Abstract
The role of Notch in the immune system was first described in the late 90s. Reports revealed that Notch is one of the most conserved developmental pathways involved in diverse biological processes such as the development, differentiation, survival and functions of many immune populations. Here, we provide an extended view of the pleiotropic effects of the Notch signaling on the innate lymphoid cell (ILC) biology. We review the current knowledge on Notch signaling in the regulation of ILC differentiation, plasticity and functions in diverse tissue types and at both the fetal and adult developmental stages. ILCs are early responder cells that secrete a large panel of cytokines after stimulation. By controlling the abundance of ILCs and the specificity of their release, the Notch pathway is also implicated in the regulation of their functions. The Notch pathway is therefore an important player in both ILC cell fate decision and ILC immune response.
Keywords: Notch pathway, Development of lymphoid cells, ILC, Immune functions, Polarization of immune subsets
The Notch gene is conserved through evolution with a signaling pathway controlling numerous developmental decisions (cell fate, homeostasis, survival) in different tissues during the embryonic and adult life (reviewed in Ref. [1]). Notch signaling is one of the few pathways allowing the communication between two neighboring cells to manage multiple developmental issues in a repeated manner. The Notch pathway occurs during the development and differentiation of numerous immune cell populations and could also interfere to regulate their survival and functions [[2], [3], [4]]. The role of the Notch pathway on T cell biology has been largely uncovered and reviewed [[5], [6], [7]]. Devoid of specific antigen receptors, innate lymphoid cells (ILCs) are separated into several subsets with secretive abilities highly similar to those of T-helper cell subsets [8]. A recent consensus nomenclature subdivided ILCs into 5 groups constituted of cytotoxic NK cells, fetal lymphoid-tissue inducer (LTi) cells and helper ILC1s, ILC2s, and ILC3s with effector programs respectively corresponding to Th1, Th2 and Th17 programs [9]. Roughly, helper ILCs are separated based on the expression of the transcription factors: T-bet, GATA-3 and RORγt. ILC1s are defined by their ability to produce interferon-γ (IFNγ) and ILC2s are able to produce Th2 related cytokines, such as IL-5 IL-13 and amphiregulin. ILC3s are related to the Th17 immunity with cells expressing the retinoic acid related orphan receptor RORγt and the affiliated IL-17 and IL-22 cytokines. ILCs are important players of the immune response and participate to maintain the tissue health as they rapidly respond to disruption of tissue homeostasis or infection by secreting large amounts of cytokines. They could also be implicated in the initiation of different inflammatory diseases [10]. Mostly detected in the mucosal area, ILCs seed peripheral tissues early in life during both the fetal and neonatal stages [11,12]. In the adult, ILCs are then slowly replaced by newly generated precursors from BM hematopoiesis [12].
Studies underpinning the role of Notch on ILCs are recent and still under active consideration. Here, current understanding of the Notch biology on ILC development, differentiation and plasticity are summarized. The aim is to define a possible “consensus” of Notch signaling functions on both fetal and adult specific subset of ILCs.
The Notch signaling pathway
Notch signaling was first defined in the context of cell fate decisions during Drosophila melanogaster development [13]. Despite well evolutionarily conserved cellular machinery, the Notch signaling pathway is more complex in mammals due to the multiplication of molecules such as Notch receptors and ligands, with all receptors capable of interacting with variable affinity to any ligands [14]. In mammals, some of the pathway genes have maintained redundancy, others have acquired novel functions, while some were considered new members as they had no obvious ortholog in Drosophila. Vertebrates express four different Notch receptors (Notch 1–4), which can engage five known ligands (Delta-like 1, 3, 4, Jagged 1, and 2). Upon recognition of Notch ligands by the Notch extracellular domain and after serial proteolytic cleavages by γ-secretase, the Notch intracellular domain (NICD) translocates from the membrane to the nucleus, displaces histone deacetylase (HDAc), and binds to the transcription factor like recombination signal binding protein for immunoglobulin Jk region (RBP-JK) considered as a member of the co-repressor complex (reviewed in Ref. [15]). Then, histone acetyltransferases (HAc) are recruited for the formation of an activation complex with members of the MAML family. The co-factors NICD/RBP-JK/MAMl mediate the signaling cascade of the canonical Notch pathway recruiting co-activators, and enabling the transcription of numerous targets [[15], [16], [17]]. Genome-wide expression and chromatin immunoprecipitation (ChIP) assays proved that very large number of genes can be directly regulated by Notch [18,19]. This is partly explained by the ability of Notch to combine with other signaling pathways such as NF-Κb or TGFβ to enlarge the extent of the possible Notch targeted genes.
Indeed, NICD, RBP-JK and HAc, could activate genes such as the basic helix-loop-helix transcription factor genes Hes or Hey in many tissues and other genes such as Tcf7, Gata3 and Tbx21, all of them important during immune cell development. Overall, the regulation of the canonical Notch pathway activation is finely tuned. The NICD is rapidly degraded and the amplification of the signal is depending on the dose, duration and environmental context.
Distribution of Notch ligands through the body
Most Notch ligands have an agonistic role along the Notch pathway except for Delta-like 3 (Dll3) which probably functions as a natural antagonist [20]. To consider the Notch's signal effects in different tissues, the characterization of the Notch ligand distribution through the body is determinant. In situ hybridization of E12 through E17 murine fetal liver evidenced widespread expression of Jag1 and Dll1 ligands [21]. Micro-niches of stromal cells expressing Dll1 in fetal liver were also confirmed by a comparative E18 fetal thymic and hepatic immunostaining for CD31 and Dll1 [22].
Dll4 is indispensable for T-cell development and specific of the T cell environment in the thymus [23]. Dll4 was defined as a nonredundant Notch1 ligand in the thymus as Dll1 was not detected on thymic epithelial cells using Dll1-lacZ reporter knock-in mice [24]. Even if Jagged (Jag) ligands were found not implicated in T cell development, Jag1 and Jag2 were expressed in the thymus with a progressive decrease of Jag1 mRNA in the developing thymus [25].
Jag1 and Jag2 are mostly expressed in bone marrow (BM) sinusoidal endothelium and perisinusoidal Nestin-GFPlow cells [26]. Jag2 expression is especially enriched in BM endothelial cells. After myeloablation, surface levels of Jag1 and Jag2 are increased by BM endothelial cells and their co-expression appeared important in case of hematopoietic reconstitution [27,28]. In the BM, the expression of notch ligands like Dll is very low compared to Jag [26]. Immunohistochemical (IHC) analyses of Dll4 in paraffin-embedded BM sections uncovered expression of Dll4 restricted to mature and immature endothelium [29]. IHC and the use of Dll4-LacZ reporter mice allowed the detection of Dll4 expression by splenic white pulp arteriolar endothelium and smaller blood vessels of the red pulp [29,30]. Dll1 and Dll4 were also found to be respectively expressed by splenic and lymph node fibroblasts and were demonstrated as crucial for the immune differentiation process of peripheral lymphocytes [31]. Indeed, conditional ablation of Dll4 from Ccl19-expressing lymph nodes fibroblasts dramatically reduced the production of high affinity antibodies as the differentiation of both follicular helper T cells and germinal center B cells was impaired. Specific genetic ablation of Dll1 from Ccl19-expressing splenic fibroblasts validated its importance in the differentiation of marginal zone B cells and Esam+ dendritic cells.
