Abstract
Intracellular delivery of enzymes is essential for protein-based diagnostic and therapeutic applications. Protein-spherical nucleic acids (ProSNAs) defined by protein core and dense shell of oligonucleotides have been demonstrated as a promising vehicle-free enzyme delivery platform. In this work, we reported a crosslinking strategy to vastly improve both delivery efficiency and intracellular sensor performance of ProSNA. By assembling individual ProSNA with lactate oxidase (LOX) core into a nanoscale particle, termed as crosslinked SNA (X-SNA), the enzyme delivery efficiency increased up to 5–6 times higher. The LOX X-SNA was later demonstrated as a ratiometric probe for quantitative detection of lactate in living cells. More importantly, X-SNA probe showed significantly improved sensor performance with signal-to-noise ratio 4 times as high as ProSNA when detecting intracellular lactate.
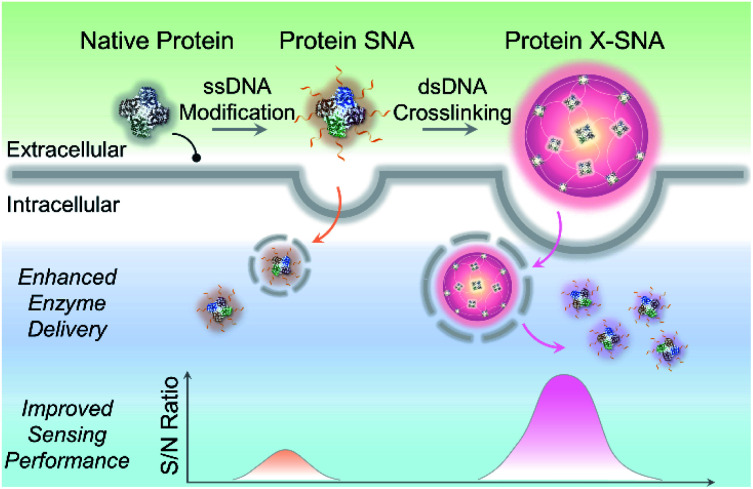
By crosslinking protein spherical nucleic acid (SNA) into a supramolecular architecture X-SNA, the intracellular enzyme delivery efficiency was significantly enhanced, showing 3–4 times higher signal-to-noise ratio in detecting intracellular lactate.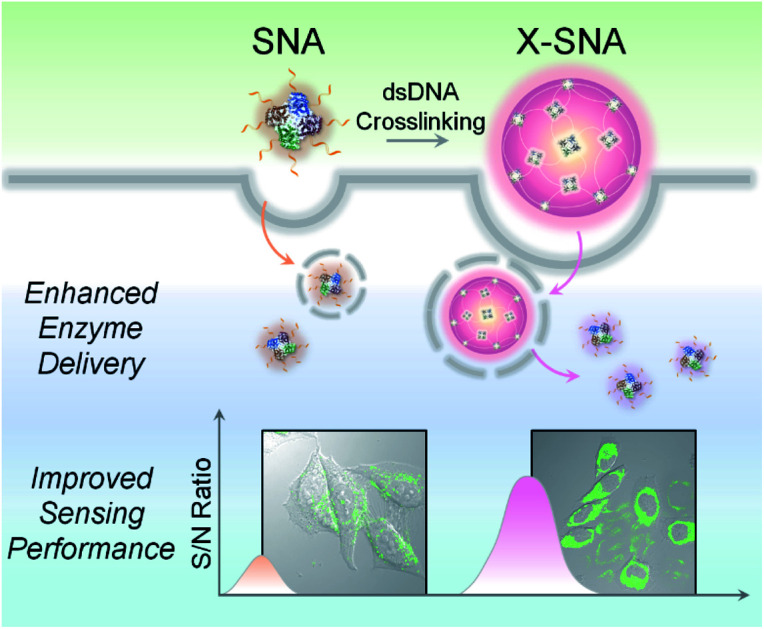
Introduction
Delivery of functional proteins to replace malfunctional ones in live cells or as intracellular probes to monitor metabolic pathways plays a key role in protein-based therapies and diagnosis.1–8 However, efficient delivery of native proteins which are normally membrane impermeable remains a major challenge due to their inherent biological sensitivity, surface charge, and large sizes.9 To meet the challenge, a variety of methods that can tune protein-membrane interfaces have been developed to facilitate protein internalization, including the use of supercharged proteins,10,11 cell-penetrating peptides,12 liposomes,13–15 virus-like particles,16 transfection agents,17 and nanoparticles.18–21 While these methods have been proven effective in different application scenarios, the use of exogenous delivery vehicles may lead to potential ill-responses of cells including cytotoxicity and immunogenicity, especially those based on virus capsids. Methods involving genetic fusion also suffer from reduced protein activity and stability.22
Recently, protein with highly oriented DNA strands on surface, known as protein spherical nucleic acid (ProSNA), has been reported as a transfection agent-free delivery system that protects protein activity, facilitates its cellular uptake, and serves as intracellular probe for live-cell analysis.23–27 SNA structure engages in cell-surface receptor-mediated endocytosis to promote protein transfection.28,29 Although ProSNA has featured advantages over its native structure, but still leaves much to be desired. One way to further improve cellular uptake is to increase the DNA density on protein surface, but that alone shields protein core and thus results in inhibition of enzymatic activity.30 The use of metal–organic framework to load proteins in SNA structure also can improve the loading and uptake, but the introduction of heavy metal ions may affect cellular processes.31–33 Indeed, a vehicle-free protein delivery system that possesses higher delivery efficiency while retaining its native structure and functionality is still sought after.
