Keywords: bioinformatics, differential expression, factor, gene, immune response, injury, pathways, protein, spinal cord
Abstract
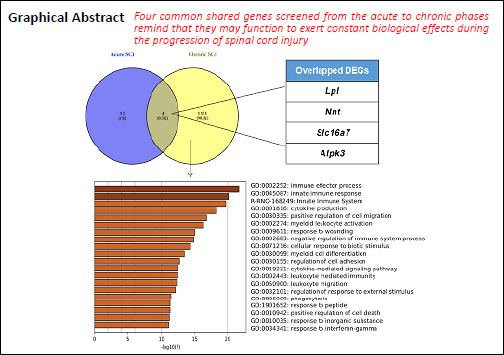
Complex pathological changes occur during the development of spinal cord injury (SCI), and determining the underlying molecular events that occur during SCI is necessary for the development of promising molecular targets and therapeutic strategies. This study was designed to explore differentially expressed genes (DEGs) associated with the acute and chronic stages of SCI using bioinformatics analysis. Gene expression profiles (GSE45006, GSE93249, and GSE45550) were downloaded from the Gene Expression Omnibus database. SCI-associated DEGs from rat samples were identified, and Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed. In addition, a protein-protein interaction network was constructed. Approximately 66 DEGs were identified in GSE45550 between 3–14 days after SCI, whereas 2418 DEGs were identified in GSE45006 1–56 days after SCI. Moreover, 1263, 195, and 75 overlapping DEGs were identified between these two expression profiles, 3, 7/8, and 14 days after SCI, respectively. Additionally, 16 overlapping DEGs were obtained in GSE45006 1–14 days after SCI, including Pank1, Hn1, Tmem150c, Rgd1309676, Lpl, Mdh1, Nnt, Loc100912219, Large1, Baiap2, Slc24a2, Fundc2, Mrps14, Slc16a7, Obfc1, and Alpk3. Importantly, 3882 overlapping DEGs were identified in GSE93249 1–6 months after SCI, including 3316 protein-coding genes and 567 long non-coding RNA genes. A comparative analysis between GSE93249 and GSE45006 resulted in the enrichment of 1135 overlapping DEGs. The significant functions of these 1135 genes were correlated with the response to the immune effector process, the innate immune response, and cytokine production. Moreover, the biological processes and KEGG pathways of the overlapping DEGs were significantly enriched in immune system-related pathways, osteoclast differentiation, the nuclear factor-κB signaling pathway, and the chemokine signaling pathway. Finally, an analysis of the overlapping DEGs associated with both acute and chronic SCI, assessed using the expression profiles GSE93249 and GSE45006, identified four overlapping DEGs: Slc16a7, Alpk3, Lpl and Nnt. These findings may be useful for revealing the biological processes associated with SCI and the development of targeted intervention strategies.
Chinese Library Classification No. R446; R744; Q811.4
Introduction
Spinal cord injury (SCI) refers to severe trauma to the spinal cord and is characterized by high incidence and morbidity (Shi et al., 2020; Wang et al., 2020). SCI is an overwhelming neurological disorder and has devastating physiological symptoms, which can vary widely from pain, to paralysis, to incontinence (van den Berg et al., 2010; Singh et al., 2014; Jazayeri et al., 2015). Traumatic SCI can be classified into primary injury and secondary injury. Primary injury refers to the mechanical and direct damage incurred against the spinal cord (Stahel et al., 2012), whereas secondary injury refers to the pathological responses induced by the immediate injury. SCI results in complex consequences that involve various changes in biological processes, including the breakdown of the blood-spinal cord barrier, neuroinflammation, oxidative stress, necrosis, apoptosis, glial scar formation, ischemic dysfunction, and functional problems, such as neuropathic pain, autonomic dysfunction, muscle atrophy, and incontinence of the bladder, rectum, and anus (Wyndaele and Wyndaele, 2006; Kotipatruni et al., 2011; Wanner et al., 2013; Anwar et al., 2016).
Despite advances in spinal cord surgical techniques and the efforts devoted to the treatment of SCI and patient care, no effective therapeutic solutions for this destructive neurological disease currently exist (Calvert et al., 2019; Lavis and Goetz, 2019; Thomaz et al., 2019). Owing to the complexity of changes that occur in gene expression and signaling pathways following SCI, understanding the underlying molecular mechanisms associated with pathological events represents a critical step in the development of promising molecular targets and therapeutic strategies. Some studies have attempted to understand this process by identifying differentially expressed genes (DEGs), using microarrays or RNA-sequencing analysis, to reveal the pathways and molecular targets that are activated during acute or chronic SCI (Nesic et al., 2002; van den Berg et al., 2010; Byrnes et al., 2011; Chen et al., 2013; Duran et al., 2017; Du et al., 2018; Guo et al., 2019; Liu et al., 2020). The gene expression profiles GSE45006, GSE93249, and GSE45550 were derived from rat SCI models across different time points after injury and have been used to analyze DEGs and their associated biological processes, based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (Wen et al., 2016; Zhang and Wang, 2016; Duran et al., 2017; Liu et al., 2017; Ma et al., 2017; Zhang et al., 2017). Previous studies have focused on temporal gene expression, molecular pathway changes, and changes in the transcriptional regulatory network and molecular mechanisms associated with SCI over time. In addition, comprehensive investigations have examined changes in the expression of both coding and long non-coding genes during the acute and chronic stages of SCI using RNA-sequencing analysis (Duran et al., 2017; Shi et al., 2017; Zhou et al., 2018).
