Abstract
Objective
There is a lack of data evaluating performance of antigenic test (AT) for SARS-CoV-2 diagnosis (Ag-RDT) in clinical practice, especially in asymptomatic subjects. The main objective of this study was to evaluate the diagnostic performance of AT compared to Reverse Transcription Polymerase Chain Reaction (RT-PCR) for SARS-CoV-2 diagnosis.
Methods
StudyCov is a monocentric cross-sectional study. A SARS-CoV-2 screening facility was set up in the Bordeaux University health campus from October 28th to November 20th 2020. Students willing to have a RT-PCR test (ARGENE SARS-CoV-2 R-GENE, BioMérieux, France) for SARS-CoV-2 diagnosis were also offered the Abbott Panbio™ SARS-CoV-2 antigenic rapid test. All participants attending the screening facility with an AT in addition to RT-PCR and having signed an informed consent were included in the study. The main objective was to assess performance of AT as compared with RT-PCR in the recruited population. Secondary objectives dealt with the analysis of the main objective stratified by current symptoms and risk exposure. A sensitivity analysis with different RT-PCR cycle thresholds was included.
Results
RT-PCR and AT results were available for 692 subjects. Overall sensitivity and specificity of AT tests were respectively 63.5% (95% confidence interval (CI): 49.0 – 76.4) and 100% (95% CI: 99.4 – 100). In the asymptomatic sub-group, they were respectively 35.0% (95% CI: 15.4% - 59.2%) and 100% (95% CI: 99.3 - 100).
Conclusions
This study shows the poor sensitivity of AT in asymptomatic subjects, specificity being however excellent. The performance results fall below the World Health Organization recommendation of 80% sensitivity and question using AT in general population, especially when asymptomatic.
Keywords: Rapid antigen detection test, Sars-cov-2, Rt-pcr, Point-of-care testing, COVID19
1. Introduction
Rapid identification of SARS-CoV-2 cases is a cornerstone in the management of the COVID-19 pandemic. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) is the reference test for SARS-CoV-2 diagnosis. In addition, various antigenic tests (AT) have been developed. They provide test results in about 15 min, which is much shorter than for RT-PCR tests.
Some studies have compared both RT-PCR and AT performances. Regulatory health agencies performed meta-analyses of the latter in order to establish testing recommendations [1], [2], [3], [4]. Concerning AT, the Haute Autorité de Santé (HAS) - the French National Authority for Health - concluded to an homogeneous good specificity of 98.7% (95% confidence interval (95% CI): 97.3 - 99.4). However, sensitivity was extremely heterogeneous, from 17 to 97%, depending on the study and the targeted population, with a pooled sensitivity of 90% (95% CI: 73 - 96) [4]. Therefore, the World Health Organization (WHO) recommended a minimum sensitivity of 80% and a minimum specificity of 97% for AT effective use [3]. However, to date, few studies have been performed in the general population and in asymptomatic people. This is of concern as some countries are promoting the use of AT without solid knowledge about their actual performance [4], [5], [6], [7].
From October 28th to November 20th 2020, a screening center has been operated in the Bordeaux University health campus (France). Students willing to be tested for SARS-CoV-2 by RT-PCR were also offered AT. The objective of this cross-sectional study was to assess the diagnosis performance of a nasopharyngeal antigenic rapid test compared to RT-PCR in this population of students and in the subgroup of asymptomatic subjects.
2. Methods
This study is a monocentric diagnosis cross-sectional retrospective study. All participants older than 18 years and tested with both AT and RT-PCR at the screening facility of the Bordeaux University health campus were included in the study. The index test was the nasopharyngeal Abbott Panbio™ SARS-CoV-2 Ag rapid test. The reference test was the RT-PCR of N and RdRp genes (ARGENE SARS-CoV-2 R-GENE, BioMérieux, France) with a maximum cycle threshold (Ct) of 45, the current standard of care in the Bordeaux University Hospital. Nasopharyngeal samplings for both tests were performed simultaneously, in each nostril. AT and RT-PCR readings were blinded to each other thanks to different logistic tracks. Testing and reading were performed by trained teams and the reading was carried out by a single reader following the manufacturer's instructions.