Herein, we report a straightforward strategy to significantly improve the intracellular delivery efficiency of proteins through controlled crosslinking of SNA structures (Fig. 1). We termed these supermolecular structures as crosslinked SNAs (X-SNAs). The central idea is that crosslinking individual proteins into nanoassemblies of well-defined sizes using long double-stranded DNAs (dsDNAs) can increase local protein concentration to facilitate uptake, while still space neighboring proteins a few nanometers apart to protect bio-functionalities. The enhanced protein uptake in turn can improve the signal-to-noise ratio of the X-SNA nanoprobes, allowing the development of better sensors for intracellular diagnosis.
Fig. 1. Scheme showing X-SNA enhances intracellular enzyme delivery efficiency and improves sensing performance compared to un-crosslinked protein SNA.
Results and discussion
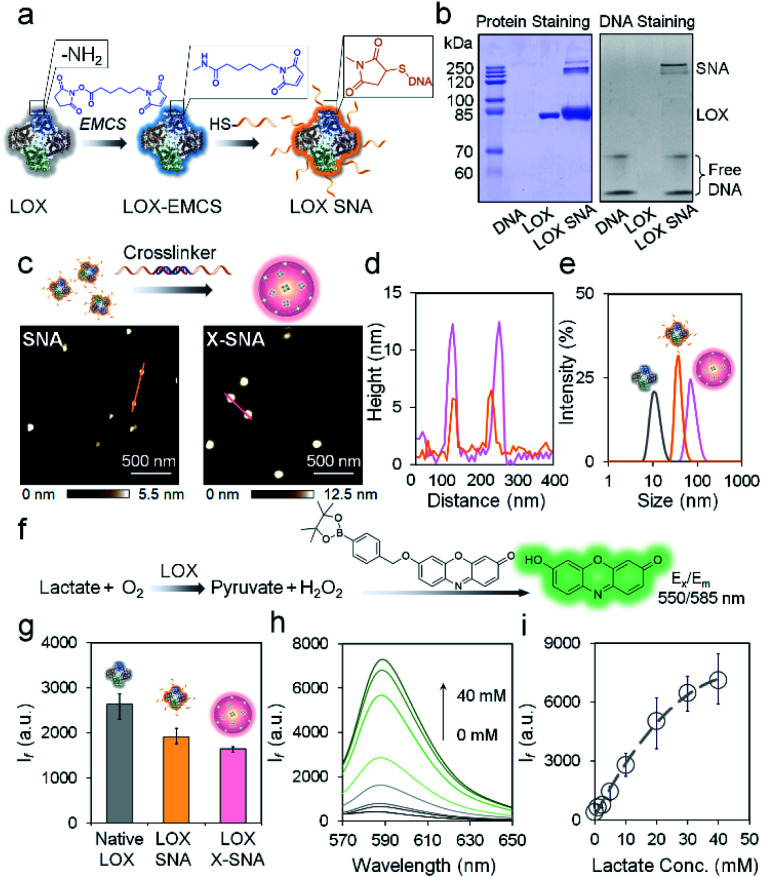
The key hypothesis of our strategy is that the X-SNA structure can promote protein delivery while maintaining its catalytic activity. To demonstrate that this is the case, tetrameric enzyme l-lactate oxidase (LOX) which is a member of alpha hydroxy acid-oxidation enzymes family was chosen as the model system. LOX is chosen for the following reasons. First, monomeric LOX has a molecular weight of ca. 80 kDa with its tetramer size of ca. 5 × 10 × 10 nm.34 Due to its relatively large size, LOX does not efficiently travel across cellular membranes and can serve as an ideal model for evaluating the intracellular delivery of functional enzymes. Second, lactate can serve as a potential biomarker owing to its abnormal accumulation in cancer cells.35,36 However, current detection of cellular lactate limits to extracellular environment or cell lysis,37,38 and intracellular detection is rarely reported.39 Thus, the efficient delivery of its oxidase, which does not exist in native mammalian cell, will allow the development of intracellular lactate probes by detecting generated H2O2. Native LOX was functionalized with nucleic acids to prepare ProSNA structure as previously reported.40,41 Briefly, surface amine groups on LOX were conjugated with 5′-end thiolated DNA strands using N-(ε-maleimidocaproyloxy)succinimide (EMCS) as a bifunctional linker (Fig. 2a). To demonstrate the successful immobilization of nucleic acids on LOX, formed LOX SNAs were characterized using denaturing polyacrylamide gel electrophoresis (SDS-PAGE). Coomassie brilliant blue and SYBR green II were used to stain protein and oligonucleotide respectively, followed by photographic and fluorescence imaging. As shown in Fig. 2b, LOX SNA exhibited clear multiple bands around 200 kDa in both LOX and DNA-staining images, while unreacted protein and free DNA showed distinct lower bands. The decreased electrophoretic mobility of LOX SNA can be attributed to the introduction of negatively charged DNA strands and increased molecular weight, suggesting the covalent attachment of multiple oligonucleotides onto LOX surface (Fig. 2b). The formation of LOX SNA was further confirmed by high performance liquid chromatography (HPLC) and UV-vis spectroscopy (Fig. S1†). The level of DNA modification was estimated using absorbance at 260 nm and 280 nm, with ca. 12 strands of DNA per tetrameric LOX and ca. 4 pmol cm−2 surface DNA density (Fig. S2†).42
Fig. 2. (a) Schematic illustration of the functionalization of LOX with DNA. (b) Protein-staining image (left) and fluorescence (right) image of a native PAGE gel showing the LOX SNA. (c) AFM height images of as-synthesized LOX SNA (left) and X-SNA constructs (right). (d) Corresponding cross-sectional analysis of SNA (orange) and X-SNA (pink). (e) DLS characterization of native LOX (gray), SNA (orange), and X-SNA (pink). (f) Signal transduction process of the lactate detection using LOX. The generated H2O2 turned on the fluorescence of indicator. (g) Histogram of fluorescence intensity showing the catalytic activity of native LOX, SNA, and X-SNA based on the reaction depicted in (f). (h) Fluorescence spectra of X-SNA probe responding to lactate concentrations from 0–40 mM. (i) Calibration profile of X-SNA probe showing the increase of lactate concentration resulted in the increase of fluorescence intensity. Error bars indicate standard deviations of three independent measurements.
