Graphical abstract
Keywords: 8-Anilinonaphthalene-1-sulfonate, Fluorescent probe, Binding assay, SARS-CoV-2, Protease inhibitor, Flavonoids
Highlights
-
•
ANS binds the SARS-CoV-2 main protease and exhibits increased fluorescence.
-
•
ANS binding is competitive with baicalein that binds in the active site.
-
•
FMO-RIMP2/PCM calculation predicts that rutin is an inhibitor of the protease.
-
•
The ANS binding and kinetic assays confirm rutin as a protease inhibitor.
-
•
The ANS binding assay could be used to identify novel protease inhibitors.
Abstract
The 3C-like main protease of SARS-CoV-2 (3CLPro) is responsible for the cleavage of the viral polyprotein. This process is essential for the viral life cycle. Therefore, 3CLPro is a promising target to develop antiviral drugs for COVID-19 prevention and treatment. Traditional enzymatic assays for the identification of 3CLPro inhibitors rely on peptide-based colorimetric or fluorogenic substrates. However, the COVID-19 pandemic has limit or delay access to these substrates, especially for researchers in developing countries attempting to screen natural product libraries. We explored the use of the fluorescent probe 8-anilinonaphthalene-1-sulfonate (ANS) as an alternative assay for inhibitor identification. Fluorescence enhancement upon binding of ANS to 3CLPro was observed, and this interaction was competitive with a peptide substrate. The utility of ANS-based competitive binding assay to identify 3CLPro inhibitors was demonstrated with the flavonoid natural products baicalein and rutin. The molecular nature of ANS and rutin interaction with 3CLPro was explored with molecular modeling. Our results suggested that ANS could be employed in a competitive binding assay to facilitate the identification of novel SARS-CoV-2 antiviral compounds.
1. Introduction
In addition to vaccines, antiviral drug development is another frontier to combat the Corona Virus Disease 2019 (COVID-19). COVID-19 is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus produces polyproteins that require further processing by proteases to liberate functional proteins that are required for the viral life cycle [1]. Inhibition of the 3C-like protease (3CLPro), or the main protease, is a known approach to inhibit SARS-CoV-1 and Middle East Respiratory Syndrome (MERS) virus replication [2]. Therefore, there has been significant interest in the scientific community to identify inhibitors of SARS-CoV-2 3CLPro, thereafter referred to as 3CLPro, since the beginning of the outbreak [3], [4]. Several research groups have reported promising results in using protease inhibitors to control SARS-CoV-2 infection [3], [4], [5]. Novel computational methodologies have also been developed to screen for inhibitor of 3CLPro [6]. Still, the discovery of novel inhibitors is necessary to overcome the high mutation rate of the RNA virus [7].
Library of compounds isolated from natural sources offers a valuable source for 3CLPro inhibitor discovery. We have been interested in the exploration of local natural products as potential 3CLPro inhibitors. Traditional assays for protease inhibitor identification employ colorimetric or fluorogenic peptide substrates. This approach has been quite successful in identification of 3CLPro inhibitors [3], [4]. The peptide substrates could readily be synthesized by existing peptide synthesis methods. However, such services are not available in many developing countries with rich natural product resources, thus investigators are required to order synthetic peptides from abroad. With the COVID-19 pandemic, which results in laboratory and business closure, we have experienced a significant import delay of peptide substrates. Therefore, we seek to develop a new assay for 3CLPro inhibitor identification that could be used for natural product and chemical library screening. The enzyme inhibition activity could later be confirmed with a standard peptide-based assay. The assay should also be relatively inexpensive so that investigators with limited funding could employ the assay. Because the native cleavage sequences of 3CLPro (TSAVLQ-SGFRK and SGVTFQ-GKFKK, with - indicating the cleavage site)[8] contain a few hydrophobic amino acids and the active site of 3CLPro has positively charged histidine residues such as H41, H163, H164, and H172, we envision that the fluorescent probe 8-anilinonaphthalene-1-sulfonate (ANS) could be utilized in the desired assay.
ANS is a water-soluble molecule that exhibits increased fluorescence upon burial into a pocket in a protein [9]. ANS-based binding assay has been utilized to investigate protein–ligand interactions and protein unfolding processes [10], [11], [12], [13]. ANS has previously been employed to characterize the folding process of SARS-CoV-1 3CLPro, in which ANS binds to the exposed hydrophobic region when the protein is unfolded [14]. However, the interaction of ANS and the native 3CLPro has not yet been explored. If ANS could bind in the active site of 3CLPro, an increase in ANS fluorescence should be observed. Inhibitors of 3CLPro should be able to compete with ANS and resulted in a reduction in fluorescence. This equilibrium binding approach is also more convenient and requires less sophisticated equipment to perform than the time-sensitive enzyme kinetic screens or end point assays. In this work, we demonstrated that ANS could bind in the active site of 3CLPro by competition with the fluorogenic peptide substrate E(EDANS)TSAVLQSGFRK(DABCYL). In addition, ANS could compete with baicalein that is known to bind in the active site [15]. We also demonstrated the utility of the ANS-based binding assay for the identification of rutin as a 3CLPro inhibitor, as predicted by several computational studies [16], [17], [18], [19], [20], [21], [22]. The ANS-based competitive binding assay will be valuable in a cost-effective screen for novel 3CLPro inhibitors, especially when access to a peptide substrate is limited.
