Abstract
Ethnopharmacological relevance
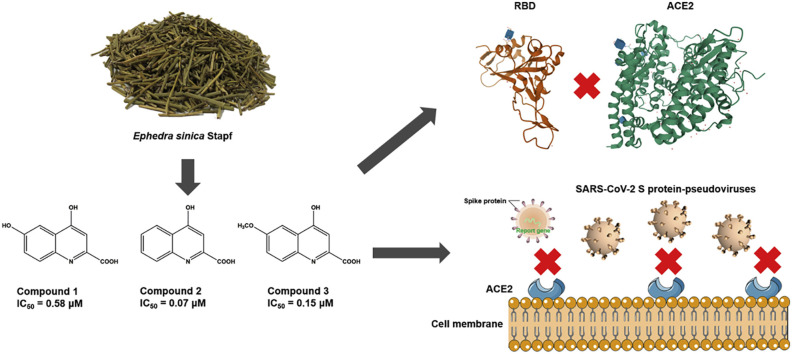
Ephedra sinica Stapf is a widely used folk medicine in Asia to treat lung diseases, such as cold, cough and asthma. Many efforts have revealed that some traditional Chinese medicine (TCM) prescriptions containing Ephedra sinica could effectively alleviate the symptoms and prevent the fatal deterioration of COVID-19.
Aim of the study
The present study aims to discover active compounds in Ephedra sinica disrupting the interaction between angiotensin-converting enzyme 2 (ACE2) and the SARS-CoV-2 spike protein receptor-binding domain (SARS-CoV-2 RBD) to inhibit SARS-CoV-2 virus infection.
Materials and methods
The ethanol extracts of Ephedra sinica were prepared. Activity guided isolation of constituents was carried out by measuring the inhibitory activity on ACE2-RBD interaction. The structures of active compounds were identified by HPLC-Q-TOF-MS/MS and NMR. To testify the contribution of main components for the inhibitory activity, different samples were prepared by components knock-out strategy. The mechanism of compounds inhibiting protein-protein interaction (PPI) was explored by competition inhibition assays, surface plasmon resonance (SPR) assays and molecular docking. SARS-CoV-2 S protein-pseudoviruses were used to observe the viropexis effect in cells.
Results
Ephedra sinica extracts (ESE) could effectively inhibit the interaction between ACE2 and SARS-CoV-2 RBD (IC50 = 95.01 μg/mL). Three active compounds, 4,6-dihydroxyquinoline-2-carboxylic acid, 4-hydroxyquinoline-2-carboxylic acid and 4-hydroxy-6-methoxyquinoline-2-carboxylic acid were identified to inhibit ACE2-RBD interaction (IC50 = 0.58 μM, 0.07 μM and 0.15 μM respectively). And knock-out the three components could eliminate the inhibitory activity of ESE. Molecular docking calculations indicated that the hydrogen bond was the major intermolecular force. Finally, our results also showed that these compounds could inhibit the infectivity of SARS-CoV-2 S protein-pseudoviruses to 293T-ACE2 (IC50 = 0.44–1.09 μM) and Calu-3 cells.
Conclusion
These findings suggested that quinoline-2-carboxylic acids in Ephedra sinica could be considered as potential therapeutic agents for COVID-19. Further, this study provided some justification for the ethnomedicinal use of Ephedra sinica for COVID-19.
Keywords: COVID-19, SARS-CoV-2, ACE2, Ephedra sinica, Quinoline-2-carboxylic acid, PPI inhibitor
Major chemical compounds studied in this article: 4,6-dihydroxyquinoline-2-carboxylic acid (PubChem CID: 440752); 4-hydroxyquinoline-2-carboxylic acid (PubChem CID: 3845); 4-hydroxy-6-methoxyquinoline-2-carboxylic acid (PubChem CID: 22017520)
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by a novel coronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020; Zhu et al., 2020). Since the outbreak of the disease at the end of 2019, about 200 countries and regions have reported this epidemic. In addition, more than 130 million people worldwide have been infected with this virus. It poses a huge threat to people's lives and health (Wang, D. et al., 2020a). Therefore, it is urgent to produce vaccines or to develop new drugs to combat this extremely dangerous coronavirus disease (Riva et al., 2020).
It was reported that the spike (S) protein of coronavirus benefits the virus to enter target cells (Xia et al., 2019). Many researches showed that SARS-CoV-2 bound to angiotensin-converting enzyme 2 (ACE2) via the receptor binding domain (RBD) in the homotrimeric spike glycoprotein (Hoffmann et al., 2020; Lan et al., 2020; Shang et al., 2020). Then the SARS-CoV-2 S protein was proteolytically activated by human proteases to mediate its entry into the cells (Gioia et al., 2020; Walls et al., 2020). Targeting the interaction between the SARS-CoV-2 S protein and the human ACE2 receptor is currently considered to be a promising therapeutic strategy (Wang, Q. et al., 2020b).
Since the pandemic, traditional Chinese medicine has been widely used and played an important role in the prevention and treatment of COVID-19. Ephedra sinica Stapf is a widely used folk medicine in Asia to treat lung diseases, such as cold, cough and asthma (Miao et al., 2020). And Ephedra sinica has been used against acute airway diseases for thousands of years in ancient China (Eng et al., 2019). Many efforts have revealed that some TCM prescriptions containing Ephedra sinica could effectively alleviate the symptoms and prevent the fatal deterioration of COVID-19 such as Lianhuaqingwen capsule/granule (Hu et al., 2020; Xiao et al., 2020), Qingfei Paidu decoction (Lee et al., 2021), Huashi Baidu granule (Huang, 2021) and Jinhua Qinggan granule (Liu, Z et al., 2020b). However, the active components and mechanisms of Ephedra sinica remain obscure. At present, many researchers have attempted to find the active components by virtual screening (Xia et al., 2021; Zhang et al., 2020). However, the microconstituents or unconventional active molecules may be neglected by using this method.
The present study aims to discover active compounds disrupting the protein-protein interaction (PPI) of ACE2-RBD to inhibit SARS-CoV-2 virus infection from Ephedra sinica extracts (ESE). Activity guided isolation of constituents was carried out by measuring the inhibitory activity on ACE2-RBD interaction. The structures of active compounds were identified HPLC-Q-TOF-MS/MS and NMR. Further, we investigated whether ESE and these active compounds (quinoline-2-carboxylic acids) could inhibit the infection of SARS-CoV-2 pseudoviruses to 293T-ACE2 and Calu-3 cells.
