Graphical abstract
Keywords: SARS-CoV-2 mutations, Mutation combination, Viral evolution, Proteome variations, Evolutionary dynamics
Abstract
The knowledge about SARS-CoV-2 proteome variations is important to understand its evolutionary tactics and in drug/vaccine design. An extensive analysis of 125,747 whole proteome reveals 7915 recurring mutations (involving 5146 positions) during December2019-November 2020. Among these, 10 and 51 are highly and moderately recurring mutations respectively. Ever since the pandemic outbreak, ∼50% new proteome variants evolve every month, resulting in 5 major clades. Intriguingly, ∼70% of the variants reported in January 2020 are due to the emergence of new mutations, which sharply declines to ∼ 40% in April 2020 and thenceforth, declines steadily till November 2020(∼10%). An exactly opposite trend is seen for variants evolved with cocktail of existing mutations: the lowest in January 2020(∼20%) and the highest in November 2020(80%). This leads to a steady increase in the average number of mutations per sequence. This indicates that the virus has reached the slow pace to accept new mutations. Instead, it uses a mutation combination strategy for survival.
1. Introduction
The SARS-CoV-2 pandemic is wreaking havoc worldwide by infecting over 87 million people with a mortality rate of more than 1 million as on January 7, 2020 (https://www.worldometers.info/coronavirus/). Although multiple factors such as age [1] weather [2] and health profile [3], [4] etc. play an important role in causing the infection, the evolution in the viral genome/proteome is of yet another major concern. Indeed, independently emerged SARS-CoV-2 variants in UK (VUI 202012/01(B.1.1.7)), South Africa (501Y.V2 (B.1.351)), Brazil (P.1(B.1.1.28.1)) and Uganda (A.23.1) [5] are suggested to be more transmissible compared with the existing variants [6]. SARS-CoV-2, a positive stranded RNA virus (30 kb size), is equipped with 10 open reading frames (ORFs) which are translated into 26 proteins that are vital for its adaptation and survival. Although the virus is equipped with a sophisticated proofreading mechanism, an earlier investigation has identified the existence of 9 and 27 high and moderate significant mutations respectively across the globe until the month of May 2020 [7]. Among these, D614G mutation in spike protein has now conquered the wild type and has proven to enhance the viral fitness and transmissibility [8], [9].
As many western countries have already reached the second wave of SARS-CoV-2 infections and started implementing the second phase of lockdown [10] it is important to understand the viral evolution to aid in the development of successful antiviral therapy for the prevention and the treatment of the infection. To this end, the variations acquired by SARS-CoV-2 proteome have been investigated by analyzing 125,747 [collected by the November 2020 and deposited in GISAID (https://www.gisaid.org) on or before December 10, 2020.] [11] whole proteome obtained from 121 countries (See Methods for details, Table S1). The analysis reveals that SARS-CoV-2 has explored 7915 recurring (occurs in at least 3 out of 125,748 sequences) mutations during December 2019-November 2020. Among them, 10 of them are highly recurring mutations (percentage frequency (PF) more than 10%) and have been explored by SARS-CoV-2 during the pre-lockdown period. By using these mutations, SARS-CoV-2 has evolved into 5 clades and their 6 major subclades during December 2019-November 2020. Further, 51 mutations recur at a moderate percentage (between 1 and 10%) while the rest of them occur below 1%. Interestingly, the evolution of SARS-CoV-2 proteome with new mutations is in declining phase after March 2020, whereas, the proteome with cocktail of existing mutations (henceforth, variants) is in growing phase after March 2020. This indicates that the virus would already have explored majority of the favorable mutations and thus, continues its evolution using cocktail of exiting mutations.
2. Results
To analyze the viral evolution strategy, the mutations acquired by the SARS-CoV-2 proteome during December 2019 to November 2020 have been analyzed and are discussed below.
2.1. Pre-lockdown emergence of 10 highly recurring SARS-CoV-2 mutations
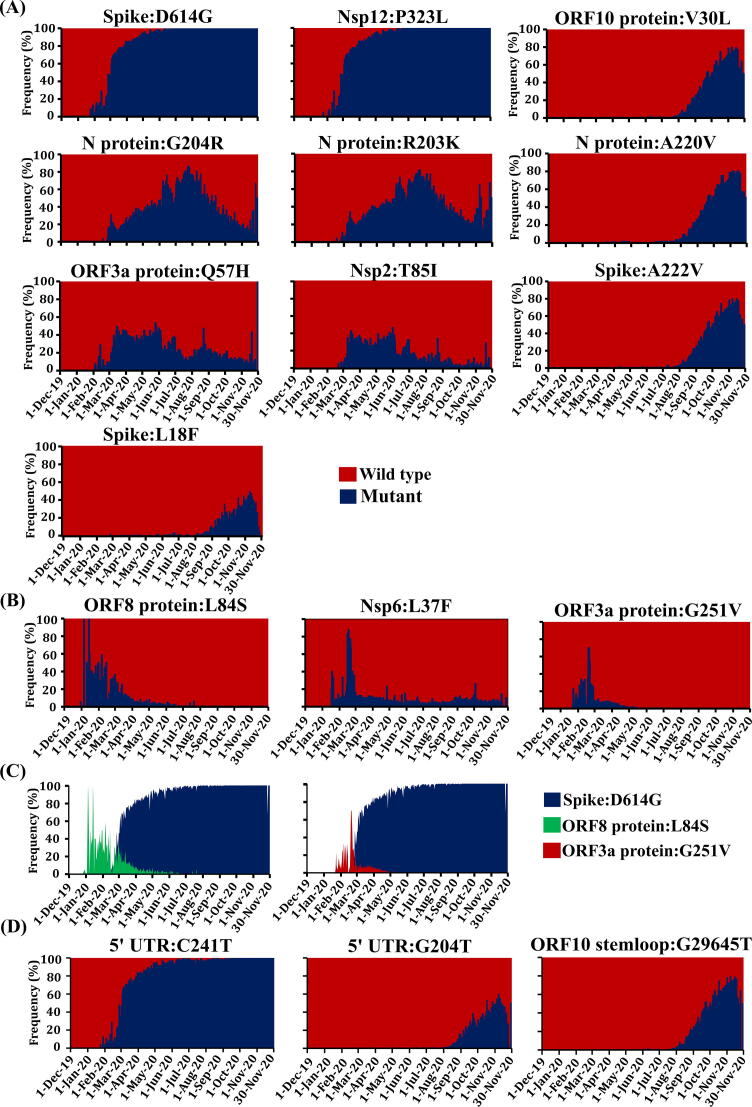
The analysis of 125,747 proteome has revealed that until the month of November 2020, 7326 amino acid positions of 26 SARS-CoV-2 proteins have undergone changes for 15,369 times (Table S2). Among these, 7915 mutations are found to occur in more than 3 sequences (viz., percentage frequency above 0.002%). Spike:D614G, Nsp12:P323L, N protein:R203K-G204R, ORF3aprotein:Q57H, Nsp2:T85I, Spike:A222V, ORF10protein:V30L, Nprotein:A220V and Spike:L18F are the 10 highly recurring (HR) mutations whose overall percentage frequency (PF) is greater than 10% during December 2019 to November 2020. Among these, the spike protein D614G mutation, which has emerged in early January 2020, has surged to nearly 100% in the month November 2020 (Fig. 1A). Nsp12:P323L, which occurs most of the time along with Spike: D614G, has also attained ∼ 100% PF during November 2020 (Fig. 1A, Fig. S1). Intriguingly, the virus has tried this combination in China during the very early stage of pandemic (Fig. 2, Table S3) (GISAID ID: EPI_ISL_422425, January 2020, Table S4). Subsequently, this combination is seen in more than 60% of the SARS-CoV-2 sequences that are collected until March 2020 (Fig. 1A) [8]. However, Germany has reported the first independent occurrence of D614G during early January (GISAID ID: EPI_ISL_406862, Table S4). Followed by these, the N protein co-mutations (N protein: R203K-G204R) occur with a PF of 33% since its first emergence in UK in February (Fig. 1 A, Fig. 2) (GISAID ID: EPI_ISL_466615). Similarly, ORF3a protein:Q57H and Nsp2:T85I that occur for the first time in Saudi Arabia (EPI_ISL_489996) and France (EPI_ISL_418218) respectively in the month of February (Fig. 2, Table S3). Although they occur greater than 10% during the entire pandemic period, their occurrence has reduced significantly after June 2020.