The majority of SCI patients are in the chronic phase of SCI owing to the lack of a cure (Fehlings and Perrin, 2006). However, the pathological changes that are induced during the early stages of SCI might contribute to the development or aggravation of secondary injuries. A major barrier to the identification of ideal therapeutic strategies remains our limited knowledge regarding the coordinated pathophysiological processes induced by SCI. Therefore, determining both the differences and similarities in the molecular changes that characterize acute and chronic SCI is necessary to develop a better understanding of the underlying molecular mechanisms and to develop potential therapeutic strategies. In this study, the expression profiles GSE45006, GSE93249. and GSE45550 were used to perform a comparative analysis of the SCI-related DEGs that change over time and during different phases.
Materials and Methods
Microarray data preprocessing
The gene expression profiles GSE45006, GSE45550, and GSE93249 were downloaded from the Gene Expression Omnibus database of the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/). These expression profiles were based on the GPL1355 platform (GSE45006 and GSE45550) of the Affymetrix Rat Genome 230 2.0 Array and the GPL14844 platform (GSE93249) of Illumina HiSeq 2000. GSE93249 included 11 rat spinal cord samples, including 3 non-injured spinal cord samples in the sham group and 8 SCI samples in the SCI group, representing the chronic phase at 1 month (m1, n = 3), 3 months (m3, n = 3), and 6 months (m6, n = 2) after injury. GSE45550 included 30 rat spinal cord samples, including 6 non-injured spinal cord samples in the sham group and 24 SCI samples representing the acute and subacute phases, at 3 days (d3, n = 8; acute phase), 8 days (d8, n = 8; acute phase), and 14 days (d14, n = 8; subacute phase) after injury. GSE45006 included 24 rat spinal cord samples, including 4 non-injured spinal cord samples in the sham group and 20 SCI samples at 1 day (d1, n = 4; acute phase), 3 days (d3, n = 4; acute phase), 7 days (d7, n = 4; acute phase), 14 days (d14, n = 4; subacute phase), and 56 days (d56, n = 4; chronic phase) post-injury. After the data associated with GSE45006, GSE93249 and GSE45550 were downloaded, the microarray data were pre-processed as follows: the data were converted into expression measures, followed by background correction, quantile normalization, log2 conversion, batch correction, and probe summarization. Finally, the gene expression matrix was obtained.
The identification of DEGs between SCIs and sham samples
The DEGs between SCI and sham samples were screened using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) online tools (Davis and Meltzer, 2007). The false discovery rate (FDR) was obtained by adjusting the raw P-values with the Benjamini–Hochberg method (Madar and Batista, 2016). The fold-change of gene expression (log2 FC) and the FDR were used to select significant DEGs. Genes with log2 FC > 1.5 and FDR < 0.05 were selected as significant DEGs.
GO and KEGG pathway enrichment analyses of the DEGs
GO analysis has been widely used to identify functional enrichment patterns in studies examining large-scale genetic sequencing. The KEGG database contains known genes and their associated pathways. The biological significance of DEGs associated with SCI can be assessed by performing GO and KEGG pathway enrichment analyses. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) provides a comprehensive set of functional annotation tools. DAVID was used to identify DEG-associated pathways by calculating P-values (Huang da et al., 2009). The raw P-values were adjusted using the Benjamini-Hochberg method, and FDR < 0.05 was selected as the cut-off criterion.
Statistical analysis
SPSS 22.0 (IBM, Armonk, NY, USA) was used for all statistical analyses. Comparisons among multiple groups were analyzed using a one-way analysis of variance, followed by Dunnett’s post hoc test. For experiments involving two groups, an unpaired Student’s t-test was performed. A value of P < 0.05 was considered statistically significant. Data are expressed as the mean ± standard deviation (SD).
Results
The DEGs identified between acute SCI and sham groups
The gene expression profile GSE45550 was used to analyze DEGs between the acute SCI and sham groups. Three time points (3, 8, and 14 days after injury) were examined in the experimental groups. The results identified approximately 2411, 463, and 184 DEGs at 3, 8, and 14 days post-injury, respectively. In addition, only 66 overlapping DEGs were identified across the three time points examined between 3 and 14 days post-injury (Figure 1A). The gene expression profile GSE45006 was also examined to further assess DEGs although this profile had a much wider time frame (1, 3, 7, 14, and 56 days post-injury). The results identified approximately 2712 overlapping DEGs during days 1 to 14 post-injury (Figure 1B). Only 2418 DEGs were identified at 56 days post-injury. The above results suggested that the data obtained from GSE45550 and GSE45006 were different.
Figure 1.
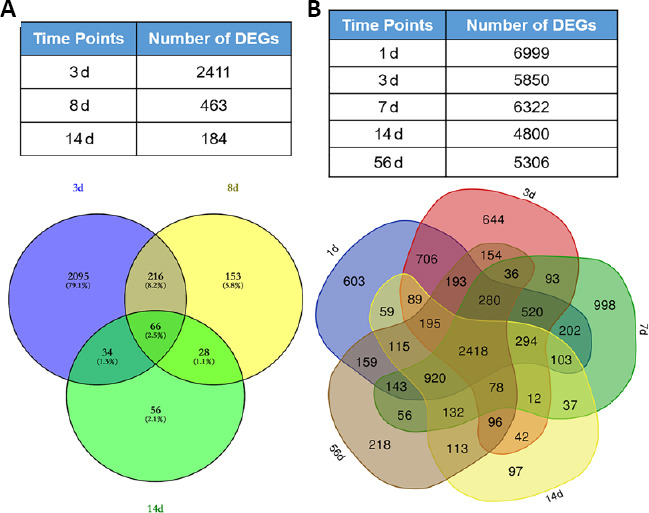
DEGs associated with acute SCI at different time points.
Deregulated transcripts (fold change ≥ 1.5) at each time point were examined for overlapping and unique genes using a Venn diagram. Overlapping areas represent common genes between different time points. (A) The numbers of DEGs in GSE45550 at different time points were obtained, and day 3 DEGs were compared with those for day 7 and day 14. (B) The numbers of DEGs in GSE45006 at different time points were obtained, and day 1 DEGs were compared with those for day 7, day 14, and day 56. DEG: Differentially expressed gene; GSE45006, GSE45550: gene expression profiles, obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; SCI: spinal cord injury.