2. Results
The interaction of ANS with 3CLPro was first explored by monitoring the increased fluorescence in an ANS-3CLPro mixture compared to the solution with no 3CLPro (Fig. 1A). When excited at 345 nm, the increased fluorescence with an emission maximum at 455 nm was observed. Therefore, ANS could be shielded from the aqueous solvent when interacting with 3CLPro. The excitation and emission wavelengths at 345 and 455 nm, respectively, were used in subsequent experiments. The saturation binding curve suggested a simple binding interaction with the apparent dissociation constant (KD) of 57 ± 7 µM (Fig. 1B).
Fig. 1.
A) Chemical structure and fluorescence emission spectrum of ANS (50 µM) bound to 3CLPro. B) Saturation binding curve to ANS to 3CLPro.
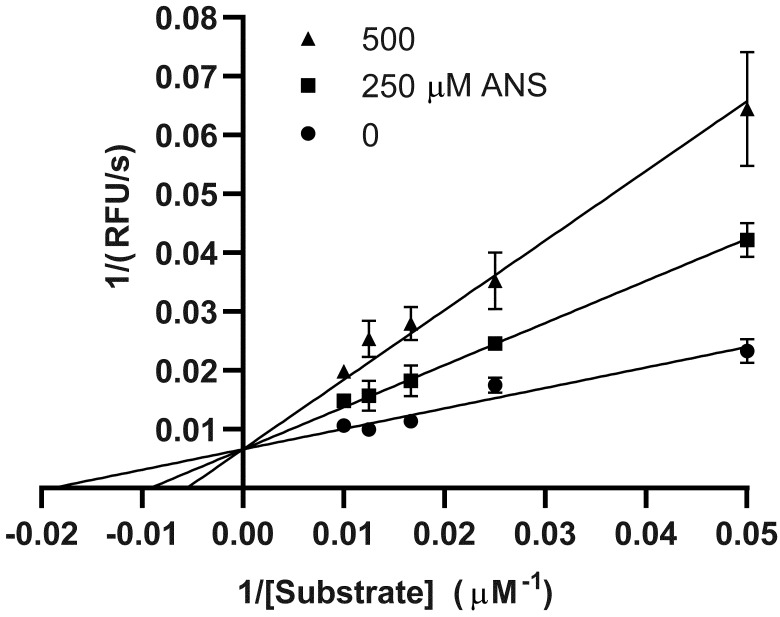
To determine whether ANS could bind in the active site of 3CLPro, inhibition experiments were performed using the fluorogenic peptide substrate E(EDANS)TSAVLQSGFRK(DABCYL) (Fig. 2). The non-linear fit of the kinetic data revealed the Michaelis constant (KM) for the fluorogenic substrate to be 51 ± 9 µM. The addition of ANS and the non-linear fit of the resulting kinetic data to the competitive inhibition model gave the inhibitory constant (KI) of 188 ± 24 µM.
Fig. 2.
Lineweaver–Burk plot of an initial rate enzyme kinetic experiment of 3CLPro at different substrate concentrations with ANS presented at 0, 250, and 500 µM. Trend lines were created with parameters derived from non-linear fit of the original data.
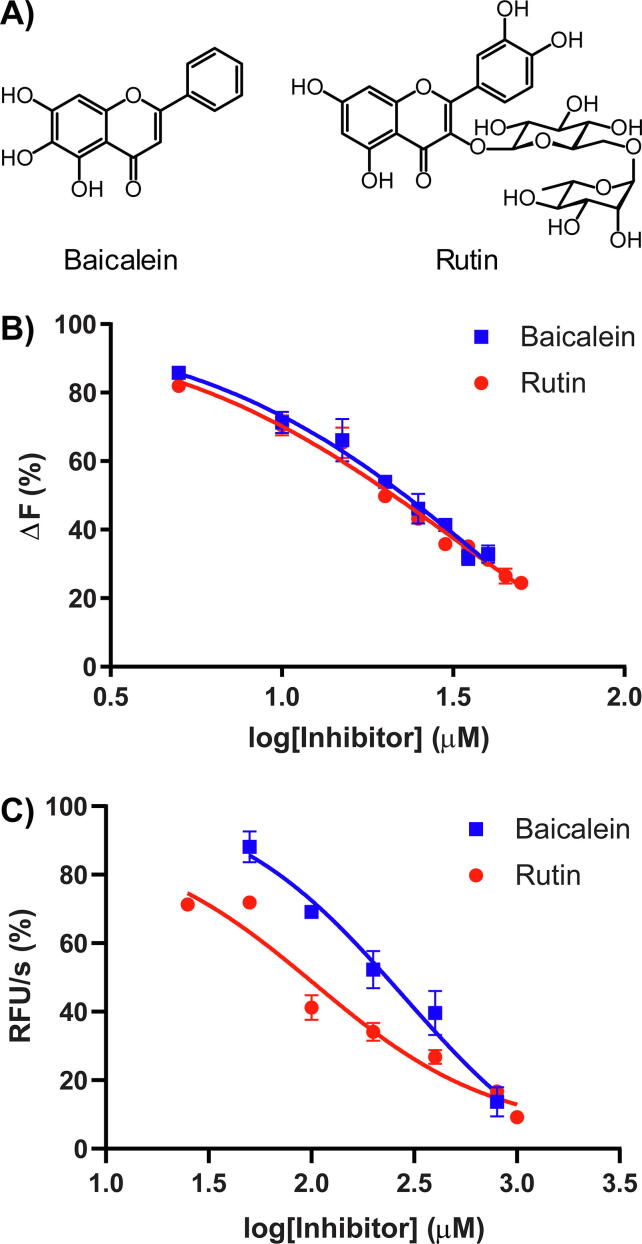
To demonstrate the utility of ANS as a fluorescent probe to identify a 3CLPro inhibitor, we performed competitive binding and competitive inhibition experiments with baicalein and rutin (Fig. 3A). Baicalein is a flavonoid natural product known to inhibit 3CLPro [15]. X-ray crystallography revealed that baicalein binds in the active site of 3CLPro. Thus, the reduction of ANS fluorescence by baicalein should be evidence that ANS binds in the active site of 3CLPro. Indeed, the reduction of ANS fluorescence was observed as baicalein was titrated (Fig. 3B). For the ANS competitive binding assay, the half-maximal inhibitory concentration (IC50) of baicalein was 42 ± 2 µM, and the corresponding KI obtained from the Cheng-Prusoff equation was 15.2 ± 0.7 µM. Inhibition of 3CLPro by baicalein was also confirmed in an activity assay using the fluorogenic peptide substrate E(EDANS)TSAVLQSGFRK(DABCYL) (Fig. 3C). For the enzyme activity inhibition assay, IC50 for baicalein was 280 ± 1 µM, and the corresponding KI obtained from the Cheng-Prusoff equation was 188 ± 1 µM.