The main objective was sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of AT compared to RT-PCR in the whole study population. RT-PCR was considered positive when both nucleocapsid (N) and RNA-dependent RNA polymerase (RdRp) genes were detected. Secondary objectives were performances of AT compared to RT-PCR stratified by current symptoms and risk situation exposure (over the previous week). Risk situation exposure was defined as attending a party, practicing team sports or having contact with another person outside home without a mask over the previous week. Sensitivity analysis with different RT-PCR Ct was included. We also compared the Ct of N and RdRp SARS-CoV-2 gene amplifications among subjects with a positive SARS-CoV-2 RT-PCR test stratified by AT results and current symptoms.
Subjects with missing RT-PCR or AT results were excluded from the analysis. Sensitivity, specificity, PPV and NPV confidence intervals were computed using the exact binomial method. Ct values for N and RdRp viral genes were compared using Wilcoxon ranked test. Analyses were performed using R v4.0.0 and the epiR package [8].
3. Results
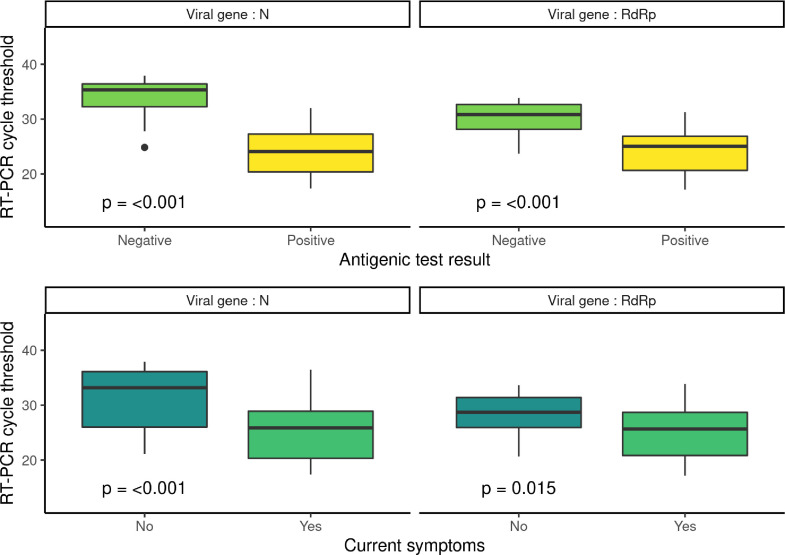
Subjects’ characteristics are described in Table 1 . There was no false-positive case when considering RT-PCR positive if both genes were detected. When considering a subject as positive if both genes were detected, one of which with a RT-PCR Ct ≤ 30, 3 subjects proved false-positive. Table 2 shows the performance of AT compared to RT-PCR by subgroup and by positive SARS-CoV-2 RT-PCR definition. Overall specificity and sensitivity were respectively 100 (95% CI: 99.4 – 100) and 63.5 (95% CI: 49.0 – 76.4). In the asymptomatic group, they were respectively 100% (95%CI: 99.3 – 100.0) and 35.0% (95% CI: 15.4 – 59.2). Results were similar when considering asymptomatic patients exposed to a risk situation. Fig. 1 shows the association between AT result, current symptoms and Ct.
Table 1.
Subjects characteristics. StudyCov study.