The Notch pathway is decisive for the maintenance of the intestinal epithelium homeostasis [32,33]. Numerous studies have been performed to locate the populations of intestinal epithelial cells (IECs) expressing Dll ligands [[34], [35], [36], [37]]. It has been shown that Dll1+ and Dll4+ IECs are distinct along the crypt–villus axis with Dll1+ IECs residing exclusively within the crypt while Dll4+ IECs were located both to crypt and villus of the small intestine [[35], [36], [37]]. Paneth cells were selectively expressing Dll4 while goblet cells co-expressed Dll1 and Dll4 despite a clear over-representation of Dll4 [37]. In case of inflamed colonic mucosa such as DSS-induced colitis models, reports concerning the Notch pathway implication could be contradictory. IHC studies showed an increased number of Dll4+ cells while crypt Dll1+ cells were lost [37]. However, a significant increase of Jag1, Dll1 and Dll4 mRNA expression was detected using qRT-PCR from proximal and distal colonic mucosa [38]. Finally, the wide expression of Notch ligands through the body suggests that environmental changes in numerous tissues could regulate immune cell development and functions.
Methods to study and manipulate the Notch signaling pathway
Both in vitro and in vivo manipulations are used to study the effect of the Notch signaling pathway on immune cell populations. In vitro cell cultures could be performed directly in liquid media or using a co-culture system with a layer of stromal cell lines. Indeed, for inducing the differentiation of hematopoietic progenitors in culture, the OP9 stroma cell lines are often used as they were derived from mouse bone marrow stromal cells. Devoid of DLL1 or DLL4 expression, OP9 stromal cell lines were transduced to express these Notch ligands in order to promote the T cell differentiation [39]. Both DLL1 and DLL4 can induce T cell differentiation with different efficiencies. Studies also used TSt4 Tet-off stromal cell lines expressing DLL1 or DLL4 for which DLL expression can be down-regulated by doxycycline (Dox) in a dose-dependent manner [40].
The canonical Notch signaling pathway could be fully abrogated irrespectively of the Notch receptor–ligand interactions by the use of pharmacological agents such as gamma-secretase inhibitors (GSIs), which inhibit the NICD release. GSIs could be used for in vitro and in vivo studies. Neutralizing mAbs against specific Notch receptors or ligands could be a more discriminatory tool. For in vivo studies, different genetic models for Notch loss or gain of function exist. A clear-cut abrogation of the canonical Notch signaling pathway could be obtained by the conditional elimination of RBP-JK or the conditional expression of a dominant negative MAML (dnMAML). Conditional inactivation of specific Notch receptors or Notch ligands are largely used mouse models. Alternatively, mice with a Notch gain-of-function in specific populations were generated by the selective induction of the constitutive NICD expression. Since the IL-7R is driving lymphopoiesis and marks all lymphoid progenitors, Il7rCre mouse were selected to specifically delete genes from the earliest stage of lymphoid progenitors. Il7rCre mice were coupled with RBP-JKfl/fl and Notch2fl/fl mouse strains to inactivate [41,42] or with Rosa26loxP-Stop-loxP-NICD to constitutively activate the Notch signaling [41].
The implication of the notch signaling pathway is quite difficult to draw due to numerous parameters involved in its regulation. Multiple Notch receptors could be expressed by the same cell and could then be simultaneously engaged increasing the complexity of the downstream activation of the pathway. Furthermore, the diverse ligands could be presented by different subsets of cell implicating divergent interactomes that may lead to alternative fate. On top of the extended possible combination of Notch receptors-ligands, differences could also arise from differential glycosylation of Notch receptors, a critical factor for ligand selectivity and signal activation [43,44]. Hence, it should be kept in mind that an apparently homogeneous population may convey different information despite the activation of the same canonical signaling pathway.
Notch and ILC development
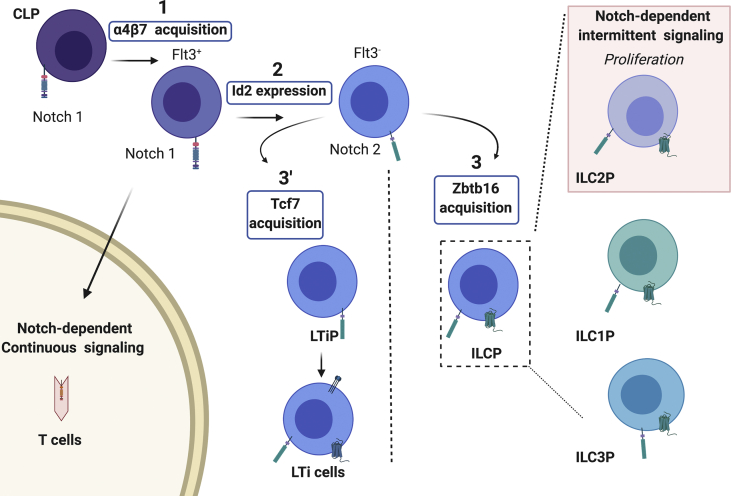
Lineage tracing, cell transfers and single cell analyses have determined that ILC1s, ILC2s and ILC3s, but not LTi cells were originated from a common ILC dedicated precursor (ILCP) characterized by the expression of the transcription factor PLZF (encoded by Zbtb16) [[45], [46], [47], [48]]. In the embryo, ILCs were mostly represented by LTi cells due to their crucial role for the formation of secondary lymphoid (lymph nodes and Peyer's patches) [49]. In the adult, an equivalent fraction of fetal LTi cells was found and named LTi-like cells (please see the review by Van de Pavert for LTi and LTi-like cell origin). Derived from the LTiP, LTi cell commitment was induced before that of helper ILC subsets. Roughly, the common lymphoid precursor (CLP) gave rise to a mix of ILCP, LTi precursor (LTiP) and NK precursors (NKP), all of them were comprised in the α4β7+ fraction of lymphoid precursors, called αLP. High levels of Id2 expression by lymphoid precursors correlated with ILC commitment. The further CXCR6 upregulation correlated with the loss of both T and B lineage potential [50]. Numerous other transcription factor genes of the early transcriptional network include Nfil3, Tox, Gata3, Tcf7 and Zbtb16 are all required for the development of several ILC lineages. More selective expression of specific genes for individual ILC lineages, such as Tbx21 for ILC1, Rora, Bcl11b and Gfi1 for ILC2 and Rorc for ILC3 are expressed later on.
Notch2 is upregulated by lymphoid precursors oriented toward the ILC lineage
While T cell development is linked to Notch1, the development of ILC is especially related to Notch2. Notch2 signaling was already shown to be essential to marginal zone B cells using either conditional Notch2 KO (Mx-Cre) mice or mice with a specific depletion of RBPJ in all B cells [51,52]. It is also needed for the development of CD11b+ dendritic cells (DC) in the spleen and intestine using mouse models deleting Notch1 and Notch2 specifically in DC (CD11c-Cre) [53,54]. Notch1 and Notch2 transcripts were both detected from lymphoid progenitor fractions with an inverse abundance along the developmental ILC path. Indeed, the progressive loss of both Notch1 expression and T cell potential coincided with the acquisition of Id2, CXCR6 and Notch2 expression by lymphoid precursors in both fetal liver and adult bone marrow [22,50]. It was shown using Id2 reporter mouse strains that committed Id2+ ILC precursors were not able to express RORγt in the bone marrow and required to leave the bone marrow to end up their differentiation in the peripheral organs [50]. Hence, peripheral Id2+ lymphoid precursors were isolated from spleen and intestinal lamina propria from which only Notch2 transcripts were detected [50].