The overlapping DEGs associated with the acute phase of SCI
Owing to the observed differences between the GSE45550 and GSE45006 profiles, a contrast analysis was performed between these two gene expression profiles for each similar point. More DEGs were identified in GSE45006 than in GSE45550, and approximately 1263, 195, and 75 overlapping DEGs were identified between the two profiles at 3, 7/8, and 14 days post-injury, respectively (Figure 2A–C). The above data indicated that the number of common DEGs significantly decreased with time following injury occurrence. The samples included in GSE45550 were obtained from 3 days to 14 days post-injury, whereas the samples included in GSE45006 covered a much wider time frame (1, 3, 7, 14, and 56 days post-injury). Therefore, GSE45006 was utilized to further determine overlapping DEGs associated with acute SCI over time. As shown in Figure 2D, approximately 16 overlapping DEGs were identified from 1 to 14 days post-injury, including Pank1, Hn1, Tmem150c, Rgd1309676, Lpl (lipoprotein lipase), Mdh1, Nnt (nicotinamide tide transhydrogenase), Loc100912219, Large1, Baiap2, Slc24a2, Fundc2, Mrps14, Slc16a7 (solute carrier family 16 member 7), Obfc1, and Alpk3 (alpha kinase 3).
Figure 2.
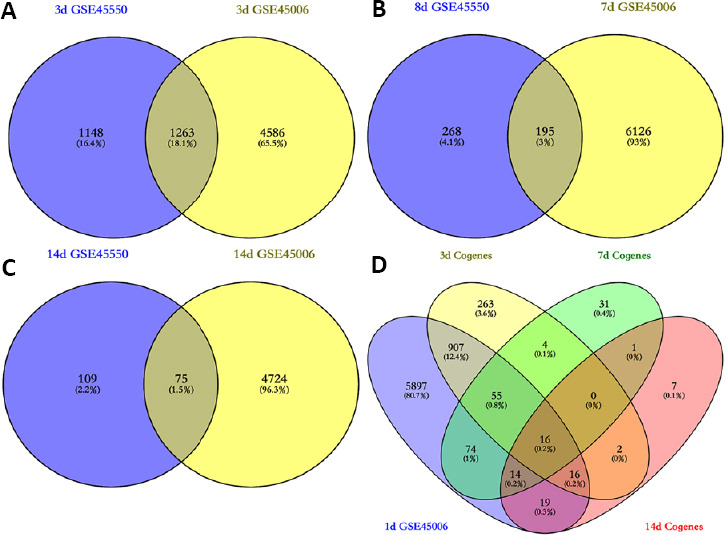
The overlapping DEGs associated with acute SCI across different time points.
(A–C) Comparative analyses of the same time points between GSE45550 and GSE45006 were performed. The overlapping DEGs were identified at 3 (A), 7/8 (B), and 14 (C) days. (D) GSE45006 was utilized to further examine overlapping DEGs during acute SCI, across time points of 1, 3, 7, and 14 days. DEG: Differentially expressed gene; GSE45006, GSE45550: gene expression profiles, which were obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; SCI: spinal cord injury.
The DEGs identified between chronic SCI and sham groups
The gene expression profile GSE93249 was used to screen DEGs between chronic SCI and sham groups. Three time points (1, 3, and 6 months) post-SCI were adopted in the experimental groups. The results indicated approximately 8096, 9683, and 7077 DEGs identified at 1, 3, and 6 months, respectively. As shown in Figure 3A, approximately 3882 overlapping DEGs were associated with chronic SCI across all time points from 1 to 6 months. Further analysis indicated that these DEGs contained both coding and long non-coding genes, including 3316 protein-coding genes and 567 long non-coding RNAs (Figure 3B and C).
Figure 3.
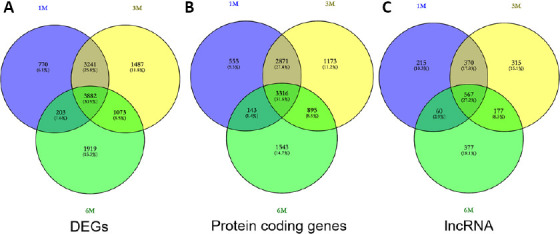
DEGs between chronic SCI and sham samples.
(A) The gene expression profile GSE93249 was used to analyze DEGs associated with chronic SCI at three time points, (B and C) The overlapping DEGs across 1-, 3-, and 6-month time points after SCI were obtained. Among these DEGs, both (B) coding and (C) long non-coding genes were identified. DEG: Differentially expressed gene; GSE93249: a gene expression profile, which was obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; SCI: spinal cord injury.