Fig. 1.
Month wise occurrence of key recurrent SARS-CoV-2 mutations. (A) Month wise occurrence of top 10 recurrent SARS-CoV-2 mutations. (A: Last column) Emergence of new highly recurrent (HR) mutations that occur after August 2020. Note that the emergence of N protein:A220V, ORF10 protein:V30L, Spike:A222V and Spike:L18F is seen only after July 2020. (B) Decrease in the percentage frequency of some of the mutations which were highly recurring before March 2020. Note that the occurrence of ORF8 protein:L84S and ORF3a protein:G251V is completely reduced after May 2020 and the occurrence of NSP6:L37F is reduced from HR to moderately recurrent (MR) after March 2020. (C) Diagram illustrating the rise (before Spike:D614G becomes dominant) and fall (after Spike:D614G takes over the wild type) of ORF8 protein:L84S and ORF3a protein:G251V. (D) Emergence of new mutations in non-coding region that occur at significantly high recurrence (HR). Note that 5′UTR:C241T mutation started occurring after Feb 2020 and 5′UTR:G204T and ORF10 stemloop:G29645T mutations started occurring after August 2020.
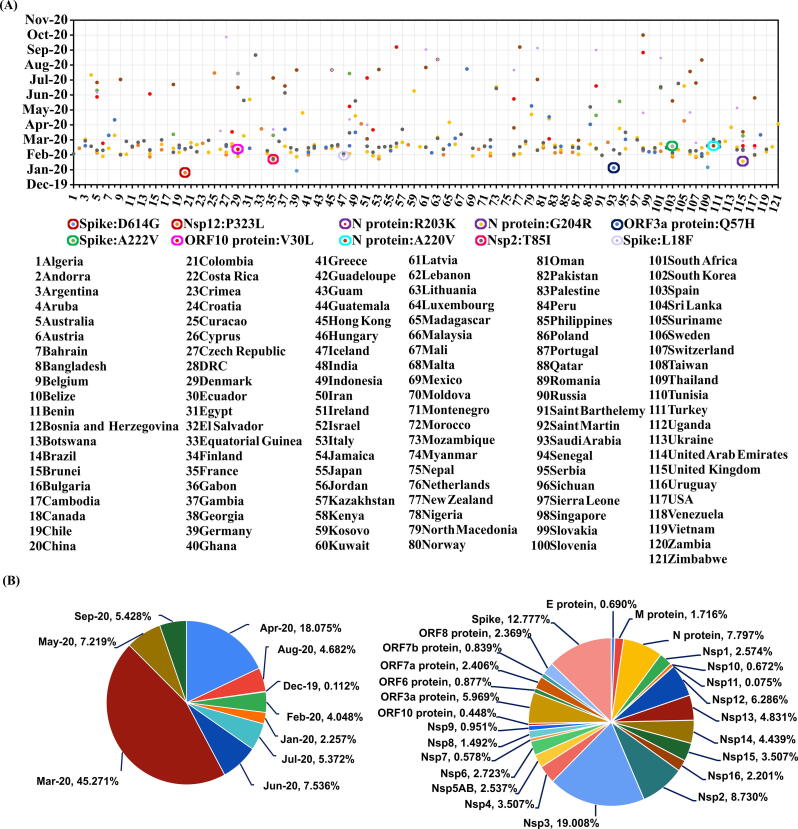
Fig. 2.
Diagram illustrating (A) the date of first incidence corresponding to 10 highly recurrent mutations in 121 countries considered in the current investigation and (B) persistence of recurrent mutations. A) The countries are number coded (refer numbers below the figure) and are indicated in the X–axis. The circles indicate the country that acquires any of the 10 mutations (differentiated by colors) before the other 121 countries. (B) Pie chart elucidating (Left) month wise and (Right) protein wise persistent mutation.
Unlike the above mutations, which occur (with the overall PF) above 10% even during pre-lockdown period, the hydrophobic trio mutations N protein:A220V, ORF10 protein: V30L and Spike: A222V have picked up only from the month of August 2020 (Fig. 1A) even though they are first seen in Tunisia in March 2020 along with Spike:D614G and Nsp12:P323L (EPI_ISL_683329). They have started spreading widely in Europe from the month of August 2020 with the PF of 11% and increased steeply and has attained the percentage frequency of 70% in November (Fig. 1 (A)). Note that their individual appearances are also seen in Europe: N protein:A220V in Portugal (EPI_ISL_511521), ORF10 protein:V30L in Denmark (EPI_ISL_444822) and Spike:A222V in Spain (EPI_ISL_467173). Similarly, Spike: L18F occurs with a high recurrence after September 2020 (with an overall PF of 12%) (Fig. 1A), although it has appeared for the first time in March 2020 in Iceland (EPI_ISL_417765).
In contrast to above, Nsp6:L37F, which occurs with a high recurrence during the early stage of the pandemic, sustains at moderate recurrence after March 2020 (Fig. 1B). A similar trend is observed for ORF8protein:L84S and ORF3a protein: G251V, which have a high recurrence before March 2020, but has subsequently reached the declining phase (Fig. 1B). Surprisingly, the surging time period of Spike:D614G and Nsp12:P323L mutation overlaps with ORF8protein:L84S and ORF3aprotein:G251V diminishing period (Fig. 1C). Strikingly, ORF8protein:L84S is one of the two characteristics mutations of the lineage A. Even though ORF8protein:L84S (GISAID ID: EPI_ISL_412982) and ORF3a protein:G251V (EPI_ISL_447919) have occurred along with Spike:D614G and Nsp12:P323L during the month of February 2020, they are rarely seen together. This suggestive of evolutionarily disadvantage of Spike:D614G and Nsp12:P323L occurring in lineage A or vice-a-versa and ORF3a protein:G251V of lineage B occurring together with Spike:D614G and Nsp12:P323L.
Fig. 2 summarizes the pre-lockdown (https://www.bbc.com/news/world-52103747) emergence, dissemination and surge of the 10 highly recurring mutations of SARS-CoV-2 proteome.