Fig. 3.
A) Chemical structures of baicalein and rutin. B) Reduction in 3CLPro-bound ANS fluorescence upon titration of baicalein and rutin. ANS concentration was at 100 µM. C) Inhibition of 3CLPro enzymatic activity by baicalein and rutin. The fluorogenic substrate concentration was at 25 µM.
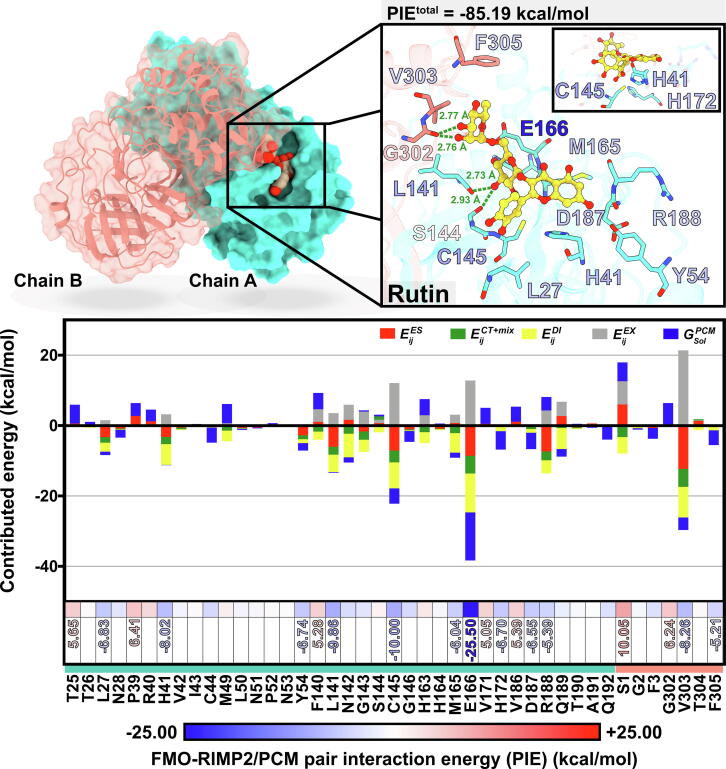
Similar to baicalein, rutin is a flavonoid previously predicted computationally to bind and inhibit 3CLPro [18], [19], [20], [22], [23]. We have also independently predicted binding of rutin within the active site of 3CLPro using a molecular docking (Fig. 4). The binding energy was −10.0 kcal/mol. Enhanced binding interactions were found for rutin binding (hydrophobic contacts with the 13 residues in chain A and 3 residues in chain B as well as four hydrogen bonds at the disaccharide rutinose (2.7–2.9 Å). This is somewhat inconsistent with the previous in silico studies using only one chain of protease (Table 1). Our results suggest that the termini of the other chain in the 3CLPro dimer are important in the interactions with rutin.
Fig. 4.
FMO-RIMP2/PCM pair interaction energy (PIEtotal) and energy components (electrostatic intyeraction (), charge transfer with higher-order mixed terms energies () dispersion (), exchange-repulsion (), and the PCM solvation effect ()) for rutin interacting with individual residues of dimeric SARS-CoV-2 3CLPro. Rutin orientation and interactions at the active site of SARS-CoV-2 3CLPro are shown above, where the green dashed line represents the hydrogen bonding. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Predicted binding mode of rutin toward SARS-CoV-2 3CLPro.