| AT test | |||
|---|---|---|---|
| Variables | Overall | Negative | Positive |
| n | 695 | 658 | 33 |
| Age (mean (SD)) | 22.95 (5.52) | 23.00 (5.62) | 21.79 (2.06) |
| Sex = Male (%) | 236 (34.0) | 223 (33.9) | 11 (33.3) |
| Type of education (%) | |||
| General first year | 28 (4.0) | 28 (4.3) | 0 (0.0) |
| Physiotherapy | 27 (3.9) | 25 (3.8) | 2 (6.1) |
| Medicine | 401 (57.7) | 379 (57.6) | 19 (57.6) |
| Nursing | 17 (2.4) | 16 (2.4) | 1 (3.0) |
| Pharmacy | 97 (14.0) | 93 (14.1) | 4 (12.1) |
| Other | 125 (18.0) | 117 (17.8) | 7 (21.2) |
| Current symptoms = Yes (%) | 132 (19.0) | 105 (16.0) | 26 (78.8) |
| Symptoms over the previous 2 weeks = Yes (%) | 94 (13.5) | 80 (12.2) | 14 (42.4) |
| Risk situation over the previous week = Yes (%) | 380 (54.7) | 356 (54.1) | 23 (69.7) |
| Antigenic test * = Positive (%) | 33 (4.8) | 0 (0.0) | 33 (100.0) |
| Ct N gene ⁎⁎ (mean (SD)) | 28.48 (6.57) | 34.65 (3.66) | 24.18 (4.30) |
| Ct RdRp gene ⁎⁎ (mean (SD)) | 26.41 (4.66) | 30.28 (2.80) | 24.19 (4.03) |
| N gene detected ⁎⁎ = yes (%) | 56 (8.1) | 23 (3.5) | 33 (100.0) |
| RdRp gene detected ⁎⁎ = yes (%) | 52 (7.5) | 19 (2.9) | 33 (100.0) |
| Both genes detected ⁎⁎ = yes (%) | 52 (7.5) | 19 (2.9) | 33 (100.0) |
*Missing antigenic test for 4 patients.
⁎⁎Missing RT-PCR for 6 patients, of which 3 also missed AT.
Both PCR and antigenic test were available for 688 patients.
Table 2.
Antigenic test performance compared to RT-PCR test for SARS-CoV-2 diagnosis by RT-PCR Ct. AT performance is shown among different subgroups depending on RT-PCR positivity definition. 95% CI are computed using the exact binomial method.
| RT-PCR max CT | Se [95% CI] | Sp [95% CI] | NPV [95% CI] | PPV [95% CI] |
|---|---|---|---|---|
| Overall | ||||
| 2 Genes | 63.5 [49.0; 76.4] | 100.0 [99.4; 100.0] | 97.1 [95.5; 98.3] | 100.0 [89.4; 100.0] |
| N Gene | 58.9 [45.0; 71.9] | 100.0 [99.4; 100.0] | 96.5 [94.8; 97.8] | 100.0 [89.4; 100.0] |
| RdRp Gene | 63.5 [49.0; 76.4] | 100.0 [99.4; 100.0] | 97.1 [95.5; 98.2] | 100.0 [89.4; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [75.3; 100.0] | 97.0 [95.5; 98.2] | 100.0 [99.4; 100.0] | 39.4 [22.9; 57.9] |
| 2 Genes + Ct ≤ 30 | 76.9 [60.7; 88.9] | 99.5 [98.7; 99.9] | 98.6 [97.4; 99.4] | 90.9 [75.7; 98.1] |
| 2 Genes + Ct ≤ 33 | 70.2 [55.1; 82.7] | 100.0 [99.4; 100.0] | 97.9 [96.4; 98.8] | 100.0 [89.4; 100.0] |
| Currently symptomatic | ||||
| 2 Genes | 81.2 [63.6; 92.8] | 100.0 [96.3; 100.0] | 94.3 [88.0; 97.9] | 100.0 [86.8; 100.0] |
| N Gene | 78.8 [61.1; 91.0] | 100.0 [96.3; 100.0] | 93.3 [86.7; 97.3] | 100.0 [86.8; 100.0] |
| RdRp Gene | 81.2 [63.6; 92.8] | 100.0 [96.3; 100.0] | 94.3 [88.0; 97.9] | 100.0 [86.8; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [73.5; 100.0] | 88.2 [81.0; 93.4] | 100.0 [96.5; 100.0] | 46.2 [26.6; 66.6] |
| 2 Genes + Ct ≤ 30 | 96.2 [80.4; 99.9] | 99.0 [94.8; 100.0] | 99.0 [94.8; 100.0] | 96.2 [80.4; 99.9] |