2. Materials and methods
2.1. Materials and reagents
Dried stems of Ephedra sinica Stapf were obtained from Inner Mongolia Autonomous Region and authenticated by Professor Boyang Yu of the School of Traditional Chinese Pharmacy, China Pharmaceutical University in April 2019. A voucher specimen (No.20190413) was deposited at the herbarium of Jiangsu Key Laboratory of TCM Evaluation and Translational Research, China Pharmaceutical University. 4,6-dihydroxyquinoline-2-carboxylic acid, 4-hydroxyquinoline-2-carboxylic acid and 4-hydroxy-6-methoxyquinoline-2-carboxylic acid isolated from ESE in our laboratory were used as reference substances. The purity of each compound was determined to be >98% by HPLC.
SARS-CoV-2 S protein RBD (Cat.No.SPD-C52H3), human ACE2 protein (Cat.No.AC2-H5257), biotinylated SARS-CoV-2 S protein RBD (Cat.No.SPD-C82E9), biotinylated human ACE2 (Cat.No.EP-105A011), streptavidin-HRP (Cat.No.EP-105A003) were purchased from Acrobiosystems (Beijing, China). 3,3′,5,5′-Tetramethylbenzidine (TMB) (Cat.No.Z724742) was purchased from Sigma-Aldrich (St. Louis, MO, USA). COVID-19-GFP pseudovirus with SARS-CoV-2 S protein (Cat.No.20200424) was purchased from GENEWIZ (Jiangsu, China).
2.2. Preparation of herb extracts
Dried stems of Ephedra sinica Stapf were milled to powder. 2.0 g of the powder was weighed out and extracted in 100 mL of ethanol-water (70:30, v/v) at 25 °C for 30 min with ultrasound. The extract was filtered, evaporated to dryness and redissolved in 200 mL deionized water to a final crude drug concentration of 10 mg/mL. The extracts were stored at −20 °C in small aliquots.
2.3. Biotinylated binding assay
The biotinylated binding assay was performed as described previously (Ho et al., 2007). Briefly, 96-well microplates (Corning, catalog # 9018) were coated at 4 °C overnight with 100 μL of 0.5 μg/mL SARS-CoV-2 RBD, rinsed with 300 μL washing buffer (0.05% Tween 20 in phosphate-buffered saline, pH 7.2) and blocked with 300 μL blocking buffer (2% bovine serum albumin in washing buffer) by incubating at 37 °C for 1 h. The absorbed protein in each well was incubated with 50 μL of 0.12 μg/mL biotinylated ACE2 and 50 μL sample at 37 °C for 1 h. Following three washes, 100 μL of streptavidin-HRP was added to each well and incubated at 37 °C for 1 h. After washed with washing buffer three times, 200 μL TMB substrate working solution was added to each well and incubated at 37 °C for 20 min. After adding 50 μL 1 M sulfuric acid, the absorbance was measured at 450 nm using a UV/Vis microplate spectrophotometer. To optimize the concentration of biotinylated ACE2 to get a complete blocking curve, we performed the assay with biotinylated ACE2 at five concentrations from 0.03 μg/mL to 0.48 μg/mL. Here, soluble recombinant human ACE2 was utilized as positive control. A dilution series of soluble recombinant human ACE2 (50 μL) was mixed with five constant concentrations of biotinylated ACE2 (50 μL) respectively in the absorbed protein wells at 37 °C for 1 h. The following HPR binding and TMB colorimetric methods were as same as above.
2.4. Surface plasmon resonance
Surface plasmon resonance (SPR) measurements were performed at 25 °C using a BIAcore T200 instrument. SARS-CoV-2 RBD and ACE2 were immobilized on a CM5 sensor chip respectively, and a blank channel was employed as a negative control for each assay. The ligands were injected in two-fold serial dilutions over SARS-CoV-2 RBD or ACE2 immobilized on the biosensor chip. The 1:1 binding model was used to assess binding kinetics. KD values were calculated with a kinetics model by BIAcore T200 analysis software.
2.5. HPLC-Q-TOF-MS/MS
HPLC-Q-TOF-MS/MS was performed with an Agilent 1260 HPLC system coupled with an Agilent 1260 diode array detector (DAD) and a Q-TOF 6530 mass spectrometer equipped with an electron spray ionization (ESI) interface. A Grace visionHT C18 column (5 μm, 4.6 × 250 mm) was used for chromatographic separations. The mobile phase was water-formic acid (A; 100:0.1, v/v) and acetonitrile (B) and a gradient elution program of 0–10 min, 5% B; 10–50 min, 5%–20% B; 50–60 min, 20%–25% B; 60–75 min, 25–35% B; 75–76 min, 35%–5% B; 76–85 min, 5% B. The flow rate was 1.0 mL/min, the detection wavelength was 254 nm and the column temperature was maintained at 35 °C. The ESI-MS positive-mode conditions were set as previously described (Guo et al., 2019).
2.6. Molecular docking
Small molecule optimization, protein preparation, binding-site definition and molecular docking were all performed in Schrödinger Maestro chemical simulation software. The structures of the compounds were drawn by ChemBiodraw 14.0. LigPrep module was used for ligand preparation. The pH was set at 7.0 ± 2.0, and the tautomeric form was determined by the Epik program. The ligand was minimized using Optimized Potentials for Liquid Simulations (OPLS)-2005 force field in the Macro Model software module. The protein structure used in the docking studies was spike proteins of SARS-CoV-2 (PDB ID = 6VSB) (Wrapp et al., 2020), which was downloaded from the Research Collaboratory for Structural Bioinformatics protein data bank and prepared with Protein Preparation Wizard workflow. The RBD of the crystal structure was taken as the docking target, and receptor binding motif (RBM) was defined as the binding site. The potential compounds were flexibly docked into the binding site with standard precision docking mode. The docking score, glide emodel, hydrogen bond and hydrophobic interactions between active site residues of RBM and compounds were recorded.
2.7. Cell culture
Human airway epithelial cells (Calu-3) and 293T cells transfected with ACE2 (293T-ACE2) were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). Calu-3 cells were grown in minimum essential medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution. 293T-ACE2 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution. Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2.
2.8. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cell viability was monitored by MTT colorimetric assay. Cells were cultivated in 96-well culture plates. After incubation at 37 °C for 24 h, various amounts of compounds were added into each well and incubated for another 24 h. Then cells were suspended in 100 μL of 10% FBS-MEM or FBS-DMEM containing 500 μg/mL MTT. After a 4 h incubation at 37 °C, 150 μL dimethyl sulfoxide was added to dissolve the MTT formazan. The absorbance value was measured at 570 nm using a microplate reader.