| Model | PDB ID | Software | Hydrogen bonding residues | Reference |
|---|---|---|---|---|
| dimer | 6LU7 | Autodock Vina | L141, S144, G302 (Chain B) | This study |
| monomer | 6LU7 | Autodock Vina | T26, Y54, L141, M165, E166 | [18] |
| monomer | 5R82 | Autodock Vina | T45, G143, E166 | [17] |
| monomer | 6LU7 | Autodock Vina | D178, R188, T190 | [20] |
| monomer | 6LU7 | Autodock Vina | N.A. | [21] |
| monomer | 6LU7 | Glide | N142, G143, C145, T190 | [22] |
| monomer | 6LU7 | LeDock | F140, E166, T26, L141, S144, C145, H163 | [19] |
To gain further insight into the interaction between rutin and 3CLPro at the atomic level, the fragment molecular orbital calculation with a high level of theory (FMO-RIMP2/PCM) was applied on the rutin-3CLPro complex. The total pair interaction energy (PIEtotal) and energy contribution from the residues within the 7-Å sphere of rutin were plotted in Fig. 4, where the negative and positive values represent the ligand stabilization and destabilization, respectively. Only the residues with energy contribution ≤ -5.0 kcal/mol and ≥ 5.0 kcal/mol were discussed. The PIEtotal value of −85.19 kcal/mol indicates that the complexation between rutin and dimeric 3CLPro was likely stable. Among the 44 fragments (amino acid residues in the FMO calculation), there are 12 residues in both chains (chain A: L27, H41, Y54, L141, C145, M165, E166, H172, D187, R188; and chain B: V303, F305), contributed to rutin binding with energy stabilization in a range of −5.2 to −25.5 kcal/mol. E166 provides most significantly to the high binding affinity of rutin in terms of electrostatic, charge transfer, dispersion, and solvation effect. The catalytic dyad H41 and C145 play a vital role in binding with the quercetin moiety of rutin. Although rutin is likely to be a non-covalent inhibitor, it shares corresponding binding patterns with the reported inhibitors of SARS-CoV-2 3CLPro [24].
Despite predictions from other research groups, including us, no experimental demonstration of rutin inhibitory activity against 3CLPro has been reported. To experimentally confirm that rutin could bind 3CLPro, we performed the ANS-based competitive binding assay. Titration of rutin into a solution of ANS-bound 3CLPro resulted in the reduction of ANS fluorescence. IC50 of rutin was 31 ± 1 µM, and the corresponding KI obtained from the Cheng-Prusoff equation was 11.3 ± 0.4 µM for the ANS competitive binding assay (Fig. 3B). Enzyme inhibition was also confirmed using the fluorogenic peptide substrate (Fig. 3C). For the enzyme activity inhibition assay, IC50 for rutin was 104 ± 1 µM, and the corresponding KI obtained from the Cheng-Prusoff equation was 69 ± 1 µM.
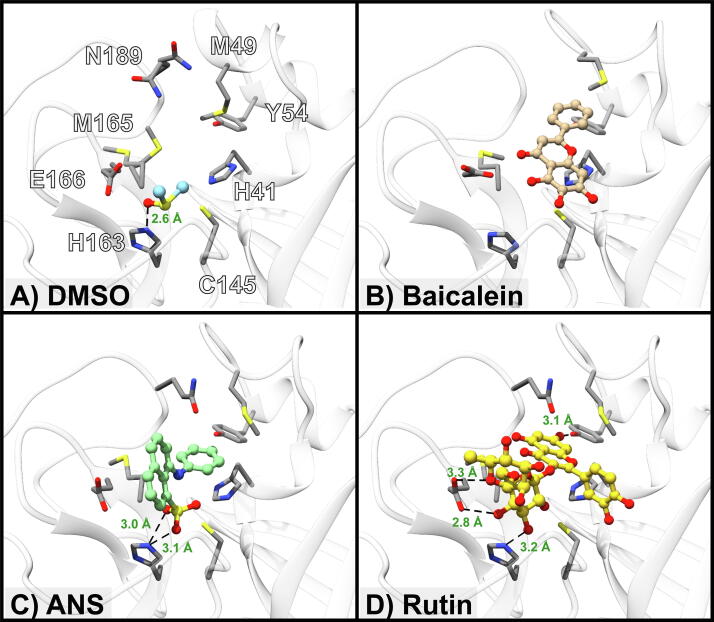
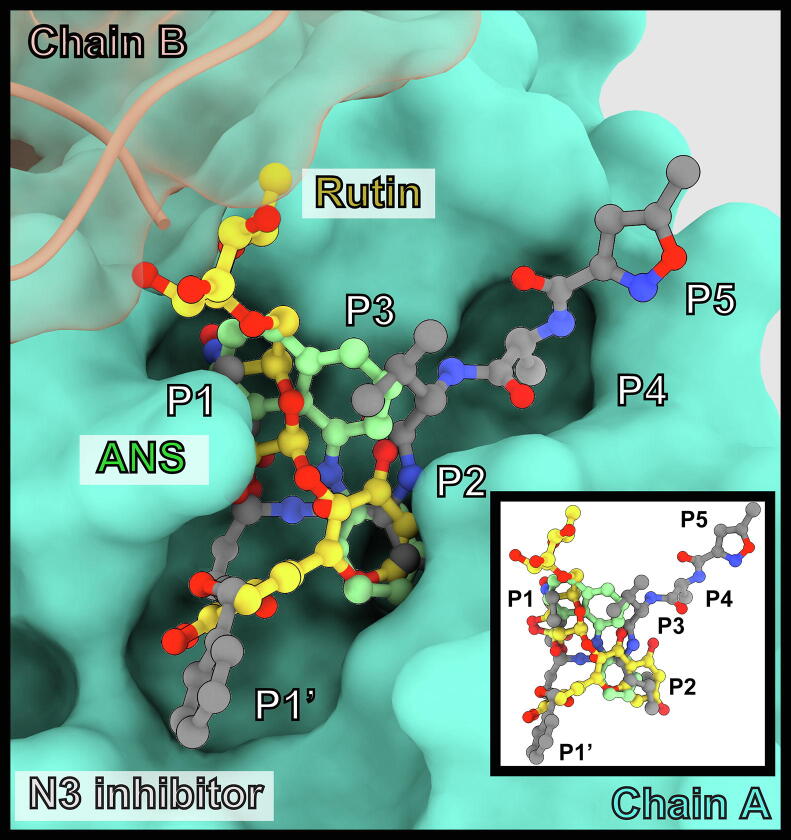
To obtain structural information for interaction between ANS and rutin, we attempted to crystallize these compounds with 3CLPro. Crystals that diffracted to around 1.5 Å were readily obtained (Table 2). However, ANS or rutin was not observed in the active site, but a molecule of DMSO was present instead (Fig. 5A). We initially speculated that DMSO might interfere with ANS binding to a certain extent. However, the ANS saturation binding curve (Fig. 1B) was obtained in the presence of 1% DMSO as in all the assay buffers. The omission of DMSO for the saturation binding experiment yielded a virtually identical curve (data not shown). The crystal structure of baicalein is already available for comparison (Fig. 5B) [15]. To obtain an ANS-bound 3CLPro structure, we performed molecular docking (Fig. 5C). The binding energy was −6.9 kcal/mol. ANS showed hydrophobic contacts with 9 residues (H41, M49, F140, L141, C145, H164, M165, E166, and Q189) in chain A, including a hydrogen bond between its sulfonate group with catalytic H163 (3.1 Å), and with the N-terminal serine residue of chain B. The structure of the rutin-3CLPro complex from docking mentioned previously was displayed in the same orientation for comparison (Fig. 5D). From our models and the crystal structure of the baicalein-3CLPro complex, ANS, baicalein, and rutin occupy an overlapping space in the active site of 3CLPro, which is also the space occupied by the peptidomimetic covalent inhibitor N3 (Fig. 6) [3].