| 2 Genes + Ct ≤ 33 | 86.7 [69.3; 96.2] | 100.0 [96.4; 100.0] | 96.2 [90.5; 99.0] | 100.0 [86.8; 100.0] |
| Currently asymptomatic | ||||
| 2 Genes | 35.0 [15.4; 59.2] | 100.0 [99.3; 100.0] | 97.6 [96.0; 98.7] | 100.0 [59.0; 100.0] |
| N Gene | 30.4 [13.2; 52.9] | 100.0 [99.3; 100.0] | 97.1 [95.3; 98.3] | 100.0 [59.0; 100.0] |
| RdRp Gene | 35.0 [15.4; 59.2] | 100.0 [99.3; 100.0] | 97.6 [96.0; 98.7] | 100.0 [59.0; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [2.5; 100.0] | 98.9 [97.7; 99.6] | 100.0 [99.3; 100.0] | 14.3 [0.4; 57.9] |
| 2 Genes + Ct ≤ 30 | 38.5 [13.9; 68.4] | 99.6 [98.7; 100.0] | 98.5 [97.2; 99.4] | 71.4 [29.0; 96.3] |
| 2 Genes + Ct ≤ 33 | 41.2 [18.4; 67.1] | 100.0 [99.3; 100.0] | 98.2 [96.7; 99.1] | 100.0 [59.0; 100.0] |
| Risk situation | ||||
| 2 Genes | 67.6 [49.5; 82.6] | 100.0 [98.9; 100.0] | 96.9 [94.5; 98.4] | 100.0 [85.2; 100.0] |
| N Gene | 63.9 [46.2; 79.2] | 100.0 [98.9; 100.0] | 96.3 [93.8; 98.0] | 100.0 [85.2; 100.0] |
| RdRp Gene | 67.6 [49.5; 82.6] | 100.0 [98.9; 100.0] | 96.9 [94.5; 98.4] | 100.0 [85.2; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [69.2; 100.0] | 96.5 [94.0; 98.1] | 100.0 [99.0; 100.0] | 43.5 [23.2; 65.5] |
| 2 Genes + Ct ≤ 30 | 81.5 [61.9; 93.7] | 99.7 [98.4; 100.0] | 98.6 [96.7; 99.5] | 95.7 [78.1; 99.9] |
| 2 Genes + Ct ≤ 33 | 71.9 [53.3; 86.3] | 100.0 [98.9; 100.0] | 97.5 [95.3; 98.8] | 100.0 [85.2; 100.0] |
| No risk situation | ||||
| 2 Genes | 55.6 [30.8; 78.5] | 100.0 [98.7; 100.0] | 97.3 [94.8; 98.8] | 100.0 [69.2; 100.0] |
| N Gene | 50.0 [27.2; 72.8] | 100.0 [98.7; 100.0] | 96.7 [94.0; 98.4] | 100.0 [69.2; 100.0] |
| RdRp Gene | 55.6 [30.8; 78.5] | 100.0 [98.7; 100.0] | 97.3 [94.8; 98.8] | 100.0 [69.2; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [29.2; 100.0] | 97.7 [95.4; 99.1] | 100.0 [98.8; 100.0] | 30.0 [6.7; 65.2] |
| 2 Genes + Ct ≤ 30 | 66.7 [34.9; 90.1] | 99.3 [97.6; 99.9] | 98.7 [96.6; 99.6] | 80.0 [44.4; 97.5] |
| 2 Genes + Ct ≤ 33 | 66.7 [38.4; 88.2] | 100.0 [98.8; 100.0] | 98.3 [96.2; 99.5] | 100.0 [69.2; 100.0] |
| Currently asymptomatic AND Risk situation over the previous week | ||||
| 2 Genes | 36.4 [10.9; 69.2] | 100.0 [98.7; 100.0] | 97.6 [95.1; 99.0] | 100.0 [39.8; 100.0] |
| N Gene | 30.8 [9.1; 61.4] | 100.0 [98.7; 100.0] | 96.9 [94.3; 98.6] | 100.0 [39.8; 100.0] |
| RdRp Gene | 36.4 [10.9; 69.2] | 100.0 [98.7; 100.0] | 97.6 [95.1; 99.0] | 100.0 [39.8; 100.0] |
| 2 Genes + Ct ≤ 23 | 100.0 [2.5; 100.0] | 99.0 [97.1; 99.8] | 100.0 [98.7; 100.0] | 25.0 [0.6; 80.6] |
| 2 Genes + Ct ≤ 30 | 44.4 [13.7; 78.8] | 100.0 [98.7; 100.0] | 98.3 [96.1; 99.4] | 100.0 [39.8; 100.0] |
| 2 Genes + Ct ≤ 33 | 40.0 [12.2; 73.8] | 100.0 [98.7; 100.0] | 98.0 [95.6; 99.2] | 100.0 [39.8; 100.0] |
Se: sensitivity - Sp: specificity - NPV: negative predictive value - PPV: positive predictive value.