To form the X-SNA construct, a 36-bp dsDNA crosslinker was applied to assemble LOX SNAs (Table S1†). The dsDNA crosslinker was designed with oligo-T10 spacer and 30-base-long 3′ sticky end which is complementary to DNAs on LOX. The relatively rigid and extended dsDNA crosslinker connects proteins with enough interparticle distance, avoiding densely packing of proteins that may inhibit its catalytic activity. LOX SNAs and dsDNA crosslinkers were mixed in 1 : 1 stoichiometry ratio, salt-aged, and then purified by low speed centrifugation. Atomic force microscopy (AFM) and dynamic light scattering (DLS) were employed to study the size and morphology of assembled X-SNA construct. AFM images showed the obtained X-SNAs to be roughly spherical with ca. 80 nm diameter and 12.5 nm height. The un-crosslinked SNAs appeared smaller than the X-SNAs with ca. 30 nm diameter and 5 nm height (Fig. 2c and d). The reduction in particle height observed in z axis can be possibly attributed to tip tapping on particles during AFM measurements. The increased size of X-SNA after crosslinking was further confirmed by dynamic light scattering (DLS) (Fig. S3†). Native LOX, SNA, and X-SNA showed an average hydrodynamic size of ca. 12 nm, 44 nm, and 78 nm, respectively (Fig. 2e).
We next studied whether oligonucleotides modification and intermolecular crosslinking block the LOX active site and inhibit its catalytic functionality. A fluorescence assay which measured H2O2 generated in the lactate oxidation reaction was employed to test LOX activity (Fig. 2f). A H2O2-responsive fluorescent indicator was synthesized and characterized as previously reported (Fig. S4†).43 The indicator is not fluorescent, but upon the addition of H2O2, the release of boronate ester produces oxidized phenoxazine-based dye molecule with fluorescence emission at 585 nm and pKa of ca. 8.75. The performance of the indicator was systematically studied to optimize the experimental conditions (Fig. S5†). To evaluate LOX activity in different constructs, native protein, SNA, and X-SNA with the same protein concentration of 5 nM were incubated with 1 mM l-lactate. After 30 min incubation to enable complete reaction, H2O2 indicator of 1 mM was then added into the solution and fluorescence emission was measured. As shown in Fig. 2g and S6,† both X-SNA and SNA were able to efficiently catalyze oxidization of lactate and remained ca. 70% activity of native enzyme. Further studies of enzymatic activity in cell lysate showed comparable results (Fig. S7†). We further tested the performance of X-SNA at different lactate concentrations in 1× PBS buffer. In the presence of lactate with a concentration increasing from 0 mM to 40 mM, fluorescence intensity centred at 585 nm increased (Fig. 2h). The response curve started to reach plateau around 40 mM, demonstrating that even nanomolar X-SNA can efficiently convert millimolar lactate into pyruvate to generate detectable H2O2 (Fig. 2i). More importantly, the lactate concentration in tumor cells is reported to be around 10–20 mM,44,45 which falls into the dynamic range of the assay, suggesting the potential of using X-SNA to quantify intracellular lactate.
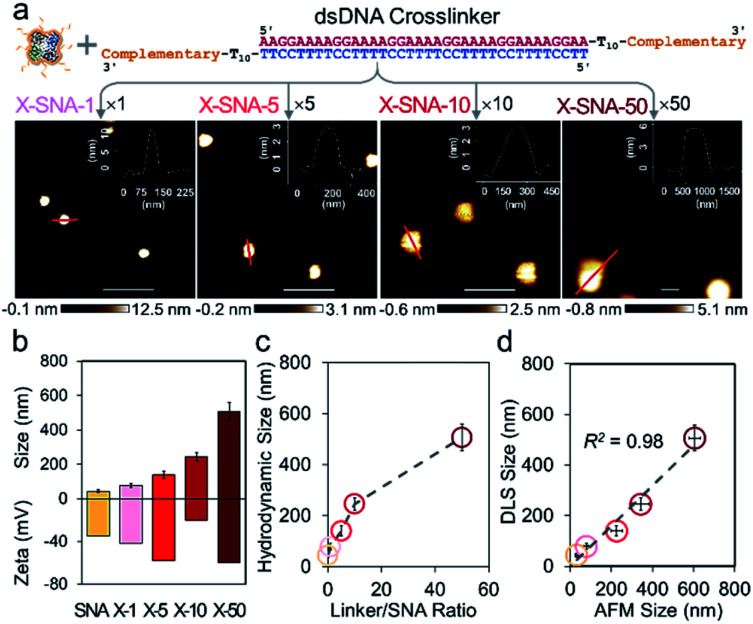
To further explore the DNA-mediated crosslinking approach, we then tried to prepare LOX X-SNAs of different sizes. The oligonucleotides on protein surface offer a preeminent orthogonal binding motif to guide programmable assembly of LOX. By systematically tuning the stoichiometry of hybridization reaction via varying SNA : linker ratio from 1 : 1 to 1 : 50, the sizes of X-SNAs can be controlled. As shown in Fig. 3a, four different sized LOX X-SNAs were successfully prepared, named as X-SNA-1, X-SNA-5, X-SNA-10, and X-SNA-50. AFM and DLS were used to characterize the formed X-SNAs. In AFM images, all four X-SNAs were observed roughly spherical with diameters controlled from ca. 80 nm to 600 nm (Fig. S8†). DLS measurements produced comparable results with AFM images (Fig. 3b and S9†), showing the hydrodynamic sizes of X-SNAs increased as the linker-to-SNA ratio increased (Fig. 3c). Zeta-potential measurements indicated X-SNAs are all electronegative with surface charge ranging from ca. −20 mV to −60 mV, supporting the DNA coverage (Fig. 3b). The correlation of AFM and DLS results showed a linear relationship between two sets of measured sizes with a R2 value of 0.98, indicating the reliability of both characterizations. The AFM-observed X-SNA size was generally larger than its hydrodynamic size, which could be attributed to the deformation of soft assemblies under tapping mode. We also tried to synthesize X-SNA with hydrodynamic size around 300–400 nm, but no uniform-sized particles were obtained. To gain more structural details of X-SNA, we used a simplified primitive cubic packing model to estimate the number of proteins in each X-SNA structure (Fig. S10†). The number of LOX in a ca. 80 nm X-SNA is calculated to be ca. 10 per particle. The number of proteins per particle increases as the size of X-SNA increases, with 500 nm one containing more than 1000 protein molecules.
