Significance
Hemagglutinin (HA) is the major influenza virus surface protein and a prime antigen candidate for vaccine development using recombinant subunit approaches. Adjuvants often are used to enhance the immunogenicity of recombinant proteins. Here, we show that a next-generation vaccine adjuvant system that seamlessly sequesters and presents antigens on the surface of immunogenic liposomes improves the functional immunogenicity of HA in mice and ferrets. The potential for antigen dose sparing and multivalent presentation is demonstrated using this approach.
Keywords: adjuvant, cobalt porphyrin, liposomes, influenza vaccine, nanoparticle vaccine
Abstract
Recombinant influenza virus vaccines based on hemagglutinin (HA) hold the potential to accelerate production timelines and improve efficacy relative to traditional egg-based platforms. Here, we assess a vaccine adjuvant system comprised of immunogenic liposomes that spontaneously convert soluble antigens into a particle format, displayed on the bilayer surface. When trimeric H3 HA was presented on liposomes, antigen delivery to macrophages was improved in vitro, and strong functional antibody responses were induced following intramuscular immunization of mice. Protection was conferred against challenge with a heterologous strain of H3N2 virus, and naive mice were also protected following passive serum transfer. When admixed with the particle-forming liposomes, immunization reduced viral infection severity at vaccine doses as low as 2 ng HA, highlighting dose-sparing potential. In ferrets, immunization induced neutralizing antibodies that reduced the upper respiratory viral load upon challenge with a more modern, heterologous H3N2 viral strain. To demonstrate the flexibility and modular nature of the liposome system, 10 recombinant surface antigens representing distinct influenza virus strains were bound simultaneously to generate a highly multivalent protein particle that with 5 ng individual antigen dosing induced antibodies in mice that specifically recognized the constituent immunogens and conferred protection against heterologous H5N1 influenza virus challenge. Taken together, these results show that stable presentation of recombinant HA on immunogenic liposome surfaces in an arrayed fashion enhances functional immune responses and warrants further attention for the development of broadly protective influenza virus vaccines.
Influenza virus is a persistent global health concern, mainly because the efficacy of current vaccines is suboptimal. The narrow breadth of protection and rapid waning of vaccination-induced antibodies along with the circulation of variant viruses necessitates annual reformulation and readministration of seasonal influenza virus vaccines. Seasonal influenza epidemics result in a global total of 3 to 5 million severe infections each year, with 290,000 to 650,000 deaths (1). The viral envelope contains two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), of which there are many antigenically distinct subtypes (H1-18, N1-11) (2, 3). Both proteins are subject to antigenic drift; the selection of advantageous mutations that alters antigenicity and promotes escape from preexisting immunity. In addition to seasonal epidemics, novel forms of the virus have arisen throughout history to cause pandemics with high morbidity and mortality rates. These strains generally arise by antigenic shift, a process through which two different influenza virus strains reassort their genomes within host cells. This results in an influenza virus strain to which the human population is largely naive (4, 5). The influenza virus's capacity to circumvent immune recognition through antigenic change has been a significant barrier to the development of long-lasting immunity and highly efficacious vaccines.
Currently, seasonal influenza vaccination relies on a global viral surveillance system and predictive models that determine which strains will be incorporated into the seasonal vaccine each year. However, seasonal vaccine efficacy varies significantly depending upon the degree of matching between the predicted and circulating strains and does not provide significant protection when variant seasonal or pandemic strains emerge. Most seasonal influenza vaccines are still produced in embryonated chicken eggs, which necessitates a protracted vaccine production timeline and can result in egg-adapted vaccine strains that are antigenically dissimilar from circulating strains (6, 7). To address this, future vaccine strategies should be developed that can either yield a more rapid production process or protect against a more extensive repertoire of influenza virus strains, ideally including strains that are yet to arise. Both approaches favor the application of recombinant influenza virus antigens, with molecular structures that can be precisely controlled and modified. Recombinant antigens have proven safe and effective in the Flublok vaccine (8), and several studies have assessed epitope-based vaccine strategies based on conserved protein sequences in the stalk region (9, 10) and head region (11) of recombinant HA antigens. However, for many experimental recombinant vaccines to achieve a sufficient degree of immunogenicity, recombinant antigens must be administered in conjunction with an adjuvant to increase the magnitude of the immune response that they elicit. While regulatory agencies have approved several adjuvanted influenza vaccines for clinical use (12, 13), existing adjuvants may not be optimum for recombinant proteins. New adjuvants are being developed including nanoparticle-based carriers for recombinant antigens (14, 15). Liposomes, in particular, have been developed as vaccine adjuvants (16) and have been assessed with recombinant HA in preclinical studies (17–19). In this study, to enhance liposome immunogenicity, a synthetic monophosphoryl lipid A (MPLA) variant, phosphorylated hexaacyl disaccharide (PHAD), was incorporated into the lipid membrane. MPLA is used as a component of AS01, a liposomal adjuvant currently included in the Shingrix vaccine for Herpes Zoster (20, 21).
We have developed an adjuvant system based on liposomes containing cobalt-porphyrin phospholipid (CoPoP). CoPoP provides a methodology that results in biostable binding due to the sequestration of His-tagged antigens directly within the bilayer (22). The CoPoP contained in the liposomal membranes results in rapid binding of His-tag modified recombinant antigens at room temperature (RT) with straightforward coincubation. When used in this way for immunization, CoPoP particles have been shown to enhance antibody responses to recombinant antigens derived from several pathogens (23–26). CoPoP has entered human clinical trials as a component of a SARS-CoV-2 vaccine (ClinicalTrials.gov Identifier: NCT04783311). In this study, we assess whether CoPoP can contribute to a platform for influenza virus vaccines. Recombinant HA trimers with trimerizing foldon domains from T4 bacteriophage fibritin were used to generate antigen associated with CoPoP liposome surfaces with conformation putatively replicating the trimers formed by native HA complexes (27, 28).
Results
His-Tagged HA Trimers Bind to CoPoP Liposomes to Form Particles.
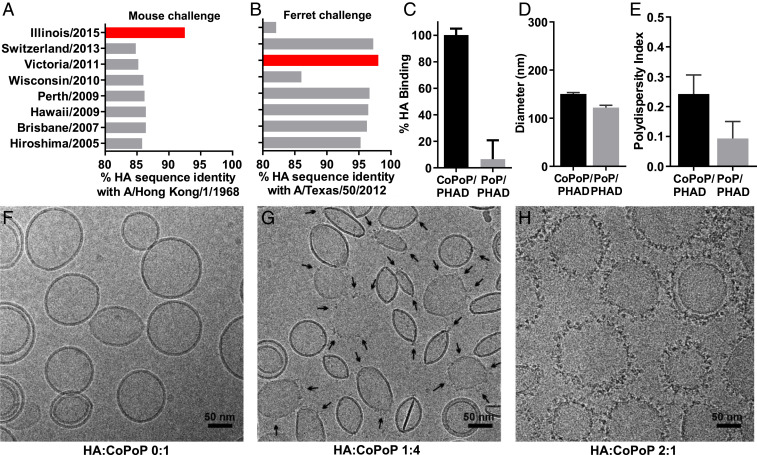
To examine CoPoP liposome application for influenza vaccines, test antigens were selected for particleization. Based on the sequence comparison of representative characterized HA trimer antigens (formed with trimerizing foldon domains) available through the International Reagent Resource (IRR) program (www.internationalreagentresource.org), H3 HA from A/canine/Illinois/11613/2015 (H3N2) was selected as the recombinant antigen to produce liposomal vaccines for testing against a mouse-adapted H3N2 influenza virus strain, A/Hong Kong/1/1968. The HA from these two strains possess relatively high sequence identity (92.5%), intended to provide a good degree of matching against the readily available mouse-adapted H3N2 virus (Fig. 1A). However, at the same time, this also represents an antigenic mismatch, which frequently results in reduced efficacy of the vaccine-induced immune response to the challenge virus strain. This strategy was also replicated when selecting A/Victoria/361/2011 H3 HA as a heterologous antigen for the ferret study that used A/Texas/50/2012 H3N2 virus challenge (Fig. 1B and SI Appendix, Fig. S1). The majority of sequence divergence exists in the variable head domain region, as defined by the sequence between Cys52 and Cys277 of the HA protein (29). Antigens possessing the highest head region sequence identity correspond with the greatest overall protein identity (SI Appendix, Table S1). The stalk region showed relatively little sequence variance, with no stalk region possessing <89% identity with any other antigen (SI Appendix, Table S2).
Fig. 1.
CoPoP/PHAD liposomes convert His-tagged HA trimers into particles. Sequence identity of representative recombinant H3 HA trimers available from the IRR with HA from the heterologous H3N2 mouse or ferret challenge strains used in this study. HA from A/canine/Illinois/11613/2015, shown in red, was selected for mouse vaccination and A/HongKong/1/1968 challenge (A) while HA from A/Victoria/361/2011 was selected for ferret immunization and A/Texas/50/2012 challenge (B). (C) Binding of His-tagged H3 HA trimers from Illinois/2015 to CoPoP/PHAD or PoP/PHAD (no Cobalt) liposomes after incubation at RT for 1 h. Particle diameter (D) and polydispersity (E) of liposomes after incubation with Illinois/2015 HA trimer. (F) Cryo-TEM images of CoPoP/PHAD liposomes before antigen binding or following incubation with Illinois/2015 H3 HA trimers at a HA-to-CoPoP mass ratio of (G) 1:4 (select HA trimers indicated by black arrows) or (H) 2:1. (C–E) Data show mean ± SD from n = 3 samples.