2.2. Highly recurring mutations in the non-coding regions
The changes in the SARS-CoV-2 genome hashave also been examined which indicate that 19,270 nucleotide positions have undergone changes for 27,200 times during the evolution. More specifically, the 6 non-coding regions of SARS-CoV-2 genome, namely, 5′UTR, ORF1ab stemloop 1 &2, ORF10 stemloop 1&2 and 3′UTR have exhibited 1,124 variations (Table S5). Out of those, 632 mutations are occurring at least thrice (PF above 0.002). Notably, C241T (PF = 92%) of 5′UTR mostly occurs along with Spike:D614G and Nsp12:P323L (Fig. 1A & D) although it has been found to occur independently in some incidences. The three mutations are first seen together in China in January 2020 (EPI_ISL_451345). Other highly recurring mutations in the non-coding regions are G204T in 5′ UTR and G29645T in ORF10 stem loop 2 (which is an equivalent of ORF10:V30L), which are on the significant rise from August 2020 with the overall PF of 14% and 23.7% respectively (Fig. 1D).
2.3. Emergence of moderately recurring 51 SARS-CoV-2 proteome mutations and 4 non-coding region mutations
In addition to the above-discussed proteome mutations, 51 variations (including substitutions and deletions, Fig. S2) are found to occur with a moderate percentage frequency (between 1 and 10%) (Table S2). Interestingly, 45 of them have the first incidence before March 2020 (Fig. S2, Table S3), while the remaining 6 mutations (N protein: A376T, Nsp13:K218R, Nsp12:E254D, Nsp12:A656S; Nsp9:M101I and Nsp12:V720I) have emerged after April 2020. Among these, 13, 33 and 5 are present in the structural, non-structural and accessory proteins respectively.
It is noteworthy that following moderately recurring mutations co-occur with the overall PF more than 3% (Table S2, Table S6): Nsp12:V720I & Nsp9:M101I and Nsp12:A185S, Nsp12:V776L, N protein:A376T & Nsp13:K218R. Among them, N protein:S194L which has emerged in China in December 2020 (EPI_ISL_406594) appears steadily after April 2020 and has the overall PF of 5.66% (the highest among the moderately recurring mutations). Markedly, Nsp13:Y541C, Nsp13:P504L, ORF8 protein: L84S and ORF3a protein: G251V have vanished after August 2020 (PF = 0% after August 2020). Other notable deletions are Spike: H69-; Spike: V70-.
Besides, 7854 recurrent mutations (492 mutations are occurring with PF (0.1–1%)) have been observed until November 2020 (PF in the range of 0.002 to 1) (Table S2). Among these, Spike:E583D, Spike:P272L, Nsp3:T1189I, Nsp2:A318V; Nsp5AB:K90R; Spike:S98F; ORF3aprotein:V202L; ORF8protein:A65V; Nsp15:K12R; N protein:H145Y; Spike:S256L; Nsp3:H295Y; Spike:D215H; ORF3aprotein:Q38R; Nsp6:V149F; ORF3aprotein:G172R; Spike:D1163Y; N protein:P13T; Spike:A688V; Spike:G1167V; N protein:Q9H; Nsp15:T114A; Nsp16:R216C; ORF3aprotein:G172V; ORF3aprotein:V163I; Nsp3:S126L; Nsp12:T76I and Nsp14:N129D have started occurring with a moderate recurrence from September or October or November 2020 onwards. Notably, 8 of them are the Spike protein mutations.
In the non-coding region, C13536T in ORF1ab stemloop 2 (PF = 1.58), G29734C (PF = 2.2) and A29771G (PF = 1.8) in 3′UTR and C66T in 5′UTR (PF = 1.76) are moderately recurring mutations. Among these, the 3′UTR mutations are steadily rising from the month of August 2020.
2.4. Lineage a specific mutations
It is well known that lineage A and lineage B differ from each other with respect to ORF8 protein:L84S and ORF1ab:C8782T (silent nucleotide mutation) mutations [12]. Interestingly, none of the lineage A mutations are found either in high or moderate percentage recurrence. Nevertheless, 42 mutations (PF in the range of 0.38–0.002%) are found to be specific for lineage A (Table S7 and Table S4), viz., they occur very rarely in lineage B.
2.5. Mutation persistence
The mutation persistence analyses (see Methods) indicate that among the 15,369 mutations, 5,361 occur persistently during the time period considered here. Among them, 63% have emerged in the month of March 2020 and April 2020 (Fig. 2B (Left)). This supports the early emergence of not only the majority of high and moderate significant mutations, but also, the majority of low significant mutations. To our surprise, Nsp3 owns the highest persistent mutations (19%) followed by the spike protein (12.8%) (Fig. 2B (Right)). The other proteins that have more than 5% of the persistent mutations are: Nsp2 > N protein > Nsp12 > ORF3a protein. Six proteins have persistent mutations less than 1%: ORF6 protein > ORF7b protein > E protein≈ Nsp10 > Nsp7 > ORF10 protein.
2.6. Post-lockdown diminish in the emergence of new mutations and escalation in the cocktail of existing mutations
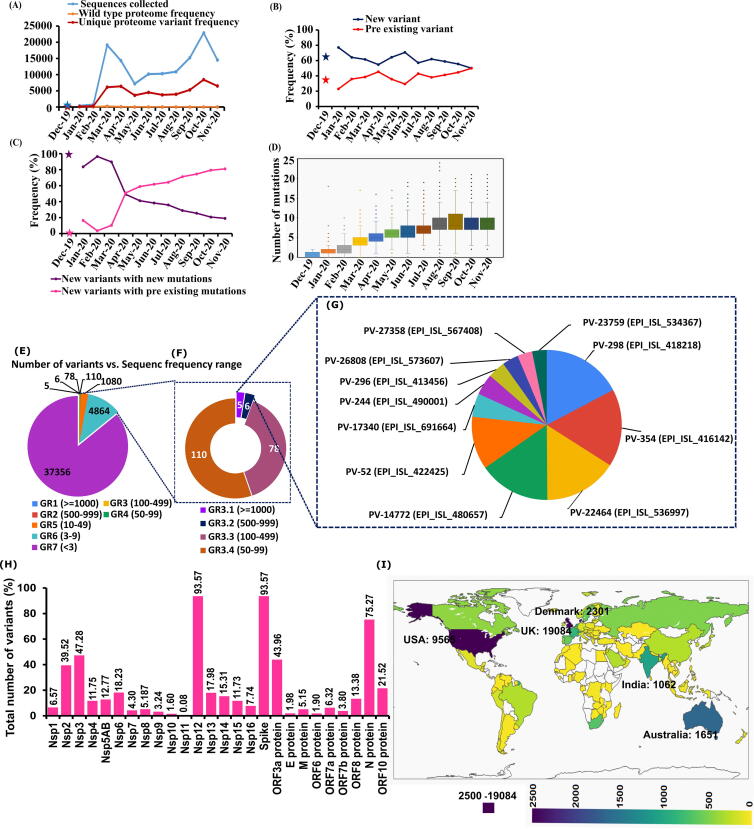
Comparative SARS-CoV-2 whole proteome analysis reports that the number of proteome variants per month follows the same trend as the number of sequences collected in a particular month. March 2020, April 2020 and November 2020 are the top 3 months having the more number of sequences with cocktail of mutations (viz., variants) (Fig. 3A, Table S8). Out of the total number of variants seen in every month, more than 50% of them are unique to a particular month (Fig. 3A, 3B, Table S8). The remaining variants are identical to the proteome variants reported in the previous month(s) (Fig. 3B). Detailed analysis indicates that the new proteome variants differ from the existing proteomes in one or more amino acid positions. Surprisingly, segregation (Fig. 3C) of the frequency of occurrence of new proteome variants into proteome with a newly emerged mutation(s) and with the cocktail of existing mutations indicates that during the month of February 2020 the evolution of new mutations is in the highest frequency (80%) which sharply declines after April 2020 (50%). Subsequently, it has entered into a steady declining phase and has reached a frequency of 20% in the month of November 2020. An exactly opposite behavior has been seen in the emergence of variants with the cocktail of existing mutations: the lowest in January 2020 (∼20%) and a sharp surge till April 2020 (60%) which is followed by a steady growth every month (∼80%) (Fig. 3C). As the result, there is a steady increase in the average number of mutations per sequence, which is nearly 0 in December 2019 and 9 in November 2020 (Fig. 3D). Indeed, a proteome with the bevy of 24 mutations are found in the month of August 2020 indicating the flexibility in the viral proteome to accommodate a greater number of mutations (EPI_ISI 570600, USA). Thus, it indicates that after examining the suitable mutations during the early stage of pandemic, the virus now tests their cocktail to continue its survival (Fig. 2, Fig. 3, Table S2, Table S6, and Table S8).