2 Genes: both genes are detected by RT-PCR regardless the Ct.
N Gene / RdRp Gene: N gene / RdRp gene is detected.
2 Genes + Ct ≤ x: both genes are detected and at least one of them is detected with a Ct ≤ x.
Fig. 1.
RT-PCR Ct of RdRp and N SARS-CoV-2 genes by antigenic test result and current symptoms among patients with positive SARS-CoV-2 RT-PCR (n = 52). Wilcoxon test is used to compare RT-PCR Ct.
Overall AT sensitivity for subjects exposed to a risk situation and sensitivity among asymptomatic exposed to a risk situation were, respectively, 67.6% (95% CI: 49.5 – 82.6) and 36.4% (95% CI:10.9 – 69.2). When considering RT-PCR as positive if both genes were detected, one of which with a Ct ≤ 30 they were respectively 81.5% (95% CI: 61.9 – 93.7) and 44.4% (95% CI: 13.7 – 78.8).
In this cohort of 692 subjects, 52 had a positive RT-PCR, 47 of which were considered likely contagious (i.e. 2 genes detected, one of which with a Ct ≤ 33) [9,10]. Using AT instead of RT-PCR, 19 (36.5%) subjects would have been missed, including 14 considered likely contagious. Among the 20 asymptomatic RT-PCR positive students, 17 were considered likely contagious. Using AT instead of RT-PCR, 13 (65%) subjects would have been missed, including 10 considered likely contagious.
4. DISCUSSION
This study highlights that AT lack sensitivity compared to RT-PCR in the context of SARS-CoV-2 diagnosis and screening. For the asymptomatic sub-group, two thirds of SARS-CoV-2 cases would be considered negative by AT even among the likely contagious ones. However, there was no false-positive in this study thus making AT reliable when positive. False negative in the asymptomatic people might be explained by high Ct, as shown in Fig. 1.
Few studies comparing AT and RT-PCR have been conducted in the general population and in asymptomatic subjects. The Abbot BinaxNOX™ was compared to RT-PCR in a walk-up population: 84% were asymptomatic and 3% had a positive RT-PCR [11]. Sensitivity and specificity were respectively 93.3% (95% CI: 68.1–99.8%) and 99.9% (95% CI: 99.4–99.9%) for a Ct < 30 case definition. Compared to our study, the swab was done in both nostrils for both tests, the population was older and the AT was different. Those differences might partly explain the performance differences. Another study compared Panbio™ SARS-CoV-2 AG Rapid Test Device with RT-PCR for emergency units-referred patients, 72.1% of which were asymptomatic [12]. Sensitivity was 73.3% (95% IC: 62.2–83.8%) which is consistent with our results.
This study is one of the first studies focusing on asymptomatic subjects. The reference test is the one used in the standard of care and both RT-PCR and AT evaluations were done blinded to each other.
As it is a retrospective study, extensive data about subjects’ characteristics such as comorbidities were not available. Included subjects are exclusively healthcare students which is relevant as it is a strategic population in contact with patients but limits the extrapolation of those findings.
StudyCov highlights lack of antigenic tests sensitivity in a student population and in a field setting for the use of AT: it falls far below the WHO performance recommendations of a minimum sensitivity of 80% [3]. It suggests the need for contextualizing AT use and its higher relevance for symptomatic cases. More studies are needed to find out if those findings are replicable in different populations. Meanwhile, this study should help to reevaluate testing policies.