Fig. 3. (a) AFM images showing four different sized X-SNA samples synthesized from different linker/SNA ratio. Inserts are corresponding cross-sectional analysis of X-SNA particles. Scale bar = 500 nm. (b) Dynamic light scattering (DLS) and zeta potential measurements of LOX SNA and four different sized X-SNA constructs. (c) Size profile showing increase of linker/SNA ratio leads to increase of the hydrodynamic size. (d) Quantitative correlation of X-SNA sizes measured by DLS and AFM images. Error bars indicate standard deviations of three independent measurements.
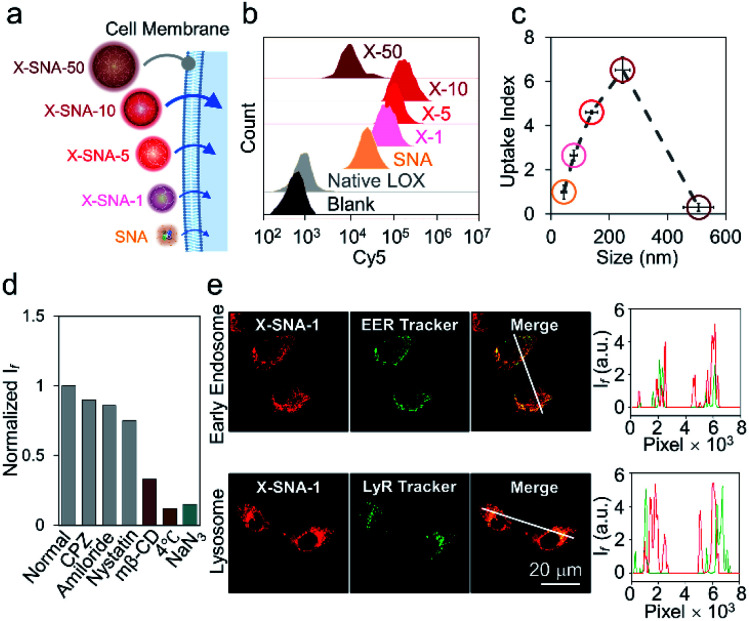
Having prepared a series of X-SNAs with controlled sizes, we next studied their intracellular delivery efficiency. Cy5 fluorophore on a T-base was labeled into the constructs to offer a handle for tracking internalization via confocal microscopy and flow cytometry. SNA, and X-SNAs of different sizes were calibrated with the same protein concentration and fluorescence emission per protein to allow quantitative analysis in cell experiments (Fig. 4a). HeLa cells were incubated with samples of 5 nM protein for 4 h and their uptake was quantified by flow cytometry. Compared to cells treated with native LOX, both SNA and X-SNAs improved LOX cellular uptake (Fig. 4b). More importantly, X-SNA constructs showed up to ca. 5–6 times higher intracellular delivery efficiency than SNA except the one of 500 nm. To further examine the effect of X-SNA size (80–500 nm) on LOX cellular uptake, we calculated an uptake index defined as the mean fluorescence ratio using SNA as the reference, representing the ability to deliver enzymes into cells. As shown in Fig. 4c, there was a general trend of increased cellular uptake as X-SNA size increased from ca. 80 to 250 nm. Specifically, 250 nm X-SNA showed 6.5 times LOX cellular uptake as much as SNA, indicating the crosslinking strategy significantly improved protein cell entry without changing its surface DNA density. However, X-SNA of the largest size 500 nm presented uptake index of ca. 0.2, suggesting poor uptake of the micrometer-sized particles. The cellular uptake abilities of different sized X-SNAs were further tested in both upright and inverted cell culture configurations using confocal microscopy (Fig. S11a†). Cells in the upright culture generally showed slightly higher fluorescence, possibly due to the gradual particle sedimentation. Importantly, the size-dependent uptake curves observed in both setups exhibited the same trend, which were consistent with flow cytometry results, confirming the effectiveness of crosslinking strategy (Fig. S11b–d†).
Fig. 4. (a) Schematic illustration of cell entry properties of constructs including SNA and four different sized X-SNAs. (b) Cell internalization properties of native LOX, SNA, and four different sized X-SNAs determined by flow cytometry. (c) Correlation curve showing the cellular uptake index of different sized constructs. (d) Flow cytometry analysis of HeLa cells incubated with 80 nm X-SNA under different inhibitor treatments. (e) Confocal images (left) and fluorescence intensity profiles (right) showing colocalization of X-SNA probes with early endosome and lysosome in HeLa cells. Error bars indicate standard deviations of three independent measurements.