The overlapping DEGs associated with the chronic phase of SCI
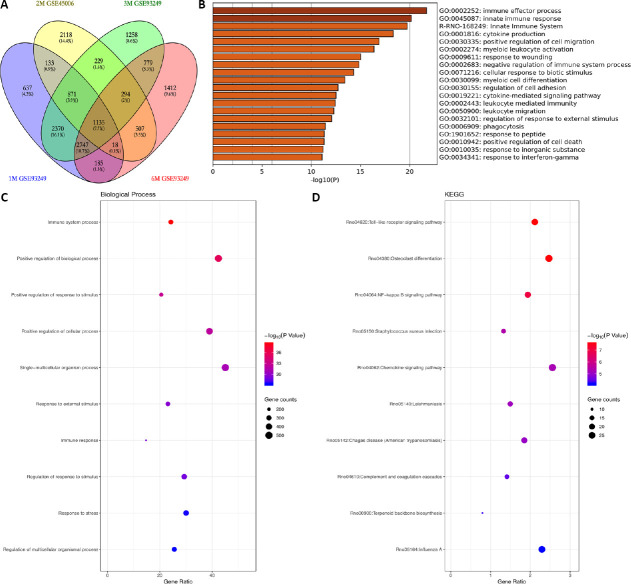
To further examine the overlapping DEGs associated with chronic SCI, the expression profiles GSE93249 and GSE45006 were compared. As shown in Figure 4A, a total of 1135 overlapping DEGs were identified between the two profiles when comparing the time points of 1, 3, 6 months, and 56 days (approximately 2 months) post-injury. The 20 most significantly enriched functions associated with the overlapping DEGs, as assessed by GO functional analysis, are shown in the heatmap in Figure 4B. The most striking functions were the immune effector process and the innate immune response. Other significant functions included cytokine production and the positive regulation of cell migration. The enriched biological process and KEGG pathways associated with the overlapping DEGs are shown in (Figure 4C and D). Significantly enriched pathways were associated with the regulation of immune responses, such as the Toll-like receptor (TLR) signaling pathway, osteoclast differentiation, the nuclear factor (NF)-κB signaling pathway, and the chemokine signaling pathway.
Figure 4.
The overlapping DEGs associated with chronic SCI across multiple time points.
The expression profiles GSE93249 and GSE45006 were used for comparative analysis to identify overlapping DEGs associated with chronic SCI. (A) The overlapping DEGs were enriched between GSE93249 and GSE45006 from 1 month to 6 months. (B) The significantly enriched functions of the overlapping DEGs are indicated in a heatmap for selected GO analysis. (C and D) The enriched biological processes (C) and KEGG pathways (D) associated with overlapping DEGs. DEG: Differentially expressed gene; GO: gene ontology; GSE93249 and GSE45006: gene expression profiles, which were obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; KEGG: Kyoto Encyclopedia of Genes and Genomes; SCI: spinal cord injury.
The overlapping DEGs associated with both acute and chronic SCI
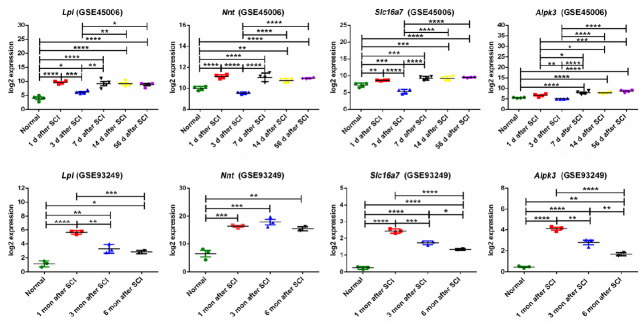
To identify common DEGs associated with both acute and chronic phases of SCI, all three expression profiles, GSE45550, GSE93249, and GSE45006, were compared. As shown in Figure 5A and B, only four overlapping DEGs were found in all three gene expression profiles at all time points (acute: 1, 3, 7/8, 14 days; chronic: 1 month, 56 days, 3, 6 months): Slc16a7, Alpk3, Lpl and Nnt. The expression of these four genes, associated with both acute and chronic spinal cord injuries, was further analyzed in GSE93249 and GSE45006 (Figure 6), and the protein-protein interaction networks associated with Slc16a7, Alpk3, Lpl and Nnt were constructed (Additional Figure 1 (1.5MB, tif) ).
Figure 5.
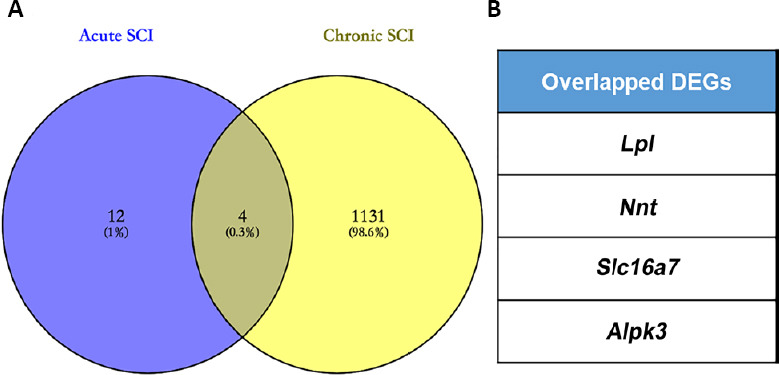
The overlapping DEGs associated with both acute SCI and chronic SCI across multiple time points.
(A) The expression profiles GSE93249 and GSE45006 were used to perform comparative analysis to determine overlapping DEGs between acute and chronic SCI. (B) The four overlapping DEGs are listed. Alpk3: Alpha kinase 3; DEG: differentially expressed gene; GSE93249 and GSE45006: gene expression profiles, which were obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; LPL: lipoprotein lipase; Nnt: nicotinamide tide transhydrogenase; SCI: spinal cord injury; Slc16a7: solute carrier family 16 member 7.
Figure 6.
The expression patterns of the overlapping DEGs.
The fold changes of Lpl, Nnt, Slc16a7, and Alpk3 expression (log2 FC) were obtained from GSE45006 and GSE93249. Data are expressed as the mean ± SD. Comparisons between results from multiple groups were analyzed using a one-way analysis of variance, followed by Dunnett’s post hoc test. For experiments involving two groups, an unpaired Student’s t-test was performed. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Alpk3: Alpha kinase 3; DEG: differentially expressed gene; log2 FC: fold change of gene expression; GSE93249 and GSE45006: gene expression profiles, which were obtained from the Gene Expression Omnibus database of the National Center of Biotechnology Information; LPL: lipoprotein lipase; Nnt: nicotinamide tide transhydrogenase; SCI: spinal cord injury; Slc16a7: solute carrier family 16 member 7.