5. OTHER information
StudyCov was conducted by the Bordeaux University Hospital, the Agence régionale de santé Nouvelle-Aquitaine (Regional Health Agency of Nouvelle-Aquitaine) and Bordeaux University without any specific funds. This study received an IRB approval (IRB00003888, 7th December 2020).
6. Transparency declaration
Conflict of interest: Pr. Dehail reports personal fees from Allergan, outside the submitted work. Other authors have nothing to disclose.
Funding
None
Access to data
Patrick Dehail has full access to the data.
CRediT authorship contribution statement
Thomas Ferté: Formal analysis, Writing – original draft, Methodology. Viviane Ramel: Methodology, Writing – original draft, Supervision. Charles Cazanave: Writing – review & editing, Conceptualization. Marie-Edith Lafon: Writing – review & editing, Resources. Cécile Bébéar: Writing – review & editing, Resources. Denis Malvy: Writing – review & editing, Supervision. Agnès Georges-Walryck: Writing – review & editing. Patrick Dehail: Methodology, Writing – original draft, Conceptualization, Project administration.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Elena Charpentier, Moufid Hajjar, Laurent Malato and Rodolphe Thiébaut for their help. We thank the Bordeaux University Health Science College medical students and staff for their technical assistance. In addition, we thank Bordeaux University hospital and the Agence régionale de santé Nouvelle-Aquitaine.
References
- 1.Canada PHA of Interim guidance on the use of rapid antigen detection tests for the identification of SARS-CoV-2 infection. Aem. 2020 https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/guidance-documents/use-rapid-antigen-detection-tests.html (accessed December 9, 2020) [Google Scholar]
- 2.CDC Information for Laboratories about Coronavirus (COVID-19) Cent Dis Control Prev. 2020 https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (accessed December 9, 2020) [Google Scholar]
- 3.World Health Organisation. Antigen-detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays (2020). https://www.who.int/publications-detail-redirect/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (accessed December 9, 2020).
- 4.Autorité de Santé Haute. Revue rapide sur les tests de détection antigénique du virus SARS-CoV-2. 2020:42. [Google Scholar]
- 5.European Center for Disease Prevention and Control, Options for the Use of Rapid Antigen Tests For COVID-19 in the EU/EEA and the UK (2020), Tech Rep n.d.:21.
- 6.Servicio Nacional de Saude. Covid-19: Estratégia Nacional De Testes para SARS-CoV-2 - INSA (2020). http://www.insa.min-saude.pt/covid-19-estrategia-nacional-de-testes-para-sars-cov-2/ (accessed December 30, 2020).
- 7.Ministry of Health Labour and Welfare. Clinical Management of Patients with COVID-19: A guide For Front-Line Healthcare Workers, version 2.1 (2020). https://www.mhlw.go.jp/content/000646531.pdf (accessed December 30, 2020).
- 8.Stevenson M., Nunes T., Sanchez J., Thornton R., Reiczigel J., Robison-Cox J., et al. epiR: an R package for the analysis of epidemiological data. R Package Version. 2013;09-43 [Google Scholar]
- 9.Société Française de Microbiologie. Avis Du 25 Septembre 2020 De La Société Française De Microbiologie (SFM) Relatif à L'interprétation De La Valeur De Ct (Estimation De La Charge virale) Obtenue En Cas De RT-PCR SARS-CoV-2 Positive Sur Les Prélèvements Cliniques Réalisés à Des Fins Diagnostiques Ou De Dépistage Version 4 (2021). https://www.sfm-microbiologie.org/wp-content/uploads/2021/01/Avis-SFM-valeur-Ct-excre%CC%81tion-virale-_-Version-def-14012021_V4.pdf (accessed January 18, 2021).
- 10.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilarowski G., Lebel P., Sunshine S., Liu J., Crawford E., Marquez C., et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. MedRxiv. 2020 doi: 10.1101/2020.11.02.20223891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linares M., Pérez-Tanoira R., Carrero A., Romanyk J., Pérez-García F., Gómez-Herruz P., et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133 doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]