The Notch signaling pathway is not necessary for early ILC specification
In order to determine whether the Notch signaling pathway was necessary for ILC commitment, sorted murine CLPs from fetal liver and bone marrow were cultured on OP9 and OP9-Dll4 stromal cell lines (+/− GSIs addition, as explained in 1.2). In both conditions, Id2+ ILCP restricted to the ILC fate were obtained suggesting that the Notch signaling was not necessary for the early specification of the ILC lineage [50]. A subsequent study also using in vitro assays proposed that Notch was needed to induce the α4β7+ Id2 committed stage from fetal liver CLPs [22]. Results from in vitro studies might vary depending on differences of culture conditions. Only mouse models for which lymphoid precursors are deficient or constitutively activated for the Notch signaling pathway could confirm the extended impact of Notch on their commitment and differentiation. Indeed, studies using mice with RBP-Jk deficient lymphoid progenitors (IL7RcrexRBP-JKf/f) confirmed that the Notch signaling was not required for the first stages of fetal ILC lineage specification [41]. The fetal liver lymphoid precursors of RBP-JK deficient mice were expressing similar levels of α4β7 and upregulated the Id2 transcription factor in similar frequencies [Fig. 1] [41]. Experiments using constitutive NICD expression (Il7rcreRosa26NIC mice) also provided evidence that a sustained strong Notch signaling directed the development of lymphoid precursors toward the T cell fate bypassing any other possible fate [Fig. 1] [41]. Contrary to their littermate control fraction, population of fetal liver αLP with a constant activation of Notch failed to express specific ILC transcripts (Id2, Rora, Rorc …) but instead expressed numerous transcripts specific of the T cell commitment (CD3e, Nrarp, Notch1 …) [41]. Hence, the duration of Notch signals could control the cell fate decision during the commitment of lymphoid precursors [Fig. 1]. The Notch intermittent activation could regulate the proliferation of fetal liver ILC2 precursors (ILC2P) [41]. The Notch pathway was determined as active in a sub-fraction of fetal liver αLP enriched in ILC2P and devoid of LTiP thanks to the use of transcriptomic analyses on single cells. Indeed, cells expressing Notch target genes were determined and their frequency were compared between RBPJk deficient and sufficient embryos. A fraction of Hes1+/Nfil3+ ILC2Ps was found reduced after the abrogation of the Notch signaling pathway. Other single cell transcriptomic studies on early fetal liver progenitors also confirmed that none of the earliest αLP stages were dependent on the Notch signaling to express ILC crucial transcription factors [55,56]. TCF-1 is absolutely required at early steps of ILC development [57,58]. The very recent identification of the regulatory element for Tcf7 expression in ILCs allowed to confirm that Notch binding was essential for T cell development, but not for ILC commitment. Hence, to initiate Tcf7 expression in T cells and ILCs, identical regulatory elements are used by different transcriptional controllers [59].
Fig. 1.
Notch implication during the early fetal stages of ILC commitment and differentiation. Notch signaling is differentially implicated in the development of the diverse subsets of fetal liver precursors. The common lymphoid progenitor (CLP) expresses the Notch1 receptor and CLPs could further differentiate into lymphoid precursors expressing the α4β7 integrin at the surface (1). Named α-lymphoid precursors (αLP), the α4β7-expressing cells comprise a mix of lymphoid precursors differentially committed. Thanks to the use of RBP-Jk deficient embryos, this first developmental step was shown to be Notch-independent and to coincide with the loss of B cell potential. Among αLP, Flt3+ cells maintain Notch1 expression and their T cell potential. For T cell development to occur, a constant Notch signal is needed. The constant Notch activation limits any other possible fate as the constitutive NICD expression restrains the Id2 expression and drives αLPs toward T cell development. Flt3- αLP precursors are Notch2+ cells, heterogeneously composed of primed ILC precursors dedicated to ILC1, ILC2, ILC3 and LTi cell lineages. The developmental step (2) concurs with the acquisition of Id2 expression and the loss of T cell potential. This step was demonstrated to be independent from the activation of the Notch pathway. While ILCPs acquire Zbtb16 expression (3), LTiPs are separated as precursors expressing Tcf7 independently from the Notch signaling pathway (3′). During the priming of ILCPs, only the fraction enriched in ILC2Ps is found sensitive to the abrogation of the Notch pathway. Hence, Notch signaling has a primordial role in enhancing the proliferation of ILC2Ps. Moreover, it was determined that Notch signaling in that context was probably intermittent but not constant. The commitment of ILCPs toward ILC1P and ILC3P branches is similar after the blockade of Notch signaling. The figure was created using BioRender.com.
The Notch signaling pathway is not necessary for fetal LTi cell development
Fetal liver CLPs generated RORγt+ ILC3s (mostly LTi cells) indifferently on OP9 or OP9-DL4 stromal cell lines [50]. The addition of GSIs to culture media did not change the fate of these fetal precursors showing that Notch, although possibly active, is not crucial to the development of fetal LTi cells [50]. Fetal liver CLPs were cultured on TSt4 Tet-off stromal cell lines with either high concentrations of Dox indicating weak Notch signaling or low concentrations of Dox for strong Notch signaling. As commonly observed, the B cell differentiation was preferentially induced in high Dox concentrations contrary to T cells. In case of intermediate concentrations of Dox, fetal liver CLPs were able to give rise to ILC2s and NK/ILC1s [40]. However, LTi cells were described as obtained under strong Notch signaling. From the data provided, LTi cells were determined as Lin−Gata3loRORγt+ and while this subset was clearly gated as an independent population in high Dox concentrations, this distinction was less clear in case of low concentrations for which Lin−Gata3+RORγt+ cells were present in a large amount [40]. While ILC3s needed strong Notch signaling, LTi cells were obtained in both medium and high Dox conditions. Mouse models confirmed that LTi cells were functional and present in normal ratio even if the Notch pathway was abrogated [Fig. 1] [40].
The absence of ILC3 commitment in the adult bone marrow is linked to the Notch pathway
The bone marrow is devoid of any RORγt expressing cells implying that ILC3 differentiation is impossible in the bone marrow in the steady state. The observation of ILC3s from bone marrow CLPs was possible but absolutely required any Dll (Dll1 or Dll4) expression by OP9 cell lines. Moreover, the use of GSIs blocked the ILC3 generation confirming that the activation of the Notch pathway was essential for the adult ILC3 differentiation [50]. The ILC3 differentiation was promoted in low dose of Dox accordingly to the need for strong Notch signaling [40]. The Notch signaling was then proposed as essential for the differentiation of adult RORγt+ ILC3s [50].
Since adult ILC3s were found in adult peripheral organs, and were reconstituted by bone marrow precursor transfers, it was postulated that bone marrow ILCPs have to circulate to encounter Notch ligands [50]. Dll1 and Dll4 could be encountered in the small intestine, which is an organ enriched in ILC subsets. Thanks to the use of reporter mice, rare Id2+CXCR6+ precursors were isolated from the small intestine suggesting that this site is a potential place for ILC3 differentiation [50]. In healthy humans, ILC progenitors have been identified in the blood while circulating ILC3s were absent [60]. It has been suggested that these ILCPs recruited to tissues could differentiate locally in response to Notch ligands and/or to other environmental stimuli [60]. Finally, Notch deficient and Notch constitutively active mouse models provided definitive proofs that the Notch signaling was absolutely required in vivo for a normal balanced ILC3 differentiation but not for the LTi branch [Fig. 2] [41,42,61].