To understand the size-dependent cellular uptake of X-SNA, we investigated its internalization mechanism using 80 nm constructs as the model system. The incubation of HeLa cells with X-SNA at 4 °C or in the presence of NaN3 led to negligible fluorescence in cells, demonstrating X-SNA relied on energy-dependent endocytosis process (Fig. 4d and S10†). To examine the endocytic pathway of X-SNA, we employed different types of biological inhibitors to inhibit established pathways by blocking surface receptors on plasma membrane.46 Confocal images of cells treated with methyl-β-cyclodextrin (MβCD), an inhibitor of caveolin-mediated endocytosis, showed significantly reduced fluorescence, indicating the suppressed X-SNA uptake (Fig. S12†). Quantitative analysis was further performed by using flow cytometry. As shown in Fig. 4d, treatment with MβCD resulted in a 60–70% decrease in the cellular uptake of X-SNA. Other inhibitors, such as chlorpromazine (CPZ, an inhibitor of clathrin-mediated endocytosis), amiloride (macropinocytosis-mediated endocytosis inhibitor), and nystatin (lipid-raft-mediated endocytosis inhibitor), showed minimized effects on X-SNA internalization. These results are consistent with previous observations that nanoscale objects are internalized by cells via a receptor-mediated endocytic pathway.47,48 In this vein, for nanoscale X-SNAs with sizes matching endocytic vesicles budded from plasma membrane, larger particles presented higher local protein concentration, resulting in significantly enhanced intracellular delivery of proteins.
Having elucidated the cell entry mechanism of X-SNA, we then studied the subcellular location of X-SNAs after endocytosis. Early endosome and lysosome were stained respectively to investigate the colocalization with X-SNAs in HeLa cell. As shown in Fig. 3e, early endosome fluorescence largely overlapped with X-SNA fluorescence, generating yellow colour. The Pearson's correlation coefficient was calculated to be ca. 0.79, suggesting a good colocalization and further confirming the receptor-mediated endocytic pathway. Lysosome fluorescence showed localized green dots while red X-SNA fluorescence spread in a larger distribution area around nucleus. Confocal z-stack images also showed clear divided green and red fluorescence at different scan depths (Fig. S13†). The calculated Pearson's correlation coefficient of lysosome and X-SNA decreased from ca. 0.28 to ca. 0.21 with the increase of the incubation time from 2 to 4 hours (Fig. S14†). These observations may possibly indicate that a part of delivered X-SNA particles released from lysosome through a slow escape process. Importantly, the ability to efficiently deliver exogenous oxidase such as LOX into cytoplasm potentially provides a general route to estimate cellular metabolite concentration by simply converting them into measurable reactive oxygen species using corresponding oxidase.
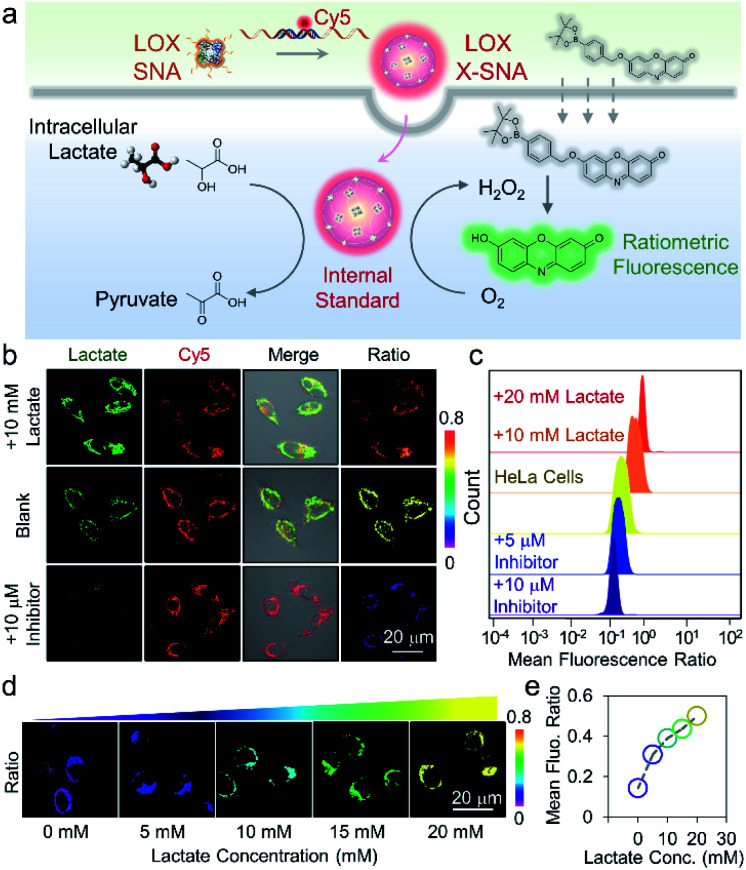
To ensure that the delivered enzymes are active in cellular milieu, we designed a ratiometric fluorescence assay using synthesized H2O2 indicator. Specifically, HeLa cells were incubated with Cy5-modified LOX X-SNA in OptiMEM for 4 h. After washing cells with PBS buffer, fresh medium containing 20 mM H2O2 indicator was added, and the cells were incubated at 37 °C for 30 min. Upon oxidation of lactate catalyzed by LOX, the released H2O2 oxidized the indicator, generating fluorescence emission at 585 nm. Meanwhile, Cy5 on X-SNA served as an internal standard, producing ratiometric signals for lactate that can be visualized and quantified by confocal microscopy and flow cytometry (Fig. 5a). Confocal z-stack images showed that X-SNA-based fluorescence assay produced green fluorescence and well-colocalized Cy5 fluorescence throughout the entire cell space, proving that the delivered enzymes remained functional and responded to lactate ratiometrically (Fig. S15†). We next treated HeLa cells with lactate medium or inhibitor oxamate to test the response of X-SNA assay to different lactate concentrations intracellularly. As shown in Fig. 5b, 10 mM lactate medium treatment increased green fluorescence in HeLa cells, while inhibitor quenched most of the emission. Intracellular Cy5 emission overlapped with green fluorescence and remained unchanged in treated cells, indicating the consistency of the internal standard. The green-to-red (G/R) fluorescence ratio of HeLa cells increased from 0.58 to 0.71 with external lactate showing more redder colour, while decreased to 0.22 with inhibitor showing more bluer colour (Fig. 5b and S16†). Flow cytometry histograms plotted in mean G/R fluorescence ratio were consistent with above-mentioned confocal images (Fig. 5c). The data fitting showed a linear relationship between the fluorescence ratio from confocal images and flow cytometry results, suggesting a good correlation of the two measurements (Fig. S17†). In addition, X-SNA treated cells showed viability up to 90%, demonstrating high biocompatibility of the nanoconstructs (Fig. S18†).