Fig. 3.
(A–D) Month wise summary of SARS-CoV-2 proteome variant distribution (Dec 2019 to Nov-2020) and (E-H) variant persistence. (A) Line plot illustrating the distribution of number of unique proteome variants (deep red color) from the total number of sequences collected (blue color) in that particular month. (B) Month wise sequence count (percentage frequency (%)) of appearance of new variants (colored dark blue) and preexisting variants (colored red). (C) Line plot illustrating the occurrence of new mutations in newly collected sequences (purple color) and without the occurrence of new mutations in newly collected sequences (pink color). (D) Box plot elucidating month wise occurrence of single amino acid mutations in SARS-CoV-2 sequences (Dec-2019 to Nov-2020). Note that the mutation average linearly increases every month in SARS-CoV-2 proteome. Due to the availability of less number of sequences in December 2019, it is indicated by “*”. (E-G) Relative percentage of occurrence of variants that are found in at least 3 (E), 50 (F) and 500 (G) sequences. (H) Pie chart representing the protein wise variant distribution. (I) Country wise distribution of SARS-CoV2 variants. Note that the variant counts have been indicated for top 5 countries. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.7. Distribution and persistence of SARS-CoV-2 proteome variants
A total of 43,499 proteome variants have been tried by the SARS-CoV-2 until November 2020 (Fig. 3E) and UK has the highest number of variants with cocktail of mutations (19084, Fig. 3I). However, only 197 variants appear at least in 50 sequences (Fig. 3E, 3F). Among them, 5, 6, 78 and 100 variants are seen at least in 1000 (Fig. 3F, 3G), 500 to 999 (Fig. 3F, 3G), 100 to 499 and 50 to 99 sequences respectively (Fig. 3F). Interestingly, only 760 variants persist during the months of October 2020 and November 2020 (Fig. S3). Not surprisingly, unlike the case of individual mutations (Fig. 2B (Left)), the variants that have emerged in August 2020 (549 sequence) and September 2020 (131 sequences) persist during October 2020 and November 2020 compared with the variants that have emerged in the previous months. The Spike, Nsp12 and N protein have contributed for above 10% of the variants (Fig. 3H). Nsp10 and E protein have taken the last 2 places in terms of variant contribution.
2.8. Suppression of clade 1 and clade 2 by spike-D614G and Nsp12- P323L
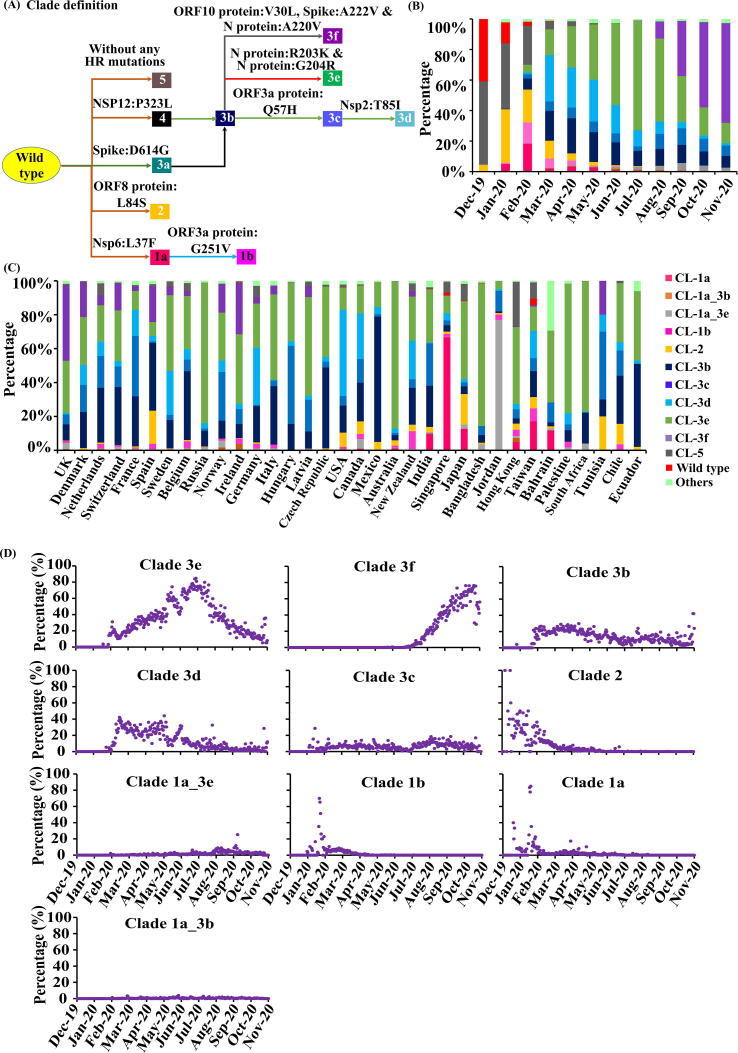
Throughout the progression of the pandemic, there have been a lot of dynamics in the proteome of the SARS-CoV-2. To understand the dynamics in the SARS-CoV-2 proteome, phylogenetic trees have been constructed using the SARS-CoV-2 proteome. Prior to the phylogenetic tree construction, a multiple sequence alignment has been carried out (as mentioned in the Methods section) and subjected to phyloproteome construction. Since phyloproteome construction depends on the probability of the amino acid(s) mutation, it will reflect in the clustering of sequences with similar mutation(s) into a new clade/sub-clade. The results indicate the dominance of wild-type proteome in December 2019, which declines drastically to 2.8% in February 2020, followed by 0% occurrence in the following months (Fig. S4). Not surprisingly, the dominant mutations reported above have led to 5 major clades as reported earlier [7]. For instance, due to D614G (highly recurrent) mutation, there is an emergence of a new clade (indicated in Fig. 4A as Spike:D614G). Similarly, the amino acid(s) responsible for the emergence of a new proteome clade or sub-clade has been indicated in Fig. 4A. Nonetheless, soon after the surge of Spike:D614G (clade-3a) and Nsp12:P323L (clade-4) (Fig. 4A & B), 5 major sub-clades have evolved during May 2020 (Fig. 4B & Fig. S5B): clade-3b (Spike:D614G and Nsp12:P323L), clade-3c (Spike:D614G, Nsp12:P323L, ORF3aprotein:Q57H), clade-3d (Spike:D614G, Nsp12:P323L, ORF3aprotein:Q57H, Nsp2:T85I) and clade-3e (Spike:D614G, Nsp12:P323L, N protein:R203K and N protein:G204R. Yet another branching in clade-3b takes place in the month of August 2020 due to the emergence of N protein:A220V, ORF10protein:V30L and Spike:A222V trio mutations (clade-3f). Consequently, merging of these clades/sub-clades (clades 3c, 3d, 3e, 3f and 1a) takes place as the virus evolves. Fig. 4B & Fig. S5 shows the month wise merging of these clades as Spike:D614G (clade-3a) and Nsp12:P323L dominate the world (Fig. 4B, Fig. S5 & Fig. 1A) (Table S9). It is noteworthy that the clade-5 doesn’t have any high recurrent mutations. Intriguingly, the clades 1b (Nsp6:L37F and ORF3aprotein:G251V) and 2 (ORF8protein:L84S) vanishes as ORF3a protein:G251V and ORF8 protein:L84S declines steeply along with the rise of Spike:D614G and Nsp12:P323L (Fig. 4B, Table S9) (Fig. 1A, 1B & 1C). However, the merging of clade 2 (ORF8protein:L84S) and clade 1b (ORF3aprotein:G251V) with clade 3 (Spike:D614G) is seen to occur very rarely, suggesting the fitness disadvantage for the virus.