2.9. Infection with SARS-CoV-2 S protein-pseudoviruses
SARS-CoV-2 S protein lentivirus-based pseudoviruses which were constructed by GENEWIZ (Jiangsu, China) were used in this study. SARS-CoV-2 spike protein can be expressed on the surface of the pseudoviruses and the virus carries GFP reporter genes, which can be used to evaluate the activity of pseudoviruses-infected cells by observing fluorescence signal (Ou et al., 2020). 96-well plates were seeded with a solution of 1 × 104 cells in 100 μL medium per well and incubated at 37 °C for 12 h. 1 μL of SARS-CoV-2 S protein-pseudoviruses (2.22 × 106 TU/mL) were mixed with various amounts of compound in 100 μL medium and incubated at 37 °C. After 1 h incubation, the mixture was added to cells in the 96-well plates. After 6 h infection, cells were incubated in fresh medium for another 48 h. Cells were harvested and the fluorescence signal was observed by the fluorescence microscope or the microplate reader with excitation of 488 nm and emission of 510 nm.
2.10. Statistical analysis
The raw data were analyzed using GraphPad Prism 6.0 software. All data are presented as mean ± SEM (standard error of the mean) of six independent assays. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. Establishment of the method for evaluating the inhibitory activity on ACE2-RBD interaction
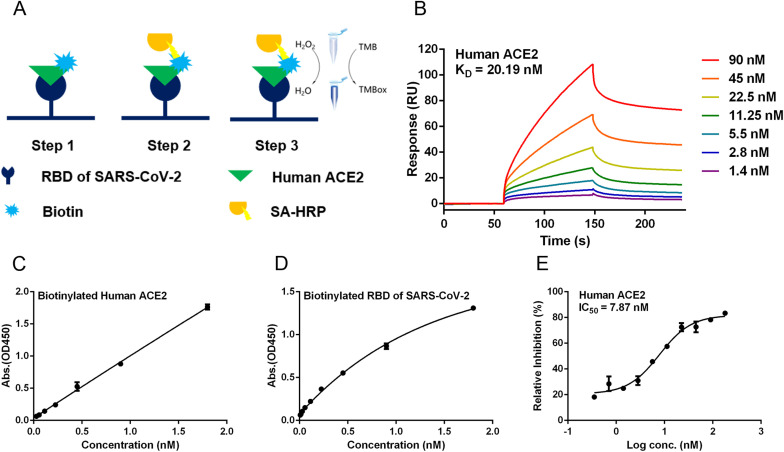
In order to measure the inhibitory activity on ACE2-RBD interaction, a biotinylated binding assay was established to evaluate the binding efficacy of ACE2 and RBD. The principle of the assay was shown as Fig. 1 A. The binding amount of the biotin-labeled ACE2 to the RBD coated on the 96-well plate could be detected directly by the color change of the TMB solution. Similarly, the binding amount of the biotin-labeled RBD to the ACE2 coated on a 96-well plate could also be tested by this method. To make sure the protein was active, the binding kinetics of ACE2 and RBD was measured by SPR, where recombinant human ACE2 was injected in two-fold serial dilutions over RBD immobilized on the biosensor chip. Similar to the results reported in the previous literature (Lei et al., 2020), SARS-CoV-2 RBD could strongly bind to ACE2 with an affinity of 20.19 nM (Fig. 1B). In order to choose reasonable test conditions, we performed the concentration-corresponding evaluation separately. As shown in Fig. 1C, the OD (Optical density) value increased with the increasement of the concentration of the biotinylated ACE2, indicating that the ACE2 bound to the immobilized RBD in a dose-dependent manner. As shown in Fig. 1D, the RBD also bound to the immobilized ACE2 in a dose-dependent manner. Previous studies have shown that soluble recombinant human ACE2 could inhibit SARS-CoV-2 infection, and related clinical trials have already begun (Monteil et al., 2020). Here, soluble recombinant human ACE2 was tested by the binding assay as positive control of this method. As expected, soluble recombinant ACE2 blocked the interaction between immobilized ACE2 and RBD with IC50 of 7.87 nM (Fig. 1E). Here, a rapid and sensitive method for evaluating the inhibitory activity on ACE2-RBD interaction was established.
Fig. 1.
Establishment of the method for evaluating the inhibitory activity on ACE2-RBD interaction. (A) Schematic diagram of the principle of biotinylated binding assay. (B) Validation of ACE2-RBD interaction by SPR. The 1:1 binding model was used to assess binding kinetics. (C) RBD was coated on a 96-well plate. (D) ACE2 was coated on a 96-well plate. (E) Soluble recombinant ACE2 blocked the interaction between immobilized ACE2 and RBD. Data are shown as mean ± SEM of six independent assays.
3.2. Activity guided isolation of constituents interrupting the interaction between ACE2 and RBD from Ephedra sinica
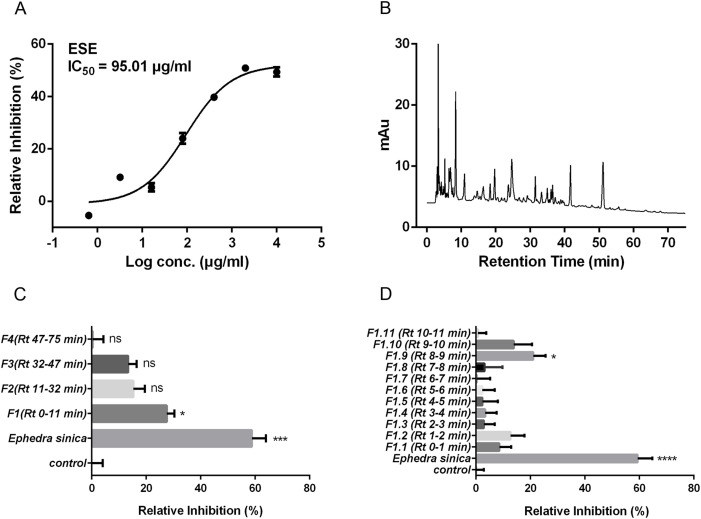
ESE was evaluated for its inhibitory efficacy on ACE2-RBD interaction. The relative inhibition ratio was determined by the biotinylated binding assay. As shown in Fig. 2 A, ESE could block the interaction between ACE2 and RBD with IC50 of 95.01 μg/mL. Then, we focused on finding biologically active molecules in ESE by activity guided isolation which has been proven to be an effective method to find biologically active molecules in natural products. Here, we performed HPLC to gradually separate the compounds in the ESE and tracked the biologically active ingredients persistently. As shown in Fig. 2B, an HPLC chromatogram of the ESE at a monitoring wavelength of 254 nm was established to identify as many compounds as possible. According to the retention time (RT) of the compound on the chromatographic column, the ESE was first divided into four fractions, namely F1 (RT 0–11 min), F2 (RT 11–32 min), F3 (RT 32–47 min), F4 (RT 47–75 min). These four fractions were tested in the binding assay for their inhibitory potential on ACE2-RBD interaction. As shown in Fig. 2C, only F1 (RT 0–11min) showed a significant inhibition effect (P < 0.05). Then F1 was sequentially separated into eleven sub-fractions based on RT, namely F1.1-F1.11. It can be clearly found that F1.9 inhibited the binding of ACE2 to RBD among these eleven sub-fractions (Fig. 2D). Then F1.9 was identified by HPLC-Q-TOF-MS/MS and NMR. The results were shown in Table 1 and Supplementary Figs. 2–4. Through comparison with literature and standard materials, it was identified as 4,6-dihydroxyquinoline-2-carboxylic acid (compound 1). However, F1.9 showed a lower inhibitory percentage than the ESE on ACE2-RBD interaction, indicating that there were other ingredients in ESE which inhibited the binding of ACE2 to RBD. Therefore, according to the HPLC-Q-TOF-MS/MS data of ESE, two derivatives of compound 1 were found and identified by HPLC-Q-TOF-MS/MS and NMR. Through comparison with literature (STARRATT and CAVENE, 1996) and standard materials, they were identified as 4-hydroxyquinoline-2-carboxylic acid (compound 2) and 4-hydroxy-6-methoxyquinoline-2-carboxylic acid (compound 3) respectively. The specific structures of three compounds (quinoline-2-carboxylic acids) were shown in Fig. 3 A.