The spontaneous binding between CoPoP liposomes and C terminus His-tagged HA trimers was assessed. Incubation of His-tagged H3 HA trimers with CoPoP liposomes yielded antigen binding of ∼100% at a mass ratio of 1:4 HA-to-CoPoP, contrasted against cobalt-lacking (and thus nonbinding in theory) PoP liposomes that resulted in minimal HA binding (Fig. 1C). Size and polydispersity assessment by light scattering indicated that liposomes maintained a size between 100 and 150 nm without the formation of large aggregates (Fig. 1 D and E). After HA trimer binding to liposomes, conservation of antigen conformation was verified by a slot blot assay of A/Victoria/361/2011 HA with four epitope-specific monoclonal antibodies (SI Appendix, Fig. S2). Imaging of the bare CoPoP/PHAD liposomes by cryo transmission electron microscopy (Cryo-TEM) confirmed the mostly unilamellar and vesicular structure of the liposomes (Fig. 1F). Individual HA trimers localized to the surface of the lipid membrane following incubation at a mass ratio of 1:4 HA-to-CoPoP, as was used in all vaccination studies (Fig. 1G). Interestingly, HA binding to liposomes appeared to be a cooperative process, as regions of high-density HA were apparent on some liposomes while other liposomes did not have any HA present. When CoPoP/PHAD liposomes were saturated with HA trimers by incubation with a mass ratio of 2:1 HA-to-CoPoP, the visibility of HA trimers on the liposomal membrane improved (Fig. 1H). The HA trimers present on the surface of CoPoP liposomes presented with a similar size compared to native influenza virus particles visualized with cryo-TEM (30).
Enhanced Immunogenicity of HA Particles Formed with CoPoP/PHAD Liposomes.
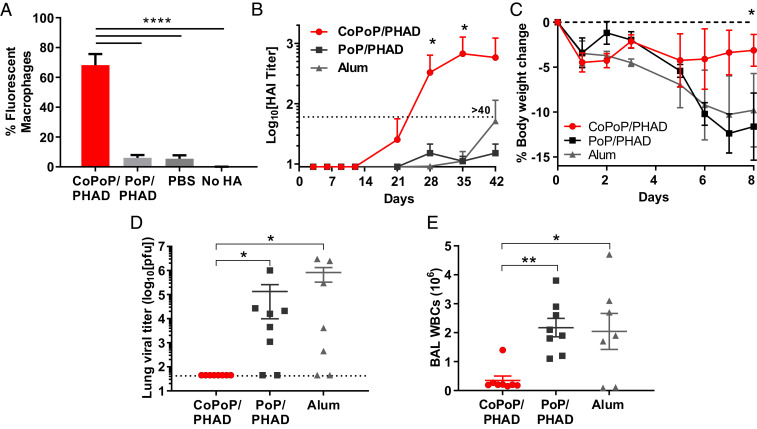
To elucidate the effects of particleization on antigen uptake, fluorescent-labeled HA trimers were incubated with liposomes and exposed to cultured murine macrophages. While nonbinding liposomes showed no enhancement in antigen delivery over soluble antigen alone, the antigen binding to the CoPoP resulted in a significant increase in antigen uptake by macrophages (Fig. 2A). Enhanced delivery to immune cells is expected to enhance the immunogenicity of the vaccine. Hemagglutinin inhibition (HAI) assays with mouse serum derived from outbred Institute of Cancer Research (ICR) mice immunized intramuscularly with 100 ng of HA trimers revealed that admixing with CoPoP/PHAD liposomes, but not nonbinding PoP/PHAD liposomes, elicited detectable HA inhibitory antibody titers after a single injection (as measured 21 d postprime) and induced sustained antibody titers above the HAI > 40 after booster injection (Fig. 2B). Mice receiving the identical vaccine regimen were then challenged with 100 plaque-forming units (pfu) (2.5 × LD50, the lethal dose that results in death of half the challenged mice) of mouse-adapted A/Hong Kong/1/1968 (H3N2). Immunization with CoPoP liposomes protected mice from morbidity as measured by weight loss (Fig. 2C). Mice immunized with HA adjuvanted with alum or PoP/PHAD liposomes lost more weight than when the CoPoP/PHAD adjuvant was used, although no mice lost more than 25% body weight. Mice were euthanized at 8 d postinfection (dpi), the time point corresponding with peak lung injury (31, 32), to assess lung viral titers and leukocyte counts of the pulmonary air spaces. Viral lung titers were below the detection limit in all animals vaccinated with HA and CoPoP/PHAD liposomes, a significant reduction over both control adjuvanted groups (Fig. 2D). Quantitative analysis of white blood cells residing in the lungs 8 dpi also showed a substantial reduction, indicating a lower severity of infection at the challenge endpoint (Fig. 2E).
Fig. 2.
Liposome presentation of HA trimers improves delivery to macrophages in vitro and outcomes of heterologous H3N2 influenza virus challenge following murine immunization. (A) Fluorescently labeled A/canine/Illinois/11613/2015 HA trimers were incubated with murine macrophages (1 μg/mL, 4 h, 37 °C) and subjected to flow cytometry to assess cellular uptake. Data show mean ± SD for n = 3 replicates. ICR mice were vaccinated intramuscularly with 100 ng A/canine/Illinois/11613/2015 (H3N2) HA trimers on days 0 and 21, prior to collecting sera and subjecting mice to challenge. (B) Day 42 Sera were assessed for HAI titer against heterologous A/Hong Kong/1/1968 (H3N2). Data show mean titer ± SD for n = 5 mice per group. Mice were challenged on day 60 with 2.5 × LD50 of mouse-adapted A/Hong Kong/1/1968 (H3N2) and euthanized 8 d postchallenge with analysis of (C) bodyweight, (D) viral titers in lung homogenates, and (E) white blood cell counts within lung tissue. Data show mean ± SEM for n = 8 mice per group. Statistical analysis was performed in B and C with one-way ANOVA and Dunnett’s comparison against alum and in D and E with one-way ANOVA with Tukey’s multiple comparisons. *P < 0.05, **P < 0.01.
Evaluation of HA Formulated with CoPoP/PHAD or Alternative Adjuvants.
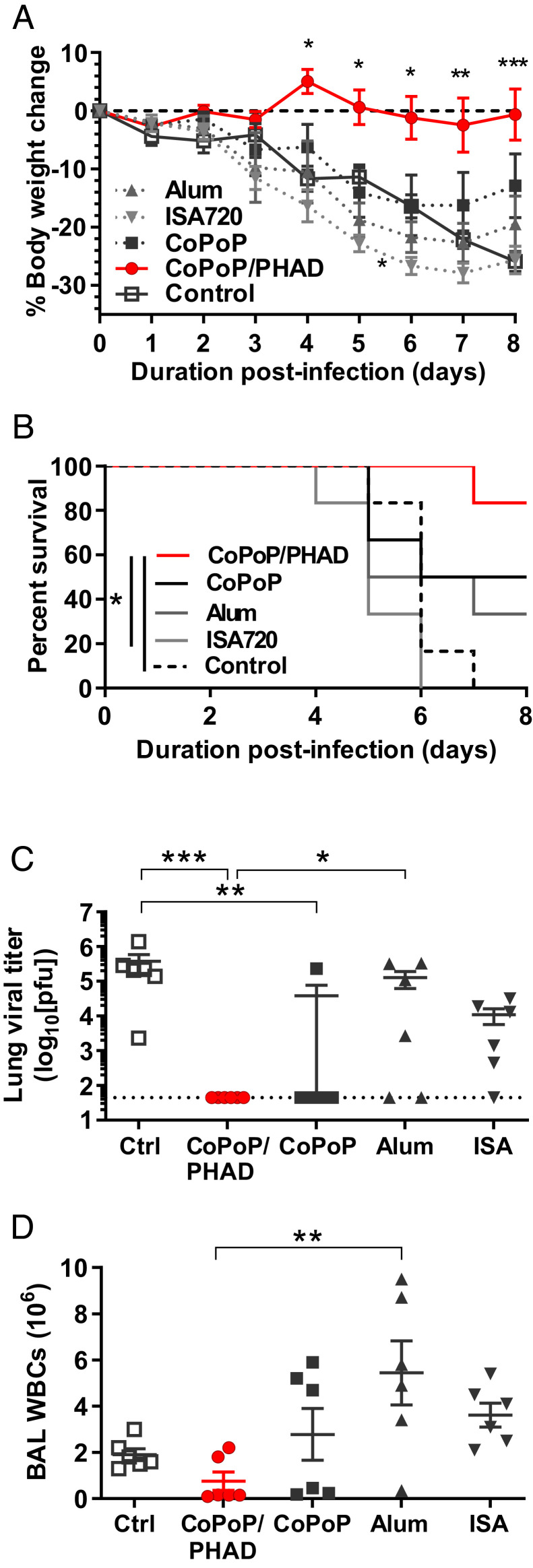
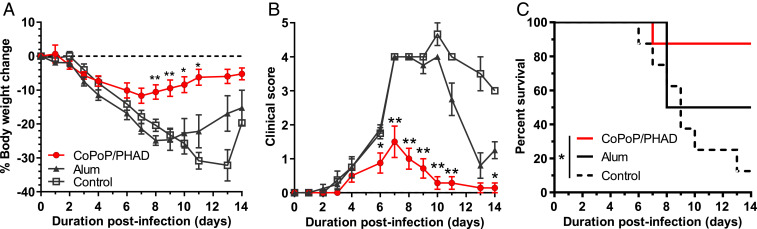
To better understand the effects of CoPoP/PHAD relative to other adjuvant systems, we performed a comparison of vaccine efficacy using CoPoP/PHAD liposomes, Montanide ISA720, and alum. Each vaccine was formulated with a 50-ng dose of His-tagged recombinant HA trimer. Intramuscular vaccination with HA admixed with CoPoP/PHAD resulted in protection from weight loss following subsequent virus infection, with significantly better protection compared to unvaccinated controls as well as vaccines formulated with alum or ISA720 adjuvants (Fig. 3A and SI Appendix, Table S3). The CoPoP/PHAD group also experienced a lower rate of severe morbidity, as indicated by weight loss, than the other groups (Fig. 3B). Mice were euthanized on day 8, and viral titers in lung homogenates and white blood cell counts in bronchoalveolar lavages (BAL) were measured as surrogates of protection. Both CoPoP/PHAD and CoPoP without PHAD significantly reduced the titer of influenza virus in the lung tissues, with most mice exhibiting titers below the limit of detection of the assay (Fig. 3C). CoPoP/PHAD mice also had the lowest white blood cell (WBC) counts in the BAL compared to all other groups, comparable to BAL WBC counts in naive mice, measured as 4.3 ± 0.3 × 105 cells (Fig. 3D).