Fig. 4.
SARS-CoV-2 proteome clades dissemination. (A) SARS-CoV-2 proteome clade definition based on highly recurrent (HR) mutations. (B) Month wise and (C) country wise distribution of top 10 clades of SARS-CoV-2 and their mergers. (B) Note that CL-3e (light green color) is majorly distributed after March 2020 to August 2020, after that it started reducing and CL-3f (violet color) is majorly distributed after August 2020. (C) The CL-3f (violet color) is majorly distributed in European countries and CL-3e (light green color) is distributed in almost all the countries. (D) Month wise percentage occurrence of top 10 clades. Note the steady increase in clade 3f from August 2020. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Further analysis indicates that some clades are found only in certain countries, as given in the Fig. 4C & Fig. S5 (Table S10). For instance, the clade 3e and 3f are majorly found among the European countries. Similarly, the following dominance is found in certain countries (overall months) clade-3d in USA, clade-1a in Singapore, clade-3e in Australia (92%), clade 1a-3e (merger of clade 1a and 3e) in Jordan and clade 3b in Mexico. Interestingly, the Spike:D614G clade or the mergers are found in less percentage in China which can be attributed to a smaller number of sequences deposited after March 2020 (after which the Spike:D614G has slowly overtaken the wild-type). Emergence of highly significant trio mutations (N protein:A220V, ORF10protein:V30L and Spike:A222V) in Europe and their merging with Spike:D614G and Nsp12:P323L (clade-3b) has led to a new subclade 3f. Our study further highlights that the origin of clade 3f (one of the widely spread proteome variants in Europe and confinement to European nations) is Tunisia, Africa (EPI_ISL_683329, March 2020) which is so far believed to be originated from Spain (July 2020) [13].
2.9. Fast evolution of SARS-CoV-2 variants through acquiring cocktail of existing mutations
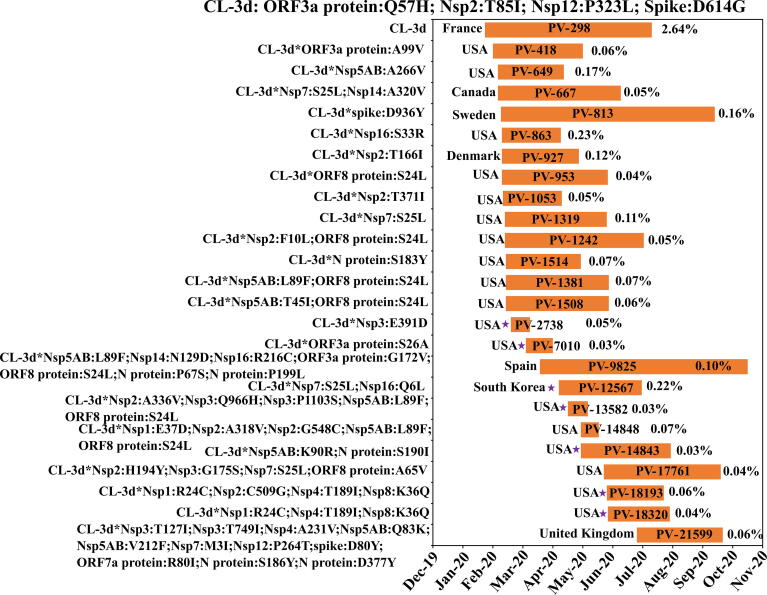
Among the 43,499 unique (proteome) variants (alternatively, 62,957 genome variants) tried by SARS-CoV-2, only 6143 (alternatively, 6914 genome variant) occurs at least 3 times, among which, the top 200 proteome variants occur with the percentage frequency above 0.04% (viz., found in 49 proteome sequences) (Figs. S6–S14, Table S4 & Table S6). Alternatively, top 200 genome variants are found above 0.03% percentage frequency (viz., found in 37 proteome sequences). Further, only 68 variants occur at a PF above 0.1%, among which, a variant of clade-3d (Nsp2:T85I, Nsp12:P323L, Spike: D614G, ORF3aprotein:Q57H), clade 3e (Nsp12:P323L, spike:D614G, N protein:R203K; N protein:G204R), clade 3f (Nsp12:P323L; spike:L18F; spike:A222V; spike:D614G; N protein:A220V; ORF 10 protein:V30L) clade 3e (Nsp2:I120F, Nsp12:P323L, spike:S477N, spike:D614G, N protein:R203K, Nprotein:G204R) and clade 3b (Nsp12:P323L, spike:D614G) appear at a moderate recurrence (PF > 1) (Fig. 5, Figs. S9, S11 & S12). These clearly indicate that the virus keeps evolving by trying different mutation combinations. Among the remaining variants, only 9 recur above 0.5%. The evolutionary dynamics analysis of the top 3 variants further reveals the emergence of various sub-clades with the inclusion of different mutations (Fig. 5, Figs. S8, S9, S10, S11 & S12). Interestingly, some of the combinations are localized to a particular country. One such example is the acquisition of additional Nsp16:Q6L and Nsp7:S27L mutations in clade 3d which is found to be specific to South Korea (PF = 0.22%) (Fig. 5). Similarly, an inclusion of Nsp2:I120F and Spike:S477N mutations into the clade-3e (Nsp12:P323L, Spike:D614G, N protein:R203K, N protein:G204R which recurs at PF = 2.56%) leads to a sub-clade (PF = 2.36%) which is specific to Australia (Fig. S11).