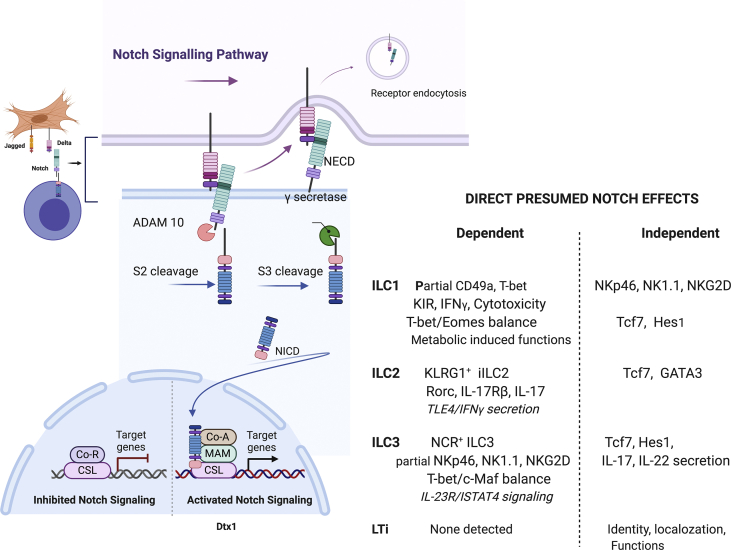
Fig. 2.
The direct Notch signaling effects on ILC biology. The Notch signaling pathway is activated by the binding to ligand presented by a neighboring cell. Here, interactions of ILC subsets with Delta (Dll) or Jagged (jag) expressing cells such as stromal fibroblastic, epithelial or dendritic cells are considered. The molecular steps involved in the Notch signaling activation are divided into the stage of Notch target gene repression and activation. In the absence of signaling, the DNA-binding protein RBP-JK recruits corepressor complexes and histone deacetylases to repress transcription of Notch target genes. Alternatively, in case of Notch ligand binding, the ADAM10 catalyzes a specific S2 cleavage step at the Notch receptor. An S3 further cleavage performed by a γ-secretase releases the intracellular domain of Notch (NICD) that migrates to the nucleus. Then, NICD interacts with RBP-JK and coactivators like Mastermind (MAML-1) and other transcription factors are recruited to form a complex resulting in the transcriptional activation of Notch target genes. On the opposite cells, the Notch ligands are eliminated from the membrane by the endocytosis of the receptors. Globally, endocytosis and membrane trafficking regulate Notch ligand and receptor availability at the cell surface. Dtx1 is a Notch target gene found as commonly activated into all ILC subsets. For each ILC subset (ILC1, ILC2, ILC3 and LTi/LTi-like), a number of target genes and effects have been proposed and the list here present those that have been experimentally shown to be Notch dependent or independent. The effects that might be related to the Notch pathway activation but not yet validated are indicated in italics. The figure was created using BioRender.com.
Notch allows the maintenance of the equilibrium and maturation of ILC3 subsets in the small intestine
In the adult intestine, ILC3s were composed of heterogeneous subsets in mice and humans [[62], [63], [64]]. CCR6 discriminates the LTi-like from other adult ILC3s, which are all CCR6- subsets. While CCR6- ILC3s are specialized in IL-22 secretion, human adult LTi-like ILC3s are preferentially secreting IL-17 while still able to produce IL-22 [63]. Contrary to the dispersion of most ILC subsets throughout the intestine, LTi-like ILC3s are clustered in cryptopatches [65]. This difference of topographical location for ILC3 subsets is probably correlated with distinct functions that result from dependency on various signaling pathway inputs such as the Notch pathway. The implication of the Notch pathway at the bifurcation of adult ILC differentiation was suspected by diverse studies [64,66,67]. Among CCR6- ILC3s, a large number of cells were positive for NKp46, the natural cytotoxicity receptor (NCR) encoded by the Ncr1 gene. It was demonstrated that NCR+ ILC3s derived from their NCR− precursors in a T-bet- and Notch-dependent manner [64,67]. Indeed, in mice with a Notch abrogated pathway due to the selective depletion of RBP-JK or Notch2 expression within the lymphoid compartment (e.g. mouse models in 1.2), NCR+ ILC3s were strongly decreased in the lamina propria, similarly to the decrease in Ahr-deficient mice [42,59,64]. AhR is a Notch-related pathway inducing Notch1, Notch2 and IL-1R expression by immune cells [68,69]. The Notch signaling was defined as one of the crucial pathways downstream of AhR responsible for the presence of intestinal NCR+ ILC3s [66]. Thanks to the use of RBP-JK depleted mice, the heterogeneity of NCR+ ILC3s was revealed as being a mix of a large Notch2-dependent and a minor Notch2-independent populations [42]. It should be noted that the disturbed homeostasis of intestinal ILC3s was equivalent when the Notch pathway was abrogated in all hematopoietic cells using a Vav-cre system [42]. Since RORγt+ NCR− ILC3s were present in RBP-JK depleted mice, Notch could not be considered as an activator of the Rorc gene expression while it is responsible for the final correct differentiation into NCR+ ILC3s.
The use of competitive bone marrow reconstitution experiments demonstrated that the Notch pathway had a direct cell-intrinsic action allowing the differentiation of NCR− precursors into NCR+ ILC3s. The Notch signaling was defined as a decisive pathway downstream of AhR in the generation of most intestinal NCR+ ILC3s [42]. However, the role of Notch was more restricted to specific subsets than the AhR as its deficiency did not impair LTi-like ILC3s. The usage of Ncr1 fate-mapping (FM) in mice defined that heterogeneity was already present among NCR− ILC3 precursors since a fraction had previously expressed the Ncr1 gene in a transitory manner [61]. Hence, it was determined that a sustained Notch signaling activation by ILC3 precursors is essential to maintain an intestinal NCR+ ILC3 identity [61]. Reciprocally, the TGFβ signaling pathway was identified as a counter effect pathway of the Notch signaling during NCR− ILC3 differentiation as it blocked the maturation of NCR+ ILC3s [61]. The use of mice with a constitutively active Notch signaling in lymphoid progenitors showed an increased number of both NCR+ ILC3s and their NCR− precursors [42]. Compatible with a role for the Notch signaling in the differentiation of NCR+ ILC3s, it confirmed that the intestinal homeostasis of ILC subsets is actively maintained by a sensitive balance between opposing pathways.
NCR+ ILC3s could be involved in type 1 inflammatory immune responses and both Notch signals and T-bet were shown to regulate the ILC3 intrinsic c-Maf expression. Reciprocally, c-Maf was able to directly restrain T-bet expression forming a negative feedback loop to maintain the CCR6- ILC3 subset equilibrium [Fig. 2]. Hence, Notch could act on the c-Maf/T-bet balance as a cell-intrinsic brake in the type 1 effector acquisition by NCR+ ILC3s [70]. During intestinal inflammation, a dysregulation of the Notch pathway might disrupt this balance and lead to pathogenic functions.