Fig. 5. (a) Scheme showing the cellular uptake of X-SNA probe and ratiometric detection of lactate in living cells. (b) Confocal images of X-SNA treated HeLa cells after incubation with lactate medium or inhibitor. Ratiometric images clearly showed colour change after treatments. (c) Showing mean fluorescence ratio of with X-SNA treated cells after treatment with either lactate medium or inhibitor, as determined by flow cytometry. (d) Confocal ratiometric images and (e) calibration curve of X-SNA treated L-02 cells in the presence of different external lactate concentrations.
To further demonstrate that the probe can respond to intracellular lactate in a quantitative manner, we chose L-02 cells with low endogenous lactate as model system and treated cells using lactate medium. As the lactate concentration increased from 0 to 20 mM in medium, cell samples showed increased green fluorescence under confocal imaging (Fig. S19†). The ratiometric colour also changed evidently from purple to yellow with fluorescent intensity ratio increasing from 0.17 to 0.58 (Fig. 5d and S20†). Quantitative flow cytometry analysis was in good agreement with the confocal images, illustrating the probe can distinguish 5 mM lactate concentration change (Fig. S21†). The calibration curve plotting mean G/R fluorescence ratio to medium lactate concentration presented a positive correlation, demonstrating that the assay can respond to intracellular lactate in a dose-dependent manner (Fig. 5e).
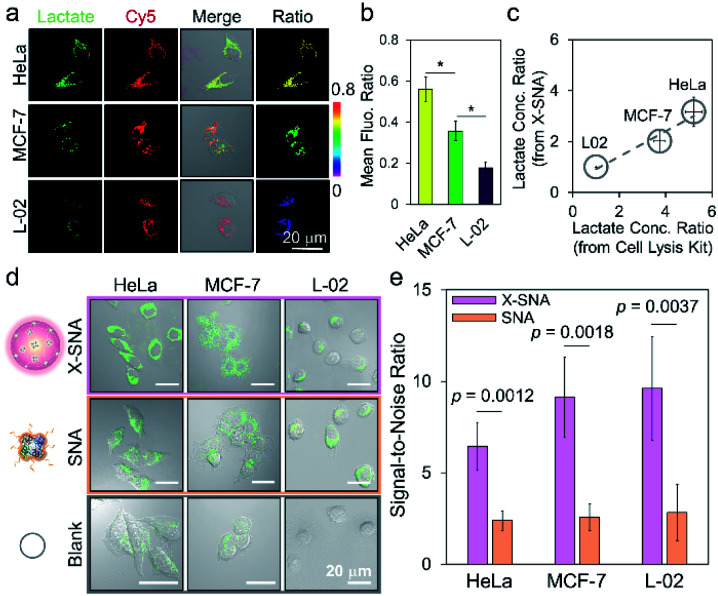
We next employed X-SNA probe to evaluate the endogenous lactate concentration in three different cell lines, including HeLa (cervical cancer), MCF-7 (breast cancer), and L-02 (normal liver) cells. All cells were cultured in medium containing 12% FBS, and then treated with X-SNA and H2O2 indicator for lactate detection. As shown in Fig. 6a and S22,† all three type of cells treated with 5 nM X-SNA probes showed Cy5 fluorescence of similar intensity, indicating that different cells showed similar X-SNA uptake. In the case of two cancer cells, HeLa cells exhibited strong green fluorescence intensity and MCF-7 showed moderate signal, indicating high and average lactate concentration. In contrast, X-SNA treated normal human hepatocytes L-02 produced faint but still visible green emission, indicating low lactate concentration. The results in confocal images were consistent with previous reports. Quantitative mean fluorescence ratio analysis from confocal images confirmed that the X-SNA probe not only can differentiate cancer cells from normal cells but also different types of cancer cells (Fig. 6b). To verify the results obtained from X-SNA probe, we used a commercial absorbance-based lactate kit to estimate lactate in cell lysis. By establishing the standard curve and measuring the absorbance of cell extract, lactate in cell lysis was calculated to be 15.9 mM for HeLa, 11.4 mM for MCF-7, and 3.1 mM for L-02, respectively (Fig. S23†). Using L-02 as reference, we plotted lactate concentration ratio obtained from X-SNA probe with cell lysis results and revealed a roughly linear relationship, suggesting a valid X-SNA assay and a correlation between intracellular and lysis lactate (Fig. 6c). X-SNA probe produced lower lactate concentration than lysis kit, possibly because nanoprobe mostly responded to cytosolic lactate rather than lactate in whole cell volume.
Fig. 6. (a) Confocal images and (b) corresponding mean fluorescence ratio of different cells treated with X-SNA for lactate detection (*p < 0.05, Student's t-test). (c) Correlation analysis studying the association between lactate concentration ratio determined by intracellular X-SNA probe and by lactate cell lysis kit. A linear fitting curve indicated the validity of X-SNA probe. (d) Confocal images of HeLa, MCF-7, and L-02 cells incubated with X-SNA or SNA in the presence of H2O2 indicator to detect intracellular lactate. (e) Corresponding histogram of the signal-to-noise ratio in three cell lines using X-DNA and SNA. P values were calculated using the Student's t-test. Error bars indicate standard deviations of three independent measurements.