Fig. 5.
Month wise evolutionary dynamics of clade 3d (CL-3d) showing the divergence of the sub-clades from its ancestors. From the month (X-axis) of first emergence to the month of last appearance of a sub-clade in a particular country is represented by a bar. The variations in the amino acid sequence are given in the Y-axis. The country of first incidence is mentioned alongside the bar. The magenta colored star indicates that the sub-clade is country specific. PV number indicates the protein variant number of the particular sub-clade (see text for details). Note that only top 25 sub-clades are shown here. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.10. Emergence of 3e and 3d sub-clade variants with cocktail of mutations and with increased transmissibility
As the world has again been thrown into a chaos in December 2020 because of the independent emergence of two variants in UK (VUI 202012/01, B.1.1.7), and South Africa (501Y.V2, B.1.351) with an increased transmissibility [14], [15]. Thus, a closer inspection has been made on these variants despite their low recurrence in the month of November 2020 (the timeline considered here for the analyses). As noted earlier [16] both the sequences have mutations in more than 10 amino acid positions and majorly in spike protein (Figs. S14 & S15). While the VUI 202012/01 (EPI_ISL_601443, September 20, 2020) has the highly recurring Spike-D614G, Nsp12:P323L, N protein:G204R and N protein: R203K, the 501Y.V2 (EPI_ISL_712081, September 8, 2020) has the highly recurring Spike:D614G, Nsp12:P323L, Nsp2:T85I and ORF3a protein:Q57H mutations. Thus, the former falls in the clade 3e and the latter falls in clade 3d. The key mutation that is common to both the variants is N501Y and is found only in the low recurrence until November 2020 (Figs. S14 & S15). Strikingly, the virus has tried these mutation combinations in the month of September 2020 itself in the respective countries, but their role in increased transmissibility was unnoticed at that time. The data collected as on January 20, 2021 shows that the former and the latter have disseminated across 35 and 10 countries respectively (Table S11). Thus, it is clear that these mutation combinations are evolutionarily advantageous.
2.11. Mutations in the SARS-CoV-2 vital motifs
To explore the impact of mutations in viral pathogenicity, survival and host interaction, the presence of mutations in 22 key functional motifs of SARS-CoV-2 that are reported earlier have been analyzed [17], [18], [19], [20], [21], [22]. Only a few mutations, which occur in a low to moderate percentage frequency, are seen in these motifs or in their vicinity (Figs. S16 & S17, Table S12). For incidence, among the N protein: S194L, N protein:S183Y and N protein:S188L mutations in SR motif , N protein: S194L is the only moderately recurring mutation. Similarly, moderately recurring H69- and V70- deletions in spike protein are seen the vicinity of GTNGTKR motif, which can jeopardize the ability of spike protein to interact with the receptors other than ACE2 [17]. The other motifs are observed to be highly conserved (except the presence of a few low recurring mutations, Figs. S16-S18) indicating their importance in viral survival.
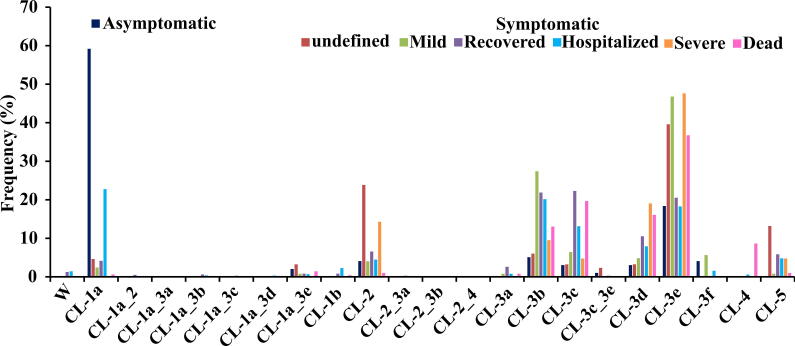
2.12. Dominance of clade 1a in asymptomatic and hospitalized patients
Analysis of the limitedly available metadata (13,343 sequences, viz., 1% total sequences considered) indicates that the clade 1a (consists of Nsp6:L37F) is dominant in asymptomatic and hospitalized patients [23] (Fig. 6, Table S13). Not surprisingly, subclades of D614G mutation are prevalent in symptomatic (with patient status is unknown), mild, severe and deceased cases as D614G has taken over the wild type.
Fig. 6.
Histogram plot illustrating the distribution of the clades in SARS-CoV-2 patients. Note that the clade distribution is shown for asymptomatic, symptomatic (undefined), mild, recovered, hospitalized, severe and dead patients. Note that the CL-1a is highly distributed in asymptomatic patients and CL-1a, CL-2, CL-3b, CL-3c, CL-3d and CL-3e clades are distributed in all kind of patients.
Further analysis of metadata to derive the relationship between the age and gender of the patients does not show any strong correlation (Fig. S20, Table S4). However, during post-lockdown, the rate of female patients infected with SARS-CoV-2 has become nearly equivalent to the male patients.
3. Discussion
Complete global proteome/genome analysis of SARS-CoV-2 would help in tracking the mutations and their implications in host immune evasion mechanisms, infectivity and mortality as well as in assessing the effectiveness of the vaccines and drugs. An earlier investigation from this laboratory [7] has reported that out of 2116 recurring mutations in the SARS-CoV-2 proteome, 7 are highly represented during the early stage of pandemic. Here, 125,747 complete SARS-CoV-2 proteome sequences collected worldwide have been analyzed to understand their spread, sustainability and their implication in viral pathogenicity and survival.
The mutation demographics indicate that only 0.14% of the total sequences are wild-type (viz., the reference sequence published in January 2020 [24], although it has disseminated across the world since its first occurrence in China in December 2019 before the implementation of the international travel ban. In fact, only 20 countries have reported the wild-type sequence and its last appearance has been seen in India in August 2020 (EPI_ISL_581502). This clearly indicates the high dynamics in the SARS-CoV-2 proteome. Although SARS-CoV-2 has equipped with a sophisticated RNA proofreading mechanism, overall 15,369 unique amino acids mutations (7326 positions) and 43,499 unique proteome variants (viz., sequences with cocktail of mutations and are different from the reference sequence) are found during December 2019-November 2020. Nonetheless, only 7915 are recurring (recurring at least in 3 sequences with a percentage frequency of at least 0.002%) mutations involving 5146 positions suggesting that they have a better evolutionary advantage compared with the rest.
Intriguingly, among the 7915 recurring mutations, 10 of them are highly recurring with the overall percentage frequency above 10 (Fig. 1A). These mutations have their first incidences during the pre-lockdown period (Fig. 2). While 6 of them are highly recurring even from the pre-lockdown period (Spike:D614G, Nsp12:P323L, N protein:R203K:G204R, ORF3aprotein:Q57H, Nsp2:T85I), some of them have picked up during the post-lockdown period indicating that they are evolutionarily advantageous. Additionally, 51 mutations are found to recur moderately (Fig. S2), among which 45 and 6 have evolved during the pre- and post- lockdown periods respectively and sustain continuously. Among the rest of 7854 less recurring mutations, 44% of them (3494) have emerged during the pre-lockdown (till April) period. Thus, the virus has identified that 3973 out of 5146 (positions of recurring mutations) amino acid positions (77% of the proteome) as evolutionarily more advantageous than the others, as they sustain with either high or moderate recurrence mutations. Further, the persistence of mutations (See Methods) indicates that most of the mutations that have emerged in the month of March 2020 are highly persistent.