Fig. 3.
CoPoP/PHAD liposomes effectively adjuvant HA trimers. Mouse challenge was used to compare CoPoP/PHAD against CoPoP formulated without PHAD (CoPoP alone), ISA720, and alum. All vaccine formulations contained 50 ng A/canine/Illinois/11613/2015 HA trimers except for the control group. Mice were immunized on day 0 and 21 and challenged on day 42 with 2.5 × LD50 influenza A/Hong Kong/1/1968 (H3N2). (A) Average body weight, (B) percentage of mice with bodyweight loss less than 20%, (C) day 8 lung influenza virus titer, and (D) BAL WBC counts are shown. Data show mean ± SEM for n = 6 mice. Statistical analysis was performed for A by one-way ANOVA with Dunnett’s comparison against control, B by log-rank Mantel–Cox test, and C and D by one-way ANOVA with Tukey’s comparisons. *P < 0.05; **P < 0.01; ****P < 0.005.
Admixing HA Trimers with CoPoP/PHAD Liposomes Confers a Dose-Sparing Effect.
Dose-sparing is a consideration for recombinant protein vaccines, especially those related to pandemic-relevant indications that could require rapid development and rollout. Mice were vaccinated with reduced antigen doses to determine the dose-sparing potential of CoPoP/PHAD liposome-adjuvanted HA. An additional adjuvant, the squalene oil-in-water emulsion AddaVax, was used to compare the CoPoP-adjuvanted vaccine results. MF59 is an established adjuvant for use in influenza vaccines, with demonstrated clinical safety and immunogenicity, and has been marketed as a component of Fluad vaccines since 1997 (33, 34). MF59 can associate proteins to the surface of micelles and activate transcription of a greater repertoire of cytokines, chemokines, and other immune factors than alum (35). AddaVax was selected as an adjuvant comparator for CoPoP/PHAD as it is a research adjuvant system based on the MF59 composition, has been shown to have comparable adjuvant qualities (36), and has the capability to induce broadly reactive antibodies against influenza virus when applied with recombinant antigens in mice and ferrets (37).
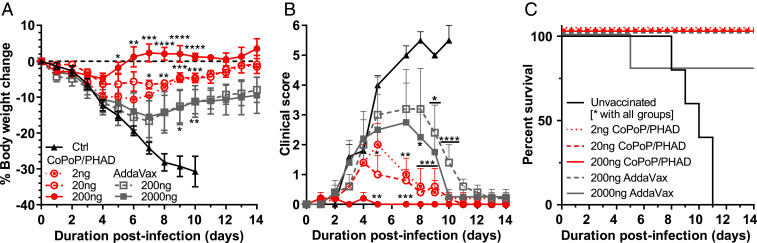
Protection was most complete when mice were immunized with HA trimer vaccine doses of 200 ng adjuvanted with CoPoP/PHAD, with complete bodyweight recovery and no observable clinical signs after 4 dpi. Mice vaccinated with lower doses of CoPoP/PHAD-HA trimers showed some morbidity; however, both the 10-fold and 100-fold reduced doses provided significant protection over unvaccinated mice. Furthermore, the application of CoPoP/PHAD showed benefits in increasing body weight protection and decreasing clinical scoring of observed infection symptoms over AddaVax-based vaccines, even when the antigen dose in AddaVax was 10-fold greater than the highest dose applied with CoPoP liposomes (Fig. 4 A and B). CoPoP/PHAD-HA trimers conferred complete survival of all vaccinated mice at all doses. In contrast, AddaVax-adjuvanted vaccines only provided complete survival at 2-µg doses, and unprotected mice all succumbed to infection (Fig. 4C). While both CoPoP/PHAD and AddaVax were able to improve the survival rates of vaccinated mice in the lethal challenge at all tested doses, the benefits shown at a 100-fold reduced dose, as well as the significant difference in protection observed against the equivalent 200-ng antigen dose with AddaVax (SI Appendix, Table S4), suggests CoPoP/PHAD could provide dose-sparing, thereby increasing vaccine doses by 10- to 100-fold and enhance vaccine efficacy.
Fig. 4.
Dose sparing potential of CoPoP/PHAD liposomes. A dose-sparing challenge was performed to test the effect of CoPoP/PHAD on promoting immunity with low A/canine/Illinois/11613/2015 H3 HA trimer antigen doses. After intramuscular immunization of ICR mice on days 0 and 21, mice were challenged on day 39 with 2.5 × LD50 influenza A/Hong Kong/1/1968 (H3N2). When compared against another adjuvant, AddaVax (squalene oil-in-water emulsion), CoPoP/PHAD provided (A) greater bodyweight protection and (B) lower clinical score based on symptoms of influenza, even with a 2-ng vaccination HA dose. (C) All dose formulations containing CoPoP achieved 100% survival, while at 200 ng with AddaVax, one mouse succumbed to infection, and all control mice succumbed before the end of the challenge. n = 5 mice per group. Data show mean ± SEM. Statistical analysis in A and B was performed by one-way ANOVA with Dunnett’s comparison against control and C with log-rank test. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
HA Trimers Admixed with CoPoP/PHAD Liposomes Induce Antibodies that Confer Protective Immunity.
To determine whether induced antibodies could mediate protection, naive mice were given a passive transfer of serum from vaccinated mice prior to challenge. Donor mice were vaccinated on day 0 and 21 with CoPoP/PHAD-HA trimers or HA trimers admixed with alum following standard procedures. On day 42, the serum from these mice was collected, pooled, and administered to naive mice, which were then challenged with A/Hong Kong/1/1968 H3N2. Serum transfer from CoPoP/PHAD-HA trimer–vaccinated mice alone significantly improved bodyweight over the course of the challenge compared to naive control mice (Fig. 5A). A substantial difference was observed in clinical scores between groups; mice receiving CoPoP/PHAD-HA trimer immune serum exhibited clinical signs of similar average severity to directly vaccinated mice, while mice receiving serum from the alum adjuvant group displayed clinical signs similar to the untreated group (Fig. 5B). The group that received transfer of CoPoP/PHAD-HA trimer postimmune sera also had greater overall survival than all other groups (Fig. 5C). These results indicate that the humoral response induced by the CoPoP/PHAD adjuvant system is sufficient to protect against influenza virus infection. Cellular immunity contributions may account for the difference between this challenge and the protection observed in challenges with directly vaccinated animals, although this was only assessed in a limited capacity. In a cursory investigation of cellular immune responses, spleen cells from mice vaccinated with HA trimers admixed with CoPoP/PHAD but not those from the alum-adjuvanted vaccine group produced detectable interferon-γ (IFN-γ) with HA restimulation (SI Appendix, Fig. S3).
Fig. 5.
Passive serum transfer protects mice influenza virus challenge. Mice were vaccinated intramuscularly with 100 ng A/canine/Illinois/11613/2015 HA trimers admixed with CoPoP/PHAD or alum, and the serum from these mice was pooled and administered to naive mice immediately prior to challenge with 2.5 × LD50 of influenza A/Hong Kong/1/1968 (H3N2) to assess antibody-mediated protection based on (A) body weight, (B) clinical score, and (C) mortality. Pooled serum (200 μL) was transferred to each recipient mouse. Data show mean ± SEM for groups of n = 8 mice. Statistical analysis in A and B was performed with one-way ANOVA with Dunnett’s comparison against control and C with log-rank test. *P < 0.05, **P < 0.01.
CoPoP/PHAD Liposome Performance in a Ferret Challenge Model.