Discussion
The pathological changes that occur after traumatic SCI are initially associated with the immediate damage, followed by secondary cascade events. Until now, most studies have concentrated on the biomolecular changes and injury-related mechanisms associated with a single phase of SCI: acute, subacute, or chronic. In this study, the time-frame for the experiments was extended from 1 day to 6 months post-injury by analyzing three gene expression profiles, allowing us to conduct a comprehensive overview of SCI.
The events associated with the SCI process can be divided into several contiguous phases, including immediate (0–2 hours), early acute (2–48 hours), subacute (2–14 days), subchronic, and chronic stages (Chen et al., 2013; Duran et al., 2017). This study aimed to provide an overall analysis of common DEGs, with persistent differential expression between the acute (1 day), subacute (7–14 days), and chronic (6 months) phases of SCI. The present study was the first to perform a comparative analysis using the gene expression profiles GSE93249, GSE45550, and GSE45006, which covered a wider time course than has been used in other studies, ranging from 1 day to 6 months post-injury. The three gene expression profiles used in this study were acquired from SCI models based on different injury positions and severity, likely causing differences in the affected signaling pathways and functional molecules. However, we hypothesized that common, shared events were likely to occur, even in different SCI models, the identification of which would provide promising candidates for therapeutic or molecular mechanism studies that may be common among all SCI injuries. DEG clusters were identified, followed by GO and pathway enrichment analyses. The results of this study may provide deep insight into the molecular mechanisms associated with SCI and facilitate the development of novel therapeutic strategies. To ensure the efficiency of DEG screening, only those molecules that were significantly differentially regulated during acute and chronic SCI were selected for our analysis procedures. Comparative analyses revealed that DEGs varied across time points in acute and chronic SCI. Our results identified 16 overlapping DEGs identified across time points in the acute and subacute phases, from 1 to 14 days post-injury. However, when DEGs were examined through 6 months post-injury, only four overlapping DEGs were identified across acute and chronic SCI: Lpl, Nnt, Slc16a7, and Alpk3. The remaining overlapping DEGs identified, in acute but not chronic SCI, were Pank1, Hn1, Tmem150c, Rgd1309676, Mdh1, Loc100912219, Large1, Baiap2, Slc24a2, Fundc2, Mrps14, and Obfc1. These 12 genes represent acute and subacute SCI-specific genes. These results suggested that early SCI DEGs are distinctly different late-stage SCI DEGs. Thus, these identified genes may represent potential and meaningful candidates for exploring the mechanisms associated with SCI development and the development of strategies to halt or slow SCI progression.
The inflammatory response induced by injuries to the spinal cord can occur within minutes. Previous studies have identified various immune responses to be the most significantly upregulated biological processes during acute SCI based on GO functional analysis, including the activation of the complement system, the induction of innate and adaptive immune responses, and antibody production (Pineau and Lacroix, 2007; de Rivero Vaccari et al., 2008). Additionally, some chemokines and cytokines produced following SCI have been shown to be involved in the inflammatory responses (Beck et al., 2010; Zhang et al., 2011; Garcia et al., 2016). In this study, one interesting molecule, peroxiredoxin-like 2A (Rgd1309676), was identified during acute and subacute SCI. Peroxiredoxin-like 2A is an antioxidant involved in the redox regulation of the cell, and it functions as a negative regulator of macrophage-mediated inflammation by inhibiting the production of inflammatory cytokines by macrophages, likely through the suppression of the mitogen-activated protein kinase signaling pathway and regulation of osteoclast differentiation (Xu et al., 2010, 2015; Guo et al., 2015). Therefore, peroxiredoxin-like 2A may function as a negative regulator that combats the excessive inflammation induced by immediate injury during the subacute stage, from days 10 to 14 post-injury. SCI has been shown to be accompanied by a low-grade, chronic inflammatory state, which may be due to the inhibition of excessive inflammation during early stages. Based on the potential function of peroxiredoxin-like 2A and its expression during acute SCI, this molecule may represent a promising and meaningful candidate for exploring SCI-associated cellular mechanisms and therapeutic targets.
Another functional protein, LPL, was identified throughout both the acute and chronic phases of SCI, in this study. LPL has been reported to respond to both SCI-induced inactivity and reloading with training (Baligand et al., 2015). LPL expression has been detected in the brain, spinal cord, and peripheral nerves. In addition to the hydrolysis of triglyceride-rich lipoproteins, LPL has been shown to play important roles in the pathophysiological responses of nerve injury. Several studies have reported the selective induction of LPL expression in response to cerebral ischemia-reperfusion injury and have suggested a protective function of LPL, which facilitates lipid uptake and reutilization in the central nervous system during demyelination events or following nerve crush injuries (Blain et al., 2004; Paradis et al., 2004; Wang et al., 2010; Bruce et al., 2018). Based on our results, LPL may fulfill biological functions during the pathophysiological responses to both acute and chronic SCI. Therefore, LPL may represent a protective factor, playing potential roles in the clearance of myelin-derived lipids after SCI.
NNT is essential for the removal of hydrogen peroxide (Meimaridou et al., 2018), and currently studies examining NNT primarily focus on adrenal diseases, heart diseases, diabetes, and other diseases (Chatterjee et al., 2014; Hershkovitz et al., 2015; Kampjut and Sazanov, 2019). SLC16A7 is a member of the monocarboxylate transporter family, and has been reported to be expressed at increased levels in prostate and colorectal cancer specimens compared with control specimens (Pinheiro et al., 2008; Pertega-Gomes et al., 2015). ALPK3 plays a key role in transferase activity, the transfer of phosphorus-containing groups, and protein serine/threonine kinase activity (Feng and Jiang, 2020). Although these three genes were identified during the acute and chronic stages of SCI, relatively few reported studies have examined these genes in nerves and the spinal cord, so their significance to the nervous system remains unclear.