As previously stated, the signal reporting process of the lactate assay relies on measurement of generated H2O2. Thus, endogenous H2O2 in cell especially in cancer cell may lead to increased background noise and affect sensing performance.49 To overcome this issue, we proposed to use X-SNA probe based on the hypothesis that the enhanced protein delivery ability of X-SNA would in turn enable an improved detection performance in living cells. To test the hypothesis, we compared the performance of X-SNA with un-crosslinked SNA in previous three cell lines to detect lactate. Cells incubated only with indicator, but no LOX were used as control groups. As shown in Fig. 6d and S24–S26,† in all three cell lines, cells incubated with X-SNA exhibited higher green fluorescence than that treated by SNA, indicating improved lactate detection ability. Visible green colour was observed in HeLa and MCF-7 control groups but negligible in L-02 control, which was attributed to the background H2O2 in cancer cells. To make a quantitative comparison, we calculated the signal-to-noise ratio of both constructs using control cells as background reference. As shown in Fig. 6e and S27,† in three tested cell lines, X-SNA showed up to ca. 9.6 signal-to-noise ratio, while un-crosslinked SNA generated signal-to-noise ratio of 2–3, suggesting the use of X-SNA can significantly improve the assay sensitivity, especially in biological environment with high background.
Conclusions
In conclusion, we have developed a straightforward strategy by crosslinking individual DNA-modified enzymes into X-SNA construct that has achieved up to 6 times higher delivery efficiency than un-crosslinked ProSNA without the use of any transfection agent. The presented results demonstrate X-SNA enters cells in a size-dependent manner and preserves most catalytic functions. The LOX-based X-SNA has been applied as intracellular lactate probe with significantly improved performance, showing up to 3–4 times higher signal-to-noise ratio than regular ProSNA. The strategy is facile and easy to create biologically active protein materials that outperform previously reported un-crosslinked method. Because of the programmability and modularity enabled by DNA crosslinker, it is possible to install a large number of functionalities into the structure such as targeting groups, imaging agents, and functional nucleic acids, leading to potential biological applications such as cellular imaging, gene regulation, and immunomodulation. Additionally, since lactate is measured through the quantification of the redox product H2O2, the idea of delivering exogenous oxidases as intracellular signal transducer provides a general strategy to develop nanoprobes which can monitor a variety of metabolites that are difficult to detect in live cells.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge National Natural Science Foundation of China (No. 21877032, No. 21527810), Hunan Province Talented Young Scientists Program (2019RS2021, 2019RS2023), National Key Research Program (No. 2019YFA0905800), and the Fundamental Research Funds for the Central Universities for financial support.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04977h
Notes and references
- Hoffman R. M. Lancet Oncol. 2002;3:546–556. doi: 10.1016/S1470-2045(02)00848-3. [DOI] [PubMed] [Google Scholar]
- Desnick R. J. Schuchman E. H. Nat. Rev. Genet. 2002;3:954–966. doi: 10.1038/nrg963. [DOI] [PubMed] [Google Scholar]
- Leader B. Baca Q. J. Golan D. E. Nat. Rev. Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- D'Astolfo D. S. Pagliero R. J. Pras A. Karthaus W. R. Clevers H. Prasad V. Lebbink R. J. Rehmann H. Geijsen N. Cell. 2015;161:674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- Sun W. J. Ji W. Y. Hall J. M. Hu Q. Y. Wang C. Beisel C. L. Gu Z. Angew. Chem., Int. Ed. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolentino M. Q. Hartmann A. K. Loe D. T. Rouge J. L. J. Mater. Chem. B. 2020;8:5627–5635. doi: 10.1039/D0TB00767F. [DOI] [PubMed] [Google Scholar]
- Zhao D. Kong Y. H. Zhao S. S. Xing H. Top. Curr. Chem. 2020;378:83–124. doi: 10.1007/s41061-020-00305-7. [DOI] [PubMed] [Google Scholar]
- Huang Z. M. Lin M. Y. Zhang C. H. Wu Z. K. Yu R. Q. Jiang J. H. Anal. Chem. 2019;91:9361–9365. doi: 10.1021/acs.analchem.9b02115. [DOI] [PubMed] [Google Scholar]
- Gu Z. Biswas A. Zhao M. X. Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/C0CS00227E. [DOI] [PubMed] [Google Scholar]
- Lawrence M. S. Phillips K. J. Liu D. R. J. Am. Chem. Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronican J. J. Thompson D. B. Beier K. T. McNaughton B. R. Cepko C. L. Liu D. R. ACS Chem. Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalik P. Elmquist A. Hansen M. Padari K. Saar K. Viht K. Langel V. Pooga M. Bioconjugate Chem. 2004;15:1246–1253. doi: 10.1021/bc049938y. [DOI] [PubMed] [Google Scholar]
- Wang M. Sun S. Neufeld C. I. Perez-Ramirez B. Xu Q. B. Angew. Chem., Int. Ed. 2014;53:13444–13448. doi: 10.1002/anie.201407234. [DOI] [PubMed] [Google Scholar]