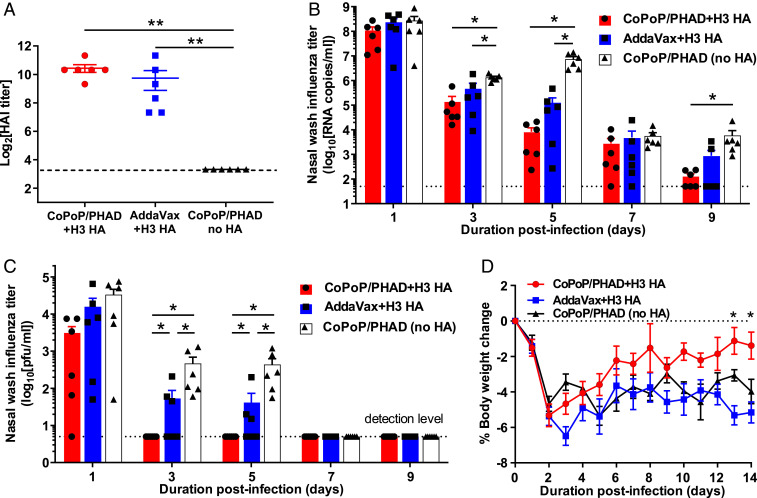
A heterologous challenge study was next performed with ferrets, a standard preclinical animal model for influenza virus research. A more recent virus challenge strain was selected to test whether the response elicited by the CoPoP/PHAD approach could provide protection outside of mouse models and in the contexts of different vaccine versus challenge strain mismatches. A/Texas/50/2012 (H3N2) was utilized as a representative seasonal H3N2 for ferret challenge due to its higher relevance to recent human H3N2 strains than the mouse-adapted H3N2 (38). Protein sequence–matching was used to determine a heterologous strain as a source for HA trimers as was done for the mouse immunogen. H3 HA trimers from A/Victoria/361/2011 (H3N2) were selected (98% sequence identity; Fig. 1A). HA from this strain was characterized, and antibodies were tested in vitro and in ferrets to ensure performance consistent with mouse challenge results (SI Appendix, Fig. S1). Ferrets were vaccinated intramuscularly with Victoria/2011 HA trimer antigens combined with CoPoP/PHAD liposomes or commercial AddaVax adjuvant or with CoPoP/PHAD liposomes without antigens as a control. HAI prior to challenge indicated that similar antibody titers were elicited by both adjuvant formulations (Fig. 6A). On day 56 postvaccination, ferrets were challenged intranasally with 106 TCID50 (the 50% tissue culture infectious dose) of influenza A/Texas/50/2012 H3N2. Nasal washes, blood samples, and body weight measurements were collected throughout the challenge. Analysis of viral load in the upper respiratory tract, assessed by qPCR assay of nasal wash samples, found that CoPoP/PHAD and AddaVax adjuvanted vaccines yielded significantly reduced viral RNA load compared to liposome alone from day 3 to day 5 postchallenge. Furthermore, the CoPoP/PHAD adjuvant with HA resulted in a significant reduction by day 9, effectively reducing viral load (as measured by qPCR) below the detection limit. Of the two adjuvants, HA combined with CoPoP/PHAD resulted in a greater average decrease in viral load at all time points (Fig. 6B). Plaque assays performed with nasal washes from ferrets revealed that infectious influenza viral titers in nasal washes were reduced to the limit of detection by day 3 postchallenge when CoPoP/PHAD adjuvant was used with HA. In contrast, detectable titers were still measured in the AddaVax ferret group at the same time point, as well as 2 d later (Fig. 6C). Bodyweight trends were similar between HA with AddaVax and CoPoP/PHAD liposome-only groups, while average body weight loss was more modest for ferrets immunized with CoPoP/PHAD-HA (Fig. 6D). In conclusion, while viral RNA was present in the lungs up to 9 d postchallenge in many animals across each group (Fig. 6B), the infectious virus titer was reduced to below the level of detection in only 3 d in CoPoP/PHAD-HA vaccinated ferrets, whereas the same reduction was not achieved in the AddaVax and control groups until 7 d postchallenge (Fig. 6C).
Fig. 6.
CoPoP/PHAD immunization vaccine efficacy in ferrets. Fitch ferrets were vaccinated intramuscularly on day 0 and 28 with 8 μg recombinant HA trimers derived from A/Victoria/361/2011 (H3N2) admixed with CoPoP/PHAD liposomes or AddaVax and challenged on day 56 with 106 TCID50 heterologous influenza A/Texas/50/2012 (H3N2). (A) Serum collected 14 d prior to viral challenge was assessed for HAI titer with homologous virus. Detection of viral load in nasal washes was determined by (B) qPCR or (C) by MDCK plaque assay. (D) Body weight of ferrets following challenge. (A) Statistical analysis by Mann–Whitney U tests, (B and C) by Mann–Whitney U test against CoPoP/PHAD without HA, and (D) by one-way ANOVA with Dunnet’s comparison against CoPoP/PHAD with no HA. * indicates P < 0.05, ** indicates P < 0.01. Data shown in mean with SD with n = 6 for each group.
CoPoP/PHAD Liposomes as a Platform to Deliver Multiple Antigens.
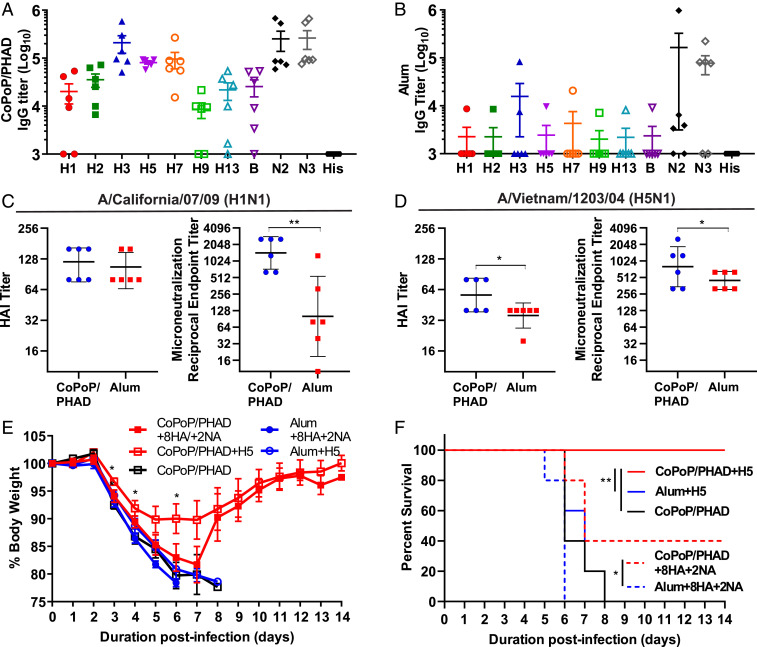
Future influenza vaccine designs might involve the copresentation of multiple antigens to enhance vaccine breadth. Due to the common use of His-tags in antigen production, a diverse repertoire of His-tagged influenza virus antigens could be displayed on CoPoP liposomes. As a proof of concept, CoPoP liposomes were first incubated with a panel of 10 influenza virus surface antigens, 7 HA, 1 B strain HA, and 2 NA from different strains (described in the methods) to form monovalent particles. Animals were divided into groups, and each group was vaccinated with one antigen from this panel. This was done to establish a baseline-expected IgG titer for each antigen and assess preexisting cross-reactivity that may occur between specific antigens. Results indicated that while some IgG cross-reactivity could be observed, off-target binding generally accounted for approximately 100-fold lower reported titer than homosubtypic binding (SI Appendix, Fig. S4A). Vaccination with recombinant NA bound to liposomes elicited antibodies capable of NA inhibition (SI Appendix, Fig. S4B). Next, to perform immunization with multivalent liposomes, a dose of 50 ng of total antigen comprising of 5 ng of each individual antigen from the panel was used. CoPoP/PHAD liposomes were incubated with an admixture of all 10 antigens prior to intramuscular murine vaccination. At 35 d postprime with a boost at day 21, serum was collected, and enzyme-linked immunosorbent assay (ELISA) was performed to determine whether antibodies were produced that could recognize each individual constituent antigen. These multivalent particles elicited detectable IgG-binding titers against all 10 antigens (Fig. 7A), although some antigens appeared more immunogenic than others. Nevertheless, the CoPoP/PHAD multiplex induced antibody titers greater than 104 against all respective antigens except H9. In stark contrast, the same 10-plex antigen admixture with alum resulted in only one animal with detectable but low-level antibody titer. For both adjuvants, the postimmune sera showed negligible reactivity against the His-tag itself (Fig. 7B).
Fig. 7.
CoPoP/PHAD liposomes for multivalent immunization. In total, 8 HA and 2 NA types of recombinant antigens were used to immunize mice with 5 ng per dose of each of the 10 antigens (50 ng total) following incubation with CoPoP/PHAD (A) or Alum (B). HAI and MNT assays were performed using sera collected from vaccinated mice. (C) Against H1N1 strain A/California/07/2009pdm09, both formulations achieved comparable HAI titers, while neutralization indicated significant neutralization by antibodies elicited with CoPoP/PHAD. (D) Against H5N1-derived strain A/Vietnam/1203/2004 (H5N1), also known as HALo, the serum antibodies from the same CoPoP/PHAD vaccinated animals significantly improved both HAI and neutralization over alum. (E) Mice challenged with 5 × LD50 HALo exhibit reduced body weight loss when vaccinated with CoPoP/PHAD bound with heterologous H5 HA from A/gyrfalcon/Washington/41088–6/2014 (H5N8) or multiplex 8HA/2NA antigens. (F) Mice vaccinated with heterologous H5 achieved 100% survival, while two of five mice in multiplex group survived to the end of the challenge period. CoPoP/PHAD with H5 or 8HA/2NA mix each provided significant increase in survival over alum adjuvanted counterparts. Data expressed in geometric mean ± geometric SD. Statistical analysis was performed for C and D with unpaired t test, E with one-way ANOVA with Dunnett’s comparison CoPoP/PHAD only, and F with log-rank test. * indicates P < 0.05, ** indicates P < 0.01.
