Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is highly contagious and has caused significant medical/socioeconomic impacts. Other than vaccination, effective public health measures, including contact tracing, isolation, and quarantine, is critical for deterring viral transmission, preventing infection progression and resuming normal activities. Viral transmission is affected by many factors, but the viral load and vitality could be among the most important ones. Although in vitro studies have indicated that the amount of virus isolated from infected individuals affects the successful rate of virus isolation, whether the viral load carried at the individual level would determine the transmissibility was unknown. We examined whether the cycle threshold (Ct) value, a measurement of viral load by RT-PCR assay, could differentiate the spreaders from the non-spreaders in a population of college students. Our results indicate that while at the population level the Ct value is lower, suggesting a higher viral load, in the symptomatic spreaders than that in the asymptomatic non-spreaders, there is a significant overlap in the Ct values between the two groups. Thus, Ct value, or the viral load, at the individual level could not predict the transmissibility. Instead, a sensitive method to detect the presence of virus is needed to identify asymptomatic individuals who may carry a low viral load but can still be infectious.
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic with serious impacts on all aspects of human life. Deterrence of viral transmission through public health measures, including contact tracing, isolation, and quarantine, is critical for infection control required to resume normal activities. Unlike two other betacoronaviruses that had caused previous local epidemics, SARS-CoV and Middle East Respiratory syndrome–CoV, SARS-CoV-2 exhibits distinct replication and transmission kinetics. It replicates more rapidly in the human upper respiratory tract, which helps its transmission through asymptomatic viral carriers and facilitates a fast spread of SARS-CoV-2. The relatively lower fatality rate (case fatality ratio) of SARS-CoV-2 (2% compared with 10% for SARS-CoV and 34% for Middle East Respiratory syndrome–CoV) may also contribute to its high transmissibility.1 SARS-CoV-2 is highly contagious, with an estimated reproductive number of 3.5,2 but significant variations exist among individuals, with some being super spreaders. This is much higher than the reproductive number for seasonal flu (1.3) and SARS-CoV (0.86 to 1.83).3 , 4
The viral load in an infected individual could affect the level of infectivity. Several studies have found that successful isolation of virus from patient samples depends on viral load as measured by the cycle threshold (Ct) value of the RT-PCR assay, which was thus suggested to correlate with infectivity.5, 6, 7, 8, 9, 10 A cutoff Ct value between 32 and 35 was proposed to guide isolation practices.5, 6, 7, 8, 9, 10 However, it was not clear whether the in vitro culture results could reflect definite viral spread in individuals and whether Ct values could actually be used to guide decisions regarding isolation and quarantine.
The effective way to block viral transmission is to identify, isolate, and treat the infected individuals, and to track down and quarantine those having close contact with the infected ones. As the infection involves more and more individuals, specific communities or regions may be forced to shut down. All social activities related to work, study, and leisure will be significantly affected, with tremendous impacts on the economy, society, and overall personal health conditions. It is thus important to better understand the dynamics of viral transmission and to examine whether certain surrogate measurements may be used to determine SARS-Cov-2 transmissibility. We therefore determined whether the Ct values, as a measurement of viral load, could be used to provide a level of prediction in a population of college students. The Ct values of the spreaders were compared with the nonspreaders. These values were found to be largely overlapping. It is thus not possible to predict viral transmissibility based on Ct values at the individual level.
Materials and Methods
Study Population
Results from undergraduate students aged <23 years were selected for this retrospective study. These students were participants in on-campus educational activities while living either on campus or off campus. They were tested twice a week in the period between September 1, 2020, and October 31, 2020. This study included only students who were tested in the Clinical Laboratory Improvement Amendments–certified Molecular Pathology Laboratory of the Department of Pathology and Laboratory, Tulane University School of Medicine, because the Ct values were obtained using the same testing method in the same laboratory for all the included subjects. Full review and approval were waived by the Tulane University institutional review board due to involvement of only secondary, deidentified data.
Sample Collection, Processing, and RNA Extraction
Nasopharyngeal swab specimens were collected following current CDC guidelines. All samples were stored at 4°C before delivery to the testing laboratory. Upon receipt, samples were inactivated at 60°C for 30 minutes in a forced-air oven (Thermo Fisher Scientific, Waltham, MA; catalog no. 151030510). RNA was extracted by using a KingFisher Flex Magnetic Particle Processor with 96 Deep-Well Head (Thermo Fisher Scientific; catalog no. 5400630) and the MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific; catalog no. A48310). An MS2 phage control was included as an extraction control in the original sample before total RNA extraction.