Table 2.
Crystallographic data collection and refinement statistics.
| Data collection statistics | |
| Wavelength (Å) | 0.97872 |
| Resolution range (Å)a | 27.57–1.45 (1.47–1.45) |
| Space group | C 2 |
| Unit cell dimensions | 113.2, 53.0, 44.7 90.0, 103.1, 90.0 |
| Total number of reflections | 339,475 (16,721) |
| Number of unique reflections | 43,879 (2,124) |
| Multiplicity | 7.7 (7.9) |
| Completeness (%) | 96.1 (93.8) |
| Mean I/σ(I) | 15.3 (2.2) |
| Wilson B factor (Å2) | 12.21 |
| Rmerge | 0.077 (1.035) |
| Rmeas | 0.082 (1.108) |
| Rpim | 0.030 (0.394) |
| CC1/2 | 0.999 (0.803) |
| Refinement Statistics | |
| Resolution range (Å)a | 27.57–1.45 (1.50–1.45) |
| R-factor | 0.1664 (0.2567) |
| R-free (5%) | 0.2010 (0.2731) |
| Number of atoms | 2,935 |
| Protein | 2,473 |
| DMSO | 16 |
| Water | 446 |
| Number of protein residues | 305 |
| RMSD for bonds (Å) | 0.005 |
| RMSD for angles (deg) | 0.822 |
| Estimated coordinate error (ML, Å) | 0.14 |
| Ramachandran favored (%) | 98.68 |
| Ramachandran outliers (%) | 0.33 |
| Average isotropic B factor (Å2) | 20.96 |
| Protein | 18.86 |
| DMSO | 27.57 |
| Water | 32.37 |
| PDB accession code | 7DJR |
Statistics for the highest-resolution shell are given in parentheses.
Fig. 5.
A) Crystal structure of 3CLPro crystallized in the presence of 5 mM ANS but has DMSO (green) bound instead of ANS. B) Crystal structure of 3CLPro with baicalein bound (PDB ID 6M2N). C-D) Conformations of ANS and rutin binding to 3CLPro by molecular docking. Potential non-covalent interactions are shown as dash lines with distances in Ångstrom (Å). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Position of ANS and rutin in the active site compared to the peptidomimetic N3 inhibitor.
3. Discussion
ANS could bind 3CLPro, which resulted in an increased fluorescence at 455 nm. Although ANS has been used previously to study the unfolding process of SARS-CoV-1 3CLPro, our results demonstrated that ANS could bind to a pocket in the native 3CLPro. To confirm that the ANS binding pocket is within the active site of 3CLPro, we first showed that ANS is a competitive inhibitor of 3CLPro. Using the fluorogenic peptide substrate, we observed a reduction in catalytic activity in the presence of ANS that could be fitted with the competitive inhibition model. Therefore, ANS binding site overlaps with the substrate-binding site.
The binding affinity of ANS towards 3CLPro could be extracted from the saturation binding curve and the competitive inhibition experiment. However, the KI value obtained from the competitive inhibition assay was higher than the KD value obtained from the saturation binding curve of ANS to 3CLPro. Several factors might have resulted in the discrepancy, including the difference in assay conditions. The major factor could be the poor solubility of the peptide substrate that limits data points at high substrate concentrations in the ANS competitive inhibition experiment. The maximum velocity estimation and KM may not be accurate. Another potential source of affinity discrepancy could be due to multiple ANS binding sites on 3CLPro. It is possible that 3CLPro possesses a high-affinity binding site for ANS that does not affect the 3CLPro catalytic activity. We cannot completely rule out this possibility with the current data. However, the saturation binding curve is evidently hyperbolic. Thus, even if there are multiple binding sites, the binding affinities are likely similar. Therefore, the KD value obtained from the saturation binding curve might reflect the binding affinity of ANS to 3CLPro more accurately than the KI value from the competitive inhibition experiment.