Strikingly, ORF3a protein:G251V (lineage B) that is having a high recurrence during pre-lockdown period has lost in the evolutionary race against Spike:D614G and Nsp12:P323L (lineage B) and their combination is evolutionarily disadvantageous. Interestingly, ORF8 protein:L84S (lineage A) also in declining phase with the surge of Spike:D614G and Nsp12:P323L. Interestingly, only a rare occurrence of Spike:D614G and Nsp12:P323L is seen in lineage A indicating their unfavorable coexistence. In support of this, the recent emergence of new variant in Uganda which possesses ORF8 protein:L84S (lineage A.23.1), lacks Spike:D614G mutation [5]. Very interestingly, 42 mutations are found to be specific to lineage A.
With the existing mutations that are (majority) acquired quite early, the virus follows the tactics of having cocktail of mutations rather than evolving new mutations (Fig. 3C). This reflects in the average number of mutations per sequence which is steadily increasing every month (Fig. 3D). Further, the variants that have emerged in the month of August 2020 and September 2020 are more persistent (Fig. S3). The most successful merging of mutations is the Spike:D614G (clade-3a) and Nsp12:P323L (clade-4) (along with 5′UTR:C241T), leading to a subclade 3b. Another notable example in line with this is the recently emerged highly transmissible variants in UK (clade-3e) and South Africa (clade-3d) with the cocktail of mutations. Interestingly, although L18F (N-terminal domain of spike) is seen quite early when it has occurred in clade 1a (EPI_ISL_417765, Iceland), its dissemination (found in 22 countries) has gained momentum only after July when the clade has started occurring along with clade 3b. Similarly, merging of clades 1a and 3e (clade 1a_3e), clades 1a and 3f (1a_3f) and clade 3f with trio mutation combinations (N protein:A220V, ORF10protein:V30L (synonymous mutation in ORF10 stemloop G29645T) and Spike:A222V has evolutionary advantages. Such a merging of highly recurring mutations has led to 5 major SARS-CoV-2 whole proteome clads (Fig. 4). Overall, the cocktail of mutations (highly and/or moderately and/or low recurring) in the whole proteome has led to increase in the number of lineages every month. Thus, highest number of unique variants (7561) have appeared in October 2020. From the sequences collected in the month of November 2020 that have been deposited by 10-December-2020, 5140 unique proteome variants have been identified. Further, the early emergence of not only the majority of high and moderate recurring mutations, but also, the majority of low recurring mutations and their persistence (Fig. 2B (Left)) may still lead to several new cocktails of existing mutations. It is possible that some of the cocktails may have more infectivity etc.
Variant persistent analysis indicates that 199 variants occur in at least 50 sequences. Nonetheless, there are 11 variants that are found to occur in more than 500 sequences indicating that these variants may be considered in the perspective of therapeutics and diagnostics. Interestingly, the protein wise persistence analyses (Fig. 2B and 3H) indicate that E protein may be more suitable for drug and vaccine development and can also be used for diagnosis as it has a low relative persistence for mutation as well as for variants.
The whole proteomic analysis of SARS-CoV-2 carried out here indicates that the virus continues to evolve by following the tactics of using the cocktail of beneficial mutations which have been mostly picked up during the early stage of pandemic. This results in the evolution of many divergent lineages under 5 major clades. Nonetheless, very few mutation combinations have turned out to be evolutionarily advantageous and have been sustained and disseminated across several countries. The outcome of this study may slightly differ since 64% of the total number of sequences used in the current investigation are from UK (47%) and USA (17%) , ∼19% of the sequences are contributed by six countries (Australia (6.7%), Denmark (6.4%), Netherlands (1.6%), Switzerland (1.5%), France (1.3%) and India (1.3%)) (Table S1) and the remaining 17% are contributed by 113 countries. As the virus keeps on examining the cocktail of mutations that can be evolutionarily favorable, more variants may keep emerging through the merging of different clades or through the inclusion of existing mutations.
4. Materials and methods
4.1. Data collection and creation of a local database
In order to analyze SARS-CoV-2 proteome collected across the world, 180,385 complete high coverage sequences (viz., sequences having less than 1% of undefined nucleotides) that were deposited on or before 10-Dec-2020 and are sequenced on or before 30-Nov-2020 were downloaded from the GISAID server (https://www.gisaid.org/). The sequence dataset was further filtered to discard the sequences with undefined nucleotides (“N”) in the coding region. The first published SARS-CoV-2 genome sequence (Genbank ID: NC_045512.2) was used as the reference sequence for both the amino acid and nucleotide mutation analyses. The sequences with nucleotide insertions compared with the wild-type sequence were separated out from the dataset. Thus, 125,747 genomic sequences with no insertions were further analyzed for any mutations in the 26 coding regions [25]. Subsequently, the coding regions of 125,747 sequences were translated into individual proteins using the in-house scripts and were subjected to mutational analysis. The in-house bash and python scripts were written to translate the SARS-CoV-2 genome into proteome and create a local repository that contains month wise and country wise SARS-CoV-2 proteome. The scripts were also used in the analysis of amino acid variations in an automated fashion. Further, these sequences were segregated based on the source country and the month of collection. Additionally, the mutations in the non-coding regions were analyzed without translation.
4.2. Metadata information
The metadata file provided by GISAID was used for collecting the information of source country, collection date, submission date, GISAID clade name, and the age and gender of the host associated to the viral sequence. These were stored in a local output file along with the assigned protein variant (PV) number, clade name used in the current study as well as with the mutations in the sequence (Table S6). It is noteworthy that only a small number of sequences were found to have patient status information.
4.3. Mutation analysis
The amino acid mutation analyses were carried out as discussed elsewhere [7]. They were categorized as highly recurrent (HR, >10%), moderately recurrent (MR, 1–10%) and low recurrent (LR,0.002–1%) mutations based on their recurrence in the 125,747 viral proteomes. Note that the mutations that don’t occur in at least 3 sequences (viz., PF = 0.002%) were not considered. Further, the first appearance and country wise distribution of HR and MR mutations were examined to understand their global dissemination. The month wise changes in the functional motifs of SARS-CoV-2 were analyzed through multiple sequence alignment (MSA) using CLUSTAL OMEGA [17], [26]. The results were presented using Weblogo3 [27].
4.4. Proteome variant analysis
Comparative proteome analyses of 125,747 sequences with the reference sequence were performed using blast program [28] and the results were analyzed using in-house scripts to identify the mutations. Based on the presence of unique nucleotide mutations with respect to the reference sequence, individual sequences were assigned a genome variant number (NV). Similarly, variant number (PV) was also assigned for SARS-CoV-2 whole proteome variants. Further, based on the presence of HR mutation(s), the variants were assigned a specific clade number (CL) (Fig. 4(A)) [25]. In order to understand the evolution of SARS-CoV-2, the frequency of occurrence of each variant was reported month wise and country wise. The variants were further classified as new or old based on their existence in the earlier months. This was to identify whether a variant was emerged for the first time in a month or exists already.