TaqPath RT-PCR COVID-19 Combo Kit Assay
The TaqPath RT-PCR COVID-19 Combo Kit Assay is US Food and Drug Administration approved under Emergency Use Authorization. Multiplex RT-qPCR was performed according to the manufacturer’s instructions (Thermo Fisher Scientific; catalog no. A47814). Viral nucleic acids were detected by using primers and probes targeting the N, S, and Orf1ab genes. A pair of primers against the extraction controls (MS2) was also included in the same reaction. RT-qPCR reactions were performed on either an ABI7500 FAST DX Real-Time PCR instrument (Thermo Fisher Scientific; catalog no. 4406985) or a QuantStudio 5 Real-Time PCR instrument (Thermo Fisher Scientific; catalog no. A34322). Positive samples were identified by using the Applied Biosystems COVID-19 Interpretive Software version 1.3 (for ABI7500 FAST DX; Foster City, CA) or Applied Biosystems COVID-19 Interpretive Software version 2.3 (for QuantStudio 5).
Analysis of Ct Values
The Ct values for the three viral genes (N, S, and Orf1ab) and extraction control were determined individually by using analytical software SDS version 1.4.1 (for ABI7500 FAST DX) or QuantStudio Design and Analysis Desktop Software version 1.5.1 (for QuantStudio 5). The averaged Ct values of the three viral genes were presented given their similarity and to minimize the bias in PCR performance for a particular gene. Selection of a Ct value of 24 or 32 as a threshold was based on the literature (Results).5, 6, 7, 8, 9, 10
Contact Tracing and Quarantine Program
Symptomatic information was collected immediately before sample collection and testing. Contact tracers received all positive results and made telephone calls to reach positive case subjects. They interviewed the positive case subjects to identify close contacts. The contact tracers also helped to establish the quarantine procedure. The information of the index cases and the contacts was recorded.
Statistical Analysis
Data are presented as means ± SD (for the age distribution), means ± SEM, or median ± interquartile (for the Ct values). Statistical significance was assessed by using a two-sided unpaired t-test for age distribution and a Mann-Whitney U test or one-way analysis of variance for Ct values with Prism Software version 9 (GraphPad Software, Inc, San Diego, CA).
Results
Colleges represent a unique environment with a dense population of primarily young students, and strict control of SARS-CoV-2 transmission is critical for their educational mission. Tulane University maintained on-campus educational activities in the fall semester of 2020. A high-throughput SARS-CoV-2 testing program was established to support the contact tracing, isolation, and quarantine efforts needed to actively restrict viral transmission throughout the campus. During the period covered in this study, the screening test was performed twice a week, with 99% of testing completed within 24 hours from collection to report. Although all students (graduate and undergraduate, on-campus and off-campus living) were screened, only data from 7440 students aged <23 years from September 1, 2020, to October 31, 2020, were included in this study for data consistency. A total of 61,982 tests for these students were performed during this period, and 602 unique positive cases were identified (Tables 1 and 2 ). Compared with all students, those who tested positive for SARS-CoV-2 were slightly younger, reflecting that more freshmen and sophomores were infected. In addition, male and female students had nearly the same proportion of infection (49.3% versus 50.7%), consistent with a meta-analysis of 90 reports.11 However, considering that male students accounted for only 37.5% of all the students screened, the male students had a higher infection rate (10.65%) than the female students (6.56%) in this cohort.
Table 1.
Sex Distribution of the Cases
| Study cohort | All cases |
Positive cases |
Infection rate, % | ||
|---|---|---|---|---|---|
| n | Proportion, % | n | Proportion, % | ||
| All | 7440 | 100 | 602 | 100 | 8.09 |
| Male | 2790 | 37.50 | 297 | 49.34 | 10.65 |
| Female | 4650 | 62.50 | 305 | 50.66 | 6.56 |
Data reflect the number of unique individual undergraduates being tested in the period between September 1, 2020, and October 31, 2020. Each individual may have been tested multiple times during this period, but each unique positive case was counted only once. Proportion of each sex in the population is calculated by dividing the total individual number by male or female individual numbers for all cases or for positive cases. The infection rate is calculated by dividing all the case numbers by the positive case number in the male or female groups or all individuals.