Fig. 2.
Activity guided isolation of constituents interrupting the interaction between ACE2 and RBD from Ephedra sinica. (A) The inhibitory efficacy of ESE on the interaction between SARS-CoV-2 RBD and ACE2. (B) The chromatogram of the ESE at a monitoring wavelength of 254 nm. (C) According to the retention time in the chromatogram, the ESE was divided into four fractions, and the relative inhibition rate was determined by the biotinylated binding assay experiment. (D) According to the retention time in the chromatogram, the ESE fraction 1 was subdivided into eleven sub-fractions, and the relative inhibition rate was determined by the biotinylated binding assay experiment. Data are shown as mean ± SEM of six independent assays. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with solvent.
Table 1.
Retention time, MS data and NMR data for identification of PPI inhibitors in Ephedra sinica Stapf.
| Compound No. | Retention time (min) | [M+H] + m/z | Formula | 1H NMR (500 MHz, DMSO) | Identification |
|---|---|---|---|---|---|
| 1 | 8.4 | 206.0437 | C10H7NO4 | 7.86 (d, J = 9.0 Hz, 1H), 7.39 (d, J = 2.5 Hz, 1H), 7.24 (dd, J = 8.9, 2.5 Hz, 1H), 6.61 (s, 1H). | 4,6-dihydroxyquinoline-2-carboxylic acid |
| 188.0337 | |||||
| 160.0386 | |||||
| 132.0426 | |||||
| 2 | 13.9 | 190.0487 | C10H7NO3 | 8.09 (d, J = 7.5 Hz, 1H), 7.96 (d, J = 8.4 Hz, 1H), 7.70 (t, J = 8.2 Hz, 1H), 7.37 (t, J = 7.5 Hz, 1H), 6.65 (s, 1H). | 4-hydroxyquinoline-2-carboxylic acid |
| 172.0373 | |||||
| 144.0433 | |||||
| 116.0490 | |||||
| 3 | 24.7 | 220.0592 | C11H9NO4 | 7.92 (d, J = 9.1 Hz, 1H), 7.47 (d, J = 2.9 Hz, 1H), 7.34 (dd, J = 9.3, 2.8 Hz, 1H), 6.60 (s, 1H), 3.85 (s, 3H). | 4-hydroxy-6-methoxyquinoline-2-carboxylic acid |
| 202.0491 | |||||
| 174.0546 | |||||
| 146.0595 |
Fig. 3.
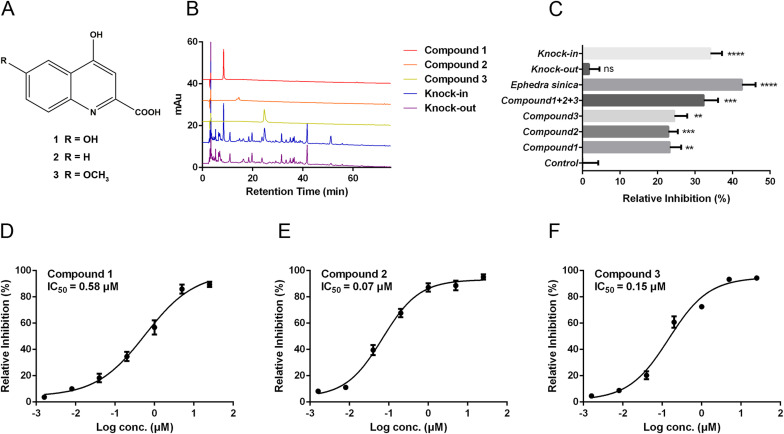
Quinoline-2-carboxylic acids were main active components in Ephedra sinica disrupting the interaction between ACE2 and RBD. (A) The structure of quinoline-2-carboxylic acids in ESE. (B) Chromatograms of compounds 1–3, the knock-out sample and the knock-in sample. (C) The inhibitory efficacy of each fractions on ACE2-RBD interaction. (D–F) The inhibitory efficacy of compounds 1–3 on ACE2-RBD interaction. Data are shown as mean ± SEM of six independent assays. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with solvent.
3.3. Quinoline-2-carboxylic acids were main active components in Ephedra sinica disrupting the interaction between ACE2 and RBD
Components knock-out technology is a good method to identify the active ingredients in natural products (Liu, X et al., 2020a). Here, we prepared compounds 1–3 knock-out samples (ESE without compounds 1–3), components knock-in samples (compounds 1–3 were added to the knock-out sample) and their HPLC chromatograms were shown in Fig. 3B. These fractions were tested in the binding assay for their inhibitory potential on ACE2-RBD interaction. As shown in Fig. 3C, compounds 1–3 blocked the interaction between ACE2 and RBD. However, the ability of knock-out sample to block the interaction between ACE2 and RBD was lost. Interestingly, this ability was restored to an equivalent level to ESE after knocking in compounds 1–3, indicating that compounds1-3 may be the main active ingredients in ESE on blocking the interaction between ACE2 and RBD. In order to evaluate the activity of disrupting ACE2-RBD interaction of different compounds, we optimize the concentration of biotinylated ACE2 as described previously (Walker et al., 2020). The biotinylated ACE2 at the concentration of 0.06 μg/mL showed a complete inhibition curve and appropriate response value (OD450) in the presence of soluble recombinant human ACE2 (Supplementary Fig. 5). As shown in Fig. 3D–F, on this optimal condition, compounds 1–3 blocked the interaction between SARS-CoV-2 RBD and ACE2 in a dose-dependent manner with IC50 of 0.58 μM, 0.07 μM and 0.15 μM with a complete blocking curve. There is no significant difference in the inhibitory activity of these three compounds.