In summary, during the present study, we performed an overall bioinformatics analysis of DEGs associated with acute, subacute, and chronic SCI through the utilization and analysis of three gene expression profiles. Approximately 16 DEGs and 1135 DEGs were associated with the acute phase and the chronic phase of SCI, respectively. However, only four overlapping DEGs, Slc16a7, Alpk3, Lpl, and Nnt, displayed persistent differential expression across the acute, subacute, and chronic phases of SCI. Thus, the twelve DEGs that were associated specifically with the acute phases of SCI may be associated with early pathological changes that occur after SCI. In contrast, the four shared genes identified in all phases of SCI may function to exert constant biological effects during SCI progression. Taken together, these data might provide some meaningful insight regarding differences in the pathogenic mechanisms and targeted therapy strategies between acute and chronic SCI. However, because these are predictive results and are based on bioinformatic analyses, the functions of key DEGs, such as peroxiredoxin-like 2A and LPL, and their roles in SCI remain to be explored and demonstrated using biological experiments. SCI models associated with different injury positions and severity are likely to result in varying DEG expression; therefore, the DEGs identified in this study should be further verified in different models in future studies.
Additional file:
Additional Figure 1 (1.5MB, tif) : The protein-protein interaction network.
The protein-protein interaction related to Lpl, Nnt, Slc16a7 and Alpk3 were constructed respectively to build a potential interaction network. Alpk3: Alpha kinase 3; LPL: lipoprotein lipase; Nnt: nicotinamide tide transhydrogenase; Slc16a7: solute carrier family 16 member 7.
Footnotes
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Giles L, Yu J, Song CP; T-Editor: Jia Y
Conflicts of interest: The authors declare no conflict of interest.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 31571236 (to YHK); Science and Technology Planning Project of Beijing of China, No D161100002816001; the National Key Research and Development Program of China, No. 2016YFC1101604 (to DYZ); the Ministry of Education Innovation Program of China, No. IRT_16R01. The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: Data were downloaded from GSE45006, GSE45550 and GSE93249 in the Gene Expression Omnibus database of the National Center of Biotechnology Information. The study does not involve ethical approval.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This study was supported by the National Natural Science Foundation of China, No. 31571236 (to YHK); Science and Technology Planning Project of Beijing of China, No D161100002816001; the National Key Research and Development Program of China, No. 2016YFC1101604 (to DYZ); the Ministry of Education Innovation Program of China, No. IRT_16R01.
References
- 1.Anwar MA, Al Shehabi TS, Eid AH. Inflammogenesis of secondary spinal cord injury. Front Cell Neurosci. 2016;10:98. doi: 10.3389/fncel.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baligand C, Chen YW, Ye F, Pandey SN, Lai SH, Liu M, Vandenborne K. Transcriptional pathways associated with skeletal muscle changes after spinal cord injury and treadmill locomotor training. Biomed Res Int. 2015;2015:387090. doi: 10.1155/2015/387090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck KD, Nguyen HX, Galvan MD, Salazar DL, Woodruff TM, Anderson AJ. Quantitative analysis of cellular inflammation after traumatic spinal cord injury: evidence for a multiphasic inflammatory response in the acute to chronic environment. Brain. 2010;133:433–447. doi: 10.1093/brain/awp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blain JF, Paradis E, Gaudreault SB, Champagne D, Richard D, Poirier J. A role for lipoprotein lipase during synaptic remodeling in the adult mouse brain. Neurobiol Dis. 2004;15:510–519. doi: 10.1016/j.nbd.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Bruce KD, Gorkhali S, Given K, Coates AM, Boyle KE, Macklin WB, Eckel RH. Lipoprotein lipase is a feature of alternatively-activated microglia and may facilitate lipid uptake in the CNS during demyelination. Front Mol Neurosci. 2018;11:57. doi: 10.3389/fnmol.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrnes KR, Washington PM, Knoblach SM, Hoffman E, Faden AI. Delayed inflammatory mRNA and protein expression after spinal cord injury. J Neuroinflammation. 2011;8:130. doi: 10.1186/1742-2094-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvert JS, Grahn PJ, Zhao KD, Lee KH. Emergence of epidural electrical stimulation to facilitate sensorimotor network functionality after spinal cord injury. Neuromodulation. 2019;22:244–252. doi: 10.1111/ner.12938. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee S, Chakraborty A, Weinberg I, Kadakia M, Wilensky RL, Sardar P, Kumbhani DJ, Mukherjee D, Jaff MR, Giri J. Thrombolysis for pulmonary embolism and risk of all-cause mortality, major bleeding, and intracranial hemorrhage: a meta-analysis. JAMA. 2014;311:2414–2421. doi: 10.1001/jama.2014.5990. [DOI] [PubMed] [Google Scholar]