Table 2.
Age Distribution of the Cases
| Study cohort | All cases, years |
Positive cases, years |
P | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| All | 20.28 | 1.31 | 19.64 | 1.11 | <0.001 |
| Male | 20.32 | 1.32 | 19.75 | 1.20 | <0.001 |
| Female | 20.25 | 1.30 | 19.54 | 1.02 | <0.001 |
Two-sided unpaired t-tests were conducted between the positive cases and all cases for all sexes, male only or female only.
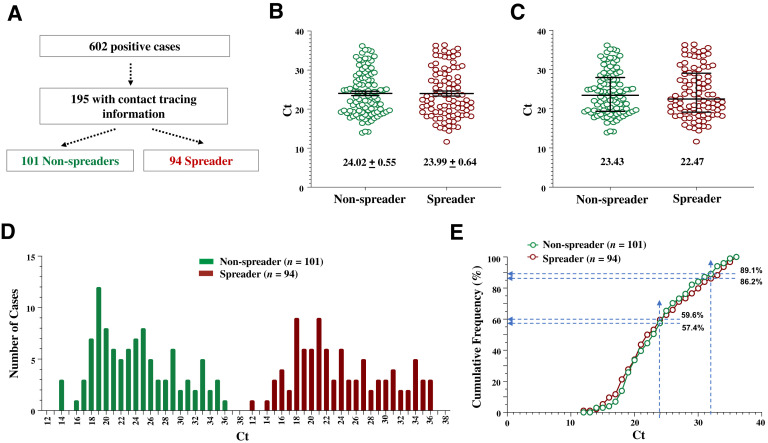
From this cohort of 602 individuals who tested positive, 195 index cases were identified with one or more reported close contacts who were then tested during their mandated 14-day quarantine period for evidence of transmission from the associated index cases (Figure 1 A). 48.2% (94 of 195) of these index cases had at least one contact who became SARS-CoV-2 positive, whereas 51.8% of the index cases (n = 101) were nonspreaders with no contacts who subsequently tested positive.
Figure 1.
The cycle threshold (Ct) values of the spreaders and the nonspreaders are largely overlapping. A: Separation of index cases into spreader and nonspreader groups. The n of the cases in each population is indicated. B and C: Scatter plots of Ct values expressed as means ± SEM (B) or median ± interquartile intervals (C). Ct values of the indicated populations are compared. D: Histogram of the distribution of Ct values. E: Cumulative frequency of Ct values. Dashed lines indicate the cumulated percentage of each population at the designated Ct value (24 or 32). At the indicated Ct value of ≤24, there is a higher percentage of spreader cases than nonspreader cases, although the differences are small. Ct values of the indicated populations are compared. P > 0.05 (U-test; B and C).
Mean Ct values of the spreaders and the nonspreaders were nearly identical (Figure 1B), but their median Ct values differed by almost one cycle (Figure 1C), suggesting that more spreaders had a lower Ct value than the nonspreaders. However, Ct distributions in these groups were similar, with the main peaks around 18 to 21 (Figure 1D), although the Ct range was slightly broader for the spreaders (12 to 36) than for the nonspreaders (14 to 36). Cumulative Ct frequencies overlapped between the spreaders and the nonspreaders, with 10.9% and 13.8% of cases having a Ct value of 32 and higher, respectively (Figure 1E); the difference, however, was not large enough to discriminate the two groups for practical use.