Both in human and mice, the downregulation of the transcription factor RORγt in NCR+ ILC3s in which the upregulation of T-bet expression was complete was described as plasticity of ILC3 into an ILC1-type named “ex-ILC3” [[71], [72], [73]]. The activation of the constitutive form of Notch by all NKp46 expressing cells did not change the phenotype of intestinal NCR+ ILC3s [42]. The activation of Notch signaling pathway or its abrogation was insufficient per se to drive ILC3 to ILC1 conversion [42].
Adult ILC2 development requires the Notch signaling
Early pioneer work on ILC2 identification first demonstrated that Notch signaling was essential to ILC2 development from murine bone marrow CLPs [74]. The in vitro differentiation of human ILC2s was also driven by the Notch signaling [75]. In vitro assays on OP9-DL1 suggested that the strength of the Notch signal was crucial as only strong Notch1 signal drove thymic progenitors towards the ILC2 fate at the expense of T cells [75]. Nevertheless, ILC2s are present in normal ratio in nude athymic mice [74,76]. To be able to enlarge that notion of strength signal to the overall ILC2 lineage, experiments have to assess diverse origin for ILC2s for which progenitors are not in competition to give rise to T or ILC2s. In murine T cells, the Notch signaling was proposed to control a distal enhancer for the initiation of the Bcl11b gene activation that might have been similarly controlled during ILC2 development [77,78]. However, recent molecular analyses of the gene network circuitry that activates the Bcl11b locus in early T cells versus ILC2s demonstrated that the protein Bcl11b is used differently [79]. Indeed, the distribution of Runx factors and Gata3 at these binding sites were different in pro-T cells and ILC2s. It underlined that even if regulatory factors are identical (e.g the Notch pathway) and similar transcription factors operates, the final regulation of the molecular program in distinct cells will also differ by targeting many different genes [79].
In conclusion, it is now widely accepted that the Notch signaling pathway is not required for the early commitment of Id2+ ILC precursors. The RBP-JK mediated Notch signaling pathway is nevertheless shown as crucial for the development and plasticity of helper ILC2s and ILC3s but it is not for the LTi lineage, neither the fetal population nor their adult LTi-Like counterparts. This differential regulation among the ILC3 branch could be related to the fact that these diverse ILC3 subsets were emerging from distinct earlier precursors, which are located in different niches (please see the review by Van de Pavert for LTi cell origin).
Regulation of ILC functions by the Notch signaling pathway
Notch can drive the acquisition of ILC3 features by ILC2s
The Notch signaling pathway was determined as important to drive the identity of KLRG1hi ILC2 subtype known as inflammatory ILC2s (iILC2s) [80]. Lineage plasticity over stability is governed by numerous unclear mechanisms and the Notch was one of the pathways defined to modulate the plastic variation toward iILC2s. iILC2s were functionally characterized as Th2 cells with complementary Th17 functions. On top of Gata3 expression, murine iILC2 expressed high amounts of Dtx1, and low levels of Rorγt, conferring to the same cell key functions of both ILC2s and ILC3s [80]. The in vivo use of GSIs or injection of selective therapeutic antibodies antagonists of Notch1 or/and Notch2, as described in 1.2, determined that the iILC2 Notch-induced plasticity was acting through both Notch1 and Notch2. It appeared from in vitro cultures on OP9-DL1 that Notch signaling was instructing the cytokine responsiveness of murine ILC2 by converting the IL-33 responsive into IL-25 responsive cells [80]. Furthermore, Notch transcriptional complex could directly bound the Rorc gene locus to stimulate its expression and Th17 capacities without affecting the expression levels of Gata3. Hence, Notch-induced IL-17 secretion did not compromise the constitutive Th2 functions of iILC2s [Fig. 2] [80].
Notch signaling could influence ILC1/NK cell identity by modulating T-bet expression
Numerous studies have shown that abrogating the Notch signaling pathway have no clear defect in the final generation of mature NK cells despite a strong decrease of the refined NKP stage in the bone marrow [81,82]. By comparing the expression of few Notch gene targets from RBP-JK deficient and competent mice, the Notch pathway was suggested to be active in ILC1 and NK cells from most tissues, except for substantial amounts of splenic Notch insensitive NK cells [83]. Next-generation sequencing of ILC subsets from the murine small intestine have pointed out that the Notch signaling together with the mTOR are major metabolic pathways used to regulate ILC1 intestinal activities [Fig. 2] [84].
The Notch pathway implication on ILC1/NK cell specification and functions was further analyzed for hepatic murine subsets. Single cell transcriptomic assays determined the heterogeneous combinatory expression of Notch1 and Notch2 by hepatic ILC1 and NK cells. Half of hepatic ILC1 and cNK populations expressed Notch receptors, and these cells either expressed Notch1 or Notch2 or both. The RBP-JK deficiency was directly correlated to the decrease of the Tbx21 gene expression and the ectopic induction of Eomes. Diverse redundant signaling pathways might regulate T-bet expression in ILC1 and NK cells with the Notch pathway being one of them for at least half of the hepatic pool [83]. Moreover, hepatic ILC1 from RBP-JK deficient mice expressed less CD49a and this decrease was specifically related to Notch1 deficiency but not Notch2. A deregulation of proliferation/survival capacities also probably affected the cNK/ILC1 ratio [Fig. 2] [83].
Notch as a possible actor for the activation/repression of ILC functions
Notch signaling was suggested as a possible way to control the IFNγ production by human ILC2s [85] as they expressed TLE4, one Notch pathway related co-repressor which has been shown to repress IFN-γ expression through epigenetic regulation [86].
Under IL-23 activation, STAT4 and STAT3 were strongly induced in murine NCR+ ILC3s for IL-22 secretion but not in LTi-like cells [87]. Strong upregulation of STAT4 is observed in activated ILC1s under IL-12 stimulation. The specific upregulation of STAT4 in response to IL-23 was found related to T-bet expression and to a delayed IFNγ secretion by NCR+ ILC3s [87]. STAT4 and IL-12 signaling were both determined as dispensable for the initial induction of T-bet expression by CCR6- ILC3s [64]. However, T-bet was shown as essential to maintain a STAT4 expression among NCR+ ILC3s but not ILC1s [87]. While T-bet is a possible direct target of the Notch signaling pathway, it was not determined whether the Notch signaling pathway could interfere along the IL23R/STAT4 axis in ILC3s.
The decrease abundance of NKp46 and NKG2D at the cell surface of Notch-depleted intestinal ILC3s suggested a partial regulation of NK cell marker expression by the Notch pathway [42]. However, this was restricted to ILC3 as RBP-JK depleted and wild-type NK cells exhibited similar NKp46 and NK1.1 expression levels [82].
Notch signaling was largely described as impacting Th1 differentiation by increasing the cytotoxicity of Th1 cells. Notch signaling has been implicated in the upregulation of KIR molecules from human NK cells [88]. DC mediated Jagged2–Notch interactions were proposed as crucial for the increase of the murine peripheral blood NK cell proliferation, cytotoxicity and IFNγ release [89]. Human NK cells could be activated by Dll4 to increase their IFNγ release respectively in presence of IL-12 for peripheral blood or IL-15 for the decidual subset [90]. However, reversed effects were observed in mouse models with a deficient Notch signaling pathway in which cNK subsets were more activated and cytotoxic [82,83]. The maintenance of the Th1 response was also observed in other studies using diverse mouse models as described in 1.2, such as Notch1/Notch2, RBP-JK deficient, and MAML1 dominant-negative mice [Fig. 2] [91,92].