3.4. Quinoline-2-carboxylic acids inhibited ACE2-RBD interaction by directly binding to SARS-CoV-2 RBD
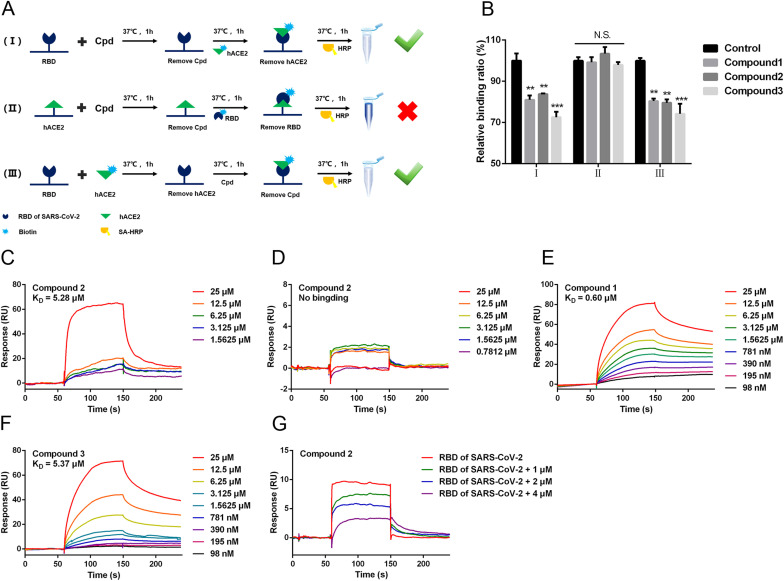
To investigate the principle of the inhibitory efficacy on ACE2-RBD interaction, we further addressed the binding of compounds 1–3 with SARS-CoV-2 RBD or ACE2. As shown in Fig. 4 A, three parallel experiments were conducted. (I) 10 μM three Compounds (Cpds) were incubated with immobilized SARS-CoV-2 RBD respectively and washed to remove the unbound part, then the relative amount of biotinylated ACE2 was determined; (Ⅱ) 10 μM Cpds were incubated with immobilized ACE2 respectively and washed to remove the unbound part, then the relative amount of biotinylated SARS-CoV-2 RBD was determined; (Ⅲ) Biotinylated ACE2 was incubated with immobilized SARS-CoV-2 RBD and washed to remove the unbound part, then 10 μM Cpds were added respectively. Finally, relative amount of biotinylated ACE2 was determined. As shown in Fig. 4B, when compounds 1–3 were incubated with immobilized SARS-CoV-2 RBD, all the relative amounts of bound ACE2 were significantly reduced compared with the solvent control group (P < 0.05). Correspondingly, when compounds were incubated with immobilized ACE2, the amounts of bound SARS-CoV-2 RBD did not change significantly compared with the solvent control group. These results indicated that the three compounds inhibited PPI by binding to SARS-CoV-2 RBD instead of ACE2. In addition, after adding compounds 1–3 into combined ACE2 and RBD (Fig. 4A Ⅲ), all the relative amounts of bound ACE2 were significantly reduced compared with the solvent control group (P < 0.05), indicating that the three compounds could also promote the dissociation of ACE2 and RBD.
Fig. 4.
Quinoline-2-carboxylic acids inhibited ACE2-RBD interaction by directly binding to SARS-CoV-2 RBD. (A) The time-of-addition experimental scheme. (B) The relative binding amount under different conditions. Compound 2 were injected in two-fold serial dilutions over SARS-CoV-2 RBD (C) or ACE2 (D) immobilized on the biosensor chip. Compound 1 (E) or 3 (F) were injected in two-fold serial dilutions over SARS-CoV-2 RBD immobilized on the biosensor chip. (G) Various amounts of compound 2 were incubated with 22.5 nM SARS-CoV-2 RBD at 37 °C for 1 h, then each mixture was injected over ACE2 immobilized on the biosensor chip. Data are shown as mean ± SEM of six independent assays. *P < 0.05, **P < 0.01, ***P < 0.001 compared with solvent.
To further verify the above results, the direct binding between the compounds and proteins were analyzed by SPR assay. Compound 2 bound to immobilized SARS-CoV-2 RBD in a dose-dependent manner. The KD value of compound 2 is 5.28 μM (Fig. 4C). While, compound 2 displayed low or no binding with ACE2 (Fig. 4D). Similarly, as shown in Fig. 4E and F, the interactions between the other two compounds and the SARS-CoV-2 RBD were concentration dependent. The KD value of compound 1 was 0.60 μM and compound 3 was 5.37 μM. To evaluate whether compound 2 interfered with the binding interface of SARS-CoV-2 RBD, we performed an SPR inhibition assay. After incubating SARS-CoV-2 RBD with gradient concentration of compound 2, the mixture was injected over ACE2 immobilized on the biosensor chip. As shown in Fig. 4G, the binding of SARS-CoV-2 RBD to ACE2 was blocked by compound 2 in a dose-dependent manner. These findings suggested that compound 1–3 could block the ACE2-RBD interaction by directly binding to SARS-CoV-2 RBD at the binding interface.