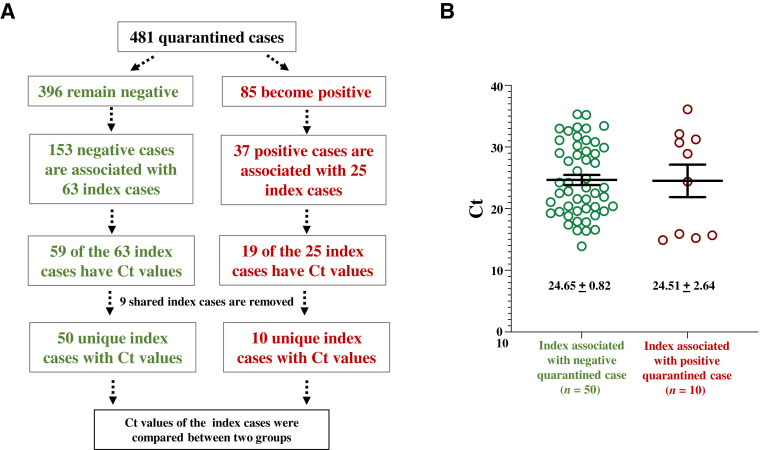
In a reverse approach, index cases were traced for 481 students undergoing quarantine at one of the three Tulane quarantine sites in September 2020 (Figure 2 A), 18% of whom (85 of 481) became positive during their quarantine period. Index cases for these 481 quarantined individuals were considered spreaders if they were linked to one or more quarantined students with a positive test result, or nonspreaders if they were associated only with individuals with negative test results. Spreaders and nonspreaders without Ct values reported were excluded from further analysis. The mean Ct values of the spreader and the nonspreader groups were similar (Figure 2B). Taken together, these index case studies suggest that Ct values alone do not predict transmission risk.
Figure 2.
Comparisons of cycle threshold (Ct) values of index cases tracked from quarantined cases. A: Diagram of the study design. Index cases with Ct values available are tracked back from their contacts in the quarantined units. B: The Ct values of spreader index cases and nonspreader index cases show a significant overlap. Data shown are means ± SEM. P > 0.05 (U-test).
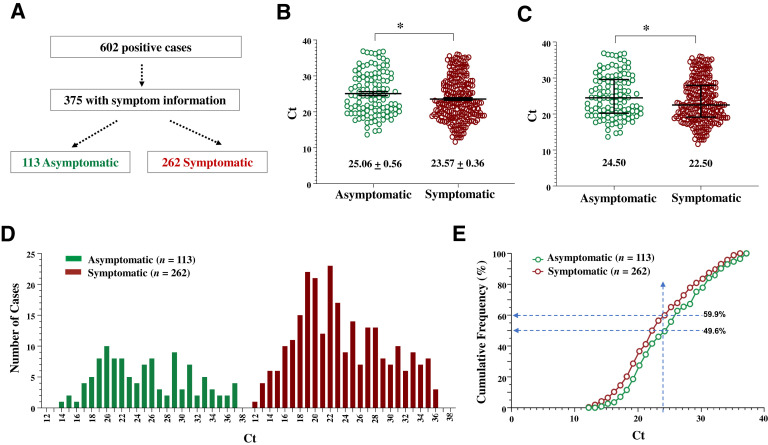
Individuals who are SARS-CoV-2 positive but asymptomatic can still be infectious12, 13, 14 and may exhibit a viral load similar to that of their symptomatic counterparts.12 , 13 , 15 Three-hundred and seventy-five positive cases were identified and evaluated for coronavirus disease 2019 (COVID-19) symptoms at testing to assess the relationship between symptom presentation and Ct values (Figure 3 A). The reported symptoms included lethargy, fever, headache, cough, runny nose, and gastrointestinal symptoms. The mean and median Ct values were significantly lower in symptomatic cases than in asymptomatic cases (Figure 3, B and C), which was also reflected by the difference in the Ct range of these groups (12 to 36 versus 14 to 37) (Figure 3D). Although both groups exhibited Ct peaks around 19 to 22, there was a noticeable rightward shift in the cumulative Ct frequency in the asymptomatic versus symptomatic population, indicative of reduced viral load in the asymptomatic group (Figure 3E). In comparison, other studies with cohorts differing in location and in constituents, including a large study involving senior citizens from nursing houses and assisted living facilities in Massachusetts, found that Ct values did not differ significantly between the symptomatic and the asymptomatic individuals; however, a faster virus clearance, as measured by Ct value, was observed in the asymptomatic cases than in the symptomatic cases.13 , 15 These findings and our studies thus suggest that infections with a higher viral load may be more likely to lead to symptom development, or that symptomatic individuals tend to have higher viral loads or to maintain their viral loads for a longer time.
Figure 3.