The impact of the Notch pathway on the developmental maturation of human tonsil-derived “ILC precursors” was recently provided using in vitro cultures on OP9-DL1 [93]. Contrary to the NK cell fate, ILC2 and ILC3 fate was only obtained when earliest stages of precursors were cultured in presence of Notch ligand Dll1 while NK cells could equally be obtained in absence or presence of Dll ligands. They also demonstrated that the presence of plate-bound Notch ligands in the absence of any stromal cells was not sufficient. Moreover, the Notch pathway was also needed to promote NKp80 acquisition from later stages of committed NK cells. Hence, the Notch pathway was needed in order to obtain ILC2s, ILC3s and functional mature human NK cells from tonsils [93].
In case of epidermal damages, it has been reported that epidermal Notch1 induction was recruiting immune subsets such as ILC3s during the wounding of the dermis [94]. The Notch signaling pathway was here indirectly responsible for ILC recruitment via CCL20 and CXCL13 expression. Notch1 was here considered as a signal of damages that could stimulate the skin repair processes by promoting IL17 secretion from recruited ILC3s [94]. Hence, on top of being an important intrinsic signaling pathway for ILC biology, Notch could also interfere on ILC functions indirectly.
Notch and the tissue homeostasis via the AhR-IL22 signaling axis
ILC3s together with T cells regulate intestinal homeostasis in case of damages by balancing immune activation and tissue repair. IL-22 is a cytokine from the IL-20 family, peculiarly known to be involved in the process of epithelial regeneration. The release of IL-22 by RORγt+ cells is induced in close proximity to epithelial damages. In case of acute or chronic injury implicating the regeneration of human tissue homeostasis, a clear induction of Notch/AhR/IL-22 axis was detected by gene expression profiling [95]. ILC3s are located in mucosal sites, where they are exposed to large amounts of commensal bacteria and possibly pathogens. Both Th17 subset of CD4+ helper T cells and ILC3s are able to secrete IL-22 in response to IL-23, which is produced by dendritic cells and macrophages after microbial metabolite recognition [96,97]. Notch is indirectly involved in the IL-22 secretion by the human T cell compartment as it promotes the differentiation of Th17 cells [98,99] and the survival of IL-17 γδT cells [100]. As discussed above, the related AhR and Notch pathways are absolutely required for the development of the intestinal IL-22 producing NCR+ ILC3s [42,50,61,101]. The innate IL-22 production in the gut mucosa was shown to require AhR [66]. However, no significant differences of IL-22, nor IL-17 secretion were noted after the in vitro stimulation of the remaining Notch deficient ILC3 subsets on OP9-DL4 [Fig. 2] [42]. Hence, the Notch pathway impacts on intestinal IL-22 production mostly via the abrogation of ILC3s maturation into NCR+ secreting cells. Finally, all these data suggest that the Notch signaling pathway may indirectly modulate the cytokine milieu in the context of tissue injury. In the case of IL-22 secretion, Notch rather regulates the abundance of ILC subsets by balancing their maturation in the tissue than modifying their secretion.
Conclusion
The Notch pathway is largely impacting the biology of ILCs from their initial development to their terminal functions. We reviewed how the Notch pathway is fine tuning the identity of ILC subsets present in different tissues and how it could balance their functions. However, more knowledge should be obtained, especially on the intrinsic molecular mechanisms. Data analyzing the implication of the Notch pathway should now be designed at the single level to avoid ambiguities on the possible resulting effects of modulating this pathway. Indeed, it is still very difficult to distinguish whether through the same receptor, the identical canonical pathway could convey a different information depending on the ligand identity and the duration/strength of the signal. An obvious difference is probably driven by the stage of the cell, which may be differentially activated (e.g. different jak–stat combinations) and via interfering signaling pathways modulating both transcription factor expression and the chromatin state. The challenge is to use the best tools to integrate data and determine a core determinant network implicating the Notch signaling steps. Finally, the Notch signaling pathway could be considered as one of the major future strategical targets to modulate the immune response of ILCs in response to the inflammation.
Conflicts of interest
The author declared no competing interests.
Acknowledgments
I am very grateful to Nathan Mackowski for his help in proofreading the review. RG work is supported by Institut Pasteur, Institut National de la Santé et de la Recherche Médicale, Université de Paris, and ANR project NASHILCCD8 (#18-CE15-0024-01), Projets ARC 2020 PJA3, Fonds AMGEN France.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Yasutomo K. Springer Singapore; Singapore: 2017. Notch signaling. [Google Scholar]
- 2.Radtke F., MacDonald H.R., Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 3.Ohishi K., Varnum-Finney B., Bernstein I.D. The notch pathway: modulation of cell fate decisions in hematopoiesis. Int J Hematol. 2002;75:449–459. doi: 10.1007/BF02982106. [DOI] [PubMed] [Google Scholar]
- 4.Vanderbeck A., Maillard I. Notch signaling at the crossroads of innate and adaptive immunity. J Leukoc Biol. 2021;109:535–548. doi: 10.1002/JLB.1RI0520-138R. [DOI] [PubMed] [Google Scholar]
- 5.Rothenberg E.V., Scripture-Adams D.D. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin Immunol. 2008;20:236–246. doi: 10.1016/j.smim.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson P.K., Zúñiga-Pflücker J.C. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–359. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Amsen D., Helbig C., Backer R.A. Notch in T Cell differentiation: all things considered. Trends Immunol. 2015;36:802–814. doi: 10.1016/j.it.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Cherrier M., Ramachandran G., Golub R. The interplay between innate lymphoid cells and T cells. Mucosal Immunol. 2020;13:732–742. doi: 10.1038/s41385-020-0320-8. [DOI] [PubMed] [Google Scholar]
- 9.Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G. Innate lymphoid cells: 10 Years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Ebbo M., Crinier A., Vély F., Vivier E. Innate lymphoid cells: major players in inflammatory diseases. Nat Rev Immunol. 2017;17:665–678. doi: 10.1038/nri.2017.86. [DOI] [PubMed] [Google Scholar]
- 11.Bando J.K., Liang H.E., Locksley R.M. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2015;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider C., Lee J., Koga S., Ricardo-Gonzalez R.R., Nussbaum J.C., Smith L.K. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity. 2019;50:1425–1438. doi: 10.1016/j.immuni.2019.04.019. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S., Muskavitch M.A., Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 15.Borggrefe T., Giaimo B.D. Springer International Publishing; Cham: 2018. Molecular mechanisms of notch signaling vol. 1066. [Google Scholar]
- 16.Oyama T., Harigaya K., Sasaki N., Okamura Y., Kokubo H., Saga Y. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development. 2011;138:5235–5246. doi: 10.1242/dev.062802. [DOI] [PubMed] [Google Scholar]
- 17.Giaimo B.D., Oswald F., Borggrefe T. Dynamic chromatin regulation at Notch target genes. Transcription. 2017;8:61–66. doi: 10.1080/21541264.2016.1265702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palomero T., Lim W.K., Odom D.T., Sulis M.L., Real P.J., Margolin A. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng A.P. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopan R., Ilagan M.X.G. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker L., Carlson A., Tan-Pertel H.T., Weinmaster G., Gasson J. The notch receptor and its ligands are selectively expressed during hematopoietic development in the mouse. Stem Cell. 2001;19:543–552. doi: 10.1634/stemcells.19-6-543. [DOI] [PubMed] [Google Scholar]