3.5. Molecular docking
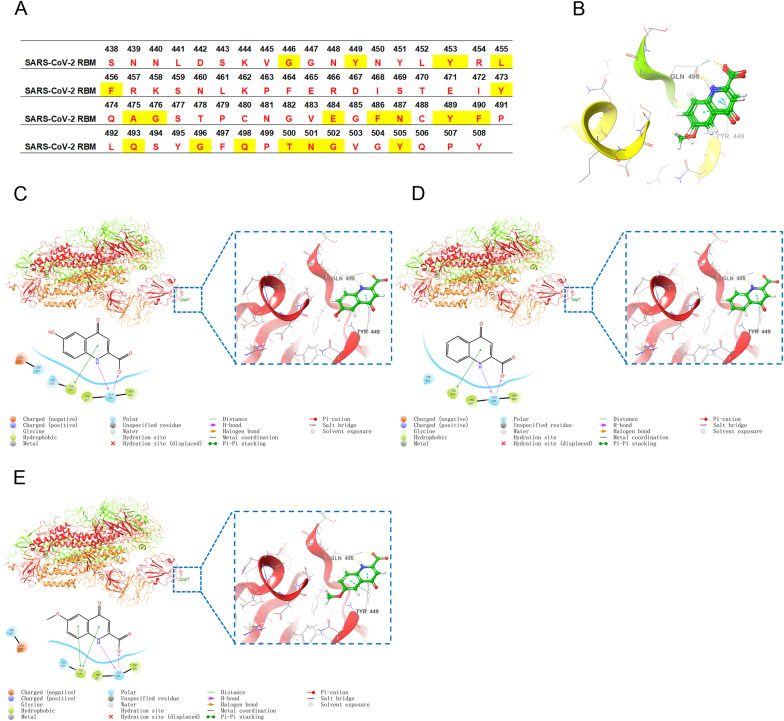
To investigate the molecular mechanism of their interference with SARS-CoV-2 RBD binding to the host cell receptor, molecular docking studies were performed. It is well known that the interface interaction between ACE2 and RBD plays a crucial role in its binding activity (Muhseen et al., 2020). The sequence of the RBM (438–508) shown in Fig. 5 A is considered particularly important for the binding of RBD to ACE2 (Yi et al., 2020). The tautomeric forms of the ligands were optimized and determined as anionic form with quinolin-4(1H)-one skeletal structure rather than quinolin-4-ol tautomer form by the Epik program. The chosen tautomeric forms were likely to be the most stable forms in aqueous solution that was consistent with literature reported (Samanta et al., 2014; Lorinczi et al., 2020). The binding affinity and binding mode of the three compounds at the RBM were investigated. Glide was used as the molecular docking tool to investigate the RBM binding conformations of the compounds because of the good reproducibility of the conformations and accuracy in molecular docking and scoring. As shown in Fig. 5B, compounds 1–3 combined with the RBM of SARS-CoV-2 RBD at similar sites, also had similar glide emodel values (−22.532, −19.446, and −21.824) and docking scores (−4.052, −4.098, and −4.206), which may explain the reason that the inhibitory efficacy on ACE2-RBD interaction of three compounds were similar and the similar affinity determined by SPR. Compound 1 built two hydrogen bond interactions with Gln498 residue. Also, it formed three hydrophobic interactions (Tyr449, Phe497, and Pro499), a Pi-pi Stacking interaction (Tyr449), three polar interactions (Ser443, Asn450, and Gln498) and a negative charged interaction (Asp442) with SARS-CoV-2 RBD (Fig. 5C). Compound 2 formed two hydrogen bond interactions (Gln498), a pi-pi stacking interaction (Tyr449), three hydrophobic interactions (Tyr449, Phe497, and Pro499) and two polar interactions (Ser443 and Gln498) with SARS-CoV-2 RBD (Fig. 5D). Compound 3 formed two hydrogen bond interactions (Gln498), two Pi-pi Stacking interactions (Tyr449), three hydrophobic interactions (Tyr449, Phe497 and Pro499), three polar interactions (Ser443, Asn450 and Gln498) and a negative charged interaction (Asp442) with SARS-CoV-2 RBD (Fig. 5E). It was worth noting that Tyr449 and Gln498 were the key ACE2 binding residues that have been previously identified (Yi et al., 2020). The combination of compounds 1–3 with these key amino acid residues may be the reason for its inhibitory activity on the interaction between ACE2 and RBD.
Fig. 5.
Molecular docking. (A) The sequence of RBM (438–508). Previously identified critical ACE2-binding residues are shaded in yellow. (B) Superimposition of the ligands and docked conformation of compounds 1–3. Three-dimensional and two-dimensional docking models of compound 1 (C), compound 2 (D) and compound 3 (E) with SARS-CoV-2 (PDB: 6VSD) generated by Schrödinger Maestro chemical simulation software.
3.6. Quinoline-2-carboxylic acids abolished the infectivity of SARS-CoV-2 S protein-pseudoviruses to 293T-ACE2 and Calu-3 cells
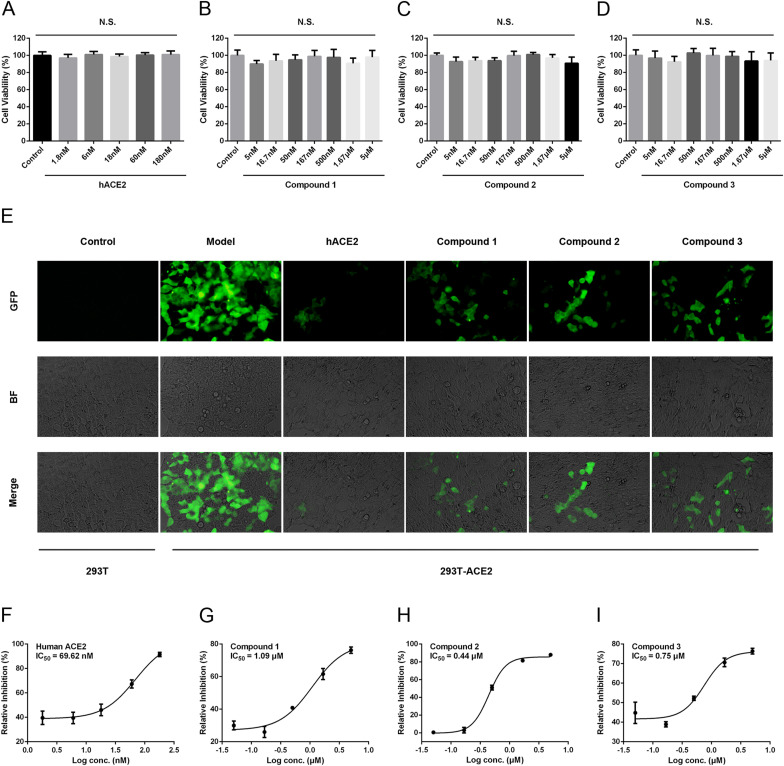
SARS-CoV-2 S protein-pseudoviruses infectivity was used to evaluate the inhibitory potential of compounds 1–3 in vitro. We first examined the cell toxicity to 293T-ACE2 cells of the compounds 1–3 and soluble human ACE2. As shown in Fig. 6 A–D, all of them had no effect on cell viability of 293T-ACE2 cells even at the highest concentration (5 μM or 180 nM). These substances shown low cytotoxicity and were safe within a certain concentration range. 293T-ACE2 cells were infected with S protein-pseudoviruses in the presence or absence of different compounds. As shown in Fig. 6E, the expression of green fluorescent protein in 293T-ACE2 cells significantly reduced caused by 5 μM compounds1-3 or 180 nM ACE2 in the images photographed by a fluorescence microscope (20 × ), indicating that they inhibited the entry of S protein-pseudoviruses into 293T-ACE2 cells. In addition, as shown in Fig. 6F, the positive control recombinant ACE2 inhibited the entry of S protein-pseudoviruses into 293T-ACE2 cells in a dose-dependent manner with IC50 of 69.62 nM. Obviously, as shown in Fig. 6G–I, compounds 1–3 also effectively inhibited the infectivity of pseudoviruses with IC50 of 1.09 μM, 0.44 μM and 0.75 μM, respectively. Similarly, Calu-3 cells were infected with S protein-pseudoviruses in the presence or absence of ESE, compound 2 or soluble human ACE2 respectively. As shown in Supplementary Fig. 1, ESE and compound 2 also abolished the infectivity of SARS-CoV-2 S protein-pseudoviruses to Calu-3 cells. These findings indicated that quinoline-2-carboxylic acids were novel compounds that were able to reduce the infectivity of SARS-CoV-2 S protein-pseudoviruses to 293T-ACE2 and Calu-3 cells.