- 9.Chen K, Deng S, Lu H, Zheng Y, Yang G, Kim D, Cao Q, Wu JQ. RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: a resource for understanding the pathology at the systems level. PLoS One. 2013;8:e72567. doi: 10.1371/journal.pone.0072567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 11.de Rivero Vaccari JP, Lotocki G, Marcillo AE, Dietrich WD, Keane RW. A molecular platform in neurons regulates inflammation after spinal cord injury. J Neurosci. 2008;28:3404–3414. doi: 10.1523/JNEUROSCI.0157-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H, Shi J, Wang M, An S, Guo X, Wang Z. Analyses of gene expression profiles in the rat dorsal horn of the spinal cord using RNA sequencing in chronic constriction injury rats. J Neuroinflammation. 2018;15:280. doi: 10.1186/s12974-018-1316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duran RC, Yan H, Zheng Y, Huang X, Grill R, Kim DH, Cao Q, Wu JQ. The systematic analysis of coding and long non-coding RNAs in the sub-chronic and chronic stages of spinal cord injury. Sci Rep. 2017;7:41008. doi: 10.1038/srep41008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehlings MG, Perrin RG. The timing of surgical intervention in the treatment of spinal cord injury: a systematic review of recent clinical evidence. Spine (Phila Pa 1976) 2006;31:S28–35. doi: 10.1097/01.brs.0000217973.11402.7f. discussion S36. [DOI] [PubMed] [Google Scholar]
- 15.Feng E, Jiang L. Peptidomic analysis of hippocampal tissue for explore leptin neuroprotective effect on the preterm ischemia-hypoxia brain damage model rats. Neuropharmacology. 2020;162:107803. doi: 10.1016/j.neuropharm.2019.107803. [DOI] [PubMed] [Google Scholar]
- 16.Garcia E, Aguilar-Cevallos J, Silva-Garcia R, Ibarra A. Cytokine and growth factor activation in vivo and in vitro after spinal cord injury. Mediators Inflamm. 2016;2016:9476020. doi: 10.1155/2016/9476020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo F, He H, Fu ZC, Huang S, Chen T, Papasian CJ, Morse LR, Xu Y, Battaglino RA, Yang XF, Jiang Z, Xin HB, Fu M. Adipocyte-derived PAMM suppresses macrophage inflammation by inhibiting MAPK signalling. Biochem J. 2015;472:309–318. doi: 10.1042/BJ20150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Lv J, Huang YF, Hao DJ, Liu JJ. Bioinformatics analyses of differentially expressed genes associated with spinal cord injury: A microarray-based analysis in a mouse model. Neural Regen Res. 2019;14:1262–1270. doi: 10.4103/1673-5374.251335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershkovitz E, Arafat M, Loewenthal N, Haim A, Parvari R. Combined adrenal failure and testicular adrenal rest tumor in a patient with nicotinamide nucleotide transhydrogenase deficiency. J Pediatr Endocrinol Metab. 2015;28:1187–1190. doi: 10.1515/jpem-2015-0075. [DOI] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Jazayeri SB, Beygi S, Shokraneh F, Hagen EM, Rahimi-Movaghar V. Incidence of traumatic spinal cord injury worldwide: a systematic review. Eur Spine J. 2015;24:905–918. doi: 10.1007/s00586-014-3424-6. [DOI] [PubMed] [Google Scholar]
- 22.Kampjut D, Sazanov LA. Structure and mechanism of mitochondrial proton-translocating transhydrogenase. Nature. 2019;573:291–295. doi: 10.1038/s41586-019-1519-2. [DOI] [PubMed] [Google Scholar]
- 23.Kotipatruni RR, Dasari VR, Veeravalli KK, Dinh DH, Fassett D, Rao JS. p53- and Bax-mediated apoptosis in injured rat spinal cord. Neurochem Res. 2011;36:2063–2074. doi: 10.1007/s11064-011-0530-2. [DOI] [PubMed] [Google Scholar]
- 24.Lavis T, Goetz LL. Comprehensive care for persons with spinal cord injury. Phys Med Rehabil Clin N Am. 2019;30:55–72. doi: 10.1016/j.pmr.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Huang X, Liang X, Zhou Y, Li H, Yu Q, Li Q. Identification of key modules and hub genes of keloids with weighted gene coexpression network analysis. Plast Reconstr Surg. 2017;139:376–390. doi: 10.1097/PRS.0000000000003014. [DOI] [PubMed] [Google Scholar]
- 26.Liu ZG, Li Y, Jiao JH, Long H, Xin ZY, Yang XY. MicroRNA regulatory pattern in spinal cord ischemia-reperfusion injury. Neural Regen Res. 2020;15:2123–2130. doi: 10.4103/1673-5374.280323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma S, Wang J, Liu L, Xia L, Tao R. Identification of temporal genes involved in the mechanisms of spinal cord injury. Spinal Cord. 2017;55:355–361. doi: 10.1038/sc.2016.183. [DOI] [PubMed] [Google Scholar]
- 28.Madar V, Batista S. FastLSU: a more practical approach for the Benjamini-Hochberg FDR controlling procedure for huge-scale testing problems. Bioinformatics. 2016;32:1716–1723. doi: 10.1093/bioinformatics/btw029. [DOI] [PubMed] [Google Scholar]
- 29.Meimaridou E, Goldsworthy M, Chortis V, Fragouli E, Foster PA, Arlt W, Cox R, Metherell LA. NNT is a key regulator of adrenal redox homeostasis and steroidogenesis in male mice. J Endocrinol. 2018;236:13–28. doi: 10.1530/JOE-16-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nesic O, Svrakic NM, Xu GY, McAdoo D, Westlund KN, Hulsebosch CE, Ye Z, Galante A, Soteropoulos P, Tolias P, Young W, Hart RP, Perez-Polo JR. DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. J Neurosci Res. 2002;68:406–423. doi: 10.1002/jnr.10171. [DOI] [PubMed] [Google Scholar]