- Zuris J. A. Thompson D. B. Shu Y. Guilinger J. P. Bessen J. L. Hu J. H. Maeder M. L. Joung J. K. Chen Z. Y. Liu D. R. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. L. Gao Y. J. Wang Y. G. Wei Q. Shi J. Y. Chen N. Li D. Fan C. H. Chem. Sci. 2018;9:5967–5975. doi: 10.1039/C8SC00367J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarczyk S. J. Sitaraman K. Young H. A. Hughes S. H. Chatterjee D. K. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16998–17003. doi: 10.1073/pnas.1101874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erazo-Oliveras A. Najjar K. La Dayani L. Wang T. Y. Johnson G. A. Pellois J. P. Nat. Methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros R. A. DeSimone J. M. Nat. Rev. Drug Discovery. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Song G. S. Chen Y. Y. Liang C. Yi X. Liu J. J. Sun X. Q. Shen S. D. Yang K. Liu Z. Adv. Mater. 2016;28:7143–7148. doi: 10.1002/adma.201602111. [DOI] [PubMed] [Google Scholar]
- Cheng L. Zhang F. R. Wang S. H. Pan X. T. Han S. C. Liu S. Ma J. J. Wang H. Y. Shen H. Y. Liu H. Y. Yuan Q. P. Angew. Chem., Int. Ed. 2019;58:7728–7732. doi: 10.1002/anie.201902476. [DOI] [PubMed] [Google Scholar]
- Dong X. Liu H. J. Feng H. Y. Yang S. C. Liu X. L. Lai X. Lu Q. Lovell J. F. Chen H. Z. Fang C. Nano Lett. 2019;19:997–1008. doi: 10.1021/acs.nanolett.8b04236. [DOI] [PubMed] [Google Scholar]
- Fu A. L. Tang R. Hardie J. Farkas M. E. Rotello V. M. Bioconjugate Chem. 2014;25:1602–1608. doi: 10.1021/bc500320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin J. D. Sprangers A. J. McMillan J. R. Mirkin C. A. J. Am. Chem. Soc. 2015;137:14838–14841. doi: 10.1021/jacs.5b09711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin J. D. Auyeung E. Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4564–4569. doi: 10.1073/pnas.1503533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D. Ebrahimi S. B. Mirkin C. A. Adv. Mater. 2020;32:1901743. doi: 10.1002/adma.201901743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi S. B. Samanta D. Mirkin C. A. J. Am. Chem. Soc. 2020;142:11343–11356. doi: 10.1021/jacs.0c04978. [DOI] [PubMed] [Google Scholar]
- Samanta D. Ebrahimi S. B. Kusmierz C. D. Cheng H. F. Mirkin C. A. J. Am. Chem. Soc. 2020;142:13350–13355. doi: 10.1021/jacs.0c06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. C. Giljohann D. A. Daniel W. L. Zheng D. Prigodich A. E. Mirkin C. A. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H. J. Hao L. L. Narayan S. P. Auyeung E. Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. R. Hayes O. G. Winegar P. H. Mirkin C. A. Acc. Chem. Res. 2019;52:1939–1948. doi: 10.1021/acs.accounts.9b00165. [DOI] [PubMed] [Google Scholar]
- Chen T. T. Yi J. T. Zhao Y. Y. Chu X. J. Am. Chem. Soc. 2018;140:9912–9920. doi: 10.1021/jacs.8b04457. [DOI] [PubMed] [Google Scholar]
- Chen Y. J. Li P. Modica J. A. Drout R. J. Farha O. K. J. Am. Chem. Soc. 2018;140:5678–5681. doi: 10.1021/jacs.8b02089. [DOI] [PubMed] [Google Scholar]
- Wang S. Z. Chen Y. J. Wang S. Y. Li P. Mirkin C. A. Farha O. K. J. Am. Chem. Soc. 2019;141:2215–2219. doi: 10.1021/jacs.8b12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y. Yorita K. Matsuoka T. Kita A. Fukui K. Morimoto Y. Biochem. Biophys. Res. Commun. 2006;350:249–256. doi: 10.1016/j.bbrc.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Heiden M. G. V. Cantley L. C. Thompson C. B. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorie O. N. Dellinger P. Crit. Care Clin. 2011;27:299–326. doi: 10.1016/j.ccc.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Mongersun A. Smeenk I. Pratx G. Asuri P. Abbyad P. Anal. Chem. 2016;88:3257–3263. doi: 10.1021/acs.analchem.5b04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendlein M. Pappa A. M. Ferro M. Lopresti A. Acquaviva C. Mamessier E. Malliaras G. G. Owens R. M. Adv. Mater. 2017;29:1605744. doi: 10.1002/adma.201605744. [DOI] [PubMed] [Google Scholar]
- San Martin A. Ceballo S. Ruminot I. Lerchundi R. Frommer W. B. Barros L. F. PLoS One. 2013;8:e57712. doi: 10.1371/journal.pone.0057712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H. Zhang C. L. Ruan G. Zhang J. J. Hwang K. Liu Y. Anal. Chem. 2016;88:1506–1510. doi: 10.1021/acs.analchem.5b04031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trads J. B. Torring T. Gothelf K. V. Acc. Chem. Res. 2017;50:1367–1374. doi: 10.1021/acs.accounts.6b00618. [DOI] [PubMed] [Google Scholar]; Li H. Zhang B. H. Lu X. G. Tan X. Y. Jia F. Xiao Y. Cheng Z. H. Li Y. Silva D. O. Schrekker H. S. Zhang K. Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4340–4344. doi: 10.1021/acs.accounts.6b00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Zhang B. H. Lu X. G. Tan X. Y. Jia F. Xiao Y. Cheng Z. H. Li Y. Silva D. O. Schrekker H. S. Zhang K. Mirkin C. A. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4340–4344. doi: 10.1073/pnas.1801836115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L. Wei L. Wang R. Y. Zhang C. Y. Zhang J. Tan T. W. Xi Z. Chem.–Eur. J. 2015;21:15167–15172. doi: 10.1002/chem.201502832. [DOI] [PubMed] [Google Scholar]
- Zheng X. T. Yang H. B. Li C. M. Anal. Chem. 2010;82:5082–5087. doi: 10.1021/ac100074n. [DOI] [PubMed] [Google Scholar]
- Hirschhaeuser F. Sattler U. G. A. Mueller-Klieser W. Cancer Res. 2011;71:6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- Ding H. M. Li J. Chen N. Hu X. J. Yang X. F. Guo L. J. Li Q. Zuo X. L. Wang L. H. Ma Y. Q. Fan C. H. ACS Cent. Sci. 2018;4:1344–1351. doi: 10.1021/acscentsci.8b00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault S. D. Walkey C. Jennings T. Fischer H. C. Chan W. C. W. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- Tan X. Y. Lu X. G. Jia F. Liu X. F. Sun Y. H. Logan J. K. Zhang K. J. Am. Chem. Soc. 2016;138:10834–10837. doi: 10.1021/jacs.6b07554. [DOI] [PubMed] [Google Scholar]
- Giorgio M. Trinei M. Migliaccio E. Pelicci P. G. Nat. Rev. Mol. Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.