4.5. Mutation or variant persistence analyses
A mutation or variant was considered to be persistent, if it has emerged anytime during December 2019-Septemeber 2020 and recurred in October 2020 or November 2020. The same methodology was used to quantify the month wise and protein wise persistence of mutations/variants.
4.6. Construction of phyloproteomic tree
The month wise phyloproteomes were created as explained earlier [7] by considering the proteome sequences (viz., the non-coding regions were excluded) that were collected in a particular month. Finally, the non-redundant whole proteomes were alone used in the construction of the phylogenetic tree (Table S14). At first, the selected sequences were subjected to alignment using MAFFT [29] and the phyloproteomic tree was constructed using the maximum likelihood method in IQ-TREE software [30]. Itol tool [31] is used to visualize, analyze and create the phylogram.
All the graphs were created using Microsoft Excel 16 and Inkscape software was used for creating the figures.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
General.
The authors thank IIT Hyderabad for computational resources.
Funding
The authors thank Indian Institute of Technology Hyderabad for the computational resources and financial support from BIRAC-SRISTI GYTI (PMU_2017_010) and BIRAC-SRISTI (PMU2019/007). LPPP and CS thank MHRD for fellowship. PPU thank CSIR for fellowship.
Author contributions
PPU collected the data. LPPP did majority of the coding part and generated the data for plotting. CS wrote coding for plotting and plotted the graphs. LPPP, CS and PPU analyzed the data. LPPP, CS, PPU and TR wrote the manuscript. TR designed and supervised the project.
Data and material availability
The nucleotide sequences of the SARS-CoV-2 genomes used in this analysis are available, upon free registration, from the GISAID database (https://www.gisaid.org/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.05.054.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mallapaty S. The coronavirus is most deadly if you are older and male - new data reveal the risks. Nature. 2020;585:16–17. doi: 10.1038/d41586-020-02483-2. published online EpubSep (10.1038/d41586-020-02483-2) [DOI] [PubMed] [Google Scholar]
- 2.Gupta S., Raghuwanshi G.S., Chanda A. Effect of weather on COVID-19 spread in the US: a prediction model for India in 2020. Sci Total Environ. 2020;728:138860. doi: 10.1016/j.scitotenv.2020.138860. published online EpubAug 1 (10.1016/j.scitotenv.2020.138860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGurnaghan Stuart J., Weir Amanda, Bishop Jen, Kennedy Sharon, Blackbourn Luke A.K., McAllister David A. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diab Endocrinol. 2021;9 doi: 10.1016/S2213-8587(20)30405-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukhari Q., Massaro J.M., D'Agostino R.B., Sr., Khan S. Effects of weather on Coronavirus pandemic. Int J Environ Res Public Health. 2020;17 doi: 10.3390/ijerph17155399. published online EpubJul 27 (10.3390/ijerph17155399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel Lule Bugembe, My V.T. Phan, Isaac Ssewanyana, Patrick Semanda, Hellen Nansumba, Beatrice Dhaala, et al., A SARS-CoV-2 lineage A variant (A.23.1) with altered spike has emerged and is dominating the current Uganda epidemic. MedRxiv 2021. https://doi.org/10.1101/2021.02.08.21251393).
- 6.Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021 doi: 10.1016/S0140-6736(21)00298-1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponoop Prasad Patro L., Sathyaseelan Chakkarai, Uttamrao Patil Pranita, Rathinavelan T. Global variation in the SARS-CoV-2 proteome reveals the mutational hotspots in the drug and vaccine candidates. BioRxiv. 2020 doi: 10.1101/2020.031.230987). [DOI] [Google Scholar]
- 8.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827 e819. doi: 10.1016/j.cell.2020.06.043. published online EpubAug 20 (10.1016/j.cell.2020.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. published online EpubOct 26 (10.1038/s41586-020-2895-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies N.G., Barnard R.C., Jarvis C.I., Russell T.W., Semple M.G., Mark Jit W.J. Association of tiered restrictions and a second lockdown with COVID-19 deaths and hospital admissions in England: a modelling study. Lancet Infect Dis. 2020;2020 doi: 10.1016/S1473-3099(20)30984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveillance Bullet Europeen sur les maladies transmissibles = Eur Commun Dis Bullet 2017;22; published online EpubMar 30 (10.2807/1560-7917.ES.2017.22.13.30494). [DOI] [PMC free article] [PubMed]
- 12.Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. published online EpubNov (10.1038/s41564-020-0770-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1038/s41586-021-03677-y. published online EpubNov 27 (10.1101/2020.10.25.20219063) [DOI] [PubMed] [Google Scholar]
- 14.Volz Erik, Mishra Swapnil, Meera Chand R.J., Jeffrey C Barrett, Geidelberg Lily. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. MedRxiv. 2020 [Google Scholar]
- 15.Tegally Houriiyah, Wilkinson Eduan, Giovanetti Marta, Iranzadeh Arash, Fonseca Vagner, Giandhari Jennifer. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020 [Google Scholar]
- 16.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. Jama. 2021 doi: 10.1001/jama.2021.1612. published online EpubFeb 11 (10.1001/jama.2021.1612) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behloul N., Baha S., Shi R., Meng J. Role of the GTNGTKR motif in the N-terminal receptor-binding domain of the SARS-CoV-2 spike protein. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198058. published online EpubSep (10.1016/j.virusres.2020.198058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolakaki E., Giannakouros T. SR/RS motifs as critical determinants of coronavirus life cycle. Front Molcular Biosci. 2020;7:219. doi: 10.3389/fmolb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadasivan J., Singh M., Sarma J.D. Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals. J Biosci. 2017;42:231–244. doi: 10.1007/s12038-017-9676-7. published online EpubJun (10.1007/s12038-017-9676-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobhy H. The potential roles of protein functional motifs in coronavirus infection. Preprints. 2020 doi: 10.20944/preprints202004.0171.v1. [DOI] [Google Scholar]
- 21.Sobhy H. Systems biology approach to characterize potential SARS-CoV-2 pathways based on protein functional motifs. Preprints. 2020 doi: 10.20944/preprints202004.0171.v2. [DOI] [Google Scholar]
- 22.Hassan S.S., Choudhury P.P., Roy B. SARS-CoV2 envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112:3890–3892. doi: 10.1016/j.ygeno.2020.07.001. published online EpubNov (10.1016/j.ygeno.2020.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R., Chen J., Hozumi Y., Yin C., Wei G.W. Decoding asymptomatic COVID-19 infection and transmission. J Phys Chem Lett. 2020;11:10007–10015. doi: 10.1021/acs.jpclett.0c02765. published online EpubDec 3 (10.1021/acs.jpclett.0c02765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. published online EpubMar (10.1038/s41586-020-2008-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. published online EpubMay (10.1016/j.apsb.2020.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 27.Crooks Gavin E., Hon Gary, John-Marc Chandonia, Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004 doi: 10.1101/gr.849004. http://www.genome.org/cgi/doi/10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. published online EpubOct 5 (10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 29.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. published online EpubJul 15 (10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. published online EpubJan (10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucl Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. published online EpubJul 8 (10.1093/nar/gkw290) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.