Antibodies elicited by multivalent CoPoP/PHAD vaccination were tested for neutralizing efficacy. HAI and microneutralization (MNT) assays were performed against two influenza virus strains: A/California/07/2009 (H1N1) and an attenuated A/Vietnam/1203/2004-derived H5N1 (H5N1 HAlo) (39). The H1 HA antigen used in the multiplex formulation was homologous with the H1N1 strain used for the assay. Simultaneously, the H5 HA was derived from a strain heterologous to H5N1 HAlo virus. In homologous neutralization, the highly multiplexed CoPoP and alum-adjuvanted antigen mixture produced comparable HAI titers; however, a significant disparity was observed in MNT between these groups, with the CoPoP/PHAD formulation yielding superior neutralization titers (Fig. 7C). In heterologous neutralization assays, the CoPoP/PHAD formulation achieved both higher HAI titer and higher neutralization titer than the alum formulation, by a statistically significant margin (Fig. 7D). These results indicate that CoPoP/PHAD liposome vaccines can induce neutralizing antibodies against multiple strains when multiplexed with many heterologous antigens.
Multivalent CoPoP/PHAD liposomes were tested with influenza virus challenge to determine whether the elicited multistrain immune response could confer protection. With the standard formulation and vaccination schedule, mice were then challenged on day 35 postprime with influenza H5N1 HALo strain. Mice were vaccinated with the 10-antigen multiplex or an equal dose containing only HA from A/gyrfalcon/Washington/41088–6/2014 (H5N8), the H5 HA component of the multiplex formula. Body weight of mice in challenge indicated best protection was achieved by single-antigen vaccination with heterologous H5 (with just 86.6% protein sequence identity); however, mice vaccinated with the multivalent formulation which survived showed comparable body weight protection and recovery with single-antigen formulation (Fig. 7E). Comparison of survival rates show that complete survival was only achieved with CoPoP/PHAD with H5 only, but vaccination with multivalent CoPoP/PHAD was the only other formulation able to preserve survival to the termination of the 14-d challenge; furthermore, multivalent CoPoP/PHAD achieved a significantly higher survival rate than mice vaccinated with alum multiplex (Fig. 7F). Considering that the multivalent formulation for this study was designed to maximize the breadth of recognition over protective efficacy, these results represent an encouraging preliminary examination of multivalent vaccine potential in CoPoP/PHAD adjuvant formulations. This study indicates that a refined selection of influenza antigens in multivalent CoPoP/PHAD could be determined, which would optimally induce broad immunity across various influenza virus strains, including both seasonal and potential pandemic strains.
Discussion
Presentation of HA antigens on nanoparticle carriers is an intriguing concept with potential benefits to influenza vaccines, with several proposed mechanisms for increased immunogenic effects (40–43). CoPoP/PHAD liposomes possess distinct properties that could provide further advantages over alternative nanoparticle vaccine approaches, including ease and flexibility of single- and multivalent-antigen binding, coformulation with lipid adjuvants such as the toll-like receptor agonist PHAD within the particle membranes, facile tuning of antigen density, and the lack of exogenous protein scaffolding.
Recombinant HA trimers adjuvanted with CoPoP/PHAD outperformed vaccines adjuvanted with alum. Furthermore, compared to experimental adjuvants such as ISA720 and AddaVax, CoPoP/PHAD performed favorably, exhibiting an advantage over these adjuvants in most experiments with marginal to significant favorable outcomes. Of note, experiments that compared binding and nonbinding liposomes (but both containing PHAD) strongly indicate that the antigen binding character of CoPoP/PHAD, when contrasted to PoP/PHAD, is a dominant factor in contributing to the potent immune response to the antigen. Likewise, the coformulation of CoPoP with PHAD outperformed the application of CoPoP liposomes without incorporated receptor agonist, indicating the capability for the incorporation of lipid adjuvant is beneficial for the particles. The CoPoP adjuvant is beneficial for hapten-like antigens that benefit from enhanced delivery to antigen-presenting cells like macrophages after conversion to particles (24, 44). Despite their much larger size compared to haptens, HA trimers also displayed enhanced uptake by murine macrophages in vitro following conversion into particle format. Taken together, the full combination of properties that can be achieved with CoPoP/PHAD contribute toward a particle with substantial immunogenicity, efficacy, and dose-sparing benefits.
The efficacy of HA trimer–based CoPoP/PHAD influenza virus vaccines in animal models is supported by the results of these preclinical challenge studies. It is strongly indicated that with ideal dosage application, CoPoP/PHAD protects against body weight loss associated with infection in mice and reduces both the severity and duration of clinical symptoms. Even when conditions were not ideal, such as dose reduction to as low as 2 ng HA, CoPoP/PHAD provided a significant benefit over untreated control and could perform comparably to the AddaVax adjuvant with a 1,000-fold higher antigen dose. CoPoP/PHAD achieves a robust humoral response, as confirmed by the protection observed in mice treated with a passive transfer of serum. CoPoP/PHAD effectively enhanced the response in ferrets against a contemporary human influenza virus strain A/Texas/50/2012. As an adjuvant, CoPoP/PHAD demonstrated the ability to induce protection against a contemporary human strain of influenza virus with a recombinant HA antigen from a strain that is homosubtypic, yet heterologous. This indicates an application for CoPoP/PHAD within the existing influenza vaccine production framework, allowing it to be used in conjunction with recombinant antigens based on seasonal predicted strains. Future work should assess whether heterosubtypic protection can be conferred using recombinant HA particles formed with CoPoP/PHAD. In addition, future work should also address the potential toxicity of HA trimers adjuvanted with CoPoP/PHAD. Prior toxicological assessment of CoPoP itself or CoPoP/PHAD complexed with recombinant protein antigens indicated that these formulations were well tolerated and did not result in local reactogenicity or observed toxicity in mice (23, 24).
The unique capabilities of CoPoP/PHAD have the potential for application in future vaccines spanning the diversity of influenza virus subtypes. CoPoP/PHAD was shown to bind with and induce detectable antibody titers against 10 distinct influenza virus antigens and was also shown to provide protection in challenge studies. The repertoire of antigens CoPoP/PHAD can bind and adjuvant includes potentially pandemic strains, such as H2, H5, and H7. We have confirmed that this broad antibody response was not due to the common His-tag, as neither CoPoP/PHAD nor alum adjuvants elicited binding antibodies against this short region. This evidence supports the potential application of CoPoP/PHAD as a flexible adjuvant for various pandemic influenza vaccines. Inclusion of recombinant NA also yielded significant antibody production and NA inhibition. NA antibodies are being recognized as an important component of the immune response to influenza virus infection. By directing an antibody response against conserved epitopes present on NA, CoPoP liposomes could easily be explored for testing the impact of inducing NA antibodies in vaccines with broad protective potential (45). Overall, the results in this preliminary application of CoPoP/PHAD show a strong indication of its adjuvant efficacy with facile stoichiometric control of antigen display and are encouraging for the future vaccine applications of such an immunostimulatory particle.
Materials and Methods
Influenza Virus Antigen Selection.
A panel of His-tagged antigens was constructed from those available through the catalog of the IRR (Influenza Division, World Health Organization Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention). HA antigens from H3N2 influenza virus strains included: A/Brisbane/10/2007 (H3N2), catalog FR-61; A/Hiroshima/52/2005 (H3N2), FR-63; A/Hawaii/07/2009 (H3N2), FR-401; A/Perth/16/2009 (H3N2), FR-472; A/Wisconsin/12/2010 (H3N2), FR-991; A/Victoria/361/2011 (H3N2), FR-1059; A/Switzerland/9715293/2013 (H3N2), FR-1416; and A/canine/Illinois/11613/2015 (H3N2), FR-1478. Sequences provided by the IRR catalog were compared with H3N2 challenge strains using online sequence alignment tools available from the National Center for Biotechnology Information’s Protein BLAST (Basic Local Alignment Search Tool).
In H3N2 challenge vaccine formulations for mice and ferrets, recombinant His-tagged H3 HA trimers from influenza virus strain A/canine/Illinois/11613/2015 (H3N2), FR-1478 or A/Victoria/361/2011 (H3N2), FR-1059 were obtained from the IRR. These have been assessed to be in ≥90% trimeric form by gel filtration chromatography. For generating multivalent liposomes, antigens from the following strains were included: HA of A/California/07/2009 (H1N1), FR-559; A/Japan/305/1957 (H2N2), FR-700; A/canine/Illinois/11613/2015 (H3N2), FR-1478; A/gyrfalcon/Washington/41088–6/2014 (H5N8), FR-1418; A/Netherlands/219/2003 (H7N7), FR-71; A/Hong Kong/33982/2009 (H9N2), FR-845; A/shorebird/Delaware/68/2004 (H13N9), FR-846; HA1 domain of B/Phuket/3073/2013 (Yamagata Lineage), FR-1417; and NA of A/Canada/444/2004 (H7N3), FR-478 and A/canine/Illinois/11613/2015 (H3N2), FR-1479.