- 22.Cherrier M., Sawa S., Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hozumi K., Mailhos C., Negishi N., Hirano K., Yahata T., Ando K. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205:2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch U., Fiorini E., Benedito R., Besseyrias V., Schuster-Gossler K., Pierres M. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felli M.P., Maroder M., Mitsiadis T.A., Campese A.F., Bellavia D., Vacca A. Expression pattern of Notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand–receptor interactions in intrathymic T cell development. Int Immunol. 1999;11:1017–1025. doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- 26.Saçma M., Pospiech J., Bogeska R., de Back W., Mallm J.P., Sakk V. Haematopoietic stem cells in perisinusoidal niches are protected from ageing. Nat Cell Biol. 2019;21:1309–1320. doi: 10.1038/s41556-019-0418-y. [DOI] [PubMed] [Google Scholar]
- 27.Ramalingam P., Poulos M.G., Butler J.M. Regulation of the hematopoietic stem cell lifecycle by the endothelial niche. Curr Opin Hematol. 2017;24:289–299. doi: 10.1097/MOH.0000000000000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo P., Poulos M.G., Palikuqi B., Badwe C.R., Lis R., Kunar B. Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J Clin Invest. 2017;127:4242–4256. doi: 10.1172/JCI92309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S.U., Maeda M., Ishikawa Y., Li S.M., Wilson A., Jubb A.M. LRF-mediated Dll4 repression in erythroblasts is necessary for hematopoietic stem cell maintenance. Blood. 2013;121:918–929. doi: 10.1182/blood-2012-03-418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan J.B., Xu K., Cretegny K., Visan I., Yuan J.S., Egan S.E. Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity. 2009;30:254–263. doi: 10.1016/j.immuni.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Fasnacht N., Huang H.Y., Koch U., Favre S., Auderset F., Chai Q. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. 2014;211:2265–2279. doi: 10.1084/jem.20132528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellegrinet L., Rodilla V., Liu Z., Chen S., Koch U., Espinosa L. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akiyama J., Okamoto R., Iwasaki M., Zheng X., Yui S., Tsuchiya K. Delta-like 1 expression promotes goblet cell differentiation in Notch-inactivated human colonic epithelial cells. Biochem Biophys Res Commun. 2010;393:662–667. doi: 10.1016/j.bbrc.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 34.Stamataki D., Holder M., Hodgetts C., Jeffery R., Nye E., Spencer-Dene B. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PloS One. 2011;6 doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim T.H., Li F., Ferreiro-Neira I., Ho L.L., Luyten A., Nalapareddy K. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu H., Okamoto R., Ito G., Fujii S., Nakata T., Suzuki K. Distinct expression patterns of Notch ligands, Dll1 and Dll4, in normal and inflamed mice intestine. PeerJ. 2014;2:e370. doi: 10.7717/peerj.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imaeda H., Andoh A., Aomatsu T., Uchiyama K., Bamba S., Tsujikawa T. Interleukin-33 suppresses Notch ligand expression and prevents goblet cell depletion in dextran sulfate sodium-induced colitis. Int J Mol Med. 2011;28:573–578. doi: 10.3892/ijmm.2011.718. [DOI] [PubMed] [Google Scholar]
- 39.Mohtashami M., Shah D.K., Kianizad K., Awong G., Zúñiga-Pflücker J.C. Induction of T-cell development by Delta-like 4-expressing fibroblasts. Int Immunol. 2013;25:601–611. doi: 10.1093/intimm/dxt027. [DOI] [PubMed] [Google Scholar]
- 40.Koga S., Hozumi K., Hirano K., Yazawa M., Terooatea T., Minoda A. Peripheral PDGFRα+gp38+ mesenchymal cells support the differentiation of fetal liver–derived ILC2. J Exp Med. 2018;215:1609–1626. doi: 10.1084/jem.20172310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chea S., Schmutz S., Berthault C., Perchet T., Petit M., Burlen-Defranoux O. Single-cell gene expression analyses reveal heterogeneous responsiveness of fetal innate lymphoid progenitors to notch signaling. Cell Rep. 2016;14:1500–1516. doi: 10.1016/j.celrep.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Chea S., Perchet T., Petit M., Verrier T., Guy-Grand D., Banchi E.G. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci Signal. 2016;9:ra45. doi: 10.1126/scisignal.aaf2223. [DOI] [PubMed] [Google Scholar]
- 43.Rana N.A., Haltiwanger R.S. Fringe Benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jafar-Nejad H., Leonardi J., Fernandez-Valdivia R. Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology. 2010;20:931–949. doi: 10.1093/glycob/cwq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishizuka I.E., Chea S., Gudjonson H., Constantinides M.G., Dinner A.R., Bendelac A. Single cell analysis defines the divergence between the innate lymphoid cell and lymphoid tissue inducer lineages. Nat Immunol. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker J.A., Clark P.A., Crisp A., Barlow J.L., Szeto A., Ferreira A.C.F. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity. 2019;51:104–118. doi: 10.1016/j.immuni.2019.05.002. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W., Cherrier D.E., Chea S., Vosshenrich C., Serafini N., Petit M. An Id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity. 2019;50:1054–1068. doi: 10.1016/j.immuni.2019.02.022. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasal D.N., Bendelac A. Multi-transcription factor reporter mice delineate early precursors to the ILC and LTi lineages. J Exp Med. 2021;218 doi: 10.1084/jem.20200487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mebius R.E., Rennert P., Weissman I.L. Developing lymph nodes collect CD4+CD3− LTβ+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 50.Possot C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A. Notch signaling is necessary for adult, but not fetal, development of RORγt + innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 51.Saito T., Chiba S., Ichikawa M., Kunisato A., Asai T., Shimizu K. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 52.Tanigaki K., Han H., Yamamoto N., Tashiro K., Ikegawa M., Kuroda K. Notch–RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 53.Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berthault C., Ramond C., Burlen-Defranoux O., Soubigou G., Chea S., Golub R. Asynchronous lineage priming determines commitment to T cell and B cell lineages in fetal liver. Nat Immunol. 2017;18:1139–1149. doi: 10.1038/ni.3820. [DOI] [PubMed] [Google Scholar]
- 56.Cumano A., Berthault C., Ramond C., Petit M., Golub R., Bandeira A. New molecular insights into immune cell development. Annu Rev Immunol. 2019;37:497–519. doi: 10.1146/annurev-immunol-042718-041319. [DOI] [PubMed] [Google Scholar]
- 57.Harly C., Kenney D., Ren G., Lai B., Raabe T., Yang Q. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat Immunol. 2019;20:1150–1160. doi: 10.1038/s41590-019-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Q., Li F., Harly C., Xing S., Ye L., Xia X. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harly C., Kenney D., Wang Y., Ding Y., Zhao Y., Awasthi P. A shared regulatory element controls the initiation of Tcf7 expression during early T cell and innate lymphoid cell developments. Front Immunol. 2020;11:470. doi: 10.3389/fimmu.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim A.I., Li Y., Lopez-Lastra S., Stadhouders R., Paul F., Casrouge A. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell. 2017;168:1086–1100. doi: 10.1016/j.cell.2017.02.021. e10. [DOI] [PubMed] [Google Scholar]