- 31.Paradis E, Clavel S, Julien P, Murthy MR, de Bilbao F, Arsenijevic D, Giannakopoulos P, Vallet P, Richard D. Lipoprotein lipase and endothelial lipase expression in mouse brain: regional distribution and selective induction following kainic acid-induced lesion and focal cerebral ischemia. Neurobiol Dis. 2004;15:312–325. doi: 10.1016/j.nbd.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Pertega-Gomes N, Vizcaino JR, Felisbino S, Warren AY, Shaw G, Kay J, Whitaker H, Lynch AG, Fryer L, Neal DE, Massie CE. Epigenetic and oncogenic regulation of SLC16A7 (MCT2) results in protein over-expression, impacting on signalling and cellular phenotypes in prostate cancer. Oncotarget. 2015;6:21675–21684. doi: 10.18632/oncotarget.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineau I, Lacroix S. Proinflammatory cytokine synthesis in the injured mouse spinal cord: multiphasic expression pattern and identification of the cell types involved. J Comp Neurol. 2007;500:267–285. doi: 10.1002/cne.21149. [DOI] [PubMed] [Google Scholar]
- 34.Pinheiro C, Longatto-Filho A, Scapulatempo C, Ferreira L, Martins S, Pellerin L, Rodrigues M, Alves VA, Schmitt F, Baltazar F. Increased expression of monocarboxylate transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch. 2008;452:139–146. doi: 10.1007/s00428-007-0558-5. [DOI] [PubMed] [Google Scholar]
- 35.Shi LL, Zhang N, Xie XM, Chen YJ, Wang R, Shen L, Zhou JS, Hu JG, Lü HZ. Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing. BMC Genomics. 2017;18:173. doi: 10.1186/s12864-017-3532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X, Liu JS, Wan R, Wang YS. Research progress in mesenchymal stem cells for treating spinal cord injury. Zhongguo Zuzhi Gongcheng Yanjiu. 2020;24:4081–4087. [Google Scholar]
- 37.Singh A, Tetreault L, Kalsi-Ryan S, Nouri A, Fehlings MG. Global prevalence and incidence of traumatic spinal cord injury. Clin Epidemiol. 2014;6:309–331. doi: 10.2147/CLEP.S68889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahel PF, VanderHeiden T, Finn MA. Management strategies for acute spinal cord injury: current options and future perspectives. Curr Opin Crit Care. 2012;18:651–660. doi: 10.1097/MCC.0b013e32835a0e54. [DOI] [PubMed] [Google Scholar]
- 39.Thomaz SR, Cipriano G, Jr, Formiga MF, Fachin-Martins E, Cipriano GFB, Martins WR, Cahalin LP. Effect of electrical stimulation on muscle atrophy and spasticity in patients with spinal cord injury - a systematic review with meta-analysis. Spinal Cord. 2019;57:258–266. doi: 10.1038/s41393-019-0250-z. [DOI] [PubMed] [Google Scholar]
- 40.van den Berg ME, Castellote JM, Mahillo-Fernandez I, de Pedro-Cuesta J. Incidence of spinal cord injury worldwide: a systematic review. Neuroepidemiology. 2010;34:184–192. doi: 10.1159/000279335. [DOI] [PubMed] [Google Scholar]
- 41.Wang GY, Cheng ZJ, Yang BH, Li Hp, He XJ. Olfactory ensheathing cell transplantation promotes the ultrastructure repair at the lesion site of rat models of spinal cord injury. Zhongguo Zuzhi Gongcheng Yanjiu. 2020;24:699–703. [Google Scholar]
- 42.Wang X, Sun W, Xu E. The expression and activity of brain lipoprotein lipase is increased after acute cerebral ischemia-reperfusion in rats. Neuropathology. 2010;30:131–139. doi: 10.1111/j.1440-1789.2009.01061.x. [DOI] [PubMed] [Google Scholar]
- 43.Wanner IB, Anderson MA, Song B, Levine J, Fernandez A, Gray-Thompson Z, Ao Y, Sofroniew MV. Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J Neurosci. 2013;33:12870–12886. doi: 10.1523/JNEUROSCI.2121-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen T, Hou J, Wang F, Zhang Y, Zhang T, Sun T. Comparative analysis of molecular mechanism of spinal cord injury with time based on bioinformatics data. Spinal Cord. 2016;54:431–438. doi: 10.1038/sc.2015.171. [DOI] [PubMed] [Google Scholar]
- 45.Wyndaele M, Wyndaele JJ. Incidence, prevalence and epidemiology of spinal cord injury: what learns a worldwide literature survey. Spinal Cord. 2006;44:523–529. doi: 10.1038/sj.sc.3101893. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Morse LR, da Silva RAB, Wang D, Battaglino RA. A short report: PAMM, a novel antioxidant protein, induced by oxidative stress. Redox Biol. 2015;6:446–453. doi: 10.1016/j.redox.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Morse LR, da Silva RA, Odgren PR, Sasaki H, Stashenko P, Battaglino RA. PAMM: a redox regulatory protein that modulates osteoclast differentiation. Antioxid Redox Signal. 2010;13:27–37. doi: 10.1089/ars.2009.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Wang Y. Identification of molecular pathway changes after spinal cord injury by microarray analysis. J Orthop Surg Res. 2016;11:101. doi: 10.1186/s13018-016-0437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, Trivedi A, Lee JU, Lohela M, Lee SM, Fandel TM, Werb Z, Noble-Haeusslein LJ. Matrix metalloproteinase-9 and stromal cell-derived factor-1 act synergistically to support migration of blood-borne monocytes into the injured spinal cord. J Neurosci. 2011;31:15894–15903. doi: 10.1523/JNEUROSCI.3943-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang YH, Song J, Wang LG, Shao J. Identification of key genes and pathways associated with spinal cord injury. Mol Med Rep. 2017;15:1577–1584. doi: 10.3892/mmr.2017.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H, Shi Z, Kang Y, Wang Y, Lu L, Pan B, Liu J, Li X, Liu L, Wei Z, Kong X, Feng S. Investigation of candidate long noncoding RNAs and messenger RNAs in the immediate phase of spinal cord injury based on gene expression profiles. Gene. 2018;661:119–125. doi: 10.1016/j.gene.2018.03.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The protein-protein interaction related to Lpl, Nnt, Slc16a7 and Alpk3 were constructed respectively to build a potential interaction network. Alpk3: Alpha kinase 3; LPL: lipoprotein lipase; Nnt: nicotinamide tide transhydrogenase; Slc16a7: solute carrier family 16 member 7.