Generation of CoPoP/PHAD Liposomes.
CoPoP was synthesized as previously described (25), and CoPoP/PHAD liposomes were prepared by ethanol injection and nitrogen-pressurized lipid extrusion (23). The composition of the liposomes included 1,2-dipalmitoylsn-glycero-3-phosphocholine (DPPC, Corden no. LP-R4-057), cholesterol (PhytoChol, Wilshire Technologies), and synthetic MPLA PHAD (Avanti catalog no. 699800P). Lipids were dissolved in 60 °C ethanol for 10 min, followed by the addition of 4 mL 60 °C phosphate-buffered saline (PBS) for another 10 min at 60 °C. Liposomes were then passed through 200-, 100-, and 80-nm stacked polycarbonate filters in a lipid extruder (Northern Lipids) with nitrogen pressure. After extrusion, liposomes were dialyzed with PBS to remove ethanol. The final liposome concentration was adjusted to 320 μg/mL CoPoP and passed through a 0.2-μm sterile filter. Liposome stock was stored at 4 °C. The liposome formulation had a mass ratio of [DPPC:CHOL:PHAD:CoPoP] [4:2:1:1].
Antigen–Liposome Binding Assay.
The binding of the antigen to liposomes was determined using a bicinchoninic acid assay (BCA, Fisher catalog no. PI23235) and separation assay. Briefly, a 50-µL volume of antigen–liposome solution was prepared by incubating proteins and liposomes for 1 h at RT and diluted to 200 µL with PBS. Samples were centrifuged at 20,000 × g for 90 min at 4 °C, and the supernatant was extracted from the resulting liposome pellet. BCA reagent, 200 μL, was added, and the samples were then incubated in a water bath at 60 °C for 10 min. A volume of 100 µL of each sample is transferred to each of three wells of a 96-well plate, and the absorbance is measured at 562 nm. The percentage binding is calculated as the ratio of the absorbance of the antigen–liposome supernatant and the liposome-only reference sample. A higher percentage corresponds with a higher protein concentration in the supernatant and thus a lower binding.
Liposome Characterization and Imaging.
The size and polydispersity of CoPoP and PoP liposomes were determined by dynamic light scattering with a NanoBrook 90 plus PALS brand instrument after 200-fold dilution in PBS. For cryo-EM, sample vitrification for cryo-EM was performed using a Vitrobot Mark IV (Thermo Fisher Scientific). For CoPoP/PHAD liposomes with high-density HA (2:1 mass ratio of protein to CoPoP), any excess protein was removed by brief incubation with Ni-NTA resin. Before samples were applied to the EM grids (C-flat 2/2–2Cu-T), grids were washed with chloroform for 2 h and treated with negative glow discharge in air at 5 mA for 15 s. For all samples, a volume of 3.6 μL was applied to the holey carbon grids and manually blotted using the Vitrobot blotting paper (Standard Vitrobot Filter Paper, Ø55/20 mm, Grade 595). Right after blotting, a new drop of the sample was applied to the EM grid and blotted again using the standard routine with the two blotting pads in the Vitrobot Mark IV (Thermo Fisher Scientific) for 3 s and with a blot force +1 before they were plunged into liquid ethane. The Vitrobot was set at 25 °C and 100% relative humidity. Grids were loaded into a Tecnai F20 electron microscope operated at 200 kV using a Gatan 626 single-tilt cryo-holder. The data acquisition was performed using SerialEM software using an FEI Tecnai G2 F20 microscope at 200 kV equipped with a TVIPS XF416 brand camera. Images were collected at a magnification of 50,000×, which produced images with a calibrated pixel size of 2.145 Å. Images were collected with a total dose of ∼50 e−/Å2 using a defocus ranging from −2 μm to −2.50 μm. Images were cropped and prepared using Adobe Photoshop.
Cellular Uptake Assay.
RAW 264.7 macrophage cells acquired from American Type Culture Collection were cultured with Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were passaged and incubated at 37 °C in 5% CO2 for 24 h until 70 to 80% confluence was achieved then exposed to H3 HA trimer antigen from A/canine/Illinois/11613/2015 (H3N2) labeled with DY490-NHS-Ester fluorescent label with the indicated conditions. Cells were incubated with antigen for 4 h, then harvested, centrifuged at 500 × g for 5 min at 4 °C, and washed with PBS three times, with centrifugation repeated for each wash. Then, cells were fixed with 2% paraformaldehyde in 0.5% BSA (bovine serum albumin)-PBS for 10 min at 4 °C, followed by centrifugation to remove the fixation buffer. Cells were resuspended in PBS containing 0.5% BSA and 0.01% sodium azide. The cells were assessed with a BD LSRFortessa TM X-20 flow cytometer, with an initial side scatter versus forward scatter gate, then gated for fluorescein isothiocyanate positive cells.
Antibody Binding Assessment with Slot Blot.
A nitrocellulose membrane was prewetted in PBS and subjected to a slot blot device with wells filled with H3 HA trimers from A/Victoria/361/2011 with indicated adjuvant. Samples were allowed to flow through the membrane by gravity over a 10-min period. The membrane was removed and blocked using 5% BSA in PBS for 30 min at RT. The membrane was cut into strips and incubated with 1,000× diluted mouse monoclonal antibodies from IRR: FR-1123 clone 6F6, FR-1124 clone 4D7, FR-1125 clone 3D7, and FR-1126 clone 1B2 in PBS for 1 h at RT. Membrane strips were washed with PBS for 5 min twice, followed by incubation with horseradish peroxidase (HRP)–labeled secondary goat anti-mouse antibody. The membrane was washed twice with PBS again, treated with substrate mixture from VisiGlo HRP substrate kit (VWR catalog no. 89424–016), and imaged using a Bio-Rad ChemiDoc Imager.
IFN-γ Quantification.
Isolated spleens from vaccinated mice were dissociated and filtered through a 70-µm cell strainer. The plunger from a sterile 3-mL syringe was used to dissociate tissue through the strainer, and 5 mL cold PBS was used to flush cells into a 50-mL tube. Cells were centrifuged at 500 × g for 5 min, and the supernatants were discarded. Red blood cells were lysed with 5 mL lysis buffer for 5 min, then 35 mL PBS was added. The cells were pelleted by centrifugation, resuspended in Roswell Park Memorial Institute 1640 supplemented with 10% FBS, 1% pen/strep, 2 mM glutamine, 1 mM sodium pyruvate, nonessential amino acids, and 50 µM β-mercapethanol and seeded into 96-well plates (2.5 × 105 cells/well). Cells were stimulated with 10 µg/mL recombinant H3 HA from A/canine/Illinois/11613/2015 and cultured for 72 h in 5% CO2/95% air at 37 °C in a humidified chamber. Following incubation, 50 µL media from each well was collected, and IFN-γ concentration was determined by an ELISA kit (Thermo Fisher BMS606TEN).
Preparation of Vaccines.
Viral antigens bearing His-tags were diluted from stock to 80 µg/mL in PBS. Appropriate volumes of 80 µg/mL working solution are mixed with an equal volume of 320 µg/mL CoPoP or PoP liposomes for a mass ratio 1:4 antigen to liposomes in the final solution. This solution was incubated for 3 h at RT (approx. 25 °C). After incubation, the solution was diluted with PBS as appropriate for assays or vaccinations.
CoPoP liposomes with HA antigen but without PHAD were prepared using methods identical to CoPoP/PHAD formulation. Alum vaccines were prepared by diluting 1 mg/mL recombinant antigen in PBS and combining with an equal volume of Alhydrogel 2% (Aluminum Hydroxide Gel Adjuvant, Brenntag Biosector A/S) for an alum dose of 50 μg. Alum–antigen mixtures were resuspended by pipetting prior to injection. For the comparative challenge study, ISA720 (Montanide ISA 720 VG, Seppic Inc.) was combined with His-tagged H3 HA from A/canine/Illinois/11613/2015 in PBS in a 7:3 volume ratio and vortexed 30 min. AddaVax vaccines were prepared by combining equal volumes of desired antigen quantity suspended in PBS with AddaVax adjuvant ready-to-use sterile emulsion (InvivoGen, catalog no. vac-adx-10), with 25 μL Addavax injected.
Mouse Immunization.
ICR (CD-1) outbred albino mice (Envigo, 6-wk-old female) were vaccinated with intramuscular injection of 50 µL test vaccine on days 0 and 21. CoPoP liposomal vaccines were prepared as described in the Liposome Preparation section, and other adjuvants were prepared following relevant protocols for use in mice.
Serum Antibody ELISA.