- 61.Viant C., Rankin L.C., Girard-Madoux M.J.H., Seillet C., Shi W., Smyth M.J. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci Signal. 2016;9:ra46. doi: 10.1126/scisignal.aaf2176. [DOI] [PubMed] [Google Scholar]
- 62.Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F. Lineage relationship analysis of RORγt + innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 63.Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K.M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klose C.S.N., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d'Hargues Y. A T-bet gradient controls the fate and function of CCR6 − RORγt + innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 65.Eberl G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., Littman D.R. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 66.Lee J.S., Cella M., McDonald K.G., Garlanda C., Kennedy G.D., Nukaya M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rankin L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kerkvliet N.I. AHR-mediated immunomodulation: the role of altered gene transcription. Biochem Pharmacol. 2009;77:746–760. doi: 10.1016/j.bcp.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevens E.A., Mezrich J.D., Bradfield C.A. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127:299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tizian C., Lahmann A., Hölsken O., Cosovanu C., Kofoed-Branzk M., Heinrich F. c-Maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. ELife. 2020;9:e52549. doi: 10.7554/eLife.52549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vonarbourg C., Mortha A., Bui V.L., Hernandez P.P., Kiss E.A., Hoyler T. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cella M., Otero K., Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U S A. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crellin N.K., Trifari S., Kaplan C.D., Satoh-Takayama N., Di Santo J.P., Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 74.Wong S.H., Walker J.A., Jolin H.E., Drynan L.F., Hams E., Camelo A. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gentek R., Munneke J.M., Helbig C., Blom B., Hazenberg M.D., Spits H. Modulation of signal strength switches notch from an inducer of T cells to an inducer of ILC2. Front Immunol. 2013;4:334 doi: 10.3389/fimmu.2013.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L., Zhang J.A., Dose M., Kueh H.Y., Mosadeghi R., Gounari F. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng K.K., Yui M.A., Mehta A., Siu S., Irwin B., Pease S. A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. ELife. 2018;7 doi: 10.7554/eLife.37851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hosokawa H., Romero-Wolf M., Yang Q., Motomura Y., Levanon D., Groner Y. Cell type–specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J Exp Med. 2020;217 doi: 10.1084/jem.20190972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang K., Xu X., Pasha M.A., Siebel C.W., Costello A., Haczku A. Cutting edge: notch signaling promotes the plasticity of group-2 innate lymphoid cells. J Immunol. 2017;198:1798–1803. doi: 10.4049/jimmunol.1601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ribeiro V.S.G., Hasan M., Wilson A., Boucontet L., Pereira P., Lesjean-Pottier S. Cutting edge: thymic NK cells develop independently from T cell precursors. J Immunol. 2010;185:4993–4997. doi: 10.4049/jimmunol.1002273. [DOI] [PubMed] [Google Scholar]
- 82.Chaves P., Zriwil A., Wittmann L., Boukarabila H., Peitzsch C., Jacobsen S.E.W. Loss of canonical notch signaling affects multiple steps in NK cell development in mice. J Immunol. 2018;201:3307–3319. doi: 10.4049/jimmunol.1701675. [DOI] [PubMed] [Google Scholar]
- 83.Perchet T., Petit M., Banchi E.G., Meunier S., Cumano A., Golub R. The notch signaling pathway is balancing type 1 innate lymphoid cell immune functions. Front Immunol. 2018;9:1252. doi: 10.3389/fimmu.2018.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gury-BenAri M., Thaiss C.A., Serafini N., Winter D.R., Giladi A., Lara-Astiaso D. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166:1231–1246. doi: 10.1016/j.cell.2016.07.043. e13. [DOI] [PubMed] [Google Scholar]
- 85.Björklund Å.K., Forkel M., Picelli S., Konya V., Theorell J., Friberg D. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 86.Bandyopadhyay S., Valdor R., Macian F. Tle4 regulates epigenetic silencing of gamma interferon expression during effector T helper cell tolerance. Mol Cell Biol. 2014;34:233–245. doi: 10.1128/MCB.00902-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mikami Y., Scarno G., Zitti B., Shih H.Y., Kanno Y., Santoni A. NCR+ ILC3 maintain larger STAT4 reservoir via T-BET to regulate type 1 features upon IL-23 stimulation in mice. Eur J Immunol. 2018;48:1174–1180. doi: 10.1002/eji.201847480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Felices M., Ankarlo D.E.M., Lenvik T.R., Nelson H.H., Blazar B.R., Verneris M.R. Notch signaling at later stages of NK cell development enhances KIR expression and functional maturation. J Immunol. 2014;193:3344–3354. doi: 10.4049/jimmunol.1400534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kijima M., Yamaguchi T., Ishifune C., Maekawa Y., Koyanagi A., Yagita H. Dendritic cell-mediated NK cell activation is controlled by Jagged2-Notch interaction. Proc Natl Acad Sci. 2008;105:7010–7015. doi: 10.1073/pnas.0709919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manaster I., Gazit R., Goldman-Wohl D., Stern-Ginossar N., Mizrahi S., Yagel S. Notch activation enhances IFNgamma secretion by human peripheral blood and decidual NK cells. J Reprod Immunol. 2010;84:1–7. doi: 10.1016/j.jri.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Amsen D., Blander J.M., Lee G.R., Tanigaki K., Honjo T., Flavell R.A. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 92.Tu L., Fang T.C., Artis D., Shestova O., Pross S.E., Maillard I. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nalin A.P., Kowalski J.J., Sprague A.C., Schumacher B.K., Gerhardt A.G., Youssef Y. Notch regulates innate lymphoid cell plasticity during human NK cell development. J Immunol. 2020;205:2679–2693. doi: 10.4049/jimmunol.2000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Z., Hodgkinson T., Gothard E.J., Boroumand S., Lamb R., Cummins I. Epidermal Notch1 recruits RORγ+ group 3 innate lymphoid cells to orchestrate normal skin repair. Nat Commun. 2016;7:11394. doi: 10.1038/ncomms11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weidenbusch M., Rodler S., Song S., Romoli S., Marschner J.A., Kraft F. Gene expression profiling of the Notch-AhR-IL22 axis at homeostasis and in response to tissue injury. Biosci Rep. 2017;37 doi: 10.1042/BSR20170099. BSR20170099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Korn T., Bettelli E., Oukka M., Kuchroo V.K. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 97.Sonnenberg G.F., Fouser L.A., Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 98.Mukherjee S., Schaller M.A., Neupane R., Kunkel S.L., Lukacs N.W. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol. 2009;182:7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keerthivasan S., Suleiman R., Lawlor R., Roderick J., Bates T., Minter L. Notch signaling regulates mouse and human Th17 differentiation. J Immunol. 2011;187:692–701. doi: 10.4049/jimmunol.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shibata K., Yamada H., Sato T., Dejima T., Nakamura M., Ikawa T. Notch-Hes1 pathway is required for the development of IL-17-producing γδ T cells. Blood. 2011;118:586–593. doi: 10.1182/blood-2011-02-334995. [DOI] [PubMed] [Google Scholar]
- 101.Lee C.C., Lin C.L., Leu S.J., Lee Y.L. Overexpression of Notch ligand Delta-like-1 by dendritic cells enhances their immunoregulatory capacity and exerts antiallergic effects on Th2-mediated allergic asthma in mice. Clin Immunol Orlando Fla. 2018;187:58–67. doi: 10.1016/j.clim.2017.10.005. [DOI] [PubMed] [Google Scholar]