Fig. 6.
Quinoline-2-carboxylic acids abolished the infectivity of SARS-CoV-2 S protein-pseudoviruses to 293T-ACE2 cells. After incubating with recombinant ACE2 (A), compound 1 (B), compound 2 (C) and compound 3 (D) for 24 h, cell viability was detected by MTT assay. (E) SARS-CoV-2 S protein-pseudoviruses entered into 293T-ACE2 cells. Images were photographed at × 20 magnification using the fluorescence microscope. (F–I) The fluorescence signal was measured with a fluorescent microplate reader. Data are shown as mean ± SEM of six independent assays.
4. Discussion
TCMs as great resource of bioactive natural products are widely used to treat COVID-19 (Xian et al., 2020). However, the active ingredients and molecular mechanisms still need to be clarified. In the present study, we confirm for the first time that quinoline-2-carboxylic acids in Ephedra sinica can disrupt ACE2-RBD interaction and inhibit the entry of pseudoviruses.
We initially performed biotinylated binding assay to determine whether ingredients from TCMs could disrupt ACE2-RBD interaction (Fig. 1A). We found that the ESE displays significant inhibitory activity (Fig. 2A). We initially used activity guided isolation and components knock-out technology to identify inhibitors of ACE2-RBD interaction from ESE (Fig. 2, Fig. 3C). In this study, we demonstrated that 4,6-dihydroxyquinoline-2-carboxylic acid, 4-hydroxyquinoline-2-carboxylic acid and 4-hydroxy-6-methoxyquinoline-2-carboxylic acid were main active components in Ephedra sinica blocking ACE2-RBD interaction (Fig. 3D–F). However, the relative inhibition ratio of the mix of compounds 1–3 was lower than ESE (Fig. 3C), that means there are other components disrupting the interaction. That is consistent with the literature (Lv et al., 2021).
It is worth noting that ESE and the active ingredients not only prevent the ACE2-RBD interaction, but also promote the dissociation of the S protein that has bound the receptor (Fig. 4A Ⅲ, B). It means that quinoline-2-carboxylic acids are possible to play crucial roles in the treatment and prevention of viruses’ invasion at the same time. Interestingly, 4-hydroxyquinoline-2-carboxylic acid (kynurenic acid) is one of the metabolites in human, the latest research showed that the content of kynurenic acid in serum of patients with COVID-19 was higher than that in negative controlled patients (Thomas et al., 2020). In our study, it was confirmed that kynurenic acid could disrupt ACE2-RBD interaction, which may be related to self-antiviral mechanism.
Traditionally, ephedrine and pseudoephedrine were considered to be the main active ingredients in Ephedra sinica but the serious side effects of these compounds restricted the clinical use of ephedra (Anderson, 2018; Wei et al., 2019). Here we proved that three trace components in Ephedra sinica not only inhibited the PPI between ACE2 and RBD but also effectively blocked the invasion of pseudovirus in vitro. We put forward a new point of view for the rational use of Ephedra sinica and also prove that activity guided isolation is a good strategy for the discovery of bioactive microconstituents in natural products.
We further demonstrated that quinoline-2-carboxylic acids inhibited PPI by directly binding to SARS-CoV-2 RBD instead of ACE2 (Fig. 4C and D). Based on the molecular docking and SPR assay, we speculated that these compounds might bind to interface between ACE2 and RBD that contributes to the disruption of the PPI. The structure of quinoline-2-carboxylic acid is important for the study on the inhibitory activity. The amidogen and carboxyl groups have two strong hydrogen interactions with Gln 498 (Fig. 5). These interactions might verify that quinoline-2-carboxylic acids have potent inhibitory activity against SARS-CoV-2.
The entry of virus into the cells is a critical step in the process of virus infection. However, SARS-CoV-2 related researches were greatly limited by the regulation that the safety level of the laboratory for direct research using virus strains need to achieve 3 or higher. The study on pseudoviruses made the conduct of researches safer and much more efficient. The surface of the pseudoviruses expresses the SARS-CoV-2 Spike protein and the viruses carry both the GFP and Luciferase reporter genes (Chen et al., 2018). Therefore, the activity of the pseudoviruses-infected cells can be evaluated by observing the fluorescence signal and detecting the luciferase activity. We confirm that quinoline-2-carboxylic acids have the ability to suppress the entrance of SARS-CoV-2 S protein-pseudoviruses into 293T-ACE2 and Calu-3 cells (Fig. 6, Supplementary Fig. 1). Thus, quinoline-2-carboxylic acids could be good inhibitors to block SARS-CoV-2 infection of human ACE2-expressing cells.
5. Conclusions
Three quinoline-2-carboxylic acids including 4,6-dihydroxyquinoline-2-carboxylic acid, 4-hydroxyquinoline-2-carboxylic acid and 4-hydroxy-6-methoxyquinoline-2-carboxylic acid in Ephedra sinica were identified as novel active constituents which blocked both the binding of SARS-CoV-2 RBD to ACE2 and the infectivity of SARS-CoV-2 S protein-pseudoviruses to 293T-ACE2 and Calu-3 cells. These findings suggested that quinoline-2-carboxylic acids could be considered as potential therapeutic compounds in the treatment of COVID-19. Further, this study provided some justification for the ethnomedicinal use of Ephedra sinica for COVID-19.
Authors' contributions
Jie Mei, Xiufeng Liu and Boyang Yu designed the experiments. Jie Mei, Yatong Zhou, Xinping Yang and Fan Zhang carried out the experiments. Jie Mei, Xiufeng Liu and Boyang Yu analyzed and interpreted the data. Jie Mei, Xiufeng Liu and Boyang Yu wrote and revised the manuscript. All authors approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
This research was supported by Double First Class University Plan (CPU2018GF06 and CPU2018GY32) and National Major Science and Technology Projects of China (2017ZX09101002-002-003).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jep.2021.114303.