ELISA was performed on vaccinated mouse serum with an initial 200-fold dilution in PBS with 0.1% Tween-20 and a fivefold serial dilution out to a maximum dilution factor of 6.25 × 105. ELISA plates were coated with 100 μL/well of 1 µg/mL antigen in coating buffer (5.3 g/L NA2CO3 and 4.2 g/L NAHCO3 in distilled water, pH 9.6) and incubated overnight, at least 18 h. Plates were then washed three times and incubated with 1% BSA for 2 h at 37 °C. BSA solution was removed, and 100 µL serum dilutions were added, followed by a 1-h incubation at 37 °C. Plates were washed three times, and 100 µL of 1 µg/mL goat anti-mouse secondary antibody with HRP (Goat Anti-Mouse IgG Antibody [H&L] [HRP], GenScript catalog no. A00160) was added and incubated an additional 30 min at 37 °C. Plates were then washed six times, and 100 µL 3,3′,5,5′-Tetramethylbenzidine (TMB) solution (TMB One Component Microwell Substrate, SouthernBiotech catalog no. 0410-01) was added and allowed to develop for 5 min. To stop the reaction, 100 µL 1 N HCl was used and absorbance was read at 450 nm.
HAI Assay.
To prevent nonspecific HAI by serum components, 20 µL serum was incubated with 60 µL receptor destroying enzyme (RDE II, by Denka Seiken, from Hardy Diagnostics catalog no. 370013) at 37 °C for 18 h (46). Samples were then incubated at 56 °C for 30 min for heat-inactivation of enzymes before being diluted to a final volume of 200 µL with sterile PBS, resulting in an initial 10-fold dilution of serum. Samples were transferred to 96-well plates, and twofold serial dilutions were performed in PBS, with a dilution range from 1/10 to 1/1,280. Each sample was treated with either H3N2 A/Hong Kong/1/1968 at a concentration of 4 hemagglutinating units (HAU) per 25 µL as determined by titration or a virus-free control of sterile PBS. Serum and virus were incubated for 1 h at RT prior to the addition of 1% chicken red blood cell (RBC) solution. Samples were incubated 1 h after the inclusion of RBCs, and HAI titer was determined by highest dilution with complete inhibition of hemagglutination, as determined by virus-free reference control.
NA Inhibition Assay.
Neuraminidase inhibition measurements were performed using the Enzyme-Linked Lectin Assay method. Nunc 96-well ELISA plates were incubated 24 h at 4 °C with 25 µg/mL fetuin (Sigma-Aldrich no. F3004) in PBS. Serum samples were treated with RDE II as described in the HAI assay. Samples were transferred to uncoated standard 96-well plates and underwent twofold dilutions ranging from 1/20 to 1/2,560, then treated with A/Hong Kong/1/1968 (H3N2) at a concentration of 4 HAU per 25 µL as determined by titration or a virus-free control of sterile PBS. Plates were incubated 2 h in darkness. Fetuin plates were washed three times with PBS with 0.05 % Tween 20 (PBST), and serum–virus mixture was transferred to the fetuin-coated plates. These plates were incubated 18 h at 37 °C. After incubation plates were washed six times with 100 µL PBST and incubated with 1 µg/mL peanut-agglutinin horseradish peroxidase conjugate (PNA-HRP) in PBST for 2 h at RT, with samples kept in darkness. Plates were then washed again three times with PBST and treated with 100 µL TMB and allowed to develop for 10 min. A total 100 µL HCl was used to stop the reaction, and absorbance was read at 450 nm.
MNT Assays.
A549 cells were seeded at 40,000 cells/well on a clear flat-bottom 96-well plate 24 h prior to infection. Mouse sera was inactivated using trypsin-periodate and incubated at 56 °C for 30 min. On a second 96-well plate, the sera were diluted twofold down each column to create a 10- to 1280-fold dilution series using DMEM supplemented with 100 U/mL Penicillin and 100 μg/100μL Streptomycin, 25 nM Hepes, and L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)–treated trypsin (0.25 μg/mL) (Sigma, T1426). Two columns were each reserved for virus-only (positive) and media-only (negative) controls. A total of 500 pfu A/Vietnam/1204/2005 H5N1 HAlo of A/California/07/2009 H1N1 virus was added to all wells (except those serving as negative controls) and incubated at RT for 30 min. The plate containing A549 cells was washed three times with 1× PBS, and the contents of plate 2 (containing serum/virus mixtures) were transferred and incubated for 1 h at 37 °C, 5% CO2. The plate was then washed three times with 1× PBS and supplemented with DMEM containing 0.2 M L-Glutamine, 100 U/mL Penicillin, and 100 μg/100 μL Streptomycin overnight (∼18 h) at 37 °C, 5% CO2, followed by fixation with prechilled 80% acetone for 30 min at −20 °C. Cell-based ELISA was then performed to detect the presence of influenza virus nucleoprotein (NP) to measure viral replication. Plates were blocked with 5% nonfat milk in PBS and quenched with 10% hydrogen peroxide in PBS. Biotin-conjugated anti-NP (EMD Millipore MAB8257B) and HRP-conjugated streptavidin (EMB Millipore 18-152) were used as primary and secondary antibodies, respectively. Plates were then washed three times with PBST before adding 100 μL SigmaFAST o-Phenylenediamine dihydrochloride HRP substrate (Sigma-Aldrich), followed by 100 μL 3 M HCl to stop the HRP substrate reaction. Absorbance at 490 nm was measured, and microneutralization-50 (MNT50) titers were calculated.
Influenza Virus Challenge in Mice.
Prior to challenge, all mice were subjected to a standard vaccination regimen involving a primary vaccination followed by a second booster vaccination at 21 d postprime. Mice were challenged 35 d postprime with an intranasal inoculation with a mouse-adapted strain of H3N2 influenza virus (A/Hong Kong/1/1968, 50 µL, 100 pfu, 2.5 × LD50). This strain was obtained through BEI Resources: Influenza A Virus, A/Hong Kong/1/1968–1 Mouse-Adapted 12A (H3N2), NR-28622. Mice were monitored daily with body weight measurements and clinical score assessments. Animals that exhibited a body weight loss of 25% or greater were euthanized (20% for the H5N1 challenge). Initial challenge sets of mice were euthanized after 8 d and BAL performed to assess the amount of leukocyte extravasation. Briefly, following induction of anesthesia with 3% isoflurane in 100% O2, a longitudinal incision was made and 0.5 to 1.0 mL blood was collected from the abdominal aorta. Following 45 min of coagulation at RT, the blood was centrifuged at 5,000 × g for 5 min at 4 °C and the serum collected and stored at −80 °C. Next, following exposure of the trachea by blunt dissection, a 22-ga catheter was secured in the trachea with a suture and BAL performed with 5 × 1 mL instillations and collection of normal saline using two 5-mL syringes and a three-way stopcock. The collected fluid was centrifuged at 1,500 × g for 3 min at 4 °C, and the supernatant was stored at −80 °C. The pellet was resuspended in PBS and the cells counted using a Coulter Counter (MultiSizer 3, Beckman Coulter) (47). The lungs were also extracted and homogenized (Bullet Blender, NextAdvance), then centrifuged at 500 × g for 5 min at 4 °C. The supernatant was collected and quick frozen in ethanol and dry ice and stored at −80 °C until the influenza virus plaque assay in Madin–Darwin Canine Kidney (MDCK) cells was performed. In the subsequent set of experiments, mice were euthanized after 14 d in order to observe the full time course of influenza virus infection, in which body weights and clinical scores were collected daily. For the clinical score, one point was assessed for each of the following traits: piloerection, labored breathing, hunched posture, lethargy, abnormal gait, and emaciation (>10% weight loss) for a maximum score of 6 (48). In the multiplex vaccine challenge, mice were challenged with an intranasal inoculation of H5N1 HALo virus (A/Vietnam/1203/2004-derived, 50 µL, 150 pfu, 5 × LD50). These mice were euthanized if body weight loss exceeded 20% from initial weight.
Influenza Virus Plaque Assay in MDCK Cells.
MDCK cells (ATCC) were grown to 70 to 80% confluency in DMEM + 0.1mM nonessential amino acids + 1mM sodium pyruvate + 50 U/mL penicillin and 50 µg/mL streptomycin + 20 µg/mL gentamycin + 10% fcs in 6-well tissue culture plates in 37 °C + 5% CO2. The medium was removed and the cells rinsed 2× with 0.3% BSA in DMEM, then 100 µL 10-fold serial dilutions (0.3% BSA in DMEM as diluent) of virus samples were adsorbed for 1 h at 37 °C + 5% CO2 (300 µL diluent was added to each well during the adsorption to prevent drying of cell sheet). The inoculum was removed, the cells rinsed once with PBS, and then overlaid with 2 mL L-15 medium + 1 µg/mL TPCK-treated trypsin + 0.5% agarose. The plates were incubated for 48 h then stained with 0.3% crystal violet + 5% isopropanol + 5% ethanol in H2O for 20 min following removal of the overlay and 30-min fixation in 90% ethanol. The cells were rinsed with H2O and the plaques enumerated.
Influenza Virus Challenge in Ferrets.