To confirm that ANS indeed binds in the substrate-binding site, we investigated whether baicalein could compete with ANS binding. Baicalein had already been crystallized in complex with 3CLPro [15]. Thus, the binding site is known. Baicalein had also been shown to inhibit 3CLPro in enzymatic inhibition assays [15] including one independently reported by us. Therefore, baicalein could be used to validate the ANS utility in inhibitor identification. Indeed, baicalein could compete with ANS for 3CLPro binding. The KI value of baicalein from the ANS competitive binding assay (11.3 µM) is comparable with the KD value previously obtained from an isothermal titration calorimetry experiment (4.03 µM) [15]. Therefore, the ANS-based competitive binding assay yielded similar results to other techniques used to identify 3CLPro inhibitors.
We next applied the ANS-based competitive binding assay to show that rutin is an inhibitor of 3CLPro. Rutin has been predicted computationally by multiple research groups to be a 3CLPro inhibitor (Table 1). We have shown that rutin is competitive with both ANS and the fluorogenic peptide substrate. Therefore, we have both demonstrated the inhibitory activity of rutin and also shown that ANS could be used to identify novel inhibitors.
To investigate the molecular details of the interaction between ANS and rutin with 3CLPro, we attempted to crystallized this compound with 3CLPro. However, only the DMSO-bound structure was obtained. Therefore, we employed computational methods to investigate the 3CLPro-ligand interactions. Both ANS and rutin binds in the active site overlapping the known baicalein[15] or N3 inhibitor binding site[3]. The sulfonate group of ANS interacts with a histidine residue, H163, as expected. H163 is not a part of the catalytic dyad but forms the S1 pocket for substrate binding and is also recognized by the γ-lactam moiety of the N3 inhibitor. Rutin interacts mainly with the S1 and S2 pockets, and also interact with residues from the neighboring 3CLPro subunit.
Compared with the crystal structure of DMSO-bound 3CLPro, other ligand-bound structures required movement of the M49 side chain and distortion of the corresponding helix. This required conformational change may explain why we did not observe any ligands in the DMSO-bound, tightly packed C2 crystal form. The baicalein-bound crystal was in the P1 space group, in which the M49-containing helix is more solvent-exposed. Therefore, further crystal engineering may be required to obtain new crystal forms for the ANS- and rutin-bound 3CLPro structures. In addition, because it appeared that the readily obtained C2 crystal form has ligand bias, it may be beneficial for future investigators to screen for other crystal forms to be used in other inhibitor discovery approaches.
In conclusion, ANS could be used in a competitive binding assay to identify ligands that interact with 3CLPro in the active site. Using baicalein as a model compound with a known binding location and 3CLPro inhibitory activity, we showed that ANS binding was competitive with baicalein. Thus, ANS likely binds in the active site of 3CLPro. We also applied this methodology to demonstrated that rutin, a compound predicted to bind and inhibit 3CLPro, was indeed binding competitively with ANS and could inhibit 3CLPro activity. These results demonstrated the utility of ANS in the identification of potential 3CLPro inhibitors. This inhibitor identification strategy could be employed when researchers have limited access to peptide-based protease assays.
4. Materials and methods
4.1. Protein expression, ANS binding assay, and enzyme kinetics
SARS-CoV-2 3CLPro with no tags at the termini was expressed, purified, and stored exactly as previously described for SARS-CoV-1 3CLPro [25]. The gene was synthesized and codon-optimized for expression in Escherichia coli based on the amino acid sequence in GenBank accession number NC_045512. 3CLPro concentrations were determined using the absorbance value at 280 nm and the extinction coefficient of 32,890 M−1 cm−1.
All assays were performed with BioTek Synergy H1 microplate reader using PBS with 1 mM DTT and 1% DMSO as the reaction buffer. The volume was fixed at 100 µL. ANS binding assay was performed with 3CLPro at 5 µM. The excitation and emission wavelengths used were 345 and 455 nm, respectively. For enzyme kinetics, the fluorogenic substrate E(EDANS)TSAVLQSGFRK(DABCYL) (Biomatik) was used with 0.2 µM of 3CLPro. The excitation and emission wavelengths employed were 340 and 490 nm, respectively. GraphPad Prism 8 (San Diego, California USA, www.graphpad.com) was used for graphing and non-linear fit. [26]
4.2. X-ray crystallography
3CLPro was crystallized as described previously (100 mM MES pH 6.5, 15% PEG 4,000, and 5% DMSO)[27] but in the presence of 5 mM ANS. Microseeding was required to obtain crystals suitable for data collection. Crystals were cryoprotected with the crystallization buffer with 5 mM ANS but the PEG 4,000 concentration raised to 35%. Diffraction data were collected at the Life Sciences Collaborative Access Team beamline 21-ID-F (Advanced Photon Source, Argonne National Laboratory). The data were indexed and integrated with XDS [28]. Space group determination and scaling were performed with AIMLESS [29]. Molecular replacement phasing was accomplished with Phaser [30] using a previously reported SARS-CoV-2 3CLPro crystal structure (PDB ID 5RE9) as a search model. Refinement and model adjustments were performed with phenix.refine [31], [32] and COOT [33] respectively. Structure figures were created with UCSF Chimera [34]. The crystal structure and the associated experimental data were deposited at the Protein Data Bank under the accession code 7DJR.