The cycle threshold (Ct) values of the symptomatic individuals are lower than those of the asymptomatic individuals as a population. A: The separation of positive cases into symptomatic and nonsymptomatic groups. The n of the cases in each population is indicated. B and C: Scatter plots of Ct values expressed as means ± SEM (B) or median ± interquartile intervals (C). D: The histogram of the distribution of Ct values. E: The cumulative frequency of Ct values. Dashed lines indicate the cumulated percentage of each population at the Ct value of 24. At this Ct value and below, there is a higher percentage of symptomatic cases (59.9%) than asymptomatic cases (49.6%). Ct values of the indicated populations are compared. ∗P < 0.05 (U-test).
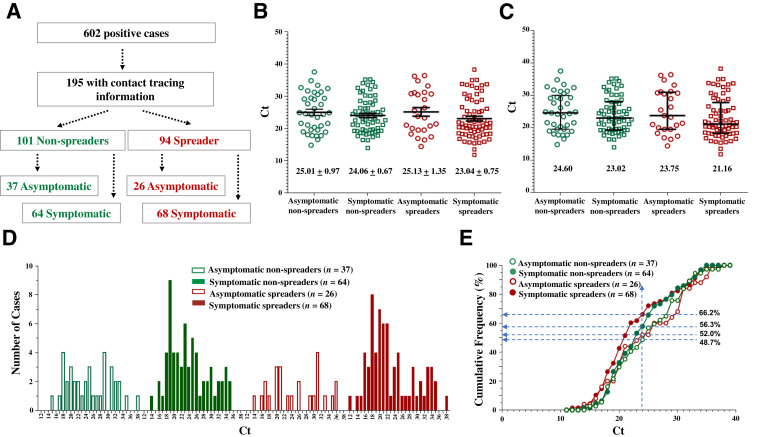
All 195 index cases with contact tracing information had data recorded regarding symptoms. The spread group and the nonspreader group was further divided based on symptom presentation (Figure 4 A). The symptomatic spreaders had the lowest mean and median Ct values, differing by 2 cycles for the mean and 3.5 cycles for the median compared with the asymptomatic nonspreaders, who had the highest mean and median Ct values (Figure 4, B and C). The Ct distribution indicated that the symptomatic groups (spreaders and nonspreaders) and the spreader groups (with or without symptoms) tended to include more individuals with lower Ct values (<24) (Figure 4, D and E). This finding suggests that SARS-CoV-2 spreaders tend to have higher viral loads and are more likely symptomatic.
Figure 4.
The symptomatic spreaders exhibit the lowest cycle threshold (Ct) values as a population. A: Separation of positive cases into spreaders and nonspreaders, with or without symptoms. The n of the cases in each population is indicated. B and C: Scatter plots of Ct values expressed as means ± SEM (B) or median ± interquartile intervals (C). Ct values of the indicated populations are compared. D: The histogram of the distribution of Ct values. E: The cumulative frequency of Ct values. Dashed lines indicate the cumulated percentage of each population at the Ct value of 24. At this Ct value and below, there is a higher percentage of symptomatic spreader cases (66.2%) than asymptomatic nonspreader cases (48.7%). The percentage of cases of the other groups falls between the two. Ct values of the indicated populations are compared. P > 0.05 (one-way analysis of variance; B and C).
Discussion
The current study compared Ct values for the first time between spreaders and nonspreaders of SARS-CoV-2 infection in a college student population. Although the mean Ct values of the spreaders, particularly the symptomatic spreaders, were lower than those of the nonspreaders, there was a significant overlap among individuals, whether they were spreaders or nonspreaders. It is thus practically not feasible to predict who would be spreaders based on the viral load as detected from the nasal swab.
Ct values are not reported in current public health practice despite the fact that they may be informative of viral burden. Our study supports this practice and indicates that, due to the broad spread and overlap in Ct values across the spectrum of symptom presentation and transmissibility, reporting of Ct values at the individual level, such as by setting a cutoff value at 32,5, 6, 7, 8, 9, 10 would provide little diagnostic value for differential case management. At the population level, Ct values may be useful, particularly in association with the symptomatic presentation, to indicate the likelihood of transmission. These values may thus have epidemiologic or surveillance importance.
Detection of SARS-CoV-2 may need to be both sensitive and rapid, which might not always be achieved by all methods. A rapid but less sensitive method should be used more frequently to identify individuals whose virus level may be increasingly elevated over the course of infection and thus presumably become more infectious. However, our results suggest that individuals with a low viral load could still be infectious. Thus, a sensitive and robust SARS-CoV-2 diagnostic testing method is needed to effectively control viral transmission by maximizing the ability to identify and quarantine those infected with a low level of virus.
