., Aref A.R., Ahadi A.M., Molaei F., Alahari S.K. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syn N., Wang L., Sethi G., Thiery J.P., Goh B.C. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol. Sci. 2016;37:606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Qian M., Wang S., Guo X., Wang J., Zhang Z., Qiu W., Gao X., Chen Z., Xu J., Zhao R. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene. 2020;39:428–442. doi: 10.1038/s41388-019-0996-y. [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L., Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 17.Hsu Y.L., Hung J.Y., Chang W.A., Jian S.F., Lin Y.S., Pan Y.C., Wu C.Y., Kuo P.L. Hypoxic Lung-Cancer-Derived Extracellular Vesicle MicroRNA-103a Increases the Oncogenic Effects of Macrophages by Targeting PTEN. Mol. Ther. 2018;26:568–581. doi: 10.1016/j.ymthe.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Zhou J., Li X., Wang X., Lin Y., Wang X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 2018;435:80–91. doi: 10.1016/j.canlet.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh C.H., Tai S.K., Yang M.H. Snail-overexpressing Cancer Cells Promote M2-Like Polarization of Tumor-Associated Macrophages by Delivering MiR-21-Abundant Exosomes. Neoplasia. 2018;20:775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C., Wei C., Wang S., Shi D., Zhang C., Lin X., Dou R., Xiong B. Elevated CD163+/CD68+ Ratio at Tumor Invasive Front is Closely Associated with Aggressive Phenotype and Poor Prognosis in Colorectal Cancer. Int. J. Biol. Sci. 2019;15:984–998. doi: 10.7150/ijbs.29836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z., Shi K., Yang S., Liu J., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichmüller S.B., Osen W., Mandelboim O., Seliger B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J. Natl. Cancer Inst. 2017;109:djx034. doi: 10.1093/jnci/djx034. [DOI] [PubMed] [Google Scholar]
- 23.Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 24.Hilliard A., Hilliard B., Zheng S.J., Sun H., Miwa T., Song W., Göke R., Chen Y.H. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 25.Sheedy F.J., Palsson-McDermott E., Hennessy E.J., Martin C., O’Leary J.J., Ruan Q., Johnson D.S., Chen Y., O’Neill L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 26.Burow D.A., Umeh-Garcia M.C., True M.B., Bakhaj C.D., Ardell D.H., Cleary M.D. Dynamic regulation of mRNA decay during neural development. Neural Dev. 2015;10:11. doi: 10.1186/s13064-015-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneda M.M., Cappello P., Nguyen A.V., Ralainirina N., Hardamon C.R., Foubert P., Schmid M.C., Sun P., Mose E., Bouvet M. Macrophage PI3Kγ Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016;6:870–885. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneda M.M., Messer K.S., Ralainirina N., Li H., Leem C.J., Gorjestani S., Woo G., Nguyen A.V., Figueiredo C.C., Foubert P. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhen Y., Fang W., Zhao M., Luo R., Liu Y., Fu Q., Chen Y., Cheng C., Zhang Y., Liu Z. miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene. 2017;36:275–285. doi: 10.1038/onc.2016.201. [DOI] [PubMed] [Google Scholar]
- 30.Statement of Retraction. Artif. Cells Nanomed. Biotechnol. 2020;48:797. doi: 10.1080/21691401.2020.1769277. [DOI] [PubMed] [Google Scholar]
- 31.Massagué J., Obenauf A.C. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Micalizzi D.S., Maheswaran S., Haber D.A. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F., Wang S., Fang Y., Zheng L., Zhi X., Cheng B., Chen Y., Zhang C., Shi D., Song H. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application. Oncotarget. 2017;8:3029–3041. doi: 10.18632/oncotarget.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayan D., Young A., Teng M.W.L., Smyth M.J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer. 2017;17:709–724. doi: 10.1038/nrc.2017.86. [DOI] [PubMed] [Google Scholar]
- 35.Frid M.G., Brunetti J.A., Burke D.L., Carpenter T.C., Davie N.J., Reeves J.T., Roedersheimer M.T., van Rooijen N., Stenmark K.R. Hypoxia-induced pulmonary vascular remodeling requires recruitment of circulating mesenchymal precursors of a monocyte/macrophage lineage. Am. J. Pathol. 2006;168:659–669. doi: 10.2353/ajpath.2006.050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y., Dang W., Zhang S., Yue W., Yang L., Zhai X., Yan Q., Lu J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z., Yang Q., Zhang B., Wu W., Yuan F., Zhu Z. miR-106b Promotes Metastasis of Early Gastric Cancer by Targeting ALEX1 in Vitro and in Vivo. Cell. Physiol. Biochem. 2019;52:606–616. doi: 10.33594/000000043. [DOI] [PubMed] [Google Scholar]
- 38.Shi D.M., Bian X.Y., Qin C.D., Wu W.Z. miR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. OncoTargets Ther. 2018;11:571–585. doi: 10.2147/OTT.S152611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li N., Miao Y., Shan Y., Liu B., Li Y., Zhao L., Jia L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017;8:e2796. doi: 10.1038/cddis.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao L.J., Yan S., Zhuang Q.F., Mao Q.Y., Xue D., He X.Z., Chen J.P. miR-106b promotes proliferation and invasion by targeting Capicua through MAPK signaling in renal carcinoma cancer. OncoTargets Ther. 2019;12:3595–3607. doi: 10.2147/OTT.S184674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng L., Zhang Y., Liu Y., Zhou M., Lu Y., Yuan L., Zhang C., Hong M., Wang S., Li X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G.J., Li J.S., Zhou H., Xiao H.X., Li Y., Zhou T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J. Exp. Clin. Cancer Res. 2015;34:73. doi: 10.1186/s13046-015-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishida N., Nagahara M., Sato T., Mimori K., Sudo T., Tanaka F., Shibata K., Ishii H., Sugihara K., Doki Y., Mori M. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin. Cancer Res. 2012;18:3054–3070. doi: 10.1158/1078-0432.CCR-11-1078. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J.X., Song W., Chen Z.H., Wei J.H., Liao Y.J., Lei J., Hu M., Chen G.Z., Liao B., Lu J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang M., Zhao S., Jiang Z., Wang S., Sun P., Quan J., Yan D., Wang X. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuhashi S., Manirujjaman M., Hamajima H., Ozaki I. Control Mechanisms of the Tumor Suppressor PDCD4: Expression and Functions. Int. J. Mol. Sci. 2019;20:2304. doi: 10.3390/ijms20092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Yang H.S. The role of Pdcd4 in tumour suppression and protein translation. Biol. Cell. 2018 doi: 10.1111/boc.201800014. Published online May 28, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 49.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 50.Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Nedaeinia R., Manian M., Jazayeri M.H., Ranjbar M., Salehi R., Sharifi M., Mohaghegh F., Goli M., Jahednia S.H., Avan A., Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48–56. doi: 10.1038/cgt.2016.77. [DOI] [PubMed] [Google Scholar]
- 52.Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.