4.3. Prediction of ligand–protein binding and interactions
The 3D structures of the ANS and rutin were built by GaussView 6.0.16 and then optimized with DFT-B3LPY/6–31 g(d) basis set using gaussian 16 [35]. The 3CLPro protein covalently bonded with the N3 inhibitor was retrieved from the protein databank (PBD ID 6LU7) [3]. The prepared protein structure without inhibitor binding was taken from our previous studies [24], [36]. To construct the initial structure of the ligand-3CLPro complex, the optimized structures of deprotonated ANS and rutin were separately docked into the binding site of N3 inhibitor using AutoDock Vina 1.1.2 according to the standard procedure [37]. The ligand orientation with the highest binding affinity was chosen for ligand–protein analysis and further ab initio fragment molecular orbital (FMO) calculation [38], [39], [40].
To provide a detailed insight into the binding of rutin to 3CLPro, the pair interaction energy (PIE) calculation using the second-order Møller–Plesset perturbation theory (MP2) with the resolution-of-the-identity (RI) approximation and polarizable continuum model (PCM) solvation effect (FMO-RIMP2/PCM) was carried out by GAMESS software [41], [42] with the 32 cores computer cluster using a generalized distributed data interface (GDDI). The complex was divided into small fragments (one residue/ligand per fragment) called a monomer, and then all fragments were performed in a parallel manner by the molecular orbital (MO) calculation [43]. Each pair interaction of monomers (I and J) was computed the PIE by a summation of several energy contributions among the clustered residues to identify the essential interacted residue for ligand binding by the following equation:
| (1) |
where is electrostatic interaction, is charge transfer with higher-order mixed terms energies dispersion, is exchange-repulsion, and is the polarizable continuum model (PCM) solvation effect [44], [45], [46].
CRediT authorship contribution statement
Peerapon Deetanya: Methodology, Investigation, Writing - review & editing. Kowit Hengphasatporn: Methodology, Investigation, Writing - original draft, Writing - review & editing, Visualization. Patcharin Wilasluck: Methodology, Investigation, Validation. Yasuteru Shigeta: Methodology, Investigation, Resources, Funding acquisition. Thanyada Rungrotmongkol: Methodology, Investigation, Writing - review & editing, Resources, Funding acquisition. Kittikhun Wangkanont: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing, Project administration, Funding acquisition, Supervision, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). This project is partly funded by the National Research Council of Thailand. KW is partially supported by the Institute for the Promotion of Teaching Science and Technology (IPST) under the Research Fund for DPST Graduate with First Placement [Grant no. 08/2559] and the Chulalongkorn University grant to the Center of Excellence for Molecular Biology and Genomics of Shrimp (GCE 6302823006-1), and to the Molecular Crop Research Unit (GRU 6407023008-1). PD is partially supported by and the 90th Anniversary of the Chulalongkorn University Scholarship. KH and TR are grateful for computational resources supported by NSTDA Supercomputer Center (ThaiSC). YS and KH are supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number JP20ae0101047h0001. KH and YS are also partly supported by CREST program “Precise arrangement toward functionality”, Grant No. JPMJCR20B3. The authors would like to thank Dr. Warinthorn Chavasiri for the gift of baicalein, and Dr. Supot Hannongbua for comments and criticism on the manuscript.
Author Contributions
KW conceived of the study and was in charge of the overall direction. KW and TR procured funding for this study. PD and PW purified 3CLPro and performed ANS binding assay and enzyme kinetics. PD and KW crystallized and determined the 3CLPro structure. KH, YS, and TR performed docking and computational characterization. The manuscript was written with contributions from all authors. All authors have given approval to the final version of the manuscript.
References
- 1.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, NY). 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boras B, Jones RM, Anson BJ, Arenson D, Aschenbrenner L, Bakowski MA et al. 2020. Discovery of a Novel Inhibitor of Coronavirus 3CL Protease as a Clinical Candidate for the Potential Treatment of COVID-19. bioRxiv.2020.2009.2012.293498.