Although limited by its retrospective nature, this study has the advantage of being less affected by host and environmental factors of viral transmission, as the college student population is generally in good health with few underlying susceptibilities, with most individuals living and interacting in a shared and relatively confined social environment (ie, campus). Data were further restricted to those from current undergraduate students (<23 years old, with an average age of 20.3 years) (Table 2), making the population more homogeneous to reduce the influence of age. Transmissibility is not only affected by the viral load of the spreaders and the environment in which transmission takes place but also by factors that emphasize the susceptibility of the population such as age, sex, and basic health conditions. Therefore, it is interesting to note that although male and female students had nearly the same proportion of infected individuals, consistent with a meta-analysis of 90 reports,11 male students had a higher infection rate (10.65%) than female students (6.56%) in this cohort. The sex disparity of COVID-19 has been well recognized in terms of severity of the disease, with male subjects being more likely to develop severe conditions.11 The effect of sex needs to be further dissected out to determine how a specific sex may lead to differences in the spread and development of COVID-19.
In conclusion, this study determined that Ct values of spreaders may be lower at the population level than that of nonspreaders; however, the large overlap in values at the individual level prevents their use as a differential diagnostic tool to guide isolation and quarantine practice. Thus, a sensitive and robust diagnostic method is necessary to restrict viral transmission from those carrying a low level of virus.
Author Contributions
D.T., R.M.G., X.-M.Y., P.D., and L.H. designed the study; D.T., E.M.K., J.C.D, N.O., R.M.G., S.Y., and S.R. collected and analyzed the data; X.-M.Y., D.T., and Z.L. wrote the manuscript; T.Y.H., R.M.G., and P.D. edited the manuscript; D.J.E., M.L., M.L.L., S.T., and P.N. provided administrative and operational support; J.T., N.T., and N.W. provided IT support.
Footnotes
Supported by Tulane University.
Disclosures: None declared.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Choi S., Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health. 2020;42:e2020011. doi: 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggerstaff M., Cauchemez S., Reed C., Gambhir M., Finelli L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis. 2014;14:480. doi: 10.1186/1471-2334-14-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowell G., Castillo-Chavez C., Fenimore P.W., Kribs-Zaleta C.M., Arriola L., Hyman J.M. Model parameters and outbreak control for SARS. Emerg Infect Dis. 2004;10:1258–1263. doi: 10.3201/eid1007.030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jefferson T., Spencer E.A., Brassey J., Heneghan C. Viral cultures for COVID-19 infectivity assessment—a systemic review. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1764. [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Hoang V.T., Colson P., Raoult D., La Scola B. Correlation between 3790 quantitative polymerase chain reaction–positives samples and positive cell cultures, including 1941 severe acute respiratory syndrome coronavirus 2 isolates. Clin Infect Dis. 2021;72:e921. doi: 10.1093/cid/ciaa1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25:2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L., Kumar S., Tran T., Ko D., Sivaruban T., Ngo C., Toi C., O'Sullivan M.V., Sintchenko V., Chen S.C.-A., Maddocks S., Dwyer D., Kok J. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1579. [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chau N.V.V., Thanh Lam V., Thanh Dung N., Yen L.M., Minh N.N.Q., Hung L.M., Ngoc N.M., Dung N.T., Man D.N.H., Nguyet L.A., Nhat L.T.H., Nhu L.N.T., Ny N.T.H., Hong N.T.T., Kestelyn E., Dung N.T.P., Xuan T.C., Hien T.T., Thanh Phong N., Tu T.N.H., Geskus R.B., Thanh T.T., Thanh Truong N., Binh N.T., Thuong T.C., Thwaites G., Tan L.V., Oxford University Clinical Research Unit COVID-19 Research Group The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin Infect Dis. 2020;71:2679–2687. doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 15.Lennon N.J., Bhattacharyya R.P., Mina M.J., Rehm H.L., Hung D.T., Smole S., Woolley A., Lander E.S., Gabriel S.B. Comparision of viral levels in individuals with or without symptoms at time of COVID-19 testing among 32,480 residents and staff of nursing homes and assisted living facilities in Massachusetts. Open Forum Infectious Diseases. 2020;70:S848–S849. [Google Scholar]