The ferret challenge was conducted by BIOQUAL, Inc. Fitch ferrets were vaccinated via intramuscular route on days 0 and 28 and challenged with human influenza A/Texas/50/2012 (H3N2). The challenge virus was generated in embryonated chicken eggs and delivered under Ketamine/Dexmedetomidine sedation in 1 mL total volume divided evenly over the two nostrils. Body weight, temperature, and clinical score were monitored daily for the 14-d challenge period. Nasal washes were collected as indicated postchallenge for lung influenza virus titer analysis. Collected nasal wash was centrifuged for 10 min at 450 × g at 4 °C and stored at −80 °C. Ferret serum prior to the challenge was analyzed by HAI using 4 HAU/25 µL challenge strain A/Texas/50/2012 (H3N2) following methods comparable to those discussed with mouse serum, though using a dilution range from 1/20 to 1/2,560. To determine viral road, qPCR was used. RNA was extracted from ferret nasal wash according to a Qiagen kit. The following sequences for the influenza virus NP primer/probe were used: H3N2 Primers: H3N2-U: GGGTTAATAATCACTCACCG; H3N2-D: CTCAGTTGCATTCTGGCG; and H3N2 Probe: H3N2-P: CAAAATCATGGCGTCCCAAGGC. These primer–probe sets cover highly conserved regions in NP. A total of 47 μL master mix and 3 μL sample RNA were added to the wells of a 96-well plate. All samples were tested in triplicate. The plate was placed in a 7500 Sequence detector and run using the following program: 48 °C for 30 min, 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, and 1 min at 60 °C. The number of copies of RNA per mL were calculated by extrapolation from the standard curve and multiplying by the reciprocal of the 0.5 mL extraction volume.
Ethics.
Experimental protocols involving mice, including immunizations and subsequent challenge studies, were reviewed and approved by the University at Buffalo, Veterans Affairs Western New York Healthcare System, and McMaster Institutional Animal Care and Use Committees (IACUC). Experiments performed in ferrets were conducted in accordance with standard operating procedures of BIOQUAL, Inc. under an IACUC-approved protocol.
Supplementary Material
Acknowledgments
This study was supported by grants from the NIH; R41AI149954 (B.A.D.), R01HL151498 (B.A.D.), and HHSN272201700015I, Task Order HHSN27200004 (BIOQUAL), and the Canadian Institutes of Health Research (CIHR) (M.S.M). M.S.M. was supported by a CIHR New Investigator Award from the Government of Ontario. This material is the result of work supported with resources and the use of facilities at the Veterans Affairs Western New York Healthcare System, Buffalo, NY. A.Z. is supported by a Physicians' Services Incorporated Research Trainee Fellowship and a CIHR Canada Graduate Scholarship. We thank staff at the Facility for Electron Microscopy Research (FEMR) at McGill University for help in microscope operation. FEMR is supported by the Canadian Foundation for Innovation, the Quebec government, and McGill University. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of Centers for Disease Control and Prevention nor the US Department of Veterans Affairs nor the US government.
Footnotes
Competing interest statement: J.F.L. and W.-C.H. hold interest in POP Biotechnologies.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025759118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.WHO , Influenza (seasonal). https://www.who.int/health-topics/influenza-seasonal#tab=tab_1. Accessed 1 June 2020.
- 2.Bouvier N. M., Palese P., The biology of influenza viruses. Vaccine 26 (suppl. 4), D49–D53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y., Wu Y., Tefsen B., Shi Y., Gao G. F., Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 22, 183–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamblin S. J., Skehel J. J., Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 285, 28403–28409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrat F., Flahault A., Influenza vaccine: The challenge of antigenic drift. Vaccine 25, 6852–6862 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Horimoto T., Kawaoka Y., Molecular changes in virulent mutants arising from avirulent avian influenza viruses during replication in 14-day-old embryonated eggs. Virology 206, 755–759 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Zost S. J., et al., Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U.S.A. 114, 12578–12583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox M. M., Izikson R., Post P., Dunkle L., Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vaccines 3, 97–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein D. I., et al., Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 20, 80–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steel J., et al., Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1, e00018-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter D. M., et al., Design and Characterization of a COBRA HA vaccine for H1N1 influenza viruses. J. Virol. 90, 4720–4734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Center for Disease Control and Prevention , 2019-2020 U.S. Flu season: Preliminary burden estimates. https://www.cdc.gov/flu/about/burden/preliminary-in-season-estimates.htm. Accessed 1 December 2020.
- 13.Tregoning J. S., Russell R. F., Kinnear E., Adjuvanted influenza vaccines. Hum. Vaccin. Immunother. 14, 550–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sia Z. R., Miller M. S., Lovell J. F., Engineered nanoparticle applications for recombinant influenza vaccines. Mol. Pharm. 18, 576–592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly H. G., Kent S. J., Wheatley A. K., Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Rev. Vaccines 18, 269–280 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Henriksen-Lacey M., Korsholm K. S., Andersen P., Perrie Y., Christensen D., Liposomal vaccine delivery systems. Expert Opin. Drug Deliv. 8, 505–519 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Vu M. N., et al., Hemagglutinin functionalized liposomal vaccines enhance germinal center and follicular helper T cell immunity. Adv. Healthc. Mater. 10.1002/adhm.202002142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babai I., et al., A novel liposomal influenza vaccine (INFLUSOME-VAC) containing hemagglutinin-neuraminidase and IL-2 or GM-CSF induces protective anti-neuraminidase antibodies cross-reacting with a wide spectrum of influenza A viral strains. Vaccine 20, 505–515 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Barnier Quer C., Elsharkawy A., Romeijn S., Kros A., Jiskoot W., Cationic liposomes as adjuvants for influenza hemagglutinin: More than charge alone. Eur. J. Pharm. Biopharm. 81, 294–302 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Didierlaurent A. M., et al., Adjuvant system AS01: Helping to overcome the challenges of modern vaccines. Expert Rev. Vaccines 16, 55–63 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Del Giudice G., Rappuoli R., Didierlaurent A. M., Correlates of adjuvanticity: A review on adjuvants in licensed vaccines. Semin. Immunol. 39, 14–21 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Federizon J., et al., Experimental and computational observations of immunogenic cobalt porphyrin lipid bilayers: Nanodomain-enhanced antigen association. Pharmaceutics 13, 98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W.-C., et al., A malaria vaccine adjuvant based on recombinant antigen binding to liposomes. Nat. Nano. 13, 1174–1181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W.-C., et al., SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv. Mater. 32, e2005637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao S., et al., Functionalization of cobalt porphyrin-phospholipid bilayers with his-tagged ligands and antigens. Nat. Chem. 7, 438–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Federizon J., et al., Immunogenicity of the Lyme disease antigen OspA, particleized by cobalt porphyrin-phospholipid liposomes. Vaccine 38, 942–950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens J., et al., Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303, 1866–1870 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Krammer F., et al., A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7, e43603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hai R., et al., Influenza viruses expressing chimeric hemagglutinins: Globular head and stalk domains derived from different subtypes. J. Virol. 86, 5774–5781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris A., et al., Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 103, 19123–19127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tait A. R., Davidson B. A., Johnson K. J., Remick D. G., Knight P. R., Halothane inhibits the intraalveolar recruitment of neutrophils, lymphocytes, and macrophages in response to influenza virus infection in mice. Anesth. Analg. 76, 1106–1113 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Penna A. M., Johnson K. J., Camilleri J., Knight P. R., Alterations in influenza A virus specific immune injury in mice anesthetized with halothane or ketamine. Intervirology 31, 188–196 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Ott G., Radhakrishnan R., Fang J.-H., Hora M., “The adjuvant MF59: A 10-year perspective” in Vaccine Adjuvants: Preparation Methods and Research Protocols, O’Hagan D. T., Ed. (Springer, 2000), pp. 211–228. [Google Scholar]
- 34.O’Hagan D. T., Rappuoli R., De Gregorio E., Tsai T., Del Giudice G., MF59 adjuvant: The best insurance against influenza strain diversity. Expert Rev. Vaccines 10, 447–462 (2011). [DOI] [PubMed] [Google Scholar]
- 35.O’Hagan D. T., Ott G. S., De Gregorio E., Seubert A., The mechanism of action of MF59 - an innately attractive adjuvant formulation. Vaccine 30, 4341–4348 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Goff P. H., et al., Adjuvants and immunization strategies to induce influenza virus hemagglutinin stalk antibodies. PLoS One 8, e79194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., et al., Broad cross-reactive IgG responses elicited by adjuvanted vaccination with recombinant influenza hemagglutinin (rHA) in ferrets and mice. PLoS One 13, e0193680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorquera P. A., et al., Insights into the antigenic advancement of influenza A(H3N2) viruses, 2011-2018. Sci. Rep. 9, 2676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steel J., et al., Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83, 1742–1753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolova V., Westendorf A. M., Buer J., Überla K., Epple M., The potential of nanoparticles for the immunization against viral infections. J. Mater. Chem. B Mater. Biol. Med. 3, 4767–4779 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyay S., Chen J.-Y., Chen H.-W., Hu C. J., Nanoparticle vaccines adopting virus-like features for enhanced immune potentiation. Nanotheranostics 1, 244–260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López-Sagaseta J., Malito E., Rappuoli R., Bottomley M. J., Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 58–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyoglu-Barnum S., et al., Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao S., et al., An engineered biomimetic MPER peptide vaccine induces weakly HIV neutralizing antibodies in mice. Ann. Biomed. Eng. 48, 1991–2001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y.-Q., et al., Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173, 417–429.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lingel A., Bullard B. L., Weaver E. A., Efficacy of an adenoviral vectored multivalent centralized influenza vaccine. Sci. Rep. 7, 14912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald B. A., et al., Halothane modulates the type i interferon response to influenza and minimizes the risk of secondary bacterial pneumonia through maintenance of neutrophil recruitment in an animal model. Anesthesiology 123, 590–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonville C. A., et al., Respiratory dysfunction and proinflammatory chemokines in the pneumonia virus of mice (PVM) model of viral bronchiolitis. Virology 349, 87–95 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.