- 6.Jukič M., Janežič D., Bren U. Ensemble docking coupled to linear interaction energy calculations for identification of coronavirus main protease (3CL(pro)) non-covalent small-molecule inhibitors. Molecules (Basel, Switzerland) 2020;25(24) doi: 10.3390/molecules25245808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushal N., Gupta Y., Goyal M., Khaiboullina S.F., Baranwal M., Verma S.C. Mutational Frequencies of SARS-CoV-2 genome during the beginning months of the outbreak in USA. Pathogens (Basel, Switzerland) 2020;9(7) doi: 10.3390/pathogens9070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J., Worrall L.J., Vuckovic M., Rosell F.I., Gentile F., Ton A.-T. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat Commun. 2020;11(1):5877. doi: 10.1038/s41467-020-19662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stryer L. Fluorescence spectroscopy of proteins. Science (New York, NY). 1968;162(3853):526–533. doi: 10.1126/science.162.3853.526. [DOI] [PubMed] [Google Scholar]
- 10.Schonbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. PNAS. 2000;97(12):6345–6349. doi: 10.1073/pnas.120120397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kane C.D., Bernlohr D.A. A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene 8-sulfonic acid. Anal Biochem. 1996;233(2):197–204. doi: 10.1006/abio.1996.0028. [DOI] [PubMed] [Google Scholar]
- 12.Pearce M.C., Rubin H., Bottomley S.P. Conformational change and intermediates in the unfolding of alpha 1-antichymotrypsin. J Biol Chem. 2000;275(37):28513–28518. doi: 10.1074/jbc.M004310200. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharjee C., Das K.P. Thermal unfolding and refolding of beta-lactoglobulin. An intrinsic andextrinsic fluorescence study. Eur J Biochem. 2000;267(13):3957–3964. doi: 10.1046/j.1432-1327.2000.01409.x. [DOI] [PubMed] [Google Scholar]
- 14.Chang H.P., Chou C.Y., Chang G.G. Reversible unfolding of the severe acute respiratory syndrome coronavirus main protease in guanidinium chloride. Biophys J. 2007;92(4):1374–1383. doi: 10.1529/biophysj.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal P.K., Agrawal C., Blunden G. Rutin: A potential antiviral for repurposing as a SARS-CoV-2 main protease (Mpro) inhibitor. Nat Prod Commun. 2021;16(4) 1934578X21991723. [Google Scholar]
- 17.Al-Zahrani A.A. Rutin as a promising inhibitor of main protease and other protein targets of COVID-19. Silico Study. 2020;15(9) 1934578X20953951. [Google Scholar]
- 18.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X., Cai X., Song X., Li C., Zhao J., Luo W. Possible SARS-coronavirus 2 inhibitor revealed by simulated molecular docking to viral main protease and host toll-like receptor. Future Virol. 2020 doi: 10.2217/fvl-2020-0099. [DOI] [Google Scholar]
- 20.Huynh T., Wang H., Luan B. Structure-based lead optimization of herbal medicine rutin for inhibiting SARS-CoV-2's main protease. PCCP. 2020;22(43):25335–25343. doi: 10.1039/d0cp03867a. [DOI] [PubMed] [Google Scholar]
- 21.Peterson L. COVID-19 and flavonoids. In silico molecular dynamics docking to the active catalytic site of SARS-CoV and SARS-CoV-2 main protease. SSRN. 2020 doi: 10.2139/ssrn.3599426. [DOI] [Google Scholar]
- 22.Xu Z., Yang L., Zhang X., Zhang Q., Yang Z., Liu Y. Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening. Strategy. 2020;7(247) doi: 10.3389/fmolb.2020.556481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman F., Tabrez S., Ali R., Alqahtani A.S., Ahmed M.Z., Rub A. Molecular docking analysis of rutin reveals possible inhibition of SARS-CoV-2 vital proteins. J Tradition Complem Med. 2021 doi: 10.1016/j.jtcme.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somboon T., Mahalapbutr P., Sanachai K., Maitarad P., Lee V.S., Hannongbua S. Computational study on peptidomimetic inhibitors against SARS-CoV-2 main protease. J Mol Liq. 2021;322 doi: 10.1016/j.molliq.2020.114999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J Mol Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowd J.E., Riggs D.S. A comparison of estimates of michaelis-menten kinetic constants from various linear transformations. J Biol Chem. 1965;240:863–869. [PubMed] [Google Scholar]
- 27.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun. 2020;11(1):5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR et al. 2016. Gaussian 16 Rev. C.01. Wallingford, CT.
- 36.Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020;59(18):1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 37.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitaura K., Sugiki S.-I., Nakano T., Komeiji Y., Uebayasi M. Fragment molecular orbital method: analytical energy gradients. Chem Phys Lett. 2001;336(1):163–170. [Google Scholar]
- 39.Pham B.Q., Gordon M.S. Development of the FMO/RI-MP2 fully analytic gradient using a hybrid-distributed/shared memory programming model. J Chem Theory Comput. 2020;16(2):1039–1054. doi: 10.1021/acs.jctc.9b01082. [DOI] [PubMed] [Google Scholar]
- 40.Tsukamoto T., Mochizuki Y., Watanabe N., Fukuzawa K., Nakano T. Partial geometry optimization with FMO-MP2 gradient: Application to TrpCage. Chem Phys Lett. 2012;535:157–162. [Google Scholar]
- 41.Fedorov D.G. The fragment molecular orbital method: theoretical development, implementation in GAMESS, and applications. WIREs Comput Mol Sci. 2017;7(6) [Google Scholar]
- 42.Steinmann C., Jensen J.H. Fragmentation John Wiley & Sons; 2017. Effective Fragment Molecular Orbital Method. [Google Scholar]
- 43.Fedorov D.G., Kitaura K. Pair interaction energy decomposition analysis. J Comput Chem. 2007;28(1):222–237. doi: 10.1002/jcc.20496. [DOI] [PubMed] [Google Scholar]
- 44.Boonyasuppayakorn S., Saelee T., Visitchanakun P., Leelahavanichkul A., Hengphasatporn K., Shigeta Y. Dibromopinocembrin and dibromopinostrobin are potential anti-dengue leads with mild animal toxicity. Molecules (Basel, Switzerland). 2020;25(18) doi: 10.3390/molecules25184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maruyama K., Sheng Y., Watanabe H., Fukuzawa K., Tanaka S. Application of singular value decomposition to the inter-fragment interaction energy analysis for ligand screening. Comput Theor Chem. 2018;1132:23–34. [Google Scholar]
- 46.Tokiwa T., Nakano S., Yamamoto Y., Ishikawa T., Ito S., Sladek V. Development of an analysis toolkit, AnalysisFMO, to visualize interaction energies generated by fragment molecular orbital calculations. J Chem Inf Model. 2019;59(1):25–30. doi: 10.1021/acs.jcim.8b00649. [DOI] [PubMed] [Google Scholar]