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- RBD
receptor-binding domain
- COVID-19
corona virus disease 2019
- PPI
protein-protein interaction
- ACE2
angiotensin-converting enzyme 2
- SPR
surface plasmon resonance
- ESE
Ephedra sinica extracts
- S
spike;
- TCM
traditional Chinese medicine;
- HPLC
high performance liquid chromatography
- NMR
nuclear magnetic resonance spectroscopy
- DAD
diode array detector
- ESI
electron spray ionization
- RBM
receptor binding motif
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
- RT
retention time;
- IC50
the 50% inhibitory concentration
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Anderson S.D. Repurposing drugs as inhaled therapies in asthma. Adv. Drug Deliv. Rev. 2018;133:19–33. doi: 10.1016/j.addr.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Chen Q., Tang K., Zhang X., Chen P., Guo Y. Establishment of pseudovirus infection mouse models for in vivo pharmacodynamics evaluation of filovirus entry inhibitors. Acta Pharm. Sin. B. 2018;8(2):200–208. doi: 10.1016/j.apsb.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng Y.S., Lee C.H., Lee W.C., Huang C.C., Chang J.S. Unraveling the molecular mechanism of traditional Chinese medicine: formulas against acute airway viral infections as examples. Molecules. 2019;24(19):3505. doi: 10.3390/molecules24193505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia M., Ciaccio C., Calligari P., De Simone G., Sbardella D., Tundo G., Fasciglione G.F., Di Masi A., Di Pierro D., Bocedi A., Ascenzi P., Coletta M. Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem. Pharmacol. 2020;182:114225. doi: 10.1016/j.bcp.2020.114225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Fu R., Qian Y., Zhou Z., Liu H., Qi J., Zhang B., Yu B. Comprehensive screening and identification of natural inducible nitric oxide synthase inhibitors from Radix Ophiopogonis by off-line multi-hyphenated analyses. J. Chromatogr. A. 2019;1592:55–63. doi: 10.1016/j.chroma.2019.01.029. [DOI] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007;74(2):92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Guan W.J., Bi Y., Zhang W., Li L., Zhang B., Liu Q., Song Y., Li X., Duan Z., Zheng Q., Yang Z., Liang J., Han M., Ruan L., Wu C., Zhang Y., Jia Z.H., Zhong N.S. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2020;153242 doi: 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.Q. Efficacy and safety assessment of severe COVID-19 patients with Chinese medicine: a retrospective case series study at early stage of the COVID-19 epidemic in wuhan, China. J. Ethnopharmacol. 2021:113888. doi: 10.1016/j.jep.2021.113888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee D.Y.W., Li Q.Y., Liu J., Efferth T. Traditional Chinese herbal medicine at the forefront battle against COVID-19: clinical experience and scientific basis. Phytomedicine. 2021;80:153337. doi: 10.1016/j.phymed.2020.153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11(1):2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jiang N., Xu X., Liu C., Liu Z., Zhang Y., Kang W. Anti-hepatoma compound determination by the method of spectrum effect relationship, component knock-out, and UPLC-MS(2) in scheflera heptaphylla (L.)Frodin harms and its mechanism. Front. Pharmacol. 2020;11:1342. doi: 10.3389/fphar.2020.01342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Li X., Gou C., Li L., Luo X., Zhang C., Zhang Y., Zhang J., Jin A., Li H., Zeng Y., Li T., Wang X. Effect of Jinhua Qinggan granules on novel coronavirus pneumonia in patients. J. Tradit. Chin. Med. 2020;40(3):467–472. doi: 10.19852/j.cnki.jtcm.2020.03.016. [DOI] [PubMed] [Google Scholar]
- Lorinczi B., Csampai A., Fulop F., Szatmari I. Synthesis of new c-3 substituted kynurenic acid derivatives. Molecules. 2020;25(4):1–14. doi: 10.3390/molecules25040937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Wang S., Liang P., Wang Y., Zhang X., Jia Q., Fu J., Han S., He L. Screening and evaluation of anti-SARS-CoV-2 components from Ephedra sinica by ACE2/CMC-HPLC-IT-TOF-MS approach. Anal. Bioanal. Chem. 2021;19:1–10. doi: 10.1007/s00216-021-03233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao S., Zhang Q., Bi X., Cui J., Wang M. A review of the phytochemistry and pharmacological activities of Ephedra herb. Chin. J. Nat. Med. 2020;18(5):321–344. doi: 10.1016/s1875-5364(20)30040-6. [DOI] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhseen Z.T., Hameed A.R., Al-Hasani H.M.H., Tahir Ul Qamar M., Li G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: integrated computational approach. J. Mol. Liq. 2020;320:114493. doi: 10.1016/j.molliq.2020.114493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586(7827):113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A., Guchhait N., Bhattacharya S.C. Photophysical aspects of biological photosensitizer Kynurenic acid from the perspective of experimental and quantum chemical study. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2014;129:457–465. doi: 10.1016/j.saa.2014.03.079. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starratt A.N., Cavene S. Quinoline-2-carboxylic acids from Ephedra species. Phytochemistry. 1996;42(5):1477–1478. doi: 10.1016/0031-9422(96)00126-4. [DOI] [Google Scholar]
- Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A., Spitalnik S.L., D'Alessandro A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.N., Chokkalingam N., Reuschel E.L., Purwar M., Xu Z., Gary E.N., Kim K.Y., Helble M., Schultheis K., Walters J., Ramos S., Muthumani K., Smith T.R.F., Broderick K.E., Tebas P., Patel A., Weiner D.B., Kulp D.W. SARS-CoV-2 assays to detect functional antibody responses that block ACE2 recognition in vaccinated animals and infected patients. J. Clin. Microbiol. 2020;58(11):1–13. doi: 10.1128/JCM.01533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):1–12. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Du H., Shao C., Zhou H., Lu Y., Yu L., Wan H., He Y. Screening of antiviral components of ma huang tang and investigation on the ephedra alkaloids efficacy on influenza virus type A. Front. Pharmacol. 2019;10:961. doi: 10.3389/fphar.2019.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/s0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhong Z., Gao B., Vong C.T., Lin X., Cai J., Gao H., Chan G., Li C. The important herbal pair for the treatment of COVID-19 and its possible mechanisms. Chin. Med. 2021;16(1):25. doi: 10.1186/s13020-021-00427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., Xu H. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm. Sin. B. 2020;10(7):1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Tian J., Zhou Y., Xu X., Min X., Lv Y., Peng M., Zhang Y., Yan D., Lang S., Zhang Q., Fan A., Ke J., Li X., Liu B., Jiang M., Liu Q., Zhu J., Yang L., Zhu Z., Zeng K., Li C., Zheng Y., Wu H., Lin J., Lian F., Li X., Tong X. Efficacy of Huoxiang Zhengqi dropping pills and Lianhua Qingwen granules in treatment of COVID-19: a randomized controlled trial. Pharmacol. Res. 2020;161:105126. doi: 10.1016/j.phrs.2020.105126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17(6):621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.H., Wu K.L., Zhang X., Deng S.Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.